Abstract
The bacterial pathogen Vibrio cholerae can occupy both the human gut and aquatic reservoirs, where it may colonize chitinous surfaces that induce the expression of factors for three phenotypes: chitin utilization, DNA uptake by natural transformation, and contact-dependent bacterial killing via a type VI secretion system (T6SS). In this study, we surveyed a diverse set of 53 isolates from different geographic locales collected over the past century from human clinical and environmental specimens for each phenotype outlined above. The set included pandemic isolates of serogroup O1, as well as several serogroup O139 and non-O1/non-O139 strains. We found that while chitin utilization was common, only 22.6% of the isolates tested were proficient at chitin-induced natural transformation, suggesting that transformation is expendable. Constitutive contact-dependent killing of Escherichia coli prey, which is indicative of a functional T6SS, was rare among clinical isolates (only 4 of 29) but common among environmental isolates (22 of 24). These results bolster the pathoadaptive model in which tight regulation of T6SS-mediated bacterial killing is beneficial in a human host, whereas constitutive killing by environmental isolates may give a competitive advantage in natural settings. Future sequence analysis of this set of diverse isolates may identify previously unknown regulators and structural components for both natural transformation and T6SS.
INTRODUCTION
Vibrio cholerae, the bacterium responsible for the diarrheal disease cholera, can occupy a range of freshwater and marine environments, where it commonly associates with abiotic chitinous material and the biotic surfaces of algae, invertebrates, plants, and fish (1). When water carrying V. cholerae is ingested, cells that survive passage through the acidic stomach may gain access to the small intestine and bind to its mucus layer. Isolates carrying the CTX prophage can secrete cholera toxin (CT), which is responsible for the potentially fatal diarrhea that also aids in transmission from the host.
Over 200 O serogroups have been described, with each defined as a group of bacteria that share a surface antigen. Although only the O1 and O139 serogroups carrying the CTX prophage are responsible for major cholera epidemics, other serogroups may be associated with isolated cases of gastroenteritis but so far have not been shown to spread globally (2). The pandemic O1 CTX+ isolates are further divided into two biotypes, Classical and El Tor, on the basis of several biochemical and phage susceptibility tests (3–6). Seven cholera pandemics have been described. The O1 Classical biotype was responsible for the sixth and likely prior pandemics but was displaced by the O1 El Tor biotype in the seventh pandemic, which began in Southeast Asia in 1961 (7). In 1992, an El Tor mutant, serotype O139, became responsible for some regional cholera outbreaks and continues to coexist with O1 El Tor, although in a minor capacity (8–10).
When inhabiting aquatic environments, V. cholerae can degrade the chitinous surfaces of copepods, zooplankton, and crabs to soluble (GlcNAc)n oligosaccharides that can be imported and utilized as a carbon source (11). Liberated chitin oligosaccharides [(GlcNAc)2–6] may also act as an extracellular signal recognized by membrane-bound receptors that triggers a signaling cascade for the expression of genes encoding a DNA uptake apparatus for natural transformation (12–17). Transformation is one mode of horizontal gene transfer that can promote rapid gene exchange, allowing bacteria to quickly adapt to their ever-changing environment, but this DNA can also be used for repair and nutrition (18). Although successful natural transformation has been studied extensively in a small number of V. cholerae reference strains, little is known regarding the broader prevalence of transformation ability among members of the species. In fact, contemporary V. cholerae isolates from the recent Haiti outbreak were impaired for natural transformation (19, 20), highlighting the need for a more comprehensive understanding of the transformation proficiency of V. cholerae.
It was recently discovered that growth on chitin also induces the expression of a type VI secretion system (T6SS) in V. cholerae (21, 22). The type VI apparatus, which is structurally analogous to a phage tail spike, can penetrate adjacent cells and deliver toxic effectors that cause contact-dependent lysis (23). These toxic effectors can be used to target either prokaryotic or eukaryotic prey cells (24). Liberated DNA from lysed prey cells may then serve as the genetic material for natural transformation (21) or as an alternative nutrient source. T6SS was originally discovered in 2006 in a non-O1/non-O139 environmental isolate of V. cholerae, V52, in an attempt to understand virulence mechanisms in nonpandemic CTX− strains that cause isolated cases of gastroenteritis (25). Subsequently, genomic analyses of sequenced genomes of V. cholerae have identified three major T6SS gene clusters that encode one T6SS (25–27). It has been proposed that contact-dependent T6SS-mediated killing ability provides a competitive advantage, allowing V. cholerae to persist in both the human gut and the environment. However, despite extensive analyses documenting the presence of T6SS genes in all sequenced V. cholerae genomes (27, 28), a broader survey of contact-dependent killing has not been performed.
In V. cholerae clinical O1 El Tor isolates C6706 and A1552, genes involved in chitin utilization, natural transformation, and the T6SS are under the control of several positive regulatory factors. TfoX, induced by growth on chitin (12), and the transcription factor CytR, controlled by nucleoside starvation, act as positive regulators of all three phenotypes in V. cholerae C6706 (22). Quorum sensing (QS) also controls natural transformation and type VI secretion in V. cholerae by upregulating the expression of transcription factor HapR in response to the accumulation of secreted autoinducer signals at high cell density (12, 21, 29, 30). HapR directly activates the transcription of the gene encoding the transcriptional regulator QstR (31). Expression of TfoX and QstR by chitin at high cell density or from a heterologous promoter is sufficient to induce transformation and T6SS-mediated killing (21, 22, 31). Although DNA uptake and T6SS are both controlled in A1552 and C6706 by the presence of chitin, suggesting coupling of these phenotypes, it is not known whether these activities are also coordinately regulated in other members of the species.
In this study, we analyzed a set of 53 patient-derived and non-patient-derived V. cholerae isolates collected between 1910 and 2011. Each isolate was characterized for chitinase activity, natural transformation, and constitutive contact-dependent killing of Escherichia coli consistent with a functional T6SS. We designated patient-derived isolates “clinical” and non-patient-derived isolates “environmental,” which are represented by the letters C and E, respectively, in Table 1. These isolates include serogroups O1 (both Classical and El Tor biotypes), O139, and non-O1/non-O139, which were further differentiated by CTX+ or CTX− status. The majority of the isolates we tested possessed chitinase activity, while >50% were deficient in chitin-induced natural transformation. Clinical isolates were largely unable to engage in constitutive contact-dependent killing of E. coli prey cells in a standard killing assay, but nearly all of the environmental isolates tested displayed constitutive bacterial killing. The diversity of the set of isolates tested in transformation proficiency and bacterial killing suggests that transformation may be dispensable in various settings. In contrast, constitutive contact-dependent antagonism, like that mediated by a T6SS, appears to be valuable in environmental habitats distinct from a human host.
TABLE 1.
V. cholerae isolates from numerous locations, sources, and years, with serogroup and CTX statusa
| Strain | Location | Source | C/E | Yr | Serogroup | CTX | Chi | TF | Killing |
|---|---|---|---|---|---|---|---|---|---|
| Wild-type C6706 | Peru | Patient | C | 1991 | O1 El Tor | + | + | + | − |
| NCTC8457 | Saudi Arabia | Patient | C | 1910 | O1 El Tor | + | − | + | + |
| MZO-2 | Bangladesh | Patient | C | 2001 | O14 | − | + | + | + |
| 2010EL-1749 | Cameroon | Patient | C | 2010 | O1 El Tor | + | + | + | + |
| MAK757 | Celebes | Patient | C | 1937 | O1 El Tor | + | + | − | + |
| CA401 | India | Patient | C | 1953 | O1 Classical | + | + | − | − |
| O395 | India | Patient | C | 1965 | O1 Classical | + | + | − | − |
| MO10 | India | Patient | C | 1992 | O139 El Tor | + | + | − | − |
| 3541-04 | GA, USA | Patient | C | 2004 | O75 CII | + | + | − | − |
| 3558-04 | AL, USA | Patient | C | 2004 | O75 CI | + | + | − | − |
| 3582-05 | Pakistan | Patient | C | 2005 | O1 El Tor | + | + | − | − |
| 3500-05 | India | Patient | C | 2005 | O1 El Tor | + | + | − | − |
| 3546-06 | India | Patient | C | 2006 | O1 El Tor | + | + | − | − |
| 3554-08 | Nepal | Patient | C | 2008 | O1 El Tor | + | + | − | − |
| 2011EL-1141 | Afghanistan | Patient | C | 2008 | O1 El Tor | + | + | − | − |
| 3566-08 | NJ, USA | Patient | C | 2008 | O141 | + | + | − | − |
| 2009V-1085 | Sri Lanka | Patient | C | 2009 | O1 El Tor | + | + | − | − |
| 2011EL-1938 | Orissa, India | Patient | C | 2009 | O1 El Tor | + | + | − | − |
| 2009V-1096 | India | Patient | C | 2009 | O1 El Tor | + | + | − | − |
| 2011EL-1137 | South Africa | Patient | C | 2009 | O1 El Tor | + | + | − | − |
| 2009V-1046 | Pakistan | Patient | C | 2009 | O1 El Tor | + | + | − | − |
| Nepal 25 | Nepal | Patient | C | 2010 | O1 El Tor | + | + | − | − |
| Nepal 14 | Nepal | Patient | C | 2010 | O1 El Tor | + | + | − | − |
| 2011EL-1941 | Kolkata, India | Patient | C | 2010 | O1 El Tor | + | + | − | − |
| 2010V-1014 | Pakistan | Patient | C | 2010 | O1 El Tor | + | + | − | − |
| 2010EL-1786 | Artibonite, Haiti | Patient | C | 2010 | O1 El Tor | + | + | − | − |
| 2011EL-1939 | India | Patient | C | 2011 | O1 El Tor | + | + | − | − |
| 2012V-1001 | United States | Patient | C | 2011 | O1 El Tor | + | + | − | − |
| 2011V-1043 | FL, USA | Patient | C | 2011 | O75 CI | + | + | − | − |
| 3223-74 | Guam | Storm drain | E | 1974 | O1 | − | + | + | + |
| 2631-78 | LA, USA | Moore swab | E | 1978 | O1 | − | + | + | + |
| 2633-78 | Brazil | Sewage | E | 1978 | O1 | − | + | + | + |
| E8498 | LA, USA | Water | E | 1978 | O75 CII | + | + | + | − |
| VC22 | FL, USA | Oyster | E | 1981 | O1 | − | + | + | + |
| 2479-86 | LA, USA | Moore swab | E | 1986 | O1 | − | + | + | + |
| TP | CA, USA | Water | E | 2000 | NAg | − | + | + | + |
| VC56 | AL, USA | Oyster | E | 2009 | O1 | − | − | + | + |
| 3225-74 | Guam | Storm drain | E | 1974 | O1 | − | + | − | + |
| 3272-78 | MD, USA | Bay | E | 1977 | O1 | − | + | − | + |
| 1074-78 | Brazil | Sewage | E | 1978 | O1 | − | + | − | + |
| 2559-78 | LA, USA | Crab | E | 1978 | O1 | + | + | − | + |
| 692-79 | LA, USA | Canal | E | 1979 | O1 | − | + | − | + |
| 2740-80 | U.S. Gulf Coast | Water | E | 1980 | O1 | − | + | − | + |
| VC48 | FL, USA | Oyster | E | 1981 | NAg | − | + | − | + |
| 2512-86 | LA, USA | Moore swab | E | 1986 | O1 | + | + | − | + |
| 2497-86 | LA, USA | Moore swab | E | 1986 | O1 | − | + | − | + |
| 1496-86 | LA, USA | Moore swab | E | 1986 | O1 | − | + | − | − |
| 2523-87 | LA, USA | Moore swab | E | 1987 | O1 | − | + | − | + |
| 857 | Bangladesh | Water | E | 1996 | O1 | − | + | − | + |
| SIO | CA, USA | Water | E | 2000 | NAg | − | + | − | + |
| 3568-07 | Mexico | Queso fresco | E | 2007 | O141 | + | + | − | + |
| VC53 | AL, USA | Oyster | E | 2009 | NAg | − | − | − | + |
| HE46 | Centre, Haiti | Gray water | E | 2011 | NAg | − | + | − | + |
Clinical (C) and environmental (E) isolates were found positive or negative for the following phenotypes: chitinase activity (Chi), natural transformation (TF), and constitutive contact-dependent bacterial killing (Killing).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were obtained from various laboratories (see Table S1 in the supplemental material for detailed descriptions). Isolates were previously characterized in the respective laboratories to determine their serogroups and the presence or absence of the CT gene (ctxA). The methods used at the Centers for Disease Control and Prevention were summarized by Talkington et al. (19, 32, 33). The V. cholerae clinical and environmental isolates were collected from a variety of locations between 1910 and 2011. E. coli MG1655 Cmr was used as the prey in bacterial killing assays. All strains were grown in LB liquid medium or on LB agar at 37°C with the appropriate antibiotics added at the following concentrations when needed: kanamycin at 50 μg/ml and chloramphenicol at 10 μg/ml for V. cholerae and 25 μg/ml for E. coli. Specific assay conditions are described below.
Construction of a plasmid for heterologous qstR and tfoX expression.
A previously constructed plasmid containing a kanamycin resistance gene and the tfoX gene under the control of the ptac promoter (29) was digested at the BamHI restriction enzyme site located 5′ of the tfoX gene. A fragment that carries the qstR gene, including its native ribosome binding site, was amplified from the C6706 chromosome with primers pQT_1 and pQT_2 (see Table S2 in the supplemental material). Gibson assembly (New England BioLabs) was used to introduce this PCR product into the BamHI-digested plasmid so that both tfoX and qstR were under the control of the same isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptac promoter. The resulting plasmid, designated pQT, was confirmed for the qstR insertion by sequencing with primers pQT_3 and pQT_4 (see Table S2). E. coli S17-λpir cells transformed by electroporation with pQT were used to introduce the plasmid into V. cholerae strains of interest by conjugation.
Construction of the suicide vector used in allelic exchange to delete vasK in isolate 692-79.
The suicide vector, pRE118, containing an R6K origin of replication, a kanamycin resistance gene, and the sacB gene conferring sucrose sensitivity, was used for allelic exchange (34). Plasmid pRE118 was digested at the KpnI and XhoI restriction enzyme sites. The genome sequence of 692-79 was used to design primers to PCR amplify 500 bp upstream (vasK_1 and vasK_2) and downstream (vasK_3 and vasK_4) of the vasK gene (see Table S2 in the supplemental material). Gibson assembly (New England BioLabs) was used to combine these two PCR products and the digested plasmid. The resulting plasmid was verified via sequencing with flanking vasK_1 and vasK_4 primers (see Table S2). E. coli S17-λpir cells transformed by electroporation with the plasmid were used to introduce the suicide vector into isolate 692-79 via conjugation. LB agar plates containing kanamycin were used to screen for transconjugants containing the chromosomally integrated plasmid. These colonies were then streaked onto LB agar plates containing 10% sucrose to select for candidates that had lost the plasmid by homologous recombination. Isolated colonies unable to grow when restreaked onto LB agar plates with kanamycin were selected as presumptive vasK deletion mutants. Deletion of vasK was confirmed by PCR with internal vasK_5 and vasK_6 primers. The resulting 692-79 ΔvasK mutant was used to verify the necessity of T6SS in contact-dependent bacterial killing.
Chitinase plate assay.
Colloidal chitin was prepared from practical-grade chitin (Sigma) derived from shrimp shells as previously described (35, 36). Colloidal chitin plates were prepared by mixing 2% (wt/vol) colloidal chitin with LB medium buffered to pH 7.0 with 0.1 M phosphate buffer. Strains were incubated overnight at 37°C in LB broth and diluted to an optical density at 600 nm (OD600) of 1.0, and 10 μl of each suspension was plated onto the colloidal chitin agar. After incubation at 37°C for 96 h, the presence or absence of a zone of chitin clearing for each colony was recorded by comparison to positive (C6706) and negative (C6706 CytR−) controls.
Natural transformation chitin assay.
The standard chitin-induced transformation assay was used to quantify the transformation frequency (TF) of V. cholerae as described previously in detail (12, 37). Briefly, cells grown in the presence of chitin (crab shell fragments) were exposed for 24 h to genomic DNA carrying a kanamycin resistance cassette and then plated on selective and nonselective media to determine the TF (37). Three independent experiments were performed with each isolate in triplicate, and the mean TF ± the standard deviation for one representative experiment were reported.
Bacterial killing assay.
The killing assay (38) was modified as described previously in detail (22). Briefly, V. cholerae predators and Cmr E. coli prey strains were grown overnight in LB medium with shaking at 37°C to an OD600 of ∼1.0. Predator V. cholerae (clinical and environmental isolates) and prey (Cmr E. coli) cells were mixed at a ratio of 10:1, and 50 μl of each suspension was spotted onto sterile Whatman cellulose gridded filters (GE Healthcare) placed on LB plates. For qstR tfoX inducible killing assays with isolates carrying pQT, IPTG (Fisher BioReagents) was added to a final concentration of 200 μM to each 50-μl suspension before samples were spotted onto filters. A prey-alone control was also prepared in the same ratio with fresh LB medium to determine total prey counts in the absence of a predator. After incubation at 37°C for 3 h, cells were removed from filters by vortexing in LB medium, serially diluted, and plated on LB agar supplemented with chloramphenicol to determine the number of CFU per milliliter of surviving E. coli prey.
Contact dependence assay.
The contact dependence assay described in reference 38 was modified slightly to parallel the standard killing assay described above. Briefly, each V. cholerae isolate (predator), as well as an E. coli control, was plated as a lawn on LB agar plates and incubated overnight. A sterile 0.22-μm filter (Pall Life Sciences) was placed on top of each confluent lawn, and then 50 μl of E. coli prey was spotted on top of the filter, allowing the E. coli cells access to nutrients but keeping them physically separated from the plated V. cholerae predator and the E. coli control. After incubation at 37°C for 3 h, cells were removed from filters by vortexing in LB medium and serially diluted and dilutions were spot plated on LB agar supplemented with chloramphenicol to determine the approximate number of CFU of surviving E. coli prey per milliliter.
RESULTS
V. cholerae clinical and environmental strains.
Strains were chosen to include various locations, sources, years of isolation, and serogroups as outlined in detail in Table 1. Serogroups of clinical and environmental isolates, as well as the presence or absence of the CT gene (ctxA), were obtained from previous publications (19, 33) or determined by methods described by Talkington et al. (32). Isolates that did not fall into major serogroup O1, O139, O75, O14, or O141 were deemed nonagglutinating and designated NAg. The majority of the 53 isolates are in the O1 serogroup and were acquired from various locations in various years. All of the isolates had comparable growth kinetics and grew to similar ODs (data not shown); therefore, any differences seen were not attributed to growth defects. Clinical O1 El Tor strain C6706 served as the reference strain in all assays (39). On the basis of the Pearson correlation matrix obtained by principal-component analysis (PCA) (see Table S3 in the supplemental material), differences in location, year, and serogroup do not appear to be correlated with differences in chitinase activity, natural transformation ability, or constitutive bacterial killing, as described below.
Chitinase activity is common in V. cholerae.
In clinical O1 El Tor isolate C6706, chitin degradation, natural transformation, and T6SS-mediated killing all require chitin as an inducing signal (22). Therefore, to characterize the set of isolates, we first sought to determine qualitatively whether each isolate was defective or proficient in chitin utilization. Specifically, we observed degradation of colloidal chitin, which requires the expression of two major chitinases, chiA-1 and chiA-2, that are both under TfoX and CytR control in V. cholerae C6706 (22). As expected, a zone of clearing was visible around a colony of C7606 on colloidal chitin agar, indicative of chitinase activity (Fig. 1) (35). Clearing was more pronounced for a C6706 strain that constitutively expressed the chitin-responsive TfoX regulator (TfoX*) (39), while a C6706 strain lacking the cytR gene (CytR−) was unable to degrade chitin by this method (Fig. 1) (22, 39). Each clinical and environmental isolate was then assessed for the ability to degrade chitin in this assay. The majority of the isolates degraded colloidal chitin and showed a visible zone of clearing. Only one clinical isolate (NCTC8457) and two environmental isolates (VC56 and VC53) were unable to produce a detectable zone of clearing comparable to that of the C6706 CytR− strain and are designated negative in Table 1. However, these three isolates were capable of growing (∼1.0E+07 CFU/ml) in minimal medium containing a chitin crab shell fragment under the conditions used in a transformation assay described below, though they reached a lower stationary-phase cell density than C6706 (∼1.0E+09 CFU/ml). Thus, although unable to degrade colloidal chitin, each negative isolate in Table 1 appeared to possess chitinase activity sufficient to utilize chitin for growth.
FIG 1.
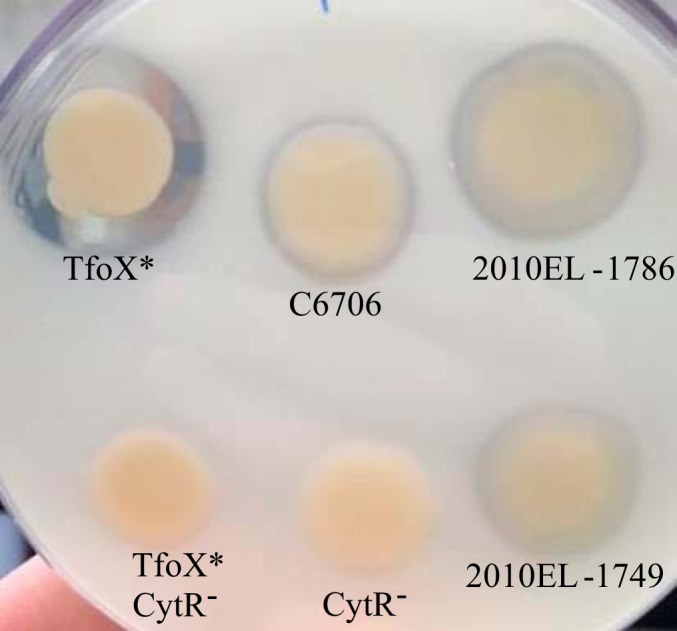
Chitin agar plate assay. V. cholerae isolate C6706 and isogenic derivatives constitutive for TfoX (TfoX*) and lacking cytR (CytR−), as well as all clinical and environmental isolates, were assayed for the ability to degrade chitin, which results in a visible zone of clearing on LB agar plates containing 2% colloidal chitin. Shown are C6706, the TfoX* derivative, and isolates 2010EL-1786 and 2010EL-1749, which produce a detectable zone of clearing; as well as two CytR− strains that do not produce a zone of clearing.
Natural transformation proficiency in V. cholerae is rare.
Since the discovery of natural competence and transformation in V. cholerae in 2005, most studies have focused on a small number of clinical strains, while less is known regarding the overall prevalence of natural transformation ability more broadly in this Vibrio species (12). To address this, each isolate was tested for the ability to take up DNA and recombine it onto the chromosome via chitin-induced natural transformation, by methods previously described (37). Isolates with a transformation frequency (TF) of ≤1.0E−08 were deemed severely impaired in transformation, as described previously (29), and are designated negative in Table 1. Proficient isolates are designated positive in Table 1, and the corresponding values are shown in Table 2.
TABLE 2.
Transformation frequencies of proficient V. cholerae clinical and environmental isolatesa
| Strain | C/Eb | Mean TF ± SD |
|---|---|---|
| Wild-type C6706 | C | 1.00E−05 ± 8.50E−06 |
| NCTC8457 | C | 5.42E−07 ± 2.04E−07 |
| MZO-2 | C | 4.79E−06 ± 1.94E−06 |
| 2010EL-1749 | C | 4.53E−06 ± 3.49E−07 |
| 3223-74 | E | 1.04E−05 ± 3.29E−07 |
| 2631-78 | E | 1.77E−06 ± 5.74E−07 |
| 2633-78 | E | 1.07E−06 ± 4.56E−07 |
| E8498 | E | 2.51E−06 ± 3.17E−07 |
| VC22 | E | 1.75E−07 ± 8.19E−08 |
| 2479-86 | E | 7.61E−06 ± 1.40E−06 |
| TP | E | 2.62E−06 ± 8.91E−07 |
| VC56 | E | 1.08E−07 ± 1.18E−08 |
Data shown are for biological triplicates from one experiment representative of three performed.
C, clinical; E, environmental.
Of the 29 clinical isolates tested, only 4 (13.8%) were proficient in natural transformation, with TFs ranging from 1.00E−05 to 5.42E−07 (Tables 1 and 2). As shown before (12, 39), O1 El Tor reference strain C6706 had an expected TF of 1.0E−05. The TF of O1 El Tor isolate NCTC8457, obtained in Saudi Arabia in 1910, was 5.42E−07, and that of O14 MZO-2, isolated in Bangladesh in 2001, was 4.79E−06. Contemporary clinical O1 El Tor isolate 2010EL-1749, collected in 2010 in Cameroon, was also transformable with a frequency of 4.53E−06. Of the 24 isolates tested that were derived from environmental sources, 8 (33.3%) were proficient at natural transformation, with a range spanning from 1.04E−05 to 1.75E−07 (Table 2). The set of proficient environmental isolates included samples obtained over a 30-year period from 1974 to 2009 at locations within and outside the United States and included O1 and non-O1 serogroup isolates (Tables 1 and 2). Similar results were obtained when we attempted to transform deficient isolates with their own genomic DNA carrying a kanamycin resistance cassette (data not shown).
Constitutive contact-dependent bacterial killing is common among environmental isolates.
Four environmental isolates, V52, 2740-80, DL4211, and DL4215, have been reported previously to display constitutive T6SS-mediated killing of E. coli under standard killing assay conditions in the absence of chitin (24, 25, 40). However, in clinical isolates C6706 and A1552, transcription of the genes encoding the T6SS requires activation by the QstR and TfoX transcription factors, which are induced at high cell density and in the presence of chitin (12, 21, 39). Chitin induction is sufficient to induce killing of prey by A1552, as observed by fluorescence microscopy (21). Nevertheless, chitin induction is insufficient for killing of E. coli prey above the limit of detection by A1552 and C6706 in a standard quantitative 3-h killing assay. Only genetic manipulation that places the TfoX regulator under the control of a nonnative constitutive promoter allows significant bacterial killing in this assay (21, 22).
On the basis of these and other studies, it was hypothesized that clinical pandemic isolates like C6706 tightly control T6SS, while nonpandemic isolates from environmental sources may express T6SS constitutively to compete with other microbes under conditions outside a human host (24). Because this “pathoadaptive” hypothesis (41) has not been extensively tested experimentally in killing assays, we examined each of the 53 isolates for killing of E. coli prey in the absence of chitin to determine constitutive bacterial killing activity indicative of a T6SS. Standard killing assays were performed by exposing each isolate of V. cholerae to Cmr E. coli to permit enumeration of surviving E. coli prey cells following the 3 h of exposure to the V. cholerae predator, as described previously (22, 38). Although bactericidal activity was not directly measured in these assays, we use the term “killing” as it was used to describe similar results in V. cholerae killing assays (21, 22, 38, 42).
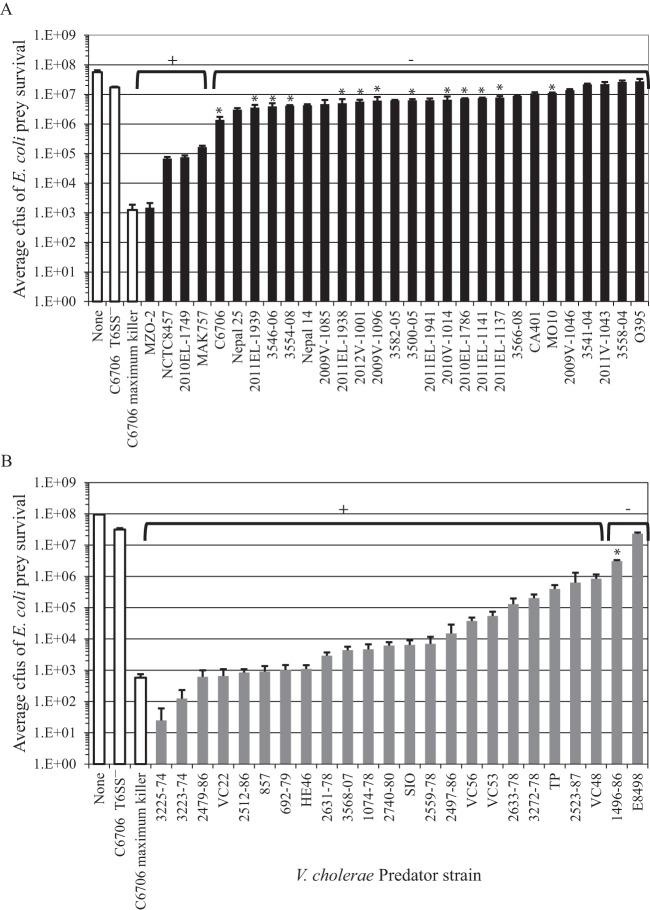
A CytR+ C6706 O1 El Tor strain that was genetically manipulated to express TfoX, HapR, and QstR constitutively (here designated the C6706 maximum killer) decreased prey survival by 10,000-fold compared to a T6SS-deficient C6706 ΔvasK mutant strain (here designated C6706 T6SS−), consistent with previous experiments (22) (Fig. 2, white bars). The modest <10-fold reduction in the survival of prey in the presence of the C6706 T6SS− derivative compared to that of the prey alone is most likely due to competition for nutrients and has also been observed in prior studies that used the standard killing assay (22, 38). Twenty-five of the 29 clinical isolates tested displayed little or no constitutive killing, with surviving E. coli counts within 10-fold of those recorded when E. coli was exposed to the C6706 T6SS− strain (Fig. 2A, below negative bracket), including the original O1 El Tor C6706 isolate as expected, because of its requirement for chitin induction. In Table 1 and Fig. 2, a plus sign denotes a >10-fold reduction of E. coli prey from the level recorded when E. coli was exposed to C6706 T6SS−, and a minus sign indicates a reduction of <10-fold. A single clinical isolate, MZO-2, reduced E. coli survival by ∼10,000-fold, similar to the C6706 maximum killer; three additional clinical isolates, NCTC8457, 2010EL-1749, and MAK757, reduced E. coli survival more modestly by ∼100-fold (Fig. 2A, below positive bracket).
FIG 2.
Bacterial killing assay results. Cmr E. coli prey cells were incubated with the V. cholerae predator strains indicated at a ratio of 1:10 on membrane filters on LB agar to measure bacterial killing. Prey-alone (None), negative (C6706 T6SS−), and positive (C6706 maximum killer) controls are represented by white bars in both panels. Shown are average prey survival values ± standard deviations after triplicate encounters of E. coli prey in contact with V. cholerae clinical isolates (panel A, black bars) and environmental isolates (panel B, gray bars). The results of one experiment representative of the three performed are shown. Isolates considered constitutive for bacterial killing as defined in the text are indicated by the positive bracket, while nonconstitutive isolates are indicated by the negative bracket. Asterisks indicate isolates that demonstrated killing when TfoX and QstR expression was induced.
In sharp contrast, the environmental isolates tested covered a 100,000-fold range of constitutive E. coli prey-killing abilities (Fig. 2B). Only two environmental isolates tested, E8498 and 1496-86, displayed little or no constitutive killing of E. coli prey (Fig. 2B, negative bracket), with the majority of the isolates (22 of 24) showing >10-fold more killing than that recorded with the C6706 T6SS− strain (Fig. 2B, positive bracket). Of these 24 isolates, 14 killed E. coli with an efficiency within 10-fold of that of the C6706 maximum killer, providing an at least 10,000-fold decrease in the prey, suggesting that these strains are efficient constitutive killers (Fig. 2B). Two isolates, 3225-74 and 3223-74, showed >100,000-fold killing, exceeding values obtained with the C6706 maximum killer (Fig. 2B). These results are consistent with previous observations, by the same standard methods described here (43), that environmental isolates, like V52 and 2740-80, constitutively kill E. coli cells. Thus, on the basis of the isolates tested here, constitutive killing appears to be common among V. cholerae environmental isolates but rare among clinical isolates.
Killing by the T6SS requires a physical association of V. cholerae predator cells with E. coli prey cells for the delivery of toxic effectors; thus, we tested whether the constitutive killing we observed was contact dependent. In contrast to the standard killing assay, where predator and prey cells are mixed prior to plating to ensure contact, E. coli cells were dispensed onto a 0.22-μm filter that was placed on a confluent lawn of V. cholerae cells to physically separate the predator and prey cells, similar to a method described previously (38). E. coli prey levels were highest when the filter was directly placed on the agar surface, and only modest (∼3-fold) reductions in E. coli survival were observed when the agar surface was first seeded with isogenic E. coli or a C6706 T6SS− mutant. However, when the C6706 maximum killer and 4 clinical and 24 environmental isolates that had displayed constitutive 10- to 100,000-fold killing of E. coli in the standard assay (Fig. 2 and Table 1, positive isolates) were incubated under these conditions, prey survival was unaltered (<2-fold) (see Fig. S1 in the supplemental material). The physical barrier of the filter allowed E. coli growth but not killing, demonstrating that the constitutive killing documented by our standard bacterial killing assay was contact dependent.
To demonstrate that constitutive contact-dependent bacterial killing observed with one of the environmental isolates was T6SS mediated, an in-frame vasK deletion mutant of isolate 692-79 was constructed via allelic exchange. The 692-79 isolate and the isogenic ΔvasK mutant (692-79 T6SS−) were tested for contact-dependent killing of E. coli along with the C6706 maximum killer and the isogenic ΔvasK mutant (C6706 T6SS−) as controls. As described previously, E. coli prey survival was highest in the absence of a predator, decreased ∼10,000-fold in the presence of the C6706 maximum killer, and decreased <10-fold in the presence of C6706 T6SS− (Fig. 3, white bars). Similarly, a >10,000-fold reduction in E. coli prey survival was observed when it was incubated with 692-79 but abolished with 692-79 T6SS−, confirming for this isolate that T6SS was responsible for the observed loss of E. coli prey viability (Fig. 3, gray bars).
FIG 3.
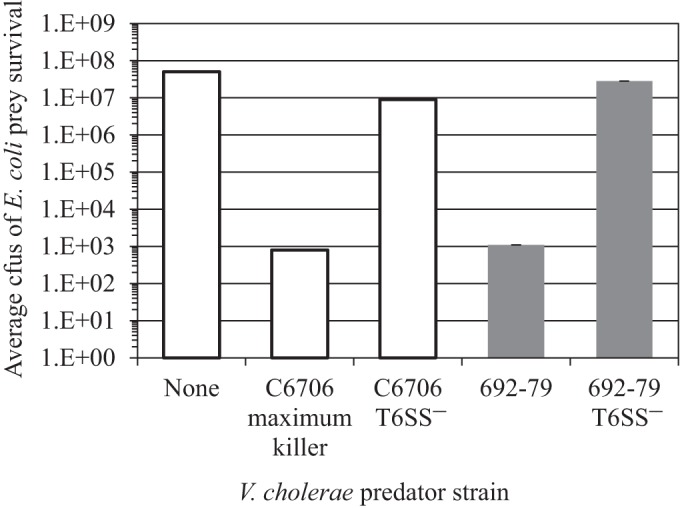
Contact-dependent bacterial killing by environmental isolate 692-79 is mediated by a T6SS. Cmr E. coli prey cells were incubated with the V. cholerae predator strains indicated at a ratio of 1:10 on membrane filters on LB agar to measure bacterial killing. Prey-alone (None), positive (C6706 maximum killer), and negative (C6706 T6SS−) controls are represented by white bars. Isolate 692-79 and the isogenic ΔvasK mutant (692-79 T6SS−) are represented by gray bars. Shown are average prey survival values ± standard deviations for triplicate predator-prey encounters. The results of one experiment representative of the three performed are shown.
Additional isolates are capable of killing under conditions that mimic chitin induction.
Chitin induction is insufficient to induce T6SS-mediated killing of E. coli by C6706 above the limit of detection in a 3-h killing assay, as described above. Thus, in order to mimic chitin signaling in isolates that did not show constitutive killing (negative in Fig. 2 and Table 1), a plasmid expressing both qstR and tfoX from a heterologous IPTG-inducible promoter, designated pQT, was introduced into each of these 27 isolates and killing assays were performed. This determined whether any of these isolates were indeed capable of contact-dependent bacterial killing by induction. The C6706 T6SS− (ΔvasK) mutant did not kill E. coli regardless of pQT induction and served as a negative control (Table 3). Relative to the T6SS− control, the C6706 maximum killer showed an ∼10,000-fold reduction in E. coli levels irrespective of pQT induction. E. coli prey levels were similarly reduced when C6706 was induced with pQT, with only a modest (<10-fold) effect observed in the absence of induction, as expected. Likewise, induction with pQT was capable of restoring bacterial killing activity in C6706 ΔqstR, ΔtfoX, and ΔqstR tfoX mutants (data not shown). Relative to the T6SS− control, 13 of 25 clinical isolates and 1 of 2 environmental isolates that were initially scored negative for constitutive contact-dependent bacterial killing (Fig. 2, asterisks) showed ≥10-fold reductions in E. coli levels only after pQT induction (Table 3). Thus, 14 additional isolates were capable of contact-dependent bacterial killing in response to conditions that mimic chitin induction, consistent with a functional T6SS.
TABLE 3.
Fold reduction of prey survival when exposed to V. cholerae isolates with and without pQT
| Strain | C/Ea | Fold reduction of prey survivalb |
|
|---|---|---|---|
| No induction | With induction | ||
| C6706 T6SS− | C | 1.0 | 1.0 |
| C6706 maximum killer | C | 13,760.0 | 15,855.0 |
| C6706 | C | 9.5 | 8,094.1 |
| 3554-08 | C | 4.2 | 557.1 |
| 1496-86 | E | 5.6 | 264.6 |
| 2011EL-1141 | C | 2.3 | 200.3 |
| MO10 | C | 1.5 | 155.0 |
| 2012V-1001 | C | 3.0 | 124.6 |
| 3546-06 | C | 4.4 | 76.3 |
| 2011EL-1938 | C | 3.4 | 43.3 |
| 2009V-1096 | C | 2.8 | 35.6 |
| 2011EL-1137 | C | 2.2 | 31.9 |
| 2010V-1014 | C | 2.6 | 26.3 |
| 3500-05 | C | 2.7 | 20.4 |
| 2011EL-1939 | C | 4.8 | 16.5 |
| 2010EL-1786 | C | 2.4 | 11.0 |
C, clinical; E, environmental.
Only isolates that killed at >10-fold the level of C6706 T6SS− when induced are shown.
DISCUSSION
To understand the diversity present in this set of 53 environmental and clinical isolates in the absence of their genome sequences, the serogroups and CTX status of all isolates were obtained from previous publications (19, 33) or determined in this study by methods we have described previously (32). Most isolates belonged to the O1 serogroup, consistent with the context of their isolation. The majority of the clinical isolates were obtained from patients during epidemics, which are typically caused by O1 strains that carry the CT-encoding genes. Many of the environmental isolates were acquired from locations believed to be the sources of cholera outbreaks, increasing the likelihood that these isolates were also of the O1 serogroup. Principal-component analysis (PCA) of the entire data set, as well as clinical or environmental isolate data alone, was performed by using the following variables: year of isolation, serogroup, CTX status, chitinase activity, TF, and fold reduction of E. coli prey in killing assays. The Pearson correlation matrix did not show any significant correlation between the serogroup, a proxy for genetic relatedness, and any of the three phenotypes studied here when clinical and environmental isolate data were tested together or separately (see Table S3 in the supplemental material). For example, looking at isolates of serogroup O141 in Table 1, 3566-08 and 3568-07 are both deficient in transformation, but one is capable of constitutive contact-dependent bacterial killing while the other is not. The only significant correlation observed when testing clinical and environmental isolates together was between the clinical or environmental designation and CTX status (see Table S3, clinical and environmental, −0.778 bold values). This correlation is expected because clinical isolates are defined as causing disease in patients by CTX. There were no significant correlations between any variables when environmental isolates were tested alone (see Table S3, environmental only).
Chitin is the most abundant polymer in the ocean; therefore, utilization of this carbon source is likely advantageous for the proliferation of many aquatic microorganisms (44). Consistent with this, most of the V. cholerae isolates tested possessed chitinase activity, determined by the production of a zone of clearing on a plate containing chitin colloid, which requires the secreted chitinases ChiA-1 and ChiA-2 (22, 35). Only three isolates were unable to produce a zone of clearing, but each was still capable of utilizing a chitin crab shell fragment during transformation assays. Indeed, two of these three isolates, NCTC8457 and VC56, were also proficient at natural transformation, which requires chitin degradation for (GlcNAc)2–6 signal production. The ubiquity of chitin utilization among all of the isolates tested here bolsters the argument that the use of chitin as a carbon source is critical to V. cholerae survival and proliferation in its natural aquatic environment.
Natural transformation has been demonstrated in only a limited set of V. cholerae isolates (12, 14, 17, 29, 30). Analysis of V. cholerae isolates obtained during the Haiti epidemic that followed the 2010 earthquake confirmed the source of the outbreak and also revealed that these contemporary isolates were severely impaired in transformation, in contrast to C6706, A1552, and others isolated in 1991 (19). This study prompted us to determine the prevalence of natural transformation among a larger set of V. cholerae isolates. Here, we showed that transformation proficiency appears to be slightly more common among isolates from environmental sources than among clinical isolates. This perhaps suggests that V. cholerae isolates occupying aquatic reservoirs may maintain the genes necessary for transformation in order to take up DNA as an alternative nutrient source in relatively nutrient-poor environmental settings compared to the nutrient-rich gut of a human host (18, 45–47). As a whole, transformation proficiency was rare among all of the isolates tested (22.6%), but our collection of strains contained a majority of O1 isolates, suggesting that transformation may simply be uncommon within the O1 serogroup.
The inability to take up DNA and recombine it onto the chromosome may result from loss-of-function mutations in competence regulators, apparatus components, or recombination genes. Indeed, two of the transformation-deficient clinical isolates, MAK757 and CA401, were previously shown to carry mutations in the gene encoding the QS regulator HapR, which is required for natural competence (33). These isolates were complemented for transformation with a plasmid carrying the C6706 hapR gene under the control of a constitutive promoter (data not shown). A third clinical isolate deficient in transformation, 2012V-1001, carries an S50Y missense mutation in the DNA binding domain of HapR (48) on the basis of the publicly available genome sequence and independent sequence analysis and its transformation ability was also restored by the same hapR-encoding plasmid (data not shown).
It is also possible that transformation-deficient isolates have acquired a factor that impaired DNA uptake. Recently, Dalia et al. showed that the transformation deficiency of clinical strains isolated during the 2010 Haiti outbreak appeared to result from of a gain of function (20). Specifically, the Haiti isolates discussed in reference 19 carry a large integrative and conjugative element (VchInd5) on chromosome 2 that encodes a constitutively expressed periplasmic DNase. That DNase, IdeA, can degrade extracytoplasmic DNA, reducing the opportunity for uptake and subsequent recombination onto the chromosome and causing a severe decrease in natural transformation (20). Sequence analysis of clinical isolates from Bangladesh showed an increase in the prevalence of ideA from 0 to 60% of that in the genomes obtained from 2001 to 2011, with the most dramatic increase after 2005, suggesting a major acquisition event (20). We identified ideA and 1 kb of its flanking sequences with 100% identity in the published genomes of all of the clinical isolates collected after 2005 and tested here. Interestingly, 2010EL-1749, isolated in 2010, was noteworthy in that it carries the ideA gene, yet the strain remains transformation proficient. Further study may reveal differences in the expression of ideA in this isolate.
Genomic analysis of the T6SS of many members of the species V. cholerae has been conducted, but T6SS-mediated killing has been demonstrated in only a small group of isolates (21, 24, 25, 40). On the basis of results from a small number of isolates, it was proposed that constitutive T6SS activity is prevalent among environmental strains because of constant exposure to predators, while clinical pandemic strains tightly regulate T6SS (24). Consistent with this “pathoadaptive” hypothesis, we showed that only 4 of 29 clinical isolates were constitutively capable of killing E. coli prey in a contact-dependent manner indicative of a T6SS, while 22 of 24 environmental isolates killed prey under laboratory conditions without chitin induction. The only clinical isolate constitutively capable of killing as effectively as the C6706 maximum-killing strain was MZO-2, which is a CTX− nonpandemic O14 isolate, as observed in the PCA of clinical isolates alone (see Table S3, clinical only, −1.000 bold values). These results further validate the pathoadaptive hypothesis. In fact, all but one of the CTX− isolates exhibited constitutive bacterial killing, but of the 39 pandemic O1 or O139 isolates, only 5 showed both CTX and constitutive killing (Table 1). It is possible that nonpandemic clinical isolates like MZO-2 have maintained constitutive killing activity to overcome deficiencies resulting from a lack of CT.
It is important to note that although wild-type pandemic isolate C6706 does not express T6SS under laboratory conditions in the absence of chitin, expression of T6SS genes has been described in vivo (49, 50). Indeed, T6SS was shown to be important in the infection of both mice and rabbits (51, 52). It was recently shown that mucins, the main protein component of the mucus layer in the small intestine, which V. cholerae colonizes, are capable of causing this activation (53). Therefore, while pandemic isolates may not display constitutive contact-dependent bacterial killing, they can still upregulate this mechanism during colonization of both chitin in the environment and of the small intestine in a human host. In fact, 13 of 25 clinical isolates, including C6706, that did not show constitutive killing were induced by qstR and tfoX expression, which mimics chitin signaling (Table 3). These results suggest that these isolates have a fully functional yet regulated T6SS.
Genetic manipulation of the environmental isolates characterized here remains challenging. Environmental isolate 692-79, but not an in-frame isogenic ΔvasK mutant constructed by allelic exchange, exhibited contact-dependent killing of E. coli (Fig. 3). Thus, the observed killing of E. coli prey by 692-79 is indeed mediated by a T6SS. Additionally, sequencing results confirm that the 24 environmental isolates characterized here, like numerous sequenced V. cholerae isolates described previously (27), contain conserved T6SS-encoding genes. However, limited success with genetic manipulation of the majority of the environmental isolates characterized here suggests that studies of the T6SS of these isolates will require methods beyond those used in this survey.
The molecular mechanism by which environmental strains are T6SS constitutive while clinical pandemic strains do not express T6SS constitutively is still unclear. It has been proposed (24) that constitutive T6SS-mediated killing by environmental isolates may be advantageous because of constant competition from other bacteria and potential predators in the environment, while pandemic strains utilize other virulence mechanisms for competition in the human gut and therefore may not require a constitutively active T6SS. Our results support this hypothesis. It is intriguing to speculate that in nutrient-poor environmental settings, the DNA and nucleotides released from neighboring cells may be consumed by V. cholerae via T6SS activity. Perhaps constitutive expression of the T6SS is favored in bacteria that occupy niches like marine settings, where DNA may be a valuable food source, but costly for enteric pathogens like pandemic V. cholerae that have become better adapted to living in the human gut, where preferential nutrients are abundant.
As stated previously, when the complete set of data from both clinical and environmental isolates tested here was analyzed by PCA, there was no significant correlation between year, location of isolation, or serogroup and possession of each of the three phenotypes. However, by PCA of clinical isolates alone, a statistically significant correlation between constitutive bacterial killing and TF was found (see Table S3, clinical only, 0.681 bold values). Among the four clinical isolates that are capable of constitutive bacterial killing to some degree, three were also transformation proficient (Table 1). This relationship is not surprising because of our knowledge about their coordinate expression and regulation. NCTC8457 was collected >100 years ago and has retained its ability to incorporate DNA onto its chromosome and constitutively kill bacteria in a contact-dependent manner, suggesting that these phenotypes may provide an adaptive advantage. All but one of the more contemporary clinical isolates, collected in 2005 or later, have lost the ability to transform DNA and no longer constitutively kill, suggesting that they are relatively poor competitors in the environment. It is interesting to speculate that V. cholerae strains adapt to different niches and express genes for contact-dependent killing and transformation differently.
Chitinase activities are likely to provide a major growth advantage in aquatic environments, consistent with their prevalence in this set of isolates. On the other hand, more complex behaviors like transformation and T6SS-mediated bacterial killing are metabolically expensive; therefore, the prevalence and regulation of these phenotypes are highly variable, as described here. These survey data will inform future sequence analyses and genomewide association studies to help identify previously unknown regulators and structural components for both transformation and the T6SS.
Supplementary Material
ACKNOWLEDGMENTS
We thank Taylor Griswold for assistance with genome analysis of the sequenced clinical isolates and Hammer lab members for discussions and critical manuscript review. We also thank Jacob Thomas for constructing the pQT plasmid and the 692-79 ΔvasK mutant.
We thank Cheryl Bopp, Jun Zhu, Andy DePaulo, Douglas Bartlett, and Rita Colwell for providing us with the isolates tested in this survey.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00351-16.
REFERENCES
- 1.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A 99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukerjee S. 1963. The bacteriophage-susceptibility test in differentiating Vibrio cholerae and Vibrio El Tor. Bull World Health Organ 28:333–336. [PMC free article] [PubMed] [Google Scholar]
- 4.Han GK, Khie TS. 1963. A new method for the differentiation of Vibrio comma and Vibrio El Tor. Am J Hyg 77:184–186. [DOI] [PubMed] [Google Scholar]
- 5.Finkelstein RA, Mukerjee S. 1963. Hemagglutination—a rapid method for differentiating Vibrio cholerae and El Tor vibrios. Exp Biol Med 112:355–359. doi: 10.3181/00379727-112-28043. [DOI] [Google Scholar]
- 6.Barrett TJ, Blake PA. 1981. Epidemiological usefulness of changes in hemolytic activity of Vibrio cholerae biotype El Tor during the seventh pandemic. J Clin Microbiol 13:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JA, Salles CA, Panigrahi P, Albert MJ, Wright AC, Johnson RJ, Morris JG Jr. 1994. Vibrio cholerae O139 synonym bengal is closely related to Vibrio cholerae El Tor but has important differences. Infect Immun 62:2108–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB. 2003. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci U S A 100:1304–1309. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A 101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Roseman S. 2004. The chitinolytic cascade in vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A 101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto S, Morita M, Izumiya H, Watanabe H. 2010. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene 457:42–49. doi: 10.1016/j.gene.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. 2011. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol 193:1953–1965. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, Izumiya H. 2014. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol Microbiol 91:326–347. doi: 10.1111/mmi.12462. [DOI] [PubMed] [Google Scholar]
- 17.Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 5:e01028-13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Bernardy EE, Hammer BK, Miyashiro T. 2013. Competence and natural transformation in vibrios. Mol Microbiol 89:583–595. doi: 10.1111/mmi.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalia AB, Seed KD, Calderwood SB, Camilli A. 2015. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc Natl Acad Sci U S A 112:10485–10490. doi: 10.1073/pnas.1509097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 22.Watve SS, Thomas J, Hammer BK. 2015. CytR is a global positive regulator of competence, type VI secretion, and chitinases in Vibrio cholerae. PLoS One 10:e0138834. doi: 10.1371/journal.pone.0138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, Pukatzki S. 2012. Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One 7:e48320. doi: 10.1371/journal.pone.0048320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata ST, Unterweger D, Rudko SP, Pukatzki S. 2013. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog 9:e1003752. doi: 10.1371/journal.ppat.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyata ST, Kitaoka M, Wieteska L, Frech C, Chen N, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system: evaluating its role in the human disease cholera. Front Microbiol 1:117. doi: 10.3389/fmicb.2010.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonova ES, Hammer BK. 2011. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol Lett 322:68–76. doi: 10.1111/j.1574-6968.2011.02328.x. [DOI] [PubMed] [Google Scholar]
- 30.Suckow G, Seitz P, Blokesch M. 2011. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol 193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Gerner-Smidt P. 2011. Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis 17:2122–2129. doi: 10.3201/eid1711.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joelsson A, Liu Z, Zhu J. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun 74:1141–1147. doi: 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 35.Bhowmick R, Ghosal A, Chatterjee NS. 2007. Effect of environmental factors on expression and activity of chitinase genes of vibrios with special reference to Vibrio cholerae. J Appl Microbiol 103:97–108. doi: 10.1111/j.1365-2672.2006.03253.x. [DOI] [PubMed] [Google Scholar]
- 36.Hsu SC, Lockwood JL. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watve SS, Bernardy EE, Hammer BK. 2014. Vibrio cholerae: measuring natural transformation frequency. Curr Protoc Microbiol 35:6A.4.1–6A.4.12. doi: 10.1002/9780471729259.mc06a04s35. [DOI] [PubMed] [Google Scholar]
- 38.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antonova ES, Bernardy EE, Hammer BK. 2012. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol 86:1215–1231. doi: 10.1111/mmi.12054. [DOI] [PubMed] [Google Scholar]
- 40.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, Uhlin BE, Wai SN. 2012. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun 80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redfield RJ. 1993. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered 84:400–404. [DOI] [PubMed] [Google Scholar]
- 46.Dubnau D. 1999. DNA uptake in bacteria. Annu Rev Microbiol 53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 47.Stewart GJ, Carlson CA. 1986. The biology of natural transformation. Annu Rev Microbiol 40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- 48.De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2007. Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J Bacteriol 189:5683–5691. doi: 10.1128/JB.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma AT, Mekalanos JJ. 2010. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A 107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bachmann V, Kostiuk B, Unterweger D, Diaz-Satizabal L, Ogg S, Pukatzki S. 2015. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.