Abstract
Phosphorylated phosphatidylinositol lipids are crucial for most eukaryotes and have diverse cellular functions. The low-abundance signaling lipid phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) is critical for cellular homeostasis and adaptation to stimuli. A large complex of proteins that includes the lipid kinase Fab1/PIKfyve, dynamically regulates the levels of PI(3,5)P2. Deficiencies in PI(3,5)P2 are linked to some human diseases, especially those of the nervous system. Future studies will likely determine new, undiscovered regulatory roles of PI(3,5)P2, as well as uncover mechanistic insights into how PI(3,5)P2 contributes to normal human physiology.
Keywords: Fab1, PIKfyve, Vac14, Fig4, lysosome, neurodegeneration
Introduction
Phosphorylated phosphatidylinositol lipids are crucial for most eukaryotes and have diverse cellular functions. They regulate multiple pathways, including organization of the cytoskeleton, cellular motility, endocytosis, and provide spatial and temporal control for membrane trafficking. The inositol ring of phosphatidylinositol can be phosphorylated and dephosphorylated on its 3,4 or 5 hydroxyl groups by several lipid kinases and lipid phosphatases. These interconversions serve as a network for the synthesis of several phosphorylated phosphatidylinositol species. Seven phosphoinositide lipid species have been identified in mammalian cells, and four in the yeast, S. cerevisiae. Early evidence that these lipids function in signal transduction came from the finding that phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) is cleaved to generate the second messengers inositol-1,4,5 triphosphate and diacylglycerol, which mobilize Ca2+ from the endoplasmic reticulum [1]. One way that phosphatidylinositol lipids regulate diverse pathways is via the recruitment of effector proteins to specific membrane domains. Importantly, by functioning at confined membrane regions, interconversion between phosphorylated phosphoinositide lipids provides spatial and temporal regulation of downstream pathways.
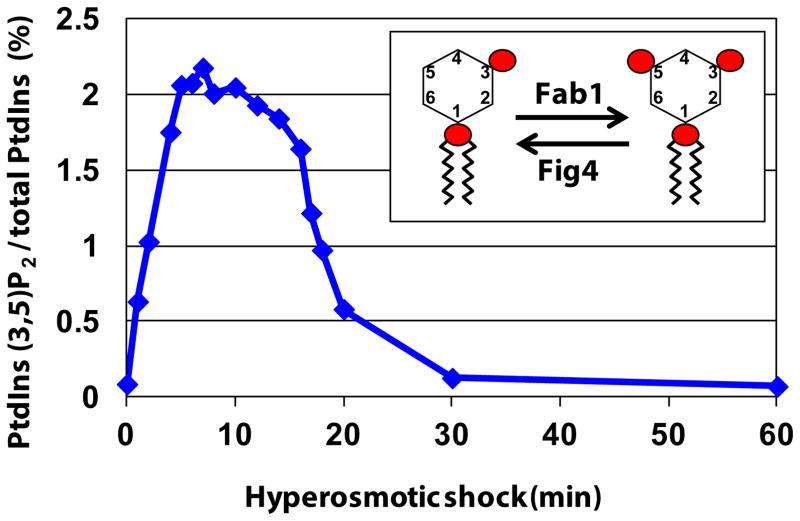
Phosphatidylinositol-3.5-bisphosphate (PI(3,5)P2) is among the more recently identified phosphoinositide lipids [2, 3]. PI(3,5)P2 is relatively low abundance, approximately, 0.05 % ~ 0.1 % of total phosphatidylinositol lipids. Physiological signals including insulin, growth factors in mammalian cells, and hyperosmotic shock in yeast and plant cells, cause an acute elevation of PI(3,5)P2 [2, 4–7]. These observations suggest that PI(3,5)P2 functions as a signaling molecule in cellular homeostasis and in adaptation. Moreover, in yeast, there is a dramatic and transient elevation of PI(3,5)P2 during hyperosmotic shock. Within 5 min of exposure to hyperosmotic media, there is a 20-fold increase and then a rapid return to the normal, low levels within 30 min [8] (Figure 1). These findings indicate that the synthesis of PI(3,5)P2 is tightly regulated and that upstream pathways are part of this regulation. However, these upstream pathways and many of the downstream pathways specific for PI(3,5)P2 are poorly understood. Here, we review studies that shed light on the regulation of PI(3,5)P2 and discuss the significance of PI(3,5)P2 as a signaling molecule, including its roles in animal physiology and human disease.
Figure 1. The synthesis of PI(3,5)P2 is tightly regulated.
The graph indicates the levels of PI(3,5)P2 during hyperosmotic shock in yeast. PI(3,5)P2 levels transiently change in response to specific stimuli. A prolonged single stimulus, introduction of yeast into hyperosmotic media, causes a transient elevation of PI(3,5)P2. Within 5 minutes, PI(3,5)P2 levels rise over 20-fold, plateau for 10 minutes, then rapidly return to basal levels. That levels of PI(3,5)P2 are tightly controlled suggests that there are multiple layers of regulation. Data modified from [8].
The PI(3)P 5-kinase Fab1/PIKfyve functions within a regulatory complex
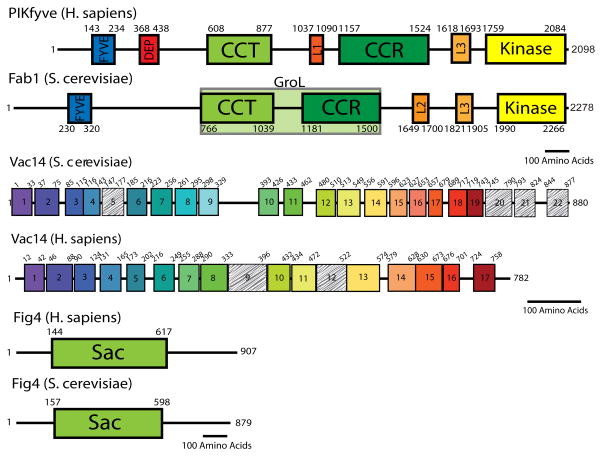
Fab1/PIKfyve is the sole lipid kinase that synthesizes PI(3,5)P2 from phosphatidylinositol-3-phosphate (PI(3)P) [6, 9, 10]. Fab1 was identified in the budding yeast S. cerevisiae [11] and was shown to function as a vacuolar PI(3)P 5-kinase [9, 12]. Mammalian Fab1/PIKfyve [4] and Arabidopsis FAB1A and FAB1B [13], are homologues of yeast Fab1. Fab1 and PIKfyve are larger than other PI 5-kinases and are composed of 2278 and 2098 amino acids, respectively. In addition to its kinase domain, Fab1/PIKfyve possesses many regulatory domains [14] (Figure 2). The FYVE domain binds to PI(3)P [4, 15]. The CCT (chaperone containing TCP1) domain has homology with TCP-1/Cpn60 chaperones and the CCR (conserved cysteine rich) domain contains conserved cysteines and histidines. The CCT and CCR domain are proposed to associate with regulatory proteins [14]. The DEP (Disheveled, Egl-10, Pleckstrin) domain is found in mammals, chordates and insects and is of unknown function. Furthermore, our analysis indicates three additional conserved regions that are either conserved in all metazoans, all fungi, or in all eukaryotes, respectively (Figure 2). The existence of these conserved domains suggests that Fab1 is highly regulated, and that this regulation is conserved.
Figure 2. Domain architecture of Fab1/PIKfyve, Vac14, and Fig4.
Boundaries of each domain were determined using a combination of Jpred4 secondary structure prediction and ClustalW multiple sequence alignment [72, 73]. For Vac14, the above techniques were used in addition to tailored HHpred alignments of select predicted HEAT repeats [74]. Fab1/PIKfyve contains previously described domains (FYVE, DEP, CCT, CCR, and Kinase); we identify three additional areas of predicted secondary structure which have structural and sequence conservation in all species (L3), in all fungi (L2), or in metazoans (L1). Vac14 is composed of tandem HEAT repeats. Colored boxes indicate homology of HEAT repeats between yeast and human Vac14. Hashed boxes indicate degenerate sequences which may be bona fide HEAT repeats. Fig4 contains a single Sac domain, which is conserved in some lipid phosphatases. Black lines represent 100 amino acids.
Indeed, yeast Fab1 has several modulators of its lipid kinase activity including Vac7, Vac14, Fig4 and Atg18. Fab1 and its regulators localize on the vacuole membrane [9, 16–18]. Vac7 and Vac14 were first identified as novel vacuolar proteins required for vacuole inheritance and morphology [16, 19]. Deletion of Vac7, Vac14 or Fab1 increases vacuole size and causes a defect in the synthesis of PI(3,5)P2 under basal conditions as well as during hyperosmotic shock [9, 16, 20], which provides an indication that Vac7 and Vac14 positively regulate Fab1 lipid kinase activity. The connection between Fig4 and Fab1 came from a yeast genetic screen for mutants that suppress the temperature sensitivity of a vac7Δ mutant [21]. Fig4 has a Sac1 phosphatase domain (Figure 2) that is found in several lipid phosphatases including Inp51, 52 and 53 [21]. Although Fig4 functions as a PI(3,5)P2 specific phosphoinositide phosphatase in vitro [17] and in vivo [8, 21], paradoxically, deletion of Fig4 causes a defect in the acute synthesis of PI(3,5)P2 during hyperosmotic shock [8]. This suggests that Fig4 has dual roles for the synthesis and turnover of PI(3,5)P2. Atg18 is a regulator of autophagy [22]. However, deletion of Atg18 results in an enlarged vacuole, similar to mutants with defects in the levels of PI(3,5)P2. Unexpectedly, the atg18Δ mutant has increased levels of PI(3,5)P2 both under basal conditions and during hyperosmotic shock [18]. These observations indicate that Atg18 negatively modulates Fab1 activity and that dynamic changes in PI(3,5)P2 levels may modulate vacuolar membrane fission and/or fusion. Note that Atg18 binds PI(3)P and PI(3,5)P2 with high affinity via two sites [18] and associates with the vacuole membrane through binding to PI(3,5)P2. Association of Atg18 with the vacuole membrane is required for its regulation of Fab1 [18, 23]. Atg18 may also have a separate role in vacuole membrane fission [21].
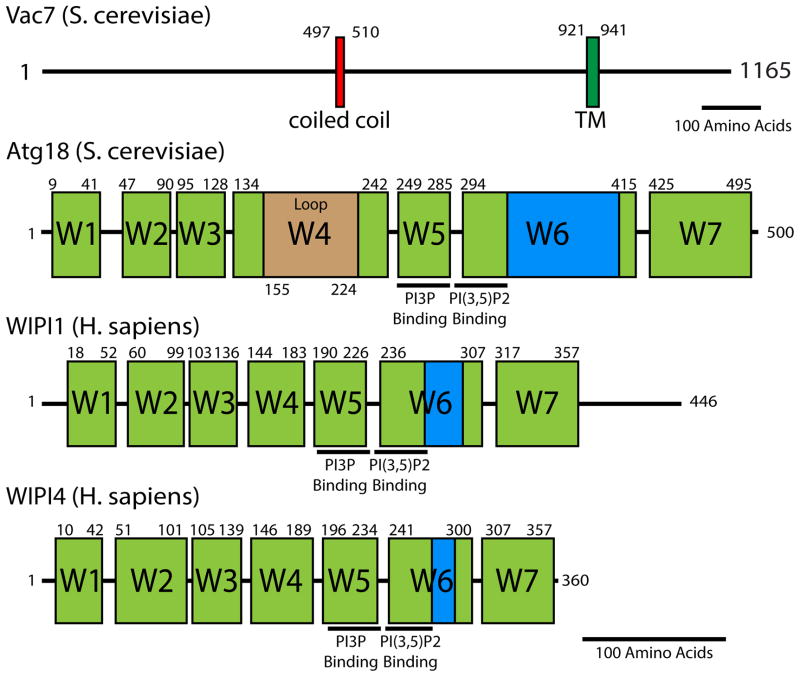
In mammalian cells, Vac14 and Fig4 are evolutionally and functionally conserved, and are also referred to as ArPIKfyve [24] and Sac3 [25] respectively. Similar to yeast, mammalian Fab1/PIKfyve, Vac14, and Fig4 are localized on early and late endosomes as well as lysosomes [25–27]. Vac7 homologues have only been observed in fungi and it is not clear whether functional homologues are present in other species. Atg18 belongs to a large family of proteins known as PROPPINS, which are found in most eukaryotes [18, 28]. PROPPIN proteins have predicted beta-propeller folds and the presence of an FRRG motif required for phosphoinositide binding. Mammals have four PROPPINS, WIPI-1,2,3, and 4 which have homology with yeast Atg18 [29–31] (Figure 3). Moreover, like Atg18, WIPI-1,3 and 4 bind PI(3)P and PI(3,5)P2 with high affinity [31]. WIPI-2 has not been tested thus far due to instability. Despite the similarities between Atg18 and WIPI-1,2,3 and 4, it is not clear whether WIPI proteins regulate PI(3,5)P2 levels in mammalian cells.
Figure 3. Domain architecture of Vac7 and Atg18.
Vac7 is found only in some fungi. Atg18 is similar to mammalian WIPI1 and WIPI2; however, it is not known if these WIPI proteins regulate PIKfyve. For Vac7, boundaries of each domain were determined using a combination of Jpred4 secondary structure prediction and ClustalW multiple sequence alignment. Vac7 contains a coiled-coil domain (80% certainty using COILS) and a transmembrane domain. Atg18, WIPI1, and WIPI4 domain boundaries were determined from ClustalW multiple sequence alignment with Hsv2—a related protein where high-resolution structures are available [31, 75, 76]. Seven WD40 blades are depicted in green. A hydrophobic lipid-associated region is highlighted in blue. Beige Atg18 Loop is a predicted unstructured region between beta sheet 2 and 3 of blade 4. WIPI2 (not depicted) is structurally similar to WIPI1 and both contain an unstructured C-terminal tail with 31% similarity to each other. WIPI3 (not depicted) is structurally similar to WIPI4. Residue pockets predicted to bind PI(3)P and PI(3,5)P2 are highlighted for ATG18, WIPI1, and WIPI4. That these regions are conserved indicates that Atg18, WIPI1, WIPI4 as well as their paralogs likely interact with phospholipids in a similar manner. Black lines represent 100 amino acids.
Several studies suggest that the levels of PI(3,5)P2 are tightly controlled by multiple layers of regulation. Fab1 activity is regulated in part via formation of a large protein complex that includes Fab1, Vac14, and Fig4 [32, 33]. The CCT and CCR domain of Fab1 are responsible for its association with Vac14 and Fig4 [32, 33]. Moreover, Vac7 and Atg18 are a part of the protein complex [33]. Similarly, mammalian Fab1/PIKfyve, Vac14 and Fig4 reside within a complex [25]. Vac14 is predicted to be entirely composed of HEAT repeats, each containing two anti-parallel helices connected by a short loop [20, 33]. Secondary structure prediction in conjunction with multiple sequence alignments indicate that there are likely 18–22 HEAT repeats in yeast Vac14 and 15–17 HEAT repeats in human Vac14 (Figure 2). Yeast Vac14 serves as a hub of the complex and through distinct HEAT repeats directly binds Fab1, Fig4, Vac7 and Atg18 [33]. The importance of Vac14 in mammals is underscored by the finding that a knock-out of Vac14 in mice causes perinatal lethality [34]. Candidate interacting proteins were isolated from Vac14-3xFLAG expressed in HEK293 cells. These include Rab9 and Rab7, which regulate endo-lysosomal membrane dynamics and trafficking pathways [35]. Thus, Vac14 may serve roles beyond its function as a scaffold for proteins within the Fab1/PIKfyve complex.
The roles, effectors, and downstream pathways of PI(3,5)P2
An obvious phenotype of yeast mutants defective in the synthesis of PI(3,5)P2 is an increase in vacuole size [9, 11, 16]. Similarly, the inhibition of Fab1/PIKfyve activity causes abnormal, enlarged endo-lysosomal compartments in mammalian cells and mouse tissues [6, 34, 36–38] as well as in the worm, C. elegans [39] and in the plant, A. thaliana [40]. These observations suggest that PI(3,5)P2 is required for normal vacuole/endo-lyososmal functions.
Under limited nutrients and/or stress, autophagy provides a degradation pathway for cytosolic content as well as organelles. PI(3,5)P2 has at least two roles in the regulation of autophagy. A decrease of PI(3,5)P2 levels in yeast causes defects in vacuolar degradation of autophagosomes that are delivered to the vacuole lumen [41]. Second, inhibition of Fab1 activity causes a loss of TORC1 activity and a concomitant increase in autophagy (discussed in below) [41]. These observations suggest that autophagy is a downstream pathway of PI(3,5)P2. Autophagy is similarly impaired by the inhibition of Fab1/PIKfyve function in mammalian cells [38, 42–44], in C. elegans [39] and in Drosophila [45]. Thus, the linkage between PI(3,5)P2 and autophagy may be conserved.
Recent studies revealed that the target of rapamycin complex 1 (TORC1) is regulated by PI(3,5)P2 in yeast and cultured adipocytes [5, 41]. TORC1 is a major regulator of cell growth and metabolic processes in many organisms and its activity is tightly regulated by the availability of nutrients. PI(3,5)P2 is required for mammalian TORC1 (mTORC1) activity upon insulin stimulation. This regulation may occur via Raptor, a component of mTORC1, that binds PI(3,5)P2 in vitro [5]. Moreover, in yeast, PI(3,5)P2 is required for TORC1 dependent regulation of autophagy as well as nutrient dependent endocytosis. The Raptor homologue, Kog1, binds PI(3,5)P2 in vitro. Furthermore, a major downstream target of TORC1, Sch9, which is a similar to mammalian S6 kinase, binds PI(3,5)P2. PI(3,5)P2 is required for the recruitment of Sch9 to the vacuole membrane and for its phosphorylation by TORC1 [41].
PI(3,5)P2 is likely required for the maintenance of intracellular osmolarity and proper pH. During hyperosmotic shock in yeast, the volume of vacuoles is transiently decreased likely due to transport of water, ions and osmolytes out of vacuole to help minimize changes in cytosolic osmolarity [8]. One possible role of PI(3,5)P2 for the maintenance of intracellular osmolarity and pH is through regulation of the vacuolar proton-translocating ATPase (V-ATPase) as well as ion channels. In yeast, PI(3,5)P2 is required for V-ATPase stability and for its reassembly after glucose starvation [46]; this occurs via the interaction between PI(3)P2 and the V0 subunit [46]. Similarly, in plants, PI(3,5)P2 is required for vacuolar acidification [47]. In mammalian cells PI(3,5)P2 regulates Ca2+ channel currents on endo-lysosome membranes via the TRPML1 channel [48, 49] and TPC sodium selective channels [50]. Importantly, TRPML1 directly binds PI(3,5)P2 [51]. Similarly, PI(3,5)P2 regulates the yeast homologue of TRPML1, Yvc1, which regulates Ca2+ currents during hyperosmotic shock [49]. Together these findings suggest that PI(3,5)P2 directly regulates the activity of selected ion channels upon physiological stimulation.
Another possible role for PI(3,5)P2 in homeostatic response to intracellular osmolarity is that PI(3,5)P2 functions in the fission of the vacuole membrane. In yeast, during hyperosmotic shock, vacuole fission would decrease vacuole volume with either no or a smaller decrease in the vacuole membrane surface area [8]. Within 30 min following hyperosmotic shock, vacuole size returns to its original size [8]. These observations suggest that PI(3,5)P2 regulates membrane dynamics via regulation of fission and/or fusion of the vacuole membrane.
Similar to most phosphatidylinositol lipids, PI(3)P [52] and PI(3,5)P2 are involved in membrane trafficking and protein sorting. PI(3,5)P2 has been proposed to be required for cargo selection into mutivesicular bodies (MVB). Many receptor proteins on the cell surface are down-regulated by the delivery to the vacuole/lysosome through the MVB pathway. The ESCRT machinery is required for this sorting [53–55]. PI(3,5)P2 is required for the MVB sorting of some proteins to the vacuole lumen in yeast [56]. Vps24, a component of ESCRT-III binds PI(3,5)P2 and is a candidate effector protein [57]. However, precisely how PI(3,5)P2 regulates cargo sorting is not yet known.
PI(3,5)P2 is also required for retrograde traffic from the yeast vacuole to the Golgi [18, 58]. Similarly in mammalian cells, PI(3,5)P2 is required for retrograde traffic from early endosomes to the trans Golgi network. SNX1 and SNX2 are required for this retrograde traffic and directly bind PI(3,5)P2 [36, 59]. In plants, SNX1 also binds PI(3,5)P2 and in addition binds to Fab1 [60]. Thus in both plants and animals, SNX1 and SNX2 are candidate effector proteins of PI(3,5)P2.
In addition, PI(3,5)P2 has been shown to selectively regulate transcription in yeast via Tup1 and Cti6 [61].This raises the possibility that PI(3,5)P2 may regulate some transcription pathways in mammalian cells. PI(3,5)P2 is also required for polarized cell growth in a moss via direct binding to a class II formin [62]. In Arabidopsis, PI(3,5)P2 is required for pollen tube growth [63]. Together these studies suggest that PI(3,5)P2 is crucial for many cellular events and has fundamental roles beyond vacuolar/endo-lysosomal function.
Roles for PI(3,5)P2 and PI(5)P in animal physiology and human disease
Similar with yeast, mammalian Fab1/PIKfyve generates all of the PI(3,5)P2 pools from PI(3)P. In addition, Fab1/PIKfyve is responsible for most of the pools of PI(5)P [6, 10]. The generation of PI(5)P occurs either via direct synthesis from PI by PIKfyve [10] and/or by the conversion of PI(3,5)P2 to PI(5)P by PI(3)P-phosphatases [6]. Myotubularin-related (MTMR) proteins are likely candidates [64, 65].
Although the distinct role of PI(3,5)P2 and/or PI(5)P in mammals is poorly understood, recent findings suggest that the activity of Fab1/PIKfyve is critical in mammalian cells, and has additional functions beyond the known roles shared with yeast. Mouse models of Fig4 and Vac14 deletions [34, 37] and a hypomorphic mutation in Pikfyve [6] exhibit neonatal lethality. Moreover, these mutants have spongiform neurodegeneration that may be due to enlarged endo-lysosomal compartments. A whole body knock-out of Pikfyve in mice results in early embryonic lethality [10, 66]. These observations suggest that PI(3,5)P2 is crucial for development and is especially critical in the nervous system. Indeed, the activity of Fab1/PIKfyve is required for the regulation of synaptic strength [27]. PI(3,5)P2 plays a role in postsynaptic weakening during chemical long-term depression and during homeostatic down scaling [67]. Notably, the levels of PI(3,5)P2 dynamically change during the initiation of long-term depression and become elevated during homeostatic downscaling [67].
PI(3,5)P2 deficiencies are linked to human diseases, especially those of the nervous system. For instance, mutations predicted to have a modest effect on the ability of cells to dynamically regulate PI(3,5)P2 levels underlie a severe form of Charcot Marie-tooth syndrome (CMT4J), a peripheral neuropathy, as well as some cases of amyotrophic lateral sclerosis (ALS) and primary lateral sclerosis (PLS) [37, 68]. Recently, a homozygous missense mutation in Fig4 was found to be the causal allele in a consanguineous family with multiple neurological problems: polymicrogyria, epilepsy, and abnormal behavior [69]. Mutations with more severe deficiencies in the regulation of PI(3,5)P2 underlie additional neurological diseases. For instance, a homozygous null mutation in FIG4 causes Yunis-Varón syndrome, which results in infant mortality and severe pathological effects on multiple tissues, including the brain [70]. Based on studies in mice [37], this mutation is predicted to lower PI(3,5)P2 to 1/3 of its normal levels Although mechanisms whereby mutations in the PI(3,5)P2 synthesis pathway cause diseases are not known, the Fig4 I41T allele in CMT4J [37] causes a defect in its association with Vac14 and Fab1/PIKfyve and results in a defect of Fab1/PIKfyve activity [37, 71]. Analysis of the causative mutations linked with the synthesis of PI(3,5)P2 may provide insights into new treatments for some diseases.
Conclusion
To date, it has been shown that PI(3,5)P2 plays fundamental roles in several cellular events. Fab1/PIKfyve binds PI(3)P and converts PI(3)P to PI(3,5)P2. Recruitment of Fab1/PIKfyve likely causes local depletion of PI(3)P and an increase in the levels of PI(3,5)P2 on endosomal membranes. These changes in PI(3)P and PI(3,5)P2 will also change the local concentration of effector proteins that bind PI(3)P or PI(3,5)P2. Thus PI(3)P and PI(3,5)P2 likely provide spatial and temporal control of diverse pathways.
Acknowledgments
Due to space limitations, we apologize to friends and colleagues for omission of some citations. We thank Drs. Yui Jin, Sai Giridharan, and Beth Strunk for helpful discussions. This review was supported by R01 GM50403 and R01 NS064015 to LSW.
Abbreviations
- PI(3)P
phosphatidylinositol 3-phosphate
- PI(3,5)
P2, phosphatidylinositol 3,5-bisphosphate
- PI(4,5)
P2, phosphatidylinositol 4,5-bisphosphate
- v-ATPase
vacuolar proton translocating ATPase
- TORC1
target of rapamycin complex 1
- mTORC1
mammalian target of rapamycin complex 1
- MVB
Multivesicular bodies
- CMT4J
Charcot-Marie Tooth syndrome 4J
References
- 1.Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 2.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 3.Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323(Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 5.Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, Saltiel AR, Meisler MH, Russell MW, Weisman LS. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci U S A. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meijer HJG, Divecha N, vac den Ende H, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl 3,5-bisphosphate in plant cells. Planta. 1999;208:294–298. [Google Scholar]
- 8.Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell. 2006;5:723–731. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J Biol Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T, Matsuzawa T, Takegawa K, Sato MH. Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol. 2011;155:797–807. doi: 10.1104/pp.110.167981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 16.Bonangelino CJ, Catlett NL, Weisman LS. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol Cell Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YX, Zhao H, Harding TM, Gomes de Mesquita DS, Woldringh CL, Klionsky DJ, Munn AL, Weisman LS. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dove SK, McEwen RK, Mayes A, Hughes DC, Beggs JD, Michell RH. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- 21.Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell. 2002;13:1238–1251. doi: 10.1091/mbc.01-10-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr, Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sbrissa D, Shisheva A. Acquisition of unprecedented phosphatidylinositol 3,5-bisphosphate rise in hyperosmotically stressed 3T3-L1 adipocytes, mediated by ArPIKfyve-PIKfyve pathway. J Biol Chem. 2005;280:7883–7889. doi: 10.1074/jbc.M412729200. [DOI] [PubMed] [Google Scholar]
- 25.Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 26.Cabezas A, Pattni K, Stenmark H. Cloning and subcellular localization of a human phosphatidylinositol 3-phosphate 5-kinase, PIKfyve/Fab1. Gene. 2006;371:34–41. doi: 10.1016/j.gene.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, McCartney AJ, Zolov SN, Ferguson CJ, Meisler MH, Sutton MA, Weisman LS. Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P(2) and PI(5)P. EMBO J. 2012;31:3442–3456. doi: 10.1038/emboj.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 29.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 30.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 31.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulze U, Vollenbroker B, Braun DA, Van Le T, Granado D, Kremerskothen J, Franzel B, Klosowski R, Barth J, Fufezan C, Wolters DA, Pavenstadt H, Weide T. The Vac14-interaction network is linked to regulators of the endolysosomal and autophagic pathway. Mol Cell Proteomics. 2014;13:1397–1411. doi: 10.1074/mcp.M113.034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, Stenmark H, Cullen PJ. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, Parker PJ. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitley P, Hinz S, Doughty J. Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol. 2009;151:1812–1822. doi: 10.1104/pp.109.146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, Weisman LS. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell. 2014;25:1171–1185. doi: 10.1091/mbc.E14-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson CJ, Lenk GM, Jones JM, Grant AE, Winters JJ, Dowling JJ, Giger RJ, Meisler MH. Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum Mol Genet. 2012;21:3525–3534. doi: 10.1093/hmg/dds179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin S, Harper CB, May LM, Coulson EJ, Meunier FA, Osborne SL. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One. 2013;8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 46.Li SC, Diakov TT, Xu T, Tarsio M, Zhu W, Couoh-Cardel S, Weisman LS, Kane PM. The signaling lipid PI(3,5)P(2) stabilizes V(1)-V(o) sector interactions and activates the V-ATPase. Mol Biol Cell. 2014;25:1251–1262. doi: 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bak G, Lee EJ, Lee Y, Kato M, Segami S, Sze H, Maeshima M, Hwang JU, Lee Y. Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell. 2013;25:2202–2216. doi: 10.1105/tpc.113.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, Cheng X, Zhang Y, Weisman LS, Delling M, Xu H. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zhang X, Dong XP, Samie M, Li X, Cheng X, Goschka A, Shen D, Zhou Y, Harlow J, Zhu MX, Clapham DE, Ren D, Xu H. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci U S A. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 53.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 54.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 55.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 56.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 57.Whitley P, Reaves BJ, Hashimoto M, Riley AM, Potter BV, Holman GD. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J Biol Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 58.Bryant NJ, Piper RC, Weisman LS, Stevens TH. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 60.Hirano T, Munnik T, Sato MH. Phosphatidylinositol 3-phosphate 5-kinase, FAB1/PIKfyve mediates endosome maturation to establish endosome-cortical microtubule interaction in Arabidopsis. Plant Physiol. 2015 doi: 10.1104/pp.15.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han BK, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Gisbergen PA, Li M, Wu SZ, Bezanilla M. Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol. 2012;198:235–250. doi: 10.1083/jcb.201112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serrazina S, Dias FV, Malho R. Characterization of FAB1 phosphatidylinositol kinases in Arabidopsis pollen tube growth and fertilization. New Phytol. 2014;203:784–793. doi: 10.1111/nph.12836. [DOI] [PubMed] [Google Scholar]
- 64.Vaccari I, Dina G, Tronchere H, Kaufman E, Chicanne G, Cerri F, Wrabetz L, Payrastre B, Quattrini A, Weisman LS, Meisler MH, Bolino A. Genetic interaction between MTMR2 and FIG4 phospholipid phosphatases involved in Charcot-Marie-Tooth neuropathies. PLoS Genet. 2011;7:e1002319. doi: 10.1371/journal.pgen.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oppelt A, Lobert VH, Haglund K, Mackey AM, Rameh LE, Liestol K, Schink KO, Pedersen NM, Wenzel EM, Haugsten EM, Brech A, Rusten TE, Stenmark H, Wesche J. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013;14:57–64. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takasuga S, Horie Y, Sasaki J, Sun-Wada GH, Kawamura N, Iizuka R, Mizuno K, Eguchi S, Kofuji S, Kimura H, Yamazaki M, Horie C, Odanaga E, Sato Y, Chida S, Kontani K, Harada A, Katada T, Suzuki A, Wada Y, Ohnishi H, Sasaki T. Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc Natl Acad Sci U S A. 2013;110:1726–1731. doi: 10.1073/pnas.1213212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCartney AJ, Zolov SN, Kauffman EJ, Zhang Y, Strunk BS, Weisman LS, Sutton MA. Activity-dependent PI(3,5)P2 synthesis controls AMPA receptor trafficking during synaptic depression. Proc Natl Acad Sci U S A. 2014;111:E4896–4905. doi: 10.1073/pnas.1411117111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baulac S, Lenk GM, Dufresnois B, Ouled Amar Bencheikh B, Couarch P, Renard J, Larson PA, Ferguson CJ, Noe E, Poirier K, Hubans C, Ferreira S, Guerrini R, Ouazzani R, El Hachimi KH, Meisler MH, Leguern E. Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology. 2014;82:1068–1075. doi: 10.1212/WNL.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campeau PM, Lenk GM, Lu JT, Bae Y, Burrage L, Turnpenny P, Roman Corona-Rivera J, Morandi L, Mora M, Reutter H, Vulto-van Silfhout AT, Faivre L, Haan E, Gibbs RA, Meisler MH, Lee BH. Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92:781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lenk GM, Ferguson CJ, Chow CY, Jin N, Jones JM, Grant AE, Zolov SN, Winters JJ, Giger RJ, Dowling JJ, Weisman LS, Meisler MH. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 74.Kippert F, Gerloff DL. Highly sensitive detection of individual HEAT and ARM repeats with HHpred and COACH. PLoS One. 2009;4:e7148. doi: 10.1371/journal.pone.0007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci U S A. 2012;109:E2042–2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]