Abstract
Nitrate-dependent ferrous iron [Fe(II)] oxidation (NDFO) is a well-recognized chemolithotrophic pathway in anoxic sediments. The neutrophilic chemolithoautotrophic enrichment culture KS originally obtained from a freshwater sediment (K. L. Straub, M. Benz, B. Schink, and F. Widdel, Appl Environ Microbiol 62:1458–1460, 1996) has been used as a model system to study NDFO. However, the primary Fe(II) oxidizer in this culture has not been isolated, despite extensive efforts to do so. Here, we present a metagenomic analysis of this enrichment culture in order to gain insight into electron transfer pathways and the roles of different bacteria in the culture. We obtained a near-complete genome of the primary Fe(II) oxidizer, a species in the family Gallionellaceae, and draft genomes from its flanking community members. A search of the putative extracellular electron transfer pathways in these genomes led to the identification of a homolog of the MtoAB complex [a porin-multiheme cytochrome c system identified in neutrophilic microaerobic Fe(II)-oxidizing Sideroxydans lithotrophicus ES-1] in a Gallionellaceae sp., and findings of other putative genes involving cytochrome c and multicopper oxidases, such as Cyc2 and OmpB. Genome-enabled metabolic reconstruction revealed that this Gallionellaceae sp. lacks nitric oxide and nitrous oxide reductase genes and may partner with flanking populations capable of complete denitrification to avoid toxic metabolite accumulation, which may explain its resistance to growth in pure culture. This and other revealed interspecies interactions and metabolic interdependencies in nitrogen and carbon metabolisms may allow these organisms to cooperate effectively to achieve robust chemolithoautotrophic NDFO. Overall, the results significantly expand our knowledge of NDFO and suggest a range of genetic targets for further exploration.
INTRODUCTION
Iron is the fourth most abundant element in the earth's crust. Microbes are important participants in iron redox cycling by oxidizing ferrous iron [Fe(II)] and reducing ferric iron [Fe(III)]. Both aqueous and solid-phase Fe(II) compounds can serve as an electron donor for chemolithotrophic Fe(II)-oxidizing bacteria (FeOB) under both oxic and anoxic conditions. Most FeOB are affiliated with Proteobacteria, and based on their physiology, they can be grouped into acidophilic aerobic, neutrophilic microaerophilic, neutrophilic anaerobic phototrophic, and neutrophilic anaerobic nitrate-reducing Fe(II) oxidizers (1).
Nitrate-dependent Fe(II) oxidation (NDFO) under anoxic circumneutral pH conditions is a well-recognized chemolithotrophic pathway in anoxic sediments, contributing to microbe-mediated iron redox cycling (2, 3). In 1996, Straub et al. (4) obtained the first chemolithoautotrophic NDFO enrichment culture from a freshwater sediment. Although a diverse group of bacteria is capable of NDFO (1, 5), this enrichment culture (referred to here as culture KS) is the most robust chemolithoautotrophic NDFO system, as it has been grown repeatedly under autotrophic conditions through numerous transfers over a period of years, and it is one of the few autotrophic NDFO systems that can completely oxidize Fe(II) in the uncomplexed form coupled to complete denitrification to generate N2. Hence, this culture has been used as a model system for physiological and geomicrobial studies of NDFO (6–9).
Based on 16S rRNA gene clone libraries, Blöthe and Roden (6) determined that culture KS was dominated by a bacterium in the family Gallionellaceae, closely related to Sideroxydans lithotrophicus ES-1, a neutrophilic microaerophilic chemoautolithotrophic FeOB (10). In addition, phylotypes related to heterotrophic nitrate-reducing Comamonas badia, Parvibaculum lavamentivorans, and Rhodanobacter thiooxidans were also detected in the clone libraries. Blöthe and Roden (6) were able to isolate the three nitrate-reducing heterotrophs and found that they were capable of only partial (12 to 24%) Fe(II) oxidation. Moreover, the authors suggested that the Sideroxydans-related bacterium in the family Gallionellaceae was the primary Fe(II) oxidizer in culture KS. However, this primary Fe(II) oxidizer has not been grown in pure culture, despite extensive efforts to isolate it, suggesting an important role(s) of the flanking community members in the physiology of this primary Fe(II) oxidizer.
A number of mechanistic models have been proposed for NDFO (11), yet the pathway(s) and genes utilized for electron transfer in chemolithoautotrophic NDFO still largely remain unknown. At circumneutral pH, oxidized (ferric) iron forms poorly soluble oxide minerals, which if present within the cell would be detrimental to the bacterium. It was suggested that NDFO likely occurs outside the cell through extracellular electron transfer (EET) (12), given the culture's capability to oxidize solid-phase Fe(II) using nitrate (8, 9). Previously, homologs of the multiheme cytochrome c (MtrA) and associated porin (MtrB) originally discovered in Shewanella oneidensis in Fe(III) reduction (13) were proposed for S. lithotrophicus ES-1 as its EET conduit and were referred to as MtoA and MtoB (for metal oxidation), respectively (14). MtoAB homologs are also present in a relative of ES-1, Gallionella capsiferriformans ES-2 (15, 16), another neutrophilic microaerophilic chemoautolithotrophic FeOB in the family Gallionellaceae (10). Given the close phylogenetic relatedness between the primary Fe(II) oxidizer in culture KS and S. lithotrophicus ES-1 and G. capsiferriformans ES-2, it seems reasonable to speculate that such a genetic system might also be present in the primary Fe(II) oxidizer in culture KS. In addition, it is also of interest whether the flanking members are equipped with the genetic machinery for EET, since heterotrophs isolated from this culture were able to partially, although not completely, oxidize Fe(II) (6).
Although a great deal has been learned about culture KS, it has remained difficult to unambiguously elucidate the metabolism and physiology at the organism level based on experiments conducted on the enrichment, due to the nature of the mixed culture. Metagenomic sequencing of microbial community DNA and reconstruction of draft genomes are powerful tools for use in inferring metabolic potential and physiology of member organisms, as has been demonstrated in both engineered and natural microbial communities (17, 18). Here, we employed two versions of culture KS, with independent transfer histories by two laboratories, for metagenomic analyses. We obtained a near-complete genome from the Fe(II) oxidizer and draft genomes from the abundant flanking species. The genome information allowed us to infer metabolic potential, search putative electron transfer pathways involved in NDFO, and explore the roles of the flanking community members and potential interspecies interactions. Along with these, we discuss a number of interesting genetic features observed in the genomes and provide insights into their potential physiological roles. We acknowledge that the discussions are based on in silico analyses and caution the reader that they provide starting points rather than final answers as to the metabolic functions of the culture.
MATERIALS AND METHODS
Enrichment cultures.
Two versions of culture KS were used in our metagenomic analyses. One has been maintained at the University of Tuebingen (Germany), where A. Kappler obtained it from K. L. Straub to study NDFO (referred to here as KS-Tueb), and the other has been independently maintained at the University of Wisconsin, Madison (WI, USA), since 1998, when E. E. Roden obtained it from K. L. Straub to study nitrate-dependent oxidation of solid-phase Fe(II) (8) (referred to here as KS-Mad). A notable difference between these two cultures is that KS-Tueb has been transferred with a 1% (vol/vol) inoculum, while KS-Mad has been transferred with a 10% inoculum. For our analyses, both KS-Tueb and KS-Mad cultures were grown in the year 2012 in a defined bicarbonate-buffered anoxic medium containing 10 mM FeCl2, 4 mM KNO3 or NaNO3, vitamins, and trace elements, as described previously (6). The growth medium and incubation conditions were largely the same, except for some minor differences, including temperature (28°C for KS-Tueb and 30°C for KS-Mad), pH (6.9 to 7.1 for KS-Tueb and 6.8 for KS-Mad), nitrate source (NaNO3 for KS-Tueb and KNO3 for KS-Mad), N2/CO2 ratio in the headspace (90/10 for Tueb and 80/20 for KS-Mad), and transfer volume (1% for KS-Tueb and 10% for KS-Mad, as described above).
DNA extraction.
The enrichment culture was vigorously shaken for at least 5 min to detach microbial cells from the iron(III) oxyhydroxides. The mixture was then allowed to settle for 10 min so that iron(III) oxyhydroxides settled down and microbial cells remained in the supernatant portion. A total of 4 ml of supernatant with microbial cells was collected and centrifuged at 10,000 × g for 5 min at room temperature to collect a cell pellet. DNA was extracted from the cell pellet using the PowerSoil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA), according to the manufacturer's instructions. Duplicate DNA extractions were performed and pooled for shotgun metagenome library construction.
Metagenome sequencing, assembly, and annotation.
Shotgun Illumina sequencing was conducted separately on the KS-Mad and KS-Tueb community DNA. For KS-Mad, an Illumina library was constructed using the Illumina Nextera tagmentation protocol, without modifications, with insert sizes ranging from 100 to 530 bp. The sequencing of KS-Mad was performed on the Illumina HiSeq 2000 platform to generate paired-end (2 × 100-bp) reads, totaling 11.2 Gbp of raw sequences. For KS-Tueb, an Illumina library was created using the Ovation Ultralow DR kit from NuGEN (San Carlos, CA, USA) without modifications, with target insert sizes of 280 to 320 bp. KS-Tueb was sequenced on the MiSeq platform to generate paired-end (2 × 250-bp) reads, totaling ∼1.5 Gbp of raw sequences.
Metagenome assembly was performed using the CLC Genomics Workbench (version 6.02; CLC bio, Inc., Cambridge, MA, USA). For the KS-Mad metagenome, after quality trimming and length filtration of the raw reads, the assembly was performed using the k-mer of 63 with scaffolding. For the KS-Tueb metagenome, because of the longer read length (250 bp, compared to 100 bp for KS-Mad), it was expected that the majority of paired reads would overlap. Therefore, after cutting the adaptor sequences, we merged paired-end reads into longer fragments prior to other read quality control (QC) procedures in order to maximize the overlap and thus the fragment sizes used in the assembly. After merging, quality trimming and length filtration were performed, and assembly was conducted using the k-mer of 63 with scaffolding. After assembly, the average fold coverage of each contig was estimated by mapping reads from the corresponding metagenome back to its contigs using CLC Genomics Workbench with 90% coverage and 95% identity cutoffs. All contigs were uploaded to the Integrated Microbial Genomes with Microbiomes (IMG/MER) database (http://img.jgi.doe.gov/mer) for gene prediction and functional annotation (19).
Analysis of essential single-copy genes.
To identify essential single-copy genes, all predicted protein sequences in the metagenome were searched against the hidden Markov models (HMMs) constructed for a total of 107 essential single-copy genes, which are conserved in 95% of all sequenced bacteria (20), by performing hmmsearch (HMMER 3.0, http://hmmer.janelia.org), as described by Albertsen et al. (17). Protein sequences from the identified essential genes were searched against the NCBI refseq_protein database using the BLASTP program, and the BLASTP results were imported to MEGAN (21) for taxonomic assignments using the lowest common ancestor algorithm, as described by Albertsen et al. (17). Essential genes assigned to the same taxonomic group were on contigs with comparable coverage, indicating that they indeed belong to the same organism. The analysis of essential single-copy genes clearly indicated the major organism bins and their expected coverage, and this information greatly facilitated the downstream taxonomic binning.
Taxonomic binning and draft genome recovery.
Taxonomic binning was performed using a combination of methods, including contig fold coverage (indicative of organism abundance) and G+C content, sequence homology, tetranucleotide frequency, and differential fold coverage between metagenomes, in order to maximize the binning accuracy and genome retrieval. For sequence homology-based binning, all protein sequences were searched against the NCBI refseq_protein database using the BLASTP program, and the search results were imported to MEGAN (21) for taxonomic assignment using the lowest common ancestor algorithm. The putative taxonomy of a contig was determined as the consensus taxon of proteins in that contig (i.e., the taxon to which the majority of proteins on that contig are assigned). An initial binning was performed on contigs >3 kbp based on homology and contig coverage and G+C content. Tetranucleotide frequencies were then calculated and analyzed using an emergent self-organizing map (ESOM) to verify the initial binning results and resolve ambiguities (22). Due to the community commonality and difference between the KS-Mad and KS-Tueb metagenomes, after the initial binning, contig differential coverage between the two metagenomes was also used to improve the binning results (17). Differential coverage is obtained by mapping reads from one metagenome to its own contigs (self-mapping coverage) and mapping reads from the other metagenome to contigs of this metagenome (cross-mapping coverage). All read mapping was performed with the CLC Genomics Workbench read mapping tool. The contig differential coverage method confirmed the initial binning results and also recruited contigs <3 kbp to some of the bins.
Phylogenetic tree construction.
We constructed a phylogenetic tree of conserved phylogenetic markers using the PhyloPhlAn analysis pipeline (24) to facilitate the taxonomic assignments of the six recovered draft genomes. For this analysis, we also included genomes of their close relatives and genomes of some related Fe(II) oxidizers. All protein sequences translated in silico from these genomes were input to PhyloPhlAn for extracting and individually aligning the conserved phylogenetic marker proteins. The alignments were then concatenated for phylogenetic tree construction using the FastTree algorithm, with default settings, by PhyloPhlAn.
To construct phylogenetic trees for Cyc2-like proteins and RubisCO, we included all identified Cyc2-like proteins and RubisCO in both metagenomes, as well as related reference genes in the public database. The protein sequences were aligned with MUSCLE (25) and trimmed to exclude columns that contain gaps for >30% of the included sequences. Maximum likelihood phylogenetic trees were constructed using PhyML 3.0 (26), with the LG substitution model (27) and the gamma distribution parameter estimated by PhyML. Bootstrap values were calculated based on 100 replicates. Trees were visualized with Dendroscope (version 3.2.10) with a midpoint root.
Other bioinformatic analyses.
Protein cellular location was predicted using CELLO version 2.5 (http://cello.life.nctu.edu.tw) (28) and PSORTb version 3.0 (http://www.psort.org/psortb) (29). The β-barrel structure of outer membrane proteins was predicted by PRED-TMBB (http://bioinformatics.biol.uoa.gr//PRED-TMBB) (30).
Metagenome and draft genome accession numbers.
All contigs are available through IMG (http://img.jgi.doe.gov/m/), with taxon object identification (ID) 3300002009 and 3300002247 for the KS-Mad and KS-Tueb metagenomes, respectively. A total of four draft genomes/bins were recovered in KS-Mad, and two were recovered in KS-Tueb. These draft genome sequences were submitted to IMG/ER (img.jgi.doe.gov/er) for annotation with the isolate microbial genome annotation pipeline (23) and are available with IMG taxon object IDs of 2565956535 to 2565956540.
RESULTS AND DISCUSSION
Community taxonomic composition.
Due to the simplicity of the enrichment communities, >94% of the total reads were assembled into contigs, and the assembled contigs totaled 21.6 Mbp and 15.0 Mbp for the KS-Mad and KS-Tueb metagenomes, respectively (see sequencing and assembly statistics summary in Table S1 in the supplemental material).
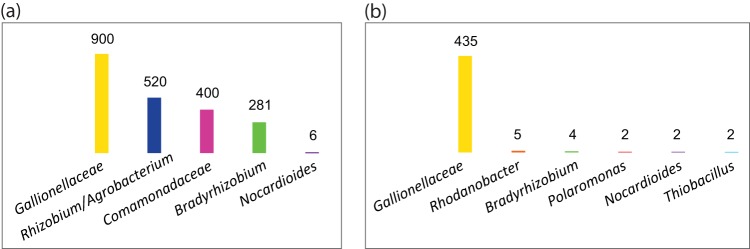
We first assessed the community taxonomic composition of the two cultures using essential single-copy genes that are conserved in most bacteria for housekeeping functions (20). Essential single-copy genes recovered in the metagenomes were assigned to different bins based on their taxonomic assignment by MEGAN using BLASTP search results and their fold coverage patterns in the metagenomes, and these bins were further assigned to the level of family or genus based on the lowest common ancestor (see Fig. S1 in the supplemental material). Using the average fold coverage of essential single-copy genes within each bin (see Fig. S1 in the supplemental material), we estimated the rank abundance of taxa in the two enrichments (Fig. 1). As expected, the most abundant bacterium in both cultures belongs to the family Gallionellaceae, with most of their essential single-copy genes having the best match to the Gallionella or Sideroxydans genus (see Fig. S1 in the supplemental material). In KS-Mad, Gallionellaceae (accounting for 42% of the total community) was flanked by the Rhizobium/Agrobacterium group (27%), Comamonadaceae (18%), Bradyrhizobium (13%) (Fig. 1a), and a very low level (<0.1%) of Nocardioides, which was detected only by its 16S rRNA gene fragment in the metagenome (see Table S2 in the supplemental material). In contrast, KS-Tueb was more strongly dominated by Gallionellaceae (accounting for 96% of the total community), flanked by Rhodanobacter (1%), Bradyrhizobium (1%), and very low levels (<1%) of Nocardioides, Polaromonas, and Thiobacillus (Fig. 1b).
FIG 1.
Rank abundance of taxa found in KS-Mad (a) and KS-Tueb (b) metagenomes. The abundance is approximated by the average fold coverage of essential single-copy genes within a taxon in the metagenome, except for Nocardioides in KS-Mad, for which the abundance is approximated by the fold coverage of its 16S rRNA gene.
Among these taxa, Gallionellaceae, Bradyrhizobium, and Nocardioides were shared between the two communities. Although species in Comamonadaceae were present in both metagenomes, their genes were affiliated with Acidovorax/Alicycliphilus/Comamonas in KS-Mad, whereas they were affiliated with Polaromonas in KS-Tueb (see Fig. S1 in the supplemental material). The two cultures exhibited some differences in their community structure (Fig. 1), despite the fact that they were propagated from the same original enrichment and their growth media were largely the same. The community differences might be partly attributed to the potential difference in cell affinity to iron(III) oxyhydroxides, as DNA extraction might have caused some biases against those with higher affinities to iron(III) oxyhydroxides. However, to a large extent, the difference may be due to the different inoculating volumes (i.e., 1% for KS-Tueb and 10% for KS-Mad). Inactive cells in the inoculum may decay and provide organic carbon for cells capable of heterotrophic denitrification (which can be performed by several flanking community members in culture KS based on their genetic potential, as discussed below). Therefore, the higher inoculating volume in KS-Mad might have resulted in a higher proportion of heterotrophic flanking populations in the community.
Previously, Blöthe and Roden (6) constructed 16S rRNA gene clone libraries from KS-Mad in 2005 and 2007, respectively, and determined that KS-Mad was dominated by a bacterium with 95% identity to S. lithotrophicus ES-1 within the Gallionellaceae family, followed by three phylotypes that were 94, 96, and 99% identical to Comamonas badia, Parvibaculum lavamentivorans, and Rhodanobacter thiooxidans, respectively. We also compared all 16S rRNA genes in the metagenomes against the clone libraries (6), and the 100% identity between the metagenome and clone library rRNA genes was found only for Gallionellaceae (both KS-Mad and KS-Tueb versions) and Rhodanobacter (KS-Tueb), whereas other pairwise comparisons had <96% identity (see Table S2 in the supplemental material).
Overall, the community comparisons indicate that the Gallionellaceae sp. was conserved with time and with different transfer histories between laboratories, while the flanking community was subject to change. This confirms that the Gallionellaceae sp. is the primary player in nitrate-driven Fe(II) oxidation (6).
Draft genome recovery.
We recovered four draft genomes/bins in KS-Mad and two in KS-Tueb, and their assembly and annotation statistics are summarized in Table 1. Specifically, for the KS-Mad metagenome, contigs >3 kbp were binned to taxonomic groups based on a combination of contig G+C content, fold coverage (see Fig. S2a in the supplemental material), and homology to reference genomes, and the results were confirmed with contig tetranucleotide frequency (see Fig. S2b in the supplemental material). Four major bins were obtained from the abundant species (see Fig. S2 and further discussion in Text S1 in the supplemental material). From the consensus of RDP classification and BLASTN search of their 16S rRNA gene sequences, taxonomic assignment based on essential single-copy genes (see Fig. S1 in the supplemental material), and the phylogenetic tree constructed with conserved phylogenetic markers (see Fig. S3 in the supplemental material), the four bins were assigned to the families Gallionellaceae and Comamonadaceae and the genera Bradyrhizobium and Rhizobium/Agrobacterium (for simplicity referred to here as Rhizobium), respectively (Table 2).
TABLE 1.
Draft genomes recovered from the metagenomes
| Draft genome | No. of contigs | Total length (bp) | G+C content (%) | Coverage in KS-Mad culture (×)a | Coverage in KS-Tueb culture (×)a | Recovered essential single-copy genesb | No. of protein-coding genes | No. of tRNAs | % genes with function prediction |
|---|---|---|---|---|---|---|---|---|---|
| Gallionellaceae sp. (KS-Tueb) | 45 | 2,644,639 | 60 | 892 | 467 | 106/106 | 2,480 | 43 | 81.85 |
| Gallionellaceae sp. (KS-Mad) | 52 | 2,639,060 | 60 | 892 | 467 | 106/106 | 2,489 | 43 | 81.85 |
| Bradyrhizobium sp. | 66 | 7,164,947 | 64 | 285 | 4 | 105/105 | 6,831 | 50 | 77.49 |
| Comamonadaceae sp. | 17 | 3,410,349 | 66 | 402 | 0 | 106/106 | 3,175 | 42 | 86.66 |
| Rhizobium composite bin | 611 | 4,909,323 | 62 | 469 | 0 | 79/105 | 5,664 | 37 | 73.63 |
| Rhodanobacter sp. | 2,874 | 3,256,089 | 68 | 0 | 3 | 89/105 | 4,894 | 27 | 71.19 |
Coverage in KS-Mad was obtained by mapping KS-Mad reads to KS-Mad assembly, and coverage in KS-Tueb was obtained by mapping KS-Tueb reads to KS-Tueb assembly.
The first number is the number of recovered essential single-copy genes, and the second number is the expected number of essential single-copy genes if the genome is complete.
TABLE 2.
Summary of taxonomic classification of draft genomes
| Draft genome | Classification of essential single-copy genes by MEGANa | Inference from phylogenic tree of conserved marker genesb | Classification of 16S rRNA sequences using RDP Classifierc | Top BLASTN hits to NCBI 16S rRNA database | Top BLASTN hit to clone libraries of Blöthe and Roden (6) |
|---|---|---|---|---|---|
| Gallionellaceae sp. (KS-Tueb) | Gallionellaceae | Gallionellaceae | Sideroxydans | 96% to Sideroxydans lithotrophicus ES-1, 94% to Gallionella capsiferriformans ES-2, 94% to Gallionella sp. strain SCGC AAA018-N21 | 100% to clone F30F68 (OTU1) |
| Gallionellaceae sp. (KS-Mad) | Gallionellaceae | Gallionellaceae | Sideroxydans | 95–96% to S. lithotrophicus ES-1, 94% to G. capsiferriformans ES-2, 94% to Gallionella sp. SCGC AAA018-N21 | 100% to clone F30F68 (OTU1), 100% to clone F29F67 (OTU1) |
| Bradyrhizobium sp. | Bradyrhizobium | Bradyrhizobium | Bradyrhizobium | 99% to Bradyrhizobium valentinum LmjM3, 99% to Bradyrhizobium jicamae PAC68, 99% to Bradyrhizobium pachyrhizi PAC48 | 89% to Parvibaculum sp. strain MBNA2 (OTU3) |
| Comamonadaceae sp. | Comamonadaceae | Comamonadaceae | Comamonas | 97% to Comamonas badia IAM 14839, 95% to Comamonas nitrativorans 23310, 94% to Alicycliphilus denitrificans BC | 95% to Comamonas sp. strain MPI12 (OTU2) |
| Rhizobium composite bin | Rhizobium/Agrobacterium group | Rhizobium/Agrobacterium group | Rhizobium | 98% to Rhizobium rosettiformans W3, 98% to Rhizobium daejeonense L61, 97% to Agrobacterium fabrum C58 | 90% to Parvibaculum sp. MBNA2 (OTU3) |
| Rhodanobacter sp. | Rhodanobacter | Rhodanobacter | Rhodanobacter | 99% to Rhodanobacter denitrificans 2APBS1, 99% to Rhodanobacter thiooxydans LCS2, 98% to Rhodanobacter caeni MJ01 | 100% to Rhodanobacter sp. strain MBNA3 (OTU4) |
Classification is based on MEGAN lowest common ancestor using BLASTP search of essential single-copy genes (see Fig. S1 in the supplemental material).
Classification is based on the phylogenetic tree constructed using conserved phylogenetic marker genes (see Fig. S3 in the supplemental material).
Classification is from Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/) Classifier, with a confidence level of 80%.
For KS-Tueb, three initial bins were obtained, with total lengths of 2.6 Mbp, 520 kbp, and 430 kbp, and were classified to Gallionellaceae, Bradyrhizobium, and Rhodanobacter, respectively. The remaining organisms were present at very low abundances (Fig. 1b) and did not assemble well. The KS-Tueb Bradyrhizobium sequences are identical to the Bradyrhizobium bin from KS-Mad, indicating that they are the same strain. Therefore, Bradyrhizobium from KS-Mad is used as the representative of Bradyrhizobium in culture KS, and no additional effort was made to recover Bradyrhizobium from KS-Tueb. Due to the commonality and difference in community composition, particularly the predominance of Gallionellaceae in both metagenomes and the lack of Rhodanobacter in KS-Mad (Fig. 1), we also tried to bin sequences <3 kbp based on their differential coverage in combination with their G+C content and sequence homology. In this way, we expanded the Rhodanobacter draft genome in KS-Tueb to 3.3 Mbp, and a number of contigs <3 kbp were also unambiguously recruited to the two Gallionellaceae genomes.
The recovery of most (or all) essential single-copy genes (Table 1) indicates that we have recovered most of their genomes, especially those for Gallionellaceae, Comamonadaceae, and Bradyrhizobium. The KS-Mad and KS-Tueb Gallionellaceae genomes are nearly identical, except for a few minor differences (see Text S2 in the supplemental material for details). As we cannot unambiguously determine whether the Gallionellaceae sp. in KS belongs to either Sideroxydans or Gallionella or represents a novel genus, we tentatively refer to it as Gallionellaceae sp. in this paper (see Text S3 in the supplemental material for details on the taxonomy of Gallionellaceae sp. and the genome-wide comparison to ES-1 and ES-2).
Extracellular electron transfer in Fe(II) oxidation.
The high degrees of genome completeness allow us to reconstruct metabolisms and search for putative genes in Fe(II) oxidation. Table 3 summarizes some key metabolic pathways in the six draft genomes recovered from culture KS, along with ES-1 and ES-2 for comparison.
TABLE 3.
Summary of key metabolic pathways in the genomesa
| Function | ES-1 | ES-2 | Gallionellaceae (KS-Tueb) | Gallionellaceae (KS-Mad) | Bradyrhizobium | Comamonadaceae | Rhizobium | Rhodanobacter |
|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | ||||||||
| CBB reductive pentose phosphate cycle | + | + | + | + | + | − | + | + |
| Glycolysis (Embden-Meyerhof pathway) | + | + | + | + | + | + | + | + |
| Pyruvate oxidation to acetyl-CoA through pyruvate dehydrogenase | + | + | + | + | + | + | + | + |
| Citrate cycle (TCA cycle, Krebs cycle) | + | + | + | + | + | + | (+) | + |
| Pentose phosphate pathway | + | + | + | + | + | +/−b | (+) | + |
| Glyoxylate cycle | − | − | − | − | + | + | − | + |
| Nitrogen metabolism | ||||||||
| Nitrogen fixation, nitrogen → ammonia | + | − | − | − | − | − | − | − |
| Ammonification, nitrate/nitrite → ammonia | + | − | − | − | + | − | + | − |
| Nitrification, ammonia → nitrite | − | − | − | − | − | − | − | − |
| Denitrification | ||||||||
| Nitrate → nitrite | − | − | + | + | + | + | (+) | + |
| Nitrite → nitric oxide | + | − | + | + | + | + | + | (+) |
| Nitric oxide → nitrous oxide | + | + | − | − | + | + | (+) | + |
| Nitrous oxide → nitrogen gas | − | − | − | − | + | + | + | (+) |
| Complete denitrification | No | No | No | No | Yes | Yes | Yes | Yes |
| Hydrogen metabolism | ||||||||
| Group 1, uptake [NiFe]-hydrogenase | + | − | + | + | − | − | + | − |
| Group 3, bidirectional cytoplasmic [NiFe]-hydrogenase | + | + | + | + | + | − | − | − |
| Group 4, H2-evolving, energy-conserving [NiFe]-hydrogenases | + | + | + | + | + | − | + | + |
| Sulfur metabolism | ||||||||
| Assimilatory sulfate reduction | + | + | + | + | + | + | + | + |
| Dissimilatory sulfate reduction | + | − | − | − | − | − | − | − |
| Thiosulfate oxidation by SOX complex | + | − | − | − | + | + | + | − |
| Phosphorus metabolism | ||||||||
| Phosphonate transport system | − | + | − | − | + | − | + | + |
| Phosphonoacetate degradation | + | + | + | + | + | + | + | + |
| High-affinity phosphate transport system | + | + | + | + | + | + | + | + |
| Low-affinity phosphate transporters | + | + | − | − | + | + | − | + |
| Polyphosphate accumulation and degradation | + | + | + | + | + | + | (+) | + |
| Alkaline phosphatase | − | − | − | − | + | − | + | − |
| Oxidative phosphorylation | ||||||||
| Complex I, NADH:quinone oxidoreductase | + | + | + | + | + | + | + | + |
| Complex II, succinate dehydrogenase | + | + | + | + | + | + | + | + |
| Complex III, cytochrome bc1 complex | + | − | + | + | + | + | + | + |
| Alternative complex III (ACIII) | + | + | − | − | − | − | − | − |
| aa3-type cytochrome c oxidase | − | − | + | + | + | + | + | + |
| Cytochrome bd ubiquinol oxidase | + | + | − | − | − | − | + | + |
| cbb3-type cytochrome c oxidase | + | + | + | + | + | + | + | + |
| F-type ATPase | + | + | + | + | + | + | + | + |
| V-type ATPase | + | − | − | − | − | − | − | − |
+, a complete pathway is present; −, a pathway is absent; (+), the missing gene(s) in that pathway was found in the metagenome and can be assigned to that genome based on fold coverage and sequence homology.
+/−, the oxidative phase of the pentose phosphate pathway is present, but the nonoxidative phase is absent.
At circumneutral pH, electron transfer likely occurs in the periplasm, with reaction products being pumped to the outside, or it occurs outside the cell (i.e., via extracellular electron transfer [EET]) to avoid intracellular mineral precipitation (7). EET is also employed by Fe(III) reducers in order to transfer electrons to poorly soluble iron minerals. The redox active enzymes in iron oxidation and reduction are often c-type cytochromes (cytc) (16, 31) and, to a lesser extent, multicopper oxidases (MCO) (32). For EET to occur, these redox active enzymes need to be able to access extracellular iron.
There are two general models for EET in Gram-negative bacteria. In one model, the redox active enzyme is secreted as an outer membrane or extracellular protein in order to directly interact with extracellular iron. Examples include outer membrane multiheme cytc (e.g., OmcB, OmcC, OmcE, and OmcS) (33, 34) and outer membrane multicopper oxidases (including OmpB and OmpC) in Fe(III)-reducing Geobacter sulfurreducens (32, 35), as well as an outer membrane cytc (Cyc2) in the acidiphilic Fe(II) oxidizer Acidithiobacillus ferrooxidans (36).
Therefore, we first searched for genes encoding outer membrane/extracellular cytc and found such a gene in the Gallionellaceae and Rhodanobacter genomes (see Fig. S4 in the supplemental material). In Gallionellaceae sp., this protein is 37 to 44% identical to Cyc2-like proteins previously found in the ES-1 and ES-2 genomes (15) and is only ∼30% identical to Cyc2 from A. ferrooxidans at the N terminus, whereas the C terminus does not share any significant homology. In Rhodanobacter sp., this protein is ∼23% identical to A. ferrooxidans Cyc2 over the entire length of the protein. Notably, sequences homologous to Gallionellaceae or Rhodanobacter Cyc2-like protein are mostly from bacteria known for Fe(II) oxidation, including Mariprofundus spp., Rhodopseudomonas spp., Ferriphaselus sp., Chlorobium ferrooxidans, etc. (see Fig. S4 in the supplemental material), and these proteins are all predicted to be outer membrane/extracellular proteins with a conserved heme-binding motif at the N terminus. An EET system involving outer membrane Cyc2, periplasmic Cyc1, and inner membrane alternative complex III (ACIII) was recently identified in the marine neutrophilic microaerophilic Fe(II) oxidizer Mariprofundus ferrooxydans PV-1 in Fe(II) oxidation (37). Indeed, Cyc2 and ACIII are present in all neutrophilic microaerophilic Fe(II) oxidizers with complete genomes available, including those of ES-1 and ES-2 (38). However, despite the close phylogenetic relatedness to ES-1 and ES-2, Gallionellaceae sp. does not possess the ACIII gene cluster, nor do any of the flanking members in culture KS. Therefore, if Cyc2-like protein is involved in EET in Gallionellaceae sp. and Rhodanobacter sp., they may employ another system in the inner membrane, such as the canonical complex III (bc1 complex), which is present in both Gallionellaceae sp. and Rhodanobacter sp., to perform the role provided by ACIII.
Homologs of Rhodobacter strain SW2 FoxE protein, a cytc involved in phototrophic Fe(II) oxidation but not associated with the outer membrane (39), were not found in any of the draft genomes.
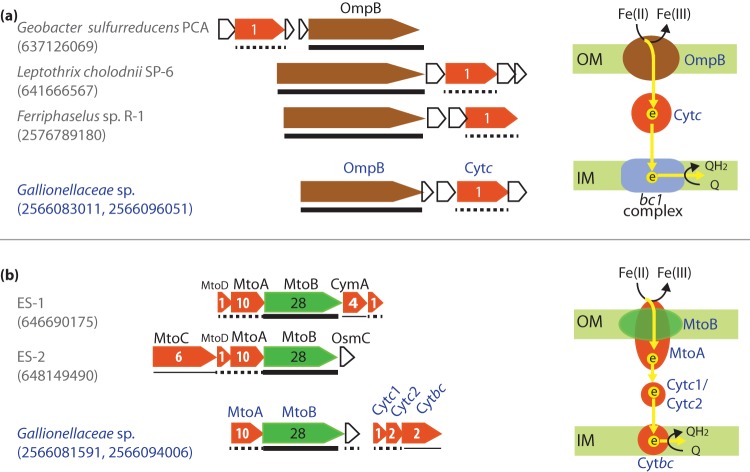
Besides cytc, we also found an outer membrane MCO in the Gallionellaceae genome (Fig. 2a). It shares homology to G. sulfurreducens outer membrane protein B (OmpB), which was loosely associated with the cell outer surface (40) and was required for the reduction of poorly soluble Fe(III) but not for dissolved Fe(III) (32). OmpB is a unique MCO, as it contains Fe(III)-binding motifs (EXXE) and a fibronectin type III domain, which is often involved in bacterial adhesion, thus possibly playing a role in attaching to Fe(III) (oxyhydr)oxides (32). Likewise, Gallionellaceae sp. OmpB-like protein has a fibronectin III domain and two iron-binding motifs. Notably, OmpB is homologous to manganese oxidase (MofA) from Leptothrix discophora (41), and we also found OmpB homologs in Fe(II)-oxidizing Leptothrix cholodnii SP-6 and Ferriphaselus sp. strain R-1 (Fig. 2a). In these cases, ompB is clustered with a gene encoding a periplasmic cytc-like protein (Fig. 2a), which might be able to relay electrons to the periplasm, supporting the hypothesized role of OmpB in EET.
FIG 2.
Gene clusters encoding putative EET enzymes, including outer membrane multicopper oxidase (MCO) homologous to OmpB in G. sulfurreducens (a) and multiheme cytc (MtoA) with a porin (MtoB) homologous to MtoAB in ES-1 and ES-2 (b). The corresponding hypothesized electron flow is schematically represented on the right side, with electron flows indicated by yellow arrows, and enzymes labeled in the same color as their coding genes in the gene cluster from Gallionellaceae sp. on the left. Note that in panel a, bc1 complex genes are not in the same gene cluster that contains ompB but are likely involved in the process. For each gene cluster, the IMG gene object IDs (OID) for OmpB and MtoB are listed in parentheses. Porins (MtoB) are labeled with green, cytc are labeled with red, and MCO are labeled with brown. Numbers in cytc-coding genes indicate the number of heme-binding sites, and numbers in the porin-coding genes indicate the number of transmembrane regions. Predicted cellular locations of porins, cytc, and MCO are indicated by different line types under the gene, with thick solid lines for outer membrane (OM), dashed lines for periplasm, and thin solid lines for inner membrane (IM), respectively.
In another general model of EET, instead of being an extracellular or outer membrane protein, the redox active enzyme is secreted to the periplasm and embedded into a porin on the outer membrane to directly or indirectly interact with extracellular iron. The genes involved in this model were first identified in Shewanella oneidensis (13). In this Fe(III) reducer, an inner membrane tetraheme cytc (CymA) transfers electrons directly or indirectly to MtrA, a decaheme cytc embedded in MtrB, which is a porin with 28 transmembrane motifs forming a sheath for MtrA in the outer membrane. MtrA then shuttles electrons to the outer membrane decaheme cytc (MtrC), which further transfers electrons to extracellular iron minerals (16, 42). Homologs of MtrAB complex were also identified in several FeOB, including PioAB in Rhodopseudomonas palustris TIE-1 (43) and MtoAB in ES-1 and ES-2 (14–16). This EET system was thus referred to as the porin-cytochrome model (42). Besides the homologous MtrAB, PioAB, and MtoAB systems, recently, a different porin-cytochrome c protein complex (Pcc) was identified in G. sulfurreducens as an EET conduit in Fe(III) reduction (44). Further, this Pcc system was identified in a number of bacteria performing redox reactions involving insoluble substrates or products, which would also require EET (45).
We searched both metagenomes for homologs of Pcc described by Liu et al. (44) but did not find any match in the KS organisms. Homologs of MtoAB in ES-1 and ES-2 were found in Gallionellaceae sp., although they are only 37% and 39% identical to MtoA and 22% and 23% identical to MtoB in ES-1 and ES-2, respectively. In Gallionellaceae sp., MtoA is a periplasmic cytc with 10 heme-binding sites, and MtoB is an outer membrane β-barrel with 28 transmembrane regions, likely forming a porin (Fig. 2b), consistent with their hypothesized role in EET. For ES-1 and ES-2, Shi et al. (16) suggested that a periplasmic cytc (MtoD) encoded in the same operon as mtoAB relays electrons from MtoA to an inner membrane cytc (CymA in ES-1 and MtoC in ES-2) encoded in the same operon (Fig. 2b). Unlike ES-1 and ES-2, the Gallionellaceae sp. mto operon contains only mtoAB and a gene encoding a small hypothetical protein. However, downstream of this operon is another three-gene operon encoding two periplasmic cytc (tentatively referred to as Cytc1 and Cytc2) and an inner membrane protein fused with a cytochrome b561 domain at the N terminus and a diheme cytc at the C terminus (tentatively referred to as Cytbc) (Fig. 2b). Given their clustering with mtoAB in the genome and their predicted cellular locations, cytochromes encoded by this operon might cooperate with MtoAB in EET: periplasmic Cytc1 and Cytc2 relay electrons from outer membrane MtoA to inner membrane Cytbc (Fig. 2b).
We also performed a broader search for gene clusters potentially encoding porin-periplasmic cytc or porin-periplasmic MCO complexes that were not previously recognized, and we found interesting cases in Gallionellaceae sp., Bradyrhizobium sp., Comamonadaceae sp., and Rhodanobacter sp. in culture KS (see Text S4 and Fig. S5 in the supplemental material for detailed discussion). These may provide more genetic targets to test in future studies.
Although we identified some candidate genes for EET, some of which were previously identified in other bacteria, such as Cyc2, OmpB, and MtoAB, we do not know whether any of these candidates are actually employed in NDFO by culture KS, and we cannot rule out the possibility that other yet-to-be-identified alternative mechanisms are employed instead.
Electron transfer in the inner membrane.
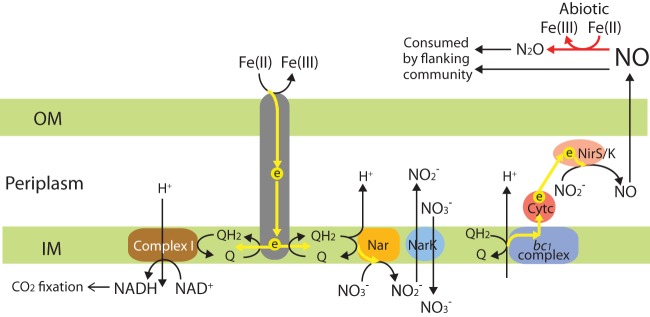
Quinols (and derivatives) are important inner membrane electron carriers in respiration. Gallionellaceae sp. and the flanking species have the genetic machinery to synthesize ubiquinone (UQ). In NDFO, how the quinone pool is reduced has remained an open question (12). Under circumneutral pH, the redox potential for the Fe(OH)3/Fe(II) couple is −0.018 V [assuming 10 mM dissolved Fe(II)], which is lower than that of the UQ/UQH2 couple (+0.113 V), and thus electron transfer from Fe(II) to UQ can probably occur without consuming energy. If Cyc2 or OmpB is utilized by Gallionellaceae sp., electrons might be transferred to the bc1 complex in the inner membrane to reduce the quinone pool (Fig. 2a), and this is further discussed below. If the MtoAB system is utilized, MtoA might transfer electrons to periplasmic Cytc1 and/or Cytc2, which relays electrons to inner membrane Cytbc encoded in the same operon downstream of the mto operon (Fig. 2b). As Cytbc contains a b-type cytochrome, which is often a subunit in quinone-active enzymes, such as complex I, bc1 complex, and [NiFe]-hydrogenase I complex (46), it is plausible that Cytbc is able to pass electrons to the quinone pool (Fig. 2b), similar to CymA in ES-1 and MtoC in ES-2 (16), although Cytbc is not homologous to CymA or MtoC.
Reduced ubiquinol could then pass electrons to inner membrane redox enzymes (such as nitrate reductase) in one direction for energy generation through proton-translocation by the quinone cycle, and in the other direction to generate reducing equivalents in the form of NAD(P)H for carbon fixation (Fig. 3). However, at cytoplasmic pH, the redox potential of the NAD(P)+/NAD(P)H couple is −0.32 V (47), which is lower than that of UQ/UQH2, hence requiring an energy-consuming “uphill” electron transport. This is probably achieved by complex I functioning in reverse at the cost of proton motive force (PMF), as previously proposed for A. ferrooxidans (48) (Fig. 3), alongside a transhydrogenase capable of interconverting NAD(P)H and NADH.
FIG 3.
A hypothesized electron transfer pathway proposed for NDFO by Gallionellaceae sp. in culture KS. The EET system for oxidizing Fe(II) and reducing the quinone pool is indicated by a gray channel in general, which may be a system involving Cyc2, OmpB, or MtoAB identified in this study, or another yet-to-be-identified alternative mechanism. Electron flows are indicated by yellow arrows, and abiotic reduction of NO by Fe(II) is indicated by red arrows. OM, outer membrane; IM, inner membrane; Nar, dissimilatory nitrate reductase complex; NarK, nitrate:nitrite antiporter; NirS and NirK, cytochrome cd1- and copper-type nitrite reductase, respectively.
The complete oxidative phosphorylation pathway (including complexes I, II, and III, cytochrome c oxidases, and F-type ATP synthase) is present in all draft genomes (Table 3). It was previously reported that ES-1 possesses a canonical complex III (bc1 complex), as well as an alternative complex III (ACIII), whereas ES-2 only has ACIII to transfer electrons between quinols and terminal oxidases (15). All draft genomes from culture KS contain bc1 complex, and none have ACIII. Notably, the Gallionellaceae genome possesses two copies of bc1 complex, with 79% amino acid identity to each other, and both copies are predicted to be located in the inner membrane. Interestingly, these two copies of bc1 complex have the best match to Thiobacillus denitrificans ATCC 25259 (68% identity) and are only remotely homologous to ES-1 (38% identity). It was previously shown that several A. ferrooxidans strains also possess two copies of bc1 complex, with one for uphill electron transport during Fe(II) oxidation, and the other for downhill electron transport during sulfur oxidation (49). Therefore, it is likely that the two copies of bc1 complex in Gallionellaceae sp. are also specialized in different functions, e.g., reducing the quinol pool using electrons obtained by the EET system (such as OmpB [Fig. 2a], Cyc2, or other alternative EET system) and oxidizing the quinol pool for downstream nitrite reduction (Fig. 3), respectively.
It appears that Gallionellaceae sp. may be able to respire oxygen, as suggested by the presence of the cbb3-type cytochrome c oxidase, which has a high affinity to oxygen (50, 51). Therefore, Gallionellaceae sp. might also be a neutrophilic microaerophilic Fe(II) oxidizer, similar to ES-1 and ES-2. Unlike ES-1 and ES-2, however, Gallionellaceae sp. lacks cytochrome bd oxidase that directly transfers electrons from quinols to oxygen. Instead, Gallionellaceae sp. possesses the aa3-type cytochrome c oxidase, which typically has a low affinity to oxygen and would require high oxygen levels (51, 52). This is not expected for neutrophilic FeOB, since the chemical oxidation of Fe(II) by oxygen can occur rapidly under neutral pH if oxygen is present at high concentrations. Therefore, the aa3-type cytochrome c oxidase might be utilized at situations other than neutrophilic Fe(II) oxidation.
Nitrogen metabolism.
Differing from ES-1, Gallionellaceae sp. in culture KS does not have nitrogen fixation genes or any ammonification pathway to convert nitrate or nitrite to ammonia. Therefore, Gallionellaceae sp. needs to rely on exogenous ammonia for growth, which was provided in the medium (4, 6), and an ammonia permease and a few amino acid transporter genes are present in its genome. Interestingly, no nitrogen fixation genes were detected in other organisms in the metagenomes, including Bradyrhizobium sp. and Rhizobium sp., whose relatives are known to be able to fix nitrogen. Instead, Bradyrhizobium sp. and Rhizobium sp. possess NirBD genes to convert nitrite to ammonia.
As a denitrifying culture, dissimilatory nitrate reductase complex (Nar) genes are present in each of the KS organisms to convert nitrate to nitrite. Despite being phylogenetically closely related to Gallionellaceae sp. in culture KS, ES-1 and ES-2 do not possess Nar, consistent with their inability to couple nitrate reduction to Fe(II) oxidation (15). In addition, Nar genes are also absent in other publicly available genomes within Gallionellaceae, including Ferriphaselus sp. strain R-1 and Ferriphaselus amnicola. This suggests that NDFO is unique to Gallionellaceae sp. in culture KS and is not a conserved feature of the Gallionellaceae family.
Nitrite, if accumulated, can rapidly react abiotically with dissolved Fe(II) (53), forming precipitates at the cell surface and leading to periplasmic and eventually cytoplasmic mineralization (54). Therefore, rapid biological reduction of nitrite may be important for achieving robust NDFO. Both the copper-type nitrite reductase (NirK) and the cytochrome cd1-type reductase (NirS) genes are present in Gallionellaceae sp., and NirS is represented by two copies with ∼80% identity to each other. Similarly, Bradyrhizobium sp. and Comamonadaceae sp. have both NirK and NirS, whereas Rhizobium sp. and Rhodanobacter sp. contain NirK only. It is interesting to note that nirS is clustered with a gene encoding a porin-like protein in the Gallionellaceae and Comamonadaceae genomes (see Fig. S5a in the supplemental material).
Unexpectedly, nitric oxide (NO) and nitrous oxide (N2O) reductase genes are missing in the Gallionellaceae genome. To exclude the possibility that the absence of these genes was due to the binning process, we searched the entire metagenomes for genes encoding both cytc-dependent (NorBC) and quinol-dependent (NorZ) NO reductases, as well as N2O reductase (NosZ). None of the hits were from Gallionellaceae sp. based on sequence homology or fold coverage. This was indeed unexpected, since this enrichment culture completely converts nitrate to nitrogen gas (6), and researchers have long assumed that the primary Fe(II) oxidizer was able to carry out the complete denitrification. It is interesting to note that its close relatives ES-1 and ES-2 both have NorBC genes.
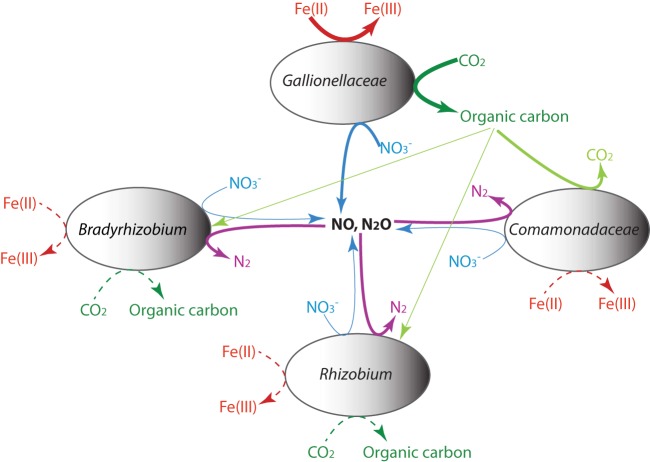
The apparent lack of NO and N2O reductase genes in Gallionellaceae sp. is surprising, since NO is toxic to cells. As the complete denitrification pathway is present in all flanking species (Table 3), it is possible that NO and N2O are consumed by these flanking populations (Fig. 3). Alternatively, as nitrite can react abiotically with Fe(II) to form N2O at near-neutral pH, with NO as an intermediate (55–57), the detoxification may also be via abiotic chemical reduction of NO, which, if true, is likely to occur outside the cell to avoid periplasmic mineralization (Fig. 3). However, chemical reduction of NO during nitrite reduction by Fe(II) was suggested to depend on the availability of reactive surface provided by Fe(III) precipitates and the sorption of Fe(II) to the reactive surface (55–57), and chemical NO reduction occurred only after a significant amount of Fe(III) precipitates had formed through Fe(II) oxidation by nitrite (56). Therefore, biological NO reduction is still important, particularly in the early growth phase, when the formation of Fe(III) minerals is limited. In addition, the molar ratio of oxidized Fe(II) to reduced nitrate was ca. 5:1 for culture KS, indicating that a complete denitrification to form N2 must have occurred (4, 6). As biological denitrification seems the only way to form N2 in this system, the flanking populations should play an important role in consuming NO and particularly N2O, regardless of whether abiotic NO reduction occurs. Therefore, Gallionellaceae sp. may partner with these microorganisms to utilize its incomplete denitrification products (Fig. 4), and this might provide an explanation as to why researchers have not been able to isolate Gallionellaceae sp., as the Gallionellaceae genome does not seem to lack pathways for amino acid or essential vitamin biosynthesis. (More discussion on the lack of NO and N2O reductase genes in Gallionellaceae sp. is provided in Text S5 and Fig. S6 in the supplemental material.)
FIG 4.
Proposed interspecies interactions among organisms in culture KS-Mad. The contribution of each reaction and interaction is qualitatively indicated by the line thickness. Dashed lines indicate biochemical transformations that potentially occurred.
Partnering of the Gallionellaceae sp. with heterotrophic flanking populations to achieve complete denitrification requires that the heterotrophs obtain a source of fixed carbon from Gallionellaceae sp. as electron donors (Fig. 4). Previous studies with an aerobic chemolithoautotrophic FeOB (strain TW2) provided evidence for release of extracellular polymeric substances (EPS), which played a role in Fe(III) complexation within oxygen-Fe(II) opposing-gradient cultures (58). Energetics calculations indicated that potential FeOB growth yield was likely severalfold in excess of the observed net growth yield for TW2 (59), suggesting that a substantial amount of energy was available for the synthesis of other cellular components (e.g., EPS). An analogous argument can be made regarding the potential for Gallionellaceae sp. to provide fixed carbon to flanking heterotrophic populations, since calculated growth yields for autotrophic Fe(II) oxidation coupled to denitrification are only ca. 10% lower than those for microaerobic oxidation. The Gallionellaceae genome contains clusters of genes encoding EPS biosynthesis, and the release of EPS may play a role in avoiding cell encrustation with Fe(III) oxides (see Text S9 in the supplemental material). The metabolism of EPS produced by Gallionellaceae sp. provides a plausible explanation for a reduction of denitrification intermediates by heterotrophic bacteria in culture KS.
Carbon metabolism.
Culture KS, as a whole, is chemolithoautotrophic, with CO2 being the sole carbon source. As has been found in ES-1 and ES-2, the Calvin-Benson-Basham (CBB) reductive pentose phosphate pathway is present in the Gallionellaceae genome. Compared to ES-1, which has both form II and form IAq ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO, the key enzyme in the CBB pathway), Gallionellaceae sp. contains a copy of form II RubisCO (see Fig. S7 in the supplemental material). Form II RubisCO has a low affinity to CO2 and therefore is adapted to high-CO2 and low-O2 environments (60). We also identified RubisCO genes in some flanking community members, including Rhizobium sp., Bradyrhizobium sp., and Rhodanobacter sp. in culture KS, and they mostly contain form IC RubisCO (see Fig. S7 in the supplemental material). Compared to form II, form IC is adapted to medium-CO2 and suboxic-O2 environments (such as soils and root nodules) (60). If these flanking members do fix CO2 in culture KS, they are also likely conducting Fe(II) oxidation coupled with denitrification in order to obtain reducing equivalents and ATP needed for CO2 fixation (Fig. 4).
Notably, RubisCO was not found in the Comamonadaceae genome. To exclude the possibility of missing this gene due to the binning process, we searched the entire metagenomes for RubisCO genes, and based on sequence homology or contig fold coverage, no RubisCO gene was assigned to Comamonadaceae species. Besides RubisCO in the draft genomes, the search process also identified form II RubisCO in Thiobacillus (see Fig. S7 in the supplemental material), for which a draft genome was not recovered due to its low abundance in the culture (Fig. 1). Other carbon fixation pathways were not identified in Comamonadaceae sp. either, suggesting that Comamonadaceae sp. is an obligate heterotroph, like its close relative Acidovorax ebreus TPSY, an anaerobic nitrate-reducing Fe(II) oxidizer that is unable to fix CO2 and lacks any known CO2 fixation pathway (61). Therefore, Comamonadaceae sp. relies on carbon fixed by autotrophs in the community for growth (Fig. 4).
Central carbon metabolic pathways, including glycolysis, the pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle, were present in all draft genomes (Table 3). Transporters for organic molecules (e.g., amino acids and carbohydrates) are abundant in Bradyrhizobium sp., Rhizobium sp., and Comamonadaceae sp., in contrast with the scarcity of these transporters in the Gallionellaceae genome (see Table S3 in the supplemental material). The abundance profile of these transporters suggests that the flanking community members are generally metabolically versatile and may utilize diverse organic carbon substrates. This is consistent with the expectation that the flanking species may utilize organic carbon fixed by Gallionellaceae sp. for complete denitrification (Fig. 4).
Whether Gallionellaceae sp. can grow with organic substrates, such as acetate, is unclear. In its genome, we found a gene encoding acetyl-coenzyme A (CoA) synthetase (ACS), which activates acetate to acetyl-CoA to be utilized in the TCA cycle. We did not find the acetate permease (ActP) gene but identified a gene annotated as a proline transporter, which belongs to the same solute:sodium symporter family as ActP. Just based on bioinformatic analysis, it is not clear whether this protein also transports acetate. In addition, a gene annotated as AMP-forming acyl-CoA synthetase belonging to the fatty acid group translocation family (TC:4.C.1) is present, and its protein is predicted to be associated with the inner membrane. This gene forms an operon with a gene encoding a major facilitator transporter and might be able to transport and activate fatty acids. Nevertheless, genes encoding key enzymes in the glyoxylate cycle, such as isocitrate lyase and malate synthase, are missing, and genes for alternate pathways for acetate assimilation (62–64) are also absent. Therefore, Gallionellaceae sp. may not be able to fulfill all assimilation needs when acetate is the sole carbon source, unless the CO2-fixing CBB cycle is also operating to supplement the assimilation needs by using the CO2, NADH, and ATP generated from acetate catabolism. In contrast, genes for the glyoxylate cycle are present in Bradyrhizobium sp., Comamonadaceae sp., and Rhodanobacter sp., likely rendering them able to grow on acetate, as has been shown in some of their heterotrophic nitrate-reducing relatives isolated from culture KS (6).
Hydrogen metabolism.
Hydrogenases identified in the KS organisms all belong to the nickel iron ([NiFe]) type, including group 1, 3, and 4 [NiFe]-hydrogenases, according to the classification by Vignais et al. (46) (Table 3). Gallionellaceae sp. has three forms of [NiFe]-hydrogenases. First, a group 1 respiratory uptake hydrogenase capable of generating PMF by oxidizing H2 in the periplasm is encoded by the hya operon (65). The small and large subunits of this hydrogenase (HyaAB) are predicted to be periplasmic proteins based on their sequences, with the small subunit containing a twin-arginine signal motif for translocation (see Fig. S8a in the supplemental material); their unique genetic arrangement with a porin-coding gene is discussed in Text S6 in the supplemental material. Second, a soluble group 3d hydrogenase, catalyzing the reversible reaction of NAD reduction, is encoded in the hoxS (hydrogen oxidation, soluble hydrogenase) operon consisting of hoxFUYH (66), which can reduce NAD+ using hydrogen in the cytoplasm, providing reducing equivalents for cell growth (see Fig. S8a in the supplemental material). Third, a group 4 H2-evolving respiratory [NiFe]-hydrogenase is clustered with formate hydrogenlyase (FHL) genes in the genome, as part of the FHL complex that catalyzes the reversible conversion of formate to CO2 and H2 (67, 68). Among the [NiFe]-hydrogenases from flanking species, Bradyrhizobium sp. possesses groups 3 and 4, Rhizobium sp. possesses groups 1 and 4, and Rhodanobacter sp. possesses group 4, whereas no hydrogenase was identified in the Comamonadaceae genome (Table 3). Experiments are under way to evaluate H2 metabolism by culture KS-Tueb and pure cultures of Bradyrhizobium, Rhodanobacter, and Nocardioides strains isolated from KS-Tueb.
Other aspects, such as sulfur and phosphorus metabolisms, strategies to avoid encrustation, and environmental sensing and chemotaxis responding to oxygen, nitrate, and nitrite, are discussed in Text S7 to S9 in the supplemental material.
Overall, our analyses suggest that culture KS microbial community composition changed over time and differed with independent transfer histories. However, the primary Fe(II) oxidizer, a bacterium in the Gallionellaceae family, remained as the predominant member. The recovered draft genomes allowed us to identify a number of putative EET genes and enabled us to infer metabolic potential and gain new insights from these organisms. For example, the genome suggests Gallionellaceae sp. has a RubisCO system with low affinity for CO2, requires ammonia in the environment for growth, might be able to use oxygen under microoxic conditions for Fe(II) oxidation besides nitrate reduction, lacks NO and N2O reductase genes, and therefore may partner with flanking populations for complete denitrification. Meanwhile, Gallionellaceae sp. may provide the flanking members with organic carbon for complete denitrification. These potential interspecies interactions and metabolic interdependencies among culture KS organisms may allow them to cooperate effectively to achieve robust chemolithoautotrophic NDFO as a whole. Therefore, our results significantly expand our knowledge of the metabolic diversity of FeOB, provide new insights into chemolithoautotrophic NDFO, and suggest a range of new targets for genomics-based analysis of the evolutionary relationships, molecular mechanisms, and environmental regulation of Fe(II)-oxidizing chemolithotrophs.
Supplementary Material
ACKNOWLEDGMENT
We thank Brandon Converse for conducting DNA extraction.
Funding Statement
This work, including the efforts of Shaomei He and Eric Roden, was funded by NASA Astrobiology Institute (NNA13AA94A). This work, including the efforts of Eric Roden, was funded by University of Wisconsin-Madison (UW) (Vilas Associateship award). This work, including the efforts of Andreas Kappler, was funded by the European Research Council under the European Union's Seventh Framework Program (FP/2007–2013)/ERC grant, agreement no. 307320 – MICROFOX.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03493-15.
REFERENCES
- 1.Hedrich S, Schlömann M, Johnson DB. 2011. The iron-oxidizing proteobacteria. Microbiology 157:1551–1564. doi: 10.1099/mic.0.045344-0. [DOI] [PubMed] [Google Scholar]
- 2.Weber KA, Urrutia MM, Churchill PF, Kukkadapu RK, Roden EE. 2006. Anaerobic redox cycling of iron by freshwater sediment microorganisms. Environ Microbiol 8:100–113. doi: 10.1111/j.1462-2920.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- 3.Roden EE. 2012. Microbial iron-redox cycling in subsurface environments. Biochem Soc Trans 40:1249–1256. doi: 10.1042/BST20120202. [DOI] [PubMed] [Google Scholar]
- 4.Straub KL, Benz M, Schink B, Widdel F. 1996. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol 62:1458–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straub KL, Schönhuber WA, Buchholz-Cleven BEE, Schink B. 2004. Diversity of ferrous iron-oxidizing, nitrate-reducing bacteria and their involvement in oxygen-independent iron cycling. Geomicrobiol J 21:371–378. doi: 10.1080/01490450490485854. [DOI] [Google Scholar]
- 6.Blöthe M, Roden EE. 2009. Composition and activity of an autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment culture. Appl Environ Microbiol 75:6937–6940. doi: 10.1128/AEM.01742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schädler S, Burkhardt C, Hegler F, Straub K, Miot J, Benzerara K, Kappler A. 2009. Formation of cell-iron-mineral aggregates by phototrophic and nitrate-reducing anaerobic Fe(II)-oxidizing bacteria. Geomicrobiol J 26:93–103. doi: 10.1080/01490450802660573. [DOI] [Google Scholar]
- 8.Weber KA, Picardal FW, Roden EE. 2001. Microbially catalyzed nitrate-dependent oxidation of biogenic solid-phase Fe(II) compounds. Environ Sci Technol 35:1644–1650. doi: 10.1021/es0016598. [DOI] [PubMed] [Google Scholar]
- 9.Shelobolina E, Xu H, Konishi H, Kukkadapu R, Wu T, Blöthe M, Roden E. 2012. Microbial lithotrophic oxidation of structural Fe(II) in biotite. Appl Environ Microbiol 78:5746–5752. doi: 10.1128/AEM.01034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerson D, Moyer C. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol 63:4784–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson HK, Clark IC, Melnyk RA, Coates JD. 2012. Toward a mechanistic understanding of anaerobic nitrate-dependent iron oxidation: balancing electron uptake and detoxification. Front Microbiol 3:57. doi: 10.3389/fmicb.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilbert M, Bonnefoy V. 2013. Insight into the evolution of the iron oxidation pathways. Biochim Biophys Acta 1827:161–175. doi: 10.1016/j.bbabio.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Beliaev AS, Saffarini DA. 1998. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol 180:6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Wang Z, Belchik SM, Edwards MJ, Liu C, Kennedy DW, Merkley ED, Lipton MS, Butt JN, Richardson DJ, Zachara JM, Fredrickson JK, Rosso KM, Shi L. 2012. Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front Microbiol 3:37. doi: 10.3389/fmicb.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4:254. doi: 10.3389/fmicb.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L, Rosso KM, Zachara JM, Fredrickson JK. 2012. Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem Soc Trans 40:1261–1267. doi: 10.1042/BST20120098. [DOI] [PubMed] [Google Scholar]
- 17.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 18.Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF. 2014. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J 8:1452–1463. doi: 10.1038/ismej.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, Huntemann M, Billis K, Varghese N, Tennessen K, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, Valas R, Novotny M, Yee-Greenbaum J, Selengut JD, Haft DH, Halpern AL, Lasken RS, Nealson K, Friedman R, Venter JC. 2012. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol 10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113–113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 27.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 28.Yu CS, Chen YC, Lu CH, Hwang JK. 2006. Prediction of protein subcellular localization. Proteins 64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 29.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. 2004. PRED-TMBB: a Web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res 32:W400-404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter K, Schicklberger M, Gescher J. 2012. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl Environ Microbiol 78:913–921. doi: 10.1128/AEM.06803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta T, Childers SE, Glaven R, Lovley DR, Mester T. 2006. A putative multicopper protein secreted by an atypical type II secretion system involved in the reduction of insoluble electron acceptors in Geobacter sulfurreducens. Microbiology 152:2257–2264. doi: 10.1099/mic.0.28864-0. [DOI] [PubMed] [Google Scholar]
- 33.Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leang C, Coppi MV, Lovley DR. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol 185:2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes DE, Mester T, O'Neil RA, Perpetua LA, Larrahondo MJ, Glaven R, Sharma ML, Ward JE, Nevin KP, Lovley DR. 2008. Genes for two multicopper proteins required for Fe(III) oxide reduction in Geobacter sulfurreducens have different expression patterns both in the subsurface and on energy-harvesting electrodes. Microbiology 154:1422–1435. doi: 10.1099/mic.0.2007/014365-0. [DOI] [PubMed] [Google Scholar]
- 36.Castelle C, Guiral M, Malarte G, Ledgham F, Leroy G, Brugna M, Giudici-Orticoni MT. 2008. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem 283:25803–25811. doi: 10.1074/jbc.M802496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barco RA, Emerson D, Sylvan JB, Orcutt BN, Jacobson Meyers ME, Ramirez GA, Zhong JD, Edwards KJ. 2015. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81:5927–5937. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M, Shapiro N, Woyke T, Chan CS. 2015. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol 6:1265. doi: 10.3389/fmicb.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croal LR, Jiao Y, Newman DK. 2007. The fox operon from Rhodobacter strain SW2 promotes phototrophic Fe(II) oxidation in Rhodobacter capsulatus SB1003. J Bacteriol 189:1774–1782. doi: 10.1128/JB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian X, Reguera G, Mester T, Lovley DR. 2007. Evidence that OmcB and OmpB of Geobacter sulfurreducens are outer membrane surface proteins. FEMS Microbiol Lett 277:21–27. doi: 10.1111/j.1574-6968.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 41.Corstjens PLAM, de Vrind JPM, Goosen T, de Vrind-de Jong EW. 1997. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol J 14:91–108. doi: 10.1080/01490459709378037. [DOI] [Google Scholar]
- 42.Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ, White G, Baiden N, Gates AJ, Marritt SJ, Clarke TA. 2012. The ‘porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol Microbiol 85:201–212. doi: 10.1111/j.1365-2958.2012.08088.x. [DOI] [PubMed] [Google Scholar]
- 43.Jiao Y, Newman DK. 2007. The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J Bacteriol 189:1765–1773. doi: 10.1128/JB.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Wang Z, Liu J, Levar C, Edwards MJ, Babauta JT, Kennedy DW, Shi Z, Beyenal H, Bond DR, Clarke TA, Butt JN, Richardson DJ, Rosso KM, Zachara JM, Fredrickson JK, Shi L. 2014. A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ Microbiol Rep 6:776–785. doi: 10.1111/1758-2229.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi L, Fredrickson JK, Zachara JM. 2014. Genomic analyses of bacterial porin-cytochrome gene clusters. Front Microbiol 5:657. doi: 10.3389/fmicb.2014.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignais PM, Billoud B, Meyer J. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 47.Karp G. 2008. Cell and molecular biology. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 48.Elbehti A, Brasseur G, Lemesle-Meunier D. 2000. First evidence for existence of an uphill electron transfer through the bc1 and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous iron-oxidizing bacterium Thiobacillus ferrooxidans. J Bacteriol 182:3602–3606. doi: 10.1128/JB.182.12.3602-3606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruscella P, Appia-Ayme C, Levican G, Ratouchniak J, Jedlicki E, Holmes DS, Bonnefoy V. 2007. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology 153:102–110. doi: 10.1099/mic.0.2006/000067-0. [DOI] [PubMed] [Google Scholar]
- 50.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol 178:1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arai H, Kawakami T, Osamura T, Hirai T, Sakai Y, Ishii M. 2014. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol 196:4206–4215. doi: 10.1128/jb.02176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabel C, Maier RJ. 1993. Oxygen-dependent transcriptional regulation of cytochrome aa3 in Bradyrhizobium japonicum. J Bacteriol 175:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picardal F. 2012. Abiotic and microbial interactions during anaerobic transformations of Fe(II) and NOx−. Front Microbiol 3:112. doi: 10.3389/fmicb.2012.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klueglein N, Zeitvogel F, Stierhof Y-D, Floetenmeyer M, Konhauser KO, Kappler A, Obst M. 2014. Potential role of nitrite for abiotic Fe(II) oxidation and cell encrustation during nitrate reduction by denitrifying bacteria. Appl Environ Microbiol 80:1051–1061. doi: 10.1128/AEM.03277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tai Y-L, Dempsey BA. 2009. Nitrite reduction with hydrous ferric oxide and Fe(II): stoichiometry, rate, and mechanism. Water Res 43:546–552. doi: 10.1016/j.watres.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 56.Kampschreur MJ, Kleerebezem R, de Vet WW, van Loosdrecht MC. 2011. Reduced iron induced nitric oxide and nitrous oxide emission. Water Res 45:5945–5952. doi: 10.1016/j.watres.2011.08.056. [DOI] [PubMed] [Google Scholar]
- 57.Sørensen J, Thorling L. 1991. Stimulation by lepidocrocite (7-FeOOH) of Fe(II)-dependent nitrite reduction. Geochim Cosmochim Acta 55:1289–1294. doi: 10.1016/0016-7037(91)90307-Q. [DOI] [Google Scholar]
- 58.Sobolev D, Roden EE. 2001. Suboxic deposition of ferric iron by bacteria in opposing gradients of Fe(II) and oxygen at circumneutral pH. Appl Environ Microbiol 67:1328–1334. doi: 10.1128/AEM.67.3.1328-1334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roden EE, Sobolev D, Glazer B, Luther GW III. 2004. Potential for microscale bacterial Fe redox cycling at the aerobic-anaerobic interface. Geomicrobiol J 21:379–391. doi: 10.1080/01490450490485872. [DOI] [Google Scholar]
- 60.Badger MR, Bek EJ. 2008. Multiple RubisCO forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 59:1525–1541. [DOI] [PubMed] [Google Scholar]
- 61.Byrne-Bailey KG, Weber KA, Chair AH, Bose S, Knox T, Spanbauer TL, Chertkov O, Coates JD. 2010. Completed genome sequence of the anaerobic iron-oxidizing bacterium Acidovorax ebreus strain TPSY. J Bacteriol 192:1475–1476. doi: 10.1128/JB.01449-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol Microbiol 61:297–309. doi: 10.1111/j.1365-2958.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- 63.Korotkova N, Chistoserdova L, Kuksa V, Lidstrom ME. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J Bacteriol 184:1750–1758. doi: 10.1128/JB.184.6.1750-1758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider K, Peyraud R, Kiefer P, Christen P, Delmotte N, Massou S, Portais JC, Vorholt JA. 2012. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium extorquens AM1 during growth on acetate. J Biol Chem 287:757–766. doi: 10.1074/jbc.M111.305219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menon NK, Robbins J, Peck HD Jr, Chatelus CY, Choi ES, Przybyla AE. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol 172:1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tran-Betcke A, Warnecke U, Bocker C, Zaborosch C, Friedrich B. 1990. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol 172:2920–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maeda T, Sanchez-Torres V, Wood TK. 2007. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Appl Microbiol Biotechnol 76:1035–1042. doi: 10.1007/s00253-007-1086-6. [DOI] [PubMed] [Google Scholar]
- 68.Sauter M, Böhm R, Böck A. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol 6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.