Abstract
Mutations disabling the TP53 tumour suppressor gene represent the most frequent events in human cancer and typically occur through a two-hit mechanism involving a missense mutation in one allele and a ‘loss of heterozygosity’ deletion encompassing the other. While TP53 missense mutations can also contribute gain-of-function activities that impact tumour progression, it remains unclear whether the deletion event, which frequently includes many genes, impacts tumorigenesis beyond TP53 loss alone. Here we show that somatic heterozygous deletion of mouse chromosome 11B3, a 4-megabase region syntenic to human 17p13.1, produces a greater effect on lymphoma and leukaemia development than Trp53 deletion. Mechanistically, the effect of 11B3 loss on tumorigenesis involves co-deleted genes such as Eif5a and Alox15b (also known as Alox8), the suppression of which cooperates with Trp53 loss to produce more aggressive disease. Our results imply that the selective advantage produced by human chromosome 17p deletion reflects the combined impact of TP53 loss and the reduced dosage of linked tumour suppressor genes.
Cancer arises through the acquisition of genetic and epigenetic changes that drive tumorigenesis. While most efforts to understand the origins of these entities have focused on the identification and functional characterization of somatic single nucleotide variants (SNVs) that activate oncogenes or inactivate tumour suppressors, the vast majority of human cancers also harbour large copy number variants (CNVs) that impact gene dosage through gain or loss of whole chromosomes or chromosome segments1. For example, recurrent segmental deletions—such as those affecting chromosome 17p—are extremely common in human cancers yet are widely considered to arise because of selection for a single ‘driver’ in the deleted region, with the adjacent gene losses reflecting ‘passenger’ events that have no effect on tumour phenotypes. However, evidence is emerging from short hairpin RNA (shRNA) screens and bioinformatic approaches that these lesions may target more than one relevant activity2–5. If true, then the impact of segmental deletions on tumour development would be fundamentally distinct from SNVs, yet, given their frequent occurrence, disproportionately understudied.
Chromosome 17p configurations in human cancer
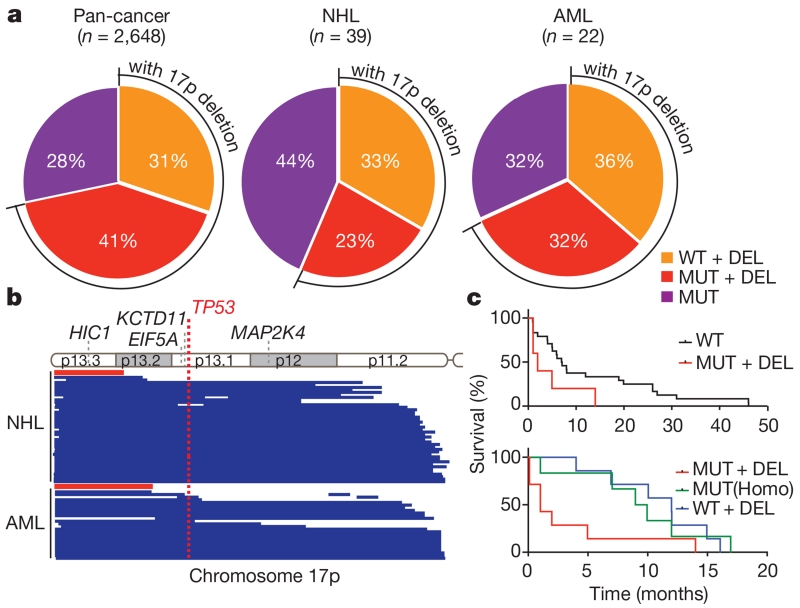
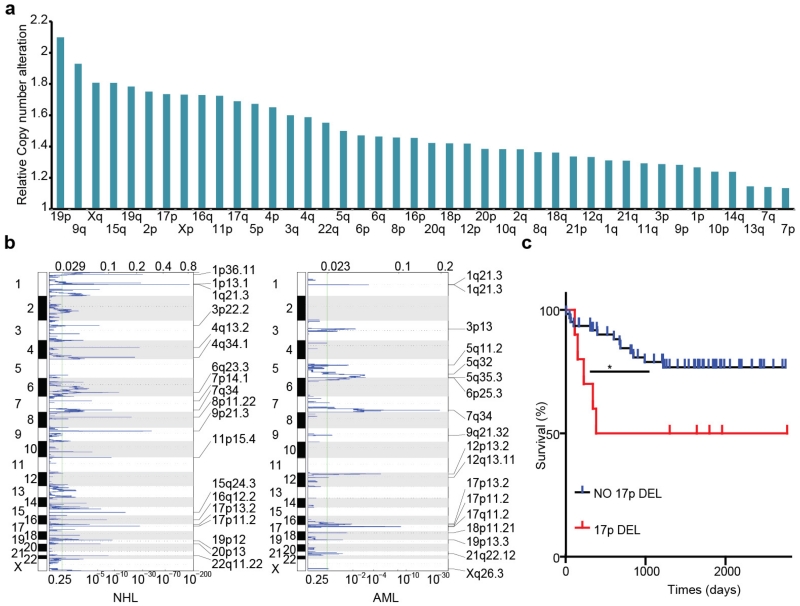
We decided to study 17p deletion as a prototype of a recurrent cancer-associated deletion owing to its high frequency and invariable inclusion of TP53, an extensively studied tumour suppressor gene that is considered to be the major driver of 17p loss6,7. To characterize better the nature and scope of these deletions, we analysed genomic data from 4,994 tumours across 18 cancer types (http://www.cbioportal.org; accessed 29 October 2014). This survey confirmed previous indications that mutation and/or deletion of the TP53 tumour suppressor gene occurs in approximately ~50% of all human cancers8. We then classified 17p-altered tumours as to whether they displayed TP53 mutation (encompassing missense, nonsense and frameshift mutations), deletion (as defined by a reduction in TP53 copy number), or both (Fig. 1a). The most common TP53 configuration involves a missense mutation together with a segmental 17p deletion, although a substantial fraction of p53-altered tumours harboured chromosome 17p deletion together with an apparently wild-type TP53 allele. Similar results were observed in blood cancers, in which TP53 mutations and/or 17p loss are less common but are linked to a particularly dismal prognosis9,10. Hence, the frequency of 17p deletion may even exceed point mutations within the TP53 gene.
Figure 1. The frequency, scope, and prognostic value of chromosome 17p alterations in human cancers.
a, The nature of TP53-containing chromosome 17p alterations in pan-human cancer (left), non-Hodgkin lymphomas (NHL; middle) and AML (right). DEL, deletion; MUT, mutations; WT, wild-type. b, The extent of chromosome 17p deletions in NHL (44 cases; top) and AML (25 cases; bottom) data sets, irrespective of TP53 deletion, as determined by single nucleotide polymorphism (SNP) array analysis. Each line represents one patient. Red bar indicates the significant copy number loss (q < 0.25) analysed by GISTIC algorithm. c, Overall survival of complex-karyotype AML patients containing both 17p deletion and TP53 mutation as compared to those without any TP53 alteration (top; P = 0.076), those with homozygous (Homo) TP53 point mutations (bottom; P = 0.059), or those with 17p deletion without any detectable TP53 mutation (bottom; P = 0.03). All are log-rank test.
Chromosome 17p deletions frequently encompass all or most of the chromosome arm (Fig. 1b). In addition to TP53, this region encodes over 300 protein-coding genes that include other established or putative tumour suppressors11–13. Unexpectedly, TP53 was not identified as a candidate tumour suppressor in non-Hodgkin lymphoma or acute myeloid leukaemia (AML) using GISTIC, an algorithm designed to pinpoint candidate drivers from CNV data by identifying ‘epicentres’ of gain or loss14 (Fig. 1b and Extended Data Fig. 1b). Moreover, AMLs harbouring a TP53 mutation together with a 17p deletion displayed a worse prognosis than those harbouring TP53 mutations alone (Fig. 1c and Extended Data Fig. 1c). Collectively, these data raise the possibility that additional genes drive selection for 17p loss during tumorigenesis.
Modelling 17p13 deletions in the mouse
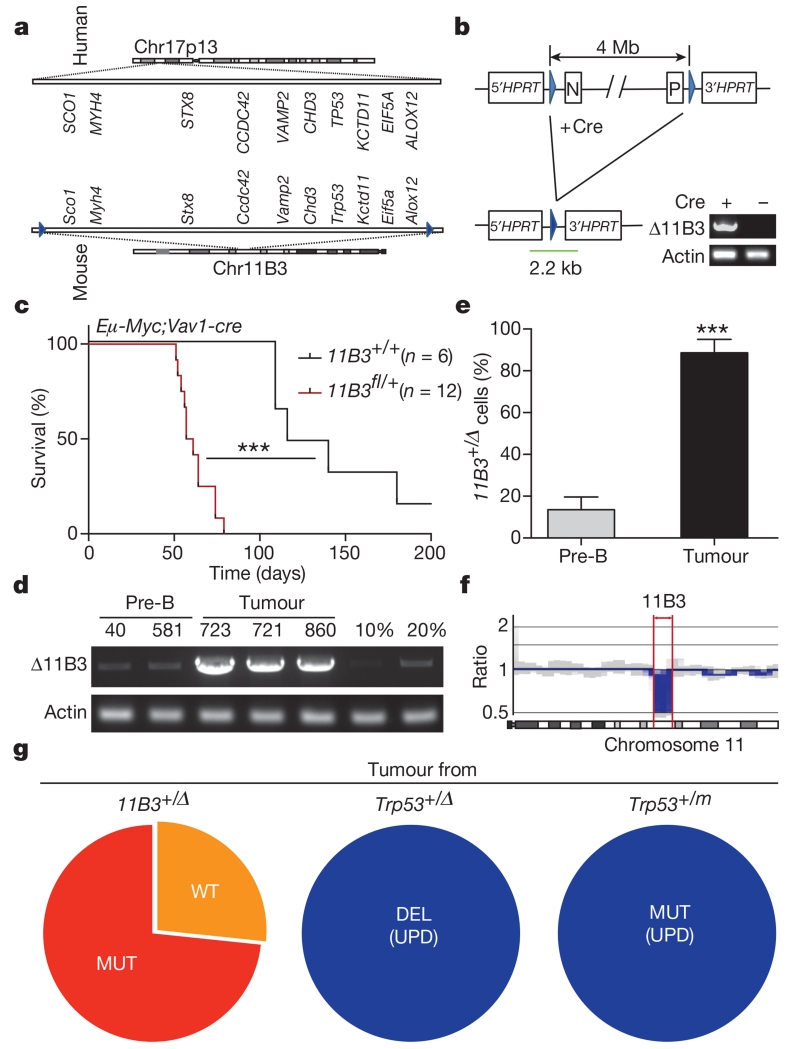
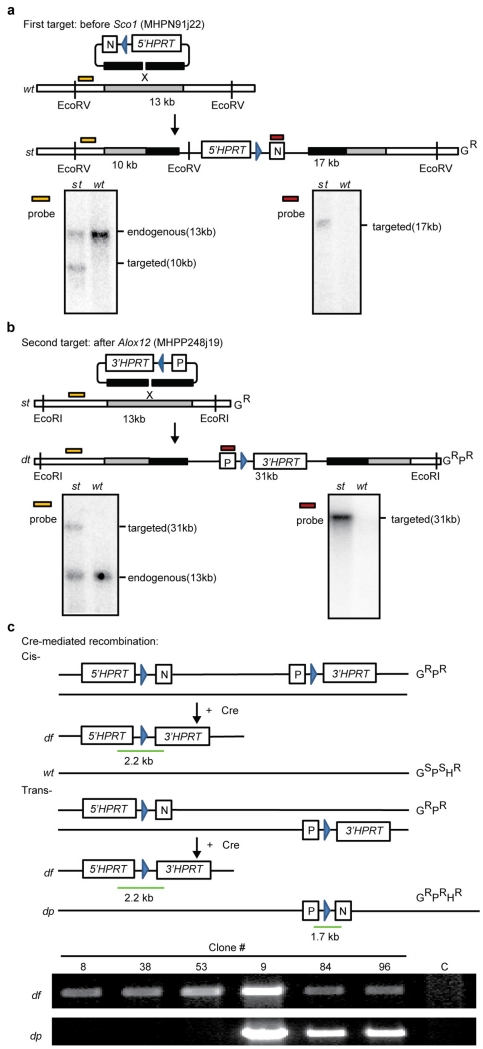
We decided to test this hypothesis in mice. To produce a strain capable of modelling somatic 17p deletions, we used the MICER chromosome engineering strategy developed previously15 to introduce loxP sites and a split HPRT gene into chromosomal regions flanking the Alox12 and Scol genes on mouse chromosome 11B3, which is a 4 Mb region on mouse chromosome 11 that is syntenic to human 17p13.1 (Fig. 2a, b and Extended Data Fig. 2a, b). Candidate clones were treated with adenovirus (Adeno)-cre to recombine the 11B3 region, selected in hypoxanthine–aminopterin–thymidine (HAT) medium, and subjected to a polymerase chain reaction (PCR) analysis to discriminate between cis and trans recombination events (Extended Data Fig. 2c). This readily identified embryonic stem (ES) cell clones harbouring loxP sites flanking 11B3 in cis and revealed that 11B3 deletion is not deleterious to ES cells (data not shown). We refer to the appropriately targeted allele above as 11B3flox (11B3fl).
Figure 2. A mouse model of human 17p13.1 deletion accelerates lymphoma development.
a, The synteny of human chromosome (Chr) 17p13.1 and mouse chromosome 11B3 (from Sco1 to Alox12b, ~4 Mb), with several representative genes listed. Blue arrowheads denote loxP sites. b, Conditional, 11B3-deletion strategy with PCR analysis (corresponding to the green bar) showing the desired deletion. c, Tumour-free survival of mice with the indicated genotype (log-rank test). d, e, The extent of 11B3 deletion in non-tumorigenic pre-B cells (Vav1-cre;11B3fl/+) and in lymphomas arising in Eμ-Myc;Vav1-cre;11B3fl/+ mice was determined by semi-quantitative PCR using mixed genomic DNA from 11B3+/Δ ES cells to 11B3fl/+ cells at different ratios (10% or 20%) (d), and qPCR (e; two-tailed t-test). Sample names correspond to the mouse identifier. Error bars represent standard deviation (s.d.). f, Copy number profile of mouse chromosome 11 as determined by low-pass whole genome sequencing of 11B3-deleted lymphoma cells obtained from c. Red arrows highlight the 11B3 region. g, Resulting Trp53 status upon LOH from tumours arising from heterozygous chromosome 11B3 deletion (left) or those originating from heterozygous Trp53 deletion (middle) or point mutation (right). UPD, uniparental disomy. ***P < 0.001.
11B3fl mice were produced by blastocyst injection and crossed to a variety of constitutive and haematopoietic-directed Cre mouse lines. We never observed viable progeny harbouring a recombined 11B3 deletion arising from 11B3fl crosses with Ella-cre or constitutive CAG-cre mice, implying that heterozygous deletion of the 11B3 interval confers embryonic lethality. However, double-mutant progeny harbouring the cre transgene and an unrecombined 11B3fl allele were produced (Extended Data Table 1), perhaps surviving owing to inefficient recombination between the two distant (~4 Mb) loxP sites.
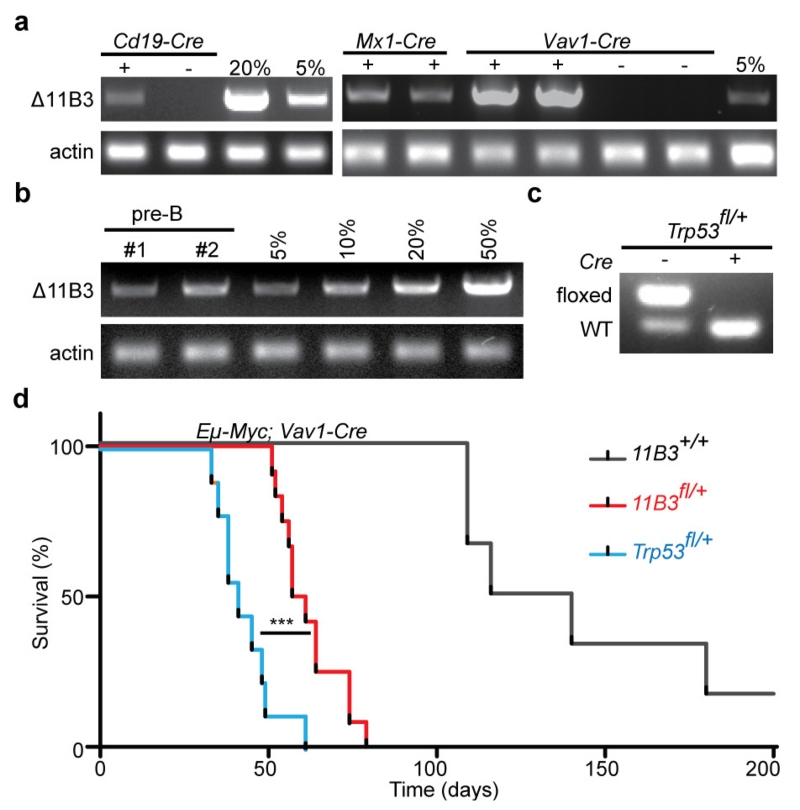
As in ES cells, somatic 11B3 deletion in the haematopoietic compartment was not an obligate cell lethal event; thus, recombined cells were detected in the peripheral blood of 11B3fl/+ mice crossed to strains harbouring Cd19-cre, Mx1-cre or Vav1-cre alleles (Extended Data Fig. 3a). Nevertheless, the apparent recombination frequency was substantially lower than observed in progeny of conditional Trp53--knockout mice crossed to the same strains (20% versus 100%; Extended Data Fig. 3b, c). While such differences can complicate direct comparisons of tumour onset between 11B3fl and Trp53fl animals, we reasoned that changes in the fraction of 11B3-deleted cells over time should indicate whether 11B3 loss provides a competitive advantage during tumorigenesis.
We first tested whether somatic deletion of 11B3 could drive cancer development in the Eμ-Myc transgenic model of non-Hodgkin lymphoma, as Trp53 deletion potently accelerates lymphomagenesis in this model16,17 and because 17p alterations are associated with adverse prognosis in the corresponding human disease18 (Extended Data Fig. 1c). Accordingly, Eμ-Myc mice were crossed to the 11B3fl allele and a Vav1-cre transgenic strain, and the resulting progeny were monitored for tumour onset (Fig. 2c). Mice harbouring the 11B3fl allele developed lymphomas much more rapidly than controls (59 versus 130 days, P < 0.001), clearly indicating that loss of the 11B3 region can promote tumorigenesis.
Paradoxically, Eμ-Myc;Trp53fl/+;Vavl-cre mice developed lymphomas even more rapidly than Eμ-Myc;11B3fl/+;Vav1-cre animals (Extended Data Fig. 3d). While this observation is consistent with a potentially negative impact of 11B3 deletion on cellular fitness, it might also reflect the nearly fivefold reduced recombination efficiency of the 11B3fl versus Trp53fl alleles. Consistent with the idea of reduced recombination efficiency, semi-quantitative and quantitative (q)PCR analysis indicated a massive enrichment in the fraction of 11B3-deleted cells in lymphomas compared with the premalignant setting (~100% versus 15%; Fig. 2d, e), which was confirmed by genome sequencing (Fig. 2f). Although lymphomas arising in the Trp53rfl/+ and 11B3fl/+ animals were both highly disseminated, 11B3-deleted lymphomas typically presented as a more mature B-cell type and showed enhanced resistance to certain chemotherapeutic agents (Extended Data Fig. 4a–c). Thus, 11B3 deletion confers a selective advantage during lymphomagenesis and produces phenotypes distinct from a Trp53-null setting.
p53 is induced after DNA damage, when it activates transcription of target genes involved in cell cycle arrest, apoptosis, and other cellular processes19,20. Strikingly, all 11B3-deleted lymphomas showed either high or undetectable levels of p53 protein, and none were capable of inducing the p53 transcriptional target p21 (also known as Cdkn1a) after treatment with the DNA-damaging chemotherapeutic drug adriamycin (ADR). Such a pattern is reminiscent of lymphoma cells that acquire inactivating Trp53 mutations21,22 and indeed, most 11B3-deleted lymphomas (75%) acquired missense or nonsense mutations in the remaining wild-type Trp53 allele (Extended Data Fig. 4f). Other 11B3-deleted lymphomas displayed no p53 activity, suggesting that each acquired some other mutational or epigenetic event that disabled the intact wild-type Trp53 allele. Regardless, the majority of 11B3-deleted lymphomas display chromosome configurations analogous to the common TP53 mutation/deletion events observed in human cancers (Fig. 1a and Fig. 2g).
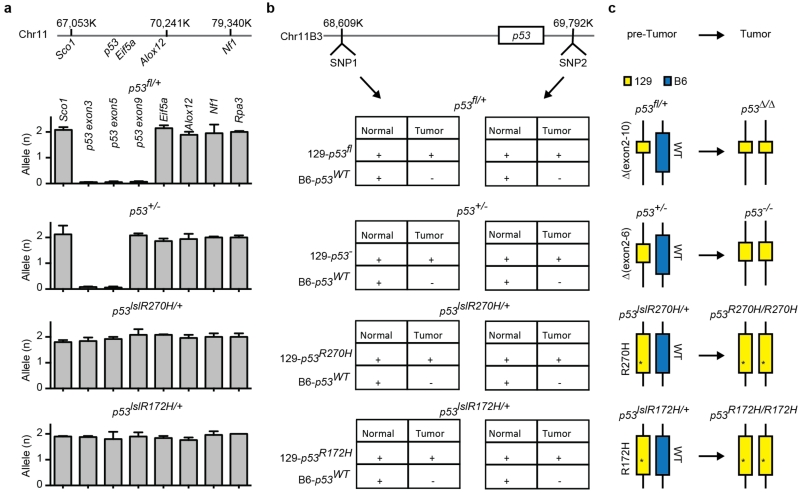
We also tested the mechanisms of loss of heterozygosity (LOH) arising in conventional Trp53-mutant mouse models. To this end, we generated Eμ-Myc strains with the following additional alleles: Trp53+/−, Vav1-cre; Trp53fl/+, Vav1-cre;Trp53LSL-R270H/+, or Vav1-cre;Trp53LSL-R172H/+, and analysed the fate of the remaining Trp53 allele upon lymphoma manifestation using a qPCR assay to detect the copy number of select regions flanking Trp53. In these models LOH clearly does not involve deletion of the remaining Trp53 locus, as the genes proximal and distal to Trp53 (for example, Eif5a and Sco1/Alox12, respectively) remained diploid in all cases (Extended Data Fig. 5a). Instead, this analysis suggests that LOH occurs through duplication of the targeted mutant Trp53 allele, a conclusion that was confirmed by analysis of polymorphic markers in this region (Fig. 2g and Extended Data Fig. 5b, c). While such 17p configurations occur in human tumours, they are less frequent than those involving large deletions (see Fig. 1a).
Role of 11B3 deletion in lymphomagenesis
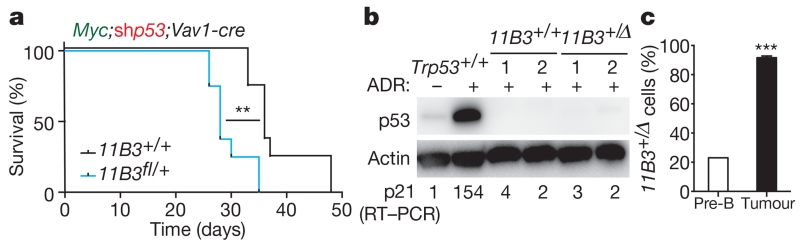
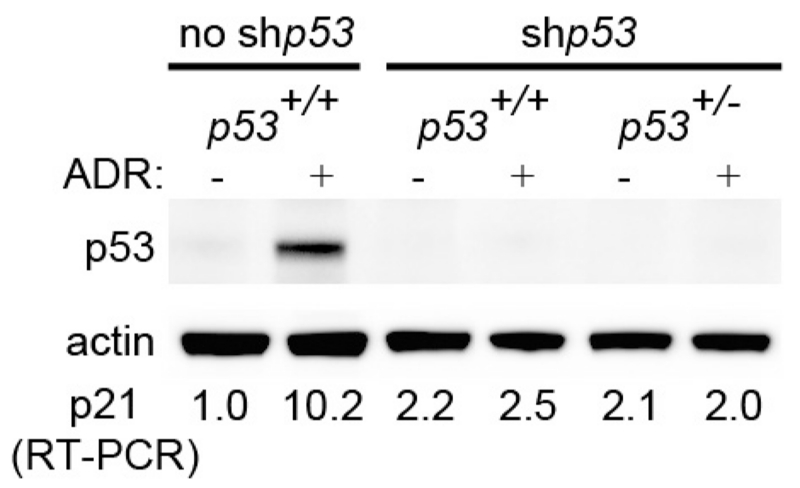
Although the data described earlier demonstrate that 11B3 deletion can contribute to tumorigenesis, the differences in recombination frequencies between the conditional 11B3 and Trp53 alleles made it impossible to assess whether other genes in the 11B3 deletion contribute to its effects. To mitigate this caveat without extensive strain intercrossing, we exploited the capability of shRNAs to suppress gene expression in trans, hypothesizing that any phenotypic differences arising between wild-type and 11B3-deleted cells expressing a potent Trp53 shRNA would be independent of Trp53. As immunoblotting and qPCR with reverse transcription (RT–qPCR) analysis confirmed that our Trp53 shRNA was equally capable of disabling Trp53 in pre-B cells harbouring one or two Trp53 alleles (Extended Data Fig. 6), we co-transduced pre-B cells derived from the bone marrow of Vav1-cre and Vav1-cre;11B3fl/+ mice with a Myc complementary DNA (linked to GFP) and the Trp53 shRNA (linked to mCherry) and studied tumorigenesis upon their transplantation into irradiated recipient mice.
Mice receiving cells harbouring the conditional 11B3 deletion and the Trp53 shRNA developed tumours more rapidly than those transplanted with cells harbouring the Trp53 shRNA alone (Fig. 3a; median onset 28 versus 36 days, P < 0.01). In both the 11B3fl/+ and 11B3+/+ settings, the disease presented as an aggressive B-cell malignancy (data not shown) in which p53 protein and activity were undetectable (Fig. 3b). Furthermore, the subsequent lymphomas were double positive for GFP and mCherry, indicating that they retained the Myc cDNA and Trp53 shRNA (data not shown). While only 15–20% of the premalignant cells underwent 11B3 recombination, almost all of the resulting lymphoma cells harboured this deletion (Fig. 3c). Thus, 11B3 deletion confers a strong selective advantage even in the absence of detectable p53 function.
Figure 3. 11B3 deletion can accelerate lymphomagenesis through p53-independent mechanisms.
a, Kaplan–Meier lymphoma-free survival curve of recipient mice receiving Vav1-cre;11B3fl/+ or Vav1-cre; 11B3+/+, transduced simultaneously with a mouse Myc cDNA and an shRNA against Trp53 (shp53). n = 8 per genotype. **P < 0.01 (log-rank test). b, The expression levels of Trp53 and p21 after ADR treatment of resulting lymphoma cells as detected by immunoblotting and RT–qPCR, respectively. p21 levels were normalized to untreated Trp53+/+ lymphomas. Shown are representative results (n = 8 per cohort). c, qPCR analysis to determine the percentage of chromosome 11B3 deletion in pre-B cells (transduced cells before transplantation; n = 3) and resulting lymphomas (n = 5) from the experiment in a. ***P < 0.001 (two-tailed t-test).
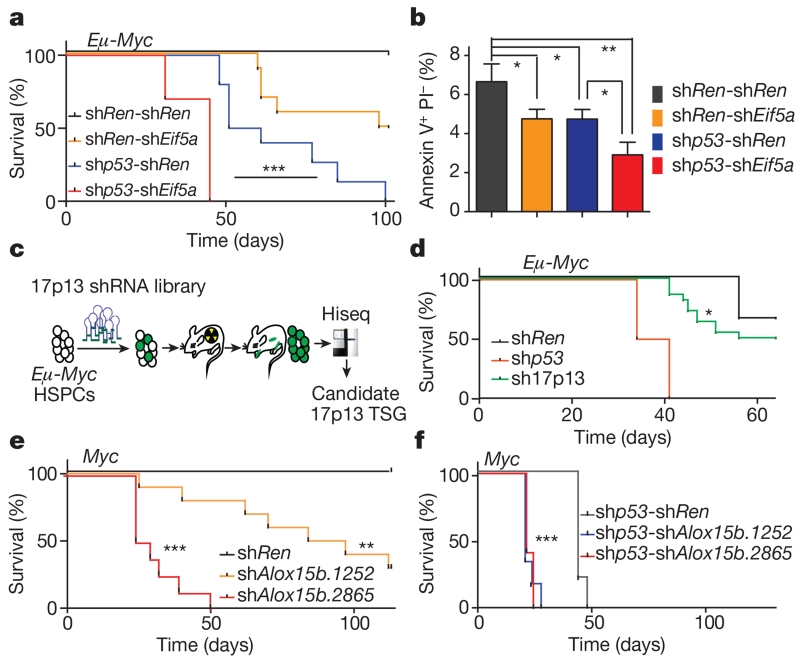
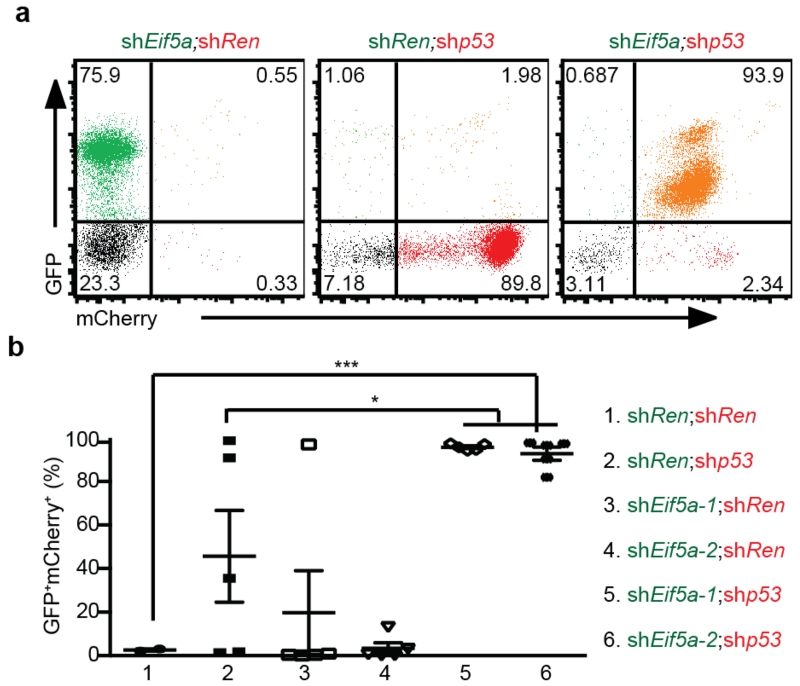
To identify genetic elements within the 11B3 interval that act together with Trp53 to suppress lymphoma development, we first took a candidate gene approach. We noted that virtually all lymphoma-associated 17p deletions encompass both TP53 and E1F5A (see Fig. 1b), the latter gene also having been identified as a tumour suppressor that promotes apoptosis11. To examine interactions between these genes during lymphomagenesis, validated shRNAs targeting Trp53 and/or Eif5a, co-expressed in tandem, were introduced into Eμ-Myc haematopoietic stem and progenitor cells (HSPCs) and then transplanted into syngeneic recipients. As expected, suppression of either Trp53 or Eif5a accelerated lymphomagenesis relative to controls, which did not develop tumours over the time of evaluation (Fig. 4a; median onset 56 and 99.5 days, respectively). Still, co-suppression of both Trp53 and Eif5a produced lymphomas even more rapidly (median onset 45 days). Apparently, premalignant cells co-suppressing Trp53 and Eif5a shRNAs have an increased selective advantage compared with cells expressing either shRNA alone, a suggestion that was confirmed by an in vivo competition assay (Extended Data Fig. 7) and apoptosis measurements in premalignant cells (Fig. 4b)16,23,24. Although our shRNA technology cannot precisely mimic the gene dosage produced by a heterozygous gene deletion, these data identify Eif5a as a second gene in the 11B3 interval that can contribute to its activity.
Figure 4. 11B3 encodes multiple genes whose attenuation cooperates with Trp53 loss to drive lymphoma.
a, Kaplan–Meier lymphoma-free survival of recipient mice transplanted with Eμ-Myc HSPCs with various GFP-linked tandem shRNAs. shp53 indicates shTrp53. n = 10 per group. b, Annexin V staining of Eμ-Myc pre-B cells transduced with the indicated tandem shRNAs constructs. n = 3. Result represents at least two independent experiments. PI, propidium iodide. c, d, In vivo shRNA screen to identify potential tumour suppressors on chromosome 17p13 (c) and resulting tumour-free survival curve of mice receiving HPSCs transduced with shRNA pools (d). Hiseq, high-throughput sequencing; TSG, tumour suppressor gene. e, f, Kaplan–Meier tumour-free survival curve of recipient mice transplanted with pre-B cells co-infected with Myc and the indicated shRNAs (e; n = 10), or infected with Myc-linked tandem shRNAs (f; n = 6). a, d–f, Log-rank test. b, Unpaired two-tailed t-test, error bars represent s.d. *P < 0.05; **P < 0.01; ***P < 0.001.
To test whether additional 17p13 genes have a role, we generated a shRNA library targeting the ~100 protein-coding genes in the 11B3 region (exclusive of Trp53 and Eif5a) and screened it for tumour-promoting activity in HSPC transplantation assays (Fig. 4c; 17p13 shRNA library detailed in Supplementary Table 1). Compared with controls, these libraries accelerated lymphomagenesis in recipient mice, implying that one or more shRNAs conferred a selective advantage (Fig. 4d). To identify such shRNAs, PCR products amplified from genomic DNA were subject to deep sequencing and the abundance of each shRNA compared to the initial pool. Forty shRNAs targeting 17 genes were enriched in tumours compared to the injected population (Supplementary Table 1).
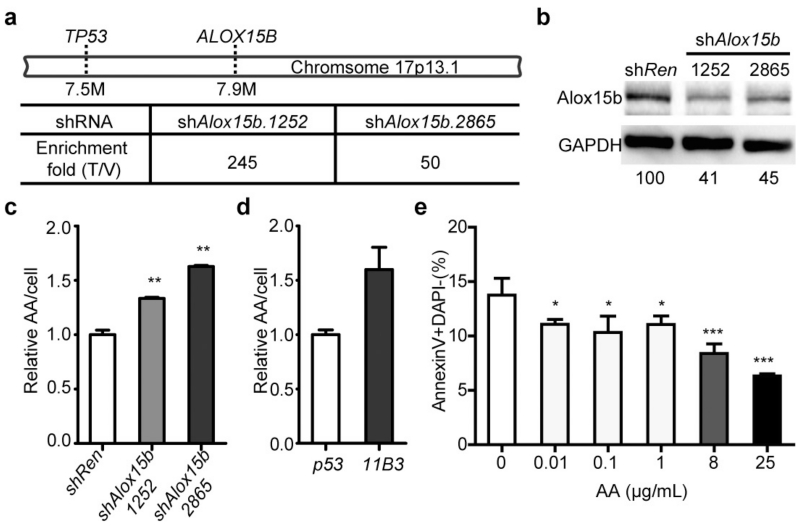
Among the potential candidates, two shRNAs targeting Alox15b (shAlox15b.1252 and shAlox15b.2865), showed a 245- and 50-fold enrichment, respectively, compared with their representation in the initial library (Extended Data Fig. 8a). We thus tested whether suppression of Alox15b alone or in combination with Trp53 could accelerate tumorigenesis as was done for Eif5a. Recipients of HSPCs targeted with Alox15b shRNAs had significantly shorter lymphoma-free survival than shRen controls (Fig. 4e), and those receiving HSPCs harbouring Alox15b-Trp53 tandem shRNA showed a further acceleration over shTrp53-shRen controls (Fig. 4f).
Partial knockdown of Alox15b by shRNAs in NIH3T3 cells resulted in accumulation of its substrate, arachidonic acid (AA) (Extended Data Fig. 8b, c), and higher AA levels were detected in 11B3-deleted lymphoma cells compared with their Trp53-null counterparts (Extended Data Fig. 8d). Interestingly, AA suppresses apoptosis in certain cancer cells25 and, in agreement, exogenous AA inhibited apoptosis of pre-B cells in a dose-dependent manner (Extended Data Fig. 8e). While a complete mechanistic understanding of its effects will require further work, these studies pinpoint Alox15b as an additional tumour suppressor in the 11B3 region. Intriguingly, shRNAs targeting two other Alox family genes adjacent to Alox15b were enriched in our screen (Alox12b, Alox3e), raising the possibility that 17p deletion coordinately attenuates the output of this gene family. Additionally, beyond protein-coding genes, the reduced dosage of non-coding RNAs not tested in our screen might contribute to the activity of 17p deletions26.
11B3 deletion contributes to AML
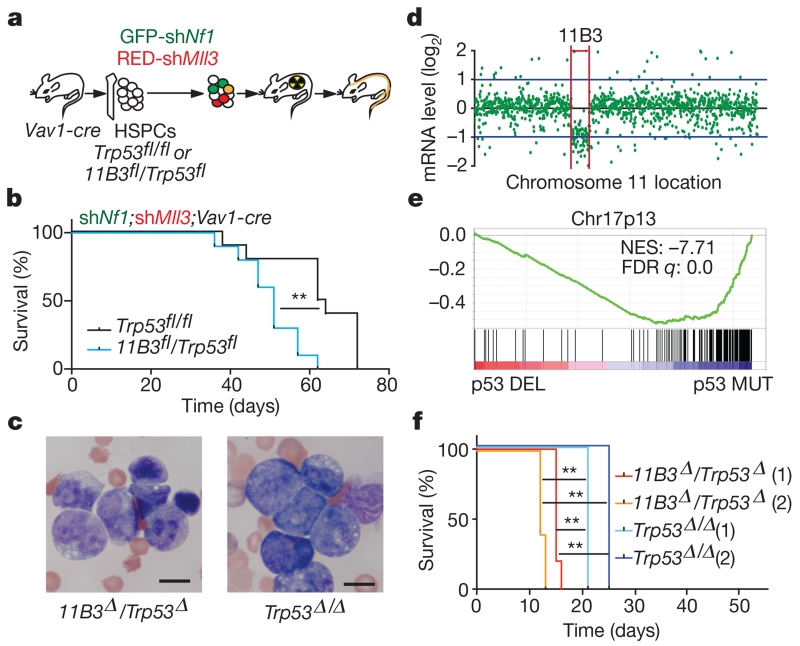
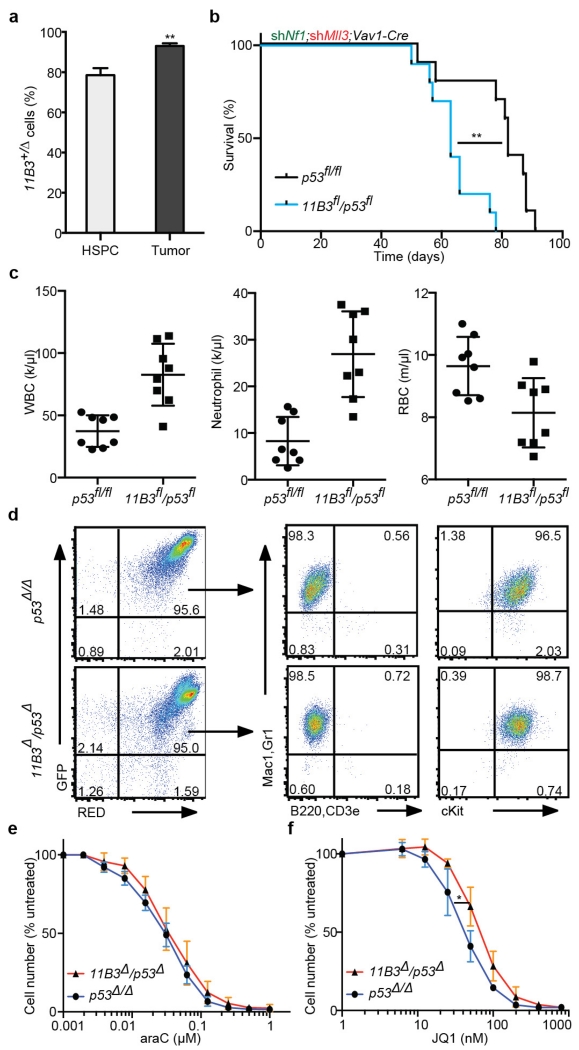
We also asked whether 11B3 loss would enhance tumorigenesis in a genetically and pathologically accurate mouse model of complex-karyotype AML, an invariably lethal leukaemia subtype9,27,28. In this disease, TP53 lesions often co-occur with NF1 loss and deletions on 5q and/or 7q, which can be approximated in mice transplanted with Trp53−/− HSPCs co-expressing shRNAs targeting Nf1 and Mll3 (also known as Kmt2c) (encoded on human 7q36)29. Taking advantage of the flexibility of this model, we compared disease onset and pathology in mice transplanted with shNf1-shMll3-transduced HSPCs derived from either Vav1-cre;Trp53fl/fl or Vav1-cre;11B3fl/Trp53fl mice (Fig. 5a).
Figure 5. Chromosome 11B3 deletion acts beyond Trp53 loss to drive myeloid leukaemia.
a, AML generation by HSPC isolation (Vav1-cre; Trp53fl/fl or Vav1-cre;11B3fl/Trp53fl) co-transduction with a GFP-linked Nf1 shRNA and an mCherry-linked Mll3 shRNA, and transplantation into sublethally irradiated recipients (n = 10 per group). b, Post-transplant, leukaemia-free survival **P < 0.01 (log-rank test). c, Blood smear of moribund mice (b) that is representative of all animals analysed in each genotype (n = 4). d, RNA-seq comparison of chromosome 11 gene-expression levels between the 11B3Δ/Trp53Δ and Trp53Δ/Δ leukaemia generated in b (n = 4). e, Gene set enrichment analysis of genes on chromosome 17p13 in chromosome 17p-deleted human AML compared to those with TP53 mutations but without 17p deletions. FDR, false discovery rate; NES, normalized enrichment score. f, Kaplan–Meier survival curve of secondary transplants from two independent primary leukaemias of each genotype in a, b. n = 5 in each cohort. **P < 0.01 (log-rank test).
These experiments revealed a clear and substantial p53-independent effect of 11B3 deletion on leukaemia development. Indeed, despite the reduced recombination frequency of the 11B3 allele (Extended Data Fig. 9a), recipients of Vav1-cre;11B3fl/Trp53fl HSPCs had a significantly decreased survival compared with recipients of Vav1-cre;Trp53fl/fl HSPCs, and succumbed to an aggressive form of AML (Fig. 5b, c and Extended Data Fig. 9b–d; median overall survival 62 versus 81 days, P = 0.002). RNA-sequencing (RNA-seq) analysis confirmed that 11B3 genes were underexpressed in 11B3-deleted AML cells compared with those in the Trp53-null setting (Fig. 5d), and gene set enrichment analysis revealed that transcriptional profiles of human 17p13 genes are downregulated in human AMLs harbouring 17p13 deletions (Fig. 5e). Moreover, 11B3-deleted AMLs were capable of serial transplantation into secondary recipients, where they promoted disease more rapidly than their Trp53-null counterparts (Fig. 5f). Finally, while the Trp53−/− AMLs are sensitive to the BET-protein inhibitor JQ1 (ref. 29), the corresponding 11B3−/Trp53− AML showed reduced sensitivity (Extended Data Fig. 9e, f). Hence, 11B3 deletions contribute p53-independent phenotypes to AML.
Discussion
This study provides compelling evidence that 17p deletion confers phenotypes beyond those achieved through TP53 loss alone. When considered in light of the established gain-of-function properties of certain missense Trp53 mutations, our results imply that the most frequent somatic event in cancer contributes multiple activities that act independently of Trp53 inactivation to drive tumorigenesis. It therefore seems likely that tumours harbouring TP53 lesions—typically considered a uniform entity—produce distinct phenotypes owing to the nature of the TP53 mutation and the extent of 17p deletion. As such, the models described here will be useful for dissecting precisely how different 17p configurations affect disease onset and ultimately impact therapy response. More broadly, our study provides direct in vivo evidence that segmental deletion events can arise owing to the selective advantage of disrupting multiple genes and provides a blueprint to dissect these common but understudied cancer-promoting lesions.
METHODS
Genomic analyses of human cancers
The data on TP53 mutations (including allele frequency) and CNVs in pan-tumours and AML are derived from The Cancer Genome Atlas (TCGA) data in the cBioPortal for Cancer Genomics (http://www.cbioportal.org/; accessed on 29 October 2014). Only sequenced samples with allele frequency information provided were included in our analysis. Considering potential normal tissue contamination, samples with TP53 mutation allele frequency above 0.6 were considered as a homozygous mutation. The SNP data were visualized in IGV and statistics for AML outcome were analysed in Prism 6. Since cBioPortal only has a few non-Hodgkin lymphoma cases available, we used published data to extract TP53 mutation and deletion information18,30–34. Clinical outcomes were annotated from follow-up data available within the Gene Expression Omnibus GSE34171 series.
CNV analysis was performed using published AML and DLBCL tumour copy number data in Affymetrix SNP Array 6.0 .cel format (http://cancergenome.nih.gov/)18,35–37 according to GISTIC2.0 (ref. 14). Specifically, the following GISTIC parameters and values were used following the latest TCGA Copy Number Portal analysis version (3 November 2014 stddata__2014_10_17; http://www.broadinstitute.org/tcga/gistic/browseGisticByTissue): core GISTIC version 2.0.22; reference genome build hg19; amplification threshold 0.1; deletion threshold 0.1; high-level amplification threshold 1.0; high-level deletion threshold 1.0; broad length cut-off 0.50; peak confidence level 0.95; cap 1.5; gene-GISTIC, true; arm-level peel-off, true; significance threshold 0.25; join segment size 8; X chromosome removed, false; maximum segments per sample 2,000; minimum samples per disease 40.
Conditional 11B3-knockout model
To create a conditional 11B3 chromosome deletion, the MICER strategy was used15. Briefly, MICER clones MHPN91j22 (centromeric to Sco1) and MHPP248j19 (telomeric to Alox12) (Sanger Institute) were introduced into AB2.2 ES cells (129S5 strain, Sanger Institute) by sequential electroporation, followed by G418 (neomycin; 180 μg ml−1) and puromycin (1 μg ml−1) selection, respectively. Successful recombination events were confirmed by Southern blotting using the hybridized probes designated in Supplementary Table 2 as described38. The cis- and trans-localizations of two loxP sites in doubly targeted ES cells were further distinguished by PCR with df-F and df-R, or dp-F and dp-R (Supplementary Table 2), respectively, after Adeno-cre infection and HAT (Gibco) selection. Correct cis-ES clones in which two loxP sites were integrated into the same allele were used to generate chimaera mice by blastocyst injection. The F1 pups were genotyped with 11B3-F and 11B3-R primers (Supplementary Table 2) and those positive backcrossed to C57BL/6 mouse strains for more than 10 generations.
Mice
All of the mouse experiments were approved by the Institutional Animal Care and Use Committee at the Memorial Sloan Kettering Cancer Center. Eμ-Myc, Vav1-cre, Ella-cre, Trp53LSL-R270H/+, Trp53LSL-R72H/+, Trp53+/−, Trp53fl/+ and Rag1−/− mice were ordered from Jackson Laboratories21,39–44 and the Arf+/− mouse strain is a gift from C. Sherr45. Eμ-Myc mice with different Trp53 alterations were monitored weekly with disease state being defined by palpable enlarged solid lymph nodes and/or paralysis. Tumour monitoring was done as blinded experiments. For lymphoma generated by transplantation, 1 million Eμ-Myc HPSCs from embryonic day (E)13.5 fetal liver or autoMACS-purified B220+ B progenitor cells isolated from 6–8-week mouse bone marrow were transduced with retroviruses, followed by tail-vein injection into sublethally irradiated (6 Gy, Cs137) C57BL/6 mice (Taconic; 6–8-week old, female, 5–10 mice per cohort)11,46. All recipient mice were randomly divided into subgroups before transplantation and monitored as described earlier. The generation of AML proceeded as previously reported29. Briefly, retrovirally infected c-Kit+ haematopoietic stem and progenitor cells were transplanted into sublethally irradiated (6 Gy, Cs137) C57BL/6 mice, followed by routine monitoring of peripheral blood cell counts and Giemsa–Wright blood smear staining. For secondary transplantation experiments, 1 million leukaemia cells were transplanted into sublethally irradiated (4.5 Gy) mice. The immunophenotypes of resulting lymphomas and leukaemias were determined by flow cytometry as previously reported using antibodies purchased from eBioscience11,29. Statistical analysis of all survival data was carried out using the log-rank test from Prism 6. No statistical methods were used to predetermine sample size.
Retroviral constructs
MSCV-Myc-IRES-GFP and MLS-based retroviral constructs harbouring a GFP or mCherry fluorescent reporter and targeting Ren, Trp53, Eif5a, Nf1 or Mll3 have all been reported before11,29,47. For the tandem shRNA experiments performed in Fig. 3, mirE-based shRNAs targeting two different genes were cloned into an MLS-based vector in an analogous fashion to what has been previously described48,49. Retrovirus packaging and infection of HSPCs was done as previously reported11,29.
Apoptosis assay
B220+ cells were isolated from the bone marrow of 6-week-old Eμ-Myc mice by autoMACS positive selection with anti-B220 microbeads (Militeny Biotech). After overnight culture, cells were infected with retroviruses carrying the indicated shRNAs. Two days after infection, 0.5 × 106 cells were washed with PBS followed by annexin V buffer (10 mM HEPES, 140 mM NaCl, 25 mM CaCl2, pH 7.4), and incubated at room temperature with Pacific Blue annexin V (BD Biosciences) and propridium iodide (PI; 1 μg ml−1; Sigma-Aldrich) for 15 min and analysed on a LSR II flow cytometer (BD Biosciences). For arachidonic acid treatment, pre-B cells were cultured out from bone marrow cells in pre-B cell medium (RPMI1640, 10% FBS, 1% penicillin/streptomycin, 50 μM β-mercap-toethanol, 3 ng ml−1 IL-7). After 3 days culture, pre-B cells were treated with a series concentration of arachidonic acid (Cayman Chemical) for 20 h, followed by annexin V staining as described earlier.
Immunoblotting
Lymphoma cells isolated from lymph nodes of diseased animals were treated with vehicle (PBS) or 1 μg ml−1 adriamycin for 4 h. Whole cell lysates were extracted in cell lysis buffer (Cell Signaling Technology) supplemented with protease inhibitors (Roche), followed by SDS–PAGE gel electrophoresis and blotting onto PVDF membranes (Millipore). Eμ-Myc;Arf−/− lymphoma cell lines were used as a positive control for p53 induction. The p53 antibody used was obtained from Novocastra (NCL-p53-505) and horseradish peroxidase (HRP)-conjugated β-actin antibody from Sigma (AC-15). Alox15b expressions were examined in NIH3T3 cells, which were infected by shRNAs targeting Ren or Alox15b and then selected by G418. Anti-Alox15b antibody is from Sigma (SAB2100110), and HRP-conjugated GAPDH antibody is from ThermoFisher Scientific (MA5-15738-HRP).
Gene expressing profiling
RNA-seq and data analysis were performed by the Integrated Genomic and Bioinformatics core at the Memorial Sloan Kettering Cancer Center. Briefly, total RNA from 11B3fl/Trp53fl;shNf1;shMll3;Vav1-cre or Trp53fl/fl;shNf1;shMll3;Vav1-cre leukaemia cells (four lines per cohort), isolated from the bone marrow of moribund mice, was isolated by Trizol extraction (Life Technologies). After ribogreen quantification (Life Technologies) and quality control on an Agilent BioAnalyzer, 500 ng of total RNA (RNA integrity number > 8) underwent polyA selection and Truseq library preparation according to instructions provided by Illumina (TruSeq RNA Sample Prep Kit v.2) with 6 cycles of PCR. Samples were barcoded and run on a Hiseq 2500 in a 50 bp/50 bp paired-end run, using the TruSeq SBS Kit v.3 (Illumina). An average of 45 million paired reads were generated per sample. At the most the ribosomal reads represented 0.1% and the percentage of mRNA bases was close to 65% on average. The output from the sequencers (FASTQ files) was mapped to the mouse genome (mm9) using the rnaStar (https://code.google.com/p/rna-star/) aligner, with the two-pass mapping methods. After mapping, the expression counts of each individual gene were computed using HTSeq (http://www-huber.embl.de/users/anders/HTSeq), followed by normalization and differential expression analysis among samples using the R/Bioconductor package DESeq (http://www-huber.embl.de/users/anders/DESeq). Gene set enrichment analysis (GSEA) was performed with Broad’s GSEA algorithm.
Quantitative PCR
A list of all primers used for PCR analysis is given in Supplementary Table 2. For detection and quantification of 11B3 recombination/deletion two methods were employed. In both cases genomic DNA (gDNA) was extracted from lymphoma or leukaemia cells using Puregene DNA purification kit (Qiagen). Initially, semi-quantitative PCR was used to detect the recombined 11B3 allele using primers df-F and df-R, generating a 2.2 kb product (Fig. 2d). The estimated frequency of recombination was determined by dropping gDNA from 11B3+/– into 11B3fl/+ at various ratios. For qPCR of the 11B3 deletion (Fig. 2e), SYBR Green PCR Master Mix (Applied Biosystems) was used and cycling and analysis was carried out on a ViiA 7 (Applied Biosystems). Primers 11B3-Q-F and 11B3-Q-R were used to detect the floxed allele, and to estimate the frequency of 11B3 deletion. Allelic frequency in UPD analysis (Extended Data Fig. 5a) was determined similarly, in this case with serial dilution of wild-type gDNA into DNase-free water to construct a standard curve. Two-tailed t-test is used for statistics analysis by Prism 6. For p21 gene expression examination by RT–qPCR, RNA was isolated with Trizol, cDNA was synthesized with SuperScript III First-Strand Synthesis System (Life Technologies) and qPCR was performed as described earlier with primers p21-Q-F and p21-Q-R.
Trp53 genomic DNA sequencing
Trp53 exons (2–10) were amplified from genomic DNAs of 11B3-deleted lymphomas by PCR (see Supplementary Table 2 for primer sequences) and subjected to Sanger sequencing. Mutations were called only if detected in sequencing reads carried out in the forward and reverse direction.
SNP analysis
SNP analysis of isolated lymphoma (tumour) or tail (normal) genomic DNAs from the same tumour-bearing mouse were carried out by Charles River laboratory. Briefly, a SNP Taqman assay with competing FAM- or VIC-labelled probes was used to detect the relevant C57BL/6 and 129S SNPs (D11Mit4 and D11NDS16) as described previously50.
Copy number profiling
Genomic DNA was extracted from freshly isolated lymphoma cells from one Eμ-Myc;11B3fl/+; Vav-cre mice. One microgram of DNA was sonicated (17 W, 75 s) on an E220 sonicator (Covaris). Samples were subsequently prepared using standard Illumina library preparation (end repair, poly A addition, and adaptor ligation). Libraries were purified using AMPure XP magnetic beads (Beckman Coulter), PCR enriched, and sequenced on an Illumina HiSeq instrument in a multiplexed format. Sequencing reads per sample were mapped using Bowtie with PCR duplicates removed. Approximately 2.5 million uniquely mappable reads were further processed for copy number determination using the ‘varbin’ algorithm51,52 with 5,000 bins, allowing for a median resolution of ~600 kb. GC content normalization, segmentation and copy number estimation was calculated as described53.
shRNA library construction and tumour sequencing
A custom shRNA library was designed to target mouse homologues (six shRNAs for one gene) to all human protein-coding genes on chromosome 17p13.1 from ALOX12 to SCO1, except TP53 and EIF5A. shRNAs were cloned into a retrovirus-based vector MLS by pool-specific PCR as previously described11. Eμ-Myc HSPCs infected with pooled shRNAs were transplanted into sublethally irradiated recipient mice. Resulting tumours were harvested, and used to extract contained shRNAs, followed by HiSeq in HiSeq 2500 (Illumina). Twenty-two oligonucleotides of shRNAs used in this study are listed in Supplementary Table 3.
LC-MS
Total lipids were extracted using Folch’s method54 and analysed by LC-MS as previously described55. Briefly, freshly harvested cells were homogenized by chloroform/methanol (2:1, v-v). After being washed by water, the lipid-containing chloroform phase is evaporated. Dried lipids were dissolved in 100 μl 95% acetonitrile (in H2O), sonicated for 3–5 min, and spiked with 10 μl of 500 ng ml−1 deuterated internal standard solution (IS; arachidonic acid-d8; Cayman Chemical, 390010). Then, 5 μl samples were injected into Acquity ultra performance liquid chromatography (UPLC) system (Waters), equipped with Acquity UPLC BEH C18 column (100 mm × 2.1 mm I.D., 1.7 μm; Waters). Samples were washed through the column with a gradient 0.1% formic acid: acetonitrile mobile elution from 35:65 (v:v) to 5:95 for 10 min. Flow rate was 0.25 ml min−1. Right after HPLC, samples were analysed in a Quattro Premier EX triple quadrupole mass spectrometer (Waters), which has electrospray negative mode and MasslynxV4.1 software. For each run, a standard curve was generated with different concentration of arachidonic acid lipid maps MS standard (Cayman Chemical, 10007268) mixed with IS (50 ng ml−1 final concentration). Arachidonic acid standard m/z is 303.2, and IS is 311.3.
In vitro drug response assays
Three Eμ-Myc lymphoma cell lines generated from Trp53fl/+;Vav1-cre or 11B3fl/+; Vavl-cre tumour-bearing mice were cultured in BCM medium (45% DMEM, 45% IMDM, 10% FBS, 2 mM glutamine, 50 μM β- mercap-toethanol, 1 × penicillin/streptomycin) in 96-well plates. Cells were treated with the indicated concentrations of 4-hydroxycyclophosphamide (Toronto Research Chemicals) or vincristine (Bedford Laboratories) for 3 days. The number of living cells was determined by PI staining and cell counting on a Guava EasyCyte (EMD Millipore). Leukaemia cell lines from Trp53Δ/Δ or 11B3Δ/Trp53Δ;shNf1;shMll3 mice were treated with cytarabine (araC; Bedford Laboratories) or JQ1 (a gift from J. Bradner) in stem cell medium (BCM medium supplemented with 1 ng ml−1 IL-3, 4 ng ml−1 IL-6 and 10 ng ml−1 SCF) and cell viability after 3 days was determined similarly. All cytokines are from Invitrogen.
Extended Data
Extended Data Figure 1. The frequency and prognostic impact of chromosome 17p deletion with the copy number loss identified by GISTIC.
a, The ratio of chromosome-copy-number-altered cases within TP53-mutated cases compared to those within wild-type cases. TP53 mutations were statistically correlated with 17p loss (P < 0.001) but also other copy number events (P < 0.001). b, Peaks of copy number loss identified by the GISTIC algorithm in NHL or AML. x axis, GISTIC q value; y axis, chromosome. q < 0.25 is considered as significant. c, Overall survival of human diffuse large B-cell lymphoma (DLBCL) patients with chromosome 17p deletion is significantly shortened compared to those with no 17p copy number variants, as annotated from the Gene Expression Omnibus GSE34171 series. *P < 0.05 (log-rank test).
Extended Data Figure 2. Generation of a chromosome 11B3 conditional knockout mouse.
a, Top, strategy to introduce 5′ HPRT gene and loxP site telomeric to Sco1 on chromosome 11B3 with MICER clone MHPN91j22. Bottom, Southern blot demonstrating correct targeting of the derived ES cells. st, single-targeted allele; wt, wild-type. Blue arrowheads denote loxP sites. b, Top, strategy to introduce 3′ HPRT gene and loxP site centromeric to Alox12 on chromosome 11B3 with MICER clone MHPP248j19. Bottom, Southern blot demonstrating correct targeting of the derived ES cells. dt, double-targeted allele. c, Top, diagram showing the expected PCR results and drug-resistance phenotypes of doubly targeted ES cells harbouring loxP sites in cis versus in trans. Gr, G418 (neomycin) resistance; Pr, puromycin resistance; Hr, HAT resistance. df, deleted allele; dp, duplicated allele. Green bar indicates the PCR product location and length. Bottom, PCR results show different ES cell clones generated in a and b.
Extended Data Figure 3. 11B3 recombination and lymphomagenesis in Eu-Myc model.
a, The extent of 11B3 deletion in peripheral blood cells, as determined by semi-quantitative PCR, in 11B3fl/+ mice crossed to Cd19-cre (left), Mx1-cre (middle) or Vav1-cre (right). Genomic DNA from 11B3+/Δ ES cells was mixed with 11B3fl/+ cells at different ratios (5% or 20%) as a standard. For Mx1-cre, 6–8-week-old mice were treated with polyinosinic:polycytidylic acid (poly(I:C)) (15 mg kg−1 every other day, 7 times) by intraperitoneal injection. b, Partial 11B3 deletion in Vav1-cre;11B3fl/+ pre-B cells as determined by semi-quantitative PCR, indicating incomplete recombination. c, Complete Trp53fl/+ recombination in Vav1-cre;Trp53fl/+ pre-B cells as determined by PCR. d, Tumour-free survival of Eμ-Myc;Vav1-cre;Trp53fl/+ (n = 9), Eμ-Myc;Vav1-cre;11B3fl/+ (n = 12) and Eμ-Myc;Vav1-cre (n = 6) mice shows that 11B3-deleted tumours have longer tumour latency than Trp53-loss-only controls. ***P < 0.001 (log-rank test).
Extended Data Figure 4. Charaterization of 11B3-deleted lymphoma.
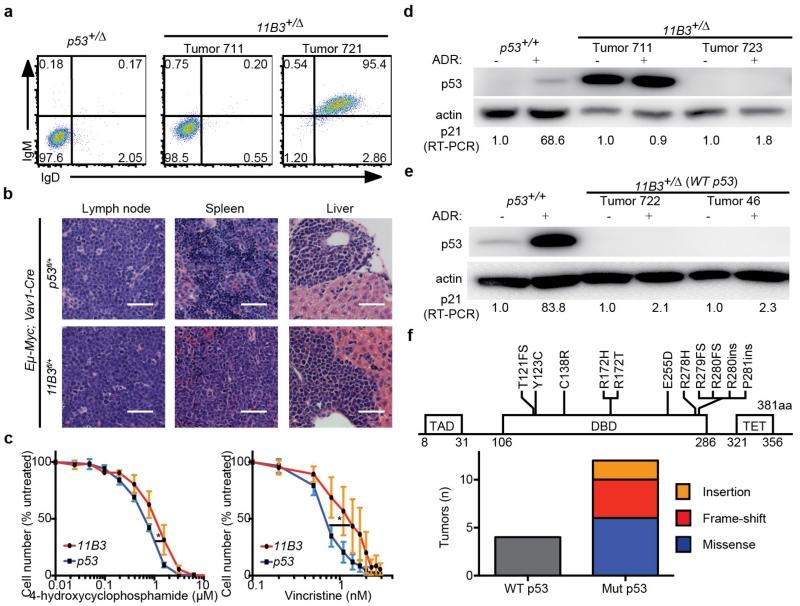
a, Immunophenotypes of B220+ Eμ-Myc lymphomas generated from Vav1-cre;p53fl/+ or Vav1-cre;11B3fl/+. 11B3-deleted lymphomas were either IgM−IgD− or IgM+IgD+ while all the Trp53-null lymphomas were IgM−IgD−. b, Haematoxylin and eosin (H&E) stainings of lymph node, spleen and liver of moribund, lymphoma-bearing mice originating from Eμ-Myc;Vav1-cre;11B3fl/+ or Eμ-Myc;Vav1-cre;Trp53fl/+ genotypes. Scale bar, 50 μm. c, 11B3-deleted lymphoma cells isolated from enlarged lymph nodes are more resistant to chemotherapy drugs 4-hydroxycyclophosphamide (left) and vincristine (right), by in vitro drug sensitivity assay. Shown are representative results of three 11B3Δ/Trp53Δframeshift (11B3) or Trp53Δ/Δ (p53) lymphoma cell lines assayed in quadruplicate. *P < 0.05 (Student’s two-tailed t-test). d, e, No functional p53 was detected in various 11B3-deleted tumours as determined by western blotting of p53 and RT–PCR analysis of p21 induction after 4-h ADR treatment. Eμ-Myc;Arf−/− (Trp53+/+) lymphomas were used as a positive control and p21 levels were normalized to untreated cells. Tumours shown in d were identified as missense (tumour 711) or frameshift mutations (tumour 723), while those in e had no detectable mutation. In total eight tumours were analysed. f, The scope of p53 mutations detected in chromosome 11B3-deleted lymphoma cells as determined by sequencing (n = 12). DBD, DNA-binding domain; FS, frameshift mutation; INS, insertion mutation; MS, missense mutation; TAD, transactivation domain; TET, tetramerization domain.
Extended Data Figure 5. Tumours in mice heterozygous for Trp53 mutations lose heterozygosity by duplicating the mutant Trp53 allele.
a, No chromosome 11B3 deletion was detected in various Trp53 heterozygous mutants. Relative allele copy number of various chromosome 11B3 genes, as determined by qPCR analysis of genomic DNA from Eμ-Myc lymphomas derived from germline mice harbouring the following additional alleles: Vav1-cre;Trp53fl/+(exon 2–10 flanked), Trp53+/− (exon 2–6 deleted), Vav1-cre;Trp53LSL-R270H/+ or Vav1-cre;Trp53LSL-R172H/+. Rpa3 on chromosome 6 was used as an endogenous normalization control. b, SNP analysis of tumour or normal tissue (tail) genomic DNA harvested from mice in a, indicating that uniparental disomy occurred during Trp53 LOH, in that C57BL/6 (B6)-derived wild-type Trp53 allele is replaced by 129-derived Trp53 mutant allele. Note that all Trp53-engineered alleles retain 129-derived SNPs; the germline wild-type Trp53 allele is C57BL/6-derived. c, Cartoon summary of the results from a and b.
Extended Data Figure 6. A Trp53 shRNA induces equivalent knockdown in cells with one or two alleles of the Trp53 gene.
Pre-B cells were isolated from Trp53+/+ or Trp53+/− bone marrow, and then transduced with GFP-linked Trp53 shRNA (shp53). GFP+ cells were sorted out by fluorescence-activated cell sorting, and treated with control wild-type pre-B cells in the present of vehicle or 1 μg ml−1 ADR for 4 h. p53 and p21 levels were detected by western blotting and RT–qPCR, respectively. Shown is the representative result of three independent experiments.
Extended Data Figure 7. In the Eμ-Myc model, Trp53 and Eif5a cooperate in tumorigenesis.
Two-colour assay for the cooperation of Trp53 and Eif5a deficiencies on lymphoma genesis. Eμ-Myc HSPCs retrovirally co-transduced with GFP- (shEif5a or shRen) and mCherry- linked shRNAs (shRen, shp53) were transplanted into sublethally-irradiated syngeneic recipients (n = 5 per group). a, b, The resulting tumours were analysed by flow cytometry (a) and the percentage of GFP+ mCherry+ lymphoma cells in each configuration was quantified (b). Error bars represent s.d. *P < 0.05, ***P < 0.001 (two tailed t-test).
Extended Data Figure 8. Alox15b deficiency promotes tumorigenesis and increases AA levels.
a, Enrichment fold of shAlox15b.1252 and shAlox15b.2865 in resulting tumours (Fig. 3i, j) compared to those in initiating shRNA libraries. b, Knockdown efficiency of shAlox15b.1252 and shAlox15b.2865 compared to control shRen in NIH3T3 cells, as detected by western blotting and quantitated by ImageJ. c, Relative levels of AA per cell are increased with Alox15b shRNAs as measured by liquid chromatography–mass spectrometry (LC-MS). NIH3T3 cells were transduced with shRen or shAlox15b. n = 3. **P < 0.01 (unpaired two tailed t-test). d, Relative levels of AA per cell in 11B3Δ/Trp53Δframeshift (11B3) lymphoma cells are higher than control cells with Trp53Δ/Δ (p53) as measured by LC-MS. n = 2. P = 0.056 (unpaired two tailed t-test). e, In vitro AA treatments reduce apoptosis, as measured by annexin V staining of pre-B cells after 20 h treatment of indicated concentration of AA. n = 4. *P < 0.05; ***P < 0.001 (unpaired two tailed t-test).
Extended Data Figure 9. 11B3 deletion accelerates leukaemogenesis beyond Trp53 loss alone and decreases sensitivity to the BET-protein inhibitor JQ-1.
a, The percentage of 11B3 deletion as determined by qPCR in premalignant c-Kit+ HSPCs (n = 2) and resulting leukaemia cells (tumour; n = 4). **P < 0.01 (unpaired two-tailed t-test). b, Overall survival of recipient mice transplanted with HSPCs from Vav1-cre;11B3fl/Trp53fl or Vav1-cre;Tr53fl/fl co-transduced with both Nf1 and Mll3 shRNAs. **P < 0.01 (log-rank test). c, Complete blood cell counts of recipient mice indicate that there are more total white blood cells (WBCs) and neutrophils, and fewer red blood cells in Vav1-cre;11B3fl/Trp53fl mice compared with the Vav1-cre;Trp53fl/fl control group at 8 weeks post-transplantation. (Note that two mice from each group died before analysis and were not included.) d, Flow cytometry analysis of GFP+mCherry+ leukaemic cells in the bone marrow of moribund mice in a shows that leukaemia cells are myeloid cells in origin and contain both shNf1 and shMll3. e, f, In vitro drug sensitivity of leukaemia cells to araC (e) and the BET-bromodomain inhibitor JQ-1 (f). Shown are representative results of three 11B3Δ/Trp53Δ and two Trp53Δ/Δ leukaemia cell lines assayed in quadruplicate. *P < 0.05 (Student’s two-tailed t-test).
Extended Data Table 1. Number and genotype of progeny resulting from crosses of 11B3ff/+;Ella-cre female mice with Ella-cre male mice.
| Progeny genotype |
11B3+/Δ | 11B3+/+ |
|---|---|---|
| Predicted | 47 | 47 |
| Observed | 0* | 69 |
No 11B3-deleted pup was produced from 11B3fl/+ female breeders with germline Ella-cre, while 25 out of 94 pups were genotyped as 11B3fl/+ without recombination.
P = 2.8 × 10−7 (Chi-squared test).
Supplementary Material
Acknowledgements
We thank J. P. Morris, L. Dow, D. Tschaharganeh, E. Manchado, T Kitzing, E. Loizou and other Lowe laboratory members for their critical discussions and technical help, C. J. Sherr for invaluable advice, L. Dai and M. Tang for liquid chromatography–mass spectrometry support, and Y. Park and S. Kim for their help in constructing the 11B3 mouse model. This work was supported by a programme project and an R01 grant from the National Cancer Institute (S.W.L.), a Center grant for Cancer Target Discovery and Development (S.W.L.), and a Memorial Sloan Kettering Cancer Center Support Grant. Y.L. was supported by an American Association for Cancer Research Millennium Fellowship in Lymphoma Research. C.C. is supported by the Thousand Young Talents Plan and the National Natural Science Foundation of China (81522003, 81570150). S.W.L. is the Geoffrey Beene Chair for Cancer Biology and a Howard Hughes Medical Institute Investigator.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions Y.L. and S.W.L. designed the experiments. Y.L., C.C., S.A., Z.X. and L.C. performed the experiments. Y.L., C.C., Z.X., T.N. and S.W.L. analysed data. Y.L., C.S. and A.A.M. designed the 11B3 model, Y.L., C.S., J.G., B.B., E.R.K., TB., B.S., TN., Q.W., N.S. and R.L.L. contributed to the human cancer genomic analysis. Y.L., C.C., C.D.R. and S.W.L. organized data and wrote the manuscript.
Author Information 17p13 shRNA library specification is provided in Supplementary Information. The raw and analysed RNA sequence data have been deposited in the Gene Expression Omnibus under accession number GSE69654.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl Acad. Sci. USA. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl Acad. Sci. USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wattel E, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]
- 10.El-Ghammaz AM, Abdelwahed E, Mostafa NN, Mansour DA. De novo deletion 17p13.1 as a predictor for disease progression in chronic lymphocytic leukemia. Clin. Exp. Med. 2015;15:493–499. doi: 10.1007/s10238-014-0317-2. [DOI] [PubMed] [Google Scholar]
- 11.Scuoppo C, et al. A tumour suppressor network relying on the polyamine–hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wales MM, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nature Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 13.Ahn YH, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor γ2 expression. Mol. Cell. Biol. 2011;31:4270–4285. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams DJ, et al. Mutagenic insertion and chromosome engineering resource (MICER) Nature Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 17.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monti S, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22:359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Hanel W, et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc. Natl Acad. Sci. USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly GL, et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28:58–70. doi: 10.1101/gad.232009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang DG, et al. Suppression of W256 carcinosarcoma cell apoptosis by arachidonic acid and other polyunsaturated fatty acids. Int. J. Cancer. 1997;72:1078–1087. doi: 10.1002/(sici)1097-0215(19970917)72:6<1078::aid-ijc24>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Balatti V, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slovak ML, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 28.Byrd JC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salaverria I, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J. Clin. Oncol. 2007;25:1216–1222. doi: 10.1200/JCO.2006.08.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio-Moscardo F, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bea S, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mestre-Escorihuela C, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 35.Rucker FG, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 36.Chigrinova E, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122:2673–2682. doi: 10.1182/blood-2013-03-489518. [DOI] [PubMed] [Google Scholar]
- 37.The Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown T. Southern blotting. Curr. Protoc. Prot Sci. 2001;13 doi: 10.1002/0471140864.psa04gs13. 4G:A.4G.1-4G:A.4G.8. [DOI] [PubMed] [Google Scholar]
- 39.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 40.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 41.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 43.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 44.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 45.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 46.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Chicas A, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson EM, et al. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nature Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 51.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baslan T, et al. Genome-wide copy number analysis of single cells. Nature Protocols. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baslan T, Hicks J. Single cell sequencing approaches for complex biological systems. Curr. Opin. Genet. Dev. 2014;26:59–65. doi: 10.1016/j.gde.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 55.Ye X, et al. Development and validation of a UPLC-MS/MS method for quantification of SKLB010, an investigational anti-inflammatory compound, and its application to pharmacokinetic studies in beagle dogs. J. Pharm. Biomed. Anal. 2011;56:366–372. doi: 10.1016/j.jpba.2011.05.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.