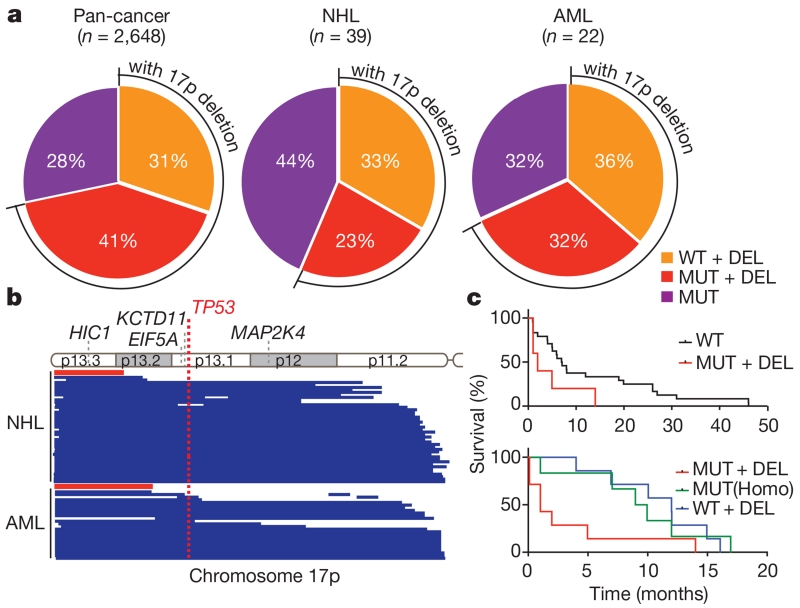
Figure 1. The frequency, scope, and prognostic value of chromosome 17p alterations in human cancers.
a, The nature of TP53-containing chromosome 17p alterations in pan-human cancer (left), non-Hodgkin lymphomas (NHL; middle) and AML (right). DEL, deletion; MUT, mutations; WT, wild-type. b, The extent of chromosome 17p deletions in NHL (44 cases; top) and AML (25 cases; bottom) data sets, irrespective of TP53 deletion, as determined by single nucleotide polymorphism (SNP) array analysis. Each line represents one patient. Red bar indicates the significant copy number loss (q < 0.25) analysed by GISTIC algorithm. c, Overall survival of complex-karyotype AML patients containing both 17p deletion and TP53 mutation as compared to those without any TP53 alteration (top; P = 0.076), those with homozygous (Homo) TP53 point mutations (bottom; P = 0.059), or those with 17p deletion without any detectable TP53 mutation (bottom; P = 0.03). All are log-rank test.