Abstract
The literature of environmental microbiology broadly discusses issues associated with microbial hazards in archives, but these publications are mainly devoted to paper documents. There are few articles on historical parchment documents, which used to be very important for the development of literature and the art of writing. These studies present a broad spectrum of methods for the assessment of biodeterioration hazards of the parchment document in question. They are based on both conventional microbiological methods and advanced techniques of molecular biology. Here, a qualitative analysis was conducted, based on genetic identification of bacteria and fungi present on the document as well as denaturing gradient gel electrophoresis profiling and examining the destructive potential of isolated microbes. Moreover, the study involved a quantitative and qualitative microbiological assessment of the indoor air in the room where the parchment was kept. The microbes with the highest destructive potential that were isolated from the investigated item were Bacillus cereus and Acinetobacter lwoffii bacteria and Penicillium chrysogenum, Chaetomium globosum, and Trichoderma longibrachiatum fungi. The presence of the B. cereus strain was particularly interesting since, under appropriate conditions, it leads to complete parchment degradation within several days.
INTRODUCTION
The incorporation charter for the city of Krakow, issued on 5 June 1257, is one of the oldest and most valuable documents stored in the National Archives in Krakow, Poland. It is also one of the oldest incorporation charters in the collections of the Polish national archives. It was a legal document that incorporated Krakow under the Magdeburg rights. It granted the city the character of a municipality with its self-government and judicial privileges. As a historical document, it is a monument of developing administration, diplomacy, and the culture of that time. The incorporation charter was written in Latin on a thick parchment, the size of which makes it one of the greatest ducal documents. The lapse of time, however, has left traces on the document. One can notice numerous cracks, breaks, and parchment defects. However, thanks to conservation and the appropriate conditions under which it has been stored, the document has been preserved in a good condition to the present day (1, 2). It is currently stored in the headquarters of the National Archives in Krakow in a specially designed packaging made of acid-free paper. In 2014, the incorporation charter for the city of Krakow was placed in the National Register of the Memory of the World UNESCO program, the main goal of which is undertaking actions aimed at saving, exhibiting, and preserving world heritage documents for contemporary and future generations.
The problem of microbial hazards of archival items is broadly addressed in the literature (3–7). Collections of written documents are a particular type of cultural heritage object, since they are carriers of broadly understood historical and cultural knowledge. Documents and other archives are usually made of organic materials, such as paper, parchment, and leather. Parchment, being the first material to carry writing, has played a crucial role in the development of culture and diplomacy. Historical parchment documents are rich sources of information, not only for history and culture but also for technology. Parchment was made of various types of hide, usually of goat, calf, or sheep. During its production, chemical agents exposed the dermis, which consisted mainly of type I collagen. Moreover, the surface layer of collagen was degraded, which led to the formation of gelatin, a polypeptide with a composition similar to that of collagen. As a result of these technological processes, the parchment product consisted of the inner collagen layer, with its typical fiber arrangement, and external gelatin layers (8–10). As with other materials that carry writing, such as paper and leather, parchment is subject to the process of biodeterioration (3, 5, 11–13). This process consists of the microbial enzymatic degradation of the organic components of these materials, which in the case of parchment is mainly collagen and in the case of paper is mainly cellulose (13, 14). Under favorable conditions, microbes capable of biodeterioration can settle onto historical objects and into environments in which they are collected and stored (15). If they grow actively, they can cause changes on the surface of documents, such as discolorations or stains, and can weaken the structure of the material and increase its brittleness until complete destruction occurs (3, 16–19). In discussions of the microbial hazards of cultural heritage objects, filamentous fungi and actinobacteria are the most important microbes, since, in comparison with bacteria, they are capable of growing actively in environments with lower temperature and relative humidity. Fungi that are most commonly isolated from archival and museum environments (both from the air and from the objects) include strains of the Alternaria, Aspergillus, Cladosporium, Chaetomium, Penicillium, Phoma, Stachybotrys, and Trichoderma genera (20). Microbial activity in an environment depends not only on species-related properties but also on external factors, such as temperature, humidity, pH or light exposure (21–23), ventilation pollutants (such as ozone and volatile organic compounds), and particulate matter (e.g., PM2.5 and PM10) (24). Recently, conservators, archivists, and museologists have emphasized the need for prevention and appropriate protection of historical objects rather than for renovation or reconstruction (13). The basis for the implementation of appropriate remedial measures is detailed research aiming at the identification of a potential microbial hazard. That is why, apart from conventional microbiological methods, researchers also apply advanced techniques of molecular biology. This is dictated by certain limitations of traditional research methods, such as limited possibilities offered by microbial culture-based techniques, their accuracy in identifying microorganisms, and the long time needed to conduct such studies (4, 25, 26).
The aim of this study was to determine the microbial hazards concerning one of the most important parchment documents stored in the National Archives in Krakow by examining microbial contamination on the parchment, determining the destructive activity of microbial isolates, and analyzing environmental conditions. Since the incorporation charter for the city of Krakow is a unique document, conventional methods were supplemented with advanced techniques of molecular biology in order to obtain more detailed information about the microbiological condition of the parchment in question. The microbiological analysis of the incorporation charter for the city of Krakow presented in this study was carried out in connection with ongoing comprehensive research of the document, including historical, conservation, and technological aspects, for better protection of such a unique document.
MATERIALS AND METHODS
The investigation involved a parchment document from the 13th century—the incorporation charter for the city of Krakow (Fig. 1). The parchment was tanned on both sides and has been partially glued with patches of parchment and paper during conservation. The size of the document is 505 mm high by 470 mm wide, with a pleat 74 mm high. It has certain defects on its upper half and left side. The parchment is creased and cracked vertically and horizontally through the center of the document. It was initially stored in a locked metal box. Subsequently, it was kept in a wooden and glass cabinet. Currently, it is stored in specially prepared packaging of acid-free paper in the storeroom of the National Archives in Krakow. The storeroom is situated on the first floor of a historical tenement, which has housed the National Archives since 1887. The room stores various collections, including paper documents (manuscripts, notes, old prints), parchments, leather bindings, and wax seals. Sampling for microbiological tests was conducted twice within a 3-month period in the autumn and winter.
FIG 1.
The incorporation charter, front and back, for the city of Krakow. The image was made available by the National Archives in Krakow.
Temperature and relative humidity monitoring.
The indoor climate of the storeroom where the incorporation charter for the city of Krakow is kept is constantly monitored, and temperature and relative humidity readings are taken twice daily (at 8 a.m. and 3 p.m.; Thermohygrometer MAX-MIN, Viking AB, Sweden). The analysis of the temperature and relative humidity changes was conducted from 8 October 2014 to 8 October 2015.
Microbiological analysis.
Samples for microbiological tests were collected with sterile swabs and FungiTape (ThermoFisher, USA). In the laboratory, the material was cultured on culture media for fungi (malt extract agar [MEA] and Sabouraud glucose agar [SGA]) and bacteria (Trypticase soy agar [TSA]). Moreover, the material was also obtained via spontaneous contamination of sterile parchment and paper samples placed on the document for the period of 3 months. Following contamination, these samples were incubated on SGA, MEA, and TSA media as well as on Weary & Canby medium without sucrose in order to isolate microbes with proteolytic properties. Because the document was glued with fragments of paper during conservation, microbes with cellulolytic properties were isolated with the use of Czapek Dox agar without sucrose. The media were incubated at a temperature of 28°C ± 2°C for 4 weeks and observed periodically. Microorganisms were then isolated until pure cultures were obtained and identified.
Molecular identification of fungi and bacteria.
Fungal DNA was isolated with the use of a commercial NucleoSpin plant II kit (Macherey-Nagel GmbH & Co. KG, Germany). Fungal growth was collected with a sterile scalpel from a 5- to 6-day culture conducted on the MEA medium and homogenized using a Minilys homogenizer (Bertin Technologies, France). The subsequent stages of isolation were conducted in accordance with the manufacturer's protocol. Bacterial DNA was extracted from 48-h cultures on broth agar using a bacterial and yeast genomic DNA purification kit (Eurx, Warsaw, Poland) in accordance with the enclosed protocol. Microbial identification was conducted on the basis of a sequence analysis of a fragment of the 16S rRNA gene in bacteria and the internal transcribed spacer (ITS) in fungi. PCR was conducted using 25 μl of a reaction mixture consisting of 1.5 μl of DNA matrix, 0.4 μmol of each primer (Table 1) (Genomed, Warsaw, Poland), 200 μl/liter of each deoxynucleoside triphosphate (dNTP; Sigma-Aldrich, St. Louis, MO, USA), 2 U of Taq DNA polymerase (Invitrogen, Thermo Fisher Scientific), 1× polymerase buffer, and 1.5 mmol of MgCl2. The PCR conditions depended on the pair of starters used. For primers U968 and L1401, the reaction program was as follows: initial denaturation at 95°C for 5 min, 34 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 1 min, and extension for 1 min at 72°C, as well as a final extension at 72°C for 10 min. In the case of fungal identification, ITS1 and ITS4 primers were used (Table 1). The amplification was conducted according to the following program: initial denaturation at 95°C for 5 min, 34 cycles of denaturation at 94°C for 50 s, annealing at 56°C for 50 s, and extension for 50 s at 72°C, as well as a final extension at 72°C for 10 min.
TABLE 1.
Primers used for microbial identification in molecular tests
| Primer | Sequence | Region | Reference(s) |
|---|---|---|---|
| Fungal primers | |||
| ITS1F | 5′-CTTGGTCATTTAGAGGAAGTAA-3′ | ITS1 | 27 |
| ITS1F-GC | 5′-CGCCCGCCGGCGGCGGCGGGCGGGGCGGGGGCA-CGGGGGGCTTGGTCATTTAGAGGAAGTAA-3′ | ITS1 | 27 |
| ITS1 | 5′-TCCGTAGGTGAACCTGCGG-3′ | ITS1 | 28, 29 |
| ITS2 | 5′-GCTGCGTTCTTCATCGATGC-3′ | ITS1 | 28 |
| ITS4 | 5′- TCCTCCGCTTATTGATATGC-3′ | ITS1 | 28, 29 |
| Bacterial primers | |||
| U968 | 5′-AACGCGAAGAACCTTAC-3′ | 16S rRNA gene | 30 |
| L1401 | 5′-CGGTGTGTACAAGACCC-3′ | 16S rRNA gene | 30 |
The PCR products obtained were separated by electrophoresis in an agarose gel with a concentration of 1.8% and visualized under UV light using a SimplySafe dye (Eurx, Poland).
The products obtained were purified and sequenced (Genomed, Warsaw, Poland). Based on the nucleotide sequence obtained, species identification was conducted using the NCBI database and the Basic Local Alignment Search Tool (BLAST) for DNA sequence analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Fungal diversity analysis.
The diversity analysis of fungi found on the surface of the incorporation chapter was conducted with the use of PCR-denaturing gradient gel electrophoresis (PCR-DGGE). This method obtains the genotypic structure of a biocenosis based on the analysis of differences in selected genomic sequences. The nested-PCR technique, which consists of two PCRs, was applied for the amplification of the fungal ITS1 region. In the first PCR, ITS1F and ITS4 primers were used, and the amplicons obtained were used as a matrix for the second PCR, which was conducted with the use of ITS1F-GC (ITS1F with a GC tail) and ITS4 primers (Table 1). Both the composition of the mix and the reaction conditions were compliant with the procedure designed in previous research (31).
The obtained PCR products underwent electrophoresis in a concentration gradient in a DCode universal mutation detection system (Bio-Rad, Hercules, CA, USA). The amplicons with a GC tail were separated in 8% polyacrylamide gel (acrylamide-to-bisacrylamide ratio, 39.5:1) containing 30% to 60% of the denaturation factor (urea). Following electrophoresis, the gel was dyed in SYBR green (1:10,000, Invitrogen). The band pattern (fingerprint) obtained was visualized under UV light and photographed. The densitometric analysis, attesting to the concentration of the products obtained, was conducted using Image J (National Institutes of Health, USA). The Shannon biodiversity index was calculated. It expresses the diversity of microbial genotypes in an investigated environment, which in this study was the surface of the document (32).
Identification of prevailing genotypes based on a DGGE profile.
Based on the DGGE profile obtained, assays of the prevailing taxons present on the object were conducted. The selected DGGE fingerprint bands were excised with a sterile scalpel. Subsequently, they were suspended and vortexed in water for molecular biology analysis (Sigma-Aldrich). The repeated reamplification of the excised products was conducted under the same conditions as the first amplification. The entire volume of the PCR product was placed on the gel, and electrophoresis was conducted. The band obtained was excised with a sterile scalpel again, and the genetic material was extracted and purified with the use of the commercial GelElute kit (Sigma-Aldrich). The product obtained underwent sequencing.
Bioluminescence analysis.
The bioluminescence test consists of the measurement of a bioluminescence level. Its formation results from the ATP released by biological matter, including microbes. Samples for investigation were collected with wet swabs from two sites on the surface of the parchment, limited within a template of 25 cm2. Subsequently, they were measured in a Hy-Lite2 system (Merck, Darmstadt, Germany), and the result was expressed in relative light units (RLU).
Verification of the biodeterioration potential of isolates.
In order to evaluate the biodeterioration potential of microorganisms isolated from the surface of the document, it was analyzed how capable these microbes were to enzymatically degrade parchment based on verification of their abilities to overgrow on parchment. Pure microbial cultures were used to prepare suspensions which were placed onto sterile parchment samples with a surface area of 4 cm2. Prior to this, the parchment samples were sterilized by incubation in 3% H2O2 for 1 h at room temperature. Next, they were rinsed in sterile water for 5 min in accordance with the procedure proposed by Strzelczyk and Karbowska-Berent (33). The inoculated parchment samples, placed on a mineral medium (Weary & Canby), were incubated at 28°C ± 2°C for 30 days and observed periodically. Bacterial and fungal growth was observed both macroscopically and microscopically (stereomicroscope, scanning electron microscope [SEM]). The proteolytic properties of microbial strains were verified in a gelatin test.
SEM.
Parchment samples with visible microbial growth were fixed in 3% glutaraldehyde (CAS no. 111-30-8) in 0.2 M cacodylic buffer for 24 h at 4°C. Subsequently, they were dehydrated in ethanol in a range of concentrations (10%, 25%, 50%, 75%, 90%, 96%) for 20 min in each concentration and in 100% ethanol four times for 30 min (33). These samples were then gold sputtered and analyzed under a microscope (model S-4700 field emission SEM; Hitachi, Tokyo, Japan).
Microbiological analysis of air.
In order to get to know the environment in which the investigated object was stored, a simplified qualitative and quantitative analysis of microbes present in the indoor air of the archival storeroom was conducted. To do this, air was collected with a MAS-100 Eco sampler (Merck) with an airflow rate of 100 liters/min for 1 min. In order to specify the count of bacteria in the air, TSA was used. For fungi, three types of media were used: MEA, SGA, and dichloran glycerol agar (DG18). The samples were incubated at 28°C ± 2°C for 72 h for bacteria and for 14 days for fungi, with periodical observations and isolations of dominant strains. The prevailing microbial strains were identified genetically according to the procedure described above.
RESULTS
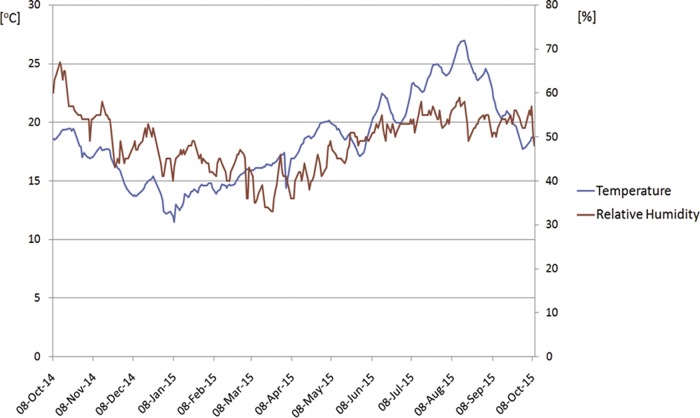
The physical parameters of the indoor climate are very important from a microbiological hazard point of view, since uncontrolled significant changes can lead to rapid microbial development. The highest temperature was noted in the summer (27°C), and the lowest was noted in the winter (11.5°C) (Fig. 2). The measurements showed relatively large temperature fluctuations (annual amplitude, 15.5°C) depending on the outdoor temperature. The relative humidity parameter seemed to be more stable. The highest value of 67% was noted in the autumn, and the lowest (33%) was noted in the winter. The difference in relative humidity was probably caused by the usage of heaters during the winter (Fig. 2).
FIG 2.
Changes of the indoor climate parameters (temperature and relative humidity) in the National Archives in Krakow, where the incorporation charter is stored.
Molecular identification of microorganisms.
Double sampling and the application of various sampling techniques as well as appropriate selection of culture media enabled 27 bacterial and 15 fungal isolates to be obtained from the surface of the document. The results of bacterial identification, conducted on the basis of the analysis of the conserved 16S rRNA gene region sequence, are presented in Table 2. The prevailing bacterial species included those of the Bacillus genus (n = 9 isolates; Bacillus cereus, Bacillus licheniformis, Bacillus megaterium, Bacillus pseudomycoides, Bacillus subtilis), the Paenibacillus genus (n = 5 isolates), and the Staphylococcus genus (n = 5 isolates). The air relative humidity in the storage room, examined for the period of conducting research, did not exceed 67%; according to the literature, the value of this parameter is not sufficient to enable active bacterial growth, but bacteria can stay in an inactive form. The identification of fungal species was made on the basis of the noncoding ITS region sequence (Table 3). Of fungal microflora, the predominant species belonged to the Penicillium genus (Penicillium chrysogenum, n = 6 isolates; Penicillium decumbens, n = 1 isolate). Isolated and assayed microorganisms, both fungi and bacteria, make up the microflora that is typical of archives (3–5, 18, 20, 22).
TABLE 2.
Identification of bacterial isolates
| Bacterial isolate | Accession no. (% similarity) |
|---|---|
| Acinetobacter lwoffii | KP282795.1 (99) |
| Bacillus sp. | JQ359106.1 (99) |
| Bacillus cereus | EU111736.1 (100) |
| Bacillus licheniformis | FJ976557.1 (99) |
| Bacillus megaterium | KJ126921.1 (99) |
| Bacillus pseudomycoides | KR063200.1 (99) |
| Bacillus subtilis | KP010381.1 (99) (4 isolates) |
| Kocuria kristinae | KF322132.1 (99) |
| Massilia sp. | KJ999603.1 (99) |
| Micrococcus luteus | KP261839.1 (98) and KM874397.1 (99) (2 isolates) |
| Micrococcus yunnanensis | KP893295.1 (99) |
| Pseudomonas psychrotolerans | LC040947.1 (99) |
| Pseudomonas stutzeri | JN613328.1 (99) |
| Paenibacillus amylolyticus | LK391509.1 (98) |
| Paenibacillus fonticola | JN638424.1 (99) |
| Paenibacillus sp. | EF156930.1 (100) (2 isolates) |
| Paenibacillus xylanilyticus | JX035945.1 (99) |
| Staphylococcus warneri | KR059861.1 (99) |
| Staphylococcus epidermidis | KP282762.1 (99) |
| Staphylococcus capitis | JF302671.1 (99) |
| Staphylococcus hominis | JQ734768.1 (99) (2 isolates) |
TABLE 3.
Identification of fungal isolates
| Fungal isolate | Accession no. (% similarity) |
|---|---|
| Alternaria tenuissima | JN624884.1 (100) |
| Aspergillus fumigatus | HM776412.1 (99) |
| Bjerkandera adusta | LN714526.1 (99) |
| Chaetomium globosum | HG530327.1 (99) and GU138648.1 (99) |
| Cryptococcus sp. | AF444449.1 (100) |
| Penicillium chrysogenum | KF999008.1 (99) (2 isolates), JF731255.1 (99) (3 isolates), and JX139710.1 (100) (1 isolate) |
| Penicillium decumbens | AY373909.1 (99) |
| Trichoderma longibrachiatum | KP281710.1 (99) and KP281708.1 (99) |
Biodiversity analysis of fungi on the surface of the document.
Since fungi play the major role in the process of biodeterioration of cultural heritage objects, mainly because of lower requirements concerning temperature and relative humidity and due to their destructive potential, an additional genetic analysis of fungal biodiversity was conducted. The PCR-DGGE method, which enables the creation of a genetic profile for the investigated environment, was used for this purpose. The advantage of this method is the ability to obtain information about the genotypic diversity of microbes present on the object without the need to conduct microbial cultures. The fact that the culture stage can be omitted is of crucial significance, since only a slight percentage of microbes can be cultured in laboratory settings. Based on the analysis, the biodiversity level was determined to be 2.12 according to the Shannon index. Moreover, 5 dominant genotypes were found on the surface of the document (Fig. 3).
FIG 3.
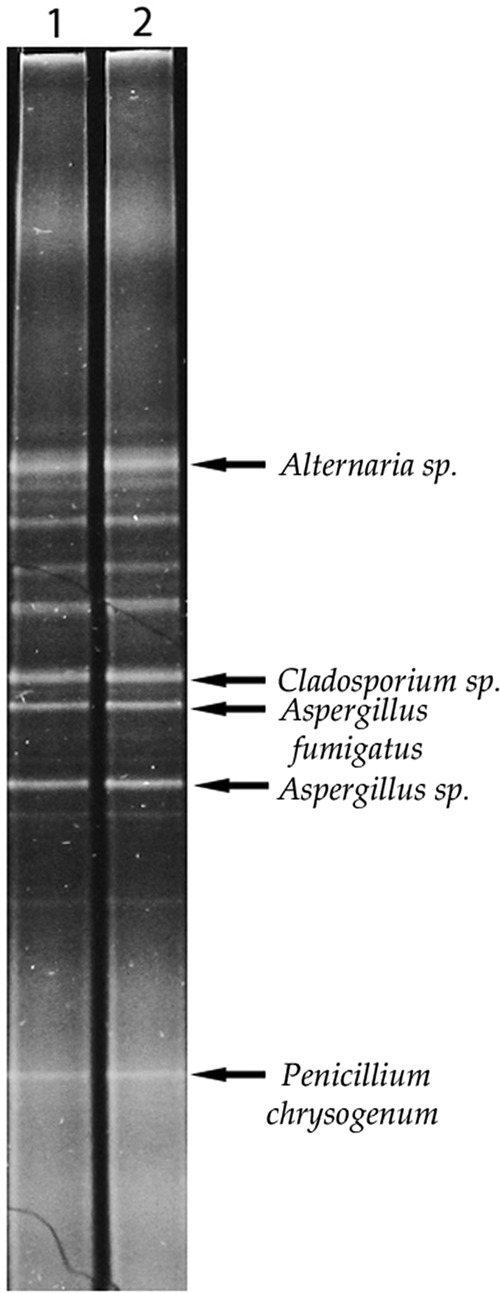
DGGE fingerprint showing fungal community structure on the surface of the document. The DGGE profile is shown in two repetitions, lanes 1 and 2. Arrows indicate the dominant genotypes.
The bands for dominant genotypes were excised from the DGGE profile and sequenced following reamplification and purification. The identification of dominating genotypes revealed the presence of previously identified fungal strains of the Aspergillus fumigatus species (HM776412.1; 98%) and Penicillium chrysogenum species (KF999008.1; 99%). However, it also enabled the identification of previously undetected species of Cladosporium (KF293973.1, 98% similarity) and Aspergillus (JQ388268, 99% similarity). The DGGE profile also enabled determination of a nonculturable strain of the Alternaria genus (HQ845239.1, 99% similarity).
Bioluminescence analysis.
The bioluminescence analysis of the tested parchment and of the inner surface of the packaging in which the document is kept measured 450 RLU for the document and 56 RLU for its packaging in the first sampling and 440 RLU and 48 RLU, respectively, for the second sampling. The bioluminescence level was stable both on the parchment and on its packaging, and it was 10 times greater on the document than on the packaging.
Verification of the biodeterioration potential.
The proteolytic properties of isolated and assayed microbes were verified in a gelatin test, the results of which are presented in Table 4 for bacterial isolates and in Table 5 for fungal isolates. Of 27 bacterial isolates tested, 60% of strains demonstrated the ability to degrade gelatin. For fungi, the test was positive in 80% of the strains examined. Considering the fact that gelatin is made of the same protein sequence as collagen, and that the only difference lies in the lack of the fibrillar structure, the ability of microbes to liquidate gelatin becomes a significant parameter. For the purpose of further verification of the microbial potential to degrade the protein material, the ability of microbes to grow on contemporary parchment was tested.
TABLE 4.
Bacterial growth on parchment and gelatin test results
| Bacterial isolate | Growtha on parchment at day: |
Gelatin test | |||
|---|---|---|---|---|---|
| 7 | 14 | 21 | 30 | ||
| Acinetobacter lwoffii | ++ | ++ | ++ | ++ | + |
| Bacillus sp. | − | − | − | − | + |
| Bacillus cereus | ++ | ++ | ++ | ++ | + |
| Bacillus licheniformis | − | − | − | − | − |
| Bacillus megaterium | − | − | − | − | + |
| Bacillus pseudomycoides | − | − | − | − | + |
| Bacillus subtilis | |||||
| Isolate 1 | +/− | + | + | ++ | + |
| Isolate 2 | +/− | + | + | + | + |
| Isolate 3 | + | ++ | ++ | ++ | + |
| Isolate 4 | + | + | + | ++ | + |
| Kocuria kristinae | − | − | − | − | − |
| Massilia sp. | − | − | − | − | − |
| Micrococcus luteus | |||||
| KP261839.1 (98%) | − | − | − | − | + |
| KM874397.1 (99%) | |||||
| Isolate 1 | − | − | − | − | − |
| Isolate 2 | − | − | − | − | − |
| Micrococcus yunnanensis | − | − | − | − | − |
| Pseudomonas psychrotolerans | − | − | − | − | + |
| Pseudomonas stutzeri | − | − | − | − | − |
| Paenibacillus amylolyticus | +/− | + | + | + | + |
| Paenibacillus fonticola | +/− | + | + | + | |
| Paenibacillus sp. | |||||
| Isolate 1 | − | − | − | − | + |
| Isolate 2 | − | + | + | ++ | + |
| Paenibacillus xylanilyticus | − | − | − | − | − |
| Staphylococcus warneri | − | − | − | − | − |
| Staphylococcus epidermidis | − | +/− | + | + | + |
| Staphylococcus capitis | − | − | − | − | − |
| Staphylococcus hominis | |||||
| Isolate 1 | − | − | − | − | + |
| Isolate 2 | − | − | − | − | − |
−, no growth; +/−, growth only on the sample edges; +, growth on the surface; ++, intensive growth with sample structural changes.
TABLE 5.
Fungal growth on parchment and gelatin test results
| Fungal strain | Growtha on parchment at day: |
Gelatin test | |||
|---|---|---|---|---|---|
| 7 | 14 | 21 | 30 | ||
| Alternaria tenuissima | − | − | − | − | − |
| Aspergillus fumigatus | + | + | + | + | + |
| Bjerkandera adusta | − | − | − | − | − |
| Chaetomium globosum HG530327.1 (99%) | − | + | + | + | + |
| Chaetomium globosum GU138648.1 (99%) | − | − | − | − | − |
| Cryptococcus sp. | − | − | + | + | + |
| Penicillium chrysogenum KF999008.1 (99%) | |||||
| Isolate 1 | − | − | + | + | + |
| Isolate 2 | − | + | + | + | + |
| Penicillium chrysogenum JF731255.1 (99%) | |||||
| Isolate 1 | − | − | − | + | + |
| Isolate 2 | − | − | − | − | + |
| Isolate 3 | − | − | + | + | + |
| Penicillum chrysogenum JX139710.1 (100%) | − | − | − | − | + |
| Penicillium decumbens | − | − | − | + | + |
| Trichoderma longibrachiatum KP281710.1 (99%) | − | − | − | + | + |
| Trichoderma longibrachiatum KP281708.1 (99%) | − | + | + | + | + |
−, no growth; +, growth on the surface.
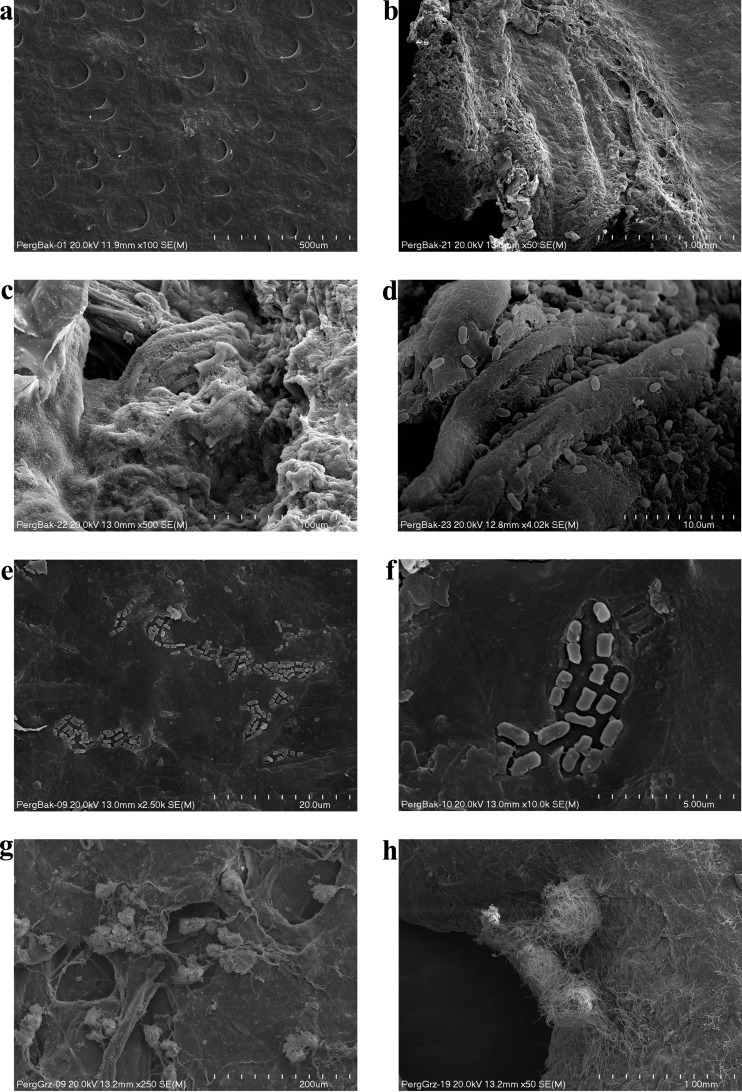
The highest growth potential among bacteria was noted in species of the Bacillus genus, particularly in 2 isolates (B. subtilis, B. cereus), in Acinetobacter lwoffii, and in one species of the Paenibacillus genus. The B. cereus isolate was characterized by particularly intensive development. Its biodeterioration activity was so high that, after only 7 days, the parchment showed numerous bright areas attesting to its defects (Fig. 4c and d). Moreover, the consistency of the parchment changed as well. It turned into a gluey substance, which prevented its fixation for an SEM analysis. The investigated bacterial strains of the Acinetobacter lwoffii, Bacillus subtilis, and Paenibacillus isolates also demonstrated high destructive potential, the evidence of which was material defects caused by bacterial growth (Fig. 4e through j). In the case of these microbes, however, the degradation process was considerably slower, and it started at the edges of the material. The SEM analysis of the surface of the parchment samples enabled the nature of the bacterial activity and the changes caused by the bacteria to be investigated in greater detail. Fig. 5 shows unaltered parchment surface (Fig. 5a) and the change of structure under the influence of Acinetobacter lwoffii, the intensive growth of the isolate, and the uncovering and degrading collagen fibers (Fig. 5b through d), as well as the slower biodeterioration caused by the B. subtilis species (Fig. 5e to f). For molds, as many as 10 of 15 investigated isolates demonstrated the ability to grow actively on parchment. Certain strains rapidly colonized parchment. Their growth could be observed as soon as after 14 days of incubation. In the case of Aspergillus fumigatus, the growth was observed from the 7th day of culture. The strains of Penicillium decumbens, Trichoderma longibrachiatum (1 isolate), and Penicillium chrysogenum (1 isolate) needed 3 weeks to colonize the material. The most intensive growth was observed for strains of the following species: P. chrysogenum, T. longibrachiatum, A. fumigatus, and Chaetomium globosum (Fig. 6). The surface and structural changes caused by fungi progressed much more slowly than those caused by bacteria. Figure 5g and h are SEM images of the growth of P. chrysogenum and C. globosum, respectively.
FIG 4.
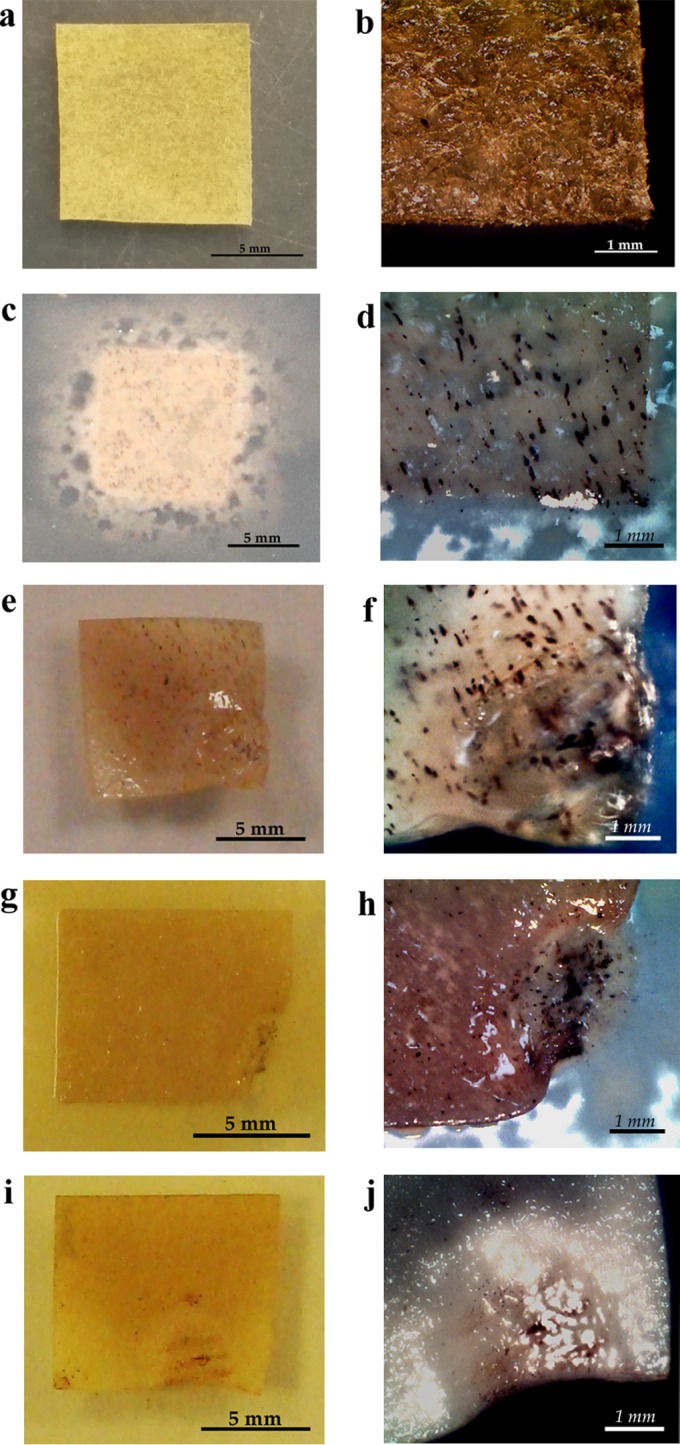
Bacterial growth on parchment. (a and b) Without bacterial growth; (c and d) B. cereus; (e and f) A. lwoffii; (g and h) B. subtilis; (i and j) Paenibacillus sp. Macroscopic images were taken with a Sony Cybershot digital camera (a, c, e, g, i); photos were taken with a Moticam 2300 3.0M digital camera with a stereoscopic magnifying glass (30×) (b, d, f, h, j).
FIG 5.
SEM microphotographs of bacterial and fungal growth on the parchment. (a) Parchment surface without microbial growth; (b to d) the structure changed by A. lwoffii; (e and f) growth of B. subtilis; (g) growth of P. chrysogenum; (h) growth of C. globosum.
FIG 6.
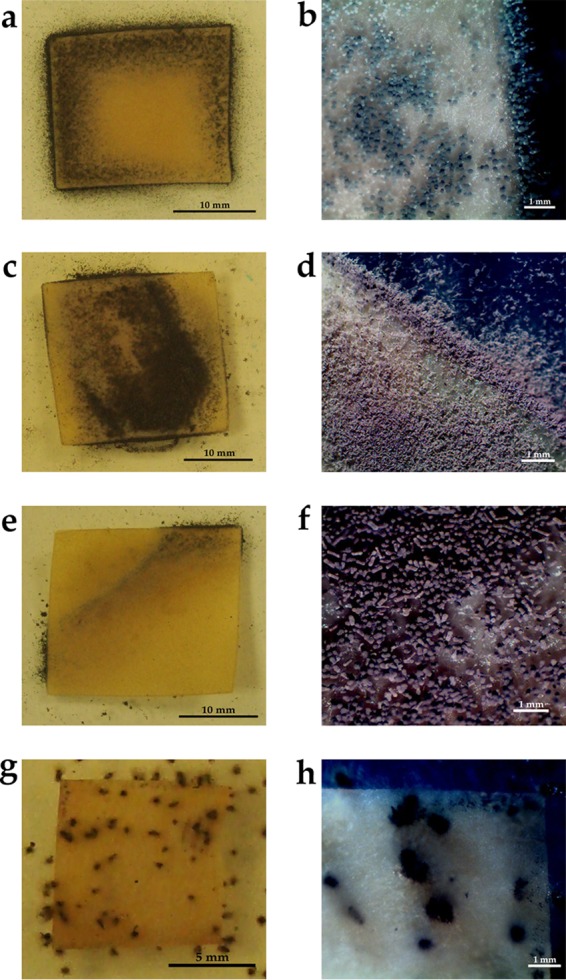
Fungal growth on parchment. (a and b) P. chrysogenum; (c and d) T. longibrachiatum; (e and f) A. fumigatus; (g and h) C. globosum. Macroscopic images were taken with a Sony Cybershot digital camera (a, c, e, g); photos were taken with a Moticam 2300 3.0M digital camera with a stereoscopic magnifying glass (40×) (b, d, f, h).
Microbiological analysis of air.
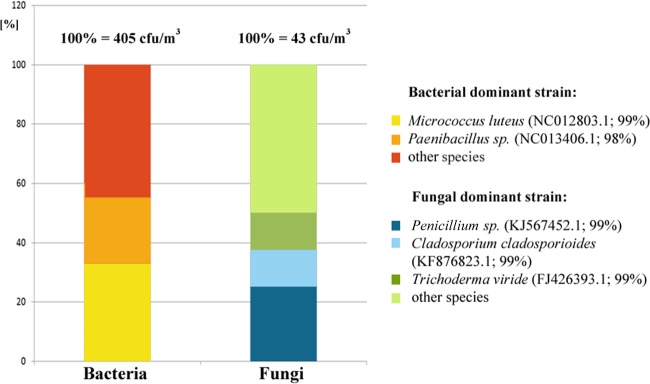
The quantitative analysis of microorganisms in the indoor air of the National Archives in Krakow revealed the presence of bacteria at the level of 405 CFU/m3 and of fungi at the level of 43 CFU/m3. The dominating fungi were those of the Penicillium, Cladosporium, and Trichoderma genera, and the prevailing bacteria belonged to the Micrococcus and Paenibacillus genera (Fig. 7).
FIG 7.
Quantitative and qualitative analyses of the indoor air in the storeroom of the National Archives in Krakow.
DISCUSSION
The most important stage in the estimation of microbial hazards, based on which remedial measures can be implemented to counteract the deterioration process, is the determination of microbial diversity on a given object and in the environment in which the object is stored. This entails the identification of individual microbial strains (34). In these studies, fungal isolates collected with various sampling methods (swab, spontaneous contamination, FungalTape) were assayed on the basis of the analysis of the ITS region sequence. A molecular analysis of this region enables species identification of most fungi with very high accuracy (34). The analysis conducted in this study revealed the presence of fungal species on the surface of the document. There were, among others, microbes of the Penicillium, Aspergillus, Trichoderma, Alternaria, and Chaetomium genera. According to literature reports, the fungal profile obtained constitutes a typical microflora of parchment documents (5, 6, 18, 35, 36). The dominating fungal isolates obtained from the surface of the document included P. chrysogenum, which has also been isolated by Krakova et al. (36), and C. globosum, previously reported by Mesquita et al. (18) and Strzelczyk (21). Moreover, A. fumigatus and Bjerkandera adusta isolates, described in the studies conducted by Krakova et al. (36), were also isolated. Particular attention should be paid to a xerophilous strain of Penicillium decumbens, which has not been isolated from parchment documents before but has been found in archival environments (37). The fungal genotype diversity analysis using the PCR-DGGE method supplemented the qualitative investigations. DGGE fingerprints, which present the structure of microbial communities in the storage environment of cultural heritage objects and on the objects themselves, have been applied in numerous microbiological studies before (20, 22, 26, 38, 39). Apart from demonstrating the genotypic structure of an environment, this method enables the identification of strains that are nonculturable in laboratory settings (20) and also, in the phylogenetic studies, provides interesting information about the community of microorganisms present in the environment or on a historical object (5–7). In the investigation of the incorporation charter for the city of Krakow, a DGGE profile was created which served as a basis for determining the biodiversity level of fungal genotypes and for identifying strains that had not been isolated in culture. The identified taxons included other strains of the Cladosporium genus, which have been isolated in numerous quantities in parchment studies (18), the Aspergillus genus, and a nonculturable strain of the Alternaria genus.
Furthermore, 27 bacterial isolates were obtained in the study, which is twice the number of fungal isolates. This fact is consistent with the results presented by Krakova et al. (36) in their study of parchment materials. Based on the analysis of the 16S rRNA gene sequence, the most dominant groups of bacteria were the Bacillus, Staphylococcus, and Paenibacillus genera, which is also consistent with the literature (36, 39). The presence of Bacillus species on parchment documents has also been described by Pinar et al. (6), who performed phylogenetic identification of the bacterial community. However, the reports found in the literature on this subject contain less information about the species of bacteria than about the species of fungi isolated from parchment documents. This is probably the result of the lesser role of bacteria in microbial hazards, particularly in terms of initiating biodeterioration. Nevertheless, it must be emphasized that in particular situations, such as heating or sewage system failures or flooding in the course of disasters, when humidity favors bacterial development, the growth of bacteria is rapid, and their destructive potential is much greater than that of fungi. Such events took place, for instance, in Florence in 1966, Petersburg in 1988, Poland in 1997 (21), and Japan in 2011 (40). The isolated bacterial strains create the microflora that is typical of archival collections (15, 35, 36) and other historical and museum objects (29, 41). The microbiological picture of the object in question was supplemented by bioluminescence analysis, which showed an average level of organic contamination on the surface of the document, compared with that of other historical objects analyzed in the author's own studies. However, ATP assay gives more reliable results about the bacterial community (7).
In order to explore the environment of the investigated document, the microbiological analysis also involved the indoor air in the room where the parchment is kept. The dominating microbes were those of the Penicillium, Cladosporium, Trichoderma, Micrococcus, and Paenibacillus genera. The quantitative levels of microbes, both fungi and bacteria, in the air were within the norms established by the Italian Ministry of Cultural Heritage, according to which the levels of heterotrophic bacteria and fungi in the indoor air of historical collection storerooms should not exceed 750 CFU/m3 and 150 CFU/m3, respectively (11). They are also consistent with research conducted in other Polish libraries and archives, where the concentrations of fungi in the air in the range of 19 to 86 CFU/m3 and of bacteria in the range of 123 to 712 CFU/m3 were described (23).
The biodeterioration process of cultural heritage objects is affected by various environmental factors, such as temperature, humidity, UV light, and the presence of organic contamination. Without a doubt, the condition of the object is of considerable importance. However, the initiation and velocity of biodeterioration primarily depend on the destructive potential of microbes that are present on the object. Usually, microbes initiate parchment breakdown from simple proteins or tannins present on its surface. The main structural material, i.e., collagen, is degraded in later stages. The hydrolysis of collagen and other protein elements can also lead to changes within nonorganic components, resulting in stains and discolorations (41). In this study, the destructive potential of isolated microbes was determined by the verification of their proteolytic properties and their ability to grow on parchment. When both of these parameters were taken into account, bacterial species of the greatest destructive potential were found to be B. cereus, A. lwoffii, B. subtilis, and one isolate of the Paenibacillus genus. The destructive potential of these strains was so high that macroscopic changes appeared even several days after the inoculation of parchment samples, leading to complete destruction, as was the case with B. cereus. The results obtained suggest that the enzymatic activity of B. cereus led to collagen degradation under aerobic conditions. The SEM analysis revealed structural changes as well as the uncovering and destruction of collagen fibers induced by bacterial growth and enzymatic activity. Structural changes caused by the biodeterioration of parchment documents and identified using SEM analysis have been previously described (5, 6). The greatest changes were observed for A. lwoffii. The microscopic image showed rapid growth of A. lwoffii bacteria and intense material degradation. The proteolytic properties of Bacillus strains have already been described in the literature (29, 35, 36). Nevertheless, the level of their destructive potential is surprising. It must be emphasized that species of the Bacillus genus have strong cellulolytic properties, which make them a threat to paper patches used to glue parchment documents and other paper archives (35). Similarly, the remaining bacterial isolates, such as Micrococcus luteus with proteolytic properties (42) or a Kocuria sp. capable of degrading polymer materials (35), can play a role in the destruction process. However, Pseudomonas psychrotolerans, Paenibacillus amylolyticus, Paenibacillus fonticola, and Paenibacillus xylanilyticus strains are rarely isolated from archival objects and probably have not been reported as found on parchment documents. However, the P. psychrotolerans, P. amylolyticus, and P. fonticola strains have proteolytic properties that have been demonstrated in a gelatin test and confirmed in the literature, which make them a potential threat to the investigated object (43–45). Similarly, P. xylanilyticus is a bacterium that produces a broad range of enzymes, including gelatinase, amylase, β-galactosidase, and xylanase, which is responsible for the breakdown of xylan, the main component of hemicellulose in paper (46).
The fungal microflora determined in this study can be a potential threat for the investigated object. Fungi of the Cladosporium, Penicillium, Aspergillus, Trichoderma, and Chaetomium genera are microbes with a high destructive potential, which has been shown in the present study and in other studies conducted in archives and museums by other authors (4–6, 13, 18, 35, 36, 39, 47). As for bacteria, the SEM images document not only the superficial growth of fungi on parchment, observed by light microscopy, but also its degradation and structural changes. The most numerous groups of isolates were fungi of the Penicillium genus, i.e., the proteolytic species of P. chrysogenum and P. decumbens, which are responsible for the destruction of leather (42). The considerable majority of isolated fungi have proteolytic and cellulolytic properties. They can therefore play a significant role in the process of enzymatic breakdown of organic materials (4, 29, 36). It is worth noting that fungi also include species that are incapable of the enzymatic breakdown of materials which were used to make historical objects. Under appropriate humidity levels, the species develop with the use of contaminants present on the surfaces of the objects. Their activity results in various metabolites which contribute to the chemical damage of the material. Moreover, the growth itself can cause mechanical damage (20).
As has already been mentioned, microorganisms play an important role in the degradation of archives, works of art, sculptures, and other historical objects (40). The detection of microorganisms, even those that may have a destructive influence on objects on which they settle, is very important information for conservators, archivists, and museologists. The disinfection of historical objects is not always possible and is usually not recommended due to the risk of damaging already weakened objects. In such situations, information about the microbiological condition of an object or collection is an indicator of further management. In the majority of cases, preventive measures to improve storage conditions are implemented in order to protect cultural heritage objects from microbial hazards. It is essential to guarantee appropriate temperature and humidity in storerooms (41, 48). According to the recommendations of the International Federation of Library Associations, the indoor temperature in archives should range from 18°C to 20°C, and the humidity should be 50% to 60%. The study presented here involved the analysis of changes of these parameters within 1 year. It indicated high temperature fluctuations. The humidity parameter was characterized by greater stability; it exceeded 60% only occasionally. A drop in humidity to the level of approximately 35% is alarming, since a decrease below 35% can lead to damage to leather bindings and stiffening of parchment documents (49, 50). It is worth emphasizing that certain xerophilous fungal species are capable of settling in environments with low humidity. They accumulate humidity while growing, thus creating a specific microclimate, enabling other fungi and bacteria to develop (12). The microflora detected in this study also includes fungi considered xerophilous, such as C. globosum, P. chrysogenum, Trichoderma spp., or Aspergillus spp. (37). The most important issue is that, despite age and technical conditions of the historical tenement in which the storeroom is located, the most important climate parameters are maintained at a satisfactory level.
The microbes isolated from the incorporation charter also include fungi and bacteria that are potentially dangerous to human health. These include Aspergillus fumigatus, a species with toxigenic properties that can cause systemic mycosis and other conditions (51), and Staphylococcus epidermidis, which can cause skin lesions that are difficult to heal and other systemic infections. The information about the presence of pathogenic microorganisms is important, considering the fact that the collections are looked after by people (conservators, archivists, or museologists). Many species that settle on cultural heritage objects and in their environments can have a negative influence on human health by the production of toxins or the induction of respiratory diseases, allergies, fungal infections, and many other troublesome and dangerous conditions. Awareness of the fact that such microbes are present in collections should make employees more sensitive and prompt them to implement appropriate means of personal protection. In such an environment, an important issue is the protection of both archival collections and worker health, which sometimes is very difficult to achieve (52–54).
The microbiological studies of the object and the environment in which it is stored revealed fungal and bacterial microflora typical of archival items and other cultural heritage objects. Certain bacteria and fungi found in this microflora are characterized by a high destructive potential, which may constitute a risk of biodeterioration of the investigated object. In order to protect such a unique and valuable document, preventive actions should be undertaken to guarantee appropriate inner climate conditions to prevent microbial development (e.g., permanent monitoring of temperature and relative humidity, providing proper ventilation with usage of air filters, passive removing of organic pollutants, cleaning strategies, and packaging strategies) (50).
ACKNOWLEDGMENTS
I thank Barbara Berska and Małgorzata Bochenek from the National Archives in Krakow for the possibility of conducting interesting studies.
This paper is part of the plan of the National Archives of Krakow to conduct summary scientific investigations for the purposes of creating a monograph concerning the incorporation charter for the city of Krakow.
The research was financed from the funds of the Polish Ministry of Science and Higher Education for young scientists, grant 089/WT-KM/02/2015/M/5089.
REFERENCES
- 1.Jelonek-Litewka K. 2003. Czy odnaleziona pieczęć Bolesława Wstydliwego jest pieczęcią oderwaną od aktu lokacyjnego miasta Krakowa. Krakowski Rocznik Archiwalny 9:31–38. [Google Scholar]
- 2.Rabiej P. 2008. Kilka uwag o dokumencie lokacyjnym Krakowa z 1257 roku (Notesullacarta di fondazione di Cracovia del 1257) Miasta, ludzie, instytucje, znaki. Ksiega jubileuszowa ofiarowana Profesor Bozenie Wyrozumskiej w 75. rocznice urodzin cur. Zenon Piech, Kraków, Towarzystwo Naukowe 951:487–500. [Google Scholar]
- 3.Zyska B. 1997. Fungi isolated from library materials: a review of the literature. Int Biodeterior Biodegradation 40:43–51. doi: 10.1016/S0964-8305(97)00061-9. [DOI] [Google Scholar]
- 4.Sterflinger K. 2010. Fungi: their role in deterioration of cultural heritage. Fungal Biol Rev 24:47–55. doi: 10.1016/j.fbr.2010.03.003. [DOI] [Google Scholar]
- 5.Piñar G, Sterflinger K, Pinzari F. 2015. Unmasking the measles-like parchment discoloration: molecular and microanalytical approach. Environ Microbiol 17:427–443. doi: 10.1111/1462-2920.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piñar G, Sterflinger K, Ettenauer J, Quandt A, Pinzari F. 2015. A combined approach to assess the microbial contamination of the Archimedes palimpsest. Microb Ecol 69:118–134. doi: 10.1007/s00248-014-0481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troiano F1, Polo A, Villa F, Cappitelli F. 2014. Assessing the microbiological risk to stored sixteenth century parchment manuscripts: a holistic approach based on molecular and environmental studies. Biofouling 30:299–311. doi: 10.1080/08927014.2013.871539. [DOI] [PubMed] [Google Scholar]
- 8.Poole JB, Reed R. 1962. The preparation of leather and parchment by the Dead Sea Scrolls community. Technol Cult 3:1–26. doi: 10.2307/3100798. [DOI] [Google Scholar]
- 9.Kennedy CJ, Wess TJ. 2008. The structure of collagen within parchment: a review. Restaurator 24:61–80. doi: 10.1515/REST.2003.61. [DOI] [Google Scholar]
- 10.Zhou P, Mulvaney SJ, Regenstein JM. 2006. Properties of Alaska pollock skin gelatin: a comparison with tilapia and pork skin gelatins. J Food Sci 71:C313–C321. doi: 10.1111/j.1750-3841.2006.00065.x. [DOI] [Google Scholar]
- 11.Cappitelli F, Fermo P, Vecchi R, Piazzalunga A, Valli G, Zanardini E, Sorlini C. 2009. Chemical-physical and microbiological measurements for indoor air quality assessment at the Ca'Granda historical archive, Milan (Italy). Water Air Soil Pollut 201:109–120. doi: 10.1007/s11270-008-9931-5. [DOI] [Google Scholar]
- 12.Montanari M, Mellonia V, Pinzari F, Innocentia G. 2012. Fungal biodeterioration of historical library materials stored in Compactus movable shelves. Int Biodeterior Biodegradation 75:83–88. doi: 10.1016/j.ibiod.2012.03.011. [DOI] [Google Scholar]
- 13.Nunes I, Mesquita N, CaboVerdea S, LeitãoBandeirac AM, Carolinod MM, Portugal A, Botelho ML. 2013. Characterization of an airborne microbial community: a case study in the archive of the University of Coimbra, Portugal. Int Biodeterior Biodegradation 79:36–41. doi: 10.1016/j.ibiod.2013.01.013. [DOI] [Google Scholar]
- 14.Cappitelli F, Sorlini C. 2005. From papyrus to compact disc: the microbial deterioration of documentary heritage. Crit Rev Microbiol 31:1–10. doi: 10.1080/10408410490884766. [DOI] [PubMed] [Google Scholar]
- 15.Guiamet P, Borrego S, Lavin P, Perdomo I, Saravia SGD. 2011. Biofouling and biodeterioration in materials stored at the historical archive of the Museum of La Plata, Argentine, and at the National Archive of the Republic of Cuba. Colloids Surf B Biointerfaces 85:229–234. doi: 10.1016/j.colsurfb.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 16.Fabbri AA, Ricelli A, Brasini S, Fanelli C. 1997. Effect of different antifungals on the control of paper biodeterioration caused by fungi. Int Biodeterior Biodegradation 39:61–65. doi: 10.1016/S0964-8305(97)00001-2. [DOI] [Google Scholar]
- 17.Arai H. 2000. Foxing caused by fungi: twenty-five years of study. Int Biodeterior Biodegradation 46:181–188. doi: 10.1016/S0964-8305(00)00063-9. [DOI] [Google Scholar]
- 18.Mesquita N, Portugal A, Videira S, Rodríguez-Echeverría S, Bandeira AML, Santos MJA, Freitas H. 2009. Fungal diversity in ancient documents: a case study on the archive of the University of Coimbra. Int Biodeterior Biodegradation 63:626–629. doi: 10.1016/j.ibiod.2009.03.010. [DOI] [Google Scholar]
- 19.Gutarowska B, Skora J, Zduniak K, Rembisz D. 2012. Analysis of the sensitivity of microorganisms contaminating museums and archives to silver nanoparticles. Int Biodeterior Biodegradation 68:7–17. doi: 10.1016/j.ibiod.2011.12.002. [DOI] [Google Scholar]
- 20.Sterflinger K, Pinzari F. 2012. The revenge of time: fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ Microbiol 14:559–566. doi: 10.1111/j.1462-2920.2011.02584.x. [DOI] [PubMed] [Google Scholar]
- 21.Strzelczyk AB. 2004. Observations on aesthetic and structural changes induced Polish historic object by microorganisms. Int Biodeterior Biodegradation 53:151–156. doi: 10.1016/S0964-8305(03)00088-X. [DOI] [Google Scholar]
- 22.Michaelsen A, Pinzari F, Ripka K, Lubitz W, Piñar G. 2006. Application of molecular techniques for the identification of fungal communities colonizing paper material. Int Biodeterior Biodegradation 58:133–141. doi: 10.1016/j.ibiod.2006.06.019. [DOI] [Google Scholar]
- 23.Karbowska-Berent J, Górny RL, Strzelczyk AB, Wlazło A. 2011. Airborne and dust borne microorganisms in selected Polish libraries and archives. Build Environ 46:1872–1879. doi: 10.1016/j.buildenv.2011.03.007. [DOI] [Google Scholar]
- 24.Pinheiro AC, Viegas C, Viegas S, Veríssimo C, Brandão J, Macedo MF. 2012. Indoor air quality in Portuguese archives: a snapshot on exposure levels. J Toxicol Environ Health A 75:1359–1370. doi: 10.1080/15287394.2012.721168. [DOI] [PubMed] [Google Scholar]
- 25.Michaelsen A, Piñar G, Montanari M, Pinzari F. 2009. Biodeterioration and restoration of a 16th-century book using a combination of conventional and molecular techniques: a case study. Int Biodeterior Biodegradation 63:161–168. doi: 10.1016/j.ibiod.2008.08.007. [DOI] [Google Scholar]
- 26.Pinheiro AC, Macedo MF, Jurado V, Saiz-Jimenez C, Viegas C, Brandão J, Rosado L. 2011. Mould and yeast identification in archival settings: preliminary results on the use of traditional methods and molecular biology options in Portuguese archives. Int Biodeterior Biodegradation 65:619–627. doi: 10.1016/j.ibiod.2011.02.008. [DOI] [Google Scholar]
- 27.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 28.White TJ, Bruns TD, Lee S, Taylor J. 1990. Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, London, United Kingdom. [Google Scholar]
- 29.Pangallo D, Krakova L, Chovanova K, Buckova M, Puskarova A, Simonovicova A. 2013. Disclosing a crypt: microbial diversity and degradation activity of the microflora isolated from funeral clothes of Cardinal Peter Pazmany. Microbiol Res 168:289–299. doi: 10.1016/j.micres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Felske A, Engelen B, Nübel U, Backhaus H. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol 62:4162–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lech T, Ziembinska-Buczynska A. 2015. Evaluation of a modified sampling method for molecular microbial communities. Genet Mol Res 14:3200–3208. doi: 10.4238/2015.April.10.32. [DOI] [PubMed] [Google Scholar]
- 32.Luczkiewicz A, Felis E, Ziembinska A, Gnida A, Kotlarska E, Olanczuk-Neyman K, Surmacz-Gorska J. 2013. Resistance of Escherichia coli and Enterococcus spp. to selected antimicrobial agents present in municipal wastewater. J Water Health 11:600–612. doi: 10.2166/wh.2013.130. [DOI] [PubMed] [Google Scholar]
- 33.Strzelczyk AB, Karbowska-Berent J. 2000. The role of Streptomycetes in the biodeterioration of historic parchment, p 158. Nicolaus Copernicus University Press, Torun, Poland. [Google Scholar]
- 34.Trovão J, Mesquita N, Paiva DS, Paiva de Carvalho H, Avelar L, Portugal A. 2013. Can arthropods act as vectors of fungal dispersion in heritage collections? A case study on the archive of the University of Coimbra, Portugal. Int Biodeterior Biodegradation 79:49–55. doi: 10.1016/j.ibiod.2012.10.015. [DOI] [Google Scholar]
- 35.Michaelsen A, Piñar G, Pinzari F. 2010. Molecular and microscopic investigation of the microflora inhabiting a deteriorated Italian manuscript dated from the 13th century. Microb Ecol 60:69–80. doi: 10.1007/s00248-010-9667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krakova L, Chovanová K, Selim SA, Šimonovičová A, Puškarová A, Maková A, Pangallo D. 2012. A multiphasic approach for investigation of the microbial diversity and its biodegradative abilities in historical paper and parchment documents. Int Biodeterior Biodegradation 70:117–125. doi: 10.1016/j.ibiod.2012.01.011. [DOI] [Google Scholar]
- 37.Montemartini-Cortea A, Ferronib A, Salvoa VS. 2003. Isolation of fungal species from test samples and maps damaged by foxing, and correlation between these species and the environment. Int Biodeterior Biodegradation 51:167–173. doi: 10.1016/S0964-8305(02)00137-3. [DOI] [Google Scholar]
- 38.Oros-Sichler M, Gomes NC, Neuber G, Smalla K. 2006. A new seminested PCR protocol to amplify large 18S rRNA gene fragments for PCR-DGGE analysis of soil fungal communities. J Microbiol Methods 65:63–75. doi: 10.1016/j.mimet.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Principi P, Villa F, Sorlini C, Cappitelli F. 2011. Molecular studies of microbial community structure on stained pages of Leonardo da Vinci's Atlantic codex. Microb Ecol 61:214–222. doi: 10.1007/s00248-010-9741-3. [DOI] [PubMed] [Google Scholar]
- 40.Sato Y, Aoki M, Kigawa R. 2014. Microbial deterioration of tsunami-affected paper-based objects: a case study. Int Biodeterior Biodegradation 88:142–149. doi: 10.1016/j.ibiod.2013.12.007. [DOI] [Google Scholar]
- 41.Sterflinger K, Piñar G. 2013. Microbial deterioration of cultural heritage and works of art: tilting at windmills? Appl Microbiol Biotechnol 97:9637–9646. doi: 10.1007/s00253-013-5283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlita A. 2004. Microbial biodeterioration of leather and its control: a review. Int Biodeterior Biodegradation 53:157–163. doi: 10.1016/S0964-8305(03)00089-1. [DOI] [Google Scholar]
- 43.Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K. 1997. Emended description of Paenibacillus amylolyticus and description of Paenibacillus illinoisensis sp. nov. and Paenibacillus chibensis sp. nov. Int J Syst Bacteriol 47:299–306. doi: 10.1099/00207713-47-2-299. [DOI] [PubMed] [Google Scholar]
- 44.Hauser E, Kämpfer P, Busse HJ. 2004. Pseudomonas psychrotolerans sp. nov. Int J Syst Evol Microbiol 54:1633–1637. doi: 10.1099/ijs.0.03024-0. [DOI] [PubMed] [Google Scholar]
- 45.Chou JH, Chou YJ, Lin KY, Sheu SY, Sheu DS, Arun AB, Young CC, Chen WM. 2007. Paenibacillus fonticola sp. nov. isolated from a warm spring. Int J Syst Evol Microbiol 57:1346–1350. doi: 10.1099/ijs.0.64872-0. [DOI] [PubMed] [Google Scholar]
- 46.Rivas R, Mateos PF, Martínez-Molina E, Velázquez E. 2005. Paenibacillus xylanilyticus sp. nov., an airborne xylanolytic bacterium. Int J Syst Evol Microbiol 55:405–408. doi: 10.1099/ijs.0.63173-0. [DOI] [PubMed] [Google Scholar]
- 47.Pinzari F, Cialei V, Piñar G. 2012. A case study of ancient parchment biodeterioration using variable pressure and high vacuum scanning electron microscopy. In Meeks N, Cartwright C, Meek A, Mongiatti A (ed), Historical technology, materials and conservation: SEM and microanalysis. Archetype Publications, International Academic Projects, London, United Kingdom. [Google Scholar]
- 48.Caneva G, Ceschin S. 2008. Ecology of biodeterioration, p 35–58. In Caneva G, Nugari MP, Salvadori O (ed), Plant biology for cultural heritage. Getty Publications, Los Angeles, CA. [Google Scholar]
- 49.ISO ISO 11799:2003. Information and documentation: document storage requirements for archive and library materials. ISO, Geneva, Switzerland. [Google Scholar]
- 50.British Standards Institution. PAS 198:2012. Specification for managing environmental conditions for cultural collections. British Standards Institution, London, United Kingdom. [Google Scholar]
- 51.Verissimo C. 2016. Fungal infections, p 27–34. In Viegas C, Pinheiro AC, Sabino R, Viegas S, Brandao J, Verissimo C (ed), Environmental mycology in public health: fungi and mycotoxins risk assessment and management. Academic Press, London, United Kingdom. [Google Scholar]
- 52.Vuong C, Otto M. 2002. Staphylococcus epidermidis infections. Microbes Infect 4:481–489. doi: 10.1016/S1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 53.Bennett JW, Klich M. 2003. Mycotoxins. Clin Microbiol Rev 16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinheiro AC. 2016. Urban settings, p 157–166. In Viegas C, Pinheiro AC, Sabino R, Viegas S, Brandao J, Verissimo C (ed), Environmental mycology in public health: fungi and mycotoxins risk assessment and management. Academic Press, London, United Kingdom. [Google Scholar]