Abstract
Human serotonin receptor 3A (5-HT3A) is a ligand-gated ion channel regulated by serotonin. A fusion protein (P9-5-HT3A) of 5-HT3A with the P9 protein, a major envelope protein of bacteriophage phi6, was highly expressed in the membrane fraction of Escherichia coli, and the expressed protein was purified to homogeneity using an affinity chromatography. P9-5-HT3A was observed as mixed oligomers in detergents. The purified P9-5-HT3A was efficiently reconstituted into proteoliposomes, and the serotonin-dependent ion-channel activity of P9-5-HT3A was observed by measuring the increased fluorescence of Fluo-3 attributed to the formation of a complex with the Ca2+ ions released from the proteoliposomes. Alanine substitution for Trp178 of 5-HT3A abolished the serotonin-dependent ion-channel activity, confirming the importance of Trp178 as a ligand-binding site. Furthermore, the ion-channel activity of the reconstituted P9-5-HT3A was effectively blocked by treatment with ondansetron, an antagonist of 5-HT3A. The bacterial expression system of human 5-HT3A and the proteoliposomes reconstituted with 5-HT3A would provide biophysical and structural analyses of 5-HT3A.
Keywords: Serotonin, Receptor, 5-HT3A, Overexpression, Purification, Reconstitution
Introduction
Neurotransmitter 5-hydroxytryptamine (5-HT) or serotonin receptors are grouped into seven families. Unlike other serotonin receptor families, which are G-protein coupled receptors, 5-HT3 is a ligand-gated ion channel [1]. Among the five subunits of 5-HT3 (5-HT3A–E), the 5-HT3A subunit has been extensively characterized both genetically and biochemically. The 5-HT3A subunit shows sequence homology to the nicotinic/GABA receptor gene super family [2], and it is most closely related to the nicotinic acetylcholine receptor (nAChR) with 30% amino acid homology [3]. The 5-HT3A subunit consists of an N-terminus extracellular domain, four transmembrane helical domains (M1–M4), and a relatively large cytoplasmic region between the M3 and M4 helixes. The 5-HT3 receptor consists of a pentamer of 5-HT3 subunits, and the 5-HT3A subunit is essential for assembly with other subtype subunits to form functional channels [2,4]. When only the 5-HT3A subunit is expressed in oocytes, it shows similar physiological characteristics to native 5-HT3 [2], suggesting that the 5-HT3A subunit could form an active homopentamer. Although a crystal structure of the 5-HT3A subunit has not been reported, electron microscopic analysis [5], molecular modeling of the 5-HT3A subunit based on a 4 Å resolution model of nAChR [6,7], and the crystal structure of the extracellular region of nAChR [8] all suggest that 5-HT3A subunit has a pentameric quaternary structure, and the ligand-binding extracellular region consists predominantly of β-sheets [9]. Furthermore, it was found that the loop between the M2 and M3 helices may have an important role in channel opening or receptor desensitization [10,11]. However, structural analysis of the open and closed states of the 5-HT3A subunit along with the principle behind this conformational change must be elucidated to understand the mechanism of ligand-gated ion channels for the purpose of drug development.
Structural and functional analyses of cell surface receptors require high-level expression in a heterologous system. Mammalian 5-HT3 is expressed at a low level in the central nervous system [12]. However, 5-HT3 is expressed at a substantially high level in mouse neuroblastoma cells, from which it can be purified and functionally reconstituted [13]. Nonetheless, structural and biochemical characterizations require a more convenient expression and purification system. The expression of membrane proteins in bacteria has been accomplished using specialized fusion partners such as Mistic [14], green fluorescent protein (GFP) [15], or maltose-binding protein and thioredoxin [16]. Recently, we had successively expressed human endothelin receptor type A (ETA), a member of the G-protein coupled receptors, in Escherichia coli using the P9 protein of bacteriophage phi6 as a fusion partner of ETA [17]. Most of the expressed P9-ETA was located in the membrane fraction, and it could be purified to homogeneity with high yield, suggesting that the P9 protein could be used as a fusion partner to assist in high-level expressions of other membrane proteins such as 5-HT3A in the membrane fraction of E. coli.
In this report, we have shown that the P9 protein was also effective in expressing human 5-HT3A at a high level in the membrane fraction of E. coli. The expressed P9-5-HT3A fusion protein was purified to homogeneity using a single-step affinity chromatography, with high yield. Furthermore, the purified P9-5-HT3A was reconstituted into proteoliposomes, which showed the serotonin-dependent release of encapsulated Ca2+ ions.
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and 1,2-dioleoyl-sn-glycero-3-{[N-(5-amino-1-carboxylpentyl)-iminodiacetic acid]succinyl} (nickel salt) (DOGS-NTA-Ni) were purchased from Avanti Polar Lipids, Inc. (USA). Fluo-3 was obtained from Invitrogen (USA). Serotonin, ondansetron, and N-lauroylsarcosine (Sarkosyl) were from Sigma (USA). Ni-NTA agarose resin was purchased from Qiagen (Germany). Bio-Beads (20–50 mesh) were from Bio-Rad (USA). All other chemicals used were of reagent grade.
Expression and purification of recombinant P9-5-HT3A
The expression vector of recombinant 5-HT3A (pP9-5-HT3A) was prepared by inserting the coding region of human 5-HT3A (amino acids 2-478; GenBank ID: CAA06442) at the C-terminus of the P9 protein from the Pseudomonas phage phi6 (GenBank ID: ABB69810.1) with a C-terminal His6 tag (amino acids: EASHHHHHH) (Fig. 1A). This sequence was inserted at the NdeI and HindIII sites of pRSETA (Invitrogen, USA) to generate the expression vector of P9-5-HT3A. The expression vector for the W178A substitution mutant of P9-5-HT3A [pP9-5-HT3A(W178A)] was prepared by site-directed mutagenesis using pP9-5-HT3A as the template DNA. The nucleotide sequences of the prepared expression vectors were confirmed by DNA sequencing. These vectors were transformed into the Rosetta2 (DE3) strain of E. coli and grown in Luria–Bertani medium containing 100 μg/ml of ampicillin and 30 μg/ml of chloramphenicol at 37 °C. Expression of the recombinant proteins was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), when the optical density of the culture at 600 nm reached 0.5–0.6. After 3 h of induction, the cells were harvested by centrifugation. The cell pellet (3 g of wet weight from 1 L culture) was resuspended in 25 ml of lysis buffer (20 mM HEPES, 150 mM KCl, pH 7.2) and lysed using a microfluidizer (M-110P; Microfluidics, USA). The insoluble materials of the lysate were then removed by centrifugation at 12,000g for 20 min, and the membrane fraction was precipitated by centrifugation at 100,000g for 1 h. The proteins in the membrane fraction were solubilized by incubation with 20 ml of lysis buffer containing 1% Sarkosyl for 1 h, and insoluble materials were removed by centrifugation at 100,000g for 1 h. The resulting supernatant was mixed with 2 ml of Ni-NTA for 1 h at 4 °C. After washing the resin with 10 ml of washing buffer (5 mM imidazole, 150 mM KCl, 20 mM HEPES, pH 7.2, 1% Sarkosyl), the proteins bound to the Ni-NTA resin were eluted using an elution buffer (200 mM imidazole, 150 mM KCl, 20 mM HEPES, pH 7.2, 1% Sarkosyl). The excess imidazole in the protein solution was removed using a desalting column.
Fig. 1.
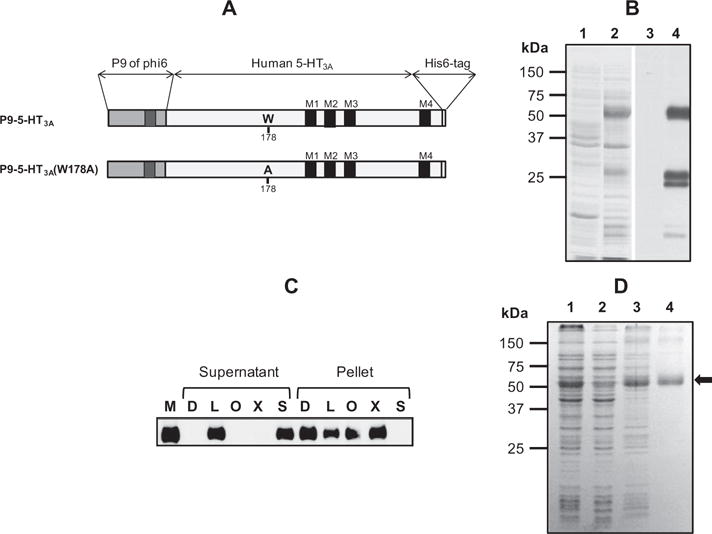
Expression and purification of P9-5-HT3A. (A) Schematic representation of wild-type and W178A mutant 5-HT3A along with the P9 and His6 tag flanking the N- and C-termini of 5-HT3A. The transmembrane regions in P9 and 5-HT3A are indicated as black rectangles, and the position of W178 or A178 is indicated. (B) The expression level of P9-5-HT3A in E. coli was examined by SDS–PAGE and western blotting. The membrane fractions of E. coli before (lanes 1 and 3) and after (lanes 2 and 4) induction with 1 mM IPTG were analyzed by SDS–PAGE, and the gel was stained with Coomassie blue dye (lanes 1 and 2) or subjected to western blotting using anti-His tag antibody (lanes 3 and 4). (C) The solubility of expressed P9-5-HT3A in various detergents was examined. The membrane fraction (M) from the lysate of cells expressing P9-5-HT3A was treated with 1% n-dodecyl β-D-maltoside (D), 1% lauryldimethylamine oxide (L), 2% n-octyl-β-d-glucopyranoside (O), 1% Triton X-100 (X), or 1% Sarkosyl (S) for 1 h at 4 °C. After ultracentrifugation at 100,000g for 1 h, the proteins in the supernatant and pellet fractions were separated by SDS–PAGE and analyzed by western blotting using anti-His tag antibody. D. SDS–PAGE analysis during the purification of P9-5-HT3A. Lane 1, cell lysate; lane 2, soluble fraction from cell lysate after ultracentrifugation; lane 3, membrane fraction; lane 4, purified protein after Ni-NTA column chromatography. Purified P9-5-HT3A is indicated as an arrow.
Reconstitution of P9-5-HT3A in liposomes
Large unilamellar liposomes (LUVs) with a diameter of 400 nm, consisting of POPC and POPS (weight ratio of POPC:POPS = 7:3) or POPC, POPS, and DOGS-NTA-Ni (weight ratio of POPC:POPS:-DOGS-NTA-Ni = 6.9:3:0.1) in buffer A (20 mM HEPES, 150 mM KCl, pH 7.2), were prepared using an extruder (Avanti, USA) as previously described [18]. Proteoliposomes were prepared by mixing 0.5 ml of LUVs (lipid concentration of 5 mg/ml) in buffer A with the same volume of P9-5-HT3A in buffer A containing 1% Sarkosyl, at a 1:1,000 protein-to-lipid molar ratio. After incubation at 4 °C for 1 h, the detergent in the mixture was removed by treatment with 100 mg of Bio-Beads for 3 h at 4 °C twice, or by dialysis against a 1,000-fold volume of buffer A for 24 h with two dialysis buffer changes. The vesicles were recovered by ultracentrifugation at 110,000g for 20 min using an Airfuge (Beckman), and any attached proteins on the surface of proteoliposomes were removed by washing 7–8 times using ultra centrifugation. The proteoliposomes were then resuspended in 200 μl of buffer A. The amount of proteins in the recovered proteoliposomes was estimated from the absorption at 280 nm, and with a BCA protein assay kit as described by the product manual (Pierce, IL, USA). The lipids in the proteoliposomes were assayed by measuring the amount of phosphates generated from acid hydrolysis, as previously described [19]. For the Ca2+-release assay, the proteoliposomes were prepared by mixing 5 mg/ml of LUVs in buffer A containing 7.5 mM CaCl2 with the same volume of P9-5-HT3A in buffer A containing 1% Sarkosyl, at a 1:4,000 protein-to-lipid molar ratio, and the proteoliposomes were prepared after using Bio-Beads to remove the Sarkosyl, as described above.
Liposomal release assay
The channel activity of the reconstituted P9-5-HT3A was examined by measuring the release of Ca2+ ions from the proteoliposomes using Fluo-3, a Ca2+ ion sensitive fluorescence dye. Fluo-3 (10 μM) and proteoliposomes (20 μg/ml) were mixed in buffer A (0.1 mL), and the release of Ca2+ ions was monitored using a Cary Eclipse Fluorescence Spectrophotometer (Varian, USA) at the excitation and emission wavelengths of 500 and 526 nm (slits, 5 nm), respectively, in a 1-cm path-length quartz cuvette at 25 °C. The extent of Ca2+ ion release was quantified on a percentage basis according to the following equation: [(Ft − F0) / (F100 − F0) × 100], where Ft is the measured fluorescence of reagent-treated LUVs at time t, F0 is the average fluorescence of the LUV suspension for the initial 1–2 min before the addition of 5-HT, and F100 is the average fluorescence value of the final 1–2 min after the complete disruption of proteoliposomes by addition of Triton X-100 (final concentration, 0.1%).
Results
P9-5-HT3A is highly expressed in the membrane fraction of E. coli as a fusion protein of the phi6 P9 protein
Phi6 is an enveloped bacteriophage that infects Pseudomonas syringae, and its P9 protein is a major envelope protein consisting of 90 amino acids with a single transmembrane helical region [20]. Previously, we have reported that human ETA could be highly expressed in the membrane fraction in E. coli when ETA was expressed as a P9 fusion protein [17]. To achieve this, the P9 protein was fused to the N-terminus of the wild-type or W178A mutant 5-HT3A along with a His6 tag at the C-terminus, as shown in Fig. 1A. The W178A mutant was prepared in order to verify the serotonin-dependent ion-channel opening activity of P9-5-HT3A, since the W178 residue, which is equivalent to W183 of mouse 5-HT3A, is critical for serotonin-dependent ion-channel opening [21,22]. These fusion proteins were expressed in E. coli Rosetta2 (DE3) cells. High-level expression of the fusion protein in the membrane fraction after induction with 1 mM IPTG was observed in the Coomassie blue-stained gel following SDS–PAGE (Fig. 1B, lanes 1 and 2). The induced protein was confirmed as P9-5-HT3A by western blot analysis using anti-His6 antibody (Fig. 1B, lanes 3 and 4). The average expression level of P9-5-HT3A was 1.5–2 mg per liter of culture, indicating that the P9 fusion system was very effective for the high-level expression of 5-HT3A in the membrane fraction of E. coli.
Expressed P9-5-HT3A was purified using a simple affinity column
To purify the expressed P9-5-HT3A in the membrane fraction, the efficiency of various detergents for the solubilization of P9-5-HT3A was examined. The membrane fraction from the lysate of cells expressing P9-5-HT3A was treated with n-dodecyl β-d-malto-side (D), lauryldimethylamine oxide (L), n-octyl-β-d-glucopyranoside (O), Triton X-100 (X), or Sarkosyl (S). After ultracentrifugation at 100,000g for 1 h, the supernatant and pellet fractions were analyzed by western blotting using anti-His tag antibody. As shown in Fig. 1C, the P9-5-HT3A in the membrane fraction was completely solubilized in 1% Sarkosyl, and about 50% was solubilized in 1% lauryldimethylamine oxide. However, P9-5-HT3A was poorly solubilized in the other detergents. The solubilized P9-5-HT3A was further purified, using a simple Ni-NTA affinity column, to a purity of >90% (Fig. 1D, lane 4). The purified P9-5-HT3A ran as a 56-kDa protein in SDS–PAGE, which was slightly lower than the calculated size (67 kDa). Membrane proteins such as GPCRs often migrate in SDS–PAGE faster than their actual size [17]. The identity of the purified protein as P9-5-HT3A was further confirmed by western blotting using anti-His6 antibody (data not shown). Finally, about 1.0 mg of purified protein was obtained from a 1 L culture. These results indicate that P9-5-HT3A was highly expressed in the membrane fraction of E. coli and was effectively purified by Ni-NTA affinity chromatography.
Purified P9-5-HT3A presents as oligomers in detergent micelles
It was well established that ligand-gated ion channels, such as nAChR, form pentamers. To investigate the oligomeric state of the purified P9-5-HT3A, the size of the Ni-NTA affinity-purified protein was analyzed by gel filtration. At least four protein peaks were observed in the chromatogram (Fig. 2A). When the proteins from these peaks were analyzed by SDS–PAGE, only the P9-5-HT3A band was observed in these peaks (Fig. 2A, upper panel), indicating that these peaks in the gel filtration chromatogram represented different oligomeric states of P9-5-HT3A. The sizes of these peaks were estimated to 78, 125, 223, and 366 kDa, respectively, based on the relative elution volumes of marker proteins (Fig. 2B). It was suggested that P9-5-HT3A tightly retains the micelles of Sarkosyl, which resulted in an increase of apparent molecular weight. Considering the predicted size of monomeric P9-5-HT3A (67.2 kDa) and the micelles of Sarcosyl (0.6 kDa), these peaks corresponded to a monomer, dimer, trimer, and pentamer, respectively. These results indicated that the purified proteins presented as a mixture of oligomers, including a pentamer.
Fig. 2.
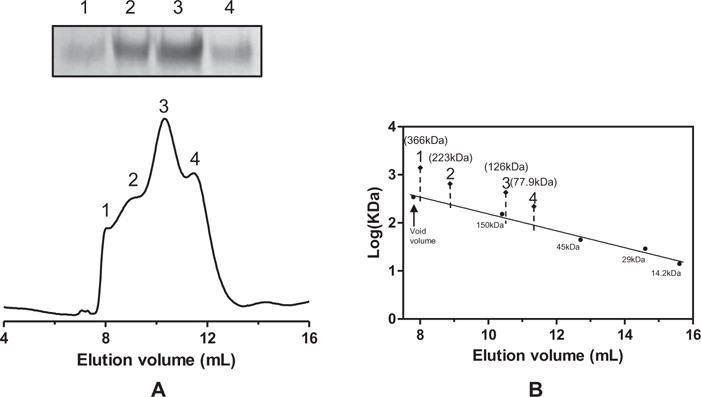
Gel filtration chromatography of P9-5-HT3A. (A) Purified P9-5-HT3A was injected onto a Superdex 200HR 10/300 GL gel filtration column equilibrated in 0.5% Sarkosyl, 50 mM HEPES, and 150 mM KCl, pH 7.2. The column was run at 0.5 ml/min, and the protein elution profile was monitored at 280 nm. The indicated fractions (1–4) were also analyzed by SDS–PAGE (upper panel). (B) The size of P9-5-HT3A was estimated using marker proteins (14.2, 29, 45, and 150 kDa).
Reconstituted P9-5-HT3A in proteoliposomes has the ligand-dependent ion-channel activity
To prepare the proteoliposomes containing the purified P9-5-HT3A, we used Bio-Beads adsorption and dialysis to remove the detergent. Additionally, the effect of NTA-Ni-containing lipids (DOGS-NTA-Ni) on the incorporation of the His-tag-containing P9-5-HT3A into proteoliposomes was examined. Between the two methods, the Bio-Beads adsorption method clearly showed a much higher recovery rate of P9-5-HT3A and lipids than the dialysis method. About 9% of proteins and 50% of lipids were recovered in the proteoliposomes prepared by the Bio-Beads method, whereas only 1.6% and 2.3% of proteins and lipids, respectively, were recovered from the dialysis method (Table 1). Furthermore, when a small portion of NTA-Ni-linked lipid (DOGS-NTA-Ni) was incorporated in the liposomes, the recovery rate of 5-HT3A in the proteoliposomes increased to 12.3%, suggesting that the specific interaction between the His6 tag of P9-5-HT3A and the NTA-Ni group in the liposomes enhanced the reconstitution of P9-5-HT3A.
Table 1.
Reconstitution efficiency of P9-5-HT3A in proteoliposomes.
| Detergent removal methods | Lipid compositiona (POPC:POPS:DOGS-NTA-Ni) | Recovery (%)b
|
|
|---|---|---|---|
| Protein | Lipids | ||
| Bio-Beads | 7:3:0 | 9.3 | 49.8 |
| 6.9:3:0.1 | 12.3 | 53.4 | |
| Dialyis | 7:3:0 | 1.6 | 2.3 |
The mass ratio of POPC, POPS, and DOGS-NTA-Ni.
The percentage of recovered P9-5-HT3A and lipids in the proteoliposomes compared with the initial amounts of P9-5-HT3A and lipids.
The expressed 5-HT3A gene in oocytes or mammalian cells showed characteristics similar to those of the native 5-HT3 receptor channel [2,23], and the permeability of divalent cations such as Ca2+ through the expressed 5-HT3 ion channel was comparable to that of monovalent cations such as Na+ or K+ [24], suggesting that a properly constituted 5-HT3A could transport Ca2+ ions. To examine whether or not the reconstituted P9-5-HT3A has ion-transport activity, the release of Ca2+ ions through P9-5-HT3A in Ca2+-encapsulated proteoliposomes was measured using Fluo-3 (Fig. 3A), a Ca2+ ion-sensitive fluorescent dye [25]. Addition of 10 μM or higher concentration of serotonin to the proteoliposome solution evoked prompt release of the Ca2+ ion, the amount of which corresponded to 7% of the total amount of encapsulated Ca2+ (Fig. 3B). In contrast, the P9-5-HT3A(W178A) mutant showed no significant Ca2+ release after treatment of up to 20 μM of 5-HT (Fig. 3C). These results indicate that the reconstituted P9-5-HT3A in proteoliposomes had serotonin-dependent ion-channel activity, and the effect of a mutation that defects this serotonin-dependent ion-channel activity of 5-HT3A [21,22] was easily observed by assaying the Ca2+ ions released from the proteoliposomes. The effective concentration of serotonin for opening the P9-5-HT3A ion channel was within 5–10 μM (Fig. S1), which was similar to the reported EC50 values for 5-HT (2–4 μM) [26,27]. The fact that the percentage of Ca2+ release relative to the total amount of Ca2+ released by detergent treatment was about 7% perhaps indicates that only 7% of the prepared proteoliposomes contained functional P9-5-HT3A. To further confirm that the reconstituted P9-5-HT3A has serotonin-dependent ion-channel activity, the inhibitory effect of ondansetron, an antagonist of 5-HT3A [28,29], on the release of Ca2+ from the proteoliposomes by 5-HT was examined. The ion-channel activity of P9-5-HT3A was not affected by the treatment of 1.0 μM of ondansetron (Fig. S2). When the proteoliposomes were incubated with 10–1000 nM of ondansetron, however, the release of Ca2+ from the proteoliposomes by 20 μM 5-HT was significantly reduced (Fig. 3D). These results indicate that P9-5-HT3A was functionally reconstituted in proteoliposomes and could bind specifically to 5-HT as well as to ondansetron.
Fig. 3.
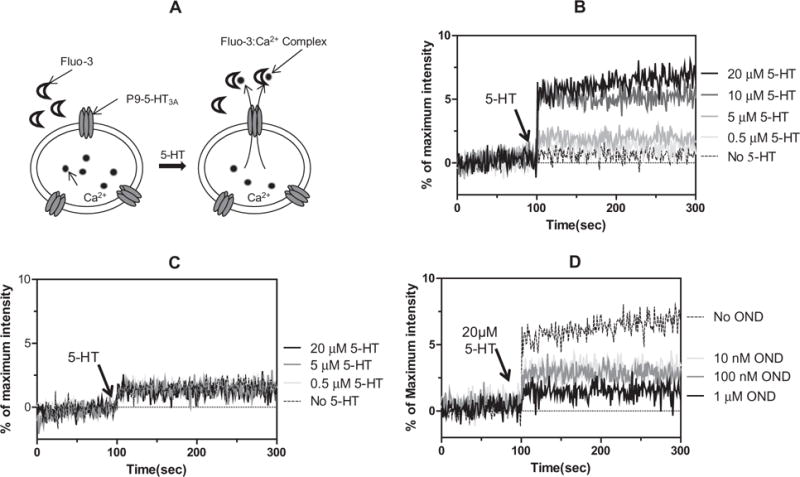
Calcium release assay of proteoliposomes containing P9-5-HT3A. (A) Schematic representation of the serotonin-dependent ion-channel opening assay with proteoliposomes reconstituted with P9-5-HT3A. The Ca2+ ions released from the P9-5-HT3A ion channel bind to Fluo-3, and the enhanced fluorescence of the Fluo-3:Ca2+ complex was measured. (B) The percentage release of Ca2+ from proteoliposomes containing wild-type P9-5-HT3A was measured by mixing different concentrations of serotonin (5-HT) at 25 °C. The time point of the addition of 0 M (dotted line), 0.5 μM (light grey line), 5 μM (grey line), 10 μM (dark grey line), or 20 μM (black line) 5-HT is indicated by the arrow. (C) Percentage release of Ca2+ from proteoliposomes containing P9-5-HT3A(W178A). The time point of the addition of 0 M (dotted line), 0.5 μM (light grey line), 5 μM (grey line), or 20 μM (black line) 5-HT is indicated by the arrow. (D) The percentage release of Ca2+ from proteoliposomes that were preincubated with 0 μM (dotted line), 10 nM (light grey line), 100 nM (grey line), or 1 μM (black line) ondansetron was measured at 25 °C. The time point of the addition of 20 μM 5-HT is indicated by the arrow.
Discussion
High-level expression of multi-transmembrane helical proteins such as channels or GPCRs has been scarcely reported. Some membrane proteins expressed as fusion proteins containing a well-folded protein such as GFP or thioredoxin have shown increased expression [15,16]. However, such an expression system might be applicable only in specific cases. In this report, we used a small single transmembrane helical protein, P9, as a fusion partner for the expression of human 5-HT3A. P9-5-HT3A showed high-level expression in E. coli, and more than 50% of the expressed protein was located in the membrane fraction. The P9 protein is a major envelope protein of bacteriophage phi6, and it was highly expressed in the membrane fraction of E. coli when using the T7 promoter (data not shown). The initial high rate of expression and insertion into the membrane might assist the high-level expression of multi-transmembrane proteins fused to the P9 protein.
The purified P9-5-HT3A appeared as a mix of oligomers (Fig. 2A) rather than a unique population of the pentamer, which was observed in 5-HT3A expressed in mammalian cells [5,30]. The ligand-dependent ion-channel activity of P9-HT3A, however, was observed when it was reconstituted into liposomes. An electrophysiological analysis indicated that Na+, K+, and Ca2+ ions are permeable through the 5-HT3 ion channel, which has a pore size of 7.6 Å [24]. When the oligomers of purified P9-5-HT3A were reconstituted into proteoliposomes, only 7% of the proteoliposomes showed serotonin-dependent Ca2+ ion release (Fig. 3B), suggesting that only a minor fraction of P9-5-HT3A, probably the pentameric form, might form functional ion channels. It had been shown that glycosylation was important for the proper folding of 5-HT3. Deglycosylation of active 5-HT3 expressed in insect cells did not reduce its ability to bind ligands but did assist in the proper folding of 5-HT3 [31]. Hence, a minor fraction of P9-5-HT3A subunits expressed in E. coli would form an active pentamer. Optimization of the folding condition, or application of an E. coli strain that can afford glycosylation, would enhance the yield of properly folded P9-5-HT3A.
The serotonin-dependent release of Ca2+ ions from the proteoliposomes reconstituted with P9-5-HT3A and the inhibition of the P9-5-HT3A ion channel by ondansetron indicated that the Ca2+ release assay could be applied to examine the activity of P9-5-HT3A. Calcium ions have shown an inhibitory effect on the 5-HT3 ion channel [32]. However, the calcium ion binding site is located in the extracellular region of 5-HT3 [11], and the calcium ions presenting only inside of the proteoliposomes could not affect the calcium-sensing site of the reconstituted P9-5-HT3A, which was located outside of the proteoliposomes. This Ca2+ release assay with the reconstituted P9-5-HT3A could be used as a model system for the structural and functional analyses of the 5-HT3 ion channel.
Supplementary Material
Acknowledgments
This work was supported by the Functional Proteomics 21 Program (FPR08B2-290). Lim was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0023759). We also thank the Research Program 2012 of Kookmin University, Korea.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.pep.2013.01.001.
References
- 1.Bradley PB, Engel G, Feniuk W, Fozard JR, Humphrey PP, Middlemiss DN, Mylecharane EJ, Richardson BP, Saxena PR. Proposals for the classification and nomenclature of functional receptors for 5-hydroxytryptamine. Neuropharmacology. 1986;25:563–676. doi: 10.1016/0028-3908(86)90207-8. [DOI] [PubMed] [Google Scholar]
- 2.Maricq AV, Peterson AS, Brake AJ, Myers RM, Julius D. Primary structure and functional expression of the 5-HT3 receptor, a serotonin-gated ion channel. Science. 1991;254:432–437. doi: 10.1126/science.1718042. [DOI] [PubMed] [Google Scholar]
- 3.Vetz P, Abdelatty F, Villarroel A, Rappold G, Weiss B, Koenen M. Organization of the murine 5-HT3 receptor gene and assignment to human chromosome 11. FEBS Lett. 1994;339:302–306. doi: 10.1016/0014-5793(94)80435-4. [DOI] [PubMed] [Google Scholar]
- 4.Miyake A, Mochizuki S, Takemoto Y, Akuzawa S. Molecular cloning of human 5-hydroxytryptamine3 receptor: heterogeneity in distribution and function among species. Mol Pharmacol. 1995;48:407–416. [PubMed] [Google Scholar]
- 5.Boess FG, Beroukhim R, Martin IL. Ultrastructure of the 5-hydroxytryptamine 3 receptor. J Neurochem. 1995;64:1401–1405. doi: 10.1046/j.1471-4159.1995.64031401.x. [DOI] [PubMed] [Google Scholar]
- 6.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 7.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR a1 bound to α-bungarotoxin at 1.94 Å resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 9.Reeves DC, Sayed MFR, Chau PL, Price KL, Lummis SCR. Prediction of 5-HT3 receptor agonist-binding residues using homology modeling. Biophys J. 2003;64:2338–2344. doi: 10.1016/S0006-3495(03)75039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lummis SCR, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis–trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 11.Paulsen IM, Martin IL, Dunn SM. Isomerization of the proline in the M2–M3 linker is not required for activation of the human 5-HT3A receptor. J Neurochem. 2009;110:870–878. doi: 10.1111/j.1471-4159.2009.06180.x. [DOI] [PubMed] [Google Scholar]
- 12.Kilpatrick GJ, Jones BJ, Tyers MB. The identification and distribution of 5-HT3 receptors in rat brain using radioligand binding. Nature. 1987;330:746–748. doi: 10.1038/330746a0. [DOI] [PubMed] [Google Scholar]
- 13.Lummis SCR, Martini IL. Solubilization, purification, and functional reconstitution of 5-hydroxytryptamine 3 receptors from N1E1-115 neuroblastoma cells. Mol pharmacol. 1991;41:18–23. [PubMed] [Google Scholar]
- 14.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 15.Drew DE, von Heijne G, Norlund P, de Gier JWL. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 16.Yeliseev AA, Wong KK, Soubias O, Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14:2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, Jung Y, Lee JY, Lee WK, Lim D, Yu YG. Purification and characterization of recombinant human endothelin receptor type A. Protein Exp Purif. 2012;84:14–18. doi: 10.1016/j.pep.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Xu Y, Zhang F, Shin YK. Constitutive versus regulated SNARE assembly: a structural basis. EMBO J. 2004;23:681–689. doi: 10.1038/sj.emboj.7600083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Arthur G. Improved procedures for the determination of lipid phosphorus by malachite green. J Lipid Res. 1992;33:1233–1236. [PubMed] [Google Scholar]
- 20.Johnson MD, III, Mindich L. Plasmid-directed assembly of the lipid-containing membrane of bacteriophage phi6. J Bacteriol. 1994;176:4124–4132. doi: 10.1128/jb.176.13.4124-4132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-p interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- 22.Thompson AJ, Price KL, Lummis SCR. Cysteine modification reveals which subunits form the ligand binding site in human heteromeric 5-HT3AB receptors. J Physiol. 2011;589:4243–4257. doi: 10.1113/jphysiol.2011.208439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussy N, Lukas W, Jones KA. Functional properties of a cloned 5-hydroxytryptamine ionotropic receptor subunit: comparison with native mouse receptors. J Physiol. 1994;481:311–323. doi: 10.1113/jphysiol.1994.sp020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J. Ion permeation through 5-hydroxytryptamine-gated channels in neuroblastoma N18 cells. J Gen Physiol. 1990;96:1177–1198. doi: 10.1085/jgp.96.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minta A, Kao JPY, Tsien RY. Fluorescent indicators for cytosolic calcium based on rhodamine and fluorescein chromophore. J Biol Chem. 1989;264:8171–8178. [PubMed] [Google Scholar]
- 26.Hussy N, Lukas W, Jones KA. Functional properties of a cloned 5-hydroxytryptamine ionotropic receptor subunit: comparison with native mouse receptors. J Physiol. 1994;3072:311–323. doi: 10.1113/jphysiol.1994.sp020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiter C, Hovius R, Costioli M, Pick H, Kellenberger S, Schild L, Vogel H. Characterization of the ligand-binding site of the serotonin 5-HT3 receptor: the role of glutamate residues 97, 224 and 235. J Biol Chem. 2003;278:22709–22716. doi: 10.1074/jbc.M301801200. [DOI] [PubMed] [Google Scholar]
- 28.Van Wijngaarden I, Hamminga D, van Hes R, Standaar PJ, Tipker J, Tulp MTM, Mol F, Olivier B, de Jonge A. Development of high-affinity 5-HT3 receptor antagonists. Structure-affinity relationships of novel 1,7-annelated indole derivatives. J Med Chem. 1993;36:3693–3699. doi: 10.1021/jm00075a026. [DOI] [PubMed] [Google Scholar]
- 29.Min KT, Wu CL, Yang J. Nondepolarizing neuromuscular blockers inhibit the serotonin-type 3A receptor expressed in Xenopus oocytes. Anesth Analg. 2000;90:476–481. doi: 10.1097/00000539-200002000-00044. [DOI] [PubMed] [Google Scholar]
- 30.McKernan RM, Gillard NP, Quirk K, Kneen CO, Stevenson GI, Swain CJ, Ragan CI. Purification of the 5-hydrozytryptamine 5-HT3 receptor from NCB20 cells. J Biol Chem. 1990;265:13572–13577. [PubMed] [Google Scholar]
- 31.Green T, Stauffer KA, Lummis SCR. Expression of recombinant homooligomeric 5-hydroxytryptamine 3 receptors provides new insights into their maturation and structure. J Biol Chem. 1995;270:6056–6061. doi: 10.1074/jbc.270.11.6056. [DOI] [PubMed] [Google Scholar]
- 32.Eisele JL, Bertrand S, Galzi JL, Devillers-Thiery A, Changeux JP, Bertrand D. Chimeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366:479–483. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


