Abstract
A new environmental bacterial strain exhibited strong antimicrobial characteristics against methicillin-resistant Staphylococcus aureus, vancomycin-resistant strains of Enterococcus faecalis and Lactobacillus plantarum, and other Gram-positive bacteria. The producer strain, designated OSY-I1, was determined to be Brevibacillus laterosporus via morphological, biochemical, and genetic analyses. The antimicrobial agent was extracted from cells of OSY-I1 with isopropanol, purified by high-performance liquid chromatography, and structurally analyzed using mass spectrometry (MS) and nuclear magnetic resonance (NMR). The MS and NMR results, taken together, uncovered a linear lipopeptide consisting of 13 amino acids and an N-terminal C6 fatty acid (FA) chain, 2-hydroxy-3-methylpentanoic acid. The lipopeptide (FA-Dhb-Leu-Orn-Ile-Ile-Val-Lys-Val-Val-Lys-Tyr-Leu-valinol, where Dhb is α,β-didehydrobutyric acid and valinol is 2-amino-3-methyl-1-butanol) has a molecular mass of 1,583.0794 Da and contains three modified amino acid residues: α,β-didehydrobutyric acid, ornithine, and valinol. The compound, designated brevibacillin, was determined to be a member of a cationic lipopeptide antibiotic family. In addition to its potency against drug-resistant bacteria, brevibacillin also exhibited low MICs (1 to 8 μg/ml) against selected foodborne pathogenic and spoilage bacteria, such as Listeria monocytogenes, Bacillus cereus, and Alicyclobacillus acidoterrestris. Purified brevibacillin showed no sign of degradation when it was held at 80°C for 60 min, and it retained at least 50% of its antimicrobial activity when it was held for 22 h under acidic or alkaline conditions. On the basis of these findings, brevibacillin is a potent antimicrobial lipopeptide which is potentially useful to combat drug-resistant bacterial pathogens and foodborne pathogenic and spoilage bacteria.
INTRODUCTION
Unregulated access to antibiotics is one of the main reasons for the spread of antibiotic-resistant pathogens and their resistance genes through migration, travel, and trade (1). It was reported that in Europe alone, 25,000 patients die annually because of bacterial infections which cannot be treated with common antibiotics (2). Examples of antibiotic-resistant bacteria are methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus spp., carbapenem-resistant Mycobacterium tuberculosis, and highly virulent multidrug-resistant Clostridium difficile strains (2–5). Therefore, the discovery and development of new antimicrobial agents are of paramount importance. Despite their natural scarcity, new antimicrobial agents can be discovered by subjecting microorganisms that potentially produce such agents to screening and isolation processes (6, 7).
The current study led to the discovery of a new strain of Brevibacillus sp. with promising antimicrobial activity. The genus Brevibacillus was established in 1996 on the basis of a genetic reclassification of strains previously recognized to be Bacillus brevis (8). Many bioactive compounds have been isolated from Brevibacillus spp. since then, including new antibacterial, antifungal, and anti-invertebrate agents (8, 9). For instance, extracellular neutral protease that controls nematode growth was obtained from a Brevibacillus sp. (10); a Brevibacillus laterosporus strain was reported to inhibit the growth of a number of fungi, including Fusarium, Aspergillus, and Alternaria (11); and a B. laterosporus strain, A60, was found to be active against Gram-negative bacteria of the genus Pseudomonas and the Gram-positive bacterium Bacillus subtilis (12). Recently, a lipopeptide antibiotic, designated tauramamide, was discovered in a culture of a B. laterosporus strain (13). A lipopeptide is generally composed of a specific lipophilic moiety attached to a peptide chain. This category includes antimicrobials that are potentially useful as antibacterial, antifungal, and antiviral agents (14–17). A series of lipopeptides synthesized by Makovitzki et al. (18) was found to interact with target pathogens, leading to cell membrane permeation and disintegration. The current study was initiated to unveil new natural antimicrobial agents effective against human pathogens, preferably drug-resistant strains, along with some foodborne pathogenic and spoilage microorganisms.
MATERIALS AND METHODS
Strain screening.
Soil and food samples were collected and screened for antimicrobial-producing bacteria. Most soil samples were collected from the vicinity of The Ohio State University campus, Columbus, OH. Fermented foods were purchased from local grocery stores; these included vegetables (kimchi, pickles, and sauerkraut), meat (salami and sausage), dairy products (yogurt, kefir, and imported cheeses), and soybean products. Soil or food subsamples (10 g each) were homogenized in 0.1% peptone water using a stomacher. Tenfold dilutions were made from the homogenate, and a 100-μl aliquot from each dilution was spread plated onto Trypticase soy agar (TSA; BD Diagnostic Systems, Sparks, MD). The plates were incubated at 37°C for 48 h. Hundreds of colonies were screened for their abilities to produce antimicrobial agents following the method described by Guo et al. (6) with slight modification. Briefly, portions of the colonies on the TSA plates were transferred by the use of sterile toothpicks onto new TSA plates and incubated at 37°C for 48 h. The incubated plates were then overlaid with soft agar medium (TSA with 0.75% agar) that had been preinoculated with Listeria innocua ATCC 33090 or Escherichia coli K-12. After further incubation at 37°C overnight, the overlaid plates were inspected for any zones of inhibition of the indicator strain. Among a few isolates showing antimicrobial activity, one isolate (designated OSY-I1) produced a clear zone of inhibition against L. innocua with a diameter of greater than 3.0 cm. Considering its strong activity against the indicator organism, this isolate was subjected to further analysis.
Cultures and media.
The new isolate, OSY-I1, was propagated on TSA, and a subculture was stocked in the Yousef laboratory's culture collection. For stock preparation, an overnight culture of OSY-I1 in Trypticase soy broth (TSB) was mixed with 80% sterile glycerol in a 1:1 ratio and stored at −80°C. Selected bacterial strains were tested for sensitivity to the newly discovered antimicrobial agent (Table 1). These bacterial strains were obtained from Yousef laboratory stock cultures at The Ohio State University, unless stated otherwise.
TABLE 1.
MICs of brevibacillin from Brevibacillus laterosporus OSY-I1, vancomycin, and nisin against selected bacteria, including antibiotic-resistant strains
| Bacterial straina | MIC (μg/ml) |
||
|---|---|---|---|
| Brevibacillin | Vancomycin | Nisin | |
| Gram-positive bacteria | |||
| Alicyclobacillus acidoterrestrisb | 1.0 | <0.5 | <0.5 |
| A. acidoterrestris ATCC 49025b | 0.5–1.0 | <0.5 | <0.5 |
| Bacillus cereus ATCC 11778 | 2.0–4.0 | 2.0–4.0 | 8.0 |
| B. cereus ATCC 14579 | 1.0 | 1.0 | 2.0 |
| Clostridium difficile A515c | 4.0–8.0 | 2.0 | 4.0–8.0 |
| C. difficile CL148d | 4.0–8.0 | 2.0 | 4.0–8.0 |
| Enterococcus faecalis ATCC 29212e | 2.0 | 2.0 | >16.0 |
| E. faecalis ATCC 51299, vancomycin resistante | 4.0–8.0 | >16.0 | >16.0 |
| Lactobacillus plantarum ATCC 8014f | 1.0 | >16.0 | <0.5 |
| Lactococcus lactis ATCC 11454g | 2.0 | <0.5 | >16.0 |
| Listeria innocua ATCC 33090h | 1.0–2.0 | 1.0 | 2.0–4.0 |
| Listeria monocytogenes OSY-8578h | 1.0–2.0 | 1.0 | <0.5 |
| L. monocytogenes Scott Ah | 1.0 | 1.0 | 4.0 |
| Staphylococcus aureus ATCC 6538 | 1.0–2.0 | 1.0 | 1.0 |
| S. aureus, methicillin resistant (MRSA)i | 1.0 | 1.0–2.0 | 2.0–4.0 |
| Gram-negative bacteria | |||
| Escherichia coli K-12 | >32 | >16 | >16 |
| E. coli O157:H7 EDL 933 | 32 | >16 | >16 |
| Pseudomonas aeruginosa ATCC 27853e | >32 | >16 | >16 |
| Salmonella enterica serovar Typhimurium DT 109 | >32 | >16 | >16 |
Unless stated otherwise, the strains were incubated in cation-adjusted Mueller-Hinton II broth (MHB; Becton, Dickinson & Co., Sparks, MD) at 37°C for 20 h.
A highly spoilage-inducing isolate provided by a food processor; the strain was cultured in yeast starch-glucose broth at 37°C for 48 h.
Provided by W. A. Gebreyes, Department of Veterinary Preventive Medicine, The Ohio State University; the strain was cultured in brain heart infusion supplemented with 5% yeast extract and incubated anaerobically at 37°C for 24 h.
Provided by J. T. Lejeune, College of Veterinary Medicine, The Ohio State University; the strain was cultured under the same conditions used with the A515 strain (see footnote c).
Incubated in TSB at 37°C for 20 h.
Incubated in de Man, Rogosa, and Sharpe broth at 30°C for 20 h. ATCC 8014 is an antibiotic-resistant strain.
Incubated in de Man, Rogosa, and Sharpe broth at 30°C for 20 h.
Incubated in TSB at 37°C for 20 h.
The MIC of oxacillin for the isolate is more than 32 μg/ml.
Strain identification.
The morphology of OSY-I1 was examined by Gram staining of cells and malachite green staining of spores. The biochemical characteristics of the isolate were examined using a commercial biochemical testing kit (API 50 CH test strips; bioMérieux, Inc., Durham, NC) following the kit manufacturer's instructions. Briefly, overnight colonies of OSY-I1 were transferred into the medium provided with the kit (API 50 CHB/E) to prepare a cell suspension with a turbidity equivalent to an optical density at 600 nm of 0.242 (McFarland standard no. 2). The prepared suspension was inoculated into the wells of the biochemical strips; this was followed by incubation at 30 or 37°C, and readings were taken after 24 and 48 h. Biochemical reactions were recorded, and the isolate's identity was determined by comparing the results to the data in the database provided by the kit manufacturer.
For genetic identification, the isolate (OSY-I1) was analyzed using the 16S rRNA gene sequencing technique (19). The genomic DNA of OSY-I1 was extracted using a commercial kit (DNeasy blood and tissue kit; Qiagen, Valencia, CA). Universal primers (16S forward primer 5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′ and 16S reverse primer 5′-CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC-3′) were used to amplify the 16S rRNA gene by the use of Taq DNA polymerase (Taq PCR core kit; Qiagen, Valencia, CA). The PCR was conducted in a thermocycler under the following conditions: an initial incubation at 94°C for 3 min and 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 2 min. The final extension step was conducted at 72°C for 10 min. The amplicon of the 16S rRNA genes was purified using a commercial DNA extraction kit (QIAquick gel extraction kit; Qiagen, Valencia, CA). The purified PCR product was then sequenced from both the 5′ and 3′ ends with an automated DNA analyzer (Applied Biosystems, Foster City, CA). The resultant 16S rRNA sequence (1,450 bp) was compared to known sequences in a national database (Ribosomal Database Project, release 11; http://rdp.cme.msu.edu) using the Seqmatch algorithm.
Isolation and purification of the antimicrobial agent from OSY-I1.
An overnight liquid culture of OSY-I1 in TSB was aliquoted (aliquots of 100 μl each) and spread plated onto TSA plates. After incubation at 37°C for 72 h, bacterial cells were scraped into centrifuge tubes by use of a microscopic slide and mixed with isopropanol in a 1:4 (wt/vol) ratio. The contents of the centrifuge tubes were then agitated at 200 rpm for 4 h, followed by centrifugation at 7,710 × g for 15 min. The supernatant was held in a chemical hood at 25°C for 48 h to allow solvent evaporation. The dry residues were then suspended in water to prepare the crude extract, and its antimicrobial activity against L. innocua ATCC 33090 was examined. The crude extract was purified with a high-performance liquid chromatography (HPLC) system (Hewlett Packard 1050; Agilent Technologies, Palo Alto, CA) equipped with an analytical reverse-phase column (particle size, 5 μm; 250 by 4.6 mm; Biobasic C18; Thermo Electron Corp., Bellefonte, PA). The purification process was achieved by elution with a linear gradient consisting of two mobile phases: HPLC-grade water with 0.1% trifluoroacetic acid (TFA) and HPLC-grade acetonitrile with 0.1% TFA. In each run, 40 μl crude extract was injected and separated by use of a linear gradient of from 0 to 66.6% acetonitrile for 20 min at a flow rate of 1 ml/min, and the effluent was monitored by a UV-visible detector set at 220 nm. Fractions were collected at a rate of one fraction per minute, and fractions from multiple runs were combined and placed into glass beakers, followed by air drying in a chemical hood. Each dry fraction was dissolved in 50% acetonitrile, and the antimicrobial activity was assayed by the spot-on-lawn method (7). Purified antimicrobial agent (obtained from fractions with antimicrobial activity) from multiple runs was pooled and reinjected into the HPLC, using the same conditions described above, to check the purity. The fraction with the strongest antimicrobial activity and the highest purity was then subjected to analysis by mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy.
MALDI-TOF MS analysis.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS analysis was conducted on a Bruker UltrafleXtreme MALDI-TOF/TOF MS (Bruker Daltonics Inc., Billerica, MA) operated in reflection positive-ion mode and accelerated at a voltage of 28 kV. The HPLC-purified sample was mixed with a matrix in a ratio of 1:5 (vol/vol). The matrix used was α-cyano-4-hydroxycinnamic acid (Bruker Daltonics Inc.), which was dissolved in 50% acetonitrile with 0.1% TFA in water. A nitrogen laser was set at a threshold level that minimized fragmentation but that was adequate to generate signals.
LC-MS/MS.
The sequence of the target antimicrobial agent was determined using capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS). A Thermo Fisher LTQ Orbitrap XL mass spectrometer equipped with a microspray source (Michrom BioResources Inc., Auburn, CA) was operated in positive-ion mode. A capillary column (Magic C18AQ [Bruker Daltonics]; 0.2 by 150 mm; particle size, 3 μm; 200 Å) was used for sample separation on an HPLC (UltiMate 3000; Thermo Scientific). The sample was dissolved in acetonitrile-H2O (50:50, vol/vol) and was loaded onto the column, bypassing the desalting trap. Mobile phase A was 50 mM acetic acid in water, and mobile phase B was acetonitrile. The flow rate was set at 2 μl/min. Typically, mobile phase B was increased from 2% to 50% in 30 min and from 50% to 90% in 5 min and then kept at 90% for another 2 min before being quickly brought back to 2% in 1 min. The column was equilibrated at 2% mobile phase B (and 98% mobile phase A) for 20 min before the injection of a subsequent sample. A spray voltage of 2.2 kV and a capillary temperature of 175°C were used to acquire the MS/MS data. The scan sequence of the mass spectrometer was based on the preview mode data-dependent TopTen method, as follows. For the analysis, the mass spectrometer was programmed for a full scan, recorded between m/z 350 and 2,000, and an MS/MS scan to generate product ion spectra to determine the amino acid sequence from consecutive scans of the 10 most abundant peaks in the spectrum. To achieve high mass accuracy during mass determination by MS, the full scan was performed in the Fourier transform (FT) mode, and the resolution was set at 60,000. The automatic gain control (AGC) target ion number for the full scan in the FT mode was set at 1 × 106 ions, the maximum ion injection time was set at 1,000 ms, and the microscan number was set at 1. MS2 was performed using the ion trap mode to ensure the highest signal intensity of the MSn spectra. The AGC target ion number for the ion trap MSn scan was set at 10,000, the maximum ion injection time was set at 50 ms, and the microscan number was set at 1. The collision-induced dissociation fragmentation energy was set to 35%.
NMR analysis.
For NMR analysis, a standard protocol was followed to determine the constituents and then their sequential arrangement (20), as has been performed previously (6, 7). The HPLC-purified dry sample (approximately 1 mg) was dissolved in 99.9% perdeuterated dimethyl sulfoxide (DMSO-d6) (Cambridge Isotope Lab, Tewksbury, MA). Experiments were conducted at 298 K on Bruker Avance-III HD-600 and -700 spectrometers (Bruker, Karlsruhe, Germany). Both spectrometers were equipped with a 5-mm (1H, 13C, 15N) triple-resonance cryoprobe (TXI and TXO, respectively) and a z-axis gradient. The following data sets were recorded: one-dimensional (1D) 1H NMR, 1D 13C NMR, 1D 13C distortionless enhancement by polarization transfer (DEPT)-90 and DEPT-135, two-dimensional (2D) 1H-homonuclear double-quantum filtered correlation spectroscopy (DQF-COSY), total correlation spectroscopy (TOCSY; 60-ms DIPSI2 [decoupling in the presence of scalar interactions-2] mixing time), nuclear Overhauser effect spectroscopy (NOESY; 200- and 400-ms mixing times), 2D heteronuclear 1H-13C heteronuclear single quantum coherence (spectroscopy) (HSQC), multiplicity-edited HSQC, heteronuclear multiple-bond correlations (HMBC), HSQC-NOESY (350-ms mixing time), HSQC-TOCSY (60-ms DIPSI2), heteronuclear multiple-quantum correlation (HMQC)-COSY (21), and 2D heteronuclear 1H-15N HSQC. All data sets were recorded using standard Bruker pulse sequences and the natural abundance. Data were processed using NMRPipe software (22) and were typically zero filled prior to the application of Window functions, followed by Fourier transform. Chemical shifts were referenced directly (1H and 13C) or indirectly (15N) to the internal residual solvent peak. The spectra for the assignments were visualized by the use of the NMRView program (23). All of the NMR spectral figures were prepared using TopSpin (version 3.2) software (Bruker, Karlsruhe, Germany).
Sensitivity to heat and pH.
Pure brevibacillin, prepared as described previously, was subjected to heat and acid treatment. For the thermal stability test, brevibacillin was diluted with DMSO and heated in a water bath at 80°C, simulating a pasteurization temperature. Samples of the antimicrobial agent solution were taken at designated intervals during heating, and changes in antimicrobial activity against MRSA as an indicator bacterium were monitored using a 2-fold dilution scheme in a 96-well microtiter plate. For the pH stability test, pure brevibacillin was dissolved in citrate buffer (0.1 M, pH 3.0), phosphate buffer (0.1 M, pH 7.0), and Tris buffer (0.1 M, pH 9.0) to achieve a concentration of 16 μg/ml. The pH range (pH 3 to 9) was chosen to encompass the pH values of most food products. The pH-adjusted buffer containing brevibacillin was then incubated at 25°C for 22 h. Treated brevibacillin buffers were then neutralized to pH 7.0 and tested for changes in antimicrobial activity using the same method used to test for the effects of heat treatment. Both the thermal and pH treatment experiments were done in triplicate.
In vitro antimicrobial activity determination.
The MICs of the HPLC-purified antimicrobial agent prepared from the OSY-I1 isolate for selected Gram-positive and Gram-negative bacteria (Table 1) were determined by the broth microdilution method following the protocol of the Clinical and Laboratory Standards Institute (24). Specifically, purified antimicrobial agent was collected from the HPLC, weighed, and dissolved in DMSO at a concentration of 3,200 μg/ml (stock solution). Working solutions were prepared from the stock solution through 2-fold serial dilution using DMSO as a diluent. The MIC study was conducted in 96-well microtiter plates. Each well contained 178 μl of medium, 20 μl of indicator bacteria (∼2.0 × 104 CFU/well), and 2 μl of diluted antimicrobial agent. Therefore, the total volume was 200 μl/well, and the DMSO concentration was 1%. In addition, vancomycin (Sigma, St. Louis, MO) and nisin (Aplin and Barrett Ltd., Trowbridge, United Kingdom), each of which was dissolved individually in DMSO, were used as positive controls, and 1% DMSO was used as a negative control. In order to test the MICs of all three antimicrobial agents under the same conditions, DMSO was used as the solvent to avoid variations in the solubilities of these agents in water. The MIC represented the lowest concentration (in micrograms per milliliter) of a certain antimicrobial agent that led to no visible growth of the indicator bacteria after incubation at 35°C for 20 h (24). The MIC experiments were done in triplicate.
Nucleotide sequence accession number.
The DNA sequence of the 16S rRNA gene of OSY-I1 (1,450 bp) has been deposited in the GenBank database under accession no. KR919625.
RESULTS
Isolation and identification of an antimicrobial-producing strain.
A total of approximately 2,500 isolates from tested samples were screened for their antimicrobial activity against L. innocua ATCC 33090 and E. coli K-12. A soil isolate showed strong activity against L. innocua, with the area of the diameter of inhibition being greater than 3.0 cm. The isolate was given the strain designation OSY-I1. The new isolate is a Gram-positive, spore-forming bacterium. Biochemical test results showed that OSY-I1 is positive for catalase and oxidase; fermentation of glucose, fructose, mannose, and mannitol; and esculin hydrolysis but that it is negative for sorbitol and lactose fermentation. The results of biochemical tests (API strips) indicated 99.9% similarity between OSY-I1 and Brevibacillus laterosporus. Genetic analysis by sequencing of the 16S rRNA gene (1,450 bp) also showed that isolate OSY-I1 shares a high degree of identity (97.7%) with B. laterosporus. Therefore, the new isolate was identified as B. laterosporus on the basis of both biochemical and genetic tests.
Extraction, isolation, and purification of the antimicrobial agent produced by OSY-I1.
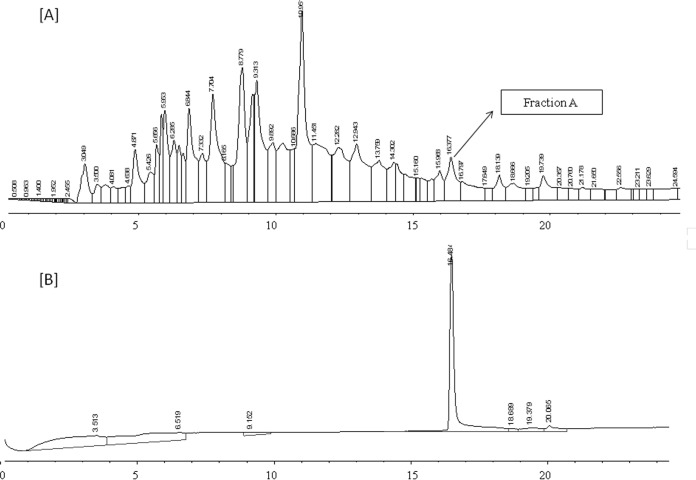
Analysis of the crude extract by HPLC (Fig. 1A) revealed that fraction A (at a retention time of 16.4 min) had activity against L. innocua ATCC 33090. The purity of the active fraction was tested by reinjection onto the HPLC, and a single peak was observed (Fig. 1B), suggesting that the compound representing the fraction was purified to homogeneity. The pure compound was designated brevibacillin.
FIG 1.
HPLC chromatograms at different stages of purification of the antimicrobial agent from Brevibacillus laterosporus OSY-I1. The antimicrobial activity of the different fractions was tested against Listeria innocua ATCC 33090. (A) Chromatogram of the crude extract; (B) chromatogram of purified antimicrobial agent after reinjection of fraction A of the crude extract.
MIC determination and stability test.
Purified brevibacillin was used for MIC determination. Targeted microorganisms included pathogenic and nonpathogenic bacteria. The antimicrobial agent was active only against the Gram-positive bacteria tested, including methicillin-resistant Staphylococcus aureus, vancomycin-resistant Lactobacillus plantarum, and vancomycin-resistant Enterococcus faecalis (Table 1). This antimicrobial agent had antimicrobial activity more potent than that of nisin for most of the tested microorganisms, and its potency was often comparable to that of vancomycin (Table 1). In the heat stability test, brevibacillin showed no sign of degradation when it was heated at 80°C for up to 60 min (data not shown). In the pH stability test, 50% and 62.5% of the antimicrobial activity remained in pH 3.0 and 9.0 buffer, respectively, compared to the amount of antimicrobial activity of brevibacillin that remained when it was incubated at pH 7.0 for 22 h.
MALDI-TOF MS and LC-MS/MS analyses.
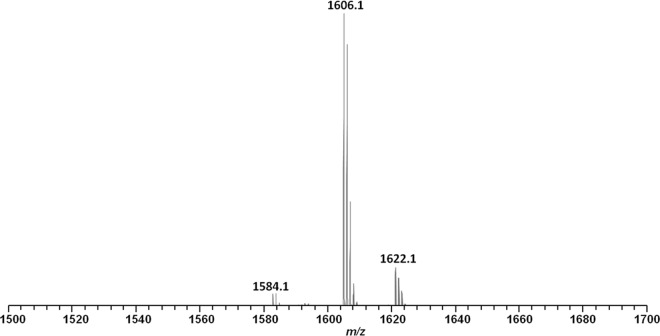
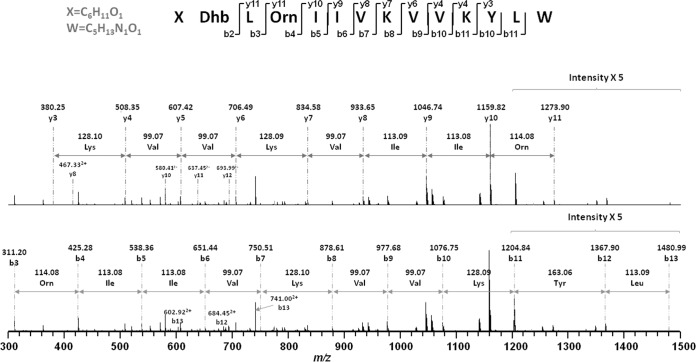
The purified active fraction was analyzed using MALDI-TOF MS to determine the molecular mass of the antimicrobial compound. As shown in Fig. 2, three peaks were measured for the fraction at m/z values of 1,584.1, 1,606.1, and 1,622.1, corresponding to the singly protonated antimicrobial agent [M + H]+, its sodium-cationized ion [M + Na]+, and its potassium-cationized ion [M + K]+. The active fraction was also subjected to LC-MS/MS analysis to deduce an accurate molecular mass and provide a preliminary peptide sequence for the antimicrobial agent. As shown in Fig. 3, the b and y ions were generated by MS/MS analysis, and the preliminary sequence was proposed to be X-Dhb-Leu-Orn-Ile-Ile-Val-Lys-Val-Val-Lys-Tyr-Leu-W (where Dhb is α,β-didehydrobutyric acid, X is C6H11O, and W is C5H13NO) from the N terminus to the C terminus. The observed m/z (z = 2) was equal to 792.54772+; the theoretical m/z was equal to 792.54682+, which indicated a mass error of 1.13 ppm; and the observed molecular mass was 1,583.0794 Da.
FIG 2.
MALDI-TOF MS analysis of the antimicrobial agent isolated from Brevibacillus laterosporus OSY-I1. The ions at m/z (z = 1) 1,584.1, 1,606.1, and 1,622.1 represent the singly protonated [M + H]+, sodium-cationized [M + Na]+, and potassium cationized [M + K]+ adducts, respectively.
FIG 3.
Fragmentation of brevibacillin by tandem MS to generate b and y ions.
Peptide sequence determination and terminal structure elucidation by NMR.
In this study, DMSO-d6 was used to dissolve the compound, as it offers higher solubility than water or CDCl3, two other commonly used NMR solvents. This choice also facilitates the observation of labile protons and their associated cross peaks in 2D NMR. The spectral assignments were performed by simultaneous analysis of multiple data sets through cross-checking in an effort to rule out alternative explanations. The representative and critical observations, particularly those regarding the elucidation of N- and C-terminal structures, are summarized in Fig. 4 to 6.
FIG 4.
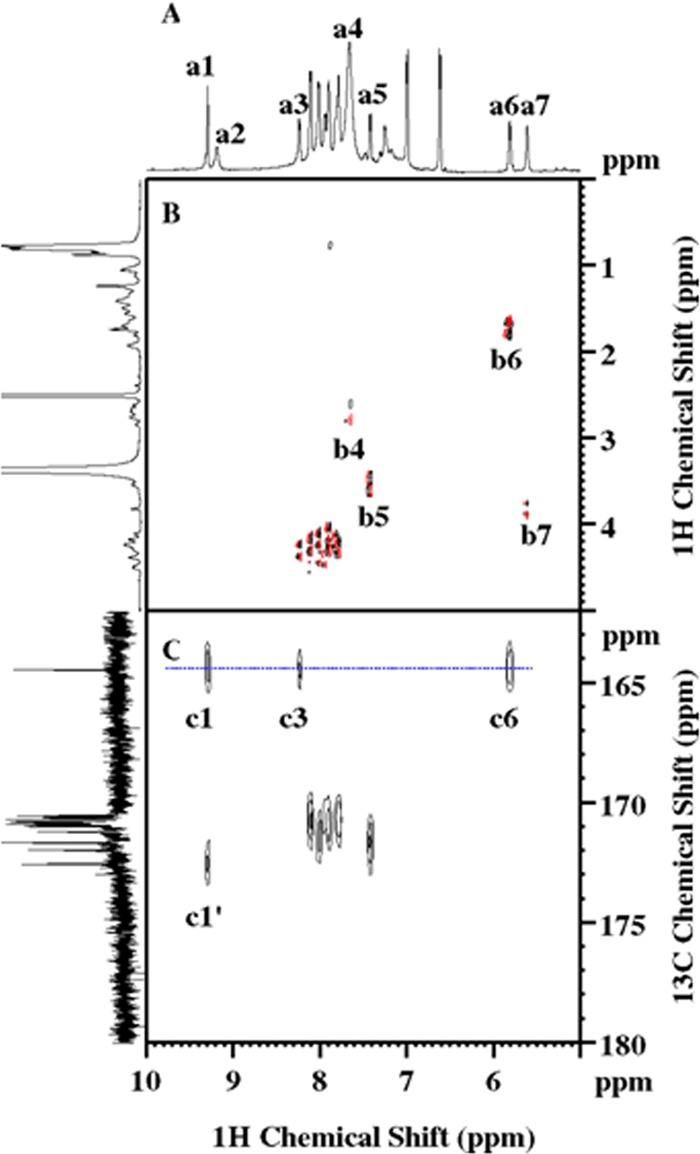
NMR spectra signifying the existence of a polypeptide and the acylated N-terminal cap. (A) 1D 1H NMR showing the spectral region from 5 to 10 ppm. The peaks of a1 to a7 are attributed to Dhb-1 HN, Tyr-11 Hη, Leu-2 HN, NH3+ (Orn-3 Hε/Lys-7 Hζ/Lys-10 Hζ), valinol-13 HN, Dhb-1 Hβ, and FA OHβ1, respectively. (B) Partial 2D 1H DQF-COSY spectrum showing the 3JHH correlations, with the following peaks being labeled: Orn Hδ-Hε or Lys Hε-Hζ (b4), valinol-13 HN-Hα (b5), Dhb-1 Hβ-Hγ1 (b6), and FA Hα-Hβ1 (b7). Also, Dhb-1 HN is unique and does not show a COSY peak. (C) 2D 1H-13C HMBC showing the 1H-13C multiple-bond correlations, including Dhb-1 C′ to Dhb-1 HN (c1), Leu-2 HN (c3), and Dhb-1 Hβ (c6) and FA C′ to Dhb-1 HN (c1′). The projection on the left of panel C is the carbonyl region of the 1D 13C NMR spectrum, recorded on a Bruker Avance-III HD-700 spectrometer equipped with a TXO cryoprobe.
FIG 5.
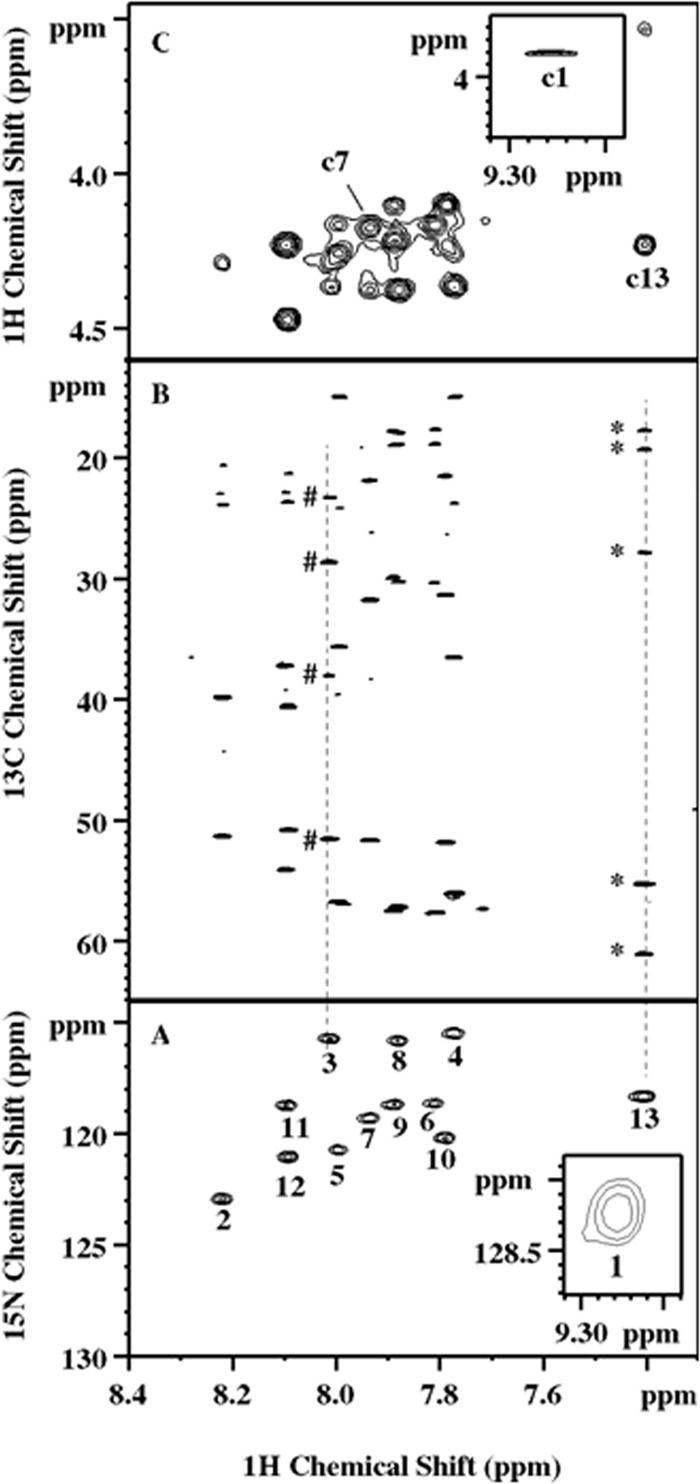
NMR spectra exemplifying the residue and sequential assignment of the lipopeptide. (A) 2D 1H-15N HSQC spectrum. (Inset) The 13 amide cross-peaks. The numbers 1 to 13 represent the order of the components in the actual final sequence. (B) 2D 1H-13C HSQC-TOCSY spectrum illustrating the identification of the side chain spin network associated with amide protons. Peaks marked by pound signs and asterisks are scalar correlated to Orn-3 HN and valinol-13 HN, respectively. Their multiplicities were determined by 2D multiplicity-edited 1H-13C HSQC as CH2, CH2, CH2, and CH for Orn-3 and CH3, CH3, CH, CH, and CH2 for valinol-13 from top to bottom, respectively, for both. (C) Partial 2D 1H-homonuclear NOESY. (Inset) Example NOE assignments of Dhb-1 HN to FA Hα (c1), Lys-7 HN to Val-6 Hα (c7), and valinol-13 HN to Leu12 Hα (c13). Ambiguities arising from a signal overlapping, for example, the region close to c7, were resolved by 2D 1H-13C HSQC-NOESY.
FIG 6.
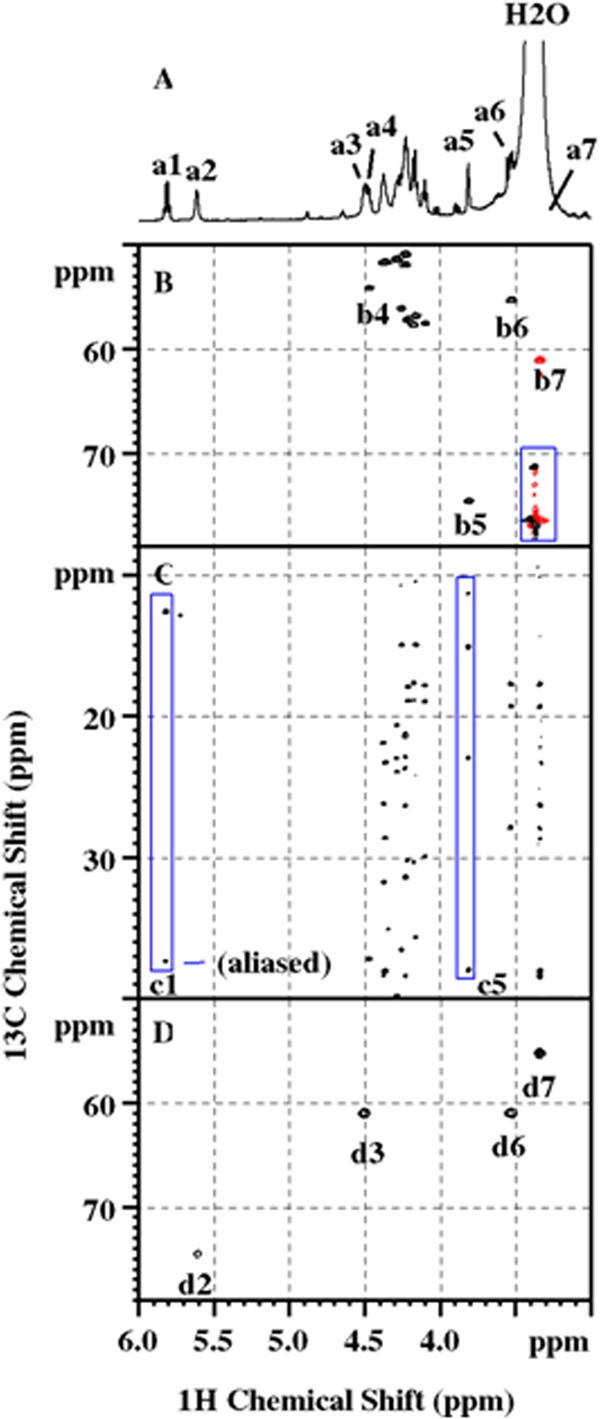
NMR data showing the critical evidence for the assignment of FA, Dhb-1, and valinol-13. (A) 1D 1H NMR showing the spectral region from 3.00 to 6.00 ppm with the following peaks marked from a1 to a7: Dhb-1 Hβ, FA OHβ1, valinol-13 OHγ1, Tyr-11 Hα, FA Hα, valinol-13 Hα, and valinol-13 Hβ1, respectively. (B) 2D multiplicity-edited 1H-13C HSQC showing the 1H-13C one-bond correlations, with the following peaks being labeled: Tyr-11 CHα (b4), FA CHα (b5), valinol-13 CHα (b6), and valinol-13 CH2β1 (b7). In this experiment, the cross-peaks of CH2 (red) and those of CH and CH3 (black) are of opposite signs. The CH and CH3 moieties can be further distinguished by 1D 13C DEPT experiments. The noise marked inside the rectangular box arose from the strong residual H2O resonance. (C) Partial 2D 1H-13C HSQC-TOCSY highlighting the spin networks associated with Dhb-1 Hβ (box c1) and FA Hα (box c5). The former consists of only 1 CH moiety (aliased in this spectrum) and 1 CH3 moiety, while the latter has five protonated carbons, including FA CHα, which is beyond the range of the spectrum shown in the figure. (D) 2D HMQC-COSY or H2BC showing the exclusive 1JCH/3JHH correlations: FA Hβ1 to 13Cα (d2), valinol-13 Hγ1 to Cβ1 (d3), valinol-13 Hα to Cβ1 (d6), and valinol-13 Hβ1 to Cα (d7).
A quick 1D 1H NMR revealed resonances between 6.5 and 9.5 ppm (Fig. 4A), the typical amide and aromatic amino acid spectral region. Subsequently, 2D 1H-15N HSQC confirmed a total of 13 amide cross peaks (Fig. 5A), signifying the existence of a peptidyl fragment. Since an equal number of carbonyl resonances was observed in 1D 13C NMR (Fig. 4C), it was plausible to assume that the polypeptide likely consists of 13 amino acids. The majority of the amides, i.e., peaks 2 through 12 in Fig. 5A, have chemical shifts of between 7.77 and 8.22 ppm in the 1H dimension. The lack of dispersion, together with chemical shifts close to random coil values, is indicative of a flexible conformation that is likely linear and unstructured in nature. Signal overlapping also occurs in the side chains and is presumably attributed to the same cause, rendering the complete and accurate assignment somewhat challenging for this molecular size. Nevertheless, the problem was alleviated by collecting high-resolution data sets as well as by exploiting heteronuclear NMR techniques, as further elaborated below.
The amide protons described above were subsequently correlated with the scalar-coupled aliphatic side chains via 2D COSY and TOCSY experiments. For example, the HN of residue 3 displays through-bond correlations in 2D 1H-13C HSQC-TOCSY (Fig. 5B) to four protonated carbons, including one CH moiety and three CH2 moieties, as determined by 2D multiplicity-edited 1H-13C HSQC. Their chemical shifts and the scalar connectivity revealed by COSY further suggest an aliphatic side chain resembling the one of an arginine residue. However, two lines of NMR evidence led to the assignment of an ornithine, in agreement with the MS observation. First, no Hε-Νε cross-peak was detected in the 2D 1H-15N HSQC described above. Second, the broad proton resonance at 7.67 ppm (Fig. 4A, a4), initially assigned to lysine Nε and H3+ groups (Fig. 4B, b4), appears from peak integration to contain close to 9 protons, indicating three lysine-like residues in this compound.
Ten other residues in the center of 2D 1H-15N HSQC were readily determined in a similar fashion, including 1 Tyr, 2 Ile, 2 Lys, 2 Leu, and 3 Val residues. The two well-dispersed peaks 1 and 13 (Fig. 5A), however, appeared to be unusual and warranted in-depth investigation. Nevertheless, the peptide sequence was subsequently deduced by analyzing the 2D 1H NOESY and 2D 1H-13C HSQC-NOESY spectra. This is exemplified by the assignment of an Hα(i)-HN(i + 1) sequential nuclear Overhauser effect (NOE) c7 between residues 6 and 7 and c13 between residues 12 and 13 (Fig. 5C). It was concluded that the sequence determined at this stage is in full agreement with the MS results presented above, and residues 1 and 13, which were not determined, are positioned at the N and C termini, respectively.
The HN of residue 13 shows scalar correlations to five protonated carbons (Fig. 5B), four of which (two CH and two CH3 moieties) form a spin network reminiscent of a valine side chain on the basis of 3JHH (3-bond proton-proton J coupling constant) correlations and the chemical shifts. The remaining methylene moiety, on the other hand, demonstrates 13C/1H chemical shifts of 61.04/3.340 ppm (Fig. 6B, b7), characteristic of a hydroxymethyl group. Its degenerate proton resonance (Fig. 6A, a7), however, is masked by the intense and broad residual water signal, hampering observation of the 3JHH correlation peaks to the vicinal protons that would otherwise provide direct evidence that would allow the covalent bonding to be unraveled. This problem was overcome by 2D 1H-13C HMQC-COSY, or the so-called H2BC approach (21), which employs the heteronuclear technique to correlate 1H and 13C spins separated by two covalent bonds via 3JHH and 1JCH while significantly suppressing unwanted signals. Figure 6D shows the reciprocal H2BC correlations, d6 and d7, between this CH2 moiety and one of the aforementioned methanetriyl groups. The latter, with 13C/1H chemical shifts of 55.24/3.530 ppm (Fig. 6B, b6), is analogous to CαH in a valine residue that is 3JHH correlated to the HN proton (Fig. 4B, b5). Moreover, a hydroxyl proton at 4.503 ppm (Fig. 6A, a3), barely resolved from Tyr-11 Hα (Fig. 6A, a4, and Fig. 6B, b4), was uncovered in the same H2BC spectrum to engage in a 1JCH/3JHH correlation with the methylene carbon described above (Fig. 6D, d3). All of these observations taken together unequivocally identified a valinol (2-amino-3-methyl-1-butanol) residue at the C terminus, which can be viewed as a modified valine residue with its carboxylic group replaced by a hydroxymethyl group.
Unlike natural amino acids, the amide proton of unknown peak 1 does not show a typical COSY peak with an Hα proton (Fig. 4B). An extremely weak peak, however, was visible in the 2D 1H homonuclear TOCSY spectrum (data not shown) between this amide proton and the protons of a methyl group at 13C/1H of 12.65/1.742 ppm. The latter, in turn, exhibited a 3JHH correlation (Fig. 4B, b6) to a CH moiety with 13C/1H chemical shifts of 117.35/5.812 ppm. It was found that the spin network consists only of these CH and CH3 moieties (Fig. 6C, box c1), and their distinctive 1H and 13C chemical shifts further imply the existence of an alkene group with one methyl substituent. Since the methanetriyl proton, described above, and the amide protons of unknown peak 1 and Leu-2 all have heteronuclear multiple-bond correlation with a carbonyl resonance at 164.43 ppm (Fig. 4C, c6, c1, and c3, respectively), the N-terminal residue was unequivocally determined to be Dhb (7).
Evidently, the peptide is acylated at the N terminus, as Dhb-1 HN has another HMBC peak that correlated with a carbonyl resonance at 172.55 ppm (Fig. 4C, c1′). It also shows several NOEs that could be rationalized only by the through-space interactions with the N-terminal cap, most notably, a CH group (Fig. 5C, c1), whose distinctive 13C/1H chemical shifts of 74.61/3.815 ppm (Fig. 6B, b5) suggest hydroxylation at this site. This group can be scalar correlated with four other protonated carbons (Fig. 6C, box c5), including 1 CH moiety, 1 CH2 moiety, and 2 CH3 moieties. On the basis of the 3JHH and 1JCH/3JHH correlations extracted from the 2D DQF-COSY and H2BC spectra, respectively, the aliphatic chain comprising these five protonated carbons was delineated to be identical to that of an isoleucine residue. Since a hydroxyl proton at 5.613 ppm (Fig. 6A, a2) shows a 3JHH correlation (Fig. 4B, b7) to the Hα and because of the 1JCH correlation with the Cα (Fig. 6D, d2), this N-terminal fatty acyl chain was unambiguously identified to be a 2-hydroxy-3-methyl-pentanoyl group.
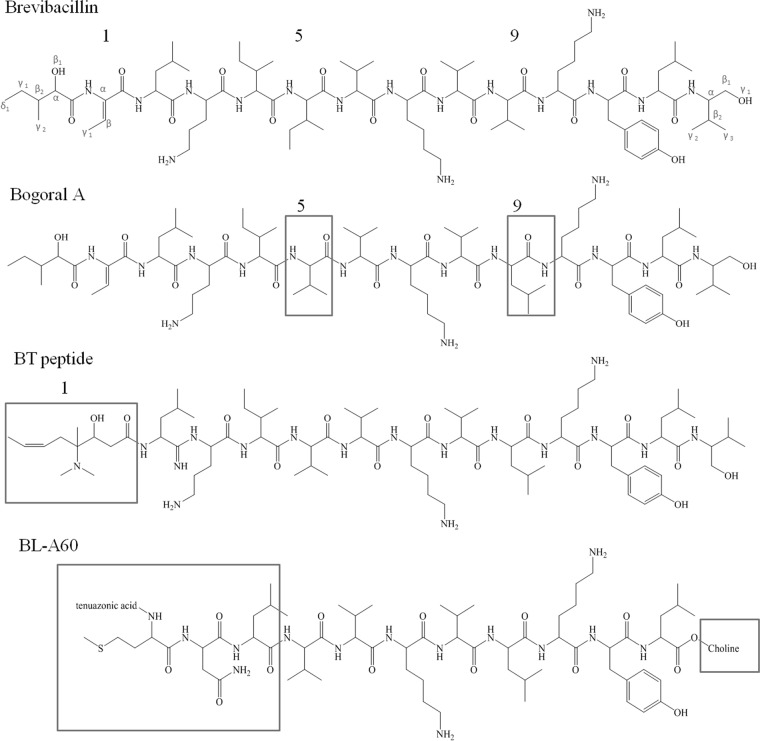
In conclusion, the structure of brevibacillin was elucidated by MS and NMR to be a linear lipopeptide (Fig. 7) comprising 13 amino acids (identified in order by the numbers after the amino acids in parentheses below) that is acylated to a C6 fatty acid (FA) tail, 2-hydroxy-3-methylpentanoic acid: FA-(Dhb-1)–(Leu-2)–(Orn-3)–(Ile-4)–(Ile-5)–(Val-6)–(Lys-7)–(Val-8)–(Val-9)–(Lys-10)–(Tyr-11)–(Leu-12)–(valinol-13). The sequence includes three modified amino acid residues, two of which are at the termini. The chemical shift assignments are summarized in Table 2. It should be noted that the stereospecific assignments are tentative with regard to the methyl groups of Val, Leu, and valinol.
FIG 7.
Chemical structures of brevibacillin and the related cationic peptide antibiotics bogorol A, BT peptide, and BL-A60 (BL-A60 is presented in reversed sequence order). The structural differences are highlighted in boxes.
TABLE 2.
Chemical shift assignments of lipopeptide in DMSO-D6 at 298.0 K
| Residue | Chemical shift (ppm) |
|||
|---|---|---|---|---|
| 15N/1HN | 13Cα/1Hα | 13Cβ/1Hβ | Others (13C/1H, C′, and OH) | |
| FA | 74.61/3.815 | OHβ1, 5.613; CHβ2, 37.94/1.700 | CH2γ1, 22.94/1.383, 1.119; CH3γ2, 15.10/0.875; CH3δ1, 11.36/0.807; C′, 172.55 | |
| Dhb-1 | 128.19/9.278 | 131.23/∼a | 117.35/5.812 | CH3γ1, 12.65/1.742; C′, 164.43 |
| Leu-2 | 122.88/8.219 | 51.29/4.288 | 39.84/1.557, 1.546 | CHγ, 23.94/1.657; CH3δ1, 22.93/0.860; CH3δ2, 20.64/0.846 |
| Orn-3 | 115.69/8.013 | 51.50/4.362 | 28.57/1.679, 1.650 | CH2γ, 23.28/1.551, 1.507; CH2δ, 38.01/2.755; Hε, 7.670 |
| Ile-4 | 115.45/7.774 | 56.03/4.265 | 36.74/1.710 | CH2γ1, 23.78/1.353, 1.009; CH3γ2, 15.01/0.768; CH3δ1, 10.72/0.752 |
| Ile-5 | 120.69/8.000 | 4.167/56.70 | 35.64/1.708 | CH2γ1, 24.13/1.427, 1.043; CH3γ2, 14.94/0.764; CH3δ1, 10.58/0.772 |
| Val-6 | 118.58/7.808 | 4.184/57.64 | 30.43/1.928 | CH3γ1, 18.88/0.806; CH3γ2, 17.57/0.802 |
| Lys-7 | 119.20/7.938 | 4.372/51.58 | 31.73/1.630, 1.482 | CH2γ, 21.85/1.267; CH2δ, 26.22/1.491; CH2ε, 38.39/2.693; Hζ, 7.670; Nζ, 33.3 |
| Val-8 | 115.77/7.883 | 4.224/57.15 | 30.23/1.907 | CH3γ1, 18.89/0.790; CH3γ2, 17.89/0.757 |
| Val-9 | 118.72/7.890 | 4.107/57.45 | 29.94/1.937 | CH3γ1, 18.93/0.775; CH3γ2, 17.83/0.774 |
| Lys-10 | 120.14/7.788 | 4.231/51.79 | 31.39/1.433, 1.321 | CH2γ, 21.58/1.040; CH2δ, 26.28/1.419; CH2ε, 38.39/2.662; Hζ, 7.670; Nζ, 33.3 |
| Tyr-11 | 118.75/8.095 | 4.471/54.06 | 37.17/2.840, 2.603 | CHδ1 and CHδ2, 129.79/6.988; CHε1 and CHε2, 114.53/6.613; Cζ, 155.81; Hη, 9.189 |
| Leu-12 | 121.00/8.095 | 4.226/50.82 | 40.56/1.400, 1.383 | CHγ, 23.65/1.331; CH3δ1, 22.87/0.818; CH3δ2, 21.21/0.764 |
| Valinol-13 | 118.27/7.405 | 3.530/55.24 | CH2β1, 61.04/3.340; CHβ2, 27.90/1.801 | OHg1, 4.507; CH3γ2, 19.31/0.814; CH3γ3, 17.71/0.786 |
∼, no 1Hα in this residue.
DISCUSSION
A new bacterial strain, B. laterosporus OSY-I1, was found to produce a potent lipopeptide with activity against Gram-positive bacteria. This species has no history of posing health hazards to animals or humans. In fact, B. laterosporus (BOD strain) has been used as a probiotic (25), and this strain has been patented (26). The product which contains the BOD probiotic strain has been commercially available since 1989 (Flora Balance). According to Chawawisit and Lertcanawanichakul (27), bioactive agents from probiotic bacteria may serve as promising candidates for the treatment of infections caused by antibiotic-resistant pathogens.
The newly discovered antimicrobial agent, brevibacillin, has a relatively novel structure, which is shown in Fig. 7. Brevibacillin is a cationic lipopeptide with three positively charged residues, which result in a net positive charge at neutral pH (28). After careful consideration, we concluded that this new compound is a member of a linear cationic antimicrobial lipopeptide family for which scattered information is available in the published literature. In 2001, Barsby et al. (29) reported the isolation of the first known compound in this family from a marine Bacillus sp. The producer organism was later reclassified as B. laterosporus by the same researchers (30). The lipopeptide was named bogorol A and is different from our brevibacillin in two amino acid resides at positions 5 and 9; isoleucine and valine in brevibacillin are replaced by valine and leucine in bogorol A, respectively. In 2005, Wu et al. (31) reported the isolation of an antimicrobial peptide, named BT peptide, from Brevibacillus texasporus. The peptide is structurally similar to bogorol A but differs in the residue at the N terminus (Fig. 7). The authors did not report this similarity, and their ambiguity about the N terminus warranted description of the compound as a lipopeptide. Seven years after the publication of the BT peptide structure, Zhao et al. (12) reported the discovery of a new antimicrobial peptide, BL-A60. Although neither the C terminus nor the N terminus was fully elucidated by the authors, the sequence of eight of the peptide's amino acid residues is similar to that of their counterparts in bogorol, except that the sequence order is reversed (Fig. 7). The BL-A60 pseudopeptide shares the highest similarity with bogorol C (30), another member of this family of antibiotics with a methionine residue at position 2, replacing the leucine residue in bogorol A (data not shown).
Brevibacillin carries three cationic amino acids: an ornithine and two lysines. These positively charged residues contribute to the hydrophilicity of the antimicrobial agent, whereas the aliphatic amino acids (Leu-3, Ile-4, Ile-5, Val-7, Val-8, Leu-9, Leu-12, Val-13) and an aromatic amino acid (Tyr-11) contribute to its hydrophobicity. In addition to its net positive charge and amphipathic nature, the lipophilic chain enhances the antimicrobial activity by increasing the hydrophobicity of the compound's N terminus. On the basis of the mechanism of action of other lipopeptides (32), it is presumed that brevibacillin may accumulate at the anionic surface of the bacterial cell membrane. The integrity of the indicator cell membrane could then be disrupted by this antimicrobial agent on the basis of its amphipathic nature. This amphipathic trait is often considered a prerequisite for the antimicrobial activity of cationic antimicrobial peptides against targeted microorganisms (33, 34). On the basis of these observations, it is presumed that brevibacillin damages the cell membrane, probably by creating holes, which depolarize the cell membrane potential and lead to the leakage of the intracellular contents. Further research is under way to confirm these hypotheses.
Brevibacillin showed strong inhibitory activity against MRSA and vancomycin-resistant strains of Enterococcus faecalis and Lactobacillus spp. This is the first time that L. plantarum ATCC 8014 has been reported to be a vancomycin-resistant species. Brevibacillin also showed potent antimicrobial activity against two Clostridium difficile strains, indicating its potency to serve as a good antibiotic with activity against anaerobic microorganisms. Brevibacillin also showed a strong inhibitory effect against foodborne spoilage and pathogenic microorganisms and MICs as low as those of nisin for many strains chosen for analysis. The antimicrobial agent was very effective against Alicyclobacillus spp., indicating the possibility of its use as a food additive in the future (35).
ACKNOWLEDGMENTS
The project was funded by the Center for Advanced Processing and Packaging Studies and by a scholarship to X. Yang from the China Scholarship Council.
We thank J. T. Lejeune (Food Animal Health Research Program, The Ohio State University) and W. A. Gebreyes (Department of Veterinary Preventive Medicine, The Ohio State University) for providing C. difficile strains.
REFERENCES
- 1.Grundmann H, Klugman KP, Walsh T, Ramon-Pardo P, Sigauque B, Khan W, Laxminarayan R, Heddini A, Stelling J. 2011. A framework for global surveillance of antibiotic resistance. Drug Resist Updat 14:79–87. doi: 10.1016/j.drup.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Cars O, Hedin A, Heddini A. 2011. The global need for effective antibiotics—moving towards concerted action. Drug Resist Updat 14:68–69. doi: 10.1016/j.drup.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Vergidis PI, Falagas ME. 2008. New antibiotic agents for bloodstream infections. Int J Antimicrob Agents 32:S60–S65. doi: 10.1016/j.ijantimicag.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Wright GD, Wright GD. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv Drug Deliv Rev 57:1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Huang E, Yuan C, Zhang L, Yousef AE. 2012. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl Environ Microbiol 78:3156–3165. doi: 10.1128/AEM.07782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Z, Kisla D, Zhang L, Yuan C, Green-Church KB, Yousef AE. 2007. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl Environ Microbiol 73:168–178. doi: 10.1128/AEM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda AK, Bisht SS, DeMondal S, Senthil Kumar N, Gurusubramanian G, Panigrahi AK. 2014. Brevibacillus as a biological tool: a short review. Antonie Van Leeuwenhoek 105:623–639. doi: 10.1007/s10482-013-0099-7. [DOI] [PubMed] [Google Scholar]
- 9.Ruiu L. 2013. Brevibacillus laterosporus, a pathogen of invertebrates and a broad-spectrum antimicrobial species. Insects 4:476–492. doi: 10.3390/insects4030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Oliveira EJ, Rabinovitch L, Monnerat RG, Passos LKJ, Zahner V. 2004. Molecular characterization of Brevibacillus laterosporus and its potential use in biological control. Appl Environ Microbiol 70:6657–6664. doi: 10.1128/AEM.70.11.6657-6664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saikia R, Gogoi DK, Mazumder S, Yadav A, Sarma RK, Bora TC, Gogoi BK. 2011. Brevibacillus laterosporus strain BPM3, a potential biocontrol agent isolated from a natural hot water spring of Assam, India. Microbiol Res 166:216–225. doi: 10.1016/j.micres.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Guo L, Zeng H, Yang X, Yuan J, Shi H, Xiong Y, Chen M, Han L, Qiu D. 2012. Purification and characterization of a novel antimicrobial peptide from Brevibacillus laterosporus strain A60. Peptides 33:206–211. doi: 10.1016/j.peptides.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Desjardine K, Pereira A, Wright H, Matainaho T, Kelly M, Andersen RJ. 2007. Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: structure elucidation and synthesis. J Nat Prod 70:1850–1853. doi: 10.1021/np070209r. [DOI] [PubMed] [Google Scholar]
- 14.Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant Gram-negative bacteria. Ann Pharmacother 33:960–967. doi: 10.1345/aph.18426. [DOI] [PubMed] [Google Scholar]
- 15.Denning DW. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- 16.Steenbergen JN, Alder J, Thorne GM, Tally FP. 2005. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 17.Huang XQ, Lu ZX, Zhao HZ, Bie XM, Lu FX, Yang SJ. 2006. Antiviral activity of antimicrobial lipopeptide from Bacillus subtilis fmbj against pseudorabies virus, porcine parvovirus, Newcastle disease virus and infectious bursal disease virus in vitro. Int J Pept Res Ther 12:373–377. doi: 10.1007/s10989-006-9041-4. [DOI] [Google Scholar]
- 18.Makovitzki A, Avrahami D, Shai Y. 2006. Ultrashort antibacterial and antifungal lipopeptides. Proc Natl Acad Sci U S A 103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38:3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wüthrich K. 1986. NMR of proteins and nucleic acids. John Wiley & Sons, New York, NY. [Google Scholar]
- 21.Nyberg NT, Duus JØ, Sørensen OW. 2005. Editing of H2BC NMR spectra. Magn Reson Chem 43:971–974. doi: 10.1002/mrc.1698. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA, Blevins RA. 1994. NMR VIEW—a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial disc susceptibility tests; approved standard. CLSI document M02-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Sanders ME, Morelli L, Tompkins TA. 2003. Sporeformers as human probiotics: Bacillus, Sporolactobacillus, and Brevibacillus. Compr Rev Food Sci Food Saf 2:101–110. doi: 10.1111/j.1541-4337.2003.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 26.O'Donnell BJ. October 1995. Method of inhibiting fungi by Bacillus laterosporus. US patent 5,455,028A.
- 27.Chawawisit K, Lertcanawanichakul M. 2008. Minimum inhibitory concentration (MIC) of crude preparations of Brevibacillus laterosporus SA14 bioactive material compared to vancomycin and oxacillin, against clinical isolates of methicillin-resistant Staphylococcus aureus. World J Microbiol Biotechnol 24:2199–2204. doi: 10.1007/s11274-008-9730-6. [DOI] [Google Scholar]
- 28.Hancock RE. 2001. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis 1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 29.Barsby T, Kelly MT, Gagné SM, Andersen RJ. 2001. Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org Lett 3:437–440. doi: 10.1021/ol006942q. [DOI] [PubMed] [Google Scholar]
- 30.Barsby T, Warabi K, Sørensen D, Zimmerman WT, Kelly MT, Andersen RJ. 2006. The bogorol family of antibiotics: template-based structure elucidation and a new approach to positioning enantiomeric pairs of amino acids. J Org Chem 71:6031–6037. doi: 10.1021/jo060667p. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Ballard J, Jiang YW. 2005. Structure and biosynthesis of the BT peptide antibiotic from Brevibacillus texasporus. Appl Environ Microbiol 71:8519–8530. doi: 10.1128/AEM.71.12.8519-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang E, Yousef AE. 2014. The lipopeptide antibiotic paenibacterin binds to the bacterial outer membrane and exerts bactericidal activity through cytoplasmic membrane damage. Appl Environ Microbiol 80:2700–2704. doi: 10.1128/AEM.03775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shai Y, Oren Z. 2001. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides 22:1629–1641. doi: 10.1016/S0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 34.Tossi A, Sandri L. 2002. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr Pharm Des 8:743–761. doi: 10.2174/1381612023395475. [DOI] [PubMed] [Google Scholar]
- 35.Grande MJ, Lucas R, Abriouel H, Ben Omar N, Maqueda M, Martínez-Bueno M, Martínez-Cañamero M, Valdivia E, Gálvez A. 2005. Control of Alicyclobacillus acidoterrestris in fruit juices by enterocin AS-48. Int J Food Microbiol 104:289–297. doi: 10.1016/j.ijfoodmicro.2005.03.010. [DOI] [PubMed] [Google Scholar]