ABSTRACT
The Epstein-Barr virus (EBV) is etiologically linked to approximately 10% of gastric cancers, in which viral genomes are maintained as multicopy episomes. EBV-positive gastric cancer cells are incompetent for progeny virus production, making viral DNA cloning extremely difficult. Here we describe a highly efficient strategy for obtaining bacterial artificial chromosome (BAC) clones of EBV episomes by utilizing a CRISPR/Cas9-mediated strand break of the viral genome and subsequent homology-directed repair. EBV strains maintained in two gastric cancer cell lines (SNU719 and YCCEL1) were cloned, and their complete viral genome sequences were determined. Infectious viruses of gastric cancer cell-derived EBVs were reconstituted, and the viruses established stable latent infections in immortalized keratinocytes. While Ras oncoprotein overexpression caused massive vacuolar degeneration and cell death in control keratinocytes, EBV-infected keratinocytes survived in the presence of Ras expression. These results implicate EBV infection in predisposing epithelial cells to malignant transformation by inducing resistance to oncogene-induced cell death.
IMPORTANCE Recent progress in DNA-sequencing technology has accelerated EBV whole-genome sequencing, and the repertoire of sequenced EBV genomes is increasing progressively. Accordingly, the presence of EBV variant strains that may be relevant to EBV-associated diseases has begun to attract interest. Clearly, the determination of additional disease-associated viral genome sequences will facilitate the identification of any disease-specific EBV variants. We found that CRISPR/Cas9-mediated cleavage of EBV episomal DNA enabled the cloning of disease-associated viral strains with unprecedented efficiency. As a proof of concept, two gastric cancer cell-derived EBV strains were cloned, and the infection of epithelial cells with reconstituted viruses provided important clues about the mechanism of EBV-mediated epithelial carcinogenesis. This experimental system should contribute to establishing the relationship between viral genome variation and EBV-associated diseases.
INTRODUCTION
Epstein-Barr virus (EBV) is one of the most widespread human pathogens. EBV infection is usually asymptomatic, but it sometimes causes severe disorders, such as EBV-related lymphoproliferative disease, B-cell lymphomas, and NK/T-cell lymphomas (1). In addition, causal relationships between EBV infection and epithelial cell-derived cancers, such as nasopharyngeal carcinomas (NPCs) and gastric cancers, have been investigated extensively (2, 3). However, the precise mechanisms underlying EBV-mediated epithelial carcinogenesis remain largely unknown.
Recent deep-sequencing studies demonstrated unexpected levels of heterogeneity in EBV genomes derived from various EBV-positive cell lines, including Burkitt's lymphoma-derived cell lines (4), spontaneously established lymphoblastoid cell lines (LCLs), Hodgkin's lymphoma cell lines, NPC-derived cell lines, a gastric cancer-derived cell line (5), and NPC biopsy samples (6). Among infected individuals, EBV-associated cancers arise in only a very small population, indicating that EBV contributes to carcinogenesis as a cofactor. An attractive hypothesis is that a specific EBV strain serves as a strong cofactor for carcinogenesis. To test this hypothesis, authentic viruses maintained in cancer cells should be isolated and further characterized; however, EBV-associated epithelial cancer cells, such as NPCs and gastric cancers, are incompetent for progeny virus production, making it difficult to reconstitute infectious viruses derived from cancer cells. A recent study demonstrated the cloning of an NPC-derived EBV strain, M81, in a bacterial artificial chromosome (BAC) vector, followed by infectious virus reconstitution (7). The study clearly demonstrated that reconstituted cancer cell-derived EBV differs significantly from B-cell-derived EBV in its enhanced epitheliotropism and its competency to enter the lytic cycle in lymphoblastoid cells.
To increase the repertoire of EBV strains derived from patients with various diseases, including cancers, we aimed to simplify the procedure for BAC cloning of EBV genomes. Genome-editing technology using clustered, regularly interspaced, short palindromic repeats (CRISPR)/Cas9 works not only for chromosomal DNAs but also for cutting EBV episomes (8, 9), the genomes of herpes simplex viruses (10, 11), and adenoviruses (10). We envisioned that transgene insertion into EBV episomes would be stimulated by cutting circular EBV episomes and simultaneously introducing a specifically designed donor plasmid into latently infected cells. This study provides the proof of concept for inserting a BAC vector sequence into a specific locus within an EBV genome via homology-directed repair. We cloned two gastric cancer cell line-derived EBV strains as EBV-BAC clones, determined their complete viral genome sequences, reconstituted infectious viruses, and clarified how viruses affect the phenotypes of stably infected epithelial cells.
MATERIALS AND METHODS
Cell culture.
SNU719 cells (12) were obtained from the Korean Cell Line Bank (KCLB 00719) and were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (PC-SM). YCCEL1 cells (13) were obtained from Sun Young Rha (Yonsei University College of Medicine, Seoul, South Korea) and were cultured in minimal essential medium supplemented with 10% FBS, nonessential amino acids, and PC-SM. HEK293 cells were cultured as described previously (14) and were used for recombinant virus production. HDK1-K4DT cells (human dermal keratinocytes immortalized by the expression of a mutant form of cyclin-dependent kinase 4/cyclin D1/human telomerase reverse transcriptase [15]) were cultured in keratinocyte serum-free medium (SFM) with supplements (catalog no. 17005-042; Invitrogen).
CRISPR/Cas9-mediated EBV-BAC cloning.
An EBV DNA sequence around a BssHII site (corresponding to nucleotides [nt] 134663 to 134668 of wild-type EBV [EBV-wt]) was chosen as a CRISPR/Cas9 target sequence according to our experience of EBV genome targeting (14). The EBV-specific CRISPR/Cas9 plasmid (pX330-sgEBV) was constructed by inserting annealed oligonucleotides (sgEBV top [CACCGCTGTGTGCGCGCCGCCAGCA] and sgEBV bottom [AAACTGCTGGCGGCGCGCACACAGC]) into the BbsI site of pX330 (catalog no. 42230; Addgene) (16). Primers EBV SacI up (CGCGGATCCGAGCTCTTCCGCTTCATCTG [with BamHI and SacI sites underlined]) and EBV SacI down (CGCGAATTCGAGCTCAGGTTTTTGCACCT [with EcoRI and SacI sites underlined]) were used to PCR amplify a SacI fragment (1,088 bp) of SNU719 EBV (or YCCEL1 EBV) spanning the target region. The PCR product was digested by BamHI and EcoRI and was cloned into pCAG-EGxxFP to obtain a reporter plasmid, pCAG-EG-ebv-FP. pCAG-EG-mebv-FP, a reporter plasmid with silent mutations within the BVRF1 coding sequence, was obtained by using primers mBVRF1 for (ATGCTCGCCGCCCGGACACAGCTGCGTCT) and mBVRF1 rev (TGTGTCCGGGCGGCGAGCATGGCAGCCGG) with a PCR-based mutagenesis protocol (17).
The PCR-amplified SacI fragment was cloned into pBluescript II-SK to make pBS-SNU719 (the EBV DNA upstream region was on the KpnI side of the vector). A unique PacI site (bPacI) was generated at the CRISPR/Cas9 target site by using inverse PCR primers BssHII inv up (TTAACGCACACAGCTGCGTCTAGC) and BssHII inv down (TTAACGCCGCCAGCATGGCAGCCG) (each with a half-PacI site [underlined]) to obtain pBS-SNU719-PacI. Primers M13-20′ (TTGTAAAACGACGGCCAGT), mBVRF1 rev, mBVRF1 for, and PacI rev (CGCGTTAATTAAAACCCCCGTGCGATGAC [with the PacI site underlined]), with pBS-SNU719 as a template, were used to obtain a PCR product with silent BVRF1 mutations. The PCR product was then digested by SpeI and PacI and was cloned into corresponding sites of pBS-SNU719-PacI to obtain pBS-SNU719-BVRF1(+)-PacI. A PacI fragment consisting of a BAC vector sequence, an enhanced green fluorescent protein (EGFP) gene, and a hygromycin resistance gene (14) was cloned into the PacI site of pBS-SNU719-BVRF1(+)-PacI to make a donor plasmid for targeting SNU719 EBV.
The upstream homology region of YCCEL1 EBV is identical to that of SNU719 EBV, while the downstream homology regions are not identical. To replace the downstream homology region of pBS-SNU719-PacI with YCCEL1 EBV DNA, a Tth111I-HpaI fragment of pBS-SNU719-PacI was replaced with the corresponding YCCEL1 EBV fragment to yield pBS-YCCEL1-PacI. A donor plasmid for targeting YCCEL1 EBV was constructed by following the strategy described above.
EBV genome targeting and screening of bacterial colonies.
SNU719 cells (or YCCEL1 cells) were plated in a well of a 6-well dish and were transfected with 0.4 μg of the donor plasmid and 4 μg of pX330-sgEBV (2 μg each for YCCEL1 cells) using the ViaFect reagent. Transfected cells were replated onto a 10-cm dish at 2 days posttransfection and were selected with hygromycin (30 μg/ml for SNU719 cells; 50 μg/ml for YCCEL1 cells). Hygromycin-resistant cells were pooled at 3 weeks posttransfection, and episomal DNAs were prepared from the pooled cells (18). Aliquots of episomal DNAs were subjected to PCR analyses using either primers rt recom for (TATCCAGTCCAGACGCTTTTC) and BAC vec down (TTGTATGCCTGCTGTGGATT) to check the upstream junction or primers lt recom for (CGGGCCATTTACCGTAAGTTA) and lt recom rev (TAAAGAACTGGCCGCTTTG) to check the downstream junction. Aliquots of episomal DNAs were used to transform ElectroMAX DH10B competent cells (Invitrogen) in order to obtain chloramphenicol-resistant bacterial colonies. The chloramphenicol-resistant colonies obtained were screened by colony direct PCR using primers BamW up (TACCAGAGGGGGCCAAGAA) and BamW down (AGGAGAGGCAGGGCCTGAA). EBV-BAC clones were identified by the presence of a 306-bp PCR product derived from EBV BamHI W fragments. Positive clones were grown, and BAC DNA preparations and restriction enzyme analyses were performed as described previously (19). Negative clones were found to contain donor plasmids themselves (data not shown).
EBV genome sequencing and annotation.
EBV-BAC clone DNAs were prepared by using a NucleoBond BAC 100 kit (Macherey-Nagel, Düren, Germany). Contaminated bacterial genomic DNAs were enzymatically digested by using Plasmid-Safe ATP-dependent DNase (Epicentre, Madison, WI) before they were subjected to sequencing. Sequencing was performed by TaKaRa Bio (Mie, Japan) on a PacBio RS instrument (Pacific Biosciences, Menlo Park, CA, USA) (20) using libraries prepared with the SMRTbell template preparation kit (version 1.0; Pacific Biosciences). A hierarchical genome assembly process was used for high-quality de novo genome assemblies (21). The contigs obtained were manually curated to remove duplicated terminal sequences and transgene sequences, and sequences of SNU719 and YCCEL1 EBV genomes were obtained. The sequences were manually annotated based on homology and positional concordance with the standard EBV-wt sequence (22), with the help of the Genome Annotation Transfer Utility (23). Nearly all open reading frames (ORFs) corresponding to those of EBV-wt were successfully annotated. Only two ORFs (EBNA3C of SNU719 EBV and EBNA2 of YCCEL1 EBV) were found to contain internal stop codons due to sequencing errors, and they were modified by referring to the results of Sanger sequencing.
Phylogenetic analysis.
The following EBV genomic sequences were obtained from GenBank: GD1 (accession number AY961628.3), Akata (KC207813.1), wt (NC_007605.1), M81 (KF373730), and Mutu (KC207814.1). Genomic DNA alignments were executed by excluding repetitive elements of the EBV genome (namely, the family of repeats, internal repeats 1 to 4, and terminal repeats). The phylogenetic tree was inferred by using the maximum likelihood method based on the Tamura-Nei model (24) with gamma distribution, which was used to model evolutionary rate differences among sites.
Recombinant EBV production, infection, and viral gene expression analyses.
Virus-producing cells were established by stably transducing EBV-BAC clone DNAs into HEK293 cells as described previously (19). Western blot analyses and TaqMan small-RNA assays (Applied Biosystems) were used to analyze viral protein and microRNA expression, respectively.
EBV infection of immortalized keratinocytes.
HDK1-K4DT cells were retrovirally transduced with CR2 as described previously (14). The HDK1-CR2 cells obtained were subsequently infected with the SNU719-BAC virus or the YCCEL1-BAC virus. Pools of stably infected HDK1-CR2 cells, designated HDK1-SNU719 or HDK1-YCCEL1 cells, were obtained by selection with hygromycin (5 μg/ml). Control HDK1 cells were obtained by infecting HDK1-CR2 cells with a retroviral vector carrying hygromycin resistance and EGFP genes (pQCXIHyg-EGFP).
Induction of Ras oncoprotein in EBV-infected keratinocytes.
HDK1-SNU719 cells, HDK1-YCCEL1 cells, and control HDK1 cells were transduced with a retroviral vector (pQCXIP-mERT2-KRASG12V) encoding an estrogen receptor (ER)-RAS fusion protein derived from LZRS ERTm RasV12, which was a gift from Paul Khavari (plasmid 21199; Addgene) (25). The infected cells were selected with puromycin (0.5 μg/ml) for 2 days to obtain pools of cells expressing the ER-RAS fusion protein. The ER-RAS-expressing cells (2.5 × 105 cells) were plated into each well of 6-well dishes, incubated overnight, and then subjected to 1 nM 4-hydroxytamoxifen (4-OHT) for 4 days to induce ER-RAS expression. Cells were photographed using inverted florescence microscopy before and after 4 days of 4-OHT treatment. Whole-cell extracts were prepared from the 4-OHT-treated cells and were subjected to Western blot analysis using an anti-K-Ras antibody (F234; Santa Cruz).
Nucleotide sequence accession numbers.
The sequences of SNU719 EBV and YCCEL1 EBV can be retrieved from DDBJ/EMBL/GenBank under accession numbers AP015015 and AP015016.
RESULTS
CRISPR/Cas9-mediated cleavage of viral episomes enables highly efficient cloning of EBV episomes from gastric cancer cells.
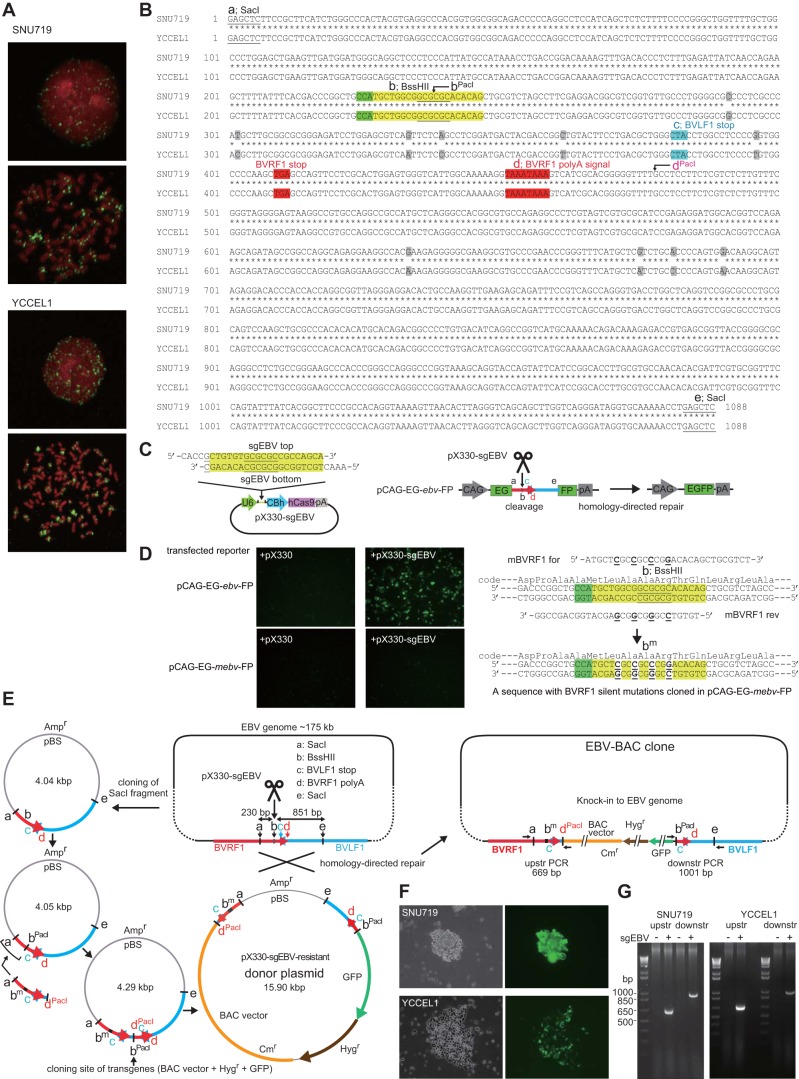
SNU719 and YCCEL1 are EBV-positive gastric cancer cell lines derived from Korean patients (12, 13). Fluorescence in situ hybridization (FISH) revealed a number of EBV genome-derived signals scattered inside interphase nuclei of SNU719 and YCCEL1 cells, or associated with mitotic chromosomes (Fig. 1A), indicating that the EBV genomes are most likely maintained as circular episomes. A conventional homologous recombination strategy to clone EBV episomes from SNU719 cells ended in failure, presumably due to the inefficiency of homologous recombination (data not shown).
Fig 1.
CRISPR/Cas9-mediated cloning of EBV episomes derived from gastric cancer cells. (A) EBV genomes in interphase nuclei of EBV-positive gastric cancer cells (SNU719 and YCCEL1) or in mitotic cells were visualized by fluorescence in situ hybridization as described previously (38). (B) Nucleotide sequences of SNU719 EBV and YCCEL1 EBV spanning the CRISPR/Cas9 target sequence and flanking regions that served as homologous arms. SacI sites (indicated by lowercase letters a and e) and a BssHII site (b) are underlined. A target and a PAM sequence are highlighted in yellow and green, respectively. A stop codon and a polyadenylation signal of BVRF1 are highlighted in red, while a stop codon (in the complementary strand) of BVLF1 is highlighted in blue. Nucleotides that are not identical in SNU719 EBV and YCCEL1 EBV are shaded. The positions of bPacI and dPacI, PacI recognition sites created for the cloning, are indicated. (C) Schematic representation of a CRISPR/Cas9 plasmid (pX330-sgEBV) and a reporter plasmid (pCAG-EG-ebv-FP). pX330-sgEBV has the indicated oligonucleotides (the BssHII site and a G nucleotide added on the 5′ end [underlined]) cloned into pX330. pCAG-EG-ebv-FP has a 1,088-bp SacI fragment (shown in panel B) inserted. The efficiency of CRISPR/Cas9-mediated DNA cleavage and homology-directed repair can be monitored by the restoration of the GFP coding sequence. (D) HEK293 cells were transfected with GFP reporter plasmids and CRISPR/Cas9 plasmids in combination as indicated and were examined for GFP expression at 2 days posttransfection. pCAG-EG-mebv-FP has a mutated insert (containing a bm sequence, shown on the right) and is resistant to pX330-sgEBV-mediated cleavage. The oligonucleotides used to generate the bm sequence are shown. (E) A donor plasmid construction strategy and an expected EBV-BAC clone obtained by homology-directed repair. An upstream homology arm (indicated as a-bm-c-dPacI) contains a CRISPR/Cas9-resistant BVRF1 coding sequence, and the BVRF1 coding sequence is recovered after homology-directed donor knock-in. The sizes of PCR products are shown to verify successful recombination at the upstream (upstr) and downstream (downstr) regions. pBS, pBluescript; Ampr, ampicillin resistance gene; Cmr, chloramphenicol resistance gene. (F) SNU719 and YCCEL1 cells were transfected with pX330-sgEBV and the donor plasmid and were selected using hygromycin. Phase-contrast (left) and GFP fluorescence (right) images of proliferating colonies were taken at 3 weeks posttransfection. (G) Detection of homologously recombined molecules in hygromycin-resistant cell pools. Note that PCR products were present only when the donor plasmid and pX330-sgEBV were cotransfected.
We therefore applied the CRISPR/Cas9-mediated genome-editing technique to increase the efficiency of homologous recombination. A DNA sequence within the BVRF1 open reading frame was chosen as a target site (Fig. 1B). A plasmid containing an hCas9 expression cassette and a single-guide RNA (sgRNA) targeting the EBV DNA (pX330-sgEBV) was constructed (Fig. 1C). The efficiency of pX330-sgEBV in cleaving the target sequence within the EBV genome was first verified using a GFP reporter plasmid in which the target EBV sequence was flanked by overlapping GFP open reading frames (26) (pCAG-EG-ebv-FP [Fig. 1C]). Massive EGFP expression was observed 48 h posttransfection when the reporter plasmid was cotransfected with pX330-sgEBV into HEK293 cells, indicating that cleavage of the EBV target sequence stimulated the reconstitution of a GFP coding sequence via homology-directed repair (Fig. 1D). The same reporter gene system was used to demonstrate that the CRISPR/Cas9 system could also work in SNU719 and YCCEL1 cells (data not shown).
We then constructed a donor plasmid to enable the insertion of transgenes (a BAC vector sequence and hygromycin resistance and GFP genes) into the EBV genome cleavage site. The experimental strategy used to construct the donor plasmid is illustrated in Fig. 1E. Minimal upstream and downstream homology regions (230-bp and 851-bp DNA fragments in Fig. 1E) were included in the donor plasmid. Since the EBV genome cleavage site is located within the BVRF1 gene (Fig. 1B and E), which encodes an essential minor capsid protein, we created a CRISPR/Cas9-resistant BVRF1 gene by introducing silent mutations and included it in the upstream homology region of the donor plasmid (Fig. 1D and E). The resistance of the mutated DNA fragment (pCAG-EG-mebv-FP) to CRISPR/Cas9-mediated DNA cleavage was verified by the lack of GFP expression, even when pX330-sgEBV was cotransfected (Fig. 1D). Thus, the donor plasmid is expected to be resistant to pX330-sgEBV.
The circular donor plasmid was cotransfected with either pX330 (without sgRNA) or pX330-sgEBV into SNU719 cells, and the transfected cells were subjected to hygromycin selection for 3 weeks. Hygromycin-resistant, GFP-positive colonies appeared when cells were transfected with both the donor plasmid and pX330-sgEBV (Fig. 1F), while very few GFP-positive cells appeared when only pX330 was cotransfected. Hygromycin-resistant colonies were pooled, and episomal DNA was prepared from them. PCR analyses provided clear evidence of successful homology-directed repair in episomal DNA samples when the donor plasmid was cotransfected with pX330-sgEBV, but not when it was cotransfected with pX330 (Fig. 1G).
We also tested a linear fragment of the donor plasmid (a 12.9-kb SacI fragment, spanning positions a through e in Fig. 1E) for homology-directed repair, but it failed to work (data not shown). Thus, only a circular donor plasmid, not a linearized one, achieved homologous recombination.
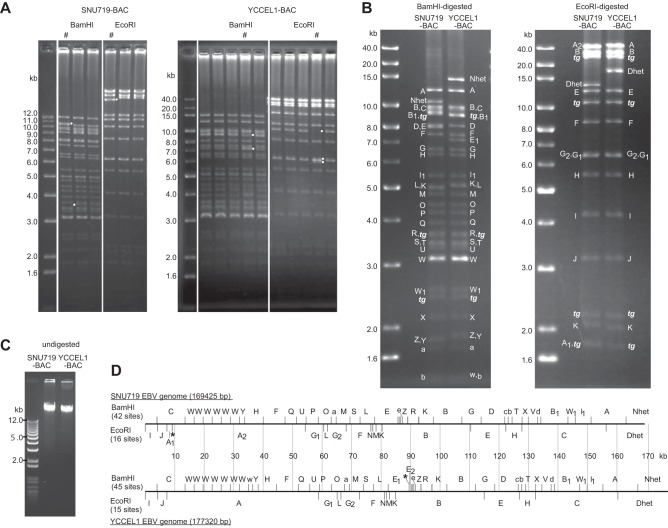
Aliquots of the episomal DNA with the evidence of homologous recombination were used to transform bacteria, and the resulting chloramphenicol-resistant colonies were PCR screened for the presence of the EBV internal repeat 1 (IR1) sequence, permitting the identification of SNU719-derived EBV-BAC clones (Fig. 2A). The frequency of obtaining EBV-BAC clones from the chloramphenicol-resistant colonies ranged from 8.8% (3/34) to 47.8% (22/46). Ten of 12 clones exhibited identical BamHI and EcoRI digestion patterns, while two exceptional clones had altered digestion patterns (Fig. 2A and data not shown). Whether these exceptional clones are artifacts resulting from the experimental procedures or represent genuine minor EBV populations of biological relevance is currently unclear.
FIG 2.
EBV-BAC clones obtained from SNU719 and YCCEL1 cells and deduced restriction enzyme maps of authentic EBV genomes without transgenes. (A) EBV-BAC clones obtained from SNU719 and YCCEL1 cells. DNAs of multiple EBV-BAC clones obtained from the cells were digested with either BamHI or EcoRI and were analyzed by agarose gel electrophoresis. Note that clones marked with pound signs (#) exhibit digestion patterns different from those of the other clones. White dots mark the individual bands that differ in the exceptional clones. (B) Restriction enzyme mapping of SNU719-BAC and YCCEL1-BAC. BAC DNA samples were digested with either BamHI or EcoRI and were analyzed by 0.8% agarose gel electrophoresis. Digested bands are labeled alphabetically, corresponding to bands derived from EBV strain B95-8. Bands derived from transgenes are indicated by tg. (C) Purified SNU719-BAC and YCCEL1-BAC DNA samples were electrophoresed through a 0.6% agarose gel. (D) Linear BamHI and EcoRI digestion maps of the SNU719 and YCCEL1 EBV genomes (without transgenes). EBV genomes are represented as if they were linearized by cleavage at the position corresponding to nucleotide 1 of EBV-wt. Note that YCCEL1 EBV has two additional BamHI W repeats, one of which is internally truncated (indicated by a lowercase “w”). The BamHI E1 fragment of YCCEL1 EBV is smaller than the corresponding fragment of SNU719 EBV (BamHI E) (Fig. 2B) due to the presence of an extra BamHI site (indicated by an asterisk). The EcoRI A fragment of SNU719 EBV is divided into two fragments (A1 and A2) by an extra EcoRI site (asterisk).
The same experimental strategy was successfully applied to the cloning of the EBV genome in YCCEL1 cells (Fig. 1F and G), indicating the versatility of this experimental strategy. In this case, 4 of 5 EBV-BAC clones exhibited identical digestion patterns (Fig. 2A).
Sequencing of gastric cancer cell-derived EBV genomes using single-molecule real-time sequencing technology.
One clone with the representative digestion pattern was derived from each cell line and selected as SNU719 EBV-BAC (SNU719-BAC) or YCCEL1 EBV-BAC (YCCEL1-BAC), respectively, and subjected to further analysis (Fig. 2B). The BamHI- and EcoRI-digested BAC clones were analyzed by Southern blotting (data not shown), and the identity of each band was determined by comparison with those of EBV strain B95-8 (Fig. 2B). SNU719-BAC and YCCEL1-BAC exhibited markedly different digestion patterns. DNAs of SNU719-BAC and YCCEL1-BAC were purified and were then subjected to DNA sequencing (Fig. 2C). The sequence of the YCCEL1 EBV genome was determined previously using a hybrid capture approach (GenBank accession no. LN827561.1) (5); however, there are a number of sequence gaps corresponding to repetitive regions within the genome. To obtain complete viral genome sequences, we employed PacBio single-molecule real-time sequencing technology (20, 21). Long reads (mean subread length, >10 kb) were obtained and were subjected to a hierarchical genome assembly process (21) to form contigs of >180 kb for both SNU719 and YCCEL1 EBV-BAC clones (Table 1). The longest contigs were manually curated, and sequences of SNU719-BAC (181,278 bp) and YCCEL1-BAC (189,173 bp) were obtained. BamHI and EcoRI digestion maps were then prepared from the sequences (data not shown). The sizes of the BamHI- and EcoRI-digested DNA fragments of EBV-BAC clones corresponded very well with the digestion maps prepared (Fig. 2B), verifying the accuracy of the assembled sequences.
TABLE 1.
Summary of sequence data obtained from PacBio sequencing
| Sample | No. of bases (megabases) | No. of subreads | Mean subread length (bases) | Qualitya | Longest contig length (bases) | Coverage (fold) |
|---|---|---|---|---|---|---|
| SNU719-BAC | 730 | 71,436 | 10,231 | 0.845 | 201,979 | 2,240 |
| YCCEL1-BAC | 710 | 66,824 | 10,635 | 0.850 | 188,893 | 1,712 |
Mean mapped subread concordance.
Transgene sequences were subsequently removed from the EBV-BAC sequences to obtain sequences of the SNU719 and YCCEL1 EBV genomes (accession numbers AP015015 and AP015016). The sizes of the SNU719 EBV and YCCEL1 EBV genomes were 169,425 and 177,320 bp, respectively. The size difference is due mainly to variations in the number of repeat sequences: 6 versus 7.6 copies of W repeats (IR1) and 5 versus 12 copies of terminal repeats for SNU719 EBV and YCCEL1 EBV, respectively (Fig. 2D). Restriction mapping data again proved that these copy numbers were accurate (data not shown).
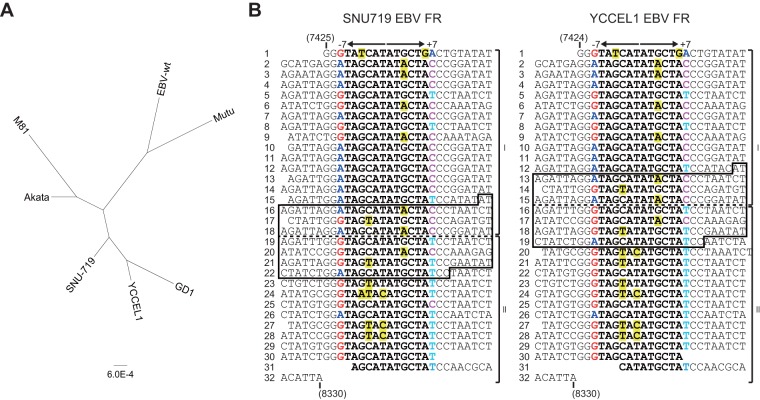
The DNA sequences of SNU719 EBV and YCCEL1 EBV were subjected to phylogenetic analyses. Five representative EBV genomes deposited in GenBank (EBV-wt [22], GD1 [27], M81 [7], Akata, and Mutu [4]) were used as reference sequences. The sequences of the SNU719 and YCCEL1 EBVs, both derived from Korean gastric cancer patients, are similar to those of GD1 and M81 (both derived from Chinese NPC patients) and to that of Akata (derived from a Japanese Burkitt's lymphoma patient), while they are distinct from EBV-wt (a strain B95.8 EBV [derived from an infectious mononucleosis patient from the United States] with its sequence gap filled with EBV strain Raji sequence [derived from an African Burkitt's lymphoma patient]) and Mutu (derived from an African Burkitt's lymphoma patient) (Fig. 3A). These results are in good agreement with the notion that different EBV strains are distributed in different geographical areas (East Asia, African countries, and Western countries) (5, 7, 28).
FIG 3.
Characterization of gastric cancer cell-derived EBV genome sequences. (A) Phylogenetic trees based on available EBV genomic DNA sequences. The tree is drawn to scale, with branch lengths corresponding to the number of substitutions per site. (B) Primary DNA sequences of the family of repeats (FR) of gastric cancer cell-derived EBVs. Sequences are aligned along the palindromic cores of each repeat unit. Nucleotides constituting the 12-bp palindromic core of each repeat unit are in boldface, and those that are mismatched to the consensus motif (TAGCATATGCTA) of the palindromic core are highlighted in yellow. Nucleotides G, A, T, and C at positions −7 and +7 are shown in different colors to emphasize the orientation of each repeat unit. At the border of subregions I and II (dashed lines), there exist 205-bp sequences (boxed) that are 100% conserved between SNU719 and YCCEL1 EBV. Nucleotide numbers in the SNU719 EBV sequence (GenBank accession no. AP015015) and the YCCEL1 EBV sequence (AP015016) are indicated.
Genome annotation revealed that the numbers of amino acids of viral latent proteins encoded by SNU719 EBV and YCCEL1 EBV differed substantially from those encoded by EBV-wt (Table 2). The reading frames of the LF3 protein were not conserved, as observed in a previous report regarding EBV strains Akata and Mutu (4). Mature microRNA sequences of SNU719 EBV and YCCEL1 EBV were found to be identical to those deposited in miRBase (www.mirbase.org).
TABLE 2.
Comparison of numbers of amino acids in EBV nuclear antigens and latent membrane proteins deduced from DNA-sequencing results in this study and previous studiesa
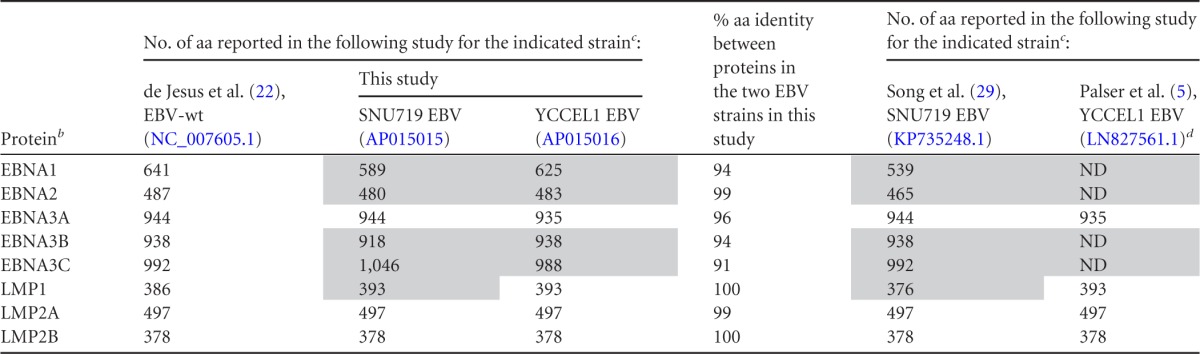
Shading indicates discrepancies between this study and previous studies.
b EBNA, EBV nuclear antigens; LMP, latent membrane proteins.
c Accession numbers are given in parentheses after EBV strain designations.
d ND, not determined.
Comparison of amino acid sequences in SNU719 EBV and YCCEL1 EBV revealed high degrees of conservation of LMP1, LMP2A, and LMP2B (Table 2). In EBNA1, the only difference resides in the lengths of internal glycine-alanine repeats, and in EBNA2, the difference is restricted to the N-terminal proline-rich domain. Thus, these proteins are also highly conserved between the strains. On the other hand, the amino acid sequences of EBNA3A, -3B, and -3C are heterogeneous between the two strains (Table 2), and the difference is not restricted to the repetitive amino acid coding regions.
The numbers of amino acids in viral latent proteins deduced from our sequencing results were compared with those deduced from previously determined EBV genome sequences, SNU719 (GenBank accession no. KP735248.1) (29) and YCCEL1 (GenBank accession no. LN827561.1) (5) (Table 2). Good concordance was found in the amino acid sequences of EBNA3A, LMP2A, and LMP2B. On the other hand, discrepancies were noticed for EBNA1, EBNA2, EBNA3B, EBNA3C, and LMP1 of the SNU719 EBV strain. A common feature of these viral proteins is that they contain repetitive amino acid coding regions. YCCEL1 sequencing data (LN827561.1) contain internal gaps corresponding to repetitive amino acid coding regions of EBNA1, EBNA2, EBNA3B, and EBNA3C. Thus, many of the discrepancies could be attributed to differences in the ability to assemble repetitive DNA sequences.
PacBio sequencing technology was not hampered by intergenic repetitive sequences. We were able to determine the sequences of the family of repeats (clustered 30-bp palindromic repeats that present the most difficulty for Sanger sequencing [19]) in both genomes. The results reveal that the region can be divided into subregions in which 30-bp units are aligned in opposite orientations (19) (Fig. 3B). The previous sequencing data of SNU719 and YCCEL1 EBV genomes contain internal gaps in the family of repeats region.
Taken together, these results demonstrate that BAC cloning of EBV episomes and subsequent PacBio single-molecule sequencing are powerful tools for obtaining complete sequences of various EBV strains.
Infection of B cells and epithelial cells with reconstituted viruses.
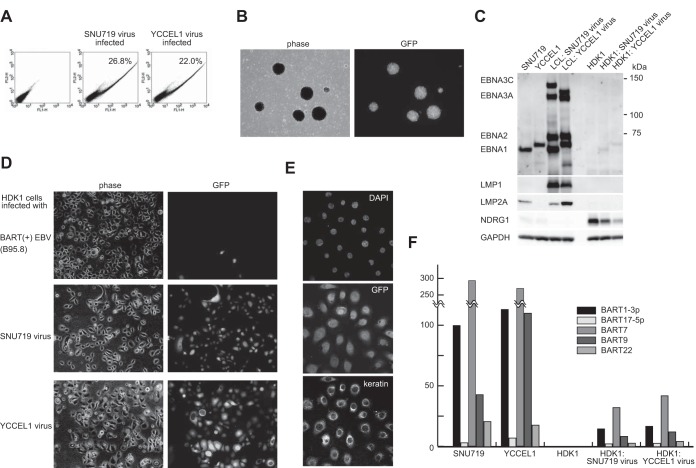
HEK293 cell clones stably transfected with either SNU719-BAC or YCCEL1-BAC were established, and EBV lytic replication was induced. GFP expression in recipient B cells was observed when culture supernatants of the induced HEK293 cells were used for infection, indicating the production of recombinant viruses (Fig. 4A). Primary B lymphocytes were then infected with the recombinant viruses, leading to the establishment of lymphoblastoid cell lines (LCLs) expressing GFP (Fig. 4B). Estimated 50% transforming doses (TD50) were approximately 104.5 TD50/ml for the SNU719-derived virus and 104.3 TD50/ml for the YCCEL1-derived virus. While the parental SNU719 and YCCEL1 cells were in type I latency (12, 13), in which only the EBNA1 protein is expressed, the established lymphoblastoid cell lines were in type III latency, with EBNA1, EBNA2, EBNA3A, EBNA3C, LMP1, and LMP2A expressed (Fig. 4C). Protein bands representing viral latent proteins of the SNU719 and YCCEL1 strains were found to migrate differently, and the results corresponded very well with the protein sizes deduced from our nucleotide-sequencing results (Fig. 4C and Table 2).
FIG 4.
Infection of B cells and epithelial cells with SNU719-BAC and YCCEL1-BAC viruses. (A) EBV-negative Akata cells were infected with the culture supernatants of HEK293-derived virus-producing cells. At 2 days postinfection, uninfected (left) and infected (center and right) cells were analyzed by fluorescence-activated cell sorting (488-nm excitation followed by FL1 and FL2 detection). Percentages of GFP-expressing cells, manifested by shifts in fluorescence intensity in the FL1 channel, are shown. (B) Phase-contrast and GFP images of a lymphoblastoid cell line established by the SNU719 virus. (C) Immunoblot analyses of latent viral gene expression in EBV-positive gastric cancer cells, LCLs, and immortalized HDK1 cells (uninfected and EBV infected). Whole-cell extracts (indicated at the top) were analyzed by immunoblotting using a human EBV-immune serum, anti-LMP1 antibody S-12, anti-LMP2A antibody 15F9, an antibody against NDRG1, and an antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) Phase-contrast and GFP images of immortalized keratinocytes (HDK1-K4DT cells) stably infected with various recombinant viruses (indicated on the left). BART(+) EBV is a derivative of EBV strain B95-8 with its BART microRNA clusters restored by using an EBV-BAC system (14). (E) Immortalized HDK1 cells stably infected with the SNU719-BAC virus were processed for immunofluorescence analysis using a pan-keratin antibody (bottom). Nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI) and GFP fluorescence are also shown. (F) The TaqMan small-RNA assay was used to determine viral microRNA expression levels in parental gastric cancer cells, uninfected HDK1 cells, and SNU719 or YCCEL1 virus-infected HDK1 cells. Relative microRNA expression levels for different cells were calculated by normalizing to hsa-miR-16 expression levels. Data are presented relative to average ebv-miR-BART1-3p expression levels in SNU719 cells, which were adjusted to 100.
We also tested whether the recombinant viruses derived from SNU719-BAC and YCCEL1-BAC could establish stable infections in epithelial cells. Either primary or immortalized primary epithelial cells were transduced with the cellular EBV receptor CR2 (CD21) and were then used as recipient cells. Although primary epithelial cells were transiently infected with the viruses, as revealed by GFP expression, GFP-positive cells never proliferated. In contrast, when immortalized human dermal keratinocytes (HDK1-K4DT cells) (15) were used as recipient cells, GFP-positive cells proliferated under hygromycin selection. Pools of 100% GFP-positive cells expressing keratins were established (Fig. 4D and E). The infected HDK1 cells expressed only traces of EBNA1, but not other viral latent proteins, indicating that the cells were in type I latency (Fig. 4C). Substantially lower levels of BamHI A rightward transcript (BART) microRNAs were expressed in infected HDK1 cells than in the parental gastric cancer cell lines (Fig. 4F). Still, downregulation of N-myc downstream regulated gene 1 (NDRG1), a previously identified BART microRNA target (14), was observed in EBV-infected cells (Fig. 4C), verifying that the microRNAs expressed were functional. Intact BAC clones were rescued from the pools of EBV-infected keratinocytes, indicating that the SNU719-BAC and YCCEL1-BAC viruses were episomally maintained (data not shown). This observation corresponds well with a previous report that telomerase-immortalized oral keratinocytes could be infected with Burkitt's lymphoma-derived EBV strain Akata, and EBV genomes were subsequently lost from the infected cells (30).
In contrast, when HDK1-K4DT cells were infected with a derivative of EBV strain B95-8 [BART(+) B95.8 virus (14)], the majority of hygromycin-resistant cells were GFP negative (Fig. 4D). The reasons underlying this phenomenon are currently unknown.
Stable infection with gastric cancer cell-derived EBV protects epithelial cells from the cytokilling activity of Ras oncoprotein expression.
SNU719 EBV-infected and uninfected control HDK1-K4DT cells, both of which were hygromycin resistant and GFP positive, grew at similar rates; therefore, the direct effects of EBV infection were obscure. Since EBV is thought to function as a cofactor in epithelial carcinogenesis, we predicted that EBV infection may result in phenotypic differences under certain circumstances, such as cellular oncoprotein expression. A gene encoding the phosphatidylinositol triphosphate (PI-3) kinase catalytic subunit (PIK3CA) is one of the most frequently mutated genes in EBV-positive gastric cancers (2). Since PI-3 kinase is a Ras effector, we examined the effect of Ras activation, which is known to induce senescence-like growth arrest in primary human fibroblasts (31) and nonapoptotic cell death in several human cancer cell lines (32). Hygromycin-resistant, EGFP-positive HDK1 cells were established by retrovirus infection and were used as non-EBV-infected control cells. EBV-infected HDK1-K4DT cells and control HDK1 cells were transduced with the active form of K-Ras (KRASG12V) fused to a mutant estrogen receptor ligand binding domain (ER–RAS). The ER-RAS fusion protein is expected to be degraded in the cytoplasm prior to 4-hydroxytamoxifen (4-OHT) treatment, and it is stabilized at the inner cytoplasmic membrane by the addition of 4-OHT, leading to Ras activation. Treating uninfected control cells with 4-OHT caused massive vacuolar degeneration and cell death within 4 days (Fig. 5A). In contrast, the majority of SNU719 EBV-infected cells survived longer than 4 days of 4-OHT treatment, with few cells undergoing vacuolar degeneration. The expression of an ER–RAS fusion protein in EBV-infected cells after 4-OHT treatment was verified by Western blotting (Fig. 5B). Essentially the same results were obtained when YCCEL1 EBV-infected HDK1 cells were subjected to the same assay (data not shown).
FIG 5.
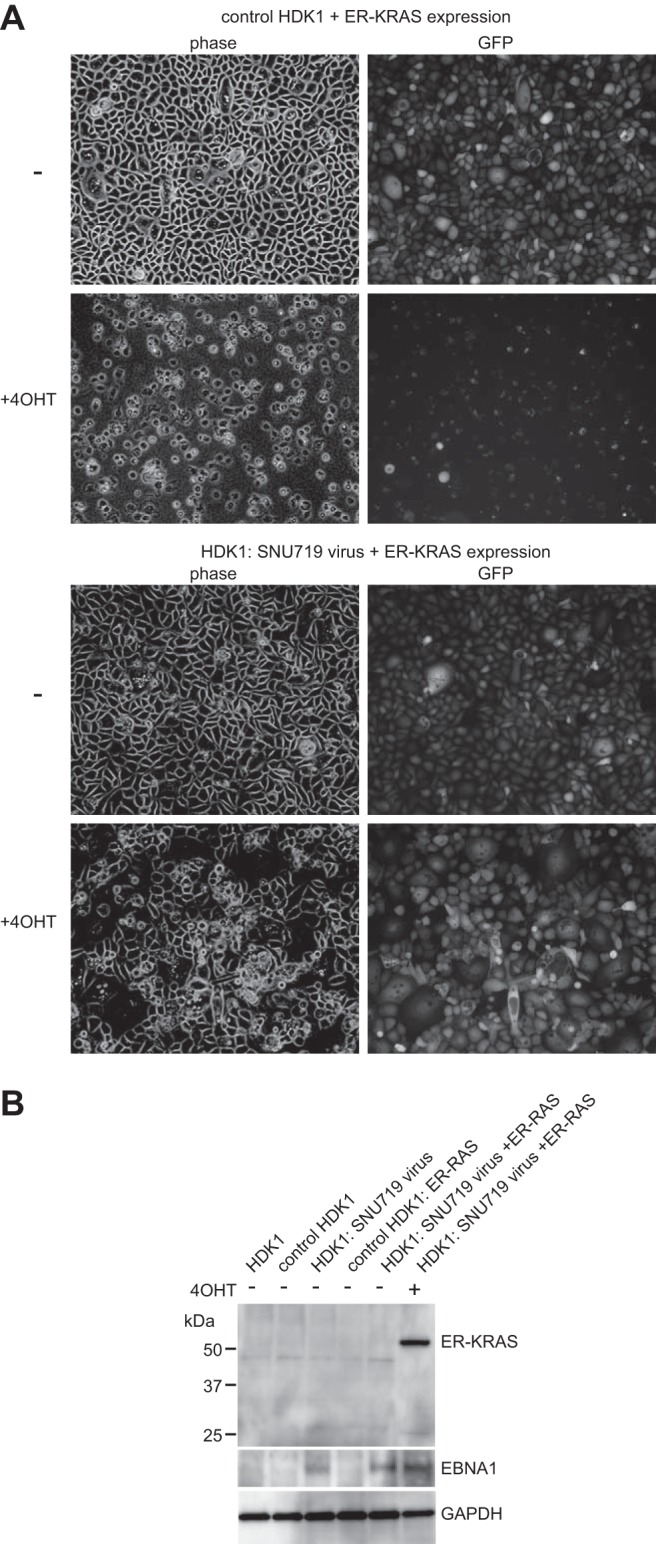
EBV infection of immortalized keratinocytes makes them resistant to oncogene-induced cell death. (A) Control HDK1 cells (EBV-negative, hygromycin-resistant, GFP-positive cells) were obtained by retrovirus infection. Control HDK1 cells and EBV-infected HDK1 cells (both GFP positive) were transduced with the ER–RAS fusion protein. Pools of transduced cells were treated with 1 nM 4-OHT for 4 days. Phase-contrast and GFP fluorescence images of untreated and 4-OHT-treated cells are shown. (B) Whole-cell extracts of uninfected and EBV-infected HDK1 cells (cultured with or without 4-OHT as indicated at the top) were analyzed by immunoblot analysis using an anti-Ras antibody and a human EBV-immune serum.
These results represent the first in vitro demonstration that gastric cancer-derived EBVs can protect epithelial cells from the cytokilling effect of Ras oncoprotein expression. The findings also imply that the limited numbers of viral gene products expressed in stably infected keratinocytes were sufficient to allow EBV to work as an important cofactor in the survival of keratinocytes following Ras activation.
DISCUSSION
Recent progress in DNA-sequencing technology has accelerated the sequencing of whole EBV genomes, and the repertoire of sequenced EBV genomes is increasing progressively. It is now clear that there are a large number of viral gene polymorphisms among EBV strains and that many of these are found in viral latent genes (4, 5). The possibility that EBV variant strains exist that are relevant to EBV-associated diseases has started to attract interest (7, 28, 33). Clearly, it will be important to sequence more disease-associated viral genomes and to reconstitute and experimentally test such viruses in order to determine whether they behave differently from nonpathogenic strains.
We used CRISPR/Cas9-mediated cleavage of EBV episomal DNA to facilitate the cloning of disease-associated viral strains. The cleavage stimulated the insertion of 12-kb transgenes via homology-directed repair. Importantly, minimal homology regions (230 bp upstream and 851 bp downstream) are necessary for this repair, making the construction of the donor plasmid extremely simple. Providing that the 1,088-bp target sequences (Fig. 1B) are identical, one can use exactly the same donor plasmid to clone multiple EBV strains. Given the high efficiency of homology-directed repair, 3 weeks of bulk selection of transduced cells was sufficient to obtain pools of drug-resistant cells, from which we successfully cloned EBV genomes. The entire cloning process could be performed within approximately 1 month.
As has been demonstrated for the sequencing of the pseudorabies virus genome (34), PacBio single-molecule sequencing technology is extremely useful for determining the sequences of repetitive regions. This technology allowed accurate determination of the numbers of W repeats (IR1), family of repeats, terminal repeats, and other repetitive sequences within EBNA protein-coding regions, as verified by the restriction enzyme digestion patterns of BAC clone DNAs and the sizes of proteins by Western blot analyses. Hence, PacBio sequencing has the clear advantage over other deep-sequencing methods of not being hampered by the repetitive sequences scattered throughout herpesvirus genomes.
Immortalized keratinocytes stably infected with recombinant SNU719 EBV and YCCEL1 EBV strains were established. In contrast, primary epithelial cells entered senescence after EBV infection. This corresponds well with a previous observation that overexpression of cyclin D1 in precancerous tissue is a prerequisite for susceptibility to stable EBV infection (35). We took advantage of an experimental system in which immortalized human keratinocytes undergo vacuolar degeneration and cell death following Ras activation. This is most likely nonapoptotic cell death, as has been demonstrated for human cancer cells (32); whether the same applies to normal human epithelial cells remains to be clarified. Importantly, stable infection with gastric cancer cell-derived EBVs made the infected keratinocytes resistant to oncogene-induced vacuolar degeneration and cell death. We speculate that this observation faithfully reflects epithelial carcinogenesis in vivo, to which EBV contributes as a cofactor. Epithelial carcinogenesis in vivo likely takes much longer than EBV-mediated B-cell transformation and involves multiple genetic and epigenetic alterations of cellular genes (36). A possible scenario is that the presence or absence of EBV infection determines the destiny of precancerous gastric epithelia that encounter additional oncogene activation. Although EBV-infected keratinocytes escaped cell death after ER-RAS expression, they failed to proliferate further. This could be due to the nonphysiological level of ER-RAS expression. There remains much room for improvement in producing a more physiological experimental system.
Whether viral sequence heterogeneity affects viral phenotypic variation remains an open question. Although not proven, it may be that only a specific EBV strain can act as a strong cofactor during epithelial carcinogenesis. One intriguing observation is that gastric cancer cell-derived EBVs, but not lymphoblast-derived EBVs, could establish stably infected keratinocytes with 100% GFP expression (Fig. 4D). Since these EBV strains have different geographical origins, the difference observed may imply that EBV strains with different biological properties are distributed in geographically different areas in the world. Alternatively, this observation raises the possibility that gastric cancer cell-derived EBVs have specific phenotypes. Final proof should be obtained by identifying sequence variations and phenotypic differences between (i) EBV maintained in B cells and (ii) EBV maintained in cancer tissue from the same individual. Such experiments are now theoretically possible, since the CRISPR/Cas9-mediated EBV targeting strategy can be applied to spontaneously outgrown lymphoblastoid cell lines.
It is also of interest to investigate whether some specific EBV strains are linked with other EBV-associated diseases. For example, chronic active EBV-associated T/NK-cell lymphoproliferative diseases are rare but are more prevalent in East Asian countries than in Western countries (37), suggesting that EBV strains with specific geological distributions may be relevant to disease etiology. Functional comparisons of EBVs derived from patients with EBV-related diseases and those derived from healthy individuals would enable testing of this hypothesis. Viral genome heterogeneity could significantly affect viral phenotypes, such as infectivity for target cells (B cells, NK/T cells, and epithelial cells), immunogenicity, and pathogenicity. The experimental system described in this study should contribute to the further clarification of the biological significance of viral strain variation and to the identification of potentially disease specific variants.
ACKNOWLEDGMENTS
We thank the Korean Cell Line Bank and Sun Young Rha for providing us with SNU719 cells and YCCEL1 cells, respectively, and Mamiko Miyata for technical assistance.
REFERENCES
- 1.Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2603–2654. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao SW, Tsang CM, Pang PS, Zhang G, Chen H, Lo KW. 2012. The biology of EBV infection in human epithelial cells. Semin Cancer Biol 22:137–143. doi: 10.1016/j.semcancer.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Lin Z, Wang X, Strong MJ, Concha M, Baddoo M, Xu G, Baribault C, Fewell C, Hulme W, Hedges D, Taylor CM, Flemington EK. 2013. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J Virol 87:1172–1182. doi: 10.1128/JVI.02517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok H, Wu CW, Palser AL, Kellam P, Sham PC, Kwong DL, Chiang AK. 2014. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol 88:10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai MH, Raykova A, Klinke O, Bernhardt K, Gartner K, Leung CS, Geletneky K, Sertel S, Munz C, Feederle R, Delecluse HJ. 2013. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep 5:458–470. doi: 10.1016/j.celrep.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Quake SR. 2014. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A 111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. 2015. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J Gen Virol 96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 10.Bi Y, Sun L, Gao D, Ding C, Li Z, Li Y, Cun W, Li Q. 2014. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog 10:e1004090. doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suenaga T, Kohyama M, Hirayasu K, Arase H. 2014. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol Immunol 58:513–522. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh ST, Seo JS, Moon UY, Kang KH, Shin DJ, Yoon SK, Kim WH, Park JG, Lee SK. 2004. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology 320:330–336. doi: 10.1016/j.virol.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Kim DN, Seo MK, Choi H, Kim SY, Shin HJ, Yoon AR, Tao Q, Rha SY, Lee SK. 2013. Characterization of naturally Epstein-Barr virus-infected gastric carcinoma cell line YCCEL1. J Gen Virol 94:497–506. doi: 10.1099/vir.0.045237-0. [DOI] [PubMed] [Google Scholar]
- 14.Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. 2015. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J Virol 89:2684–2697. doi: 10.1128/JVI.03189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egawa N, Nakahara T, Ohno S, Narisawa-Saito M, Yugawa T, Fujita M, Yamato K, Natori Y, Kiyono T. 2012. The E1 protein of human papillomavirus type 16 is dispensable for maintenance replication of the viral genome. J Virol 86:3276–3283. doi: 10.1128/JVI.06450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimada A, Dohke K, Sadaie M, Shinmyozu K, Nakayama J, Urano T, Murakami Y. 2009. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev 23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda T, Yajima M, Ahsan N, Tanaka M, Takada K. 2004. Production of high-titer Epstein-Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J Virol 78:7004–7015. doi: 10.1128/JVI.78.13.7004-7015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanda T, Shibata S, Saito S, Murata T, Isomura H, Yoshiyama H, Takada K, Tsurumi T. 2011. Unexpected instability of Family of Repeats (FR), the critical cis-acting sequence required for EBV latent infection, in EBV-BAC Systems. PLoS One 6:e27758. doi: 10.1371/journal.pone.0027758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 21.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 22.de Jesus O, Smith PR, Spender LC, Elgueta Karstegl C, Niller HH, Huang D, Farrell PJ. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J Gen Virol 84:1443–1450. doi: 10.1099/vir.0.19054-0. [DOI] [PubMed] [Google Scholar]
- 23.Tcherepanov V, Ehlers A, Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 25.Dajee M, Tarutani M, Deng H, Cai T, Khavari PA. 2002. Epidermal Ras blockade demonstrates spatially localized Ras promotion of proliferation and inhibition of differentiation. Oncogene 21:1527–1538. doi: 10.1038/sj.onc.1205287. [DOI] [PubMed] [Google Scholar]
- 26.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng MS, Li DJ, Liu QL, Song LB, Li MZ, Zhang RH, Yu XJ, Wang HM, Ernberg I, Zeng YX. 2005. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J Virol 79:15323–15330. doi: 10.1128/JVI.79.24.15323-15330.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feederle R, Klinke O, Kutikhin A, Poirey R, Tsai MH, Delecluse HJ. 2015. Epstein-Barr virus: from the detection of sequence polymorphisms to the recognition of viral types. Curr Top Microbiol Immunol 390:119–148. doi: 10.1007/978-3-319-22822-8_7. [DOI] [PubMed] [Google Scholar]
- 29.Song KA, Yang SD, Hwang J, Kim JI, Kang MS. 2015. The full-length DNA sequence of Epstein Barr virus from a human gastric carcinoma cell line, SNU-719. Virus Genes 51:329–337. doi: 10.1007/s11262-015-1248-z. [DOI] [PubMed] [Google Scholar]
- 30.Birdwell CE, Queen KJ, Kilgore PC, Rollyson P, Trutschl M, Cvek U, Scott RS. 2014. Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes. J Virol 88:11442–11458. doi: 10.1128/JVI.00972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 32.Chi S, Kitanaka C, Noguchi K, Mochizuki T, Nagashima Y, Shirouzu M, Fujita H, Yoshida M, Chen W, Asai A, Himeno M, Yokoyama S, Kuchino Y. 1999. Oncogenic Ras triggers cell suicide through the activation of a caspase-independent cell death program in human cancer cells. Oncogene 18:2281–2290. doi: 10.1038/sj.onc.1202538. [DOI] [PubMed] [Google Scholar]
- 33.Farrell PJ. 2015. Epstein-Barr virus strain variation. Curr Top Microbiol Immunol 390:45–69. doi: 10.1007/978-3-319-22822-8_4. [DOI] [PubMed] [Google Scholar]
- 34.Tombácz D, Sharon D, Olah P, Csabai Z, Snyder M, Boldogkoi Z. 2014. Strain Kaplan of pseudorabies virus genome sequenced by PacBio single-molecule real-time sequencing technology. Genome Announc 2:e00628–14. doi: 10.1128/genomeA.00628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang CM, Yip YL, Lo KW, Deng W, To KF, Hau PM, Lau VM, Takada K, Lui VW, Lung ML, Chen H, Zeng M, Middeldorp JM, Cheung AL, Tsao SW. 2012. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc Natl Acad Sci U S A 109:E3473–E3482. doi: 10.1073/pnas.1202637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X, Tang CM, Kang W, Lung RW, Li JW, Chan TF, Xing R, Lu Y, Lo KW, Wong N, To KF, Yu C, Chan FK, Sung JJ, Yu J. 2014. Integrative identification of Epstein-Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology 147:1350–1362.e4. doi: 10.1053/j.gastro.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 37.Kimura H, Ito Y, Kawabe S, Gotoh K, Takahashi Y, Kojima S, Naoe T, Esaki S, Kikuta A, Sawada A, Kawa K, Ohshima K, Nakamura S. 2012. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood 119:673–686. doi: 10.1182/blood-2011-10-381921. [DOI] [PubMed] [Google Scholar]
- 38.Kanda T, Kamiya M, Maruo S, Iwakiri D, Takada K. 2007. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J Cell Sci 120:1529–1539. doi: 10.1242/jcs.03434. [DOI] [PubMed] [Google Scholar]