ABSTRACT
Virus entry into cells is a multistep process that often requires the subversion of subcellular machineries. A more complete understanding of these steps is necessary to develop new antiviral strategies. While studying the potential role of the actin network and one of its master regulators, the small GTPase Cdc42, during Junin virus (JUNV) entry, we serendipitously uncovered the small molecule ZCL278, reported to inhibit Cdc42 function as an entry inhibitor for JUNV and for vesicular stomatitis virus, lymphocytic choriomeningitis virus, and dengue virus but not for the nonenveloped poliovirus. Although ZCL278 did not interfere with JUNV attachment to the cell surface or virus particle internalization into host cells, it prevented the release of JUNV ribonucleoprotein cores into the cytosol and decreased pH-mediated viral fusion with host membranes. We also identified SVG-A astroglial cell-derived cells to be highly permissive for JUNV infection and generated new cell lines expressing fluorescently tagged Rab5c or Rab7a or lacking Cdc42 using clustered regularly interspaced short palindromic repeat (CRISPR)-caspase 9 (Cas9) gene-editing strategies. Aided by these tools, we uncovered that perturbations in the actin cytoskeleton or Cdc42 activity minimally affect JUNV entry, suggesting that the inhibitory effect of ZCL278 is not mediated by ZCL278 interfering with the activity of Cdc42. Instead, ZCL278 appears to redistribute viral particles from endosomal to lysosomal compartments. ZCL278 also inhibited JUNV replication in a mouse model, and no toxicity was detected. Together, our data suggest the unexpected antiviral activity of ZCL278 and highlight its potential for use in the development of valuable new tools to study the intracellular trafficking of pathogens.
IMPORTANCE The Junin virus is responsible for outbreaks of Argentine hemorrhagic fever in South America, where 5 million people are at risk. Limited options are currently available to treat infections by Junin virus or other viruses of the Arenaviridae, making the identification of additional tools, including small-molecule inhibitors, of great importance. How Junin virus enters cells is not yet fully understood. Here we describe new cell culture models in which the cells are susceptible to Junin virus infection and to which we applied CRISPR-Cas9 genome engineering strategies to help characterize early steps during virus entry. We also uncovered ZCL278 to be a new antiviral small molecule that potently inhibits the cellular entry of the Junin virus and other enveloped viruses. Moreover, we show that ZCL278 also functions in vivo, thereby preventing Junin virus replication in a mouse model, opening the possibility for the discovery of ZCL278 derivatives of therapeutic potential.
INTRODUCTION
The Junin virus (JUNV) is an important member of the New World arenavirus family (clade B) and is responsible for outbreaks of Argentine hemorrhagic fever in South America, where 5 million people are considered to be at risk (1). The reservoir of JUNV is the rodent Calomys musculinus, a known healthy carrier (although infertile), from which the virus can cross the species barrier and infect humans (2). Several arenaviruses, including JUNV, are classified as category A pathogens by the Centers for Disease Control and Prevention and considered potential bioterrorism agents, requiring proactive research and the development of preventive and therapeutic antiviral treatments (3).
JUNV is an enveloped virus composed of a small segment and a large segment of ambisense single-stranded RNA encoding four proteins: the nucleoprotein (NP), the envelope glycoprotein (GP) complex, the RNA-dependent RNA polymerase (L), and the matrix protein (Z). The GP complex is first expressed as a polyprotein precursor that is cleaved into the stable signal peptide (SSP), GP1, and GP2, with SSP being involved in GP intracellular trafficking and membrane fusion (4).
Interventions for JUNV infection are currently limited. The JUNV strain Candid #1 is used as a live-attenuated vaccine in regions where JUNV is endemic and shows strong protection efficacy in humans (5). Injection of immune plasma from a previously JUNV-infected patient is currently the only option that improves a patient's prognosis (6). Therefore, safe and reliable therapies should be developed to face potential future outbreaks. The use of antiviral compounds targeting entry steps of the viral cycle represents an attractive approach to fight infections upstream of viral replication.
JUNV entry can be dissected into several steps: attachment of virus particles to the cell surface, internalization into endosomal compartments, and penetration of the viral genome into the cytosol. JUNV requires cellular entry factors, including the transferrin (Tf) receptor and the voltage-gated calcium channel, to attach to cells and further fuse with cellular membranes (7, 8). Fusion allows the release of the ribonucleoproteins (RNPs), composed of the viral genomic RNA protected by NP proteins, into the cytosol of the infected cell. Available evidence supports the possibility that the internalization of JUNV depends on clathrin-mediated endocytosis and requires subsequent endosomal acidification for membrane fusion and penetration (9–11). The early and late endosomal compartments, as identified by the small GTPases Rab5 and Rab7, respectively, have also been proposed to be important for efficient JUNV infection (12). Entry of JUNV is thought to depend in part on the integrity of the actin network, on the basis of the findings of experiments performed using long-term treatment with actin polymerization inhibitors (13, 14). One difficulty with the interpretation of these results, however, is the possibility that the inhibitory effects are indirect, given the importance of the actin cytoskeleton in many other cellular functions.
Cdc42 is a small GTPase protein known to regulate actin polymerization (15, 16). The activity of Cdc42 can be regulated by the guanine nucleotide exchange factor intersectin (17–19). A virtual in silico screen based on the three-dimensional structure of Cdc42 and intersectin identified the small molecule ZCL278 [4-{3-[2-(4-bromo-2-chloro-phenoxy)-acetyl]-thioureido}-N-(4,6-dimethyl-pyrimidin-2-yl)-benzenesulfonamide] to be an inhibitor of Cdc42 function with the potential to insert into the Cdc42 groove that interacts with intersectin (20). In cells, ZCL278 was shown to efficiently inhibit chemically induced filopodium formation, a process dependent on Cdc42 activity.
Here, we identify ZCL278 to be a potent inhibitor of JUNV cellular entry. Unexpectedly, we found that the entry of JUNV is mostly independent of Cdc42 and actin. Investigation of the step at which ZCL278 inhibits JUNV entry demonstrates that viral attachment and the internalization of JUNV particles are unaffected. In contrast, ZCL278 decreases the amount of the JUNV RNP cores released into the cytosol and prevents pH-dependent GP-mediated fusion. Moreover, we observed that the intracellular trafficking of JUNV particles is perturbed in ZCL278-treated cells, with three times more particles being associated with proteolytically active lysosomal compartments. The distribution of JUNV particles within early/late endosomes in cells treated with ZCL278 is also severely impaired compared to that in untreated cells. In the absence of infection, ZCL278 treatment of cells significantly increases the degradation of exogenous bovine serum albumin (BSA) but has no effect on the degradation of endogenous epidermal growth factor receptor (EGFR) or on endosomal/lysosomal acidity. We show that ZCL278 antiviral activity also extends to vesicular stomatitis virus (VSV), lymphocytic choriomeningitis virus (LCMV), and dengue virus serotype 2 (DV2), while poliovirus type 1 (PV1) was unaffected. Finally, ZCL278 is extremely potent in preventing JUNV replication in a mouse model, demonstrating that ZCL278 or related compounds could be considered therapeutic candidates to prevent JUNV-related diseases.
MATERIALS AND METHODS
Reagents.
ZCL278 [4-{3-[2-(4-bromo-2-chloro-phenoxy)-acetyl]-thioureido}-N-(4,6-dimethyl-pyrimidin-2-yl)-benzenesulfonamide] was purchased from Tocris Bioscience, latrunculin A (Lat A) and latrunculin B (Lat B) were purchased from EMD Millipore, and jasplakinolide (Jasp) was purchased from Enzo Life Sciences. Ribavirin, chloroquine (CQ), bafilomycin A1 (BafA), and cytochalasin D (Cyto D) were obtained from Sigma-Aldrich, and green dequenched (DQ) BSA, Tf from human serum, and Alexa Fluor 647 (A647) conjugate were obtained from Life Technologies. The CellTiter-Glo luminescent cell viability assay was obtained from Promega. The rabbit polyclonal anti-Cdc42 antibody (antibody sc-87) was purchased from Santa Cruz. The rabbit monoclonal anti-Lamp1 antibody (antibody D2D11) was purchased from Cell Signaling. The mouse monoclonal anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (antibody 6C5) was obtained from GeneTex. The mouse anti-EGFR antibody (antibody 13C9) was obtained from Genentech. The following antibodies targeting JUNV proteins have been previously characterized (21) and were provided by BEI Resources: SA02 mouse monoclonal anti-NP antibody (clone SA02-BG12; catalog number NR-2573), QB06 mouse monoclonal anti-NP antibody (clone QB06-AE05; catalog number NR-2576), LD05 mouse monoclonal anti-GP antibody (clone LD05-BF09; catalog number NR-48833), GD01 mouse monoclonal anti-GP antibody (clone GD01-AG02; catalog number NR-43776), and GB03 mouse monoclonal anti-GP antibody (clone GB03-BE08; catalog number NR-2564). Antibodies were coupled to Alexa Fluor 488 (A488), Alexa Fluor 568 (A568), or Alexa Fluor 647 carboxylic acid (succinimidyl ester) or biotin-XX (sulfosuccinimidyl ester) from Life Technologies. The plasmid pcDNA3-EGFP-Cdc42(wt) and pcDNA3-EGFP-Cdc42(T17N) were gifts from Klaus Hahn (Addgene plasmids 12599 and 12601, respectively). All oligonucleotides were synthesized by Integrated DNA Technologies, and sequencing analyses were performed by Macrogen Corp. The compound 17C9 was obtained from Dale Boger's lab and corresponds to the optically active S-enantiomer form at 10 mM resuspended in dimethyl sulfoxide (22, 23).
Cell maintenance.
Vero, HEK 293FT, Huh7, and SVG-A cells (ATCC) were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) with GlutaMAX (Life Technologies-Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals) and 100 IU ml−1 penicillin–100 mg ml−1 streptomycin (Life Technologies-Gibco). SUM159 human breast carcinoma cells (24) were cultured in DMEM–F-12 medium–GlutaMAX (Life Technologies-Gibco) supplemented with 5% heat-inactivated FBS, 100 IU ml−1 penicillin, 100 mg ml−1 streptomycin, 1 μg ml−1 hydrocortisone, and 5 μg ml−1 insulin (Sigma-Aldrich). A549 cells were cultured in DMEM–F-12 medium–GlutaMAX supplemented with 10% heat-inactivated FBS, 100 IU ml−1 penicillin, and 100 mg ml−1 streptomycin. Virus medium corresponds to DMEM-GlutaMAX supplemented with 2% heat-inactivated FBS, 100 IU ml−1 penicillin, and 100 mg ml−1 streptomycin.
Virus production.
Throughout this article, Junin virus or JUNV refers to the nonpathogenic vaccine strain Candid #1 (obtained from the S. Bavari laboratory at USAMRIID), which was the only strain used in this study. Production of JUNV was performed by inoculation of JUNV (passage 2 or 3) onto subconfluent Vero cells in a T170 flask at a multiplicity of infection (MOI) of 0.5 in 3 ml of virus medium. After 1 h, additional medium was provided, and cells were incubated for 72 h at 37°C with 5% CO2. The supernatant was harvested and cleared of cellular debris by centrifugation at 10,000 × g at 4°C for 10 min. The virus-containing supernatant was frozen at −80°C before use. The Armstrong strain of LCMV used in this study was prepared as described above for JUNV. Viruses were titrated by plaque assay on Vero cells. DV2 and PV1 were grown as described elsewhere (25).
Virus infectivity assays.
Plaque assays were performed as previously described (25, 26). For flow cytometry, cells were harvested at 16 h postinfection, fixed with 4% paraformaldehyde, and permeabilized with 0.5% bovine serum albumin (BSA) and 0.05% saponin in phosphate-buffered saline (PBS), followed by incubation with the SA02 mouse monoclonal antibody (2 μg ml−1; clone SA02-BG12; catalog number NR-2573; BEI Resources) specific for the JUNV nucleoprotein (NP) and coupled to Alexa Fluor 488 or Alexa Fluor 647. Measurement of the mean fluorescence intensity of infected cells was performed on a FACSCanto II flow cytometer with 488- and 640-nm lasers (BD Biosciences). Analysis of the percentage of infected cells was completed using FlowJo software (TreeStar Inc.).
Virus labeling.
The assay for the labeling of JUNV with A647 was performed as described previously (26). A JUNV stock was incubated for 30 min at 25°C with a nonneutralizing mouse monoclonal antibody raised against the JUNV GP (4 μg ml−1; clone LD05) coupled to Alexa Fluor 647 (Life Technologies). The mixture was gently applied on top of a 10% OptiPrep cushion (Sigma-Aldrich), and the combination was ultracentrifuged at 150,000 × g for 2 h at 4°C using an SW55 Ti rotor (Beckman Coulter). The supernatant was discarded, and the pellets were resuspended in virus medium overnight in the dark at 4°C for gentle resolubilization.
RNA interference.
Transfection of small interfering RNAs (siRNAs) was performed as described previously (27). Briefly, 10 nM siRNA (FlexiTube siRNA; Qiagen) was used for the transfection of Vero cells with the Lipofectamine RNAiMax reagent (Life Technologies). The cells were washed at 6 h posttransfection and replated 2 days later. On day 3, the cells were either infected or lysed for RNA expression analysis. The siRNA target sequences used in this study were TTCAGCAATGCAGACAATTAA for the first siRNA targeting Cdc42 (siCdc42#1) and CATCAGATTTGAAATATTTAA for the second siRNA targeting Cdc42 (siCdc42#2) and are proprietary for siAllstar.
RNA analysis.
Total RNA from cells was purified using an RNeasy minikit (Qiagen) and was used as the template for cDNA synthesis (iScript kit; Bio-Rad) according to the manufacturer's instructions. Quantitative PCR (qPCR) assays used iQ SYBR green supermix (Bio-Rad) and were performed on a MyiQ iCycler machine (Bio-Rad) as follows: an initial PCR activation step of 5 min at 95°C and 40 cycles of denaturation and combined annealing/extension for 15 s at 95°C and 30 s at 60°C, respectively. Fluorescence data collection was performed at the end of each cycle, and a melt curve that showed the specificity of the primers used and that started at 50°C and increased 0.5°C every 10 s for 90 cycles was included at the end of each PCR. Viral and cellular complementary DNAs were detected using forward and reverse primers specific for PV1 (primers 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TTGATTTTGGAGGGATCTCG-3′, respectively), as described previously (28), and GAPDH (primers 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TTGATTTTGGAGGGATCTCG-3′, respectively).
Knock-in using CRISPR-Cas9.
To generate a small guide RNA (sgRNA) containing a specific sequence targeting either Rab5c or Rab7a, a free PCR product strategy was used (29). This involved PCR amplification of the U6 promoter using a unique forward primer (5′-GCCGGTACCTGAGGGCCTATTTCCC-3′) and a reverse primer that anneals with the end of the U6 promoter, followed by the 20-nucleotide target sequence, as well as the clustered regularly interspaced short palindromic repeat (CRISPR) RNA recognized by the caspase 9 (Cas9) enzyme. The 134-base primer was purchased as an ultramers from Integrated DNA Technologies and has the following sequences: 5′-ACCTCTAGAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACNNNNNNNNNNNNNNNNNNNNCGGTGTTTCGTCCTTTCCACAAG-3′. The underlined nucleotides correspond to the complementary sequence of the targeted DNA (see Fig. 6). The PCR amplification was performed using Herculase II DNA polymerase (Agilent), and the PCR cycles were set as follows: an initial activation step of 2 min at 95°C; 30 cycles of denaturation at 95°C for 30 s, annealing at 66°C for 30 s, and extension at 72°C for 90 s; and a final extension of 3 min at 72°C. Upon electrophoresis in 1% agarose, an ∼430-bp band was obtained, and gel purification was performed using a Zymoclean gel DNA recovery kit (Zymo Research). Aliquots were kept at −20°C before use.
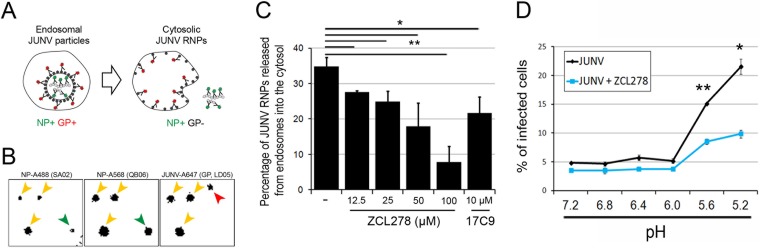
FIG 6.
ZCL278 prevents the release of JUNV RNP into the cytosol and GP-mediated fusion. (A to C) JUNV material released into the cytosol, measured using immunofluorescence labeling. (A) (Left) NP+ and GP+ objects that are considered to be JUNV particles with an intact envelope within endosomes; (right) NP+ and GP− JUNV material released from endosomes. (B, C) To detect both states, Vero cells were left untreated or treated with increasing amounts of ZCL278 and incubated with JUNV-A647 for 30 min at 37°C. Cells were stained with two noncompetitive anti-NP antibodies coupled to different fluorophores (NP-A488 and NP-A568). (B) Representative snapshots of the inverted fluorescence intensity for each individual channel. Yellow arrowheads, objects positive for all three fluorophores and thus considered to be NP+ and GP+ particles; green arrowheads, NP+ and GP− objects considered to be released events; all detected objects were classified as either NP+ and GP+ or NP+ and GP−; red arrowhead, a GP+ and NP− object (not taken into account in the following analysis) that possibly represents an endosome with which fusion already occurred. (C) Quantification of the percentage of JUNV material (NP+ and GP−) released from endosomes. The data are the averages ± SDs from two or three experiments, with each experiment corresponding to 10 fields of view with more than 100 objects counted per condition. Statistical analysis using the two-tailed Student t test was used to show significance. *, P < 0.05; **, P < 0.005. (D) Viral fusion at the plasma membrane was artificially induced by placing Vero cells on which JUNV virions were prebound at 4°C, followed by dropping the pH of the extracellular medium in the presence or absence of 50 μM ZCL278. The histogram represents the percentage of infected cells for the different conditions measured by flow cytometry. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition, and the histogram is representative of the data from two independent experiments. Statistical analysis for significance was performed using the two-tailed Student t test. *, P < 0.05; **, P < 0.005.
Donor DNA templates were generated from genomic DNA extracted from SVG-A cells by PCR amplification reactions using a QIAamp DNA minikit (Qiagen). The fragments obtained with primers F1-R1 and F3-R3 spanned ≈800 nucleotides upstream and 800 nucleotides downstream of the ATG codon of either Rab5c or Rab7a. The open reading frame encoding enhanced green fluorescent protein (EGFP) and a flexible linker with an amino acid sequence of GGSGGSGGS were obtained by PCR using primers F2-R2 from an expression plasmid vector. The PCR products and a pUC19 plasmid linearized using the SmaI restriction enzyme were annealed with each other using Gibson assembly master mix (New England BioLabs), according to the manufacturer's instructions. The reaction product was transformed into competent TOP10 cells, and sequencing of the bacterial colonies was performed.
SVG-A cells were cotransfected with 0.8 μg of the plasmid coding for Streptococcus pyogenes Cas9, 0.8 μg of the free PCR product coding for the target sgRNA, and 0.8 μg of the template donor plasmid using the Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer's instructions. Cells were expanded for 7 to 10 days and prepared for bulk sorting using a FACSAria II cell sorter (BD Biosciences) with an 85-μm nozzle. The EGFP-positive population was initially ∼1% and was enriched over 2 or 3 subsequent bulk sortings. When the EGFP-positive population reached at least 50%, sorting of single cells into 96-well plates was performed, and the subsequent monoclonal populations were expanded and screened for endogenous/edited alleles by PCR using forward primer 5′-GAGCCTGAAGTTGGGAGACC-3′ and reverse primer 5′-CATGCCCACTCACCTCCAAT-3′ for Rab5c and forward primer 5′-GCGGTCACTTCTTTGAGAAAGT-3′ and reverse primer 5′-AAGTGGCAGCACGGACAGTGT-3′ for Rab7a. We selected two clones, named EGFP-Rab5c+/+ and EGFP-Rab7a+/−, based on positive PCR identification.
Knockout using CRISPR-Cas9.
The knockout of specific genes was performed using a CRISPR-Cas9 lentiviral vector (lentiCRISPR) (30) expressing Streptococcus pyogenes Cas9 and the selection marker puromycin flanked by the long terminal repeats necessary for lentiviral production. The vector also contained a U6 promoter followed by two BsmBI restriction sites to allow the insertion of double-stranded material with a CACCG overhang at the 5′ end of one strand and overhangs of CAAA at the 5′ end and C at the 3′ end of the other complementary strand. The detailed digestion/ligation protocol is described elsewhere (30). Lentiviral production was performed by cotransfecting HEK 293FT cells with the lentiCRISPR construct containing the guide RNA (gRNA) of interest, the psPAX2 plasmid coding for the packaging lentiviral proteins, and pVSV-G coding for the GP envelope from VSV using the transfection reagent Trans-IT 2020 (Mirus Bio LLC) according to the manufacturer's instructions. The cells were washed at 6 to 10 h posttransfection, and the supernatant was harvested 3 days later. Lentiviral infection was performed by adding pure lentivirus-containing supernatant to cells in the presence of 10 μg ml−1 Polybrene (EMD Millipore) for 6 to 10 h. The cells were washed, incubated for a further 36 h in cell culture medium, and subsequently incubated with 4 μg ml−1 puromycin (InvivoGen) for 4 days. The medium was switched to normal culture medium for at least 24 h before experiments were performed.
Transferrin uptake.
The Tf uptake assay was performed as described previously (27). Briefly, cells were incubated for 10 min with 5 μg ml−1 transferrin-Alexa Fluor 647 (Tf-A647; Life Technologies) at 4°C or 37°C. After incubation, the plates were cooled on ice, rinsed with cold PBS, and harvested or first briefly incubated twice with 150 mM NaCl, 1 mM MgCl2, 0.125 mM CaCl2, 0.1 M glycine, pH 2.5 (referred to as the acid wash), to remove the surface-bound Tf-A647. The cells were resuspended in 200 μl PBS containing 1% BSA and 0.5 mM EDTA at 4°C. Measurement of the fluorescence intensity, reflecting the amount of Tf-A647 for each cell, was determined by flow cytometry using the 633-nm laser line of the FACSCanto II flow cytometer (BD Biosciences).
EGFR degradation assay.
The EGFR degradation assay was performed in a way similar to that previously described (31). Cells were serum starved for 2 h in minimal essential medium alpha (Gibco) and treated with 250 ng ml−1 epidermal growth factor (EGF) for an additional 2 h. Cells were subsequently fixed, permeabilized, stained with a mouse anti-EGFR antibody and a secondary anti-mouse IgG antibody coupled to A647, and analyzed by flow cytometry. The EGFR mean fluorescence intensity was measured by flow cytometry using a FACSCanto II flow cytometer. Cells were untreated or treated with 50 μM ZCL278 or 1 μM Jasp from 15 min prior to EGF addition to the end of the EGF incubation.
LTR staining.
Cells were incubated with 50 nM LysoTracker red (LTR) for 5 min at 20°C. The cells were detached by trypsinization and kept on ice for the following two washes using cold PBS. The LTR mean fluorescence intensity was measured by flow cytometry using a FACSCanto II flow cytometer.
Plasma membrane fusion assay.
Cells were cooled on ice for 20 min, followed by the inoculation of cold JUNV at an MOI of 5. Cells were washed in cold PBS and incubated with solutions at the pH indicated below at 4°C for an additional 30 min in the presence or absence of 50 μM ZCL278. Viral fusion was blocked by incubating the cells with 4 μg/ml neutralizing antibody for 15 min at 4°C. The cells were subsequently shifted to 37°C for 16 h, and the percentage of infected cells was determined by flow cytometry.
Immunofluorescence.
Infected cells grown on coverslips were fixed with 4% paraformaldehyde for at least 15 min, permeabilized with 0.5% BSA and 0.05% saponin in PBS (see Fig. 5) or 5% BSA and 0.1% Triton X-100 (see Fig. 6), and incubated with the following primary antibodies at the indicated concentrations: anti-NP SA02 and QB06 antibodies and anti-GP GB03 antibody at 2 μg ml−1 and anti-Lamp1 antibody at a 1:100 dilution. The acquisition of fluorescence intensity was performed on a Zeiss AxioObserver.Z1 inverted microscope mounted with a spinning disk head (Yokogawa), a QuantEM:512SC electron-multiplying charge-coupled-device camera (EMCCD; Photometrics), and 63× (see Fig. 5) or 100× (see Fig. 6) (numerical aperture, 1.4) oil objectives (Zeiss). Each acquisition corresponds to stacks spaced by 0.25 to 0.5 μm that spanned the entire cell volume. Image processing started by creating a maximum-intensity z projection for each channel, followed by segmentation using ImageJ software (version 1.48d). Binary masks were created, and the masks from NP signal images were multiplied, resulting in a new binary mask corresponding to all JUNV NP-positive (NP+) objects (total NP+ objects). This mask was then multiplied by the GP mask, generating a new mask corresponding to objects positive for both NP and GP (GP+) (NP+ and GP+ objects). The amount of total NP+ objects and NP+ and GP+ objects was measured using the Analyze Particles plug-in. The percentage of NP+ and GP-negative (GP−) objects was calculated by subtracting the number of NP+ and GP+ objects from the total number of NP+ objects. The number of virus particles that were naturally devoid of GP (NP+ and GP− objects) was estimated by spreading JUNV onto glow-discharged glass coverslips. Staining and determination of NP+ and GP− objects were performed following the same protocol described above for cells.
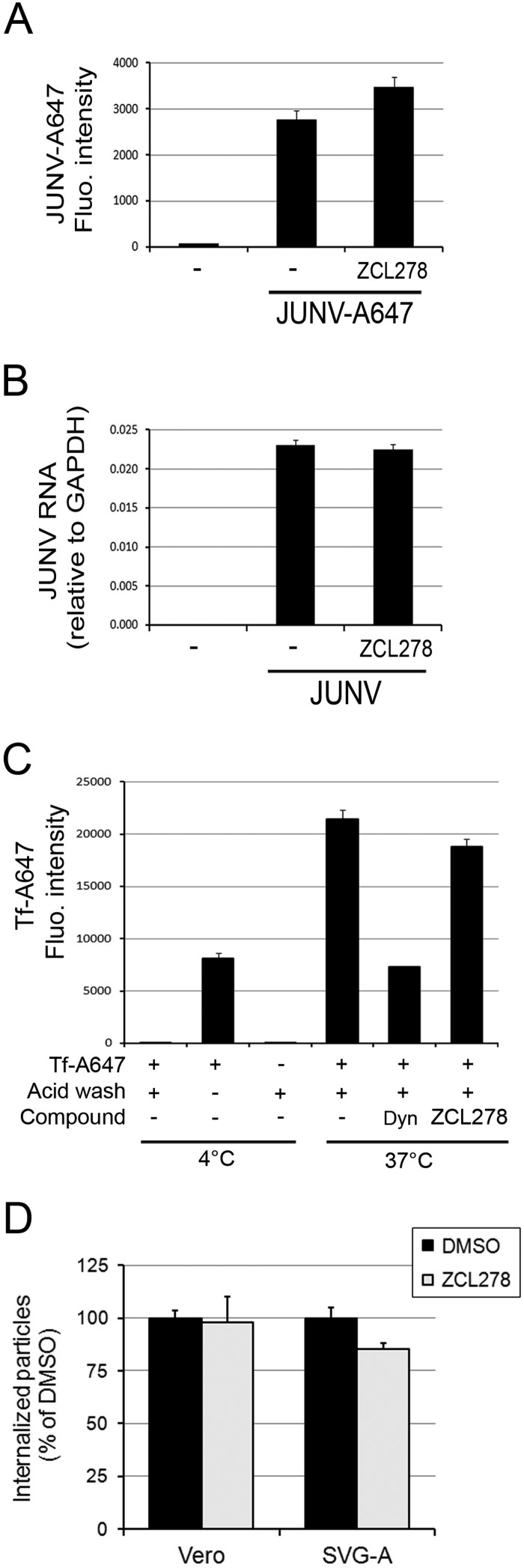
FIG 5.
ZCL278 does not affect attachment of Junin virus to the cell surface or endocytosis. (A) Vero cells were incubated with JUNV-A647 for 30 min at 4°C in the presence or absence of 50 μM ZCL278. The A647 fluorescence (Fluo.) intensity associated with the cells, which accounted for bound JUNV, was measured by flow cytometry. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (B) Vero cells were incubated with JUNV at an MOI of 5 for 30 min at 4°C. Cells were extensively washed with cold PBS and lysed for RT-qPCR analysis. The amounts indicated by the histograms are normalized to those for the GAPDH housekeeping gene, and error bars are the means ± SDs from triplicate experiments. (C) Tf-A647 uptake by Vero cells was performed in the presence or absence of 50 μM ZCL278 or 20 μM Dynasore-OH (Dyn). An acid wash was performed to remove surface-bound transferrin that was not endocytosed. The Tf-A647 mean fluorescence intensity was measured by flow cytometry. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (D) Vero cells or SVG-A cells were prepared as described in the legends to Fig. 2C and D in the presence or absence of 50 μM ZCL278, and the amount of endocytosed particles (A647 positive and A568 negative) normalized by the amount of particles at the cell surface (A647 and A568 positive) was quantified by fluorescence microscopy. A total of 641 particles in the untreated control cells were counted and 698 particles in the ZCL278-treated cells were counted. DMSO, dimethyl sulfoxide. Error bars are the means ± SDs from two individual experiments.
Wound healing assay.
Cells were plated at confluency on a single coverslip with adjacent individual chambers. The cell monolayer was then scratched using a 200-μl pipette tip. Cell migration at 37°C with 5% CO2 was monitored every 10 to 30 min for 12 h by bright-field illumination with a 10× objective. An automated stage allowed the acquisition of up to 30 fields of view over the three chambers.
Cell viability.
Cell viability was measured using a CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer's instructions. Luminescence was acquired using a FLUOstar Omega microplate reader (BMG Labtech).
Mice.
Mice were housed according to the policies of the Institutional Animal Care and Use Committee of the University of Pennsylvania, and this committee approved all experiments. C57BL/6 mice were purchased from The Jackson Laboratory and housed and bred at the University of Pennsylvania under animal biosafety level 2 conditions. Four-week-old mice received intravenous injections of gabapentin (Hi-Tech Pharmacal) or ZCL278 (Axon Medchem). At 1 h after treatment, the mice were inoculated intraperitoneally with JUNV Candid #1 (1 × 106 PFU) in no more than 1 ml with a 27 1/2-gauge needle. Daily drug treatments at the doses depicted in the legend to Fig. 8B were administered intraperitoneally. At the end of the experiment, the mice were sacrificed, their spleens were homogenized with a Dounce homogenizer and centrifuged to generate a cell pellet and supernatant, and RNA expression levels were determined as described previously (7). Briefly, JUNV RNA was quantified by real-time quantitative PCR (RT-qPCR) using a 7800HT sequence detector system (Applied Biosystems) and a specific primer pair to detect the small segment of JUNV (Candid #1 strain; primers 5′-GGGGCAGTTCATTAGCTTCATGC-3′ and 5′-CAAAGGTAGGTCATGTGGATTGTTGG-3′). All RNA quantifications were normalized to the amount of GAPDH RNA (obtained by PCR with primers 5′-CCCCTTCATTGACCTCAACTACA-3′ and 5′-CGCTCCTGGAGGATGGTGAT-3′). The Power SYBR green PCR kit (Applied Biosystems) was used to perform all RT-qPCR amplifications. The amplification conditions were 50°C for 2 min, followed by 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. At the end of the RT-qPCR run, a dissociation curve was determined to ensure that each primer pair generated a single product of amplification.
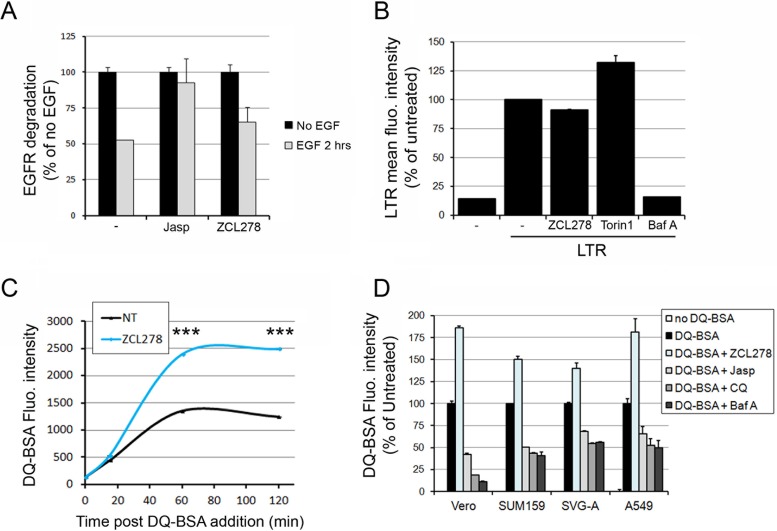
FIG 8.
ZCL278 increases the degradation of exogenous DQ-BSA but not endogenous EGFR independently of endosomal acidification. (A) The amount of EGFR in the absence (black bars) or presence (gray bars) of EGF was measured by flow cytometry in cells treated with the indicated compounds. The histogram represents the A647 mean fluorescence intensity and reflects the amount of EGFR per cell normalized to that for the control not treated with EGF for each condition. Data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (B) SUM159 cells were treated with 50 μM ZCL278, 10 μM 17C9, 250 nM Torin1, or 100 nM bafilomycin (Baf A) for 1 h 15 min at 37°C (similar to the time used during the JUNV infection experiments for which the results are depicted in Fig. 3D), and the acidic compartments were stained with LTR. The histogram represents the cellular mean fluorescence intensity of LTR measured by flow cytometry for each condition. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition. Torin1 treatment induced higher LTR fluorescence levels than the other treatments, as has been previously observed (46), while ZCL278 had no effect on LTR staining. (C) Vero cells were untreated or pretreated with 50 μM ZCL278 and subsequently incubated for 15, 60, or 120 min with 10 μg ml−1 green DQ-BSA in the presence (blue line) or absence (black line) of the compound. The cells were analyzed by flow cytometry, and the curves represent the mean fluorescence intensity of 488-nm laser-excited cells, which is proportional to the amount of DQ-BSA degraded at the indicated time points after DQ-BSA addition. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition. Statistical analysis using the two-tailed Student t test shows significance differences. ***, P < 0.0005. (D) Vero, SUM159, SVG-A, or A549 cells were pretreated for 15 min with 50 μM ZCL278 or 1 μM Jasp or pretreated for 2 h with 100 μM chloroquine (CQ) or 100 nM Baf A. The cells were then coincubated for 1 h with 10 μg ml−1 green DQ-BSA in the presence of the indicated drugs. The histogram represents the DQ-BSA mean fluorescence intensity normalized by that obtained under the untreated condition for each cell type. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition.
RESULTS
Characterization of new cell line models suitable for JUNV infection.
We sought to determine the role of specific host proteins during the entry of JUNV while being conscious to maintain physiologically relevant levels of those factors. We employed gene editing based on a CRISPR-Cas9 strategy to knock out specific genes or to obtain cells that endogenously express proteins of interest fused to fluorescent tags. To this end, we had to identify new human cellular models compatible with such techniques because the genome of Vero cells (derived from the green African monkey) has not been fully sequenced and at present is not suited for use in efficient gene-editing approaches. We had successful experiences in editing the genes of cells of the human breast cancer cell line SUM159 (27, 32, 33) and therefore evaluated SUM159 cells for permissiveness and compared them to the Vero and A549 cells routinely used in JUNV infection assays. In addition, we also tested SVG-A human astroglial cells as a potential host relevant for JUNV physiopathological studies. We found that all four cell lines support infection and produce infectious particles (Fig. 1). Vero and SVG-A cells very efficiently supported JUNV infection and spread (Fig. 1A and B), produced infectious viral particles (Fig. 1C and D), and exhibited no major cell death due to JUNV infection over time (Fig. 1E). In contrast, SUM159 and A549 cells supported JUNV replication to a lesser extent, produced fewer viral particles at day 3 postinfection, and poorly supported JUNV infection after 5 days postinfection.
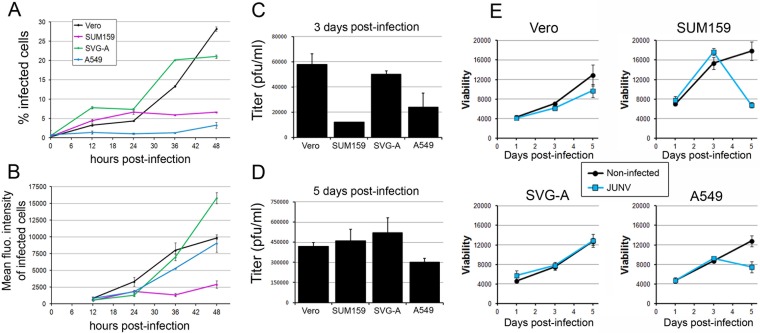
FIG 1.
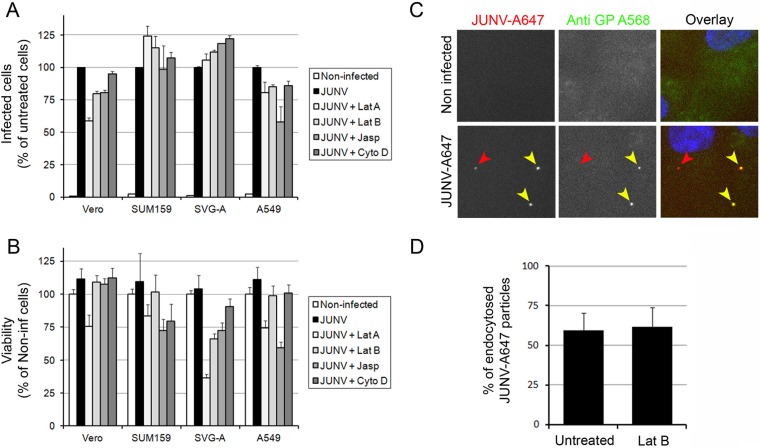
Characterization of infection of Vero, SUM159, SVG-A, and A549 cells by JUNV. (A, B) JUNV (MOI, 0.1) was inoculated onto Vero, SUM159, SVG-A, or A549 cells for 1 h at 37°C, and the cells were subsequently washed and reincubated for the indicated times postinfection. Cells were fixed and stained with an anti-NP antibody coupled to A647, and the percentage of infected cells (A) and the A647 mean fluorescence (fluo.) intensity of the infected cells (B) were measured by flow cytometry. The data correspond to the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (C, D) Vero, SUM159, SVG-A, or A549 cells infected with JUNV (MOI, 0.03) for 1 h were subsequently washed extensively and reincubated in virus medium for 24 h at 37°C. The supernatant was removed, and fresh medium was added to the cells for 48 h (3 days postinfection). The supernatant was collected, and fresh medium was added for an additional 48 h (5 days postinfection). The supernatant was collected, and plaque assays were performed to quantify the number of infectious particles released by the different cell types between days 1 and 3 postinfection (C) and days 3 and 5 postinfection (D). The data correspond to the means ± SDs from duplicate experiments. (E) Vero, SUM159, SVG-A, or A549 cells treated as described in the legends to panels C and D were used to assess cell viability using the CellTiter-Glo viability assay with noninfected cells (black line) or JUNV-infected cells (blue line). The luminescence was measured using the same luminometer parameters for each cell type, and the results are expressed as arbitrary luminescence units. The data correspond to the means ± SDs from triplicate experiments.
In conclusion, all four cell lines were permissive to JUNV infection. Moreover, we identified the SVG-A glial cell line to be a useful alternative to Vero and A549 cells to study JUNV biology; indeed, it appears to be highly permissive and potentially more physiologically relevant.
Limited role of the actin network during JUNV entry.
Next, we revisited whether the actin cytoskeleton plays a role during the early entry steps of JUNV infection in these four cell lines, as has been previously suggested (13, 14). Vero, SUM159, SVG-A, and A549 cells were treated with Lat A, Lat B, Jasp, or Cyto D, four compounds well-known to interfere with the dynamics of the actin cytoskeleton through different mechanisms. To assess the role of actin specifically during the attachment/internalization phase, cells were exposed to JUNV together with one of the compounds for only 30 min to minimize the potential perturbation of later stages (endosomal trafficking and RNP release into the cytosol). Subsequently, the cells were incubated for 16 h without virus or compounds but in the presence of an anti-GP neutralizing antibody (see Fig. S1, GD01 antibody, in the supplemental material) (21, 26) to prevent further infection posttreatment. Upon treatment, a very limited inhibitory effect on JUNV infection was observed in the four cell lines tested (Fig. 2A). The relatively high concentrations of actin-interfering drugs used in this experiment (1 μM for each drug) induced some toxicity (Fig. 2B), but no correlation between infectivity and toxicity could be established. These concentrations were used to ensure the fast and complete disruption of the actin network, and fluorescence microscopy imaging of cells transiently expressing LifeAct-RFP, which binds only to F actin, confirmed the effect of these treatments (see Fig. S2 in the supplemental material).
FIG 2.
Importance of the actin network during JUNV entry. (A, B) Vero, SUM159, SVG-A, or A549 cells were untreated or pretreated for 15 min with 1 μM latrunculin A (Lat A), 1 μM latrunculin B (Lat B), 1 μM cytochalasin D (Cyto D), or 1 μM jasplakinolide (Jasp) and subsequently infected with JUNV in the presence or absence of the drugs for an additional 30 min. Then, the cells were washed and incubated for 16 h in virus medium containing an anti-GP neutralizing antibody. (A) The cells were fixed and stained with an anti-NP antibody coupled to A647, and the percentage of infected cells was measured by flow cytometry. The data are normalized to the percentage of infected untreated cells for each cell type. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (B) Cell viability was assessed using the CellTiter-Glo viability assay with cells prepared in parallel with the cells in the assay for which the results are presented in panel A. Error bars are the means ± SDs from triplicate experiments. Non-inf, noninfected. (C, D) Vero cells were pretreated in the presence or absence of Lat B for 15 min and subsequently infected with JUNV-A647 for 30 min in the presence or absence of Lat B. The cells were then fixed and stained with an anti-GP antibody (GB03) coupled to A568 without permeabilization. (Left and middle) Fluorescence from individual channels; (right) overlay of the fluorescence from the GP-A568 (green) and JUNV-A647 (red) channels together with the DAPI (4′,6-diamidino-2-phenylindole) signal (blue). Red arrowheads, a JUNV particle that has been endocytosed (A647 positive and A568 negative); yellow arrowheads, two particles that are still at the surface of the cell (A647 and A568 positive). (D) Quantification of the amount of A647-positive and A568-negative particles (endocytosed) normalized by the amount A647- and A568-positive particles (at the cell surface). A total of 124 particles over 26 fields of view were counted for the untreated control, and 42 particles over 16 fields of view were counted for cells treated with 1 μg ml−1 Lat B. Error bars are the means ± SDs from two individual experiments.
To assess the role of the actin network specifically during JUNV internalization, we used a recently described assay (34) to monitor by immunofluorescence microscopy the location of viral particles on Vero cells infected with JUNV that was fluorescently labeled with a nonneutralizing antibody (JUNV-A647; see Fig. S1, LD05 antibody, in the supplemental material) (21, 26) in the presence or absence of Lat B. Upon fixation, the virus particles that were not internalized were detected using an anti-GP antibody coupled to Alexa Fluor 568 (A568) without prior permeabilization of the cells. As shown in Fig. 2C, this protocol allowed us to discriminate between endocytosed viruses (A647-positive cells only; red arrowhead) and virus particles remaining at the cell surface (A647- and A568-positive cells; yellow arrowheads). The images in the lower and upper panels of Fig. 2C were acquired, processed, and contrasted in the same way. Quantification of the percentage of endocytosed virions following 1 μM Lat B treatment confirmed that the actin cytoskeleton is not involved in the process of JUNV endocytosis in Vero cells (Fig. 2D).
In conclusion, our data highlight that, at best, the actin network plays a limited role during JUNV endocytosis.
Identification of ZCL278 as a potent JUNV entry inhibitor.
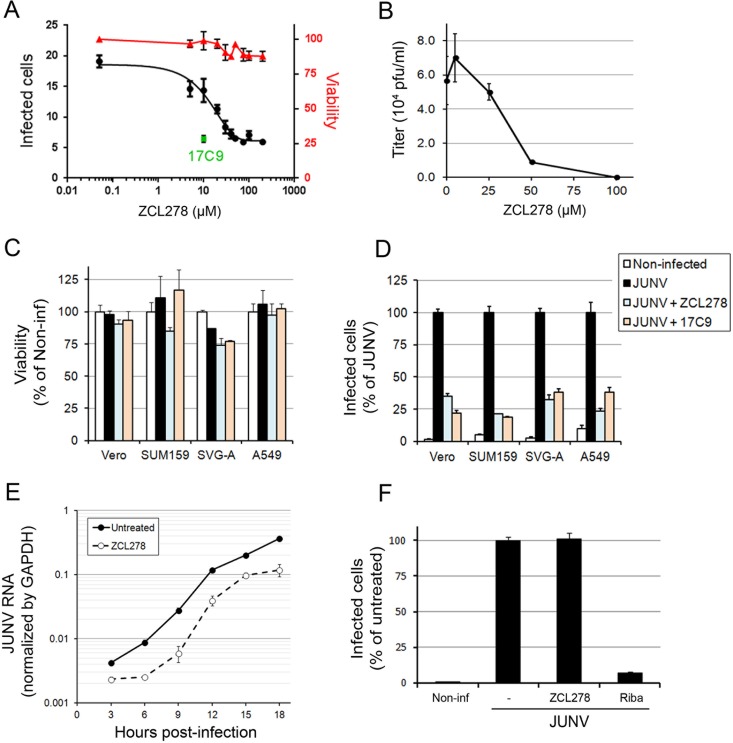
In addition to investigating the effect of the actin-interfering agents just described, we also tested the small molecule ZCL278, an inhibitor of the Cdc42 small GTPase (20). Cdc42 is an important regulator of branched actin dynamics (16). Given the absence of a significant role of actin dynamics during JUNV entry and infection, we expected that the compound would not interfere with JUNV infection; however, we found that ZCL278 possesses significant antiviral activity against JUNV (Fig. 3). Dose-response experiments were first carried out in Vero cells, and while ZCL278 was not toxic at concentrations up to 200 μM, we found that it inhibited JUNV with a 50% inhibitory concentration (IC50) of ∼14 μM, as measured by flow cytometry (Fig. 3A). ZCL278-treated cells also produced fewer infectious particles (Fig. 3B). No significant toxicity was detected in the four JUNV-infected cell lines tested using 50 μM ZCL278 (Fig. 3C), but potent antiviral activity was observed (Fig. 3D). This inhibition of JUNV infection was as robust as that observed with treatment with 10 μM 17C9, a known small-molecule inhibitor of the fusion mediated by the viral GP (22, 23). A one-step growth curve of JUNV-infected cells treated with ZCL278 showed a significant decrease in the amount of viral RNA expressed compared to the amount expressed by untreated cells (Fig. 3E). Finally, we found that treatment with ZCL278 added at 1 h postinfection for up to 16 h did not prevent JUNV replication, in contrast to the effect of a similar treatment with ribavirin (Fig. 3F), a known postentry inhibitor of replication of RNA viruses, including JUNV (35). These results indicate that ZCL278 interferes with early steps of JUNV infection prior to virus replication.
FIG 3.
Identification of ZCL278 as a potent JUNV entry inhibitor. (A) Cells were untreated or pretreated for 15 min with ZCL278 and subsequently infected with JUNV (MOI, 0.3) in the presence or absence of the compound for an additional hour. Then, the cells were washed and incubated for 16 h in virus medium containing an anti-GP neutralizing antibody (GD01). Cells were fixed and stained with an anti-NP antibody coupled to A647, and the percentage of infected cells was measured by flow cytometry. The percentage of infected Vero cells (left axis) was plotted against the ZCL278 concentration (black line). Samples treated in parallel were used to measure cell viability using the CellTiter-Glo assay. Viability (right axis), which was normalized to that of the untreated sample, is shown by the red line. 17C9 at a single concentration of 10 μM was used as a positive control, and the percentage of infected cells is shown as a green dot. Error bars are the means ± SDs from duplicate experiments. (B) Vero cells were untreated or pretreated with increasing amounts of ZCL278 for 15 min and subsequently infected with JUNV for 1 h in the presence or absence of ZCL278. After 24 h, the supernatant was harvested and the number of PFU of virus per milliliter was measured by plaque assay. Error bars are the means ± SDs from duplicate experiments. (C, D) Vero, SUM159, SVG-A, or A549 cells were treated as described in the legend to panel A. (C) Viability was determined by measuring the luminescence using the CellTiter-Glo assay. Error bars are the means ± SDs from triplicate experiments. (D) The normalized percentage of infected cells of each cell type and condition. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (E) One-step growth curve of Vero cells that were infected with JUNV at an MOI of 3 and that were pre- and cotreated with 50 μM ZCL278 as described in the legend to panel A. RNAs were extracted at the indicated times postinfection. An RT-qPCR specific for JUNV RNA was performed, and values were normalized by GAPDH values. Error bars are the means ± SDs from duplicate experiments. The differences between untreated and ZCL278-treated cells were highly significant at every time point. (F) Vero cells were first infected with JUNV for 1 h and subsequently incubated with 50 μM ZCL278 or 400 μM ribavirin (Riba) in virus medium containing an anti-GP neutralizing antibody for 15 h. Cells were fixed, stained, and analyzed by flow cytometry as described in the legend to panel A. The data were normalized to the percentage of untreated infected cells. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition.
ZCL278 inhibits JUNV entry in a Cdc42-independent manner.
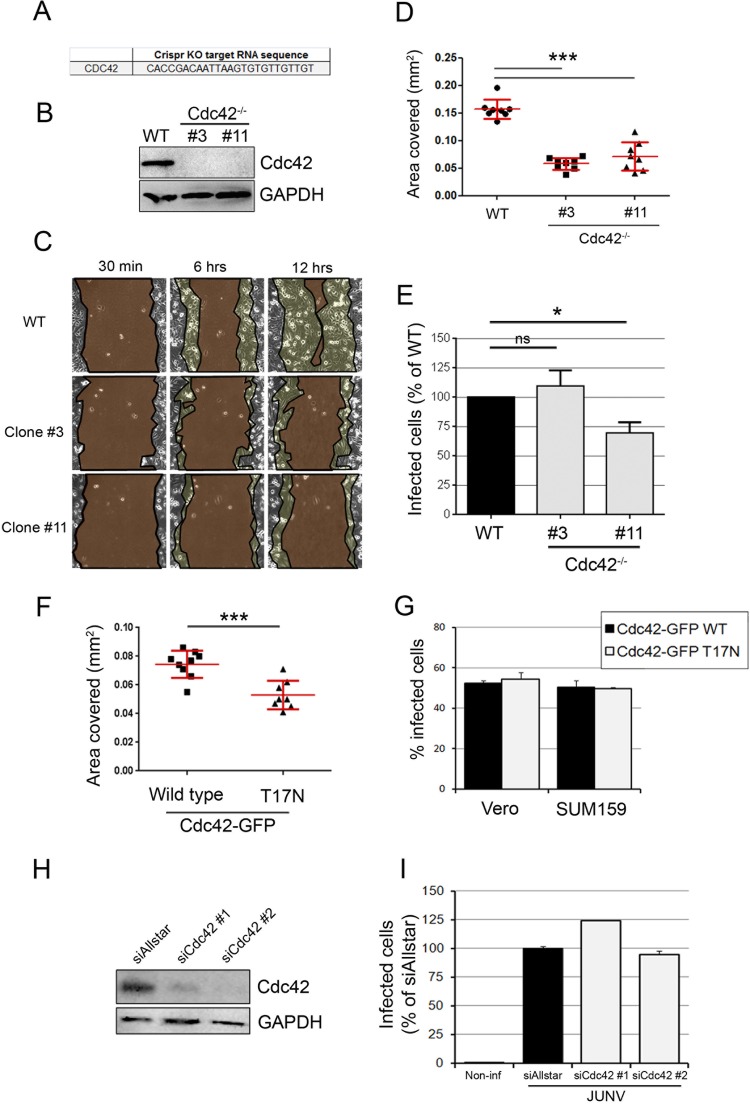
Given that it has been proposed that ZCL278 interferes with Cdc42 function (20), we investigated whether the protein Cdc42 was required for JUNV entry. We optimized a recently described knockout strategy using CRISPR-Cas9 lentiviral vectors (30) to fully eliminate the expression of Cdc42 in SUM159 cells. We designed an sgRNA targeting a region near the ATG codon of the Cdc42 gene (Fig. 4A), which directs the Cas9-mediated cleavage of the genomic double-stranded DNA at this specific site, leading to error-prone nonhomologous end joining and the induction of irreversible gene disruptions. We selected two Cdc42-knockout SUM159 monoclonal cell lines with undetectable Cdc42 protein expression, as assessed by Western blotting (Cdc42−/− clones 3 and 11; Fig. 4B). Cells from both lines exhibited shapes slightly more elongated than those of their wild-type counterparts but maintained an actin cytoskeleton rather similar to that of their wild-type counterparts (see Fig. S3A and B in the supplemental material), as previously observed when Cdc42 activity was inhibited in the absence of stimulation (20, 36). Cdc42 has a well-established role during filopodium formation and cell migration (37). To test whether the two Cdc42−/− cell line clones were functionally deficient in this respect, we set up a wound healing assay and quantified the area covered by migrating cells for 6 h postwounding (Fig. 4C and D). As expected and previously described using Cdc42-specific siRNA (38), the migration of the Cdc42−/− cell lines was strongly reduced compared to that of wild-type cells. Infection of these cells by JUNV showed that while clone 3 was as permissive as wild-type cells, a slight but significant decrease in infection could be observed in clone 11 (Fig. 4E). This difference could be explained by the fact that complete long-term depletion of Cdc42 may activate cellular compensatory mechanisms and/or cause unexpected side effects.
FIG 4.
JUNV enters cells in a Cdc42-independent manner. (A) Sequence of the gRNA targeting Cdc42 inserted into the LentiCRISPR vector. KO, knockout. (B) SUM159 cells subjected to Cdc42 knockout by transduction of a lentiviral vector were selected for 15 days postransduction, and monoclonal populations were isolated. The Cdc42 protein expression level from the wild type (WT), clone 3, and clone 11 was measured by Western blotting using an anti-Cdc42 antibody and revealed by use of a secondary antibody coupled to horseradish peroxidase. An anti-GAPDH antibody was used as a loading control. Even with a long exposure time, no Cdc42 protein could be detected in clones 3 and 11. (C) Wound healing assay of wild-type, Cdc42−/− clone 3, or Cdc42−/− clone 11 SUM159 cells that had been plated to confluence on the day before the scratch. Cells migrating to close the wound were imaged over a 12-h period. Micrographs show the same field of view at different time points after wounding for each cell type. The regions highlighted in dark brown indicate the surface area of the wound, which became smaller with time. The light brown regions delineate the cells that migrated, closing the wound, and correspond to the area covered (in millimeters squared). (D) The plot represents the area covered by the cells at 6 h after wounding compared to that covered at 30 min after wounding. Each dot represents a single movie, and eight movies were acquired per condition. Red lines, means ± SDs. Statistical analyses for significance were performed using the two-tailed Student t test to compare the results for the wild type and either clone 3 or clone 11. ***, P < 0.0001. (E) Wild-type or Cdc42−/− SUM159 cells (clones 3 and 11) were infected with JUNV for 16 h, and the percentage of infected cells was measured by flow cytometry. The data are normalized to the percentage of infected wild-type cells. Error bars are the means ± SDs from eight independent experiments, in which the results for at least 5,000 cells per condition were acquired in duplicate. Statistical analyses were performed using the two-tailed Student t test and showed no significant differences (ns) between the wild type and clone 3 (P = 0.49) and significance differences between the wild type and clone 11 (*, P = 0.013). (F) Wound healing assay, as exemplified in panel C, of SUM159 cells transfected with wild-type Cdc42-EGFP or a dominant negative mutant counterpart, Cdc42-EGFP T17N. Statistical analysis was performed using the two-tailed Student t test and showed a significant difference. ***, P < 0.0001. (G) Vero or SUM159 cells transfected to overexpress the Cdc42-EGFP wild type (black bars) or the T17N mutant (gray bars) were infected with JUNV for 16 h, and the percentage of infected cells in the EGFP-positive cell population was measured by flow cytometry. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (H, I) Cdc42 protein expression was knocked down in Vero cells by transfecting two individual siRNAs targeting Cdc42 mRNA. (H) At 2 days posttransfection, the silencing efficiency was estimated by Western blotting to be 82% (siCdc42#1) and 92% (siCdc42#2). (I) In parallel, a subset of these cells was infected with JUNV for 16 h, and the percentage of infected cells was assessed by flow cytometry. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition.
We next performed experiments that induce Cdc42 perturbations for 3 days (rather than the several weeks needed as part of the selection procedure used to isolate knockout clones). Overexpression of a dominant negative form of Cdc42 in which the threonine at position 17 is replaced by an asparagine (T17N) strongly reduces the protein's affinity for GTP (39). Cells expressing Cdc42 T17N showed a significant defect in cell migration (Fig. 4F), while they remained permissive to JUNV infection (Fig. 4G). Furthermore, depletion of Cdc42 in Vero cells by 92% and 81% using two individual siRNAs (Fig. 4H) did not reduce the level of JUNV infection (Fig. 4I). Finally, acute treatment with another small-molecule inhibitor of Cdc42 function (ML141 [40]) caused partial JUNV entry inhibition to a lesser extent than 17C9 (see Fig. S4 in the supplemental material).
From the results of these experiments, we concluded that the elimination of Cdc42 expression or long-term interference with its function did not affect JUNV infection.
ZCL278 does not inhibit clathrin-mediated endocytosis, attachment of JUNV to the cell surface, or viral particle internalization.
We next investigated which step of viral entry was inhibited by ZCL278. No significant difference in the number of JUNV-A647 particles bound to the cell surface at 4°C was observed between ZCL278-treated and untreated Vero cells, as measured by flow cytometry (Fig. 5A). These data were confirmed by quantifying by RT-qPCR the amount of JUNV RNA bound to the surface of Vero cells at 4°C (Fig. 5B). We also tested whether ZCL278 interferes with clathrin-mediated endocytosis using a Tf uptake assay that was described previously (27, 34), with uptake efficiency being assessed by measuring the mean fluorescence intensity of endocytosed Tf-A647 by flow cytometry (Fig. 5C). We observed no significant difference in Tf uptake between ZCL278-treated and untreated cells, whereas treatment with the small-molecule dynamin inhibitor Dynasore-OH (41, 42) efficiently inhibited Tf-A647 endocytosis. Of note, the results of our previous study indicated that JUNV infection does not downregulate Tf receptor expression (34). Finally, quantification of the percentage of endocytosed virions by immunofluorescence (Fig. 2D) showed that the internalization of JUNV was not impaired by the presence of ZCL278 (Fig. 5D).
From these results, we concluded that ZCL278 does not prevent the binding of JUNV particles to the cell surface or its subsequent internalization.
ZCL278 inhibits the release of JUNV ribonucleoproteins from the endosomes to the cytosol.
We investigated whether ZCL278 could interfere at a later entry step, namely, the exit of JUNV RNPs from the endosomes into the cytosol. We developed an immunofluorescence-based assay that distinguished complete viral particles located in either endosomes or the cytosol. Particles that were positive for both the NP and GP proteins of JUNV were defined as particles within endosomes, and particles that were positive for NP but negative for GP were defined as JUNV RNPs that had reached the cytosol upon exit from the endosome (see the schematic in Fig. 6A). To reduce nonspecific staining, JUNV particles were first labeled with a nonneutralizing GP-specific antibody coupled to A647 (Fig. 2C and D) because labeling of cell-free virions strongly minimized the nonspecific antibody fluorescence background otherwise associated with cells. Vero cells infected with these labeled particles were fixed and stained with two noncompetitive monoclonal mouse anti-NP antibodies coupled to either Alexa Fluor 488 or Alexa Fluor 568. The positions of these NP fluorescent signals were correlated (see Materials and Methods) as a way to increase the accuracy of identification of true NP-positive objects by minimizing the nonspecific background given by each antibody separately (Fig. 6B). Images were acquired by spinning disk confocal microscopy, and the relative amount of JUNV RNPs released into the cytosol was determined by dividing the number of NP+ and GP− objects (released RNPs) by the total number of NP+ objects (Fig. 6C). The proportion of particles lacking GP present in our viral stock was estimated to be ∼25% of the particles; this number was deducted from the values presented in Fig. 6C. ZCL278-treated cells showed a dose-dependent decrease in NP-positive objects that did not contain GP, suggesting that ZCL278 prevents viral particles from releasing their contents into the cytosol.
Next, we set up an infection assay that bypasses the endocytic step where JUNV was allowed to fuse directly at the plasma membrane using extracellular acidic pH switching. Vero cells were incubated with JUNV at 4°C to allow virus binding at the cell surface but to prevent endocytosis. After unbound viruses were removed, medium with the indicated pH was applied to cells at 4°C in the presence or absence of ZCL278 (Fig. 6D). The fusion of the JUNV GP with cellular membranes requires endosomal acidification (7, 43); therefore, a low extracellular pH would allow JUNV fusion with the plasma membrane. Fusion events were stopped by incubation with a neutralizing antibody, and cells were switched to 37°C for 16 h to allow viruses that had fused to the plasma membrane to productively infect cells (Fig. 6D). The increased percentage of infected cells at a pH of <5.6, quantified by flow cytometry, confirmed the ability of JUNV to fuse at the plasma membrane at 4°C (Fig. 6D, black line). A significant 2-fold decrease in the percentage of infected cells was observed with ZCL278 treatment (Fig. 6D, blue line).
Together, our data suggest that ZCL278 prevents the release of JUNV RNP by inhibiting GP-mediated fusion.
ZCL278 prevents the localization of JUNV particles in early and late endosomes.
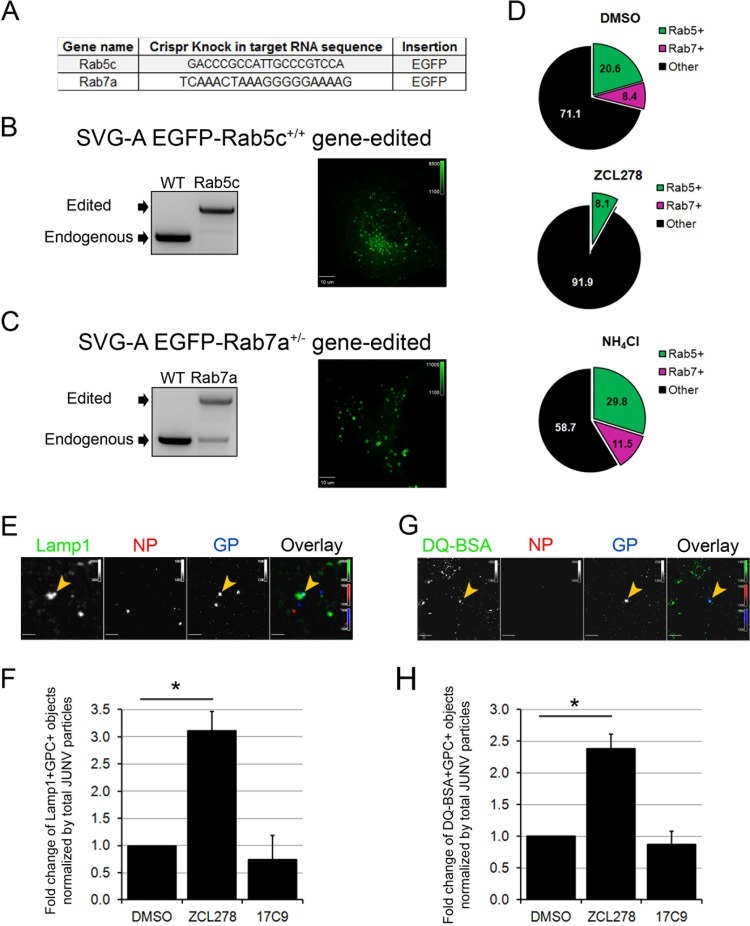
Because ZCL278 interferes with JUNV RNP release into the cytosol, we explored whether intact viral particles accumulated in early and/or late endosomes upon compound treatment. To perform this experiment, we used the CRISPR-Cas9 system to create two new gene-edited SVG-A cell lines expressing endogenous levels of EGFP-Rab5c or EGFP-Rab7a. Briefly, SVG-A cells were cotransfected with a plasmid carrying Cas9, a double-stranded linear DNA fragment encoding an sgRNA targeting a region near the ATG codon of either Rab5c or Rab7a (Fig. 7A) under the control of the U6 promoter, and a template plasmid carrying the EGFP sequence flanked by ∼800 bp upstream and downstream of the targeted region (see Materials and Methods for details). After sorting of single cells into 96-well plates by fluorescence-activated cell sorting, monoclonal populations were expanded and screened for endogenous/edited alleles. We selected two clones, one corresponding to EGFP-Rab5c+/+ and the other corresponding to EGFP-Rab7a+/−, as assessed by genomic PCR (Fig. 7B and C).
FIG 7.
JUNV particles are mostly absent from early and late endosomes but are enriched within lysosomal compartments in ZCL278-treated cells. (A) Gene names and the sequences of the target gRNA used to insert the EGFP and a linker consisting of the amino acid sequence GGSGGSGGS upstream of their ATG codons by homologous recombination in SVG-A cells. (B, C) SVG-A cells were cotransfected with a plasmid coding for Cas9, a linear PCR product coding for the specific gRNAs targeting a region near the ATG codon of either Rab5c or Rab7a under the control of the U6 promoter, and a template plasmid containing the EGFP sequence flanked by 800 bp upstream and downstream of the region targeted (see Materials and Methods for details). Monoclonal populations were expanded and screened for endogenous/edited alleles by genomic PCR with primers specific for Rab5c (B) or Rab7a (C). (Right) Snapshots of the EGFP fluorescence distribution for each cell type acquired by spinning disk confocal microscopy. The dynamics of the EGFP-positive vesicles are shown in Movies S1 and S2 in the supplemental material. (D) SVG-A cells edited for EGFP-Rab5c+/+ and EGFP-Rab7a+/− were pretreated for 15 min and then infected with JUNV-A647 for 30 min in the presence or absence of 50 μM ZCL278 or 20 mM NH4Cl. The cells were then washed, fixed, and stained with an anti-NP antibody coupled to A568. Images were acquired by spinning disk confocal microscopy, and the number of JUNV particles positive for A647 and A568 (NP+ and GP+ objects) were quantified visually using the same contrast settings. The percentages of the NP+ and GP+ objects that also contained an EGFP signal are represented in a pie chart, in which the green slice accounts for Rab5c-positive (Rab5+), NP+, and GP+ objects and the pink slice accounts for Rab7a-positive (Rab7+), NP+, and GP+ objects. Each condition represents data collected from 10 fields of view, in which more than 120 GP+ particles were quantified. (E, F) SVG-A cells were treated as described in the legend to panel D and stained with an anti-NP antibody coupled to A548 and a rabbit anti-Lamp1 antibody, detected with a secondary anti-rabbit IgG antibody coupled to A488. (E) Images were acquired by spinning disk confocal microscopy. Yellow arrowheads, Lamp1-positive and GP+ objects. (F) The number of Lamp1-positive and GP+ objects normalized by the total number of GP+ and NP+ particles. Statistical analysis for significance was performed using the two-tailed Student t test. *, P < 0.05. (G, H) SVG-A cells were prepared as described in the legend to panel D, but inoculation of JUNV-A647 was performed in the presence of green DQ-BSA. (G) A representative snapshot. Yellow arrowheads, DQ-BSA-positive GP+ objects. (H) The number of DQ-BSA-positive and GP+ objects normalized to the total amount of GP+ and NP+ particles. Statistical analysis for significance was performed using the two-tailed Student t test. *, P < 0.05.
The EGFP fluorescence in these EGFP-Rab5c+/+ and EGFP-Rab7a+/− cells showed rapidly moving punctate structures, as expected for endosomal vesicles (Fig. 7B and C; see also Movies S1 and S2 in the supplemental material). We tested whether endosomal trafficking in these cells was normal by performing an EGFR degradation assay as previously described (31). Wild-type SVG-A cells treated with EGF had significantly reduced EGFR expression levels, reflecting the expected proteolysis of the receptor in the late endosomal/lysosomal compartment (see Fig. S5A in the supplemental material). The EGFP-Rab5c+/+ and EGFP-Rab7a+/− cells showed capacities to degrade EGFR similar to those of their wild-type unedited counterparts, suggesting that the intracellular transport of EGFR from the plasma membrane to the lysosomes was not significantly affected by the gene-editing procedure. Moreover, we observed that treatment with ZCL278 slightly reduced EGFR degradation in all the cell lines, while Jasp treatment blocked its degradation, as expected (see Fig. S5B and C in the supplemental material) (44).
The EGFP-Rab5c+/+ and EGFP-Rab7a+/− cells were then infected with JUNV-A647 (generated as described in the legend to Fig. 2C) for 30 min in the presence or absence of ZCL278 or NH4Cl, a compound that neutralizes endosomal acidification. The cells were then fixed and stained with an anti-NP antibody coupled to Alexa Fluor 568, and JUNV particles positive for A647 and A568 were analyzed for the presence of an EGFP signal. The percentage of intact viral particles found in the Rab5c- or Rab7a-positive and -negative compartments was quantified in the presence or absence of ZCL278 or NH4Cl (Fig. 7D). Surprisingly, treatment with ZCL278 strongly decreased the presence of JUNV particles in Rab5c and Rab7a endosomes compared to that in control cells and NH4Cl-treated cells. Although we cannot exclude the possibility that a subset of JUNV particles was distributed in endosomes containing undetectable amounts of Rab5/7, we conclude that ZCL278 interferes with intracellular trafficking during JUNV entry.
ZCL278 increases JUNV particle accumulation in proteolytically active lysosomal compartments.
During the analysis of the results presented in Fig. 7D, we observed that the total number of A647-positive puncta (GP+) lacking NP (NP−) was significantly increased, from 38% ± 23% of GP+ and NP− particles in untreated cells to 76% ± 9% in ZCL278-treated cells. Such a phenotype could reflect the degradation of JUNV particles in the lysosomes, leading to the absence of detection of NP. The lysosome-resistant A647 signal could therefore correspond to the dye used to label the GP antibody trapped alone in a compartment in which the protein components of JUNV would have been degraded. Therefore, we next investigated whether JUNV particles could be found in proteolytically active compartments upon ZCL278 treatment. Image analysis of cells stained for NP and Lamp1, an established lysosomal marker, revealed that ZCL278 treatment significantly increased the colocalization of the A647 signal with Lamp1-positive compartments by a factor of 3 (Fig. 7E and F). To confirm that the Lamp1 compartments containing JUNV particles were proteolytically active, SVG-A cells were coincubated with JUNV-A647 and green fluorescent DQ-BSA in the presence or absence of ZCL278. The DQ-BSA reagent corresponds to the unfolded BSA protein incubated in vitro with very large amounts of FL-BODIPY dye so that upon folding, the dye self-quenches and exhibits minimal fluorescence. Upon proteolytic processing of this BSA, the dye is released and dequenches, emitting a bright fluorescent signal (45). Strikingly, we observed that ZCL278 increased JUNV colocalization with DQ-BSA (Fig. 7G and H), suggesting that a subset of endocytosed JUNV particles was retargeted to proteolytically active compartments in ZCL278-treated cells.
ZCL278 increases the degradation of DQ-BSA but does not affect EGFR degradation or endosomal acidity.
Because ZCL278-treated cells had a greater ability to prevent JUNV infection and redirect JUNV particles to lysosomes, we asked whether ZCL278 could have broader cellular effects, including an enhanced proteolytic capacity in infection-free assays. We found that ZCL278 treatment did not affect the degradation of EGFR in SUM159 cells (Fig. 8A) or SVG-A cells (see Fig. S5A and B in the supplemental material).
We also investigated whether ZCL278 treatment interfered with the acidity of the endosomal/lysosomal compartments using LysoTracker red (LTR). Cells were treated with the compounds indicated below for 75 min (comparable to the treatment time used for the JUNV infection experiments for which the results are presented in Fig. 3D), followed by LTR staining to fluorescently label all acidic compartments. ZCL278-treated cells exhibited LTR fluorescence levels similar to those of untreated controls, as measured by flow cytometry (Fig. 8B). As controls, cells treated with Baf A, a vacuolar H+-ATPase inhibitor, had a strongly decreased LTR fluorescence, as expected (46), while treatment with Torin1, a drug that induces autophagy by inhibiting mTOR (47), led to an increase in LTR fluorescence, as previously observed (48).
Because we found that JUNV was redirected toward DQ-BSA-positive compartments (Fig. 7H), we tested the effect of ZCL278 on the degradation of DQ-BSA. ZCL278 induced an almost 2-fold increase in the intensity of DQ-BSA fluorescence, as observed by flow cytometry (Fig. 8C), corresponding to an increase in dequenching of the DQ-BSA due to greater proteolytic processing. The plateau of DQ-BSA fluorescence was reached at the same time point (1 h) by untreated and ZCL278-treated cells, which suggests that the cells exhibit similar degradation kinetics but do so with a different overall capacity. This result indicates that ZCL278 increases the overall lysosomal proteolytic activity of the cell, but the reason for this increase remains unknown. We observed a similar phenotype in the four cell types studied, and compounds that prevent lysosomal degradation by interfering with endosomal trafficking (Jasp) or with the endosomal/lysosomal pH (Baf A and CQ) efficiently prevented BSA degradation (Fig. 8D).
Taken together, the results of these experiments demonstrate that ZCL278 induces a rapid and robust upregulation of proteolytic degradation of the exogenous soluble BSA added to cells in culture.
Antiviral activity of ZCL278 extends to other enveloped viruses.
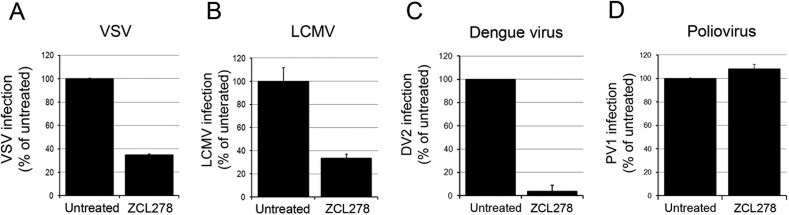
Given the suspected role of ZCL278 described above, we hypothesized that the entry of viruses from other families may also be impaired by ZCL278 treatment. We found that the cellular entry of VSV, LCMV, and DV2 was strongly inhibited (Fig. 9A to C) upon ZCL278 treatment, while that of the nonenveloped PV1 remained unaffected (Fig. 9D). These findings highlight the antiviral activity of ZCL278 against several other enveloped viruses. However, the extent of ZCL278 antiviral activity is variable according to the virus studied, and the precise mechanism of action of ZCL278 remains to be determined.
FIG 9.
The antiviral activity of ZCL278 extends to other enveloped viruses. Vero (A, B, D) or Huh7 (C) cells pre- and cotreated in the presence or absence of 50 μM ZCL278 as described in the legend to Fig. 3D were infected with VSV for 6 h at an MOI of 0.3 (A), LCMV for 16 h at an MOI of 0.3 (B), DV2 for 24 h at an MOI of 1 (C), or PV1 for 6 h at an MOI of 1 (D). The percentage of VSV- and LCMV-infected cells detected by virus-specific antibody staining was measured by flow cytometry (A, B). The amount of infectious DV2 particles released was measured by plaque assay (C), and the replication of PV1 was evaluated by RT-qPCR (D). For all histograms, the values presented are relative to those for the untreated control. Error bars represent SDs from experiments performed at least in duplicate.
ZCL278 inhibits JUNV infection in mice.
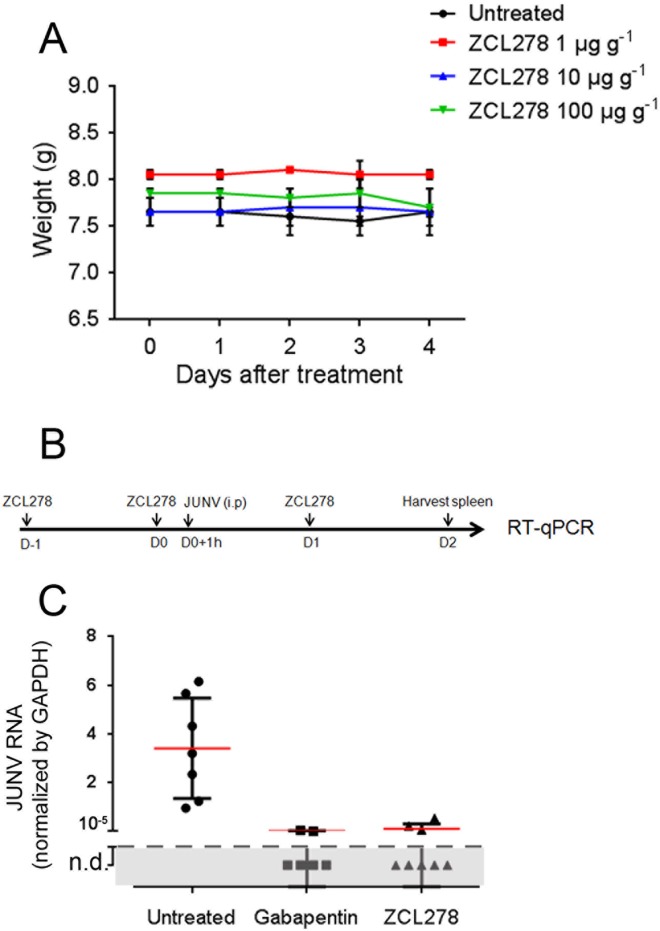
Given the antiviral activity of ZCL278 detected in our in vitro assays, we tested whether it could also prevent JUNV replication in a previously described mouse model of JUNV infection (7). First, to check for drug toxicity, C57BL/6 mice were injected intraperitoneally with the amounts of ZCL278 indicated below every 24 h for 4 days, and their weights, used as a readout of overall health, were recorded before each injection. The ZCL278 dosage had no significant effect on the body weights of the mice over the treatment period (Fig. 10A), and the mice in all treatment groups appeared healthy.
FIG 10.
ZCL278 inhibits Junin virus replication in mice. (A) C57BL/6 mice were weighed and then intraperitoneally injected with the indicated amounts of ZCL278 every 24 h for 4 days. The data are the means ± SDs for two mice per condition. (B) Mice untreated or treated with 100 μg g−1 gabapentin or 100 μg g−1 ZCL278 were inoculated intraperitoneally (i.p.) with 106 PFU of the JUNV Candid #1 strain. At the end of the experiment, the mice were euthanized, their spleens were harvested, and RNA was extracted. D, day. (C) JUNV NP RNA was detected by RT-qPCR, and the data were normalized to those for the GAPDH housekeeping gene. The result for each mouse is represented by one data point. Red lines, means; error bars, SDs (n = 7 for untreated mice, 6 for gabapentin-treated mice, and 8 for ZCL278-treated mice). P was <0.0001 for gabapentin- and ZCL278-treated mice compared to untreated mice (one-way analysis of variance). n.d., not detected.
Mice were treated with ZCL278 before and after systemic inoculation with 106 PFU of the Candid #1 strain of JUNV, according to the protocol depicted in Fig. 10B. As a positive control for viral inhibition, mice were treated with gabapentin, a small molecule that inhibits JUNV entry both in vitro and in vivo by downregulating the JUNV entry factor voltage gate calcium channel (7). Strikingly, ZCL278 reduced the JUNV RNA load in the spleen more than 33-fold, with JUNV RNA being undetectable in 5 out of 8 mice (Fig. 10C). These results are similar to those seen in gabapentin-treated mice, demonstrating that ZCL278 could abrogate JUNV replication in vivo. We concluded that ZCL278 is also a potent in vivo antiviral inhibitor of JUNV infection in the mouse model used.
DISCUSSION
Our investigation of the cellular entry pathway of the Junin virus led us to establish the absence of a significant role of actin dynamics during JUNV internalization in four cell types. Consistent with this result, we uncovered that Cdc42 plays a minimal role during JUNV infection. We identified the small molecule ZCL278 to be a novel broad-spectrum antiviral compound that inhibits not only JUNV but also VSV, LCMV, and DV2. Finally, we showed that ZCL278 is effective in blocking JUNV replication in mice.
Our work presents new cell line models for the study of JUNV infection. Vero cells are derived from the green African monkey (Chlorocebus sp.) and are commonly used for JUNV infection assays but at present are unsuitable for use in genomic manipulations due to the absence of whole-genome sequencing data from the source organism. A549 cells are human lung cancer cells previously used in JUNV infection experiments (49), but they exhibit a relatively lower permissiveness to JUNV (Fig. 1). We found that SVG-A cells, derived from a human fetal glial cell line immortalized by simian virus 40 (SV40) large T antigen expression and previously described to support both SV40 and JC virus infection (50, 51), are highly permissive to JUNV infection. This cell line represents a physiologically relevant model to study JUNV biology because JUNV-induced Argentine hemorrhagic fever manifests with neurological symptoms (1). While they are less relevant, breast cancer SUM159 cells were used here because we have previously optimized gene-editing strategies to express fluorescently labeled endogenous proteins in these cells (27, 33). Here we showed that we can also manipulate the genome of SVG-A cells by inserting a fluorescent EGFP tag into the N terminus of the endosomal small GTPases Rab5c and Rab7a (Fig. 7). These cell lines represent useful tools with which to track endosomes without the need to overexpress proteins, which is particularly important because ectopic Rab5 and Rab7 overexpression affects membrane trafficking (52, 53). Of note, we also generated SVG-A and SUM159 EGFP-Rab5a+/+ gene-edited cells, but the fluorescence intensity signal was much weaker than that of the EGFP-Rab5c+/+ gene-edited cells described here.
We found that actin microfilament integrity is not strongly required for JUNV internalization (Fig. 2). These results are in contrast to those of a previous study that showed decreased JUNV infectivity upon long-term small-molecule interference with actin dynamics (14). In that study, whereas the authors also performed their experiments in Vero cells, they used the IV4454 strain of JUNV, in contrast to the Candid #1 strain used here. Moreover, our protocol was designed to specifically assess the role of actin during early entry events by interfering with the actin organization for the first 30 min of virus internalization, with further infection being blocked with a neutralizing antibody. Therefore, from the results of our experiments, we conclude that the actin network is not required for JUNV internalization, but we cannot rule out the possibility of a role for actin at later stages of the viral cycle.
ZCL278 was developed as an inhibitor of Cdc42 function (20), and it is currently patented for its chemotherapeutic potential in the treatment of various cancers (54). Here, we show that ZCL278 also exhibits antiviral activity with an unknown mechanism of action that differs from its reported function. New small molecules in development that target the viral polymerase or the GP of JUNV have been reported (22, 35, 55–60). These drugs are very specific and exhibit inhibitory effects at relatively low concentrations, ranging from 0.5 to 10 μM. A recurrent issue when targeting viral proteins as therapy is the potential emergence of drug-resistant viruses. ZCL278 targets a cellular process and exhibits antiviral activity at higher concentrations (Fig. 3A; IC50, ∼14 μM); however, this concentration remains lower than that of other well-known broad-spectrum antipathogen drugs targeting cellular processes, such as ribavirin or chloroquine (usually used at concentrations of ≥100 μM). Future work should also assess whether virus mutants that are resistant to the antiviral activity of ZCL278 can be identified.
The relatively high IC50 required to inhibit infection suggests that either ZCL278 acts on several targets to inhibit virus entry or this small molecule is not fully fitted to interfere with a unique off target crucial for virus entry. Although ZCL278 is described to be a Cdc42 inhibitor, we found that JUNV does not require Cdc42 to infect cells (Fig. 4), at least in cells exhibiting sustained interference with Cdc42 function. However, we cannot exclude the possibility that cells may be able to compensate for some of the Cdc42 functions by upregulating other related GTPases. Another Cdc42 inhibitor was also tested and exhibited weaker antiviral activity (see Fig. S4 in the supplemental material), but the antiviral mechanism for this compound remains to be determined. In conclusion, although we cannot completely exclude the possibility that Cdc42 plays some role during JUNV entry, our experiments clearly indicate that Cdc42 is not absolutely required for JUNV infection. In this context, we suspect that ZCL278 antiviral activity may not depend on its ability to inhibit Cdc42 function.
ZCL278 affects at least two steps of JUNV entry: first, ZCL278 decreases JUNV RNP release into the cytosol and virus particle fusion when induced at the plasma membrane. This activity is independent of virus attachment or internalization (Fig. 5 and 6). A possibility is that ZCL278 directly prevents GP-mediated viral fusion by interfering with either the viral GP or its cellular receptors. Given that the GPs of the various viruses tested differ substantially, it is unlikely that ZCL278 acts on the viral proteins. Second, we found that ZCL278 strongly perturbs the intracellular endosomal trafficking of JUNV particles, on the one hand, and increases the lysosomal proteolytic activity of treated cells, on the other hand (Fig. 7 and 8). However, we did not directly correlate the antiviral activity of ZCL278 with its ability to increase lysosomal degradation. At the cellular level, how ZCL278 affects intracellular trafficking remains unclear. It could be that the altered intracellular trafficking of incoming JUNV particles may prevent them from reaching the appropriate compartment required for fusion. Alternatively, ZCL278 might activate autophagy. ZCL278, however, does not induce an increase in acidic cellular compartments (Fig. 8B), as has been previously shown for activated autophagy (48, 61) and as we have confirmed using the small molecule Torin1. ZCL278 could promote the fusion of early/late endosomes with lysosomes, for instance, by targeting SNARE proteins. Although we did not observe major differences in the distribution of Rab5c, Rab7a, or Lamp1 markers upon ZCL278 treatment, the dynamic turnover of these proteins may prevent the detection of a phenotype using fixed samples. Further investigation of the intracellular trafficking of exogenous cargos in ZCL278-treated cells should help to validate or refute this hypothesis. In conclusion, we surmise that ZCL278 alters membrane trafficking in a yet-to-be-discovered fashion.
We found that ZCL278 inhibited infection by VSV (Rhabdoviridae), LCMV (Old World Arenaviridae), and DV2 (Flaviviridae), while PV1 (Picornaviridae) infectivity remained unaffected. The potency of ZCL278 varied among the different viruses that we tested, with high antiviral activity against JUNV and DV2 and more modest inhibitory activity against LCMV and VSV being observed. Of note, investigating whether ZCL278 also inhibits pathogenic JUNV strains would be useful because their fusion properties may differ from those of the Candid #1 strain that we used (62). The viruses that we tested are single-stranded RNA viruses with either a positive or a negative orientation, and their GPs are structurally unrelated. Thus, it is unlikely that the activity of ZCL278 against different viruses comes from its ability to specifically bind to such diverse viral envelope proteins. As a common feature, the four enveloped viruses inhibited by ZCL278 require internalization and GP-mediated pH-dependent membrane fusion (11, 63–66). In contrast, poliovirus entry is thought to be pH independent (67, 68), and the viral particles release their RNA content either at the plasma membrane or in vesicles located within 100 to 200 nm of the plasma membrane (68). Hence, the antiviral effect of ZCL278 could be attributed to the intracellular perturbation of late endosomal membranes, which in turn would prevent/decrease pH-dependent GP-mediated fusion. This model requires further investigation.
Finally, we show that ZCL278 inhibits JUNV replication in vivo (Fig. 10). The low toxicity observed and the fact that ZCL278 maintains antiviral activity in mice are promising. Small-molecule chemistry should be applied to improve ZCL278's potency and, potentially, help us identify the target that confers its antiviral activity.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Dale Boger and Landon Whitby for providing the small molecule 17C9 and Nathanael Gray for the Torin1 compound. We thank Walter J. Atwood for providing the SVG-A cell line. We also thank Florian Winau and Yu Hu for providing us with LCMV.
We declare that there are no conflicts of interest.
Funding Statement
This work was supported by National Institutes of Health grants GM-075252 and U54 AI057159 (New England Regional Center of Excellence in Biodefense and Emerging Infectious Diseases [NERCE BEID]) to T.K. and U54 AI 057168 (Mid-Atlantic RCE) to S.R.R. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00103-16.
REFERENCES
- 1.Enria DA, Briggiler AM, Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res 78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez RM, Jaquenod de Giusti C, Sanchez Vallduvi MM, Frik J, Ferrer MF, Schattner M. 2011. Junin virus. A XXI century update. Microbes Infect 13:303–311. doi: 10.1016/j.micinf.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Bronze MS, Greenfield RA. 2003. Preventive and therapeutic approaches to viral agents of bioterrorism. Drug Discov Today 8:740–745. doi: 10.1016/S1359-6446(03)02778-8. [DOI] [PubMed] [Google Scholar]
- 4.Nunberg JH, York J. 2012. The curious case of arenavirus entry, and its inhibition. Viruses 4:83–101. doi: 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maiztegui JI, McKee KT Jr, Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, Enria DA, Briggiler AM, Levis SC, Ambrosio AM, Halsey NA, Peters CJ. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis 177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- 6.Maiztegui JI, Fernandez NJ, de Damilano AJ. 1979. Efficacy of immune plasma in treatment of Argentine haemorrhagic fever and association between treatment and a late neurological syndrome. Lancet ii:1216–1217. [DOI] [PubMed] [Google Scholar]
- 7.Lavanya M, Cuevas CD, Thomas M, Cherry S, Ross SR. 2013. siRNA screen for genes that affect Junin virus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci Transl Med 5:204ra131. doi: 10.1126/scitranslmed.3006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castilla V, Mersich SE, Candurra NA, Damonte EB. 1994. The entry of Junin virus into Vero cells. Arch Virol 136:363–374. doi: 10.1007/BF01321064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez MG, Cordo SM, Candurra NA. 2007. Characterization of Junin arenavirus cell entry. J Gen Virol 88:1776–1784. doi: 10.1099/vir.0.82808-0. [DOI] [PubMed] [Google Scholar]
- 11.Rojek JM, Sanchez AB, Nguyen NT, de la Torre JC, Kunz S. 2008. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J Virol 82:7677–7687. doi: 10.1128/JVI.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez MG, Forlenza MB, Candurra NA. 2009. Involvement of cellular proteins in Junin arenavirus entry. Biotechnol J 4:866–870. doi: 10.1002/biot.200800357. [DOI] [PubMed] [Google Scholar]
- 13.Candurra NA, Lago MJ, Maskin L, Damonte EB. 1999. Involvement of the cytoskeleton in Junin virus multiplication. J Gen Virol 80(Pt 1):147–156. [DOI] [PubMed] [Google Scholar]
- 14.Martinez MG, Cordo SM, Candurra NA. 2008. Involvement of cytoskeleton in Junin virus entry. Virus Res 138:17–25. doi: 10.1016/j.virusres.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Hall A. 1994. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol 10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 16.Wear MA, Schafer DA, Cooper JA. 2000. Actin dynamics: assembly and disassembly of actin networks. Curr Biol 10:R891–R895. doi: 10.1016/S0960-9822(00)00845-9. [DOI] [PubMed] [Google Scholar]
- 17.Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. 2002. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol 9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- 18.Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS. 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol 3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Fraticelli AE, Vergarajauregui S, Eastburn DJ, Datta A, Alonso MA, Mostov K, Martin-Belmonte F. 2010. The Cdc42 GEF intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J Cell Biol 189:725–738. doi: 10.1083/jcb.201002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friesland A, Zhao Y, Chen YH, Wang L, Zhou H, Lu Q. 2013. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc Natl Acad Sci U S A 110:1261–1266. doi: 10.1073/pnas.1116051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez A, Pifat DY, Kenyon RH, Peters CJ, McCormick JB, Kiley MP. 1989. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J Gen Virol 70(Pt 5):1125–1132. doi: 10.1099/0022-1317-70-5-1125. [DOI] [PubMed] [Google Scholar]
- 22.Lee AM, Rojek JM, Spiropoulou CF, Gundersen AT, Jin W, Shaginian A, York J, Nunberg JH, Boger DL, Oldstone MB, Kunz S. 2008. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J Biol Chem 283:18734–18742. doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitby LR, Lee AM, Kunz S, Oldstone MB, Boger DL. 2009. Characterization of Lassa virus cell entry inhibitors: determination of the active enantiomer by asymmetric synthesis. Bioorg Med Chem Lett 19:3771–3774. doi: 10.1016/j.bmcl.2009.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. 1999. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer 81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carocci M, Hinshaw SM, Rodgers MA, Villareal VA, Burri DJ, Pilankatta R, Maharaj NP, Gack MU, Stavale EJ, Warfield KL, Yang PL. 2015. The bioactive lipid 4-hydroxyphenyl retinamide inhibits flavivirus replication. Antimicrob Agents Chemother 59:85–95. doi: 10.1128/AAC.04177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudin R, Barteneva NS. 2015. Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat Commun 6:6022. doi: 10.1038/ncomms7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocucci E, Gaudin R, Kirchhausen T. 2014. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol Biol Cell 25:3595–3609. doi: 10.1091/mbc.E14-07-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotla S, Major SC, Gustin KE. 2009. Rapid detection and quantitation of poliovirus and rhinovirus sequences in viral stocks and infected cells. J Virol Methods 157:32–39. doi: 10.1016/j.jviromet.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma YM, Boucrot E, Villen J, Affar EB, Gygi SP, Gottlinger HG, Kirchhausen T. 2007. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem 282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 32.Gaudin R. 2015. Enhancing our single molecule experience using Crispr. Med Sci (Paris) 31:959–961. (In French.) doi: 10.1051/medsci/20153111007. [DOI] [PubMed] [Google Scholar]
- 33.Kural C, Akatay AA, Gaudin R, Chen BC, Legant WR, Betzig E, Kirchhausen T. 2015. Asymmetric formation of coated pits on dorsal and ventral surfaces at the leading edges of motile cells and on protrusions of immobile cells. Mol Biol Cell 26:2044–2053. doi: 10.1091/mbc.E15-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudin R, Kirchhausen T. 2015. Superinfection exclusion is absent during acute Junin virus infection of Vero and A549 cells. Sci Rep 5:15990. doi: 10.1038/srep15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sepulveda CS, Garcia CC, Fascio ML, D'Accorso NB, Docampo Palacios ML, Pellon RF, Damonte EB. 2012. Inhibition of Junin virus RNA synthesis by an antiviral acridone derivative. Antiviral Res 93:16–22. doi: 10.1016/j.antiviral.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 36.El-Sibai M, Nalbant P, Pang H, Flinn RJ, Sarmiento C, Macaluso F, Cammer M, Condeelis JS, Hahn KM, Backer JM. 2007. Cdc42 is required for EGF-stimulated protrusion and motility in MTLn3 carcinoma cells. J Cell Sci 120:3465–3474. doi: 10.1242/jcs.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridley AJ. 2006. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Pothula S, Bazan HE, Chandrasekher G. 2013. Regulation of Cdc42 expression and signaling is critical for promoting corneal epithelial wound healing. Invest Ophthalmol Vis Sci 54:5343–5352. doi: 10.1167/iovs.13-11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. 2004. Activation of endogenous Cdc42 visualized in living cells. Science 305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 40.Surviladze Z, Waller A, Strouse JJ, Bologa C, Ursu O, Salas V, Parkinson JF, Phillips GK, Romero E, Wandinger-Ness A, Sklar LA, Schroeder C, Simpson D, Noth J, Wang J, Golden J, Aube J. 2010. A potent and selective inhibitor of Cdc42 GTPase. Probe reports from the NIH Molecular Libraries Program. NIH Molecular Libraries Program, Bethesda, MD. [PubMed] [Google Scholar]
- 41.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 42.McCluskey A, Daniel JA, Hadzic G, Chau N, Clayton EL, Mariana A, Whiting A, Gorgani NN, Lloyd J, Quan A, Moshkanbaryans L, Krishnan S, Perera S, Chircop M, von Kleist L, McGeachie AB, Howes MT, Parton RG, Campbell M, Sakoff JA, Wang X, Sun JY, Robertson MJ, Deane FM, Nguyen TH, Meunier FA, Cousin MA, Robinson PJ. 2013. Building a better Dynasore: the Dyngo compounds potently inhibit dynamin and endocytosis. Traffic 14:1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castilla V, Palermo LM, Coto CE. 2001. Involvement of vacuolar proton ATPase in Junin virus multiplication. Arch Virol 146:251–263. doi: 10.1007/s007050170173. [DOI] [PubMed] [Google Scholar]
- 44.Stoorvogel W, Kerstens S, Fritzsche I, den Hartigh JC, Oud R, van der Heyden MA, Voortman J, van Bergen en Henegouwen PM. 2004. Sorting of ligand-activated epidermal growth factor receptor to lysosomes requires its actin-binding domain. J Biol Chem 279:11562–11569. doi: 10.1074/jbc.M308449200. [DOI] [PubMed] [Google Scholar]
- 45.Reis RC, Sorgine MH, Coelho-Sampaio T. 1998. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol 75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- 46.Shacka JJ, Klocke BJ, Shibata M, Uchiyama Y, Datta G, Schmidt RE, Roth KA. 2006. Bafilomycin A1 inhibits chloroquine-induced death of cerebellar granule neurons. Mol Pharmacol 69:1125–1136. doi: 10.1124/mol.105.018408. [DOI] [PubMed] [Google Scholar]
- 47.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petit CS, Roczniak-Ferguson A, Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cordo SM, Cesio y Acuna M, Candurra NA. 2005. Polarized entry and release of Junin virus, a New World arenavirus. J Gen Virol 86:1475–1479. doi: 10.1099/vir.0.80473-0. [DOI] [PubMed] [Google Scholar]
- 50.Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. 1985. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A 82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashok A, Atwood WJ. 2003. Contrasting roles of endosomal pH and the cytoskeleton in infection of human glial cells by JC virus and simian virus 40. J Virol 77:1347–1356. doi: 10.1128/JVI.77.2.1347-1356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. 1994. Inhibition of Rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukhopadhyay A, Barbieri AM, Funato K, Roberts R, Stahl PD. 1997. Sequential actions of Rab5 and Rab7 regulate endocytosis in the Xenopus oocyte. J Cell Biol 136:1227–1237. doi: 10.1083/jcb.136.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Lu Q, Zhou H. November 2012. Cdc42 inhibitor and uses thereof. US patent 20140194451 A1.
- 55.Bolken TC, Laquerre S, Zhang Y, Bailey TR, Pevear DC, Kickner SS, Sperzel LE, Jones KF, Warren TK, Lund SA, Kirkwood-Watts DL, King DS, Shurtleff AC, Guttieri MC, Deng Y, Bleam M, Hruby DE. 2006. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antiviral Res 69:86–97. doi: 10.1016/j.antiviral.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larson RA, Dai D, Hosack VT, Tan Y, Bolken TC, Hruby DE, Amberg SM. 2008. Identification of a broad-spectrum arenavirus entry inhibitor. J Virol 82:10768–10775. doi: 10.1128/JVI.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gowen BB, Juelich TL, Sefing EJ, Brasel T, Smith JK, Zhang L, Tigabu B, Hill TE, Yun T, Pietzsch C, Furuta Y, Freiberg AN. 2013. Favipiravir (T-705) inhibits Junin virus infection and reduces mortality in a guinea pig model of Argentine hemorrhagic fever. PLoS Negl Trop Dis 7:e2614. doi: 10.1371/journal.pntd.0002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rathbun JY, Droniou ME, Damoiseaux R, Haworth KG, Henley JE, Exline CM, Choe H, Cannon PM. 2015. Novel arenavirus entry inhibitors discovered using a minigenome rescue system for high-throughput drug screening. J Virol 89:8428–8443. doi: 10.1128/JVI.00997-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasquato A, Burri DJ, Kunz S. 2012. Current drug discovery strategies against arenavirus infections. Expert Rev Anti Infect Ther 10:1297–1309. doi: 10.1586/eri.12.117. [DOI] [PubMed] [Google Scholar]
- 60.Thomas CJ, Casquilho-Gray HE, York J, DeCamp DL, Dai D, Petrilli EB, Boger DL, Slayden RA, Amberg SM, Sprang SR, Nunberg JH. 2011. A specific interaction of small molecule entry inhibitors with the envelope glycoprotein complex of the Junin hemorrhagic fever arenavirus. J Biol Chem 286:6192–6200. doi: 10.1074/jbc.M110.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chikte S, Panchal N, Warnes G. 2014. Use of LysoTracker dyes: a flow cytometric study of autophagy. Cytometry A 85:169–178. doi: 10.1002/cyto.a.22312. [DOI] [PubMed] [Google Scholar]
- 62.Droniou-Bonzom ME, Reignier T, Oldenburg JE, Cox AU, Exline CM, Rathbun JY, Cannon PM. 2011. Substitutions in the glycoprotein (GP) of the Candid #1 vaccine strain of Junin virus increase dependence on human transferrin receptor 1 for entry and destabilize the metastable conformation of GP. J Virol 85:13457–13462. doi: 10.1128/JVI.05616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog 4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fritz R, Stiasny K, Heinz FX. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol 183:353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quirin K, Eschli B, Scheu I, Poort L, Kartenbeck J, Helenius A. 2008. Lymphocytic choriomeningitis virus uses a novel endocytic pathway for infectious entry via late endosomes. Virology 378:21–33. doi: 10.1016/j.virol.2008.04.046. [DOI] [PubMed] [Google Scholar]
- 66.Blumenthal R, Bali-Puri A, Walter A, Covell D, Eidelman O. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J Biol Chem 262:13614–13619. [PubMed] [Google Scholar]
- 67.Perez L, Carrasco L. 1993. Entry of poliovirus into cells does not require a low-pH step. J Virol 67:4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. 2007. Imaging poliovirus entry in live cells. PLoS Biol 5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.