Abstract
Diet is one of the primary drivers that sculpts the form and function of the mammalian gut microbiota. However, the enormous taxonomic and metabolic diversity held within the gut microbiota makes it difficult to isolate specific diet-microbe interactions. The objective of the current study was to elucidate interactions between the gut microbiota of the mammalian herbivore Neotoma albigula and dietary oxalate, a plant secondary compound (PSC) degraded exclusively by the gut microbiota. We quantified oxalate degradation in N. albigula fed increasing amounts of oxalate over time and tracked the response of the fecal microbiota using high-throughput sequencing. The amount of oxalate degraded in vivo was linearly correlated with the amount of oxalate consumed. The addition of dietary oxalate was found to impact microbial species diversity by increasing the representation of certain taxa, some of which are known to be capable of degrading oxalate (e.g., Oxalobacter spp.). Furthermore, the relative abundances of 117 operational taxonomic units (OTU) exhibited a significant correlation with oxalate consumption. The results of this study indicate that dietary oxalate induces complex interactions within the gut microbiota that include an increase in the relative abundance of a community of bacteria that may contribute either directly or indirectly to oxalate degradation in mammalian herbivores.
INTRODUCTION
Mammals live in a complex and largely symbiotic relationship with their gut microbiota. This microbiota harbors 150 times more genes than the host and exhibits complex interactions with the host's diet (1–3). In mammalian herbivores, diverse intestinal bacteria ferment a diet high in recalcitrant cellulose and in turn synthesize nutrients from the diet in a form more amenable to absorption by the host (4). Furthermore, mammalian herbivores harbor greater microbial diversity in their gut than either omnivores or carnivores (1). Despite the progress of research into the interactions between the mammalian gut microbiota and diet, the isolation of specific diet-microbe interactions in such a complex system has proven to be difficult (5, 6).
In addition to having a role in fermentation, microbes play an important role in the biotransformation of dietary toxins in mammalian herbivores (4, 7–10). For some toxins, such as oxalate or 3,4-dihydroxypyridine (DHP), a single species of bacteria is capable of biotransforming the toxin, and this function can be transferred to other mammals through microbial transplants (7, 8, 11, 12). For other toxins, such as creosote resin, whole microbial community transplantation into other mammals can increase tolerance (10).
Oxalate, a widely produced and ingested plant secondary compound (PSC), serves as an excellent model to study diet-microbe interactions (13). It is the simplest organic acid and is toxic to mammals (14–16). Oxalate can bind to free calcium ions in the blood and aggregate in the kidneys to form kidney stones (17). In fact, oxalate is a constituent in 80% of kidney stones in humans (17). Oxalate is not metabolized by mammalian enzymes but rather is biotransformed into formate and CO2 by gut microbes (7, 18–21). While some oxalate-degrading bacteria, such as Oxalobacter formigenes, biotransform oxalate for use as a carbon and energy source, the growth of other oxalate-degrading bacteria, such as Lactobacillus acidophilus, is inhibited by the presence of oxalate, even though these bacteria biotransform the compound when present (7, 22). Additionally, the by-products of microbial oxalate degradation, formate and CO2, may be used by a number of bacteria in the process of acetogenesis or methanogenesis, potentially benefitting other gut bacteria not directly involved in the oxalate degradation function (23). While there is no direct evidence for either acetogenesis or methanogenesis, several known acetogenic taxa, such as Clostridium, Streptococcus, and Ruminococcus, are prevalent in the N. albigula gut (24–26). These attributes constitute a unique system to isolate the interactions between dietary toxins and gut microbes, along with their contribution to the overall metabolism of the host.
The wild mammalian herbivore Neotoma albigula (white-throated woodrat) is an ideal species to study the effects of dietary oxalate. Some populations of N. albigula consume a diet composed of nearly 100% Opuntia species cactus, which contains a high oxalate content (1.5%, dry weight) (26). Neotoma albigula can degrade >90% of dietary oxalate when fed artificial diets of up to 9% oxalate (dry weight). This high level of oxalate degradation has been hypothesized to be the result of microbial metabolism (27, 28). Furthermore, N. albigula harbors a diversity of known and potentially oxalate-degrading bacteria distributed across the gastrointestinal tract, including Oxalobacter, Lactobacillus, Clostridium, and Enterococcus, among others (26). Thus, N. albigula regularly consumes large amounts of oxalate and harbors a diversity of bacteria that exhibit complex interactions with oxalate, making it an ideal species to elucidate oxalate-microbiota interactions.
The purpose of the current study was to identify the ecological and functional interactions between dietary oxalate and the gut microbiota of N. albigula. This study has two primary objectives. The first is to quantify the effect of increasing oxalate consumption on oxalate degradation in vivo. The second is to determine if the gut microbiota of N. albigula exhibits a community-level response to oxalate consumption. Given the previously identified differential responses of oxalate-degrading bacteria to the presence of oxalate, we predicted that oxalate would stimulate the growth of some microbial taxa and inhibit the growth of others, while having a neutral effect on the remaining community. Our data support the hypothesis that a specialized microbial network of bacteria is responsible for oxalate degradation in N. albigula.
MATERIALS AND METHODS
Location, collection, and diet of animals.
Six white-throated woodrats (N. albigula), were collected with Sherman live traps from Castle Valley, Utah (38.63°N, 109.41°W), in October 2012. The woodrats were immediately transported to the University of Utah Department of Biology Animal Facility and housed in individual cages (48 by 27 by 20 cm) under a 12/12-h light/dark cycle at 28°C and 20% humidity. The animals were maintained in captivity and fed high-fiber rabbit chow with 0.2% oxalate (Teklad formula 2031; Harlan, Denver, CO, USA) for 6 months prior to experimentation. All methods were approved by the Institutional Animal Care and Use Committee (IACUC) under protocol 12-12010.
To examine the interactions between dietary oxalate and gut microbes, animals were placed in a diet trial where oxalate was gradually increased over time (Table 1). The 5-day time periods for the 0.05% oxalate diet were chosen to ensure that any effect of oxalate on the microbiota was removed, while a 3-day period for each of the oxalate diet periods was chosen based on the study of Belenguer et al. (29), in which 3 days on oxalate was long enough to elicit a microbial response. Metabolic cages were used to separate urine and feces and allow for the quantification of food and water intake, which were given ad libitum. In metabolic cages, N. albigula had access to direct coprophagy (consumption of feces from the anus) but not indirect coprophagy (caching of feces to consume later). To minimize the oxalate concentration of the rabbit chow without reducing food intake, a 3:1 ratio of powdered purified rat chow (Harlan, Denver, CO, USA) to powdered rabbit chow (Harlan) was used in the study. This diet contained an oxalate concentration of 0.05%, which is herein referred to as “no oxalate” (see Table S1 in the supplemental material). The oxalate diets were prepared by mixing sodium oxalate (Fisher Scientific, Pittsburgh, PA, USA) into the powdered chow on a dry weight basis. At the end of the diet trial, all animals were returned to the no-oxalate diet to ensure that any effect on the microbiota was the result of oxalate and not some other factor. Urine and feces were collected daily in sterile 50-ml Falcon tubes for oxalate assays and microbial inventories. Additionally, we collected data on body mass, food and water intake, and fecal and urinary output daily. Using the food intake and fecal output data, we estimated the dry matter digestibility (DMD) as 1 − (dry fecal output/food consumed). These data were evaluated with a repeated-measures analysis of variance (ANOVA).
TABLE 1.
Design of the diet triala
The oxalate percentage was determined by the mass.
Oxalate assays.
Oxalate in the urine was quantified by following a modified protocol described by Ingale et al. (30). Urine samples were collected daily from each animal for the assays and pooled for each treatment period. Urine samples were acidified with 3 M HNO3 to a pH of <3 to solubilize any oxalate crystals. The acidified urine was centrifuged to remove precipitates, and the supernatant was reserved. The pH of the supernatant was brought up to 7 with NaOH. Approximately 0.1 g of CaCl2 was added and mixed to precipitate oxalate. The samples were then centrifuged and decanted. A volume of distilled water matching the total urinary volume was added to the calcium oxalate precipitate. The samples were then titrated as described below.
For fecal oxalate assays, feces for each animal were collected daily, dried at 45°C overnight, and pooled by animal at the end of each treatment period. The oxalate assays were conducted by following a modified protocol from Justice (28). Approximately 0.4 g of dried feces was ground and added to 5 ml of 6 N H2SO4 for 15 min to solubilize the oxalate. After 15 min, 25 ml of distilled water was added, and the entire solution was filtered through grade 4 Whatman filter paper. The filtrate was brought up to a pH of 7 with NaOH, and 0.1 g of CaCl2 was added to precipitate the oxalate. The samples were centrifuged and decanted. After centrifugation, a volume of distilled water equal to that recovered after filtration was added, and the samples were titrated.
The urine and fecal extracts containing calcium oxalate were titrated in 5-ml aliquots with 0.01 M KMnO4 in triplicate. The aliquots were first acidified with 1 ml of 6 N H2SO4 and heated to 70 to 90°C. The KMnO4 was then added until a pink color persisted for 30 s, and the volume of KMnO4 was recorded. These volumes were then compared to a standard curve. Standard curves were made by addition of 0 mM, 5 mM, 10 mM, 15 mM, or 20 mM sodium oxalate to the urine or feces of the woodrats consuming 0.05% oxalate. After extraction and titration, the volume of KMnO4 required to titrate the samples with no oxalate added was subtracted from the volume of all samples to account for endogenous oxalate production. With these methods, we are able to recover 102.69% ± 12.94% of the oxalate from urine and 97.47% ± 6.78% of the oxalate from feces. Both titration curves were linear, with r2 values of >0.9.
To estimate how much dietary oxalate was being degraded, we quantified the difference between the oxalate consumed and the total oxalate excreted. This estimate is conservative, given that some endogenously produced oxalate excreted in the urine and feces is not accounted for with this method. However, our estimates of total oxalate excretion on the no-oxalate diet indicate that the endogenous contribution is typically small (<10% of the oxalate consumed with a 0.5% oxalate diet). Furthermore, given that endogenous oxalate production is determined by the consumption of certain dietary precursors, it should not change under the diet regime used in this study and is unlikely to impact the conclusions drawn (15).
Microbial inventories.
We collected fresh feces for the microbial inventories on the last day of each diet treatment, which were frozen at −80°C until DNA extraction. DNA was extracted from 180 to 220 g of feces using the QIAamp DNA stool minikit (Qiagen, Germantown, MD, USA). DNA extractions were also performed on oxalate, food, and the reagents of the extraction kit to identify potential sources of contamination. Microbial inventories from a total of 36 fecal samples were generated by amplifying the V4 region of the 16S rRNA gene with the primers 515F and 806R (31). The primers contained a 12-base barcode sequence, which allowed for multiplexing of samples within a single-lane sequencing run on an Illumina MiSeq, with paired-end sequencing of 150 bp each, as previously described (32).
Sequences were analyzed using QIIME (33). Standard quality control was conducted, and sequences were demultiplexed using the default parameters in QIIME. A de novo picking strategy was used to classify the operational taxonomic units (OTU) with UCLUST (34) with a minimum sequence identity of 97%. This strategy resulted in an OTU table and phylogenetic tree, which were used in downstream analyses. Sequences identified as chloroplasts or mitochondria or those that had fewer than 10 representations across the data set were removed. Additionally, samples of microbial communities with fewer than 3,000 sequence reads total were removed from further data analysis. For the comparative analyses, the samples were rarified to the same sampling depth of 27,378 reads, which was the highest number that included all samples remaining after quality control.
We calculated the α-diversity metrics species richness (Margalef's richness index), evenness (equitability), and Shannon index. The community membership and structure were determined using unweighted and weighted UniFrac analyses, respectively, to compare levels of microbial community similarity across individuals and diet treatments. The unweighted UniFrac analysis compares the members of a community, whereas the weighted analysis also takes into consideration relative abundances (35). Comparisons were made with analysis of similarity (ANOSIM) after 999 permutations. Additionally, a repeated-measures Pearson correlation analysis between the relative abundance of an OTU and oxalate consumption was conducted for all samples and OTU. The open-source software QIIME was used for diversity, ANOSIM, and correlation metrics, with a false discovery rate (FDR) correction for the Pearson correlation. Significance was set at a P value of <0.05 for all analyses.
RESULTS
Oxalate degradation.
Body mass, food intake, DMD, water intake, and urine output did not differ significantly among the treatments (Table 2). The oxalate intake increased significantly with increasing dietary oxalate concentrations (P < 0.001), and the amount of oxalate degraded correlated significantly with oxalate consumption (Fig. 1). When the excretion of endogenous oxalate is taken into consideration (i.e., by subtracting the amount excreted on the no-oxalate diet), the oxalate degradation exceeded 90% of that consumed regardless of the concentration in the diet. Furthermore, 94 to 99% of the excreted dietary oxalate was found in the feces, indicating that little oxalate was absorbed into the blood.
TABLE 2.
Metrics associated with woodrats that were not significantly affected by oxalatea
| Metric | Mean value ± SE (g) | F | P |
|---|---|---|---|
| Body mass | 171.08 ± 12.83 | 0.06 | 1.00 |
| Food intake | 18.71 ± 0.06 | 1.66 | 0.18 |
| Fecal output | 1.907 ± 0.088 | 3.16 | 0.27 |
| DMD | 0.758 ± 0.012 | 0.88 | 0.49 |
| Water intake | 12.71 ± 2.25 | 0.74 | 0.57 |
| Urine output | 5.88 ± 0.96 | 0.39 | 0.82 |
Means were compared over the course of the experiment with a repeated-measures ANOVA (df = 5, 30). Shown is the global mean for each metric.
FIG 1.
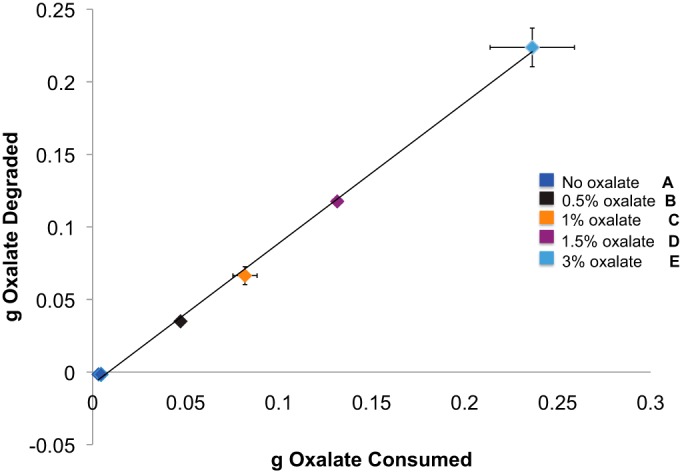
The amount of oxalate consumed is correlated with the amount of oxalate degraded (estimated from the differential between oxalate consumed and the total oxalate excreted in the urine and feces). The data were analyzed with a repeated-measures Pearson correlation (r = 0.99845, P < 0.001). The oxalate consumed also increased significantly with increasing oxalate consumption as determined by a repeated-measures ANOVA with a post hoc Holm-corrected Tukey analysis (the statistical groups are shown by bold letters).
Response of gut microbiota.
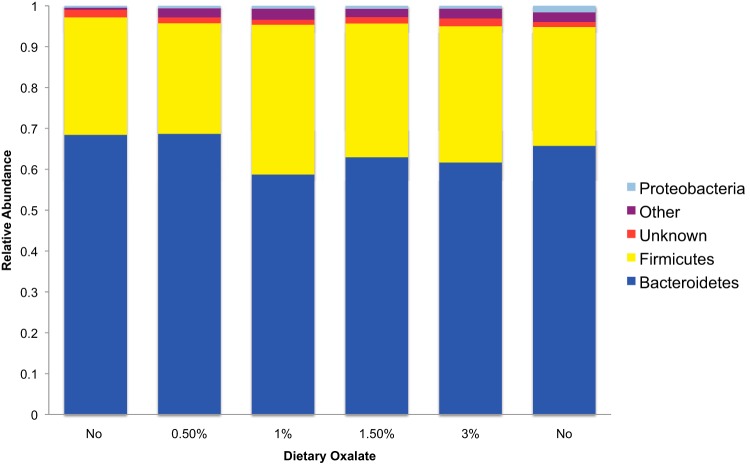
High-throughput sequencing yielded a total of 2,208,347 high-quality sequences of 150 overlapping base pairs. The data set from one animal was removed because two of the microbial inventories contained <3,000 sequence reads. Furthermore, a total of 38,723 OTU were removed from the data set, having fewer than 10 sequence reads total. The remaining inventories contained an average of 69,010 ± 5,353 sequences per sample. A rarefaction analysis concluded that the diversity at 27,378 is a good estimate of the true diversity (see Fig. S1 in the supplemental material). With a cutoff of 27.378, an additional 15 OTU were removed from the data set. When DNA was extracted from oxalate or food and used as the template for PCR with universal 16S rRNA primers, no amplification products were detected following gel electrophoresis. Similarly, the DNA extraction reagents used in the study yielded no PCR amplification of 16S rRNA, indicating that there was no detectable contamination. Across all fecal samples, sequences were assigned to 6,232 OTU. Of these OTU, 97.6% were assigned to 14 bacterial phyla with 25.3% assigned to 88 genera. The fecal microbiota showed a composition typical of woodrats and other mammals that was dominated by Bacteroidetes, particularly the family S24-7 that comprised between 49.8 to 67.2% of the microbiota (26). There were no significant differences in community membership or community structure across treatments, based on the ANOSIM (P = 0.496 and 0.691, respectively) (Fig. 2). Species richness and the Shannon index increased significantly with dietary oxalate concentration; however, levels of evenness did not differ significantly (Fig. 3). Species richness correlated significantly with oxalate consumption (Fig. 4). However, a repeated-measures ANOVA followed by a post hoc Tukey analysis revealed that only species richness with a 3% oxalate diet was significantly different from that with the no-oxalate diet (Fig. 3A). This shift in α-diversity prompted us to further investigate the microbial involvement in oxalate biotransformation.
FIG 2.
Relative abundances of the major phyla present within the gut at different dietary oxalate concentrations over time. The “Other” category contains several phyla with minor contributions to the microbiota. The columns are ordered relative to the time series of the experiment. Neither community membership nor structure changed with oxalate treatment (P = 0.496 or 0.691, respectively).
FIG 3.
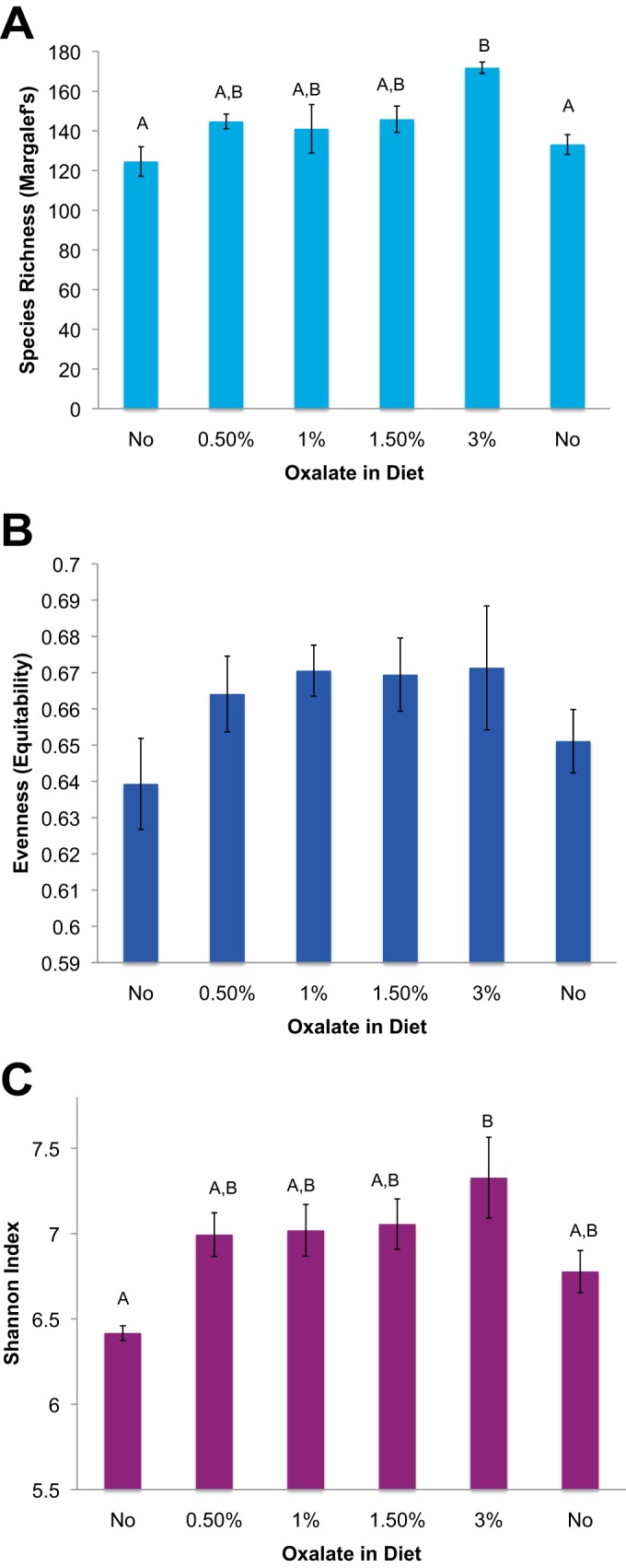
α-Diversity metrics between different dietary oxalate treatments. Species richness was determined with Margalef's richness index, species evenness was determined by their equitability, and the Shannon index is a combination of species richness and evenness. The statistical analyses were repeated -measures ANOVAs followed by a Holm-corrected paired t test. The same letters indicate statistically similar treatment groups. The order of the columns reflects the time series of the experiment. (A) Species richness as demonstrated by repeated-measures ANOVA [F(5, 30) = 2.67, P = 0.044]; (B) species evenness as demonstrated by repeated-measures ANOVA [F(5, 30) = 1.1126, P = 0.38]; (C) Shannon index as determined by repeated-measures ANOVA [F(5, 30) = 2.9928, P = 0.031].
FIG 4.
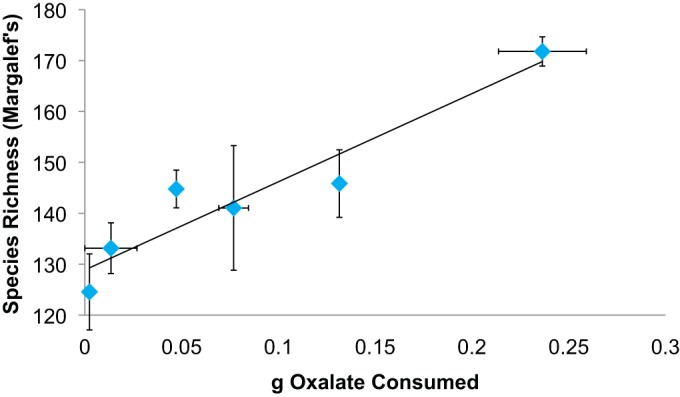
Species richness was correlated with oxalate consumption (repeated-measures Pearson correlation with a P of 0.001 and an r of 0.529). The symbols represent mean oxalate consumption and species richness for each treatment.
Of the 6,232 identified OTU, a total of 116 OTU exhibited a significant positive correlation (P < 0.05 after an FDR correction) with oxalate consumption, while 1 OTU exhibited a negative correlation (Table 3). Those OTU exhibiting a positive correlation included known oxalate-degrading bacteria: Oxalobacter, another Oxalobacteraceae sp., Clostridiales, and Lachnospiraceae, among others. The taxonomic clade with the greatest number of OTU that exhibited a positive correlation was the S24-7 family.
TABLE 3.
Microbial OTUs whose relative abundances were positively correlated with oxalate intakea
| Lowest assigned taxonomy | Taxonomic level | No. of OTU | Relative abundance with diet of: |
r | P | |
|---|---|---|---|---|---|---|
| 0% oxalate | 3% oxalate | |||||
| Oscillospira | Genus | 4 | 1 × 10−4 | 5.8 × 10−3 | 0.54 ± 0.037 | 0.009–0.037 |
| Oxalobacter | Genus | 1 | 6.39 × 10−5 | 5.24 × 10−4 | 0.54 | 0.031 |
| Clostridiales | Order | 16 | 9.13 × 10−6 | 1.03 × 10−3 | 0.52 ± 0.015 | 0.009–0.04 |
| Ruminococcus | Genus | 5 | 0 | 3.29 × 10−4 | 0.52 ± 0.015 | 0.031–0.038 |
| Allobaculum | Genus | 3 | 0 | 3.29 × 10−4 | 0.51 ± 0.017 | 0.031–0.034 |
| S24-7 | Family | 50 | 1.01 × 10−3 | 1.27 × 10−2 | 0.51 ± 0.007 | 0.009–0.041 |
| Lactobacillus | Genus | 1 | 0 | 2.44 × 10−5 | 0.51 | 0.031 |
| Oxalobacteraceae | Family | 1 | 5.48 × 10−5 | 6.09 × 10−4 | 0.51 | 0.032 |
| RF39 | Order | 1 | 0 | 1.22 × 10−5 | 0.51 | 0.031 |
| Bifidobacterium | Genus | 3 | 9.13 × 10−6 | 1.83 × 10−3 | 0.50 ± 0.022 | 0.031–0.04 |
| Unassigned | NA | 19 | 4.57 × 10−5 | 8.52 × 10−4 | 0.49 ± 0.007 | 0.031–0.044 |
| Ruminococcaceae | Family | 3 | 2.74 × 10−5 | 3.17 × 10−4 | 0.48 ± 0.02 | 0.031–0.044 |
| Lachnospiraceae | Family | 3 | 3.65 × 10−5 | 2.19 × 10−4 | 0.48 ± 0.017 | 0.031–0.043 |
| Rikenellaceae | Family | 4 | 9.13 × 10−6 | 1.87 × 10−3 | 0.48 ± 0.004 | 0.034–0.037 |
| Coprococcus | Genus | 1 | 0 | 2.44 × 10−5 | 0.48 | 0.034 |
| Proteus | Genus | 1 | 0 | 2.44 × 10−5 | 0.46 | 0.04 |
| Salinibacterium | Genus | 1 | 2.74 × 10−5 | 0 | −0.46 | 0.039 |
We tested 6,232 OTU. Comparisons were performed by Pearson correlation regression analysis with a false discovery rate (FDR) correction for multiple comparisons. For taxa with multiple OTU that were correlated with oxalate consumption, the average r values and the range of P values are given and the relative abundance refers to the group of OTU as a whole. NA, not applicable.
A subset of identified OTU were shared across all animals and treatments. A total of 103 OTU were present in all six animals on the no-oxalate diet, and 282 OTU were shared by animals on the 3% oxalate diet, including all of those present in all animals on the no-oxalate diet (see Table S2 in the supplemental material).
DISCUSSION
The current study sought to address two important gaps in gut microbiota research. First, there is a need to understand the factors that contribute to changes in the form and function of the mammalian gut microbiota, both to aid in the development of personalized therapies and to advance ecological theories (5, 6). However, studying these factors is confounded by the complexity inherent within the gut microbiota, with its immense and variable diversity and considerable microbe-microbe and microbe-host interactions (2, 6, 36). Second, there is a need to understand how oxalate affects the mammalian gut microbiota as a whole. Previous research has focused on the role of individual taxa in oxalate degradation (7, 20, 29, 37, 38). However, several oxalate-degrading taxa have now been identified from the mammalian gut, and other taxa may be affected by oxalate in obscure ways (20, 26, 39, 40). To address the gaps, we combined controlled laboratory diet trials, physiological assays, and microbial ecology to examine the taxonomic and functional response of the whole gut microbiota in a mammalian herbivore, N. albigula, which naturally consumes large amounts of oxalate in its diet, a simple compound that is metabolized exclusively by the gut microbiota (18, 26).
The microbiota of N. albigula is exceptional in its capacity to degrade oxalate. The animals exhibited no adverse effects associated with oxalate intake (Table 1), and the microbiota was capable of degrading >90% of dietary oxalate regardless of the amount of oxalate consumed, showing a strong microbial response to oxalate consumption. Studies conducted on other mammals indicate that the level of dietary oxalate degradation in N. albigula is unique (27, 28). The Norway rat (Rattus norvegicus) becomes hyperoxaluric on a 1.5% oxalate diet (41; unpublished data), whereas another study demonstrated that N. albigula can tolerate 9% oxalate with no detrimental effect (27). One potential morphological characteristic that may facilitate oxalate degradation in N. albigula is the presence of a foregut that houses a microbiota with a high potential for oxalate degradation (10, 26). In metabolic cages, N. albigula animals have access to direct coprophagy (consumption of feces from the anus), which may help to inoculate the foregut with oxalate-degrading bacteria. Given the results of the current and previous studies, the gut microbiota of N. albigula appears to have a considerable capacity for oxalate degradation, indicative of a rapid microbial response to oxalate consumption.
Our work shows that dietary oxalate affects both the diversity of the microbial community as a whole in N. albigula and the relative abundances of specific OTU. The correlation between oxalate consumption and species richness (Fig. 3A) suggests that OTU that were present below detectable limits in animals on a no-oxalate diet increased in relative abundance with higher oxalate consumption to detectable levels. Such a correlation between the consumption of (natural) dietary toxins and gut microbiota diversity has been observed in other woodrat studies and is likely indicative of a dynamic, community-wide adaptation to dietary change (42). Although there was a strong individual signature to the gut microbiota in the current study, some OTUs both were broadly distributed among animals in general and exhibited a significant correlation with oxalate consumption (Table 3; see also Table S2 in the supplemental material). The subset of microbes that increased with oxalate consumption may represent a core community of microbes essential for the function of oxalate degradation, or an “oxalate microbiome.”
A core gut microbiota has previously been associated with diverse mammalian host phenotypes (43–45). In the current study, we have identified a core set of bacteria that are commonly distributed across individuals and are responsive to oxalate, suggesting that this microbial network may be important in reducing oxalate absorption in N. albigula. Some of the bacteria in this group, such as Oxalobacter, may engage in oxalate degradation directly. Others, such as Oscillospira and Clostridiales, may benefit indirectly from oxalate degradation possibly via acetogenesis and facilitate the continued presence of those bacteria that degrade oxalate.
Strategies to utilize known oxalate-degrading bacteria as probiotic therapies to reduce urinary oxalate excretion in humans and rat models typically result in only an ephemeral reduction of urinary oxalate and a transient colonization by the probiotic bacteria (11, 37, 41, 46, 47). This is in contrast to mammals that are natural hosts to oxalate-degrading bacteria, such as the animals in the current study, which maintain those populations and their associated functions across generations and respond to increasing dietary oxalate even after long periods of time without oxalate in the diet (38, 48, 49). The transient colonization of the oxalate-degrading bacteria following probiotic treatment suggests that these transplanted bacteria are unable to integrate successfully into a foreign community, implying that there are underlying mechanisms of support for these bacteria in their natural hosts.
The S24-7 family appears to play a critical role both in the oxalate microbiome specifically and in the gut microbiota of N. albigula generally. This family comprised 43% of the OTU that exhibited a significant correlation with oxalate consumption and consistently makes up >50% of the entire gut microbiota in N. albigula (10; this study). This family is commonly found in rats, mice, goats, and humans and has also been correlated with a high-fat diet, immunoglobin A, tapeworms, etc. Thus, the S24-7 family may generally be sensitive to dietary shifts (50–54). Given the widespread distribution of this family and correlation with a number of dietary components, S24-7 represents a significant gap in our understanding of the gut microbiota form and function.
Oxalate is a simple molecular compound with characteristics that make it amenable to elucidating specific diet-microbiota interactions within the mammalian gut. In the current study, we were able to predict the identities of a subcommunity of microbes that exhibits a strong, rapid response to oxalate ingestion. Our results suggest that a distinct oxalate-metabolizing microbiome that increases in abundance when oxalate is consumed exists. Furthermore, we have shown that the methods utilized here are effective at identifying subcommunities within the mammalian gut microbiota that engage in a particular function of interest and that may be useful to manipulate in a therapeutic context.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kevin Kohl for help collecting animals, Jesse Nelson for help with diet trials, and Ky-Phuong Luong for comments on the manuscript. We thank Bob Weiss for feedback on the experiment.
We declare no conflicts of interest.
Funding Statement
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00216-16.
REFERENCES
- 1.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, et al. . 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karasov WH, Carey HV. 2009. Metabolic teamwork between gut microbes and host. Microbe 4:323–328. [Google Scholar]
- 5.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison MJ, Dawson KA, Mayberry WR, Foss JG. 1985. Oxalobacter formigenes gen. nov., sp. nov.: Oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol 141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 8.Jones R, Megarrity R. 1986. Successful transfer of DHP-degrading bacteria from Hawaiian goats to Australian ruminants to overcome the toxicity of Leucaena. Aust Vet J 63:259–262. doi: 10.1111/j.1751-0813.1986.tb02990.x. [DOI] [PubMed] [Google Scholar]
- 9.Sundset MA, Barboza PS, Green TK, Folkow LP, Blix AS, Mathiesen SD. 2010. Microbial degradation of usnic acid in the reindeer rumen. Naturwissenschaften 97:273–278. doi: 10.1007/s00114-009-0639-1. [DOI] [PubMed] [Google Scholar]
- 10.Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. 2014. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol Lett 17:1238–1246. doi: 10.1111/ele.12329. [DOI] [PubMed] [Google Scholar]
- 11.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. 2011. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest. Liver Physiol 300:G461-9. doi: 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace JR. 2008. Gut microbiology-broad genetic diversity, yet specific metabolic niches. Animal 2:661–668. doi: 10.1017/S1751731108001687. [DOI] [PubMed] [Google Scholar]
- 13.Franceschi VR, Nakata PA. 2005. Calcium oxalate in plants: formation and function. Annu Rev Plant Biol 56:41–71. doi: 10.1146/annurev.arplant.56.032604.144106. [DOI] [PubMed] [Google Scholar]
- 14.James LF, Butcher JE. 1972. Halogeton poisoning of sheep: effect of high level oxalate intake. J Anim Sci 35:1233–1238. [DOI] [PubMed] [Google Scholar]
- 15.Conyers RA, Bais R, Rofe AM. 1990. The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin Chem 36:1717–1730. [PubMed] [Google Scholar]
- 16.Massey LK, Roman-Smith H, Sutton RA. 1993. Effect of dietary oxalate and calcium on urinary oxalate and risk of formation of calcium oxalate kidney stones. J Am Diet Assoc 93:901–906. doi: 10.1016/0002-8223(93)91530-4. [DOI] [PubMed] [Google Scholar]
- 17.Moe OW. 2006. Kidney stones: pathophysiology and medical management. Lancet 367:333–344. doi: 10.1016/S0140-6736(06)68071-9. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkinson A. 1977. Oxalic acid in biology and medicine. Academic Press, New York, NY. [Google Scholar]
- 19.Allison MJ, Cook HM, Milne DB, Gallagher S, Clayman RV. 1986. Oxalate degradation by gastrointestinal bacteria from humans. J Nutr 116:455–460. [DOI] [PubMed] [Google Scholar]
- 20.Hokama S, Honma Y, Toma C, Ogawa Y. 2000. Oxalate-degrading Enterococcus faecalis. Microbiol Immunol 44:235–240. doi: 10.1111/j.1348-0421.2000.tb02489.x. [DOI] [PubMed] [Google Scholar]
- 21.Ren Z, Pan C, Jiang L, Wu C, Liu Y, Zhong Z, Ran L, Ren F, Chen X, Wang Y, Zhu Y, Huang K. 2011. Oxalate-degrading capacities of lactic acid bacteria in canine feces. Vet Microbiol 152:368–373. doi: 10.1016/j.vetmic.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Campieri C, Campieri M, Bertuzzi V, Swennen E, Matteuzzi D, Stefoni S, Pirovano F, Centi C, Ulisse S, Famularo G, De Simone C. 2001. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int 60:1097–1105. doi: 10.1046/j.1523-1755.2001.0600031097.x. [DOI] [PubMed] [Google Scholar]
- 23.Drake HL. 2012. Acetogenesis. Springer Science and Business Media, New York, NY. [Google Scholar]
- 24.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 25.Leclerc M, Bernalier A, Donadille G, Lelait M. 1997. H2/CO2 metabolism in acetogenic bacteria isolated from the human colon. Anaerobe 3:307–315. doi: 10.1006/anae.1997.0117. [DOI] [PubMed] [Google Scholar]
- 26.Miller AW, Kohl KD, Dearing MD. 2014. The gastrointestinal tract of the white-throated woodrat (Neotoma albigula) harbors distinct consortia of oxalate-degrading bacteria. Appl Environ Microbiol 80:1595–1601. doi: 10.1128/AEM.03742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirley EK, Schmidt-Nielsen K. 1967. Oxalate metabolism in the pack rat, sand rat, hamster, and white rat. J Nutr 91:496–502. [DOI] [PubMed] [Google Scholar]
- 28.Justice KE. 1985. Oxalate digestibility in Neotoma albigula and Neotoma mexicana. Oecologia 67:231–234. doi: 10.1007/BF00384290. [DOI] [PubMed] [Google Scholar]
- 29.Belenguer A, Ben Bati M, Hervás G, Toral PG, Yáñez-Ruiz DR, Frutos P. 2013. Impact of oxalic acid on rumen function and bacterial community in sheep. Animal 7:940–947. doi: 10.1017/S1751731112002455. [DOI] [PubMed] [Google Scholar]
- 30.Ingale KG, Thakurdesai PA, Vyawahare NS. 2012. Effect of Hygrophila spinosa in ethylene glycol induced nephrolithiasis in rats. Indian J Pharmacol 44:639. doi: 10.4103/0253-7613.100402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens S, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Lozupone CA, Hamady H, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel SL, Hartman PA, Allison MJ. 1987. Intestinal colonization of laboratory rats with Oxalobacter formigenes. Appl Environ Microbiol 53:2767–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppe B, Beck B, Gatter N, Von Unruh G, Tischer A, Hesse A, Laube N, Kaul P, Sidhu H. 2006. Oxalobacter formigenes: a potential tool for the treatment of primary hyperoxaluria type 1. Kidney Int 70:1305–1311. doi: 10.1038/sj.ki.5001707. [DOI] [PubMed] [Google Scholar]
- 39.Murphy C, Murphy S, O'Brien F, O'Donoghue M, Boileau T, Sunvold G, Reinhart G, Kiely B, Shanahan F, O'Mahony L. 2009. Metabolic activity of probiotics-oxalate degradation. Vet Microbiol 136:100–107. doi: 10.1016/j.vetmic.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Turroni S, Bendazzoli C, Dipalo SCF, Candela M, Vitali B, Gotti R, Brigidi P. 2010. Oxalate-degrading activity in Bifidobacterium animalis subsp. lactis: impact of acidic conditions on the transcriptional levels of the oxalyl-CoA decarboxylase and formyl-CoA transferase genes. Appl Environ Microbiol 76:5609–5620. doi: 10.1128/AEM.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieske JC, Tremaine WJ, De Simone C, O'Conner HM, Li X, Bergstralh E, Goldfarb DS. 2010. Diet, but not oral probiotics, effectively reduces urinary oxalate excretion and calcium oxalate supersaturation. Kidney Int 78:1178–1185. doi: 10.1038/ki.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohl KD, Dearing MD. 2012. Experience matters: Prior exposure to plant toxins enhances diversity of gut microbiome in herbivores. Ecol Lett 15:1008–1015. doi: 10.1111/j.1461-0248.2012.01822.x. [DOI] [PubMed] [Google Scholar]
- 43.Sekelja M, Berget I, Naes T, Rudi K. 2011. Unveiling an abundant core microbiota in the human adult colon by a phylogroup-independent searching approach. ISME J 5:519–531. doi: 10.1038/ismej.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. 2012. A crypt-specific core microbiota resides in the mouse colon. mBio 3:e00116-12. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elli M, Colombo O, Tagliabue A. 2010. A common core gut microbiota between obese individuals and their lean relatives? Evaluation of the predisposition to obesity on the basis of the fecal microflora profile. Med Hypotheses 75:350–352. doi: 10.1016/j.mehy.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 46.Sidhu H, Allison MJ, Chow JM, Clark A, Peck AB. 2001. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J Urol 166:1487–1491. doi: 10.1016/S0022-5347(05)65817-X. [DOI] [PubMed] [Google Scholar]
- 47.Knight J, Deora R, Assimos DG, Holmes RP. 2013. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis 41:187–196. doi: 10.1007/s00240-013-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allison MJ, Littledike E, James L. 1977. Changes in ruminal oxalate degradation rates associated with adaptation to oxalate ingestion. J Anim Sci 45:1173–1179. [DOI] [PubMed] [Google Scholar]
- 49.Palgi N, Ronen Z, Pinshow B. 2008. Oxalate balance in fat sand rats feeding on high and low calcium diets. J Comp Phys B Biochem Syst Environ Physiol 178:617–622. doi: 10.1007/s00360-008-0252-1. [DOI] [PubMed] [Google Scholar]
- 50.Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C, Mariette J, Bouchez O, Lluch J, Ouarné F, Monsan P, Valet P, Roques C, Amar J, Bouloumié A, Théodorou V, Burcelin R. 2012. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato T, Fukuda S, Fujiwara A, Suda W, Hattori M, Kikuchi J, Ohno H. 2014. Mutiple omics uncovers host-gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res 21:469–480. doi: 10.1093/dnares/dsu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, Antonopoulous DA, Smith D, Chang EB, Ciancio MJ. 2014. Excercise prevents weight gain and alters the gut microbiota in a mouse model of high-fat diet induced obesity. PLoS One 9:e92193. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. 2015. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos Trans R Soc Lond B Biol Sci 370:20140295. doi: 10.1098/rstb.2014.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.