ABSTRACT
5-Aminolevulinic acid (ALA), a nonprotein amino acid involved in tetrapyrrole synthesis, has been widely applied in agriculture, medicine, and food production. Many engineered metabolic pathways have been constructed; however, the production yields are still low. In this study, several 5-aminolevulinic acid synthases (ALASs) from different sources were evaluated and compared with respect to their ALA production capacities in an engineered Corynebacterium glutamicum CgS1 strain that can accumulate succinyl-coenzyme A (CoA). A codon-optimized ALAS from Rhodobacter capsulatus SB1003 displayed the best potential. Recombinant strain CgS1/pEC-SB produced 7.6 g/liter ALA using a mineral salt medium in a fed-batch fermentation mode. Employing two-stage fermentation, 12.46 g/liter ALA was produced within 17 h, with a productivity of 0.73 g/liter/h, in recombinant C. glutamicum. Through overexpression of the heterologous nonspecific ALA exporter RhtA from Escherichia coli, the titer was further increased to 14.7 g/liter. This indicated that strain CgS1/pEC-SB-rhtA holds attractive industrial application potential for the future.
IMPORTANCE In this study, a two-stage fermentation strategy was used for production of the value-added nonprotein amino acid 5-aminolevulinic acid from glucose and glycine in a generally recognized as safe (GRAS) host, Corynebacterium glutamicum. The ALA titer represented the highest in the literature, to our knowledge. This high production capacity, combined with the potential easy downstream processes, made the recombinant strain an attractive candidate for industrial use in the future.
INTRODUCTION
As the common precursor of tetrapyrroles such as porphyrin, heme, vitamin B12, and chlorophyll, 5-aminolevulinic acid (ALA) has been reported to be effective in tumor-localizing and photodynamic therapy for various diseases (1–3). ALA can also be used as a selective biodegradable herbicide and insecticide or an adversity resistance and growth-accelerating agent in agriculture (4, 5).
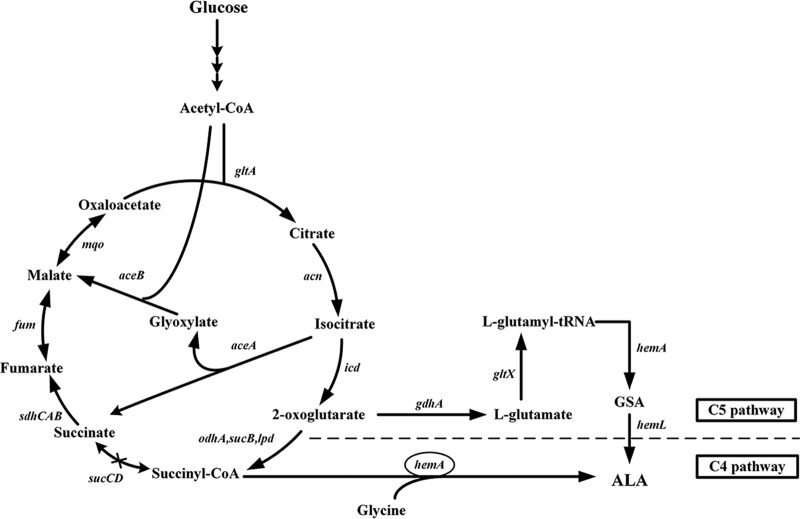
In living organisms, two kinds of metabolic pathways have been described for ALA biosynthesis (Fig. 1). One is the C5 pathway, which occurs in algae, higher plants, and many bacteria, including Escherichia coli and archaea. The C5 pathway involves the following three enzymatic activities: glutamyl-tRNA synthetase (GluRS) (encoded by gltX), a NADPH-dependent glutamyl-tRNA reductase (HemA, encoded by hemA), and a glutamate-1-semialdehyde aminotransferase (HemL, encoded by hemL). The other is the C4 pathway, which is present in birds, mammals, yeast, and purple non-sulfur-photosynthetic bacteria. In this pathway, ALA is formed through one-step catalysis by 5-aminolevulinic acid synthase (ALAS), which condenses glycine and succinyl-coenzyme A (CoA), an intermediate of the tricarboxylic acid (TCA) cycle.
FIG 1.
ALA biosynthesis pathway in C. glutamicum. The enzymes encoded by the corresponding genes are as follows: GdhA, glutamate dehydrogenase; GltX, glutamyl-tRNA synthetase; HemA, glutamyl-tRNA reductase; HemL, glutamate-1-semialdehyde aminotransferase (in the C5 pathway); HemA, 5-aminolevulinate synthase (in the C4 pathway). Cross in pathway, sucC and sucD, encoding succinyl-CoA synthetase, were deleted; circled gene, HemA in the C4 pathway was exogenously expressed.
In E. coli, the native pathway for ALA biosynthesis is the C5 pathway, which is tightly regulated by feedback inhibition of the end product heme (6). Previously, we developed a strategy to produce ALA in recombinant E. coli via the C5 pathway. Through overexpression of heterologous stabilized HemA from Salmonella arizona and HemL from E. coli, with the concomitant expression of an ALA exporter, the recombinant E. coli produced 4.13 g/liter ALA in modified minimal medium, using glucose as the sole carbon source (7). However, because the C5 pathway involves a number of enzymatic activities, utilizes ATP and NADPH as cofactors, and is dependent on tRNA-Glu, its complicated relationships with energy metabolism, oxidation-reduction states, and protein synthesis make subsequent strain improvement rather difficult (8, 9).
Recombinant E. coli expressing ALAS, the key enzyme of the C4 pathway, has attracted much attention due to its easy regulation and operation (10–13). In a recent study, a titer of 6.3 g/liter ALA was achieved in a 5-liter bioreactor with the expression of hemO from Rhodopseudomonas palustris in E. coli (14). A titer of 8.8 g/liter ALA was achieved in a 15-liter fermenter under optimized conditions, with the addition of glycine and succinate, using a recombinant strain harboring the ALAS of Rhodobacter capsulatus, i.e., E. coli Rosetta(DE3)/pET28a-R.C.hemA (15). The industrial application of ALA will be viable only when it is available in large quantities at competitive market prices. Hence, it is pressing to improve the titer of ALA and to reduce the costs of substrates and extraction to establish conditions favorable for industrial application. Moreover, E. coli has been shown to produce potentially toxic substances that are a cause of health and safety concerns (e.g., endotoxin and lipopolysaccharide) (16, 17). In view of the practical applications of ALA-based porphyrin drugs, as well as the wide use of ALA in agriculture and health care, the use of endotoxin-free Gram-positive bacteria is preferable.
As an aerobic, Gram-positive, nonsporulating, nonpathogenic bacterium with generally recognized as safe (GRAS) status, Corynebacterium glutamicum is widely used for large-scale industrial production of the flavor enhancer l-glutamate, the food additive l-lysine, and vitamins. Based on the extensive knowledge regarding its metabolism and regulation, together with the comprehensive tools available for its genetic manipulation, considerable research effort has been invested in the metabolic engineering of C. glutamicum for production of various biologically based chemicals, such as organic acids, diamines, and biofuels (18). Previously, fermentative production of ALA in C. glutamicum via the C5 pathway was demonstrated by our laboratory and the Han laboratory (19, 20). Very recently, Feng et al. reported the first attempt at using C. glutamicum for ALA production via the C4 pathway. With systematical metabolic engineering strategies, a titer of 7.53 g/liter ALA was achieved with 5-liter bioreactor fermentation (21).
In this paper, a recombinant C. glutamicum strain with high titer and productivity of ALA production via the C4 pathway, using both fed-batch fermentation and two-stage fermentation strategies, was demonstrated; 14.7 g/liter ALA was produced with a high productivity of 0.92 g/liter/h. This high production capacity, combined with the potential easy downstream process, made the recombinant strain an attractive candidate for industrial use in the future.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for cloning purposes and was propagated in Luria-Bertani (LB) medium at 37°C, with aeration. C. glutamicum ATCC 13032 and derivatives were propagated in BHIS medium (2.5 g/liter beef extract, 5 g/liter tryptone, 5 g/liter NaCl, 18.5 g/liter brain heart infusion, 91 g/liter sorbitol) at 30°C, with aeration. Kanamycin (Kan) was added at a final concentration of 25 μg/ml when necessary.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Wild type; subcloning host | TransGen Biotech |
| C. glutamicum ATCC 13032 | Wild type; biotin auxotrophic | ATCC |
| C. glutamicum CgS1 | C. glutamicum ATCC 13032 ΔsucCD | This study |
| C. glutamicum CgS1-ASV | ASV tag added to C terminus of ALAD in CgS1 | This study |
| Plasmids | ||
| pEC-XK99E | Kanr; E. coli-C. glutamicum shuttle vector | Laboratory stock |
| pEC-RS | Kanr; pEC-XK99E carrying gene for ALAS from R. sphaeroides 2.4.1 | This study |
| pEC-SB | Kanr; pEC-XK99E carrying gene for ALAS from R. capsulatus SB1003 | This study |
| pEC-GS | Kanr; pEC-XK99E carrying gene for ALAS from P. pastoris GS115 | This study |
| pEC-SC | Kanr; pEC-XK99E carrying gene for ALAS from S. cerevisiae s288c | This study |
| pEC-AT | Kanr; pEC-XK99E carrying gene for ALAS from A. tumefaciens C58 | This study |
| pEC-SB-gltA | pEC-SB carrying citrate synthase (gltA) from C. glutamicum ATCC 13032 additionally | This study |
| pEC-SB-icd | pEC-SB carrying isocitrate dehydrogenase (icd) from C. glutamicum ATCC 13032 additionally | This study |
| pEC-SB-odhA | pEC-SB carrying 2-oxoglutarate dehydrogenase (odhA-sucB-lpd) from C. glutamicum ATCC 13032 additionally | This study |
| pEC-SB-coaA | pEC-SB carrying pantothenate kinase (coaA) from C. glutamicum ATCC 13032 additionally | This study |
| pEC-SB-rhtA | pEC-SB carrying ALA exporter (rhtA) from E. coli MG1655 additionally | This study |
| pEC-SB-rhtA-coaA | pEC-SB carrying rhtA and coaA additionally | This study |
| pK-JL | pK19mobsacB derivative | 23 |
| pK-JLΔsucCD | Kanr; containing flanking regions of C. glutamicum sucCD genes | This study |
Plasmid construction.
The hemA gene of R. capsulatus SB1003, encoding ALA synthetase, was synthesized with codon optimization (GenBank accession no. KU687108). Other hemA genes were amplified by PCR with the genomic DNA of Rhodobacter sphaeroides 2.4.1 (GenBank accession no. ABA79145.1), Pichia pastoris GS115 (GenBank accession no. XM_002491600), Saccharomyces cerevisiae s288c (GenBank accession no. DAA12073.1), and Agrobacterium tumefaciens C58 (GenBank accession no. AAK88335.2) as the templates. The same ribosomal binding site (AAGGAGA) was added upstream of each gene. The PCR products with EcoRI and BamHI restriction sites at the proximal ends were cloned into the C. glutamicum expression vector pEC-XK99E digested with the same enzymes, resulting in pEC-SB, pEC-RS, pEC-GS, pEC-SC, and pEC-AT. The genes cg0949 (gltA), cg0766 (icd), cg1280-cg2421-cg0441 (odhA-sucB-lpd), and cg1132 (coaA) were amplified by PCR with the genomic DNA of C. glutamicum ATCC 13032 as the template, and rhtA was amplified by PCR with the genomic DNA of E. coli MG1655 as the template. Amplified PCR products were assembled into the vector pEC-SB digested with XbaI, resulting in plasmids pEC-SB-gltA, pEC-SB-icd, pEC-SB-odhA, pEC-SB-coaA, pEC-SB-rhtA, and pEC-SB-rhtA-coaA. The sequences of all constructed plasmids were confirmed by DNA sequencing.
Gene deletion.
The genes cg2837 (sucC) and cg2836 (sucD), encoding the two subunits of succinyl-CoA synthetase, were inactivated by in-frame deletion. A sucCD-deficient mutant of C. glutamicum ATCC 13032 was constructed with pK-JL, using the procedure described by Niebisch and Bott (22, 23). Briefly, upstream and downstream flanking regions of sucCD were amplified by PCR and assembled with SmaI-digested pK19mobsacB vector using the Gibson assembly method (24), yielding pK-JLΔsucCD. Transformation of wild-type C. glutamicum and screening for the first and second homologous recombination events were performed as described previously (25). Kanamycin-sensitive and sucrose-resistant clones were analyzed by PCR.
ALA fermentation.
For ALA batch and fed-batch fermentation, a single colony was inoculated into 5 ml BHIS medium and incubated at 180 rpm for 22 h at 30°C. The 5-ml preculture was then inoculated into a 300-ml baffled shake flask containing 50 ml modified CGXII medium; CGXII medium contained 20 g/liter (NH4)2SO4, 5 g/liter urea, 1 g/liter KH2PO4, 1 g/liter K2HPO4, 0.25 g/liter MgSO4·7H2O, 42 g/liter MOPS (3-morpholinopropanesulfonic acid), 10 mg/liter CaCl2, 10 mg/liter FeSO4·7H2O, 10 mg/liter MnSO4·H2O, 1 mg/liter ZnSO4·7H2O, 0.2 mg/liter CuSO4, 0.02 mg/liter NiCl2·6H2O, and 0.02 g/liter citrate sodium. The initial optical density at 600 nm (OD600) was controlled at about 0.5, and the glucose concentration was controlled at 40 g/liter; 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added at 0 h to induce the expression of plasmid genes. Fermentation was performed at 30°C and 180 rpm, and the pH was maintained at approximately 6.5 with 4 M NaOH throughout the fermentation. Glucose was added at 10 g/liter when its concentration was below 5 g/liter in fed-batch fermentation. The precursor glycine was added at 2 g/liter every 12 h, and 10 g/liter succinate was added when needed.
For two-stage ALA fermentation, a single colony was inoculated into 50 ml LB medium containing the appropriate antibiotics, and incubation was performed at 180 rpm for 24 h at 30°C. The preculture was then inoculated into a second culture (200 ml LB medium with 0.25 mM IPTG in a 1-liter baffled shake flask) to an OD600 of 0.4. The cells were harvested at an OD600 of 7 to 10 by centrifugation (6,000 rpm for 8 min at 4°C) and were resuspended in 50 ml solution buffer (20 mM sodium phosphate buffer unless stated otherwise) to an appropriate cell density in a 300-ml baffled shake flask. The initial glucose concentration was 40 g/liter, and glucose was added at 10 g/liter when its concentration was below 5 g/liter. The precursor glycine was added at 10 g/liter at the start, and 10 g/liter succinate was added when needed. Fermentation was performed at 30°C and 180 rpm, and the pH was maintained at approximately 6.5 with 4 M NaOH throughout the fermentation. Samples were taken at 1- to 2-h intervals for analysis.
Measurement of 5-aminolevulinic acid synthase activity in cell extracts.
Ten milliliters of fermentation broth from the 5 strains CgS1/pEC-SB, CgS1/pEC-GS, CgS1/pEC-SC, CgS1/pEC-AT, and CgS1/pEC-RS in CGXII medium at an OD600 of 7 to 8 was centrifuged (6,000 rpm at 4°C for 8 min). The cells were then washed in 50 mM Tris-HCl buffer (pH 7.5) and disrupted by sonication. The crude extracts were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatants were used as cell extracts. The assay mixture contained 50 μl of 1 mol/liter glycine, 50 μl of 2 mmol/liter succinyl-CoA, 6 μl of 1 mol/liter Tris buffer (pH 7.5), and 14 μl of 10 mmol/liter pyridoxal phosphate, as described previously (15). One unit of enzyme activity was defined as the amount of enzyme catalyzing the formation of 1 nmol ALA per min at 37°C. Protein concentrations were determined by the method of Bradford (26), using bovine serum albumin as a standard.
Analytical methods.
Optical density was measured at 600 nm with a spectrophotometer (Shimazu, Japan). Glucose, acetate, lactate, succinate, formate, malate, and citrate were quantitatively assessed by high-performance liquid chromatography (HPLC) (Shimadzu, Japan) with a refractive index detector (RID-10A; Shimadzu, Japan) and an Aminex HPX-87H ion exclusion column (Bio-Rad, USA), as described previously (27). Fumarate, isocitrate, glyoxylate, and α-ketoglutarate levels were determined by HPLC with a UV detector (SPD-20A; Shimadzu, Japan) and an Aminex HPX-87H ion exclusion column; 2.75 mM H2SO4 solution was applied to the column as a mobile phase, at a flow rate of 0.6 ml/min, at 50°C. Glycine was assessed by HPLC with a UV detector (SPD-20A) and an NH2 column (4.6 by 250 mm) packed with packing material with a 5-mm particle size (28). ALA levels were measured using modified Ehrlich's reagent after centrifugation of the cultures (8).
Nucleotide sequence accession number.
The hemA gene sequence was deposited in GenBank under accession number KU687108.
RESULTS
Construction of C. glutamicum ATCC 13032 mutant strain deficient in sucCD.
In C. glutamicum, succinyl-CoA synthetase, which catalyzes the reversible reaction of succinate + ATP + coenzyme A ↔ succinyl-CoA + ADP + phosphate, is encoded by two neighboring genes, i.e., sucC (cg2837) and sucD (cg2836). As reported previously, inactivation of sucCD would redirect succinyl-CoA from the TCA cycle to the ALA synthesis branch (29). Thus, as a first step for constructing an ALA production strain, sucCD was inactivated. In-frame deletion of the sucC and sucD genes was achieved with the use of a suicide vector, pK-JL, which enabled two rounds of positive-negative selection for homologous recombination events. The resulting strain was named CgS1. Compared with the wild-type strain, CgS1 showed a similar TCA cycle intermediate pattern (Table 2), with almost equal amounts of these intermediates, which may indicate activation of alternative pathways (e.g., the glyoxylate cycle and the α-ketoglutarate decarboxylase pathway) (30). The CgS1 mutant exhibited an only slightly reduced specific growth rate (0.31 h−1, compared with a wild-type value of 0.35 h−1), which may be beneficial for industrial use.
TABLE 2.
Determination of TCA intermediate levels in wild-type C. glutamicum and CgS1a
| Strain | Intermediate level (g/liter) |
||||||
|---|---|---|---|---|---|---|---|
| Citrate | Isocitrate | α-Ketoglutarate | Succinate | Fumarate | Malate | Glyoxylate | |
| Wild type | 1.25 ± 0.09 | 1.62 ± 0.21 | 0.63 ± 0.11 | 0.13 ± 0.03 | 0.15 ± 0.02 | 0.22 ± 0.03 | 0.34 ± 0.02 |
| CgS1 | 1.16 ± 0.1 | 1.42 ± 0.13 | 0.54 ± 0.09 | 0.13 ± 0.02 | 0.16 ± 0.02 | 0.25 ± 0.01 | 0.36 ± 0.01 |
Wild-type C. glutamicum and CgS1 strains were cultured in modified CGXII medium for 24 h, and fermentation supernatants were analyzed for related intermediates.
To assess the effects of sucCD inactivation on ALA production, we performed ALA fermentation using the wild-type strain or the mutant harboring the ALAS of R. sphaeroides 2.4.1 (Fig. 2). In the wild-type strain, a low titer of ALA (25.1 mg/liter) was achieved, perhaps due to a shortage of the succinyl-CoA precursor, which was readily converted to other TCA cycle metabolites. Succinate addition improved the ALA titer to 44.61 mg/liter. This improvement may be due to conversion of succinate to succinyl-CoA by the reversible SucCD. The sucCD inactivation led to an ALA titer 3.7-fold greater than that of the wild-type strain (92.87 mg/liter versus 25.1 mg/liter), an indication of redirection of succinyl-CoA into ALA synthesis. However, succinate addition to the mutant did not result in a further improved titer. In a sucCD deletion scenario, added succinate could turn into succinyl-CoA via the forward TCA cycle, but the decreased ALA titer indicated that this pathway was inefficient under these conditions.
FIG 2.
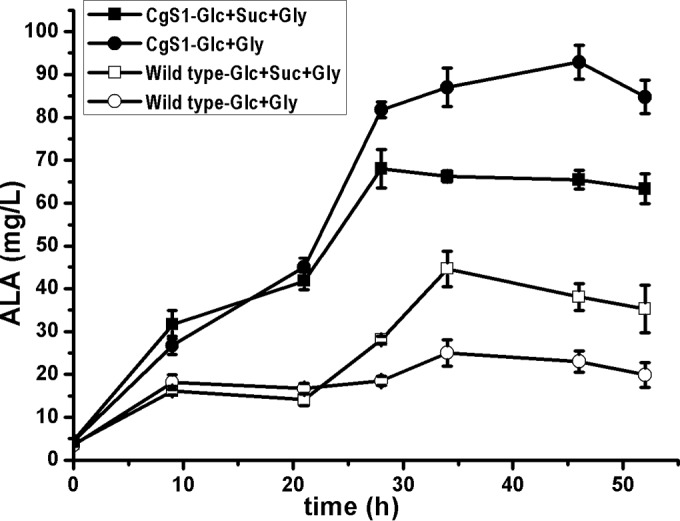
Time course of ALA production by recombinant C. glutamicum strains. Wild-type C. glutamicum and CgS1 harboring the ALAS of R. sphaeroides 2.4.1 were cultured in a 300-ml baffled shake flask containing 50 ml modified CGXII medium, at 30°C and 180 rpm, and the pH was maintained at approximately 6.5. The initial glucose (Glc) concentration was 40 g/liter. The precursor glycine (Gly) was added at 2 g/liter every 12 h, and 10 g/liter succinate (Suc) was added when needed. Results are averages from three independent experiments, with standard deviations indicated by error bars.
Screen of ALASs for ALA production in CgS1.
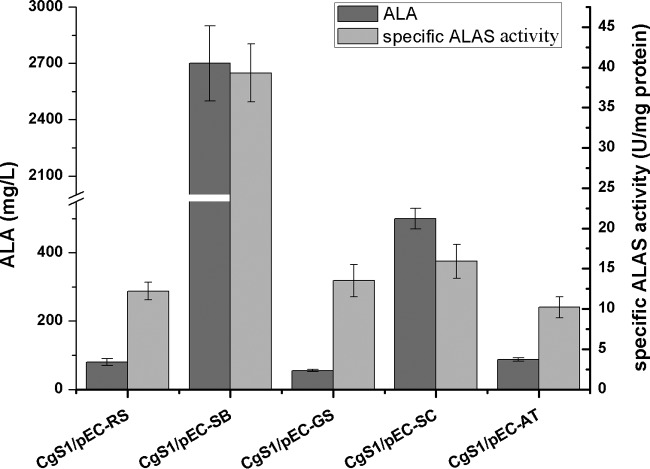
ALAS catalyzes a one-step condensation of succinyl-CoA and glycine to form ALA and is the key enzyme involved in the C4 pathway. Since C. glutamicum does not possess an ALAS naturally, a heterologous ALAS must be introduced to complement the C4 pathway. Different ALASs could show wide differences in solubility, gene expression levels, and mRNA stability, resulting in distinctive enzyme specific activities, which ultimately determine ALA production. With this in mind, four ALASs (from R. capsulatus SB1003, P. pastoris GS115, S. cerevisiae s288c, and A. tumefaciens C58) in addition to the one from R. sphaeroides 2.4.1 were selected and investigated in strain CgS1; the resulting strains harboring ALASs from different sources were named CgS1/pEC-SB, CgS1/pEC-GS, CgS1/pEC-SC, CgS1/pEC-AT, and CgS1/pEC-RS, respectively. The ALAS from R. capsulatus SB1003 displayed the best potential for ALA production, with an ALA titer of 2.7 g/liter (Fig. 3). Consistently, enzyme activity measurements confirmed that the specific ALAS activity of strain CgS1/pEC-SB was the highest (39.3 ± 3.6 U/mg protein). However, the other four enzymes displayed similar specific ALAS activities in recombinant CgS1, indicating that different metabolic perturbations may be brought about by overexpression of different ALASs.
FIG 3.
Comparison of ALASs from different sources. CgS1 strains harboring different ALASs were compared for ALA production in batch fermentation performed under the same conditions as in Fig. 2. Cell extracts of the five strains were used to determine ALAS activity as described previously (15). Results are averages from three independent experiments, with standard deviations indicated by error bars.
Fed-batch fermentation with CgS1/pEC-SB for ALA production.
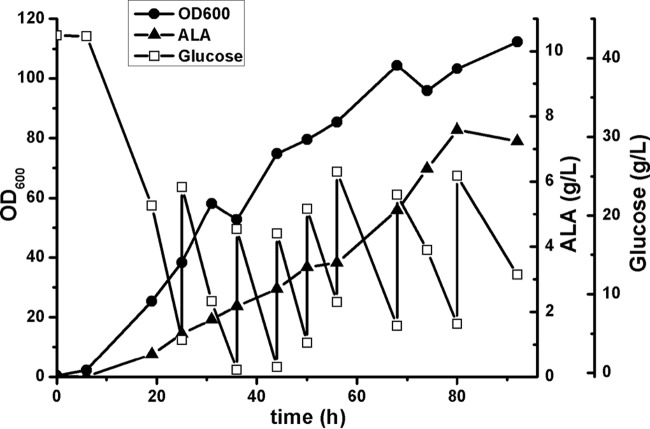
As the best potential producer, CgS1/pEC-SB was employed for fed-batch fermentation in a 1-liter baffled shake flask containing 100 ml modified CGXII medium. The OD600 gradually increased throughout the fermentation, and the ALA titer increased concomitantly, reaching 7.6 g/liter at 80 h (Fig. 4). At the end of the fermentation, 79.22 g/liter glucose was consumed. In analysis of the fermentation broth, very small amounts of by-products were found in the supernatant (total organic acid levels of less than 2 g/liter), indicating conversion of the carbon source to biomass (OD600 of 103.24) and CO2. This led to a low ALA yield of 0.096 g/g glucose.
FIG 4.
Fed-batch fermentation profile of CgS1/pEC-SB for ALA production. Fermentation was performed in a 1-liter baffled shake flask containing 100 ml modified CGXII medium, at 30°C and 180 rpm, and the pH was maintained at approximately 6.5. The initial glucose concentration was about 40 g/liter. The precursor glycine was added at 2 g/liter every 12 h. For clarity, a representative curve from three independent experiments is shown, with standard differences (relative standard deviations [RSDs]) of less than 10%.
Two-stage fermentation with CgS1/pEC-SB for improved ALA production.
Two-stage fermentation has been used to produce 2,3-butanediol, succinate, and cyclohexanone derivatives in C. glutamicum (31–34). In this strategy, a first stage of biomass production is followed by a second stage of product fermentation. The decoupling of growth and product synthesis leads to a high yield and productivity; furthermore, the simple buffer system is conducive to the downstream separation and purification of products.
After growth, cells were transferred into fermentation buffer supplied with glucose and glycine for the second stage of fermentation. As shown in Table 3, CgS1/pEC-SB showed an ALA titer of 7.49 g/liter at 15 h. Succinate addition slightly decreased the titer, indicating the inefficiency of conversion of succinate to succinyl-CoA in strain CgS1. This titer was comparable to that observed with a fed-batch fermentation strategy (7.49 g/liter versus 7.6 g/liter), but only 15 h was needed. This yielded a 4.26-fold improvement in productivity (0.5 g/liter/h) when two-stage fermentation was employed. In both cases, total organic acid by-product levels were less than 2 g/liter.
TABLE 3.
Two-stage fermentation for ALA production in an engineered C. glutamicum straina
| Strain | Substrates | ALA titer (g/liter) | ALA productivity (g/liter/h) | Yield (mol/mol) |
|
|---|---|---|---|---|---|
| ALA/glucose | ALA/glycine | ||||
| CgS1/pEC-SB | Glucose, succinate, glycine | 6.82 ± 0.11 | 0.45 ± 0.007 | 0.31 ± 0.01 | 0.69 ± 0.06 |
| Glucose, glycine | 7.49 ± 0.09 | 0.50 ± 0.006 | 0.35 ± 0.02 | 0.51 ± 0.04 | |
The results presented are means ± standard deviations from three independent experiments.
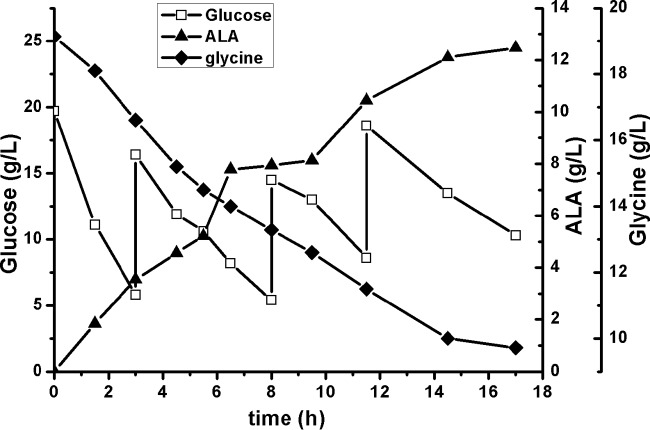
To improve the overall titer and to decrease the cost, we optimized the fermentation conditions. Using the optimized conditions, 12.46 g/liter ALA was produced within 17 h, with a productivity of 0.73 g/liter/h in the accumulation stage, which was 66% improved in comparison with the nonoptimized fermentation (Fig. 5). In total, 36.6 g/liter glucose or 9.42 g/liter glycine was consumed, giving an ALA yield of 0.47 mol ALA/mol glucose or 0.76 mol ALA/mol glycine, respectively.
FIG 5.
Two-stage fermentation profile of CgS1/pEC-SB for ALA production under optimized conditions. Fermentation was performed in a 300-ml baffled shake flask containing 50 ml 2 mM MgSO4 in 250 mM potassium phosphate buffer, at 30°C and 180 rpm, and the pH was maintained at approximately 6.5. The initial glucose and glycine concentrations were ∼20 g/liter. For clarity, a representative curve from three independent experiments is shown, with standard differences (RSDs) of less than 10%.
Metabolic flux redirection for further increases in ALA production.
Metabolic flux redirection, including enhancing product pathway flux and minimizing off-pathway and competitive pathway flux, has proven crucial for increasing target product titers in metabolic engineering. To improve ALA production in CgS1/pEC-SB, we rebalanced the pathway flux by increasing the succinyl-CoA supply, diminishing downstream drain and increasing export of ALA.
First, we tried to increase the flux through the forward TCA cycle, and thus the supply of succinyl-CoA, by overexpressing citrate synthase (gltA), isocitrate dehydrogenase (icd), or 2-oxoglutarate dehydrogenase (odhA-sucB-lpd), together with the R. capsulatus ALAS (hemA), to form a series of polycistronic plasmids, namely, pEC-SB-gltA, pEC-SB-icd, and pEC-SB-odhA, as shown in Table 1. Pantothenate kinase (coaA), which was reported previously to be important for CoA synthesis, was also overexpressed (pEC-SB-coaA) (35). As a result, gltA overexpression substantially decreased ALA production (by 30%), with decreased glucose consumption (23 g/liter versus 36 g/liter), compared with CgS1/pEC-SB; odhA-sucB-lpd overexpression increased ALA production by about 6%, icd overexpression resulted in no obvious improvement, and coaA overexpression resulted in a moderate improvement of 10% to 13.7 g/liter (Fig. 6). We then tried to diminish downstream flux by downregulating the enzymatic activity of ALA dehydratase (ALAD) (encoded by hemB, the first enzyme downstream of ALA in the heme synthesis pathway) through addition of an ASV degradation tag (AAEKSQRDYAASV) to the C terminus of ALAD, to increase its degradation rate (20). This did not improve the titer, however, which was different from our results for the C5 pathway of ALA biosynthesis (Fig. 6) (20). Finally, we overexpressed the heterologous nonspecific ALA exporter RhtA from E. coli together with the R. capsulatus ALAS (hemA) (7). This modification increased the ALA titer from 12.46 g/liter to 14.7 g/liter in 16 h, resulting in a high productivity of 0.92 g/liter/h (Fig. 6).
FIG 6.
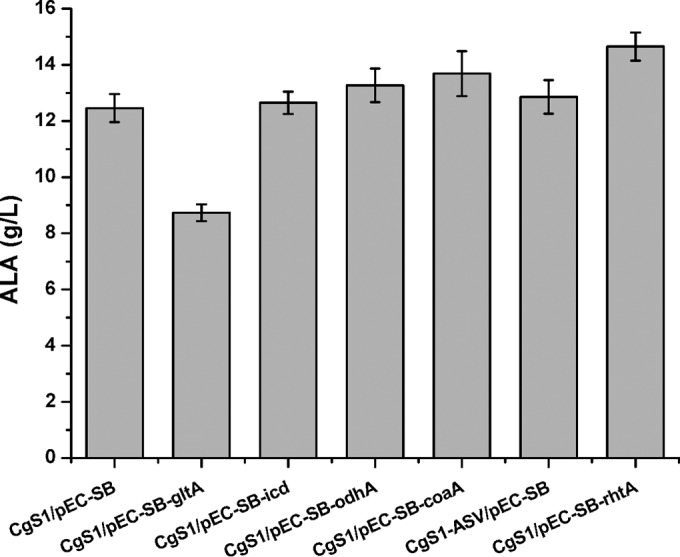
ALA production by two-stage fermentation in various recombinant C. glutamicum strains. Fermentation was performed under the same conditions as in Fig. 5. Results are averages from three independent experiments, with standard deviations indicated by error bars.
Encouraged by these results, we overexpressed coaA and rhtA together with the R. capsulatus ALAS (hemA) to construct pEC-SB-rhtA-coaA. However, this trial did not increase the titer further (data not shown).
DISCUSSION
The application of ALA in various fields has led to increasing demand for ALA production. Compared with E. coli, the most widely used host for microbial production of ALA, the GRAS organism C. glutamicum holds certain advantages concerning the broad use of ALA in agriculture and health care. In this study, two-stage high-yield ALA production in recombinant C. glutamicum via the C4 pathway was demonstrated.
In the C4 pathway, ALA is formed through one-step condensation of glycine and succinyl-CoA by ALAS. Most studies on ALA production required the addition of glycine and succinate as precursors, resulting in high production costs. Recently, Ma and colleagues investigated ALA production in sdhAB- and sucCD-deficient E. coli strains without the addition of succinate and improved ALA titers by 25.59% and 12.40%, respectively (29). We deleted sucCD from C. glutamicum ATCC 13032 in a similar way, resulting in strain CgS1. This modification also made succinate addition unnecessary (Fig. 2). Apparently, sucCD inactivation did not impair cell growth. Levels of TCA intermediate accumulation in CgS1 and the wild-type strain were essentially the same. All of these results indicated that the disturbed TCA cycle was rescued by alternative pathways in CgS1 (30).
Since C. glutamicum does not possess an ALAS naturally, a heterologous ALAS must be introduced to complement the C4 pathway. Of the five ALASs investigated, the one from R. capsulatus SB1003 displayed the best potential for ALA production. Recently, three ALASs, from Agrobacterium radiobacter, R. sphaeroides, and R. capsulatus, were purified and characterized (15). The specific activity of R. capsulatus ALAS (198.2 U/mg) was about 31.2% higher than that of A. radiobacter ALAS (151.1 U/mg) and 69.5% higher than that of R. sphaeroides ALAS (116. 9 U/mg). In vivo, the level of ALAS was tightly regulated at the level of either transcription or enzymatic activity (36–38). Thus, extensive screening of ALASs with higher activity, combined with expression optimization and protein evolution, may facilitate the identification of a better catalyzer in follow-up studies (10, 11, 39–43).
In a trial to increase the supply of succinyl-CoA, odhA-sucB-lpd and coaA overexpression increased ALA titers moderately, by 6% and 10%, respectively, while gltA overexpression substantially reduced titers. OdhA-SucB-Lpd catalyzes the reaction for succinyl-CoA synthesis directly, and CoaA increases the CoA substrate supply for this reaction; odhA-sucB-lpd and coaA overexpression improved ALA production but icd overexpression did not, which indicated an off-pathway flux through 2-oxoglutarate. Because organic acid by-products were very limited (total amounts of less than 2 g/liter) in the fermentation supernatant from CgS1/pEC-SB-gltA, we speculated that gltA overexpression may result in an imbalance between the reverse and forward TCA cycles and thus impaired glucose decomposition in a sucCD deletion scenario. Accelerated degradation of ALAD through addition of the ASV tag to the C terminus of ALAD did not increase the titer, which is different from the results reported previously for the C5 pathway. This indicated that the downstream flux was limited, compared with ALA synthesis, and the heterologous HemA may be less feedback inhibited by downstream tetrapyrroles. Heterologous expression of rhtA from E. coli proved effective for ALA production in C. glutamicum, but simple overexpression of coaA and rhtA with hemA did not result in additive effects, which indicated that further optimization for combinational expression of related genes is needed.
In the past 2 decades, much attention has been paid to cultivation process optimization, medium composition investigation, and ALAS expression and pathway optimization for ALA production (44, 45). As a widely used and efficient strategy, two-stage fermentation was employed in this study for ALA production by nongrowing cells, with related parameters being optimized by uniformity design. In two-stage fermentation, production of the target compound is decoupled from cell growth, so cell resources can be more dedicated to product synthesis. Also, potential inhibitors (e.g., heme in this ALA production case) are removed after the cell growth stage and, due to cessation of cell growth in the production stage, accumulation of these inhibitors may be much less. Finally, the simple buffer system is conducive to the downstream processes. Notably, the output of ALA here was nearly 1.67-fold greater than that from E. coli Rosetta(DE3)/pET28a-hemA (15), although the latter has been optimized and scaled up in a fermenter with added succinate. During preparation of the manuscript, production of ALA in C. glutamicum via the C4 pathway, with titers of 3.14 g/liter in a shake flask and 7.53 g/liter in a 5-liter bioreactor, was reported (21). Those authors demonstrated how metabolic engineering improved strain capacity through elimination of competitive pathways, increases in precursor supply, modification of cell permeability, and expression of an ALA exporter (21). With system biology and synthetic biology tools, CgS1 may be engineered to improve its production capacity further in the future.
In conclusion, this work demonstrated ALA production in C. glutamicum with high titers and productivity. Recombinant C. glutamicum CgS1/pEC-SB-rhtA produced 14.7 g/liter ALA with a volumetric productivity of 0.92 g/liter/h in a two-stage fermentation mode. This study paves the way for development of low-cost processes for production of ALA from glucose and glycine using a GRAS organism.
ACKNOWLEDGMENTS
This work was funded by grants from the National Natural Science Foundation of China (grant 31370085), the National High-Tech Research and Development Plan of China (grant 2012AA022104), and the Independent Innovation and Achievements Transformation of Shandong (grant 201422CX02602).
REFERENCES
- 1.Besur S, Hou W, Schmeltzer P, Bonkovsky HL. 2014. Clinically important features of porphyrin and heme metabolism and the porphyrias. Metabolites 4:977–1006. doi: 10.3390/metabo4040977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tetard MC, Vermandel M, Mordon S, Lejeune JP, Reyns N. 2014. Experimental use of photodynamic therapy in high grade gliomas: a review focused on 5-aminolevulinic acid. Photodiagnosis Photodyn Ther 11:319–330. doi: 10.1016/j.pdpdt.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa T, Kajimoto Y, Inoue Y, Ikegami Y, Kuroiwa T. 2015. Critical role of ABCG2 in ALA-photodynamic diagnosis and therapy of human brain tumor. Adv Cancer Res 125:197–216. doi: 10.1016/bs.acr.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki K, Watanabe M, Tanaka T, Tanaka T. 2002. Biosynthesis, biotechnological production and applications of 5-aminolevulinic acid. Appl Microbiol Biotechnol 58:23–29. doi: 10.1007/s00253-001-0858-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang ZJ, Li HZ, Zhou WJ, Takeuchi Y, Yoneyama K. 2006. Effect of 5-aminolevulinic acid on development and salt tolerance of potato (Solanum tuberosum L.) microtubers in vitro. Plant Growth Regul 49:27–34. [Google Scholar]
- 6.Woodard SI, Dailey HA. 1995. Regulation of heme biosynthesis in Escherichia coli. Arch Biochem Biophys 316:110–115. doi: 10.1006/abbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 7.Kang Z, Wang Y, Gu P, Wang Q, Qi Q. 2011. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab Eng 13:492–498. doi: 10.1016/j.ymben.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Wang Y, Gong K, Wang Q, Liang Q, Qi Q. 2014. Constitutive expression of RyhB regulates the heme biosynthesis pathway and increases the 5-aminolevulinic acid accumulation in Escherichia coli. FEMS Microbiol Lett 350:209–215. doi: 10.1111/1574-6968.12322. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Kang Z, Chen J, Du G. 2015. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci Rep 5:8584. doi: 10.1038/srep08584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu XX, Wang L, Wang YJ, Cai LL. 2010. d-Glucose enhanced 5-aminolevulinic acid production in recombinant Escherichia coli culture. Appl Biochem Biotechnol 160:822–830. doi: 10.1007/s12010-009-8608-x. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Fu W, Cen P. 2009. Characterization of 5-aminolevulinate synthase from Agrobacterium radiobacter, screening new inhibitors for 5-aminolevulinate dehydratase from Escherichia coli and their potential use for high 5-aminolevulinate production. Bioresour Technol 100:2293–2297. doi: 10.1016/j.biortech.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Fu W, Lin J, Cen P. 2008. Enhancement of 5-aminolevulinate production with recombinant Escherichia coli using batch and fed-batch culture system. Bioresour Technol 99:4864–4870. doi: 10.1016/j.biortech.2007.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Fu W, Lin J, Cen P. 2007. 5-Aminolevulinate production with recombinant Escherichia coli using a rare codon optimizer host strain. Appl Microbiol Biotechnol 75:777–782. doi: 10.1007/s00253-007-0887-y. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Chen J, Chen N, Sun J, Zheng P, Ma Y. 2013. Cloning of two 5-aminolevulinic acid synthase isozymes HemA and HemO from Rhodopseudomonas palustris with favorable characteristics for 5-aminolevulinic acid production. Biotechnol Lett 35:763–768. doi: 10.1007/s10529-013-1143-4. [DOI] [PubMed] [Google Scholar]
- 15.Lou JW, Zhu L, Wu MB, Yang LR, Lin JP, Cen PL. 2014. High-level soluble expression of the hemA gene from Rhodobacter capsulatus and comparative study of its enzymatic properties. J Zhejiang Univ Sci B 15:491–499. doi: 10.1631/jzus.B1300283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA, Corchero JL, Villaverde A, Schaffer L, Head SR, Souvignier C, Meredith TC, Woodard RW. 2015. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Fact 14:57. doi: 10.1186/s12934-015-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klena J, Zhang P, Schwartz O, Hull S, Chen T. 2005. The core lipopolysaccharide of Escherichia coli is a ligand for the dendritic-cell-specific intercellular adhesion molecule nonintegrin CD209 receptor. J Bacteriol 187:1710–1715. doi: 10.1128/JB.187.5.1710-1715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker J, Wittmann C. 2012. Bio-based production of chemicals, materials and fuels: Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Ramzi AB, Hyeon JE, Kim SW, Park C, Han SO. 2015. 5-Aminolevulinic acid production in engineered Corynebacterium glutamicum via C5 biosynthesis pathway. Enzyme Microb Technol 81:1–7. doi: 10.1016/j.enzmictec.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Jin H, Liu W, Wang Q, Qi Q. 2015. Engineering Corynebacterium glutamicum to produce 5-aminolevulinic acid from glucose. Microb Cell Fact 14:183. doi: 10.1186/s12934-015-0364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng L, Zhang Y, Fu J, Mao Y, Chen T, Zhao X, Wang Z. 2015. Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid. Biotechnol Bioeng doi: 10.1002/bit.25886. [DOI] [PubMed] [Google Scholar]
- 22.Niebisch A, Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch Microbiol 175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- 23.Jiang LY, Chen SG, Zhang YY, Liu JZ. 2013. Metabolic evolution of Corynebacterium glutamicum for increased production of l-ornithine. BMC Biotechnol 13:47. doi: 10.1186/1472-6750-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rittmann D, Schaffer S, Wendisch VF, Sahm H. 2003. Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch Microbiol 180:285–292. doi: 10.1007/s00203-003-0588-6. [DOI] [PubMed] [Google Scholar]
- 26.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Li M, Zhang X, Yang P, Liang Q, Qi Q. 2013. A novel whole-phase succinate fermentation strategy with high volumetric productivity in engineered Escherichia coli. Bioresour Technol 149:333–340. doi: 10.1016/j.biortech.2013.09.077. [DOI] [PubMed] [Google Scholar]
- 28.Chin D, Achari RG. 1982. Analysis of glycine in antiperspirant products by HPLC. J Soc Cosmet Chem 33:359–362. [Google Scholar]
- 29.Pu W, Chen J, Sun C, Chen N, Sun J, Zheng P, Ma Y. 2013. Deficiency of succinic dehydrogenase or succinyl-CoA synthetase enhances the production of 5-aminolevulinic acid in recombinant Escherichia coli. Sheng Wu Gong Cheng Xue Bao 29:1494–1503. (In Chinese.) [PubMed] [Google Scholar]
- 30.Kind S, Becker J, Wittmann C. 2013. Increased lysine production by flux coupling of the tricarboxylic acid cycle and the lysine biosynthetic pathway: metabolic engineering of the availability of succinyl-CoA in Corynebacterium glutamicum. Metab Eng 15:184–195. doi: 10.1016/j.ymben.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Rados D, Carvalho AL, Wieschalka S, Neves AR, Blombach B, Eikmanns BJ, Santos H. 2015. Engineering Corynebacterium glutamicum for the production of 2,3-butanediol. Microb Cell Fact 14:171. doi: 10.1186/s12934-015-0362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun JY, Lee JE, Yang KM, Cho S, Kim A, Kwon YU, Park JB. 2012. Ethambutol-mediated cell wall modification in recombinant Corynebacterium glutamicum increases the biotransformation rates of cyclohexanone derivatives. Bioprocess Biosyst Eng 35:211–216. doi: 10.1007/s00449-011-0594-z. [DOI] [PubMed] [Google Scholar]
- 33.Doo EH, Lee WH, Seo HS, Seo JH, Park JB. 2009. Productivity of cyclohexanone oxidation of the recombinant Corynebacterium glutamicum expressing chnB of Acinetobacter calcoaceticus. J Biotechnol 142:164–169. doi: 10.1016/j.jbiotec.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MJ, Kim HJ, Lee JY, Kwon AS, Jun SY, Kang SH, Kim P. 2013. Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J Microbiol Biotechnol 23:668–673. doi: 10.4014/jmb.1302.02022. [DOI] [PubMed] [Google Scholar]
- 36.Fales L, Nogaj L, Zeilstra-Ryalls J. 2002. Analysis of the upstream sequences of the Rhodobacter sphaeroides 2.4.1 hemA gene: in vivo evidence for the presence of two promoters that are both regulated by fnrL. Photosynth Res 74:143–151. doi: 10.1023/A:1020947308227. [DOI] [PubMed] [Google Scholar]
- 37.Ranson-Olson B, Zeilstra-Ryalls JH. 2008. Regulation of the Rhodobacter sphaeroides 2.4.1 hemA gene by PrrA and FnrL. J Bacteriol 190:6769–6778. doi: 10.1128/JB.00828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Q, Li T, Hou W, Zheng J, Schrum LW, Bonkovsky HL. 2011. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J Biol Chem 286:26424–26430. doi: 10.1074/jbc.M110.215772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng Q, Zhang Y, Ma C, Ma H, Zhao X, Chen T. 2015. Purification and functional characterization of thermostable 5-aminolevulinic acid synthases. Biotechnol Lett 37:2247–2253. doi: 10.1007/s10529-015-1903-4. [DOI] [PubMed] [Google Scholar]
- 40.Misumi K, Sugiura T, Yamaguchi S, Mori T, Kamei I, Hirai H, Kawagishi H, Kondo R. 2011. Cloning and transcriptional analysis of the gene encoding 5-aminolevulinic acid synthase of the white-rot fungus Phanerochaete sordida YK-624. Biosci Biotechnol Biochem 75:178–180. doi: 10.1271/bbb.100674. [DOI] [PubMed] [Google Scholar]
- 41.Choi HP, Hong JW, Rhee KH, Sung HC. 2004. Cloning, expression, and characterization of 5-aminolevulinic acid synthase from Rhodopseudomonas palustris KUGB306. FEMS Microbiol Lett 236:175–181. doi: 10.1111/j.1574-6968.2004.tb09644.x. [DOI] [PubMed] [Google Scholar]
- 42.Page MD, Ferguson SJ. 1994. Differential reduction in soluble and membrane-bound c-type cytochrome contents in a Paracoccus denitrificans mutant partially deficient in 5-aminolevulinate synthase activity. J Bacteriol 176:5919–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi C, Hong BS, Sung HC, Lee HS, Kim JH. 1999. Optimization of extracellular 5-aminolevulinic acid production from Escherichia coli transformed with ALA synthase gene of Bradyrhizobium japonicum. Biotechnol Lett 21:551–554. doi: 10.1023/A:1005520007230. [DOI] [Google Scholar]
- 44.Kang Z, Zhang J, Zhou J, Qi Q, Du G, Chen J. 2012. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv 30:1533–1542. doi: 10.1016/j.biotechadv.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Zhang G, Li X, Zhang J. 2014. Microbial production and applications of 5-aminolevulinic acid. Appl Microbiol Biotechnol 98:7349–7357. doi: 10.1007/s00253-014-5925-y. [DOI] [PubMed] [Google Scholar]