Abstract
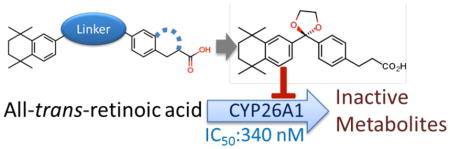
Cytochrome P450 CYP26 enzymes are responsible for all-trans-retinoic acid (atRA) clearance. Inhibition of CYP26 enzymes will increase endogenous atRA concentrations and is an attractive therapeutic target. However, the selectivity and potency of the existing atRA metabolism inhibitors towards CYP26A1 and CYP26B1 is unknown, and no selective CYP26A1 or CYP26B1 inhibitors have been developed. Here the synthesis and potent inhibitory activity of the first CYP26A1 selective inhibitors is reported. A series of non-azole CYP26A1 selective inhibitors was identified with low nM potency. The lead compound 3-{4-[2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl] phenyl}4 propanoic acid (24) had 43-fold selectivity towards CYP26A1 with an IC50 of 340 nM. Compound 24 and its two structural analogs also inhibited atRA metabolism in HepG2 cells resulting in increased potency of atRA towards RAR activation. The identified compounds have potential to become novel treatments aiming to elevate endogenous atRA concentrations and may be useful as cotreatment with atRA to combat therapy resistance.
Keywords: Cytochrome P450, retinoic acid, CYP26, retinoic acid receptor
Graphical Abstract
INTRODUCTION
Retinoic acid (RA) is the endogenous active metabolite of vitamin A (retinol). It is an essential regulator of cell growth, cell cycle and differentiation.1,2 RA exists in several isomeric forms, all-trans retinoic acid (atRA), 13-cis-retinoic acid (13-cisRA), 9,13-dicis-retinoic acid (9,13-dicisRA), and 9-cis-retinoic acid (9-cisRA).3 Of these isomers, atRA is believed to be the main biologically active form exhibiting its activity through binding predominantly to nuclear retinoic acid receptors (RARs). Through its binding to nuclear receptors and regulating gene transcription, atRA plays a critical role in many biological processes including reproduction, maintenance of skin and epithelia, regulation of the immune system and T- and B-cell function, and in fetal development.4–6 In addition, imbalance in vitamin A and RA homeostasis has been implied in development and progression of several human diseases such as acne, psoriasis and ichthyosis, type II diabetes, neurodegenerative diseases and some cancers.1 The potential of RA in the treatment of human diseases has been manifested in the success of 13-cisRA (Accutane) in treatment of acne and neuroblastoma, and the effectiveness of atRA in the treatment of acute promyelocytic leukemia (APL). However, the use and long term efficacy of atRA has been limited in all of the therapeutic areas by its high clearance that limits oral administration of this agent, and the resistance that develops towards atRA due to autoinduction of its metabolism.7 Additionally 13cis-RA via isomerization to atRA, and atRA both cause broad activation of RARs that is believed to be responsible for some of the untoward side effects of these agents. Due to these shortcomings, the full clinical and therapeutic potential of retinoids has not been reached.
The clearance of atRA is predominantly mediated by cytochrome P450 family 26 enzymes (CYP26).8,9 The CYP26 family has three isoforms: CYP26A1, CYP26B1 and CYP26C1 which are expressed in a tissue specific manner. In the human liver CYP26A1 is the predominant CYP26 enzyme responsible for majority of the hepatic clearance of atRA.10 CYP26B1 protein is not detected in human liver and the expression of this isoform appears to be restricted to extrahepatic sites.11 The biological role and expression pattern of CYP26C1 is not well characterized but this enzyme appears to be responsible for 9-cisRA metabolism and be less critical in regulating RA concentrations than CYP26A1 and CYP26B1.
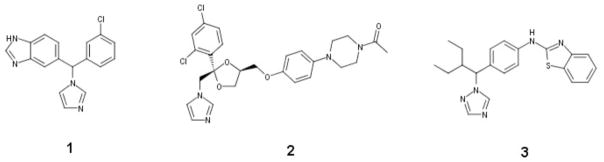
As atRA is predominantly cleared by the CYP26 enzymes, it has been proposed that inhibition of the CYP26 enzymes will increase atRA concentrations in target tissues, and inhibition of CYP26 enzymes during atRA or 13-cisRA therapy could be used to combat disease resistance.12 However, inhibitors of CYP26 that have been tested in humans (Figure 1) commonly suffer from broad spectrum CYP inhibition resulting in untoward side effects. Examples of this include the inhibition of steroid metabolism by liarozole (compound 1) and the drug-drug interactions seen with ketoconazole (compound 2).7 The lack of specificity of these inhibitors is likely due to the imidazole and triazole moieties in the compounds that are known to coordinate with the heme iron providing additional potency towards CYP inhibition. Although more selective triazole and imidazole containing CYP26 inhibitors have been developed,7 none of these compounds has yet reached human trials and therefore their drug-drug interaction potential in vivo is unknown. In addition, none of the existing inhibitors of CYP26s have been evaluated for specificity between the CYP26 enzymes and therefore the benefits and shortcomings of selective or broad CYP26 inhibition have not been established.
Figure 1.
Chemical structures of previously identified CYP26 inhibitors, 1 (liarozole), 2 (ketoconazole), and 3 (talarozole also called R11586613).
CYP26 enzymes appear to have cell type and tissue specific expression,11 and therefore it is likely that specific CYP26A1 or CYP26B1 inhibitors would be beneficial for increasing RA concentrations in selected target tissues without causing broad side effects. Specifically, since CYP26A1 appears to be the predominant CYP26 enzyme in the liver, selective inhibition of CYP26A1 is expected to decrease systemic clearance of atRA and therefore be useful for combatting therapy resistance in individuals being treated with atRA or 13cisRA. In addition, it is likely that use of CYP26A1 specific inhibitor will selectively increase RA concentrations in the liver. Notably, based on conditional knock-out mouse studies, CYP26B1 inhibition could be a liability for male reproduction as testes cell type specific CYP26B1 knock-out mice show adverse effects on the testes.14 Similarly, T-cell specific CYP26B1 knockouts have shown that CYP26B1 regulates iTreg and Th17 cell polarization,15 an effect that may be a liability for a broad CYP26 inhibitor. A CYP26A1 selective inhibitor would avoid both of these effects and likely be beneficial when systemic atRA concentrations are targeted.
The aim of this study was to identify novel non-azole inhibitors of CYP26 enzymes that would be selective for CYP26A1 and devoid of CYP26B1 inhibition. A series of novel CYP26A1 selective inhibitors was identified with low nM potency for CYP26A1 inhibition. The lead compound from the series had 43-fold selectivity towards CYP26A1 in comparison to CYP26B1 and other P450 enzymes and inhibited CYP26A1 with an IC50 of 340 nM. The lead compound and two of its structural analogs also inhibited atRA metabolism in HepG2 cells resulting in increased potency of atRA towards RAR activation in these cells. The identified compounds are the first CYP26A1 selective inhibitors discovered and have potential to become novel treatments aiming to elevate endogenous RA concentrations in circulation and in the liver. These compounds would also be useful as cotreatment with atRA to combat therapy resistance likely caused by increased systemic clearance of atRA and autoinduciton of CYP26A1, particularly in the liver.
RESULTS
Chemistry
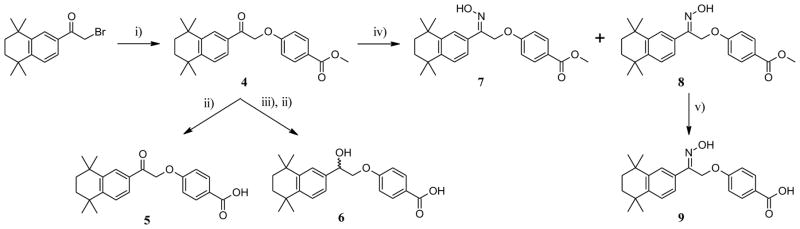
Initial coupling of 2-bromo-1-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethan-1-one with methyl-4-hydroxybenzoate in the presence of K2CO3 in methyl ethyl ketone under microwave conditions, gave ester 4 in good yield (Scheme 1). It was further hydrolyzed under basic conditions to afford carboxylic acid 5 in good yield. Reduction of the ketone moiety of compound 4 to alcohol by treatment with NaBH4, followed by basic hydrolysis at the ester position, gave carboxylic acid 6. Reaction of 4 with hydroxylamine hydrochloride in MeOH under reflux formed oxime isomers E (7) and Z (8), in moderate and good yield, respectively. Z-isomer 8 was further hydrolyzed in aqueous K2CO3 to provide carboxylic acid 9.
Scheme 1a.
aReagents and conditions: (i) Methyl-4-hydroxybenzoate, K2CO3/MEK, μW, 100°C, 2×20 min; (ii) NaOH, THF. EtOH, H2O, 80°C, 12 h; (iii) NaBH4, THF, r.t., 1 h; (iv) H2NOH.HCl/MeOH, pyridine, reflux, 6 h; (v) K2CO3 (2M)/MeOH, reflux, 80°C.
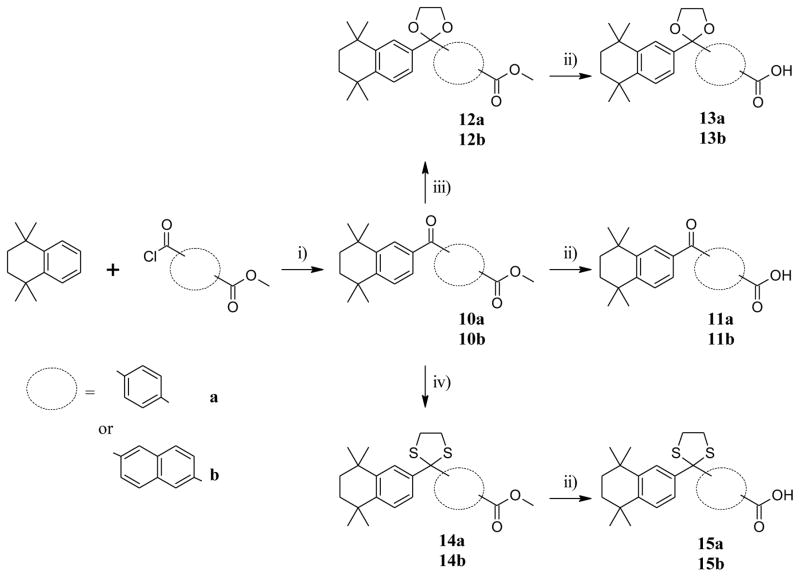
Friedel-Crafts acylation16,17 of 1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene (TTN) with methyl 4-(chlorocarbonyl)benzoate and methyl 6-(chlorocarbonyl)naphthalene-2-carboxylate in CH2Cl2 in the presence of AlCl3 yielded the expected ketones 10a and 10b (Scheme 2). The ester moieties of 10a and 10b were then hydrolyzed in basic conditions to respectively afford carboxylic acids 11a and 11b in excellent yields. Further, treatment of the ketone 10a and 10b with ethylene glycol17 resulted in the formation of ketals 12a and 12b which after basic hydrolysis, afforded the expected carboxylic acids 13a and 13b. Likewise thioketalization of 10a and 10b was achieved by reaction with ethanedithiol in the presence of boron trifluoride diethyl etherate (BF3•Et2O), giving the thioketals 14a and 14b in excellent yields which after basic hydrolysis, gave carboxylic acids 15a and 15b.
Scheme 2a.
aReagents and conditions: (i) AlCl3, CH2Cl2, r.t., overnight; (ii) K2CO3 (2M)/MeOH, reflux, 80°C,; (iii) (CH2OH)2, pTsOH, toluene, reflux, overnight; (iv) (CH2SH)2, BF3.Et2O, CH2Cl2, r.t., 0.5–1h.
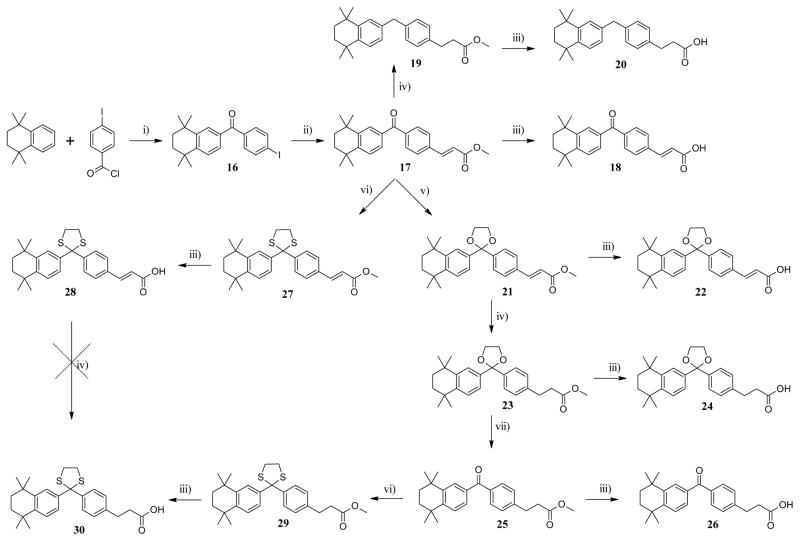
Friedel-Crafts acylation16 of TTN by 4-iodobenzoyl chloride according to the previously described procedure, afforded ketone 16 in good yield (Scheme 3). Heck18,19 coupling reaction of 16 with methyl acrylate under palladium catalysis proceeded smoothly to give alkene 17. Compound 17 was then hydrolyzed under basic conditions, to give carboxylic acid 18. Hydrogenation of 17 under Pd/C catalysis not only reduced the alkene moiety, but also the carbonyl moiety resulting in the formation of benzylbenzene 19 which upon subsequent basic hydrolysis provided the corresponding carboxylic acid 20. Ketalization of 17 was carried out according to the previously described procedure, to afford ketal 21 in moderate yield which after basic hydrolysis gave carboxylic acid 22. Hydrogenation of 21 in presence of Pd/C followed by hydrolysis gave carboxylic acid 24. The ketal moiety of 23 was cleaved with iodine in acetone under reflux according to a modified procedure of Sun et al,20 to afford ketone 25 which after saponification yielded to the carboxylic acid 26. Thioketalization of 17 followed by hydrolysis provided carboxylic acid 28. The alkene moiety of 28 was found to be resistant to hydrogenation using Pd-C or Wilkinson’s catalyst. Therefore, ketal 29 was obtained directly from the thioketalization of ketone 25 which after basic hydrolysis gave carboxylic acid 30.
Scheme 3a.
aReagents and conditions: (i) AlCl3, CH2Cl2, r.t., overnight; (ii) methyl acrylate, Pd(OAc)2, NEt3, DMF, 100°C, 6 h; (iii) LiOH aq. 1N, THF, reflux, 3 h or r.t., overnight; (iv) H2, Pd/C, EtOH/EtOAc 20 psi, 7–12 h; (v) (CH2OH)2, pTsOH, toluene, reflux, overnight; (vi) (CH2SH)2, BF3.Et2O, CH2Cl2, r.t., 0.5–1 h; (vii) I2, acetone, 4Å MS, reflux, 16 h.
Sulfone 34 was prepared in 4 steps starting from TTN. TTN was first converted to disulfide 31 (Scheme 4) by treatment with chlorosulfonic acid followed by heating in presence of zinc in hydrochloric acid.21 Disulfide 31 was then reduced and coupled with ethyl 3-(4-bromophenyl)propanoate in presence of PdCl2(dppf) and zinc,22 to give diaryl sulfide 32. Oxidation of 32 with oxone produced the expected sulfone 33 which after hydrolysis under basic condition afforded carboxylic acid 34 in good yield.
Scheme 4a.
aReagents and conditions: i) 1) Chlorosulfonic acid, 0°C to r.t., 3h, 2) Zn, EtOH, conc. HCl, reflux 45 min, r.t. overnight; ii) ethyl 3-(4-bromophenyl)propanoate, PdCl2(dppf), Zn, THF, reflux, 24h; iii) oxone, H2O, MeOH, 0°C to r.t., 12h; iv) LiOH 1N, THF, H2O, r.t. 12h.
To explore the role of the atoms of oxygen (or sulfur) from the dioxolane (dithiolane) moiety, we investigated the activity of a novel series of compounds with a sulfide moiety or a sulfoxide moiety as a linker in between the two aromatic rings. Synthesis started with preparation of iodo TTN derivative 35 from TTN (Scheme 5).23 Diaryl sulfides 36a and 36b were then prepared by nickel catalyzed coupling with the iodo derivative 32 in the presence of polymer-supported borohydride in butanol, to respectively provide the 4- and 3-mercaptophenylacetic acid methyl esters.24 Methyl esters 36a and 36b were obtained as mixtures with their butyl ester analogues (25% butyl ester). Biaryl sulfides 36a and 36b were then oxidized with oxone in methanol to afford the expected biaryl sulfones 37a and 37b. Compounds 36a, 36b, 37a and 37b were then hydrolyzed under basic conditions to afford the desired carboxylic acids 38a, 38b, 39a and 39b respectively in good to excellent yields.
Scheme 5a.
aReagents and conditions: i) I2, H2SO4, HIO4, AcOH, H2O, 70°C ii) 3- or 4-mercaptophenylacetic acid methyl ester, borohydride, polymer supported, (bpy)2NiBr2, dioxane, ButOH, 130–145°C, 3 h to 5 h; iii) oxone, H2O, MeOH, 0°C to r.t., 12 h; iv) LiOH 1N, THF, H2O, r.t. 12h.
Enzyme Inhibition and structure-activity relationship
A method was previously developed using recombinant CYP26A1 microsomes and 9-cis-RA as a substrate to test for inhibition of CYP26A1.25 Previously, 9-cis-RA was chosen as the screening substrate instead of atRA due to the lack of substrate and metabolite depletion in the screening assays with 9-cis-RA, and the lower affinity of 9-cis-RA to CYP26A1 when compared to atRA allowing screening of inhibitors at substrate concentration at or below the Km value.25 Similar to CYP26A1, atRA is a high affinity substrate of CYP26B1 and the metabolites formed by CYP26B1 are also substrates of CYP26B111 confounding kinetic inhibition measurements. To allow characterization of inhibition of CYP26B1, 9-cis-RA turnover by recombinant CYP26B1 was also characterized. 9-cis-RA was found to be a substrate of CYP26B1 and the formation of the metabolite that coeluted with 9-cis-4-OH-RA was NADPH dependent. The Km and Vmax for 9-cis-4-OH-RA formation by CYP26B1 were of 555 nM and 3.6 pmol/min/pmol P450, respectively. Based on the Km value, all compounds were evaluated for CYP26B1 inhibition at a substrate (9-cis-RA) concentration of 100 nM (concentration ≪Km to increase sensitivity and decrease the dependence of IC50 values on inhibition mechanism).
In an effort to identify lead CYP26A1 specific inhibitors, we decided to focus our screening on synthetic analogues of RA. The compounds were selected on the basis of structural similarity with atRA, the endogenous substrate of CYP26A1, and a small number of analogues were prepared to obtain an initial characterization of the structure-activity relationships (SAR) of CYP26A1 binding and selectivity. This approach was based on prior work showing that some RAR agonists bind and inhibit CYP26A1.25 The classic RAMBAs (compounds 1 and 3) and three commercially available synthetic retinoids 40 (bexarotene) (Table 1), 13a (SR1123726) (Table 2), and 15b (MM1125327) (Table 1), and two close analogues 13b and 15a, were tested for their CYP26A1 and CYP26B1 inhibition potency. This screening showed that both the RAMBAs and synthetic retinoids inhibited CYP26A1 and CYP26B1 (Table 1 and 2). The azole containing compound 3 inhibited both CYP26A1 and CYP26B1 potently whereas compound 1 was a more potent inhibitor of CYP26B1 than CYP26A1. Neither one of these compounds provided sufficient specificity and selectivity to inhibit atRA metabolism specifically by CYP26A1.
Table 1.
IC50 values and 95% confidence intervals of the IC50 for the RAMBAs and synthetic retinoids against CYP26A1 and CYP26B1 for compounds 1, 3, 40, and the naphthyl analogues 11b, 13b and 15b.
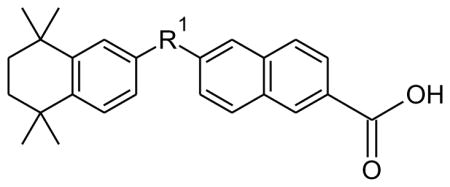
| |||
|---|---|---|---|
| Compound | R1 | Activity (μM) | |
| CYP26A1 | CYP26B1 | ||
| 1 | _ | 1.9 (1.5–2.3) | 0.018 (0.013–0.027) |
| 3 | _ | 0.0051a (0.0034–0.0068) | 0.00046a (0.000069–0.00085) |
 40 |
_ | 13.5 (8.6–21.3) | 5.9 (2.6–13.3) |
| 11b |
|
1.63 (0.95–2.9) | 0.52 (0.4–0.7) |
| 13b |
|
0.11 (0.05–0.24) | 1.03 (0.62–1.7) |
| 15b |
|
0.06 (0.04–0.10) | 1.03 (0.69–1.5) |
Inhibition constant determined based on tight binding quadratic equation.
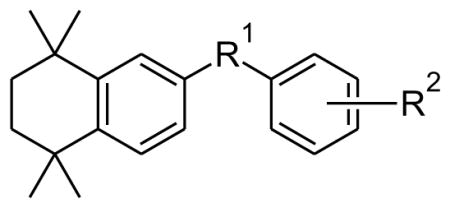
Table 2.
IC50 values and 95% confidence intervals of the IC50 for the novel CYP26 inhibitors against CYP26A1 and CYP26B1.
| |||||
|---|---|---|---|---|---|
| Cpd | R1 | R2 | Activity (μM) | ||
| CYP26A1 | CYP26B1 | ||||
| 5 |
|
|
para | 4.0 (2.5–6.3) | 0.93 (0.61–1.4) |
| 6 |
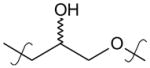
|

|
para | 2.8 (1.7–4.8) | 1.4 (0.77–2.6) |
| 9 |
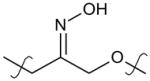
|

|
para | 3.5 (2.2–5.6) | 1.4 (0.4–4.8) |
| 11a |

|

|
para | >25 | >25 |
| 13a |

|

|
para | 7.8 (2.8–21.4) | 12.6b |
| 15a |

|

|
para | 1.3 (0.64–2.54) | 1.1 (0.1–11.7) |
| 18 |

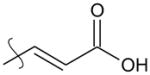
|
|
para | 4.7 | >5 |
| 20 |
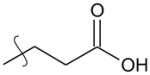
|
|
para | 1.66 | 1.7 (0.8–3.5) |
| 22 |

|

|
para | 0.24 (0.10–0.59) | >10 |
| 24 |

|

|
para | 0.34 (0.06–2.0) | 14.5 (3.5–59.7) |
| 26 |

|

|
para | >5 | >5 |
| 28 |

|

|
para | >10 | 4.4 (1.8–10.6) |
| 30 |

|

|
para | 0.27 (0.15–0.47) | 0.68 (0.2–2.4) |
| 34 |
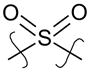
|

|
para | >5 | 6.2 |
| 38a |
|
|
para | 10.6 (5–23) | >5 |
| 38b |
|

|
para | >5 | >5 |
| 39a |

|

|
para | >5 | >5 |
| 39b |

|

|
meta | >5 | >5 |
Maximum inhibition 59% at 20μM
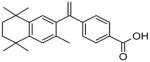
Compounds 40, 13a and 15a had IC50 values in the micromolar range and did not demonstrate significant selectivity for CYP26A1, whereas the RARγ selective antagonist 15b28 had a 0.061 μM IC50 for CYP26A1 and 1.03 μM IC50 for CYP26B1 (Table 1) demonstrating a 17-fold higher IC50 towards CYP26B1 than CYP26A1. Compound 13b, the ketal homologue of 15b displayed the same CYP26B1 inhibitory activity but exhibited a reduced CYP26A1 activity compared to the thioketal analogues. Replacement of the original dithiolane linker of 15b by a carbonyl in compounds 11b and 18, and synthesis of their shorter homologues 11a and 15a resulted in a reduction of CYP26A1 inhibitory activities and a modest enhancement of CYP26B1 inhibition for compound 11b. All these data together indicate that the geometry of the thioketal ring and the distance between the carboxylic acid and the thioketal moiety are crucial for the CYP26A1 inhibitory activity. Our subsequent research efforts to complete SAR of synthetic retinoids led to the synthesis of compounds 5, 6 and 9. We decided to incorporate polar groups in the spacer between the two aromatic moieties to explore potential hydrogen bonding interaction with CYP26A1. The distance between the carboxylic acid and the tetramethyltetrahydronaphthalene moiety was kept in the same range as for compound 15b. The naphthyl moiety was replaced by a methyleneoxyphenyl moiety to minimize potential steric hindrance that would prevent hydrogen bonding. However, such a chemical modification did not produce significant improvement in affinity towards CYP26A1. We also explored the effect of potential steric hindrance on compound 11b by replacing the naphthyl moiety by a styryl moiety (18) This change led to a dramatic loss of CYP26B1 inhibition and a 2 to 3-fold reduction in CYP26A1 activity. A similar trend was observed for the CYP26B1 activity of the styryl derivative 22 whereas CYP26A1 activity was comparable to the naphthyl analogue 13b. Surprisingly compound 28, the styryl analogue of compound 15b was not active towards CYP26A1. While 15b is a potent inhibitor of CYP26A1 it is not an ideal inhibitor to be developed further due to its high RAR affinity.28 Therefore, 15b was chosen as a lead compound for the design and synthesis of novel inhibitors that would be free of RAR binding (agonist or antagonist), and inhibition of other P450s except CYP26A1. We decided to explore the effect of the replacement of the naphthyl group bearing the carboxylic acid by a phenethyl group on CYP26 and RAR activities. Phenethyl derivative 24 displayed a lower CYP26B1 activity compared to the naphthyl analogue 13b while CYP26A1 activity was only reduced by 2 to 3-fold. CYP26A1 inhibitory activity for the thioketal analogue 30 was in the same range of compound 24 whereas CYP26B1 activity was increased. We also explored whether it is possible to improve CYP26A1 inhibitory activity and selectivity by modifying the substituents on the one-carbon between the two ring systems of compound 24. These will result in the modification of the spatial orientation of the two aromatic moieties. Such a chemical modification did not produce significant improvement in CYP26 inhibition for analogues 20, 26 and 34. In the course of our initial exploratory work on the SARs for this novel series of CYP26 inhibitors, we also explored whether it is possible to impact the spatial positioning of the carboxylic moiety by shortening the phenethyl moiety. The thioethers (38a, 38b) and the sulfones (39a, 39b) lacked inhibitory activity towards CYP26A1 and CYP26B1.
In summary, in contrast to the relatively robust nanomolar inhibition of CYP26A1 by the dithiolane and dioxolane compounds in this series, and the micromolar inhibition of CYP26A1 by majority of the remaining compounds, the inhibitory potency of the compounds (5–39) towards CYP26B1 was variable with only compounds 11b, 30 and 5 showing submicromolar potency towards CYP26B1. Interestingly, compounds 5, 6, 9, 11b, 34 were more potent inhibitors of CYP26B1 than CYP26A1 suggesting that interactions between these inhibitors and the CYP26 enzymes within the active site are different allowing development of CYP26 isoform selective inhibitors. Compounds 15b and 13b were approximately 10–20 fold more potent inhibitors of CYP26A1 than CYP26B1. The replacement of the naphthyl group in the dioxolane compound 13b with a saturated or an unsaturated two carbon chain (22 and 24) led to over 10-fold decrease in the CYP26B1 inhibition with a lower effect on CYP26A1 inhibition. Therefore, these changes increased the CYP26A1 selectivity to >40-fold between CYP26B1 and CYP26A1. In contrast, with the dithiolane compound 15b, replacement of the naphthyl with an unsaturated or a saturated two carbon chain led to either considerable decrease in CYP26A1 inhibition (28) or loss of CYP26A1 selectivity (30).
RAR activation
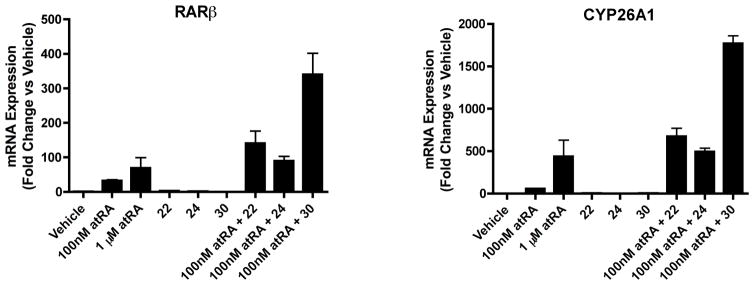
The initial lead compound in the series, 15b, is a known RAR antagonist28 and therefore it is possible that also the novel compounds identified will possess off-target effects via RAR binding. Hence, the two most promising selective inhibitors, compounds 22 and 24, as well as compound 30 which was a potent non-selective CYP26A1/B1 inhibitor, were assessed for RAR activation. We first used a luciferase-based reporter assay to screen compounds 22, 24 and 30 for RARα and RARγ activation at a concentration of 1 μM. Screen was accomplished using human RARα and RARγ reporter assay kits provided by Indigo Biosciences as reported previously. No measurable RARα and RARγ activation was detected for the three compounds (Data not shown). We then used the HepG2 cells, which have robust atRA metabolism activity and RAR mediated effects on gene transcription29 to test the effects of compounds 22, 24 and 30 on RAR activation in a cell system. The induction of the mRNA of the classic RAR target genes CYP26A1 and RARβ was evaluated after treatment with each of the novel compounds in comparison to vehicle and atRA (Figure 2). As shown, none of the three compounds resulted in activation of the pool of RARs in the HepG2 cells at a test concentration of 1 μM, a concentration at least 5 times greater than the IC50 value of these compounds towards CYP26A1.
Figure 2.
Induction of RARβ and CYP26A1 mRNA in HepG2 cells as measured by rt-PCR. HepG2 cells were treated with 100 nM or 1 μM atRA or with 1 μM compound 22, 24 or 30 alone or in combination with 100 nM atRA.
Inhibition of RA metabolism and Enhancement of RA potency
To further determine the biological activity of the lead compounds, their efficacy to increase the potency of atRA was tested in the HepG2 cells. It has been previously shown that atRA is efficiently metabolized in HepG2 cells and that the metabolism of atRA in HepG2 cells is predominantly mediated by CYP26A1.29 Therefore, RA metabolism in HepG2 cells and the activation of RARs (CYP26A1 and RARβ induction) by atRA in these cells provide a reliable test system for cellular activity of the compounds of interest. When HepG2 cells were treated with 100 nM atRA in combination with 1 μM of compounds 22, 24 and 30, the magnitude of induction of RARβ and CYP26A1 was significantly increased when compared to atRA treatment alone (Figure 2), demonstrating that all three compounds potentiate the effect of atRA in cells presumably via inhibiting atRA metabolism and increasing the cellular concentrations of atRA. Importantly the induction of RARβ and CYP26A1 following treatment with 100 nM atRA together with the inhibitors was significantly greater than the induction observed following treatment with 1 μM atRA alone demonstrating a considerable increase in the potency of atRA by the inhibitors. Furthermore these data demonstrate that these compounds do not exhibit an RAR antagonistic activity.
To test whether the increased induction of RAR target genes in the presence of the inhibitors and atRA was due to inhibition of atRA depletion in the cells, the concentrations of atRA in the media at the end of the 24-hour treatment were measured. Table 3 summarizes the effects of the lead compounds on atRA depletion in HepG2 cells. All three CYP26 inhibitors significantly increased atRA concentrations in the cell media with compound 30 having the greatest effect, approximately 9-fold increase in atRA concentrations in the media. Compound 22 increased atRA concentration 3.5-fold and compound 24 increased atRA concentration in the media by 2-fold. This increase in atRA concentrations correlated well with the observed magnitude of effect on potentiating atRA mediated RAR activation in cells with compound 30 also having the greatest effect on increasing atRA mediated induction of RARβ and CYP26A1.
Table 3.
atRA concentration in cell media after 24 hour treatment
| Treatments | atRA in cell media (nM) |
|---|---|
| Vehicle | 0.49 ± 0.33 |
| 100nM atRA | 13.2 ± 0.14 |
| 1μM atRA | 214 ± 13.6 |
| 1μM 22 | 1.09 ± 0.52 |
| 1μM 24 | 0.27 ± 0.09 |
| 100nM atRA + 1μM 22 | 45.5 ± 2.8* |
| 100nM atRA + 1μM 24 | 25.6 ± 3.4* |
| 100nM atRA + 1μM 30 | 113 ± 9* |
significantly different from the 100 nM RA treatment
Selectivity for CYP26A1
To determine the selectivity of the lead compounds towards other P450 enzymes the inhibition of CYP2D6, CYP3A4, CYP2C19 and CYP2B6 by compounds 22, 24 and 30 was evaluated. The inhibition data is summarized in Table 4. When screened at 10 μM inhibitor concentration, compounds 22 and 24 were free of any CYP3A4 inhibition while compound 30 inhibited CYP3A4 by 25%. The IC50 values for CYP3A4 inhibition for compounds 22 and 30 were 30 μM and 7.5 μM respectively. For compound 24 no concentration dependent inhibition of CYP3A4 was observed up to 100 μM with maximum 40% inhibition observed, demonstrating that compound 24 is unlikely to result in CYP3A4 inhibition at concentrations required to inhibit CYP26A1. Similarly, compound 24 was free of any significant inhibition of CYP2D6, CYP2C19 and CYP2B6 suggesting that this compound may become a novel CYP26A1 selective inhibitor. In contrast compound 22 inhibited CYP2D6 by 50% at 10 μM concentration and compound 30 inhibited CYP2B6 by 70% at 10 μM.
Table 4.
Percent inhibition for each of the tested CYP enzymes when screened with 10 μM of CYP26 inhibitors. NI indicates no inhibition observed. * shows that activity in the presence of the inhibitor was significantly different (p<0.01) from that in control.
activity in the presence of the test inhibitor (22 or 30) was significantly different from control
DISCUSSION
Inhibition of atRA metabolism has been a potential drug target for some decades now and several promising inhibitors of overall atRA metabolism have emerged from variety of drug development programs in industry and academia.7 Yet, in the development of the inhibitors of atRA metabolism or CYP26, the role of inhibition of the different CYP26 isoforms has never been addressed. In fact, the screening data in this study shows that talarozole is a potent inhibitor of both CYP26A1 and CYP26B1 while liarozole is a preferential inhibitor of CYP26B1. This difference in their inhibition profile likely contributes to some differences in their in vivo pharmacology and side effect profile. As CYP26A1 and CYP26B1 are expressed in a tissue specific manner,11 selective inhibition of either CYP26 isoform holds great promise in organ specific targeting of the retinoid system. This study is the first to test and identify CYP26 isoform specific inhibitors with the goal of identifying a CYP26A1 selective inhibitor that can be used to inhibit atRA metabolism in the liver. Compound 24 has 43-fold specificity towards CYP26A1 compared to CYP26B1 and it emerged as the lead inhibitor in this novel structural series. The selectivity of compound 24 together with compounds 15b, 13b and 22 is striking as up to now only minor differences in the biochemical structure function between CYP26A1 and CYP26B1 have been observed. Both enzymes use atRA as a substrate, bind atRA with high affinity and form very similar metabolites from atRA although at slightly different product ratios.11 This is despite the fact that the two CYP26 enzymes only share about 40% sequence similarity and hence would be expected to be structurally and functionally different. As the CYP26 enzymes have proven exceedingly difficult to express and purify in recombinant systems, no X-ray crystal structure or low-resolution structural information is available for these enzymes at present. Therefore the discovery of these novel selective inhibitors of CYP26A1 provide critical insight to the differences in the active site features and structure-activity relationships of these two enzymes and provide a lead for future development of selective inhibitors of the two enzymes.
The novel compounds in this series were developed using known RAR ligands as lead compounds and by exploring synthetically feasible modifications that could reduce RAR binding activity without compromising CYP26A1 inhibition. Replacement of the naphthyl group of the dioxolane derivative 13b by a phenethyl moiety in compound 24 appears to have a minor impact on CYP26A1 inhibitory activity while reducing CYP26B1 activity. The results in HepG2 cells show that indeed the lead compounds 22, 24 and 30 were devoid of RAR modulation while maintaining the CYP26 inhibition potency. In addition, the studies in HepG2 cells showed that compound 24 and the two other potent inhibitors in this series, compounds 22 and 30, were effective at inhibiting atRA metabolism in cell culture systems and they potentiated the activity of atRA in the cell system. To mimic the in vivo endogenous atRA concentrations in cells, the HepG2 cells were treated with 100 nM atRA and under these conditions, the lead inhibitors increased the activity of atRA by 5–100 fold and decreased atRA metabolism significantly. The HepG2 cells have been previously shown to be devoid of CYP26B1 expression although CYP26B1 expression is somewhat inducible in these cells.30 Similarly HepG2 cells do not express significant levels of the common drug metabolizing enzymes such as CYP3A4 and CYP2C enzymes. Therefore it is likely that the effect of the inhibitors on atRA activity in these cells is solely due to CYP26A1 inhibition.
In this investigation, we present a broad range of experimental data on a novel series of retinoid-based CYP26A1 inhibitors. Within this series, a SAR could be established that showed that lipophilic bulk induced by a dioxolane or a dithiolane on the spacer appears to be required for enhanced CYP26A1 inhibitory activity. Dioxolane analogues 22 and 24 were identified as selective CYP26A1 inhibitors. Classic CYP26 inhibitors have all included a triazole or imidazole functional group to increase their affinity to CYP26s via coordination to the heme iron.31,32 However, atRA itself binds to CYP26A1 with 10 nM affinity suggesting that high affinity inhibitors devoid of the imidazole or triazole group prone to broad spectrum CYP inhibition can be developed. Indeed, this study shows that nM inhibitors of both CYP26A1 and CYP26B1 free of imidazole or triazole functionality can be synthesized. These inhibitors were also free of inhibition of off-target CYPs such as CYP3A4, CYP2C19 and CYP2D6 at concentrations relevant for CYP26A1 inhibition. However, these compounds do include a ketal that may be metabolically labile and compound 22 is an unsaturated acid. These functional groups are potential in vivo liabilities of these compounds and require further study. Nevertheless, these compounds provide the first structural leads for selective CYP26A1 inhibition, and have the potential to become the first selective CYP26A1 inhibitors for testing in preclinical models. Eventually these compounds may become useful for treatment of diseases that would require increasing systemic or liver atRA concentrations or in inhibiting atRA metabolism when atRA is administered clinically to combat therapy resistance that develops due to CYP26A1 induction by atRA.
EXPERIMENTAL SECTION
General Experimental methods
Moisture sensitive reactions were performed in an inert, dry atmosphere of argon in flame dried glassware. Air sensitive liquids were transferred via syringe or cannula through rubber septa. Reagent grade solvents were used for extraction and flash chromatography. THF was distilled from Na/benzophenone under argon; dichloromethane (CH2Cl2) and chloroform (CHCl3) were distilled from CaH2 under argon. All other reagents including 2-bromo-1-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethan-1-one and 1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene and solvents which were purchased from commercial sources, were used directly without further purification. The progress of reactions was checked by analytical thin-layer chromatography (Sorbent Technologies, Silica G TLC plates w/UV 254). Flash column chromatography was performed using prepacked Biotage SNAP/ZIP cartridges on a Biotage Isolera One instrument. Microwave reactions were performed using a Biotage Initiator instrument. The solvent compositions reported for all chromatographic separations are on a volume/volume (v/v) basis. 1HNMR spectra were recorded at 400 or 500 MHz and are reported in parts per million (ppm) on the δ scale relative to tetramethylsilane as an internal standard. 13CNMR spectra were recorded at 100 or 125 MHz and are reported in parts per million (ppm) on the δ scale relative to CDCl3. Melting points were determined on a Stuart melting point apparatus from Bibby Scientific Limited and are uncorrected. LC-UV and high resolution mass spectrometry (HRMS) analyses were conducted on a Waters ACQUITY UPLC-series liquid chromatography system equipped with a diode array detector and LCT PREMIER XE™ time of flight (TOF) mass spectrometer operated on the electrospray ionization mode. Compounds were separated using a Phenomenex (Torrance, CA) NX-C18 column (50 × 4.60 mm) with 3 μm particle size and gradient elution at mobile phase flow rate of 0.4 mL/min with solvents A: water with 0.1% formic acid and B: acetonitrile. The gradient was from 90% A 10% B to 2% A 98% B over 6.6 min then held isocratic till 13 min followed by return to initial conditions by 15 min. Compound purity was assigned on the basis of UV detection data at 254-nM detection by comparing relative peak areas of the signals. All final compounds were more than 95% pure.
General procedure for ester hydrolysis using K2CO3 (method A)
To a solution of ester (0.25 mmol) in MeOH (4 mL) was added aqueous K2CO3 (0.4 mL, 2.0 M) dropwise. The reaction mixture was stirred at 80°C for 1 hour. Completion of reaction was assessed by TLC. The reaction was then cooled (0 °C), acidified to pH 2 with 1.0 N HCl, and extracted with EtOAc (3 × 10 mL). The combined organic phases were dried (MgSO4) and concentrated under reduced pressure.
General procedure for ester hydrolysis using LiOH (method B)
To a solution of the ester (0.11 mmol) in anhydrous THF (0.5 mL) was added aqueous LiOH (0.25 mL, 1.0 N) dropwise. The reaction mixture was refluxed for 2 hours or stirred overnight at room temperature. The reaction was then cooled (0 °C), acidified to pH 2 with 1.0 N HCl, and extracted with EtOAc (3 × 10 mL). The combined organic phases were dried (MgSO4) and concentrated.
General procedure for Friedel-Crafts acylation (method C)
Acyl chloride (11.7 mmol) and 1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene material (2.0 g, 10.6 mmol) were weighed into a round-bottomed flask equipped with a stir bar and 24 mL dichloromethane added. Stirring commenced and AlCl3 (3.10 g, 23.3 mmol) was added slowly. After completion of the addition, stirring was continued at room temperature overnight. The reaction was then poured into an ice solution (40 mL), acidified with a 20% HCl solution (10 mL) and the organic layer removed. Then extracted further with 2×40 mL EtOAc. The organic layers were combined and washed with water (40 mL) and brine respectively, dried over MgSO4, filtered and concentrated under reduced pressure.
General procedure for formation of dioxolane (method D)
Diarylketone (2.0 mmol) was treated with ethylene glycol (2 mL) and a catalytic amount of pTsOH (20 mg) in toluene (10 mL) at 145°C overnight using a Dean Stark trap. After cooling to room temperature, the mixture was washed with saturated NaHCO3(aq.), brine, dried over MgSO4 and concentrated under reduced pressure.
General procedure for formation of dithiolane (method E)
To a solution of diarylketone (1.14 mmol) in CH2Cl2 (2 mL) at 0°C under argon, was added a solution of (CH2SH)2 (144 μL, 1.71 mmol) in CH2Cl2 (0.5 mL) followed by BF3•Et20 (217 μL, 1.71 mmol). The resulting mixture was stirred at 0°C for 1 h and then warmed to room temperature overnight. The reaction was quenched by pouring the mixture into saturated NaHCO3, and the mixture was extracted with 20 mL CH2C12. The combined organic layers were dried (MgSO4) and concentrated.
General procedure for hydrogenation or hydrogenolysis (method F)
Alkene (515 mg, 1.36 mmol) was placed in a hydrogenation apparatus equipped with a magnetic stir bar and EtOH/EtOAc (30 mL) added followed by Pd/C (240 mg). H2 gas was introduced at 20 psi pressure and reacted at room temperature for 7 hrs. The black solution was filtered using a celite pad and concentrated under reduced pressure.
General procedure for coupling thioaryl to aryl halides (method G)
To a mixture of thioaryl (0.71 mmol,1 eq.) and aryl halides (0.71 mmol, 1 eq.) in dioxane (5 mL) and butanol (5 mL), were added the borohydride, polymer supported (650 mg, 2.5 – 5 mmol/g, 3 eq.) and (bpy)2NiBr2 (70 mg, 0.2 eq.). The mixture was heated to 145°C under an atmosphere of nitrogen. The polymer beads were removed by filtration, and the mixture was concentrated to dryness before purification by flash chromatography using the following gradient system, (cyclohexane/dichloromethane): (20/80) to (0/100).
General procedure for oxidation of sulfide to sulfone (method H)
To a solution sulfide (0.26 mmol) in THF (7 mL), was added a solution of oxone (475 mg, 0.775 mmol, 3 eq.) in water (4 mL). The reaction was stirred overnight at room temperature. Water (10 mL) was added and the mixture was extracted with dichloromethane (3×50 mL).
Methyl-4-(2-oxo-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethoxy)benzoate (4)
A mixture 500 mg (1.62 mmol) of the commercially available 2-bromo-1-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethan-1-one, 271 mg of the commercially available methyl 4-hydroxybenzoate (1.78 mmol) and K2CO3 (246 mg, 1.78 mmol) in 10 ml of methyl ethyl ketone was heated 2 times at 100°C for 20 minutes in the microwave and then filtered and concentrated on a rotary evaporator under vacuum. The product was purified by crystallization with a (heptane/ethyl acetate: 70/30) solution to give the expected compound (450 mg; 73%) as a white solid. mp = 112–114°C. 1H NMR (500 MHz, CDCl3) δ 7.98 (d, J = 8.9 Hz, 2H), 7.97 (d, J = 1.9 Hz, 1H), 7.72 (dd, J = 8.3, 1.9 Hz, 1H), 7.43 (d, J = 8.3 Hz, 1H), 6.95 (d, J = 8.9 Hz, 2H), 5.32 (s, 2H), 3.88 (s, 3H), 1.71 (s, 4H), 1.32 (s, 6H), 1.31 (s, 6H). 13C NMR (126 MHz, CDCl3) δ 193.34, 166.63, 161.72, 152.00, 145.85, 131.74, 131.58, 127.14, 126.68, 125.02, 123.32, 114.32, 70.48, 51.87, 34.80, 34.70, 34.58, 34.42, 31.73, 31.53. MS (TOF ESI+): m/z calcd for C24H29O4+ (M+H)+ calcd. 381.21, found 381.20.
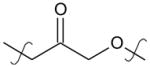
4-(2-Oxo-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethoxy)benzoic acid (5)
Compound 4 (200 mg, 0.53 mmol) was stirred at 80°C with sodium hydroxide (200 mg) in a mixture of EtOH, THF and water (10 mL, 10 mL and 1.5 mL) for 12 hours. The reaction was then cooled at r.t., acidified to pH 2 with 1.0 N HCl, and extracted with EtOAc (3 × 20 mL). The combined organic phases were dried (MgSO4) and concentrated under reduced pressure. The residue was purified by crystallization in a mixture of heptane and EtOAc (70/30) to provide 149 mg (77%) of a white solid. mp = 162–163°C. 1H NMR (400 MHz, CDCl3) δ 8.06 (d, J = 8.9 Hz, 2H), 7.98 (d, J = 1.7 Hz, 1H), 7.72 (dd, J = 8.3, 1.8 Hz, 1H), 7.43 (d, J = 8.3 Hz, 1H), 6.97 (d, J = 8.9 Hz, 2H), 5.34 (s, 2H), 1.72 (s, 4H), 1.32 (s, 6H), 1.31 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 193.30, 170.88, 162.50, 152.11, 145.95, 132.40, 131.80, 127.21, 126.77, 125.07, 122.38, 114.51, 70.56, 34.86, 34.77, 34.66, 34.47, 31.77, 31.57. HPLC (t= 8.12 min, 96%). MS (TOF ESI+) for C23H27O4+ (M+H)+ calcd. 367.20, found 367.19.
4-(2-Hydroxy-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethoxy)benzoic acid (6)
To a solution of compound 4 95 mg (0,25 mmole) dissolved in 3 mL of THF under nitrogen, was carefully added 10 mg of sodium borohydride(0,25 mmole). The mixture was stirred 3 h at room temperature. The residue was taken up in 10 mL water, and the aqueous layer was extracted with 3×10 mL EtOAc. The combined organic extract was washed with 30 mL water and 30 mL brine respectively. The organic solution was dried (MgSO4), filtered, and concentrated to give 75 mg (79%) of a colorless oil. The resulting ester (70 mg, 0.18 mmol) was stirred at 80°C with sodium hydroxide (70 mg) in a mixture of EtOH, THF and water (3mL, 3mL and 0.5 mL) for 12 hours. The reaction was then cooled at r.t., acidified to pH 2 with 1.0 N HCl, and extracted with EtOAc (3 × 10 mL). The combined organic phases were dried (MgSO4) and concentrated under reduced pressure. The residue was purified by crystallization in a mixture of heptane and EtOAc (70/30) to provide 40 mg (60%) of a white solid. mp = 156–158°C. 1H NMR (400 MHz, CDCl3) δ 8.06 (d, J = 8.8 Hz, 2H), 7.38 (d, J = 1.5 Hz, 1H), 7.34 (d, J = 8.1 Hz, 1H), 7.21 (dd, J = 8.1, 1.6 Hz, 1H), 6.97 (d, J = 8.9 Hz, 2H), 5.11 (dd, J = 8.3, 3.4 Hz, 1H), 4.23 – 4.04 (m, 2H), 1.69 (s, 4H), 1.35 – 1.22 (m, 12H).13C NMR (101 MHz, CDCl3) δ 171.39, 162.97, 145.28, 145.17, 136.35, 132.39, 126.92, 124.46, 123.41, 122.09, 114.37, 73.45, 72.63, 35.07, 34.99, 34.35, 34.19, 31.87, 31.85, 31.83. HPLC (t= 8.34 min, 97%). MS (TOF ESI+) for C23H29O4 + (M+H)+ calcd. 369.21, found 369.21.
(E)-Methyl-4-(2-(hydroxyimino)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethoxy-)benzoate (7)
The E-isomer was recrystallized from EtOAc/heptane (3/7) to give 69 mg (24%) of a white solid. mp = 146–149°C. 1H NMR (500 MHz, CDCl3) δ 8.21 (s, 1H), 7.98 (d, J = 9.0 Hz, 2H), 7.59 (d, J = 1.8 Hz, 1H), 7.40 (dd, J = 8.3, 1.9 Hz, 1H), 7.35 (d, J = 8.3 Hz, 1H), 6.98 (d, J = 9.0 Hz, 2H), 4.95 (s, 2H), 3.88 (s, 3H), 1.69 (s, 4H), 1.28 (s, 6H), 1.25 (s, 6H). 13C NMR (126 MHz, CDCl3) δ 166.79, 161.93, 153.08, 146.73, 144.85, 131.56, 127.49, 126.93, 126.48, 125.51, 123.11, 114.55, 69.32, 51.91, 34.88, 34.83, 34.34, 34.29, 31.78, 31.65. MS (TOF ESI+) for C24H30NO4 + (M+H)+ calcd. 396.22, found 396.21.
(Z)-Methyl-4-(2-(hydroxyimino)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethoxy-)benzoate (8)
A solution of compound 5 (275 mg, 0.72 mmol) in MeOH (7 mL) was treated with hydroxylamine hydrochloride (100 mg, 1.45 mmol) and pyridine (235 μL, 2.9 mmol), and the mixture was heated at reflux for 6 h. The mixture was cooled to room temperature, and the MeOH was removed in vacuo. The residue was taken up in 20 mL water, and the aqueous layer was extracted with 3×30 mL EtOAc. The combined organic extract was washed with 20 mL water and 20 mL brine respectively. The organic solution was dried (MgSO4), filtered, and concentrated. The residue was purified by flash column chromatography (heptane-ethyl acetate 100:0 v/v increasing to 70:30 v/v)to yield the 2 isomers E and Z. The Z-isomer was recrystallized from EtOAc/heptane to give 165 mg (71%) of a white solid. mp = 123–125°C. 1H NMR (500 MHz, CDCl3) δ 8.79 (s, 1H), 7.97 (d, J = 8.8 Hz, 2H), 7.60 (d, J = 1.9 Hz, 1H), 7.39 (dd, J = 8.3, 1.9 Hz, 1H), 7.29 (d, J = 8.3 Hz, 1H), 6.98 (d, J = 8.8 Hz, 2H), 5.32 (s, 2H), 3.88 (s, 3H), 1.67 (s, 4H), 1.27 (s, 6H), 1.26 (s, 6H). 13C NMR (126 MHz, CDCl3) δ 166.81, 161.81, 155.20, 146.73, 144.91, 131.59, 130.12, 126.69, 125.35, 124.01, 123.09, 114.25, 59.86, 51.91, 34.93, 34.85, 34.30, 34.29, 31.78, 31.65. MS (TOF ESI+) for C24H30NO4+ (M+H)+ calcd. 396.22, found 396.21.
(Z)-4-(2-(Hydroxyimino)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2- yl)ethoxy)benzoic acid (9)
Compound 8 (100 mg, 0.25 mmol) was hydrolysed according to method A. The residue was purified by flash column chromatography (DCM-MeOH 100:0 v/v increasing to 95:5 v/v) to provide a white solid (25 mg, 27%). mp = 208–211°C. 1H NMR (500 MHz, CDCl3) δ 7.96 (d, J = 8.9 Hz, 2H), 7.58 (d, J = 1.9 Hz, 1H), 7.48 (s, 1H), 7.39 (dd, J = 8.3, 1.9 Hz, 1H), 7.28 (d, J = 8.3 Hz, 1H), 6.99 (d, J = 8.9 Hz, 2H), 5.33 (s, 2H), 1.68 (s, 4H), 1.26 (s, 6H), 1.25 (s, 6H). HPLC (t= 8.14 min, 95%). HRMS (TOF ESI+) for C23H28NO4+ (M+H)+ calcd. 382.2018, found 382.2012.
Methyl-4-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)benzoate (10a)
Compound 10a was prepared according to method C starting from 2.0 g of the commercially available 1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene (10.6 mmol). The crude was crystallized from hot EtOAc by addition of 20 mL MeOH, resulting in white crystals after cooling. Yield 2.15 g (58%). mp = 140–142°C. 1H NMR (400 MHz, CDCl3) δ 8.15 (d, J = 8.4 Hz, 2H), 7.83 (d, J = 8.4 Hz, 2H), 7.79 (d, J = 1.8 Hz, 1H), 7.54 (dd, J = 8.2, 1.9 Hz, 1H), 7.41 (d, J = 8.2 Hz, 1H), 3.97 (s, 3H), 1.72 (s, 4H), 1.32 (s, 6H), 1.29 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 195.88, 166.40, 150.78, 145.30, 141.88, 134.15, 132.91, 129.67, 129.39, 128.89, 127.35, 126.70, 52.40, 34.81, 34.74, 34.41, 31.74, 31.60. MS (TOF ESI+) for C23H27O3+ (M+H)+ calcd. 351.20, found 351.20.
Methyl-6-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)-2-naphthoate (10b)
Compound 10b was synthesized according to method C starting from 875 mg of 1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene (4.65 mmol). The compound 10b crystallized from hot EtOAc by addition of 20 mL MeOH, resulting in white crystals after cooling (1.94 g, 69%). mp = 134–136°C. 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.30 (s, 1H), 8.13 (dd, J = 8.6, 1.6 Hz, 1H), 8.06 (d, J = 8.5 Hz, 1H), 7.98 (dd, J = 8.5, 1.6 Hz, 2H), 7.97 (d, J = 8.6 Hz, 1H), 7.87 (d, J = 1.8 Hz, 1H), 7.60 (dd, J = 8.2, 1.8 Hz, 1H), 7.44 (d, J = 8.2 Hz, 1H), 4.01 (s, 3H), 1.74 (s, 4H), 1.34 (s, 6H), 1.31 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 196.21, 166.86, 150.47, 145.26, 137.33, 134.59, 134.45, 134.13, 130.91, 130.68, 129.49, 129.33, 128.94, 127.44, 126.69, 126.66, 126.08, 52.40, 34.85, 34.78, 34.73, 34.43, 31.76, 31.63. MS (TOF ESI+) for C27H29O3+ (M+H)+ calcd. 401.21, found 401.21.
4-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)benzoic acid (11a)
Compound 10a (60 mg, 0.17 mmol) was hydrolyzed according to method A. The resultant oil was triturated with heptanes to yield compound 11 as a white solid (52 mg, 91%). mp = 194–195°C. 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 8.3 Hz, 2H), 7.87 (d, J = 8.3 Hz, 2H), 7.81 (d, J = 1.8 Hz, 1H), 7.55 (dd, J = 8.2, 1.8 Hz, 1H), 7.42 (d, J = 8.2 Hz, 1H), 1.73 (s, 4H), 1.33 (s, 6H), 1.30 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 195.84, 170.91, 150.97, 145.41, 142.74, 134.03, 131.89, 130.05, 129.75, 128.91, 127.42, 126.76, 34.81, 34.77, 34.74, 34.43, 31.76, 31.61, 26.91. HPLC (t= 8.82 min, 100%). HRMS (TOF ESI+) for C22H25O3 + (M+H)+ calcd. 337.1804, found 337.1811.
6-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)-2-naphthoic acid (11b)
Compound 10b (220 mg, 0.55 mmol) was hydrolyzed according to method A. The crude was crystalized from cyclohexane to provide compound 11b as a white solid (90 mg, 95%). mp = 224–225°C. 1H NMR (400 MHz, MeOD) δ 8.70 (s, 1H), 8.30 (s, 1H), 8.15 (d, J = 8.5 Hz, 1H), 8.08 (d, J = 8.5 Hz, 1H), 7.98 (t, J = 8.0 Hz, 2H), 7.86 (s, 1H), 7.61 (d, J = 8.2 Hz, 1H), 7.46 (d, J = 8.2 Hz, 1H), 1.75 (s, 4H), 1.35 (s, 6H), 1.32 (s, 6H). 13C NMR (101 MHz, MeOD) δ 196.84, 168.40, 150.59, 145.17, 136.98, 134.34, 134.32, 134.12, 130.94, 130.80, 129.70, 129.45, 129.30, 128.88, 127.30, 126.59, 126.33, 126.27, 34.65, 34.58, 34.26, 31.53, 31.40. HPLC (t= 9.40 min, 95%). HRMS (TOF ESI+) for C26H27O3 + (M+H)+ calcd. 387.1960, found 387.1998.
Methyl-4-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)benzoate (12a)
Compound 10a (705 mg, 2.01 mmol) was treated treated according to method D. After work-up, the residue was purified by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 60:40 v/v) to yield a white solid, 641 mg (81%). mp= 148–148°C. 1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.3 Hz, 2H), 7.61 (d, J = 8.3 Hz, 2H), 7.42 (d, J = 1.8 Hz, 1H), 7.23 (d, J = 8.3 Hz, 1H), 7.17 (dd, J = 8.2, 1.8 Hz, 1H), 4.14 – 4.07 (m, 2H), 4.07 – 3.97 (m, 2H), 3.90 (s, 3H), 1.65 (s, 4H), 1.23 (s, 12H). 13C NMR (101 MHz, CDCl3) δ 166.88, 147.43, 144.86, 144.65, 138.21, 129.61, 129.47, 126.45, 126.17, 123.82, 123.25, 109.22, 64.90, 52.07, 35.04, 34.99, 34.31, 34.09, 31.83, 31.76. MS (TOF ESI+) for C25H31O4+ (M+H)+ calcd. 395.22, found 395.23.
Methyl-6-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)-2-naphthoate (12b)
Compound 11b (570 mg, 1.42 mmol) was treated according to method D. The residue was purified by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 60:40 v/v) to yield a white solid (557 mg, 88%). mp= 182–184°C. 1H NMR (400 MHz, CDCl3) δ 8.57 (s, 1H), 8.07 (s, 1H), 8.05 (dd, J = 8.6, 1.5 Hz, 1H), 7.89 (dd, J = 8.5, 5.1 Hz, 2H), 7.67 (dd, J = 8.6, 1.6 Hz, 1H), 7.52 (s, 1H), 7.24 (d, J = 9.1 Hz, 2H), 4.19 – 4.11 (m, 2H), 4.11 – 4.02 (m, 2H), 3.97 (s, 3H), 1.65 (s, 4H), 1.25 (s, 6H), 1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 167.22, 144.81, 144.63, 142.46, 138.37, 135.12, 132.07, 130.67, 129.38, 128.63, 127.59, 126.45, 125.43, 125.35, 124.55, 123.85, 123.47, 109.51, 64.98, 52.20, 35.06, 35.00, 34.32, 34.10, 31.85, 31.77. MS (TOF ESI+) for C29H33O4 + (MH+) calcd. 445.24, found 445.23.
4-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)benzoic acid (13a)
Compound 12a (236 mg, 0.59 mmol) was hydrolyzed according to method A. The crude was crystalized from cyclohexane and washed to provide compound 13a as a white solid (161 mg, 72%). mp = 242–243°C. 1H NMR (400 MHz, CDCl3) δ 12.05 (bs, 1H), 8.07 (d, J = 8.5 Hz, 2H), 7.65 (d, J = 8.5 Hz, 2H), 7.44 (d, J = 1.8 Hz, 1H), 7.24 (d, J = 8.3 Hz, 1H), 7.18 (dd, J = 8.3, 1.9 Hz, 1H), 4.15 – 4.08 (m, 2H), 4.08 – 3.99 (m, 2H), 1.65 (s, 4H), 1.24 (s, 6H), 1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.55, 148.46, 144.93, 144.72, 138.13, 130.14, 128.67, 126.50, 126.30, 123.79, 123.24, 109.19, 64.94, 35.05, 35.00, 34.33, 34.11, 31.84, 31.77. HPLC (t= 8.85 min, 98%). HRMS (TOF MS ES+) for C24H29O4+ (MH+) calcd. 381.2066, found 381.2038.
6-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)-2-naphthoic acid (13b)
Compound 12b was hydrolyzed according to method B. The crude oil was purified by flash column chromatography (cyclohexane-EtOAc-HOAc 100:0:0.1 v/v increasing to 80:20:0.1 v/v) to provide a white solid (30 mg, 63%). mp = 222–223°C. 1H NMR (400 MHz, CDCl3) δ 8.68 (s, 1H), 8.12 (d, J = 8.6 Hz, 2H), 7.94 (dd, J = 8.5, 4.6 Hz, 2H), 7.70 (dd, J = 8.6, 1.3 Hz, 1H), 7.52 (s, 1H), 7.24 (d, J = 6.1 Hz, 2H), 4.17 (m,, 2H), 4.13 – 4.03 (m, 2H), 1.65 (s, 4H), 1.25 (s, 6H), 1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 172.09, 144.86, 144.67, 142.95, 138.32, 135.60, 132.02, 131.77, 129.58, 128.82, 126.68, 126.48, 125.60, 125.52, 124.60, 123.87, 123.46, 109.50, 65.01, 35.06, 35.00, 34.33, 34.11, 31.85, 31.77. HPLC (t= 7.74 min, 95%). HRMS (TOF ESI+) for C28H31O4 + (M+H)+ calcd. 431.2222, found 431.2222.
Methyl-4-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)benzoate (14a)
Compound 10a (400 mg, 1.14 mmol) was treated according to method E. The crude residue was purified by flash column chromatography (cyclohexane-DCM 100:0 v/v increasing to 60:40 v/v) to yield 14a as a white solid (482 mg, 99%). mp= 103–105°C. 1H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 8.7 Hz, 2H), 7.72 (d, J = 8.6 Hz, 2H), 7.45 (d, J = 2.1 Hz, 1H), 7.22 (dd, J = 8.4, 2.1 Hz, 1H), 7.17 (d, J = 8.3 Hz, 1H), 3.90 (s, 3H), 3.52 – 3.41 (m, 2H), 3.41 – 3.30 (m, 2H), 1.65 (s, 4H), 1.24 (s, 6H), 1.20 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 166.79, 150.39, 144.37, 144.03, 140.08, 129.12, 128.77, 128.37, 126.24, 125.34, 52.07, 40.23, 35.02, 34.98, 34.33, 34.02, 31.77, 31.71. MS (TOF ESI+) for C25H31O2S2+ (M+H)+ calcd. 427.18, found 427.18.
Methyl-6-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)-2-naphthoate (14b)
Compound 10b (0.83 g, 2.07 mmol) was treated according to method E. The crude was purified by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 95:5 v/v) to yield 14b as a white solid(851 mg, 86%). mp= 153–155°C. 1H NMR (400 MHz, CDCl3) δ 8.55 (s, 1H), 8.24 (d, J = 1.1 Hz, 1H), 8.06 (dd, J = 8.6, 1.5 Hz, 1H), 7.87 (d, J = 8.6 Hz, 1H), 7.84 (d, J = 8.8 Hz, 1H), 7.67 (dd, J = 8.7, 1.9 Hz, 1H), 7.53 (d, J = 2.1 Hz, 1H), 7.24 (dd, J = 8.6, 1.5 Hz, 1H), 7.18 (d, J = 8.4 Hz, 1H), 3.97 (s, 3H), 3.57 – 3.45 (m, 2H), 3.44 – 3.33 (m, 2H), 1.65 (s, 4H), 1.25 (s, 6H), 1.19 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 167.23, 145.05, 144.41, 144.06, 139.86, 134.81, 131.49, 130.38, 128.91, 128.60, 128.24, 127.64, 126.36, 126.26, 125.92, 125.63, 125.53, 52.20, 40.18, 35.05, 35.01, 34.35, 34.04, 31.77, 31.73. MS (TOF ESI+) for C29H33O2S2+ (M+H)+ calcd. 477.20, found 477.19.
4-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)benzoic acid (15a)
Compound 14a (100 mg, 0.23 mmol) in MeOH (5 mL) was hydrolyzed according to method A. The crude was crystalized from cyclohexane to provide compound 15a as a white solid (90 mg, 95%). mp = 211–212°C. 1H NMR (400 MHz, CDCl3) δ 8.03 (d, J = 8.6 Hz, 2H), 7.76 (d, J = 8.6 Hz, 2H), 7.46 (d, J = 2.0 Hz, 1H), 7.22 (dd, J = 8.4, 2.1 Hz, 1H), 7.18 (d, J = 8.3 Hz, 1H), 3.53 – 3.43 (m, 2H), 3.42 – 3.32 (m, 2H), 1.65 (s, 4H), 1.25 (s, 6H), 1.20 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.58, 151.50, 144.44, 144.10, 139.98, 129.80, 128.49, 127.83, 126.30, 126.21, 125.33, 40.28, 35.02, 34.99, 34.35, 34.03, 31.78, 31.72. HPLC (t= 9.35 min, 95%). HRMS (TOF MS ES+) for C24H29O2S2 + (MH+) calcd. 413.1609, found 413.1648.
6-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)-2-naphthoic acid (15b)
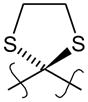
To a suspension of the ester 14b (680 mg, 1.43 mmol) in 75% aqueous MeOH (21 mL) was added KOH (0.77 g), and the mixture was stirred at 80°C for 3 h during which time the compound dissolved. The solution was cooled to room temperature, acidified with 1 N HCl, and a white solid precipitated out. The solid was filtered, washed with water and allowed to dry to yield a white solid, 621 mg (94%). mp= 270–272°C. 1H NMR (400 MHz, DMSO) δ 13.08 (bs, 1H), 8.54 (s, 1H), 8.29 (s, 1H), 8.04 (d, J = 8.6 Hz, 1H), 8.00 (d, J = 8.7 Hz, 2H), 8.00 – 7.96 (m, 1H), 7.53 (dd, J = 8.7, 1.7 Hz, 1H), 7.47 (d, J = 1.9 Hz, 1H), 7.23 (d, J = 8.4 Hz, 1H), 7.14 (dd, J = 8.4, 1.8 Hz, 1H), 3.59 – 3.47 (m, 2H), 3.44 – 3.27 (m, 2H), 1.60 (s, 4H), 1.20 (s, 6H), 1.13 (s, 6H). 13C NMR (101 MHz, DMSO) δ 167.35, 144.88, 143.82, 143.49, 140.23, 134.15, 131.02, 129.84, 129.02, 128.63, 128.45, 127.76, 126.25, 125.76, 125.41, 125.28, 124.99, 76.31, 34.41, 34.36, 33.90, 33.67, 31.55, 31.43. HPLC (t= 9.97 min, 95%). HRMS (TOF ESI+) for C28H31O2S2 + (M+H)+ calcd. 463.1765, found 463.1769.
(4-Iodophenyl)(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)methanone (16)
Compound 16 was prepared according to method C starting from 1.90 g of the commercially available 1,1,4,4- tetramethyl-1,2,3,4-tetrahydronaphthalene (10 mmol). After work-up, the crude product was purified by flash column chromatography to yield compound 16 as a yellow oil, 3.57 g (85%). 1H NMR (400 MHz, CDCl3) δ 7.84 (d, J = 8.4 Hz, 2H), 7.76 (d, J = 1.8 Hz, 1H), 7.51 (dd, J = 8.2, 2.0 Hz, 1H), 7.52 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 8.2 Hz, 1H), 1.72 (s, 4H), 1.31 (s, 6H), 1.29 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 195.71, 150.44, 145.20, 137.43, 137.31, 134.27, 131.42, 128.72, 127.21, 126.63, 99.67, 34.81, 34.73, 34.69, 34.39, 31.74, 31.60. MS (TOF ESI+) for C21H24IO+ (M+H)+ calcd. 419.09, found 419.09.
(E)-Methyl 3-(4-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)phenyl)acrylate (17)
Prepared according to the modified procedure reported by Heck et al.(9, 10) To a 50-mL round bottomed flask equipped with a magnetic stir bar was added 1.65 g (3.94 mmol) of iodide 16 and methyl acrylate (1.0 mL, 9.86 mmol). Pd(OAc)2 (9 mg, 0.04 mmol), (2-Tol)3P (48 mg, 0.16 mmol) and Et3N (2.2 mL, 15.78 mmol) were then simultaneously added and a cooled condenser attached. The solution was heated at 100°C for 6 hours. The reaction mixture was allowed to cool to room temperature and 20 mL EtOAc was added After filtration, the solution was concentrated under reduced pressure and purified by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 95:5 v/v) to yield 17 as a yellow solid (1.22 g, 82%). mp= 138–140°C. 1H NMR (400 MHz, CDCl3) δ 7.82 (d, J = 8.3 Hz, 2H), 7.79 (d, J = 1.8 Hz, 1H), 7.75 (d, J = 16.1 Hz, 1H), 7.63 (d, J = 8.3 Hz, 2H), 7.54 (dd, J = 8.2, 1.9 Hz, 1H), 7.41 (d, J = 8.2 Hz, 1H), 6.55 (d, J = 16.0 Hz, 1H), 3.83 (s, 3H), 1.72 (s, 4H), 1.32 (s, 6H), 1.30 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 195.88, 167.16, 150.58, 145.36, 143.72, 139.42, 137.90, 134.59, 130.65, 128.90, 127.90, 127.44, 126.76, 120.09, 52.01, 34.98, 34.90, 34.85, 34.55, 31.90, 31.77. MS (TOF MS ES+) for C25H29O3 (MH+) calcd. 377.21, found 377.21.
(E)-3-(4-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)phenyl)acrylic acid (18)
Compound 17 (0.82 mmol) was hydrolyzed according to method B. The resultant oil was purified by flash column chromatography (cyclohexane-EtOAc-HOAc 100:0:0.1 v/v increasing to 80:20:0.1 v/v) to provide compound 18 as a white solid (278 mg, 93%). mp = 220–221°C. 1H NMR (400 MHz, CDCl3) δ 7.85 (d, J = 16.8 Hz, 1H), 7.84 (d, J = 8.0 Hz, 1H), 7.80 (d, J = 1.8 Hz, 1H), 7.66 (d, J = 8.2 Hz, 2H), 7.54 (dd, J = 8.2, 1.8 Hz, 1H), 7.41 (d, J = 8.2 Hz, 1H), 6.57 (d, J = 16.0 Hz, 1H), 1.73 (s, 4H), 1.32 (s, 6H), 1.30 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 195.94, 171.95, 150.72, 145.91, 145.41, 139.81, 137.49, 134.51, 130.70, 128.95, 128.21, 128.03, 127.48, 126.80, 119.58, 114.86, 34.98, 34.90, 34.88, 34.57, 31.91, 31.77. HPLC (t= 10.06 min, 80%, (E)-isomer). HRMS (TOF ESI−) for C24H25O3− (M−H)− calcd. 361.1804 found 361.1817.
Methyl-3-(4-((5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)methyl)phenyl)propanoate (19)
Compound 17 (515 mg, 1.36 mmol) was treated according to method F. The crude was purified on by column chromatography (cyclohexane-ethyl acetate: 100:0 v/v increasing to 90:10 v/v) to provide a colorless oil (479 mg, 93%). 1H NMR (400 MHz, CDCl3) δ 7.19 (d, J = 8.1 Hz, 1H), 7.15 – 7.08 (Aromatic, 5H), 6.90 (dd, J = 8.1, 1.9 Hz, 1H), 3.89 (s, 2H), 3.65 (s, 3H), 2.91 (t, J = 7.9 Hz, 2H), 2.61 (t, J = 7.9 Hz, 2H), 1.66 (s, 4H), 1.25 (s, 12H). 13C NMR (101 MHz, CDCl3) δ 173.52, 144.87, 142.56, 139.40, 138.15, 138.03, 129.17, 128.40, 126.97, 126.69, 126.21, 51.70, 41.41, 35.87, 35.30, 35.25, 34.30, 34.06, 32.02, 32.00, 30.67.
3-{4-[(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)methyl]phenyl}9 propanoic acid (20)
Compound 19 (422 mg, 1.16 mmol) was hydrolyzed according to method B. The crude was purified by flash column chromatography (DCM-MeOH 100:0 v/v increasing to 95:5 v/v) to provide compound 20 as a white solid (301 mg, 77%). 1H NMR (400 MHz, CDCl3) δ 7.20 (d, J = 8.1 Hz, 1H), 7.12 (5H), 6.90 (dd, J = 8.1, 1.8 Hz, 1H), 3.89 (s, 2H), 2.92 (t, J = 7.8 Hz, 2H), 2.66 (t, J = 7.8 Hz, 2H), 1.66 (s, 4H), 1.25 (s, 12H). mp= 142–143°C. HPLC (t= 9.83 min, 95%). HRMS (TOF ESI+) for C24H31O2+ (M+H)+ calcd. 351.2324, found 351.2321.
(E)-Methyl-3-(4-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)- phenyl)acrylate (21)
Compound 17 (650 mg, 1.73 mmol) was treated according to method D. The crude was puridied by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 80:20 v/v). Compound 21 was obtained as a white solid, 385 mg (53%). mp= 155–157°C. 1H NMR (400 MHz, CDCl3) δ 7.67 (d, J = 16.0 Hz, 1H), 7.55 (d, J = 8.4 Hz, 2H), 7.48 (d, J = 8.3 Hz, 2H), 7.45 (d, J = 1.9 Hz, 1H), 7.23 (d, J = 8.2 Hz, 1H), 7.17 (dd, J = 8.2, 1.9 Hz, 1H), 6.42 (d, J = 16.0 Hz, 1H), 4.12 – 4.05 (m, 2H), 4.05 – 3.99 (m, 2H), 1.65 (s, 4H), 1.24 (s, 6H), 1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 167.56, 144.94, 144.92, 144.77, 144.62, 138.51, 134.09, 128.06, 126.86, 126.60, 123.91, 123.47, 118.03, 109.42, 77.48, 77.16, 76.84, 65.02, 51.84, 35.22, 35.16, 34.47, 34.25, 32.00, 31.93. MS (TOF ESI+) for C27H33O4+ (M+H)+ calcd. 421.24, found 421.24.
(E)-3-(4-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl)phenyl)- acrylic acid (22)
Compound 21 was hydrolyzed according to method B. T The crude was purified by flash column chromatography (DCM-MeOH 100:0 v/v increasing to 95:5 v/v) to provide compound 22 as a white solid, 50 mg (56%). mp = 241–243°C. 1H NMR (400 MHz, CDCl3) δ 7.77 (d, J = 15.9 Hz, 1H), 7.57 (d, J = 8.3 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H), 7.45 (d, J = 1.6 Hz, 1H), 7.23 (d, J = 8.3 Hz, 1H), 7.17 (dd, J = 8.2, 1.7 Hz, 1H), 6.43 (d, J = 16.0 Hz, 1H), 4.21 – 3.93 (m, 4H), 1.65 (s, 4H), 1.25 (s, 6H), 1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 172.04, 146.81, 145.43, 144.97, 144.81, 138.45, 133.74, 128.37, 126.94, 126.62, 123.92, 123.46, 117.42, 109.40, 65.04, 35.22, 35.16, 34.48, 34.26, 32.01, 31.93. HPLC (t= 9.60 min, 100%). HRMS (TOF ESI+) for C26H31O4+ (MH+) calcd. 407.2222, found 407.2228.
Methyl 3-{4-[2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl]phenyl}propano ate (23)
Compound 21 (750 mg, 1.78 mmol) was treated according to method F. Further purification by flash column chromatography (cyclohexane-dichloromethane 80:20 v/v increasing to 0:100 v/v) yielded compound 23 as a white solid, 520 mg (70%). mp= 120–122°C. 1H NMR (400 MHz, CDCl3) δ 7.38 – 7.49 (m, 3H), 7.19 – 7.24 (m, 3H), 7.10 – 7.19 (m, 3H), 3.93 – 4.13 (m, 4H), 3.66 (s, 3H), 2.93 (t, J = 7.84 Hz, 2H), 2.61 (t, J = 7.84 Hz, 2H), 1.65 (s, 4H), 1.24 (2s, 12H). 13C NMR (101 MHz, CDCl3) δ 173.3, 144.5, 144.4, 140.5, 140.2, 138.8, 128.0, 126.4, 126.3, 123.9, 123.5, 109.6, 64.8, 51.6, 35.6, 35.2, 35.1, 34.3, 34.1, 31.9, 31.8, 30.6. MS (ESI+) [M+H]+ 423.24, calc. 423.25.
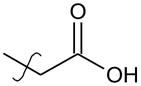
3-{4-[2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dioxolan-2-yl] phenyl}4 propanoic acid (24)
Compound 23 (250 mg, 0.60 mmol) was hydrolyzed according to method B. No further purification is needed after work-up. Compound 24 was obtained as a white solid (220 mg, 91%). mp= 219°–220°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.44 (m, 3 H), 7.19 – 7.24 (d, J=8.28 Hz 1 H), 7.17 (m, 3 H), 3.97 – 4.09 (m, 4 H), 2.94 (t, J=7.84 Hz, 2 H), 2.66 (t, J=7.78 Hz, 2 H), 1.65 (s, 4 H), 1.24 (s, 6 H),1.23 (s, 6H). 13C NMR (101 MHz, CDCl3) δ ppm 178.53, 144.53, 144.46, 140.58, 139.83, 138.78, 128.00, 126.46, 126.36, 123.88, 123.50, 109.56, 64.80, 35.40, 30.26, 35.14, 35.09, 34.33, 34.12, 31.89, 31.83. HPLC (t= 9.13 min, 99%). HRMS (TOF ESI−) for C26H31O4− (M−H)− calcd. 407.2222 found 407.2216.
Methyl-3-[4-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl) phenyl] propanoate (25)
A mixture of compound 23 (265 mg, 0.63 mmol, 1 eq.), iodine (160 mg, 0.63 mmol, 1 eq.) and 4Å molecular sieves (250 mg) in acetone (6 mL) was stirred for 8 hours under reflux according to a modified method of Sun et al.20 After work-up and purification by flash column chromatography (cyclohexane-DCM 80:20 v/v increasing to 20:80 v/v, compound 25 was obtained as a colorless oil (150 mg, 63%). 1H NMR (400 MHz, CDCl3) δ ppm 7.71 – 7.80 (m, 3 H), 7.53 (d, J=8.16 Hz, 1 H), 7.39 (d, J=8.16 Hz, 1 H), 7.31 (d, J=7.91 Hz, 2 H), 3.69 (s, 3 H), 3.04 (t, J=7.78 Hz, 2 H), 2.69 (t, J=7.72 Hz, 2 H), 1.72 (s, 4 H), 1.31 (s, 6 H), 1.30 (s, 6H). 13C NMR (101 MHz, CDCl3) δ ppm 196.25, 173.01, 149.95, 145.11, 145.01, 136.17, 134.89, 130.42, 128.71, 128.12, 127.29, 126.45, 51.72, 35.21, 34.89, 34.81, 34.66, 34.40, 31.76, 31.64, 30.86. MS (TOF ESI+) for C25H31O3+ (M+H)+ calcd. 379.23, found 379.22.
3-[4-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-carbonyl)phenyl] propanoic acid (26)
Compound 25 (50 mg, 0.132 mmol) was hydrolyzed according to method B. No further purification was needed after work-up. Compound 25 was obtained as a white solid (45 mg, 94%). mp= 153°–155°C. 1H NMR (400 MHz, CDCl3) δ 7.70 – 7.83 (m, 3H), 7.53 (dd, J = 1.82, 8.25 Hz, 1H), 7.39 (d, J = 8.25 Hz, 1H), 7.32 (d, J = 8.16 Hz, 2H), 3.05 (t, J = 7.72 Hz, 2H), 2.75 (t, J = 7.72 Hz, 2H), 1.72 (s, 6H), 1.31 (2s, 12H). 13C NMR (101 MHz, CDCl3) δ ppm 196.31, 150.01, 145.03, 144.76, 136.24, 134.84, 130.46, 128.73, 128.12, 127.30, 126.47, 35.01, 34.88, 34.80, 34.66, 34.40, 31.76, 31.64, 30.51. HPLC (t= 8.96 min, 99%). HRMS (TOF ESI−) (M−H)− calcd. 363.1960, found 363.1975.
(E)-Methyl-3-(4-(2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)- phenyl)acrylate (27)
Compound 17 (347 mg, 0.92 mmol) was treated according to method E. The crude was purified by flash column chromatography (cyclohexane-EtOAc 100:0 v/v increasing to 90:10 v/v) to provide 27 a colorless oil (380 mg, 91%). 1H NMR (400 MHz, CDCl3) δ 7.67 (d, J = 16.2 Hz, 1H), 7.66 (d, J = 8.4 Hz, 2H), 7.50 (d, J = 2.0 Hz, 1H), 7.44 (d, J = 8.4 Hz, 2H), 7.22 (dd, J = 8.4, 2.1 Hz, 1H), 7.18 (d, J = 8.3 Hz, 1H), 6.42 (d, J = 16.0 Hz, 1H), 3.80 (s, 3H), 3.50 – 3.40 (m, 2H), 3.42 – 3.32 (m, 2H), 1.66 (s, 4H), 1.25 (s, 6H), 1.21 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 167.59, 147.72, 144.48, 144.42, 144.11, 140.44, 133.28, 129.03, 127.73, 126.37, 125.58, 117.95, 51.85, 40.37, 35.20, 35.16, 34.51, 34.18, 31.94, 31.88. MS (TOF ESI+) for C27H33O2S2+ (M+H)+ calcd. 453.1922, found 453.1916.
(E)-3-(4-(2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl)phenyl)- acrylic acid (28)
Compound 27 (86 mg, 0.19 mmol) was hydrolyzed according to method B. Without any further purification, compound 28 is obtained as a white solid, 81 mg (97%). mp = 234–236°C. 1H NMR (400 MHz, CDCl3) δ 7.77 (d, J = 15.9 Hz, 1H), 7.68 (d, J = 8.4 Hz, 2H), 7.50 (d, J = 2.0 Hz, 1H), 7.47 (d, J = 8.4 Hz, 2H), 7.22 (dd, J = 8.4, 2.1 Hz, 1H), 7.18 (d, J = 8.3 Hz, 1H), 6.43 (d, J = 16.0 Hz, 1H), 3.46 (ddd, J = 12.3, 9.4, 7.4 Hz, 2H), 3.42 – 3.34 (m, 2H), 1.66 (s, 4H), 1.25 (s, 6H), 1.21 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 171.89, 148.29, 146.63, 144.52, 144.16, 140.37, 132.92, 129.11, 128.04, 126.40, 126.38, 125.57, 117.27, 40.40, 35.20, 35.16, 34.52, 34.19, 31.95, 31.89. HPLC (t= 10.02 min, 100%). HRMS (TOF ESI+) for C26H31O2S2+ (M+H)+ calcd. 439.1765, found 439.1765.
Methyl-3-{4-[2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl]phenyl}propanoate (29)
Compound 25 (142 mg, 0.375 mmol) was treated according to method E. The crude was purified by flash column chromatography (cyclohexane/EtOAc 95:5 v/v increasing to 75:25 v/v) to provide a colorless oil (116 mg, 68%). 1H NMR (400 MHz, CDCl3) δ 7.54 (d, J = 8.4 Hz, 2H), 7.52 (d, J = 2.1 Hz, 1H), 7.22 (dd, J = 8.4, 2.2 Hz, 1H), 7.16 (d, J = 8.4 Hz, 1H), 7.11 (d, J = 8.3 Hz, 2H), 3.67 (s, 3H), 3.48 – 3.29 (m, 4H), 2.93 (t, J = 7.9 Hz, 2H), 2.62 (t, J = 7.9 Hz, 2H), 1.65 (s, 4H), 1.24 (s, 6H), 1.21 (s, 6H). 13C NMR (101 MHz, CDCl3) δ ppm 173.32, 144.05, 143.64, 142.86, 140.91, 139.30, 128.50, 127.66, 126.28, 126.05, 125.54, 51.61, 40.04, 35.49, 35.10, 35.07, 34.33, 33.99, 31.78, 31.74, 30.41. MS (TOF ESI+) for C27H35O2S2O2+ (M+H)+ calcd. 455.21, found 455.21.
3-{4-[2-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-1,3-dithiolan-2-yl]phenyl}propanoic acid (30)
Compound 29 (102 mg, 0.224 mmol) was hydrolyzed according to method B. Without any further purification, compound 30 was obtained as a white solid (80 mg, 82%). mp= 165–167°C. 1H NMR (400 MHz, CDCl3) δ 7.55 (d, J = 8.3 Hz, 2H), 7.52 (d, J = 2.1 Hz, 1H), 7.22 (dd, J = 8.4, 2.2 Hz, 1H), 7.16 (d, J = 8.4 Hz, 1H), 7.12 (d, J = 7.4 Hz, 2H), 3.49 – 3.32 (m, 4H), 2.94 (t, J = 7.8 Hz, 2H), 2.67 (t, J = 7.8 Hz, 2H), 1.65 (s, 4H), 1.25 (d, J = 1.7 Hz, 6H), 1.21 (s, 6H). 13C NMR (101 MHz, CDCl3) δ ppm 177.50, 144.10, 143.68, 143.04, 140.91, 139.00, 128.58, 127.67, 126.32, 126.08, 125.57, 40.06, 35.15, 35.11, 34.35, 34.02, 31.80, 31.76, 30.11. HPLC (t= 9.67 min, 96%). HRMS (TOF ESI−) for C26H31O2S2− (M−H)− calcd. 439.1765, found 439.1743.
5,5,8,8-Tetramethyl-5, 6, 7,8-tetrahydro-naphthalene-2-disulfide (31)
5,5,8,8-Tetramethyl-5, 6, 7,8-tetrahydro-naphthalene-2-disulfide was prepared according to a procedure described by Boiteau et al.21 mp= 83–85°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.41 (d, J=1.88 Hz, 2 H), 7.28 (dd, J=8.28, 1.88 Hz, 2 H), 7.23 (d, J=8.28 Hz, 2 H), 1.65 (s, 8 H), 1.21 (s, 12 H,) 1.24 (s, 12 H). 13C NMR (101 MHz, CDCl3) δ ppm 145.79, 144.46, 134.05, 127.45, 126.89, 125.93, 34.97, 34.95, 34.45, 34.15, 31.75. MS (TOF ESI+) for C28H39S2+ (M+H)+ calcd. 439.24, found 439.25.
Ethyl-3-{4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)sulfanyl] phenyl} propanoate (32)
Compound 32 was prepared according to the method described by Fukuzawa et al.22 5, 5,8,8-Tetramethyl-5, 6, 7,8-tetrahydro-naphthalene-2-disulfide (100 mg, 0.228 mmol, 0.5eq.), PdCl2(dppf) (17 mg, 0.023 mmol, 0.05 eq.), and zinc (36 mg, 0.547 mmol, 1.2 eq.) were placed in a flask and then a solution of ethyl 3-(4-bromophenyl) propanoate (115 mg, 0.456 mmol, 1 eq.) in THF (3 mL) was added. The mixture was refluxed for 24 h and diluted with Et2O (30 mL) after cooling. The precipitate was removed by filtration and the filtrate was washed with brine and dried over CaSO4. After concentration of the organic layer, the crude was concentrated and grossly purified by column chromatography (cyclohexane-DCM 95:5 v/v increasing to 55:45 v/v). A mixture of 32 and ethyl 3-(4-bromophenyl) propanoate (30%) was obtained (125 mg). It was used in the next step without any further purification.
Ethyl 3-[4-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl)phenyl] propanoate (33)
Compound 32 (125 mg, 0.32 mmol) was treated according to method H. After drying over CaSO4 and evaporation, the crude was purified by flash column chromatography (cyclohexane-DCM 80:20 v/v increasing to 0:100 v/v). Compound 33 was obtained as a white solid (82 mg, 87%). mp= 105–106°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.88 – 7.92 (m, 1 H), 7.85 (m, J=8.16 Hz, 2 H), 7.58 (dd, J=8.41, 1.51 Hz, 1 H), 7.39 (d, J=8.41 Hz, 1 H), 7.33 (m, J=8.16 Hz, 2 H), 4.10 (q, J=7.11 Hz, 2 H), 2.99 (t, J=7.53 Hz, 2 H), 2.62 (t, J=7.53 Hz, 2 H), 1.68 (s, 4 H), 1.28 (s, 6 H), 1.25 (s, 6 H),1.19 (t, J=7.11 Hz, 3 H). 13C NMR (101 MHz, CDCl3) δ ppm 172.28, 150.80, 146.45, 146.21, 140.05, 138.55, 129.18, 127.81, 127.77, 125.89, 124.49, 60.63, 35.14, 34.69, 34.66, 34.61, 34.55, 31.70, 31.57, 30.68, 14.15. MS (TOF ESI+) for C50H65S2O4 + (2M+H)+ calcd. 857.40, found 857.40.
3-[4-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl)phenyl] propanoic acid (34)
3-Compound 33 (66 mg, 0.014 mmol) was hydrolyzed according to method B. With no further purification, compound 34 is obtained as a white solid (53 mg, 85%). mp= 211°C. 1H NMR (400 MHz, METHANOL-d4) δ ppm 7.90 (d, J=2.01 Hz, 1 H), 7.87 (m, J=8.41 Hz, 2 H), 7.65 (dd, J=8.41, 2.01 Hz, 1 H), 7.56 (d, J=8.41 Hz, 1 H), 7.48 (m, J=8.41 Hz, 2 H), 3.01 (t, J=7.59 Hz, 2 H), 2.65 (t, J=7.59 Hz, 2 H), 1.75 (s, 4 H), 1.32 (s, 6H), 1.30 (s, 6H). 13C NMR (101 MHz, METHANOL-d4) δ ppm 176.28, 152.55, 148.81,31. 147.99,87, 141.29, 140.25, 130.73, 129.42, 128.83, 127.00, 125.71, 36.03, 35.87, 35.78, 32.18, 32.02. HPLC (t= 8.44 min, 99%). HRMS (TOF ESI−) for C23H27O4S− (M−H)− calcd. 399.1630, found 399.1658.
6-Iodo-1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene (35)
6-iodo-1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene was prepared according to Christie et al.23
Methyl-2-{4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)sulfanyl] phenyl}acetate (36a)
Compound 35 (305 mg, 0.97 mmol) was treated according to method G. A colorless oil was obtained (225 mg, 60%) as a mixture of compound 36a and its butyl ester derivative (25%). 1H NMR (400 MHz, CDCl3) δ ppm 7.35 (d, J=1.88 Hz, 1 H), 7.22 – 7.25 (m, 3 H), 7.16 – 7.21 (m, 2 H), 7.09 (dd, J=8.28, 1.51 Hz, 1 H), 3.69 (s, 3 H), 3.59 (s, 2 H), 1.67 (s, 4 H), 1.24 (s, 6 H), 1.26 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 171.82, 146.10, 144.52, 135.78, 132.16, 130.95, 130.25, 129.97, 129.91, 129.10, 127.63, 52.09, 40.70, 34.97, 34.93, 34.37, 34.15, 31.75. MS (TOF ESI+) for C46H57O4S2Na+ (2M+Na)+ calcd. 759.36, found 759.35.
Methyl-2-{3-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)sulfanyl] phenyl} acetate (36b)
Compound 35 (305 mg, 0.97 mmol) was treated according to method G. A colorless oil (210 mg, 55%) was obtained as a mixture of compound 36b and its butyl ester derivative (25%). 1H NMR (400 MHz, CDCl3) δ ppm 7.34 (d, J=1.88 Hz, 1 H), 7.23 (m, 3 H), 7.14 – 7.18 (m, 1 H), 7.07 – 7.14 (m, 2 H), 3.67 (s, 3 H), 3.57 (s, 2 H), 1.68 (s, 4 H), 1.24 (s, 6 H), 1.27 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 171.62, 146.11, 144.52, 137.28, 134.83, 130.80, 130.43, 130.18, 129.16, 129.02, 128.42, 127.62, 127.31, 52.05, 40.97, 34.96, 34.93, 34.37, 34.15, 31.75, 31.71. MS (TOF ESI+) for C46H57O4S2Na+ (2M+Na)+ calcd. 759.36, found 759.35.
Methyl-2-[4-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl) phenyl]acetate (37a)
Compound 36a (105 mg, 0.27 mmol) was treated according to method H. After drying over CaSO4 and evaporation, the crude was purified by flash column chromatography (cyclohexane/dichloromethane): (80/20) to (0/100). An off-white solid was obtained (61 mg, 54%) as a mixture of compound 37a and its butyl ester derivative (5%). 1H NMR (400 MHz, CDCl3) δ ppm 7.86 – 7.93 (m, 3 H), 7.58 (dd, J=8.41, 2.01 Hz, 1 H), 7.40 (m, 3 H), 3.69 (s, 3 H), 3.67 (s, 2H), 1.68 (s, 4 H),1.29 (s, 6 H), 1.26 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 170.99, 150.99, 146.54, 141.05, 139.24, 138.36, 130.29, 130.19, 127.88, 126.00, 124.61, 52.32, 40.84, 34.74, 34.69, 34.63, 34.58, 31.72, 31.59. MS (TOF ESI+) for C46H57O8S2+ (2M+H)+ calcd. 801.34, found 801.34.
Methyl-2-[3-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl) phenyl] acetate (37b)
Compound 36b (90 mg, 0.23 mmol) was treated according to method H. After drying over CaSO4 and evaporation, the crude was purified by flash column chromatography (cyclohexane/dichloromethane): (80/20) to (0/100). Compound 37b was obtained as a thick oil (50 mg, 51%). 1H NMR (400 MHz, CDCl3) δ ppm 7.90 (d, J=2.01 Hz, 1 H), 7.88 (s, 1 H), 7.83 (dt, J=7.00, 1.65 Hz, 1 H), 7.59 (dd, J=8.41, 1.88 Hz, 1 H), 7.43 – 7.52 (m, 2 H), 7.40 (d, J=8.41 Hz, 1 H), 3.69 (s, 5 H), 1.68 (s, 4 H), 1.29 (s, 6 H), 1.26 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 170.98, 150.99, 146.54, 142.39, 138.30, 135.38, 133.94, 129.43, 128.34, 127.84, 126.44, 126.01, 124.55, 52.25, 40.70, 34.72, 34.67, 34.61, 34.55, 31.70, 31.57. MS (TOF ESI+) for C46H57O8S2+ (2M+H)+ calcd. 801.34, found 801.34.
2-{4-[(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)sulfanyl] phenyl}acetic acid (38a)
Compound 36a (50 mg, 0.136 mmol) was hydrolyzed according to method B. With no further purification, compound 38a was obtained as a white solid (43 mg, 99%). mp = 87°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.33 – 7.37 (m, 1 H), 7.15 – 7.25 (m, 5 H), 7.07 – 7.12 (m, 1 H), 3.60 (s, 2 H), 1.67 (s, 4 H), 1.27 (s, 6 H), 1.24(s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 177.38, 146.17, 144.67, 136.36, 131.30, 130.71, 130.50, 130.03, 129.97, 129.80, 129.34, 127.70, 40.52, 34.99, 34.96, 34.41, 34.19, 31.79. HPLC (t= 9.50 min, 95%). HRMS (TOF ESI−) for [(C22H26O2S)2−1]− (2M−H)− calcd. 707.3229, found 707.3204.
2-{3-[(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)sulfanyl]phenyl} acetic acid (38b)
Compound 36b (50 mg, 0.136 mmol) was hydrolyzed according to method B. With no further purification, compound 38b was obtained as a colorless oil (43 mg, 99%). 1H NMR (400 MHz, CDCl3) δ ppm 7.34 (d, J=1.88 Hz, 1 H), 7.19 – 7.24 (m, 3 H), 7.14 – 7.19 (m, 1 H), 7.07 – 7.13 (m, 2 H), 3.58 (s, 2 H), 1.67 (s, 4 H), 1.27 (s, 6 H), 1.22 (s, 6H). 13C NMR (101 MHz, CDCl3) δ ppm 177.31, 146.20, 144.63, 137.55, 134.15, 130.68, 130.50, 130.30, 129.25, 129.14, 128.58, 127.68, 127.41, 34.99, 34.98, 34.40, 34.19, 31.79, 31.75. HPLC (t= 9.45 min, 95%). HRMS (TOF ESI−) for C22H25O2S− (2M−H)− calcd. 707.3229, found 707.3286.
2-[4-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl)phenyl]acetic acid (39a)
Compound 37a (45 mg, 0.112 mmol) was hydrolyzed according to method B. With no further purification, compound 39a was obtained as white solid (31 mg, 72%). mp = 214°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.87 – 7.93 (m, 3 H), 7.58 (dd, J=8.41, 2.01 Hz, 1 H), 7.40 (m, 3 H), 3.70 (s, 2 H), 1.68 (s, 4 H), 1.29 (s, 6 H), 1.25 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 175.32, 151.05, 146.56, 141.30, 138.42, 138.23, 130.25, 127.92, 127.88, 126.01, 124.60, 34.74, 34.68, 34.62, 34.55, 31.71, 31.59. HPLC (t= 8.32 min, 99%). HRMS (TOF ESI−) for [(C22H26O4S)2−1]− (M−H)− calcd. 771.3025, found 771.3063.
2-[3-(5,5,8,8-Tetramethyl-5,6,7,8-tetrahydronaphthalene-2-sulfonyl)phenyl]acetic acid (39b)
Compound 38b (45 mg, 0.112 mmol) was hydrolyzed according to method B. With no further purification, compound 39b was obtained as white solid (41 mg, 95%). mp = 75°C. 1H NMR (400 MHz, CDCl3) δ ppm 7.89 (s, 2 H), 7.84 (d, J=6.27 Hz, 1 H), 7.56 – 7.63 (m, 1 H), 7.43 – 7.52 (m, 2 H), 7.39 (d, J=8.41 Hz, 1 H), 3.72 (s, 2 H), 1.67 (s, 4 H), 1.27 (s, 6 H), 1.25 (s, 6 H). 13C NMR (101 MHz, CDCl3) δ ppm 175.74, 151.05, 146.59, 142.48, 138.22, 134.75, 134.01, 129.49, 128.46, 127.86, 126.62, 126.04, 124.54, 34.73, 34.66, 34.61, 34.56, 31.68, 31.57. HPLC (t= 8.27 min, 99%). HRMS (TOF ESI−) for C22H25O4S− (M−H)− calcd. 385.1474, found 385.1494.
Crystallography
Compounds 7 and 8 yielded crystals of suitable quality for X-ray diffraction by slow evaporation of ethyl acetate/heptane solutions. These crystals were used to confirm the structures using X-ray data collected at 90 K, with Cu Kα radiation (λ=1.54178 Å) on a Bruker Kappa Apex-II diffractometer. Crystals of 8 are triclinic, space group P-1 with Z=2, R=0.036, CCDC 896010. Crystals of 7 are monoclinic, space group P21/c with Z=4, R=0.065, CCDC 896011. There is a conformational disorder of the six-membered ring carrying the four methyl groups. The CIFs have been deposited at the Cambridge Crystallographic Data Centre and can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif.
CYP26A1 and CYP26B1 inhibition assays
CYP26A1 and CYP26B1 were expressed in Sf9 cells and used as microsomal fractions supplemented with rat P450 reductase expressed in Escherichia coli as described previously.11,33 Incubations were performed with 5 pmol of P450 (CYP26A1 or CYP26B1) and 10 pmol of P450 reductase. The purified rat reductase was added to CYP26A1 or CYP26B1 microsomes, and allowed to incorporate into the membrane for 10 min at room temperature. The final volume of each incubation sample was then brought to 1 ml by adding 100 mM potassium phosphate (KPi) buffer, pH 7.4, 9-cis-RA, and, when appropriate, inhibitor or solvent. Compounds were dissolved in methanol or dimethyl sulfoxide, and final solvent amounts in the incubations were kept at 1%. The samples were preincubated for 5 min at 37°C before the reaction was initiated with NADPH (final concentration 1 mM). Incubation times were 1 minute and 5 minutes for CYP26A1 and CYP26B1 incubations respectively.
To determine whether the RA isomer 9-cis-RA is a substrate of CYP26B1, 9-cis-RA was incubated with CYP26B1. The formation of 9-cis-4-OH-RA by CYP26B1 was measured by HPLC as described previously for CYP26A1.25 Product formation was linear from 1 minute to 8 minutes. The Km and kcat of 9-cis-RA hydroxylation by CYP26B1 was determined by incubating 8 different concentrations of 9-cis-RA between 50 nM and 1000 nM with CYP26B1. Five minutes after reactions were initiated with NADPH the reactions were quenched with 5 ml of ethyl acetate, acitretin was added as an internal standard, samples extracted, evaporated to dryness, reconstituted in methanol and analyzed by HPLC as described previously.25 A standard curve of 9-cis-4-OH-RA was used to quantify product formation by analysis of the peak area of the primary metabolite on an HPLC. The Michaelis-Menten equation was fit to the data using GraphPad Prism (GraphPad Software Inc., San Diego, CA), and the Km and Vmax values were obtained from this fit. 9-cis-RA was then used as the substrate for subsequent assays of inhibitor potency at 100 nM concentration.
Twenty novel compounds, 1, 3 and two RAR ligands 15b and 13a were tested as inhibitors of CYP26A1 and CYP26B1. The formation of 9-cis-4-OH-RA metabolite was monitored and the percent activity remaining in the presence of the inhibitor in comparison to the solvent only control was quantified. For IC50 determination, 6–8 concentrations of the inhibitor spanning below and above the predicted IC50 were tested, and each concentration was analyzed in triplicate. The IC50 values were determined by nonlinear regression using GraphPad Prism, according to eq. 1:
| (1) |
in which 100% *(Vi/V) is the percentage of activity remaining at a given inhibitor (I) concentration, (Vi/V)max*100% is the fitted maximum percentage activity remaining, and (Vi/V)min*100% is the minimum percentage activity remaining. For compounds with IC50 values less than 100 nM, all fits were corrected for inhibitor depletion, and the Kd was determined using the Morrison equation as described previously (15)according to eq. 2:
| (2) |
in which Kd is the affinity constant of the inhibitor, [I] is the concentration of inhibitor, [E] is the concentration of enzyme, and [EI] is the concentration of the enzyme-inhibitor complex.
RAR activation and induction of RARγ and CYP26A1 in HepG2 cells
The HepG2 cells were obtained and cultured in a humidified incubator at 37 °C under a 5% atmosphere of carbon dioxide using DMEM (Dulbecco’s Modified Eagle Medium) containing 4.5g/L D-glucose, L-glutamine, and 110mg/L Sodium Pyruvate supplemented with 10% FBS and 1% penicillin. (Life Technologies, Grand Island, NY) For all treatments, cells were plated into six well plates in 2 mL media and allowed to adhere for 24 hours prior to treatments.
To test whether any of the candidate compounds activate RAR, HepG2 cells were treated for 24 hours in 2 mL media with 0.1% DMSO (vehicle control), and the test compounds at a concentration of 1 μM in 0.1% DMSO. To determine whether the test compounds inhibited atRA metabolism in the HepG2 cells and hence potentiated the RAR activation (CYP26A1 and RARγ induction) cells were treated with either vehicle control (0.1% DMSO+0.01% ethanol), 10, 100 or 1000nM atRA or 1 μM of each test compound together with 100 nM atRA. Each treatment was performed as triplicate. At the completion of each treatment, the media was collected for measurement of atRA concentration and cells were harvested for mRNA extraction. To each well, 300 μL of TRI reagent (Invitrogen, Grand Island, NY) was added and mRNA extracted according to manufacturers recommendations. mRNA concentration was quantified using the Nanodrop 2000c Spectrophotometer. (Thermofisher Sci., Waltham, MA) and cDNA generated from 1 μg mRNA using Taqman® Gene expression reagents (Applied Biosystems, Carlsbad, CA). RT-PCR was used to quantify CYP26A1 mRNA (StepOnePlus™, Applied Biosystems, Carlsbad, CA) as previously described.30 TaqMan real-time gene expression master mix and PCR primers and fluorescent probes were obtained from Applied Biosystems (Foster City, CA). Probes were labeled with the 5′-reporter dye 5-carboxyfluorescein and a nonfluorescent black hole quencher on the 3′-end. Primer and probe pairs used included: CYP26A1 (Hs00175627_m1, FAM), GAPDH (Hs99999905_m1, VIC). GAPDH was used as the housekeeping gene and all assays were done as multiplexes. All triplicate samples were analyzed in singlet. Changes in target mRNA were measured using relative quantification (fold-difference) and the ΔΔCT method.30 The effect of the inhibitors on atRA metabolism in HepG2 cells was determined using LC-MS/MS as previously described,29 using an AB Sciex QTRAP 4500 mass spectrometer coupled with an Agilent 1290 Infinity UHPLC and Agilent Zorbax C18 column (3.5 μm, 2.1 mm × 100 mm) using negative ion electrospray detection
Inhibition of CYP3A4, CYP2C19, CYP2D6 and CYP2B6
The inhibition of drug metabolizing P450 enzymes by the lead compounds was tested using midazolam (CYP3A4), (S)-mephenytoin (CYP2C19), dextromethorphan (CYP2D6) and bupropion (CYP2B6) as selective substrates as previously described.34,35 Briefly, human liver microsomes were incubated with the substrate and inhibitor in 100mM potassium phosphate buffer. All incubations were initiated with NADPH and quenched with ice cold acetonitrile at the end of the incubation period. Product formation was measured by LC-MS/MS using an AB Sciex 4500 mass spectrometer (AB Sciex, Foster City, CA) coupled to a Shimadzu UFLC XR DGU-20A5 (Shimadzu Scientific Instruments, Columbia, MD).
Supplementary Material
Acknowledgments
This work was supported by NIH grants P30NS055022 (PD, CMK), RRIA award from the Michael J. Fox Foundation for Parkinson’s Research (PD), R41AG046987 (FAD, NG), UL1 TR00042301 (PD, NI), R01GM081569 (NI, BB) and R01GM111772 (NI, ST, LP, WH), CNPq Scholar – Brasil (VGS).
ABBREVIATIONS
- CYP
Cytochrome P450
- HRMS
High resolution mass spectrometry
- RA
retinoic acid
- RAR
retinoic acid receptor
- TTN
1,1,4,4-tetramethyl-1,2,3,4-tetrahydronaphthalene
Footnotes
Supporting Information. Additional characterization data, 1H and 13C-NMR spectra, compound purity.
References
- 1.Gudas LJ. Emerging Roles for Retinoids in Regeneration and Differentiation in Normal and Disease States. Biochim Biophys Acta. 2012;1821:213–221. doi: 10.1016/j.bbalip.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Napoli JL. Physiological Insights into All-Trans-Retinoic Acid Biosynthesis. Biochim Biophys Acta. 2012;1821:152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomhoff R, Blomhoff HK. Overview of Retinoid Metabolism and Function. J Neurobiol. 2006;66:606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 4.Hogarth CA, Griswold MD. Retinoic Acid Regulation of Male Meiosis. Curr Opin Endocrinol Diabetes Obes. 2013;20:217–223. doi: 10.1097/MED.0b013e32836067cf. [DOI] [PubMed] [Google Scholar]
- 5.Maden M. Retinoic Acid in the Development, Regeneration and Maintenance of the Nervous System. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 6.Ross AC. Vitamin A and Retinoic Acid in T Cell-Related Immunity. Am J Clin Nutr. 2012;96:1166S– 72S. doi: 10.3945/ajcn.112.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson CH, Buttrick BR, Isoherranen N. Therapeutic Potential of the Inhibition of the Retinoic Acid Hydroxylases CYP26A1 and CYP26B1 by Xenobiotics. Curr Top Med Chem. 2013;13:1402–1428. doi: 10.2174/1568026611313120004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thatcher JE, Isoherranen N. The Role of CYP26 Enzymes in Retinoic Acid Clearance. Expert Opin Drug Metab Toxicol. 2009;5:875–886. doi: 10.1517/17425250903032681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross AC, Zolfaghari R. Cytochrome P450s in the Regulation of Cellular Retinoic Acid Metabolism. Annu Rev Nutr. 2011;31:65–87. doi: 10.1146/annurev-nutr-072610-145127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thatcher JE, Zelter A, Isoherranen N. The Relative Importance of CYP26A1 in Hepatic Clearance of All-Trans Retinoic Acid. Biochem Pharmacol. 2010;80:903–912. doi: 10.1016/j.bcp.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topletz AR, Thatcher JE, Zelter A, Lutz JD, Tay S, Nelson WL, Isoherranen N. Comparison of the Function and Expression of CYP26A1 and CYP26B1, the Two Retinoic Acid Hydroxylases. Biochem Pharmacol. 2012;83:149–163. doi: 10.1016/j.bcp.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njar VC, Gediya L, Purushottamachar P, Chopra P, Vasaitis TS, Khandelwal A, Mehta J, Huynh C, Belosay A, Patel J. Retinoic Acid Metabolism Blocking Agents (RAMBAs) for Treatment of Cancer and Dermatological Diseases. Bioorg Med Chem. 2006;14:4323–4340. doi: 10.1016/j.bmc.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Stoppie P, Borgers M, Borghgraef P, Dillen L, Goossens J, Sanz G, Szel H, Van Hove C, Van Nyen G, Nobels G, Vanden Bossche H, Venet M, Willemsens G, Van Wauwe J. R115866 Inhibits All-Trans-Retinoic Acid Metabolism and Exerts Retinoidal Effects in Rodents. J Pharmacol Exp Ther. 2000;293:304–312. [PubMed] [Google Scholar]
- 14.Hogarth CA, Evans E, Onken J, Kent T, Mitchell D, Petkovich M, Griswold MD. CYP26 Enzymes Are Necessary Within the Postnatal Seminiferous Epithelium for Normal Murine Spermatogenesis. Biol Reprod. 2015;93:19. doi: 10.1095/biolreprod.115.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chenery A, Burrows K, Antignano F, Underhill TM, Petkovich M, Zaph C. The Retinoic Acid-Metabolizing Enzyme Cyp26b1 Regulates CD4 T Cell Differentiation and Function. PLoS One. 2013;8:e72308. doi: 10.1371/journal.pone.0072308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm MF, Zhang L, Badea BA, White SK, Mais DE, Berger E, Suto CM, Goldman ME, Heyman RA. Synthesis and Structure-Activity Relationships of Novel Retinoid X Receptor-Selective Retinoids. J Med Chem. 1994;37:2930–2941. doi: 10.1021/jm00044a014. [DOI] [PubMed] [Google Scholar]
- 17.Dawson MI, Jong L, Hobbs PD, Cameron JF, Chao WR, Pfahl M, Lee MO, Shroot B. Conformational Effects on Retinoid Receptor Selectivity. 2. Effects of Retinoid Bridging Group on Retinoid X Receptor Activity and Selectivity. J Med Chem. 1995;38:3368– 3383. doi: 10.1021/jm00017a021. [DOI] [PubMed] [Google Scholar]
- 18.Patel BA, Ziegler CB, Cortese NA, Plevyak JE, Zebovitz TC, Terpko M, Heck RF. Palladium-Catalyzed Vinylic Substitution Reactions with Carboxylic Acid Derivatives. J Org Chem. 1977;42:3903–3907. [Google Scholar]
- 19.Heck RF, Nolley JP. Palladium-Catalyzed Vinylic Hydrogen Substitution Reactions with Aryl, Benzyl, and Styryl Halides. J Org Chem. 1972;37:2320–2322. [Google Scholar]
- 20.Sun J, Dong Y, Cao L, Wang X, Wang S, Hu Y. Highly Efficient Chemoselective Deprotection of O,O-Acetals and O,O-Ketals Catalyzed by Molecular Iodine in Acetone. J Org Chem. 2004;69:8932–8934. doi: 10.1021/jo0486239. [DOI] [PubMed] [Google Scholar]
- 21.Boiteau J, Rivier M, Feraille GA, Jomard A. WO2014016507 A1. Nouveaux Composes Inhibiteurs Selectifs Du cyp26a1 Utiles Dans Des Compositions Cosmétiques et Pharmaceutiques. 2014 Jan 30;
- 22.Fukuzawa S, Tanihara D, Kikuchi S. Palladium-Catalyzed Coupling Reaction of Diaryl Dichalcogenide with Aryl Bromide Leading to the Synthesis of Unsymmetrical Aryl Chalcogenide. Synlett. 2006;2006:2145–2147. [Google Scholar]
- 23.Christie VB, Barnard JH, Batsanov AS, Bridgens CE, Cartmell EB, Collings JC, Maltman DJ, Redfern CP, Marder TB, Przyborski S, Whiting A. Synthesis and Evaluation of Synthetic Retinoid Derivatives as Inducers of Stem Cell Differentiation. Org Biomol Chem. 2008;6:3497–3507. doi: 10.1039/b808574a. [DOI] [PubMed] [Google Scholar]
- 24.Millois C, Diaz P. Solution-Phase Synthesis of Diaryl Selenides Using Polymer-Supported Borohydride. Org Lett. 2000;2:1705–1708. doi: 10.1021/ol0058184. [DOI] [PubMed] [Google Scholar]
- 25.Thatcher JE, Buttrick B, Shaffer SA, Shimshoni JA, Goodlett DR, Nelson WL, Isoherranen N. Substrate Specificity and Ligand Interactions of CYP26A1, the Human Liver Retinoic Acid Hydroxylase. Mol Pharmacol. 2011;80:228–239. doi: 10.1124/mol.111.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendimenico GJ, Stim TB, Corbo M, Janssen B, Mezick JA. A Pleiotropic Response Is Induced in F9 Embryonal Carcinoma Cells and Rhino Mouse Skin by All-Trans-Retinoic Acid, a RAR Agonist but Not by SR11237, a RXR-Selective Agonist. J Invest Dermatol. 1994;102:676–680. doi: 10.1111/1523-1747.ep12374092. [DOI] [PubMed] [Google Scholar]
- 27.Le Q, Dawson MI, Soprano DR, Soprano KJ. Modulation of Retinoic Acid Receptor Function Alters the Growth Inhibitory Response of Oral SCC Cells to Retinoids. Oncogene. 2000;19:1457–1465. doi: 10.1038/sj.onc.1203436. [DOI] [PubMed] [Google Scholar]
- 28.Holmes WF, Dawson MI, Robert Soprano D, Soprano KJ. Induction of Apoptosis in Ovarian Carcinoma Cells by AHPN/CD437 Is Mediated by Retinoic Acid Receptors. J Cell Physiol. 2000;185:61–67. doi: 10.1002/1097-4652(200010)185:1<61::AID-JCP5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Topletz AR, Tripathy S, Foti RS, Shimshoni JA, Nelson WL, Isoherranen N. Induction of CYP26A1 by Metabolites of Retinoic Acid: Evidence That CYP26A1 Is an Important Enzyme in the Elimination of Active Retinoids. Mol Pharmacol. 2015;87:430–441. doi: 10.1124/mol.114.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay S, Dickmann L, Dixit V, Isoherranen N. A Comparison of the Roles of Peroxisome Proliferator-Activated Receptor and Retinoic Acid Receptor on CYP26 Regulation. Mol Pharmacol. 2010;77:218–227. doi: 10.1124/mol.109.059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomaa MS, Bridgens CE, Veal GJ, Redfern CP, Brancale A, Armstrong JL, Simons C. Synthesis and Biological Evaluation of 3-(1H-Imidazol- and Triazol-1-Yl)-2,2- Dimethyl-3-[4-(naphthalen-2-Ylamino)phenyl]propyl Derivatives as Small Molecule Inhibitors of Retinoic Acid 4-Hydroxylase (CYP26) J Med Chem. 2011;54:6803–6811. doi: 10.1021/jm200695m. [DOI] [PubMed] [Google Scholar]
- 32.Patel JB, Mehta J, Belosay A, Sabnis G, Khandelwal A, Brodie AM, Soprano DR, Njar VC. Novel Retinoic Acid Metabolism Blocking Agents Have Potent Inhibitory Activities on Human Breast Cancer Cells and Tumour Growth. Br J Cancer. 2007;96:1204–1215. doi: 10.1038/sj.bjc.6603705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. Expression and Functional Characterization of Cytochrome P450 26A1, a Retinoic Acid Hydroxylase. Biochem Pharmacol. 2009;77:258–268. doi: 10.1016/j.bcp.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirasaka Y, Sager JE, Lutz JD, Davis C, Isoherranen N. Inhibition of CYP2C19 and CYP3A4 by Omeprazole Metabolites and Their Contribution to Drug-Drug Interactionss. Drug Metab Dispos. 2013;41:1414–1424. doi: 10.1124/dmd.113.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sager JE, Lutz JD, Foti RS, Davis C, Kunze KL, Isoherranen N. Fluoxetine- and Norfluoxetine-Mediated Complex Drug-Drug Interactions: In Vitro to In Vivo Correlation of Effects on CYP2D6, CYP2C19, and CYP3A4. Clin Pharmacol Ther. 2014;95:653–662. doi: 10.1038/clpt.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.