ABSTRACT
Recent characterization of the bacterial community structure in beach sands has revealed patterns of biogeography similar to those observed in aquatic environments. Studies to date, however, have mainly focused on subtidal sediments from marine beaches. Here, we investigate the bacterial diversity, using Illumina-based sequencing of the V5-V6 region of the 16S rRNA gene, at 11 beaches representing those next to the Great Lakes, Florida, and the Pacific Ocean. The alpha diversity differed significantly among regions (P < 0.0001), while the within-region diversity was more similar. The beta diversity also differed by region (P < 0.001), where freshwater sands had significantly higher abundances of taxa within the Actinobacteria, Betaproteobacteria, and Verrucomicrobia than marine environments. In contrast, marine sands harbored greater abundances of Gammaproteobacteria and Planctomycetes, and those from Florida had more Deltaproteobacteria and Firmicutes. Marine beaches had significantly different phylogenetic community structures (P ≤ 0.018), but freshwater and Florida beaches showed fewer within-region phylogenetic differences. Furthermore, regionally distinct patterns in taxonomic variation were observed in backshore sands, which had communities distinct from those in nearshore sands (P < 0.001). Sample depth minimally influenced the community composition. The results of this study reveal distinct bacterial community structures in sand on a broad geographic scale but moderate regional similarity and suggest that local variation is primarily related to the distance from the shoreline. This study offers a novel comparison of the bacterial communities in freshwater and marine beach sands and provides an important basis for future comparisons and analyses to elucidate factors affecting microbial ecology in this underexplored environment.
IMPORTANCE This study presents a large-scale geographic characterization of the bacterial communities present in beach sands. While previous studies have evaluated how environmental factors influence bacterial community composition, few have evaluated bacterial communities in freshwater sands. Furthermore, the use of a consistent methodology to characterize bacterial communities here allowed a novel comparison of communities across geographic regions. We reveal that while the community composition in sands at individual beaches is distinct, beach sands within the same region harbor similar assemblages of bacteria and these assemblages differ greatly between regions. In addition, moisture, associated with distance from the shoreline, strongly influences the bacteria present in sands and more strongly influences the bacteria present than sample depth does. Thus, the data presented here offer an important basis for a broader characterization of the ecology of bacteria in sands, which may also be relevant to public health and resource management initiatives.
INTRODUCTION
Beach sands are dynamic ecosystems that harbor diverse microbial communities vital to ecosystem services, including water purification, biogeochemical cycling, and the mineralization of organic compounds (1–3). In marine systems, a beach cross section can be divided into three major compartments that vary in physical and chemical properties in large part due to tidal cycles (depicted in Fig. S1 in the supplemental material): (i) the nearshore, subtidal zone, which is permanently saturated by the overlying water column; (ii) the intertidal zone, which experiences periodic tidal wetting and drying; and (iii) the backshore, supratidal region, which does not experience tidal wetting (4). Variation also likely occurs due to depth, with a vadose zone of dry sand sitting above an ephemeral intertidal saltwater cell that often overlies fresh groundwater (5). The last two saturated zones interact with a deep saltwater wedge located below the subtidal sands, resulting in complex chemical gradients and the potential exchange of nutrients and bacteria (3, 6). Similarly, in the Great Lakes, seiche dynamics (a tide-like standing wave found in enclosed bodies of water) play a role similar to that played by the tidal cycles of marine beaches (7), and there is also frequently a continual discharge of groundwater to many lakes (8).
There are several methods by which bacterial communities in sands may be shaped by stochastic variables, as well as by transport and exchange between sand and water (4). The majority of microbial species in beach sands are thought to be autochthonous, and a cosmopolitan assemblage has been identified in a limited geographic area (3). However, contamination of beach sands by pollution in the water has also been suggested (9). Bacteria in beach sands may also enter the water column via overbeach transport, where tidal events or wave action release bacteria attached to sand particles or those residing in interstitial spaces and these bacteria then enter the water directly (10, 11). Alternatively, through-beach transport can also occur when bacteria in unsaturated sands are mobilized downward due to tidal or wave action, enter the groundwater, and are transported via subterranean discharge to the water column (3, 12). Consequently, beach sand microbes and those in the lake water or in sediments can be thought of as being in a state of dynamic equilibrium.
Recently, bacterial communities in beach sands have begun receiving increasing attention due to their relevance to public health (4, 13–18). While recreational waters have long been monitored for indicators of human fecal pollution and their presumed indication of risk to humans because of the presence of human pathogens (19, 20), beach sands have historically been neglected in this area. Over the last several years, both marine and freshwater sands have been recognized to be important reservoirs of fecal indicator bacteria (21–23) and environments that support naturalized microbial populations consisting of prokaryotes and eukaryotes (10, 24). Furthermore, the number of studies that have identified bacterial, fungal, and viral pathogens in beach sands is steadily increasing (4, 18, 22), and not all of these pathogens have been of fecal origin. Therefore, a more thorough understanding of the bacterial communities typical of beach sands is likely to offer novel insights into the ecology of these complex ecosystems and perhaps guide monitoring efforts to protect public health.
The evolution of next-generation sequencing has allowed a more comprehensive characterization of the microbial communities present in a variety of environments, including in marine waters (25), in riverine ecosystems (26, 27), in soils (28), and in and on humans and animals (29). Over the last 5 years, many studies have employed next-generation sequencing to study primarily subtidal sands and sediments (2, 30, 31). These studies have revealed that members of the rare biosphere, i.e., taxa of low abundance, fluctuate in abundance in relation to physicochemical parameters and sample depth (2, 30, 31). A recent study of intertidal sand samples collected from California beaches revealed the presence of abundant, cosmopolitan taxa that were active in through-beach transport (3). Furthermore, a biogeographic relationship in the microbial community composition has been revealed, where the communities from sands with similar characteristics or an increased anthropogenic impact were more similar to each other (3). Taken together, these studies indicate that, similar to what has been noted in soils, water and water availability play a prominent role in the distributions, types, and abundances of microbes in sand environments.
To date, nearly all of the next-generation sequencing studies of sands have focused on sand from marine beaches (2, 3, 31–35), but one recent study has focused on sand from freshwater beaches (17), and due to differences in methodology, including the sequencing platform and biases associated with the sequencing region associated with primer targets (36), the ability to compare these data sets with each other has been limited. In this study, beach sands from 11 beaches throughout the United States, Japan, and South Korea, including 4 beaches from the Great Lakes, were extensively characterized by using Illumina next-generation sequencing of the 16S rRNA gene. Generation of this data set allowed assessment of the variation in communities at global, regional, and local scales. Samples were collected from nearshore, intertidal, and backshore segments of all beaches at three depths at 10-cm intervals. We hypothesized that, due to geographic separation, each beach would harbor a unique bacterial community but that patterns in local variation would be similar at all locations. The results of this study revealed novel patterns in the bacterial community composition and structure at beaches throughout the world and further emphasized that local differences in bacterial community composition and structure due to distance from the tidal zone and sample depth are likely.
MATERIALS AND METHODS
Sample collection and processing.
Samples were collected from 11 beaches throughout the United States, Japan, and South Korea by local collaborators at each site (Table 1; see also Fig. S2 in the supplemental material). Due to geographic proximity, similarity in water type (i.e., fresh or marine), and statistical grouping based on the bacterial communities present, the sampling sites were grouped as Great Lakes, Florida, and Pacific Ocean. All samplings were performed between 29 September and 3 October 2014. For beaches influenced by tides, samples were collected at an outgoing, low tide. Samples were collected at depths of 0 to 10 cm, 10 to 20 cm, and 20 to 30 cm at the same spot using an ethanol-sterilized auger. Triplicate locations 2 m apart were sampled at each distance from the shoreline, and the sampling distances from the shoreline included (i) at the shoreline, (ii) 1 m from the shoreline, and (iii) 10 m from the shoreline (see Fig. S1 in the supplemental material). Samples were stored in 50-ml conical tubes or Whirl-Pak bags (Nasco, Fort Atkinson, WI, USA) and transported back to the lab on ice. Samples were shipped on dry ice to the University of Minnesota for DNA extraction, or DNA was extracted as described below and shipped on dry ice to the University of Minnesota. Samples were stored at −20°C prior to DNA extraction. DNA extraction was done using 250 to 300 mg of sand and PowerSoil DNA isolation kits (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) as described by the manufacturer.
TABLE 1.
Summary of beach locations
| Region | Location | Beach | Sand moisture content (%) | Latitude | Longitude |
|---|---|---|---|---|---|
| Great Lakes | Duluth, MN, USA | Minnesota Point | 7.5 ± 8.0 | 46.728 | −92.048 |
| Chicago, IL, USA | 63rd Street Beach | 26.1 ± 7.6 | 41.782 | −87.573 | |
| Burlington, Ontario, Canada | Burlington Beach | 20.7 ± 4.4 | 43.314 | −79.800 | |
| Toronto, Ontario, Canada | Marie Curtis Park | 12.0 ± 6.5 | 43.583 | −79.543 | |
| Florida | Tampa, FL, USA | Fort DeSoto | 20.2 ± 2.6 | 27.617 | −82.737 |
| Miami, FL, USA | Crandon Park | 14.6 ± 6.5 | 25.713 | −80.151 | |
| Pacific Ocean | Southern California, USA | Huntington Beach | 19.8 ± 5.3 | 33.656 | −118.000 |
| Oahu, HI, USA | Sandy Beach | 8.2 ± 5.4 | 21.286 | −157.673 | |
| Northern Japan | Otaru Dream Beach | 28.6 ± 12.6 | 43.158 | 141.208 | |
| Southern Japan | Fukiage-hama Beach | 6.9 ± 4.5 | 31.521 | 130.325 | |
| Jeju, South Korea | Jeju Beach | NDa | 33.445 | 126.294 |
ND, not determined.
Sequencing and bioinformatics.
PCR amplification, amplicon purification, sample pooling, and sequencing were performed by the University of Minnesota Genomics Center (Minneapolis, MN, USA) as previously described (37). The V5-V6 hypervariable region of the 16S rRNA gene was amplified using the BSF784/1064R primer set (25, 38), and the amplicons were gel purified. Negative controls consisting of sterile water were included to test the PCR mixtures, and amplification products were not obtained from those controls. Purified amplicons were pooled in equal amounts, and paired ends were sequenced at a read length of 150 nucleotides (nt) on a HiSeq2500 platform (Illumina, Inc., San Diego, CA, USA).
All sequence processing was performed using mothur (version 1.34.0) software (39). Sequences were paired-end joined using fastq-join software (40) and quality trimmed using a mean quality score of 35 and a 50-nt window. Sequences containing homopolymers of >8 nt, ambiguous bases, or more than 2 nt mismatches from the primer sequences were removed. High-quality sequences were aligned against the SILVA database (version 119) (41), screened to remove sequences that did not fall within the region amplified by the primers, and subjected to a 2% preclustering step (42). UCHIME software was used to identify and remove chimeric sequences (43). Sequences were normalized by random subsampling to 25,000 sequence reads per sample for statistical comparisons (44). Operational taxonomic units (OTUs) were assigned at a 97% identity using the furthest-neighbor algorithm, and taxonomic assignments were made using the Ribosomal Database Project taxonomy (version 10) described previously (45).
Statistical analyses.
Analysis of variance (ANOVA) and Spearman correlations were performed using XLSTAT (version 2015.1.01) software (Addinsoft, Belmont, MA, USA). All other diversity indices and statistics were calculated using mothur software. Shannon indices and abundance-based coverage estimates (ACEs) were calculated to assess parametric and nonparametric diversity. Comparisons between samples were performed on the basis of Bray-Curtis dissimilarity matrices (46). Beta diversity was compared using analysis of similarity (ANOSIM) (47), ordination was performed via principal coordinate analysis (PCoA), and clustering of sample groups was evaluated using analysis of molecular variance (AMOVA) (48). Determination of OTUs that significantly affected ordination was performed by the Spearman method using mothur software. Variations in the abundances of OTUs among sample groups were determined using the Kruskal-Wallis test (49), and differences in phylogenetic structure were assessed on the basis of both unweighted and weighted UniFrac distances (50). All statistics were evaluated at an α level of 0.05.
Nucleotide sequence accession numbers.
Fastq files containing the raw sequencing data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession number SRP064396.
RESULTS
The mean sand moisture content was 15.7% ± 9.7% among all samples, and the moisture content was significantly different among all beaches (P < 0.001; Table 1). Moisture content was negatively correlated with distance from the shoreline (Spearman's r = −0.483, P < 0.0001), but not sample depth (r = 0.019, P = 0.776). The ACE index was also negatively correlated with distance from the shoreline (r = −0.130, P = 0.046) and positively correlated with the moisture content (r = 0.302, P < 0.0001).
Large-scale variation in bacterial communities.
Among all samples included in the analysis (n = 258), a mean coverage of 96.7% ± 2.0% was achieved, with the coverage ranging from 89.4% to 99.9%. Mean Shannon indices for individual sites, taking all depths and distances from the shoreline together, ranged from 5.38 to 6.43, and Shannon diversity differed significantly on the basis of the sampling region, where the ranking of diversity was as follows: Florida beaches > Pacific Ocean beaches > Great Lakes beaches (P < 0.0001; Table 2). This trend was also observed using the ACE index (P < 0.0001).
TABLE 2.
Alpha diversity indices
| Beach | Mean index value ± SDa |
|
|---|---|---|
| Shannon | ACE | |
| Minnesota Point | 5.38 ± 0.41A | 1,702 ± 516A |
| 63rd Street Beach | 5.50 ± 0.41A,B,C | 1,479 ± 867A |
| Burlington Beach | 5.81 ± 0.13B,C | 2,056 ± 658A,B |
| Marie Curtis Park | 5.94 ± 0.28B,D | 2,830 ± 598B,C |
| Fort DeSoto | 6.43 ± 0.21C | 7,247 ± 2189D |
| Crandon Park | 6.43 ± 0.24C | 4,564 ± 1235E |
| Huntington Beach | 5.94 ± 0.24B,D | 2,853 ± 476B,C |
| Sandy Beach | 6.20 ± 0.40C,D | 3,750 ± 1601C,E |
| Otaru Dream Beach | 5.88 ± 0.31B,D | 3,021 ± 598B,C |
| Fukiage-hama Beach | 5.43 ± 1.02A,C | 1,313 ± 667A |
| Jeju Beach | 6.12 ± 0.11B,C,D | 6,661 ± 1135D |
Horizontal spaces divide sampling regions. The results for beaches sharing the same superscript letter (A, B, C, D, and E) did not differ significantly at an α value of 0.05.
Taking all sampling sites together, samples taken 10 m from the shoreline had significantly lower Shannon diversity indices (P ≤ 0.025) than samples taken at the shoreline and samples taken 1 m from the shoreline, for which the Shannon diversity indices did not differ significantly from each other (P = 0.992). This trend was maintained regionally within the Great Lakes and Pacific Ocean sands (see Tables S1 and S2 in the supplemental material) but not within Florida sands (see Table S3 in the supplemental material). In contrast, differences in ACE indices for samples collected at all distances were not significant (P = 0.092). Regionally, this trend was maintained in the Great Lakes and Pacific Ocean sands, but in Florida sands, ACE diversity was significantly lower for samples taken 1 m from the shoreline than samples taken either at the shoreline or 10 m from the shoreline (P = 0.041).
Differences in Shannon diversity at various depths were significant (P = 0.018), although post hoc tests revealed that only the difference between the samples obtained at 10-cm and 20-cm depths was significant (P = 0.026). Regionally, however, differences in Shannon diversity were significantly different only among Pacific Ocean sands (P = 0.042; see Table S2 in the supplemental material). Sample depth did not significantly affect ACE diversity among all samples (P = 0.324) or samples from the same region (P ≥ 0.305).
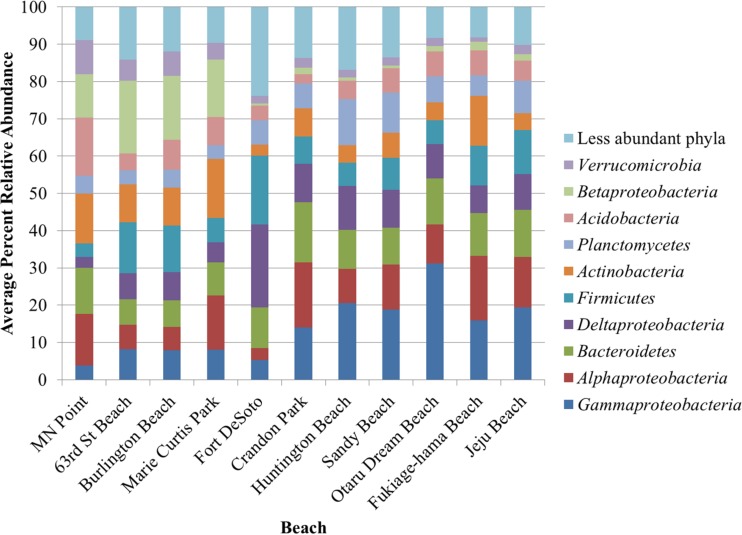
The bacterial communities found in all samples primarily comprised the phyla Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria (Fig. 1). The communities in samples collected from beaches in the same region (i.e., the Great Lakes, Florida, or Pacific Ocean) tended to be more taxonomically similar to each other than the communities in samples collected from different regions. Notably, the freshwater beaches tended to have greater relative abundances of Actinobacteria, Betaproteobacteria, and Verrucomicrobia, while the marine beaches had greater relative abundances of Gammaproteobacteria, Deltaproteobacteria, Firmicutes, and Planctomycetes. The observed differences in the phylum distribution were statistically significant (P < 0.05; Fig. 2) on the basis of Kruskal-Wallis tests of differences in the relative abundances of OTUs among regions. Furthermore, differences in the sand communities between Florida and Pacific Ocean beaches were resolved in greater detail. While Florida beach sand communities had greater abundances of Deltaproteobacteria (e.g., Desulfobacteraceae) and Firmicutes, the Pacific Ocean beach sands harbored higher proportions of Gammaproteobacteria (e.g., Thiotrichaceae) and Planctomycetes.
FIG 1.
Distribution of phyla and classes of Proteobacteria, averaged among all samples, for each beach.
FIG 2.
Family-level distribution of OTUs found to differ significantly by the Kruskal-Wallis test at an α level of 0.05.
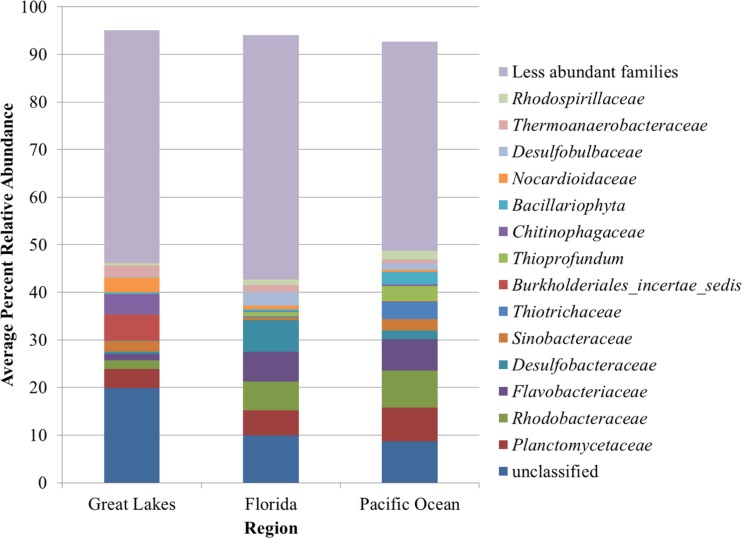
Analysis of the abundant families, which were those that had a mean relative abundance of at least 1% over the entire data set, revealed that their abundances differed significantly by region, as determined using ANOVA (Table 3), with greater differences between the abundances of these families in Great Lakes sands versus sands of the two marine regions. Among the less abundant families for which results are not depicted in Fig. 2, regional differences were also observed, where Great Lakes beaches had significantly greater abundances of families within the Alphaproteobacteria (e.g., Sphingomonadaceae), Betaproteobacteria (e.g., Comamonadaceae and Rhodocyclaceae), and Verrucomicrobiaceae. Sands from Florida and Pacific Ocean beaches had in common at comparable abundances several low-abundance families (e.g., Geobacteraceae, Syntrophobacteraceae, and Chromatiaceae) that were present at significantly greater abundances than they were in sands from Great Lakes beaches. However, sands from Florida beaches harbored families within the Bacteroidetes (e.g., Cytophagaceae) at abundances greater than those in sands from Pacific Ocean beaches. Furthermore, families within the Firmicutes differed in abundance; for example, Florida sands had greater relative abundances of Veillonellaceae, while Pacific sands had greater abundances of Bacillaceae.
TABLE 3.
Relative abundance of families from all regions
| Familya | Mean relative abundance ± SDb (%) |
||
|---|---|---|---|
| Great Lakes | Florida | Pacific Ocean | |
| Unclassified | 20.51 ± 6.26A | 10.54 ± 3.74B | 9.48 ± 2.60B |
| Planctomycetaceae | 4.04 ± 1.69A | 5.51 ± 1.65B | 7.53 ± 2.92C |
| Rhodobacteraceae | 1.90 ± 0.91A | 6.05 ± 4.66B | 7.98 ± 2.71C |
| Flavobacteriaceae | 1.40 ± 1.12A | 6.49 ± 4.00B | 6.77 ± 4.50B |
| Desulfobacteraceae | 0.46 ± 0.48A | 6.87 ± 4.86B | 1.87 ± 1.01C |
| Sinobacteraceae | 2.36 ± 1.54A | 0.61 ± 0.51B | 2.5 ± 1.51A |
| Thiotrichaceae | 0.13 ± 0.09A | 0.19 ± 0.17A | 3.67 ± 2.37B |
| Burkholderiales incertae sedis | 5.33 ± 2.35A | 0.07 ± 0.12B | 0.11 ± 0.20B |
| Chitinophagaceae | 4.81 ± 2.28A | 0.26 ± 0.44B | 0.36 ± 0.30B |
| Thioprofundum | 0.01 ± 0.06A | 0.84 ± 1.26B | 3.23 ± 1.98C |
| Bacillariophyta | 0.58 ± 0.84A | 0.60 ± 0.57A | 2.92 ± 3.62B |
| Desulfobulbaceae | 0.25 ± 0.25A | 3.07 ± 2.16B | 1.51 ± 0.64C |
| Thermoanaerobacteraceae | 2.25 ± 1.41A | 1.46 ± 0.80B | 0.88 ± 0.36C |
| Nocardioidaceae | 3.22 ± 3.37A | 0.74 ± 1.08B | 0.48 ± 1.55B |
| Rhodospirillaceae | 0.61 ± 0.37A | 1.25 ± 0.92B | 1.98 ± 0.95C |
| Geobacteraceae | 0.59 ± 0.31A | 0.96 ± 0.36B | 2.04 ± 0.91C |
| Comamonadaceae | 3.84 ± 1.01A | 0.16 ± 0.21B | 0.13 ± 0.22B |
| Sphingomonadaceae | 3.41 ± 2.98A | 0.24 ± 0.36B | 0.22 ± 0.36B |
| Verrucomicrobiaceae | 1.43 ± 0.78A | 1.04 ± 0.77B | 1.15 ± 0.52B |
| Syntrophobacteraceae | 0.95 ± 0.51A | 1.66 ± 2.21B | 1.19 ± 0.69A |
| Chromatiaceae | 0.20 ± 0.22A | 1.77 ± 0.90B | 1.56 ± 0.54B |
| Saprospiraceae | 0.32 ± 0.24A | 0.92 ± 1.01B | 1.64 ± 0.65C |
Only families present at a mean level of >1% over the entire data set are shown.
The relative abundance in samples sharing the same superscript letter (A, B, or C) do not differ significantly by ANOVA at an α value of 0.05.
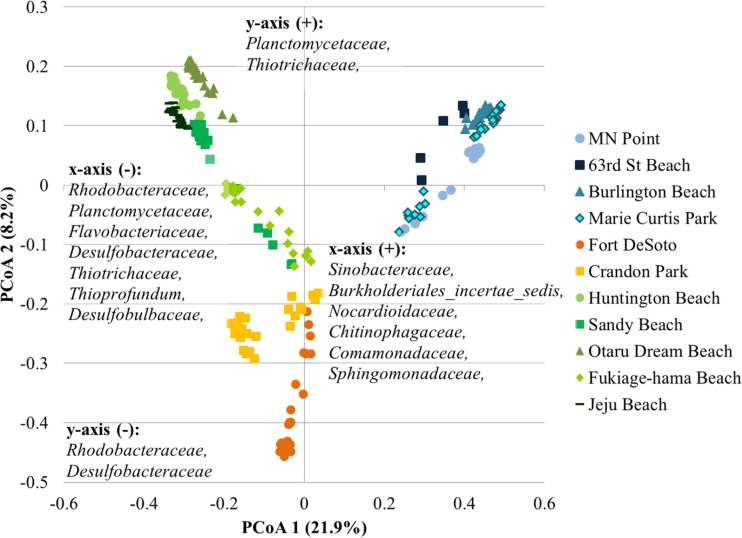
Ordination of samples by PCoA revealed the separation of samples by region (Fig. 3), and this clustering was significant by AMOVA (P < 0.001). Families within the Proteobacteria were the predominant drivers of separation of the communities among beaches regionally (Fig. 3). Furthermore, differences in bacterial community compositions (beta diversity) between regions were significant by ANOSIM (P < 0.001). Evaluation of both unweighted and weighted UniFrac distances also revealed significant differences in the phylogenetic structures of bacterial communities among regions (P < 0.001 for all analyses; see Fig. S3 in the supplemental material).
FIG 3.
Principal coordinate analysis of microbiota from all sand samples. The relationship between the ordination plot and the distance matrix had an r2 value of 0.57. The families listed represent the taxonomic assignments of OTUs that significantly affected positioning along either axis (P < 0.05) and were present at a mean relative abundance of >1.0% among all samples. Families are listed in order of declining abundance.
Regional variation in bacterial communities.
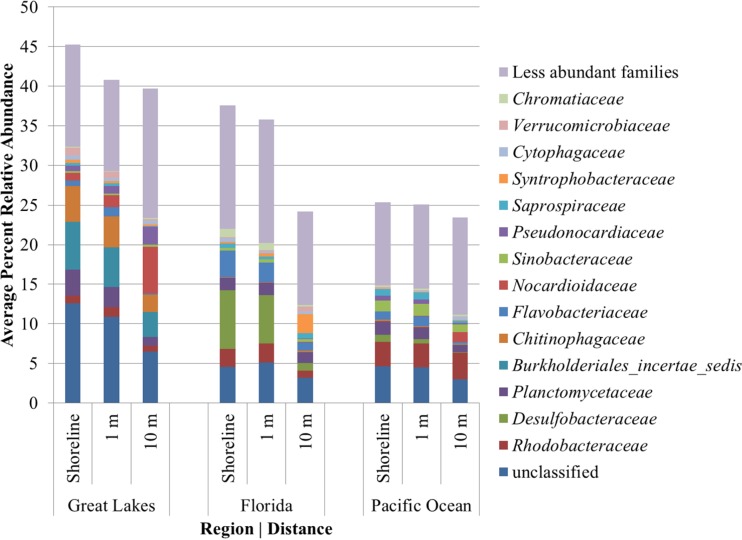
Interrogation of taxonomic shifts among the major phyla and Proteobacteria classes on the basis of sample depth and distance from the shoreline revealed little variation due to depth (discussed in greater detail below) and regionally specific variation with distance from the shoreline by ANOVA. Among the Great Lakes sand samples, distance from the shoreline was associated with an expansion of the abundance of Alphaproteobacteria (P = 0.026) and Actinobacteria (P < 0.001) and a decline in the abundance of Bacteroidetes (P < 0.001), Planctomycetes (P < 0.001), Betaproteobacteria (P = 0.009), and Verrucomicrobia (P < 0.001) (Fig. 4). The Verrucomicrobia (P = 0.001) showed higher relative abundances at a greater distance from the shoreline among the Florida beaches, with a corresponding reduction in Deltaproteobacteria (P = 0.028) and Acidobacteria (P < 0.001). Among the Pacific Ocean beaches, Firmicutes, Actinobacteria, and Betaproteobacteria had higher relative abundances at a distance of 10 m from the shoreline (P < 0.001 to 0.021) than at the other distances, with reductions in Deltaproteobacteria, Planctomycetes, Acidobacteria, and Verrucomicrobia being detected (P < 0.001 to 0.043). Notably, post hoc tests revealed no significant differences in the relative abundances of phyla between the samples taken at the shoreline and those taken 1 m from the shoreline (P ≥ 0.670).
FIG 4.
Family-level classification of OTUs for each region that differed significantly in relative abundance by distance from the shoreline by the Kruskal-Wallis test (P < 0.05).
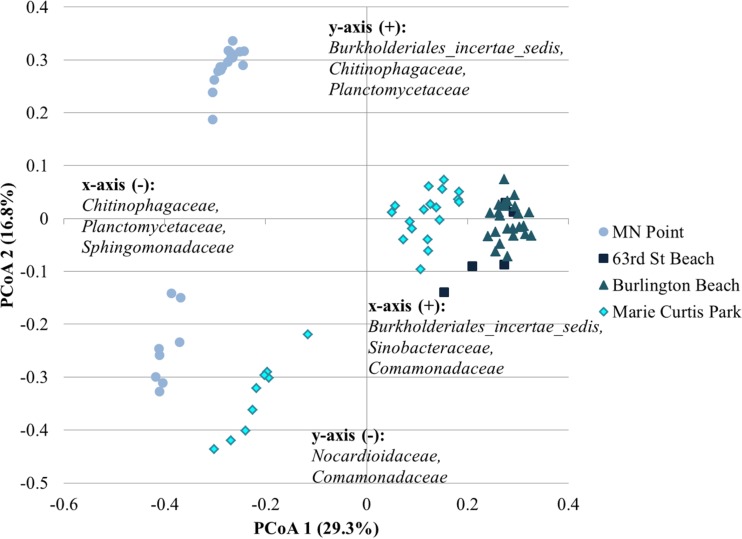
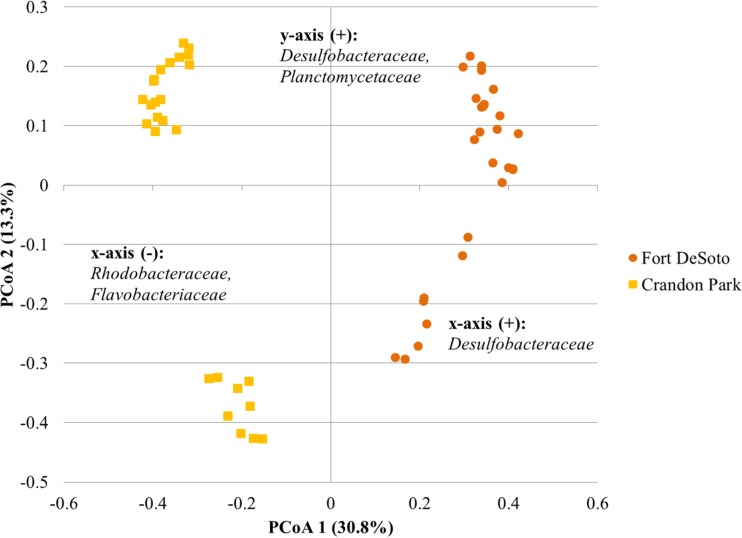
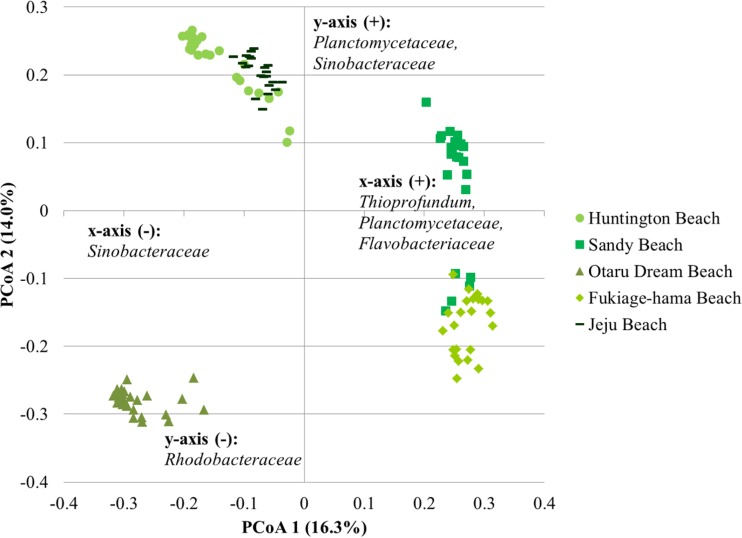
Ordination of samples within a given region revealed a significant separation of communities on the basis of the beach sampled by AMOVA (P < 0.001 for all analyses; Fig. 5 to 7). Differences in beta diversity were also significantly different among beaches within a region by ANOSIM (P ≤ 0.009, P < 0.001, and P < 0.001 for the Great Lakes, Florida, and Pacific Ocean regions, respectively). Similarly, weighted UniFrac distances indicated significant differences in the abundance-based phylogenetic structure of communities among beaches within each region (P < 0.001). Evaluation of unweighted UniFrac distances revealed a greater phylogenetic similarity of communities within the Great Lakes, with no significant differences between the communities in samples from 63rd Street Beach in Chicago, IL (USA), and those in the other samples tested (P = 0.458 to 0.553). However, the communities in sands from all other freshwater beaches differed significantly (P = 0.031 to 0.041). The phylogenetic structures of the communities in sands from the two beaches sampled in Florida were not significantly different using unweighted UniFrac analyses, although differences in UniFrac metrics approached significance (P = 0.050). In contrast, the differences in the unweighted phylogenetic structures of the communities in the sands of all beaches in the Pacific Ocean region were significant (P ≤ 0.018).
FIG 5.
Principal coordinate analysis of microbiota from sand samples collected from Great Lakes beaches. The relationship between the ordination plot and the distance matrix had an r2 value of 0.76. The families listed represent the taxonomic assignments of OTUs that significantly affected positioning along either axis (P < 0.05) and were present at a mean relative abundance of >1.0% among all samples. Families are listed in order of declining abundance.
FIG 6.
Principal coordinate analysis of microbiota from sand samples collected from Florida beaches. The relationship between the ordination plot and the distance matrix had an r2 value of 0.74. The families listed represent the taxonomic assignments of OTUs that significantly affected positioning along either axis (P < 0.05) and were present at a mean relative abundance of >1.0% among all samples. Families are listed in order of declining abundance. No families showed significantly higher relative abundances with a decrease in the y-axis coordinate.
FIG 7.
Principal coordinate analysis of microbiota from sand samples collected from Pacific Ocean beaches. The relationship between the ordination plot and the distance matrix had an r2 value of 0.44. The families listed represent the taxonomic assignments of OTUs that significantly affected positioning along either axis (P < 0.05) and were present at a mean relative abundance of >1.0% among all samples. Families are listed in order of declining abundance.
Local variation in bacterial communities.
The local variation in sand communities was assessed with respect to sample depth (10, 20, and 30 cm) and distance from the shoreline (0, 1, and 10 m) for all locations except the 63rd Street Beach, where replicate samples could not be amplified or too few sequence reads were obtained for inclusion in the analysis. As a result of insufficient replication, data for this beach were excluded from local analyses.
Ordination of samples from individual beaches revealed slightly different trends in the local variation by region (see Fig. S4 to S13 in the supplemental material). For the Great Lakes beaches, depth did not result in significant clustering of bacterial communities (P ≥ 0.633). There was, however, significant clustering of samples by distance from the shoreline (P ≤ 0.03), with each distance clustering separately. Furthermore, distance from the shoreline was associated with decreases in bacterial groups commonly associated with freshwater environments, e.g., Planctomycetaceae, Burkholderiales, and Chitinophagaceae (Fig. 4).
At the Florida beaches, communities characterized from the Crandon Park beach also followed a trend similar to that for communities characterized from the Great Lakes beaches and clustered by distance from the shoreline (P < 0.001) but not by depth (P = 0.092). In contrast, communities from Fort DeSoto saw significant clustering by both depth and distance from the shoreline (P = 0.035 and 0.003). At this beach, the sample taken 10 m from the shoreline clustered independently (P ≤ 0.005) and there was separation of the samples collected at 10-cm and 20-cm depths (P = 0.023). At both sites, distance from the shoreline corresponded to significant decreases in families, such as Desulfobacteraceae and Flavobacteriaceae, that may be associated with the water column (Fig. 4), but at Fort Desoto, where differences in communities by depth were observed, variation in the abundances of bacterial groups showed no consistent trends by depth.
Among the marine beaches, Huntington Beach and Sandy Beach communities showed independent clustering at all distances from the shoreline (P ≤ 0.042), with no clustering by depth detected (P ≥ 0.458). Similarly, clustering by depth was not significant among the remaining marine beaches (P ≥ 0.171), but communities in samples collected 10 m from the shoreline were significantly separated from those collected 1 m from or at the shoreline (P ≤ 0.002). Similar to the findings for the Florida beaches, Desulfobacteraceae and Flavobacteriaceae, as well as Planctomycetaceae, were among the most abundant families that showed significant shifts in abundance with distance from the shoreline. With few exceptions, these trends were also observed among differences in beta diversity, as assessed by ANOSIM (Table 4).
TABLE 4.
Summary of local differences in beta diversity (ANOSIM statistics) among all beaches
| Region | Beach | Beta diversity |
Post hoc results | |
|---|---|---|---|---|
| Depth | Distance from shoreline | |||
| Great Lakes | Minnesota Point | 0.982 | <0.001 | The results for all distances from the shoreline were significantly different from each other (P < 0.001) |
| Burlington Beach | 0.597 | <0.001 | The result for 10 m from the shoreline was significantly different from the results for the other distances (P < 0.001) | |
| Marie Curtis Park | 0.677 | <0.001 | The results for all distances from the shoreline were significantly different from each other (P ≤ 0.012) | |
| Florida | Fort DeSoto | 0.055 | 0.001 | The result for 10 m from the shoreline was significantly different from the results for the other distances (P ≤ 0.003) |
| Crandon Park | 0.029 | <0.001 | The difference between depths of 10 cm and 30 cm was significant (P = 0.013); the results for all distances from the shoreline were significantly different from each other (P ≤ 0.035) | |
| Pacific Ocean | Huntington Beach | 0.763 | <0.001 | The result for 10 m from the shoreline was significantly different from the other distances (P < 0.001) |
| Sandy Beach | 0.577 | <0.001 | The results for all distances from the shoreline were significantly different from each other (P < 0.001) | |
| Otaru Dream Beach | 0.604 | <0.001 | The result for 10 m from the shoreline was significantly different from the results for the other distances (P < 0.001) | |
| Fukiage-hama Beach | 0.354 | <0.001 | The result for 10 m from the shoreline was significantly different from the results for the other distances (P < 0.001) | |
| Jeju Beach | 0.324 | <0.001 | The result for 10 m from the shoreline was significantly different from the results for the other distances (P ≤ 0.001) | |
DISCUSSION
Factors shaping the bacterial community structure in intertidal beach sands have only recently begun to be examined (3, 17, 32–35), and these studies have been limited to either local or regionally scaled study areas and primarily marine beaches. Data presented here provide novel insights into the bacterial community composition of freshwater beaches determined using next-generation sequencing. Furthermore, due to the consistency in the methodology used, OTU-level statistical comparisons among geographically isolated beaches in the Northern Hemisphere were possible. As originally hypothesized, each beach harbored a community with a unique composition, even among those beaches in the same region that were closer in proximity relative to the rest of the beaches in the data set. These differences are likely due to variations in factors, such as wave action, sand grain size, and nutrient content, which were previously suggested to be associated with community homogeneity (3). Furthermore, the local variation at each beach showed similar patterns among the beaches, with a significant variation in community structure between backshore and nearshore locations being detected, as reported elsewhere (32, 33).
One of the key findings of this study was the observation of regional similarity among beaches, as was observed, for example, in the close clustering and taxonomic similarity between the microbiota at Huntington and Jeju Beaches, even though they are separated by a linear distance of nearly 10,000 km. Previously, a distance-taxon relationship among intertidal sands sampled from sites at distances over 1,350 km apart was observed and was related to sand grain size, wave climate, and nutrient content (3). Thus, the regional similarity of these parameters may explain the similar distributions of bacteria. Importantly, in this study beaches were broadly classified into regions primarily on the basis of salinity, as other metadata were not collected, but more specific groupings on the basis of physicochemical parameters may reveal more robust trends shaping bacterial communities. Unfortunately, only moisture content data and no corresponding data regarding nutrient concentrations, wave climate, or sand grain size were collected as metadata here. Significant differences in moisture content suggest corresponding variations in physicochemical parameters. These differences likely affect species distributions at a finer taxonomic resolution, thus resulting in the observed differences in beta diversity.
The taxonomic distribution observed for marine samples in this study is similar to that reported elsewhere, where the predominant phyla and classes were the Gammaproteobacteria, Alphaproteobacteria, and Bacteroidetes (34). Family-level classifications revealed greater diversity among marine beaches. The abundant, cosmopolitan community identified at California beaches was comprised of Alteromonadaceae, Bacillaceae, Flavobacteriaceae, Halomonadaceae, Planococcaceae, Pseudoalteromonadaceae, and Rhodobacteraceae (3), of which only Flavobacteriaceae and Rhodobacteraceae were identified among the abundant families found in the present study. Members of the Flavobacteriaceae, Planctomycetaceae, Saprospiraceae, and Sinobacteraceae were abundant in intertidal sands in Florida (35), and, in addition to Flavobacteriaceae, the Planctomycetaceae and Saprospiraceae were observed among the abundant families found in the present study. Similarly, Paracoccus, within the Rhodobacteraceae, was among the most abundant genera identified in sands in Hawaii (32). Notably, these families, some of which are typical marine taxa (51), were found at a significantly greater abundance in marine sands than in freshwater sands.
Several of the abundant taxa in freshwater beaches, such as the Alphaproteobacteria, including Sphingomonadaceae, Actinobacteria, and Betaproteobacteria, were previously found to be abundant in sands of Lake Michigan (17), and these lineages have been reported to be abundant and ubiquitous in freshwater (52). Therefore, it is likely that interactions between waterborne and sand communities are major drivers of low-resolution taxonomic diversity, especially among abundant taxa. Furthermore, the less frequent and less intensive water-sand interactions with distance from the shoreline likely explain why water-associated taxa show decreases in abundance in backshore sands. Subsequent variations in sand communities are then likely to occur in response to physicochemical parameters, especially among less abundant taxa, as has been previously suggested (2, 3, 17, 31).
In addition to environmental drivers of diversity and differentiation among the communities sampled, there are several methodological explanations that must be considered. As samples were collected at each beach at only a single time point, temporal variation is unlikely to be apparent in the current data set, but time has previously been reported to account for 34% of the variation in bacterial communities in subtidal sands (31). Differences in sampling dates were likely to be masked in the current work by other meteorological parameters, such as prior rain events, which are more likely to disturb bacterial communities. Furthermore, the inclusion of 10 labs in the sampling strategy used here almost certainly resulted in technical variability in the interpretation and implementation of the sampling strategy. For example, differences in moisture content at the shoreline may reflect differences among labs regarding the degree of water saturation allowed in samples, which would also have resulted in differences in the communities characterized. This discrepancy may explain why differences due to depth, when observed, showed no trends in the relative abundances of taxa. Moreover, comparisons to previous studies, as described above, may show imperfect relationships due to biases introduced by the sequencing method used and the primers selected (36).
The local variations in community structure between backshore and intertidal samples found in the current study are similar to those described in a previous report (32), but the differences in diversity observed in the current study were opposite those observed previously. Here we observed lower Shannon indices for backshore samples, associated with a decline in moisture content. Another study reported no significant difference in the Shannon index between wet and dry sand (33). These discrepancies may be explained by characteristics in the water column, as both prior studies were evaluating bacterial communities in relation to fecal indicator bacteria and impaired water quality status, and impairment has been associated with a decrease in bacterial diversity (33). Interestingly, in freshwater beaches that were not subject to strong tidal influences, the community composition in samples obtained at each distance from the shoreline tended to be unique, while the community compositions in samples obtained at the shoreline and 1 m from the shoreline were generally not distinct among marine beaches. This is similar to the findings of a previous study of the communities in freshwater sand, which saw a differentiation of the community structures between backshore and berm sand samples (17). These results may indicate that tide and wave actions serve to homogenize the bacterial communities of intertidal samples, while the more sporadic wetting of foreshore samples by wave action within the Great Lakes allows differentiation of these communities. These findings may be of particular importance to public health and, depending on the degree of tidal and/or wave activity, may affect the time required for beach sands to recover from the presence of pathogens or to achieve a decline in the level of risk from pathogens after a recent contamination event.
While sampling depth was previously shown to influence bacterial diversity and the community structure in subtidal sands (31), the differences in diversity and community composition found here did not vary. It is possible that in this study differences were not observed due to sampling during an outgoing tide, where intertidal sands would have been inundated and homogenized to the relatively shallow depth (30 cm) sampled. The finding that depth was a significant factor at Crandon Park is unusual, especially since differences were observed between 10- and 20-cm depths but not 10- and 30-cm depths. However, the differences in community structures by depth in the Fort DeSoto sands also approached statistical significance, suggesting that differences in community structure by depth may warrant further study, especially in Florida.
The results of this study expand our current understanding of bacterial communities in freshwater beach sands throughout the Great Lakes, as well as marine beach sands throughout the United States, Japan, and South Korea. This study is the first to assess geographic variability in beach sand bacterial communities on a global scale within the Northern Hemisphere. One of the key findings of this study is the high degree of regional taxonomic and, in some cases, phylogenetic similarity. This may be driven in part by interactions between the sand and water communities, since persistent core microbiomes for both freshwater and marine environments have been previously suggested (53, 54). We further show the presence of highly similar local community dynamics within the same beach, where moisture, most likely resulting from tidal cycles, is a major driver of the bacterial communities present. This study represents an important initial effort to characterize the bacterial communities in global beach sands and provides a fundamental basis for future efforts to determine factors affecting regional similarities in sand bacterial communities.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joshua Thiede for assistance with sample collection and DNA extraction. The resources of the Minnesota Supercomputing Institute were used for sequence processing and analysis. We also are grateful to the following individuals and laboratories for collection of local beach sand samples: Zachery Staley and Thomas Edge, Canada Centre for Inland Waters (Burlington, ON, Canada), for samples from Burlington Beach and Marie Curtis Park; Muruleedhara Byappanahalli and the Lake Michigan Ecological Research Station, United States Geological Survey lab (Porter, IN), for samples from the 63rd Street Beach; Valerie J. Harwood (University of South Florida, Tampa, FL) for samples from Fort DeSoto; Helena Solo-Gabriele (University of Miami, Miami, FL) for samples from Crandon Park; John Griffith and the Southern California Coastal Water Research Program lab (Costa Mesa, CA) for samples from Huntington Beach; Tao Yan (University of Hawaii, Honolulu, HI) for samples from Sandy Beach; Satoshi Ishii (Hokkaido University, Sapporo, Japan) for samples from Otaru Dream Beach; the Toshiki Uchiumi lab (Kagoshima University, Kagoshima, Japan) for samples from Fukiage-hama Beach; and Tatsuya Unno (Jeju University, Jeju, South Korea) for samples from Jeju Beach.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00247-16.
REFERENCES
- 1.de Beer D, Wenzhöfer F, Ferdelman TG, Boehme SE, Huettel M, van Beusekom JEE, Böttcher ME, Musat N, Dubilier N. 2005. Transport and mineralization rates in North Sea sandy intertidal sediments, Sylt-Rømø Basin, Wadden Sea. Limnol Oceanogr 50:113–127. doi: 10.4319/lo.2005.50.1.0113. [DOI] [Google Scholar]
- 2.Gobet A, Böer SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML, Boetius A, Ramette A. 2012. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6:542–553. doi: 10.1038/ismej.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehm AB, Yamahara KM, Sassoubre LM. 2014. Diversity and transport of microorganisms in intertidal sands of the California coast. Appl Environ Microbiol 80:3943–3951. doi: 10.1128/AEM.00513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitman R, Harwood VJ, Edge TA, Nevers M, Byappanahalli M, Vijayavel K, Brandão J, Sadowsky MJ, Alm EW, Crowe A, Ferguson D, Ge Z, Halliday E, Kinzelman J, Kleinheinz G, Przybyla-Kelly K, Staley C, Staley Z, Solo-Gabriele HM. 2014. Microbes in beach sands: integrating environment, ecology and public health. Rev Environ Sci Biotechnol 13:329–368. doi: 10.1007/s11157-014-9340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abarca E, Karam H, Hemond HF, Harvey CF. 2013. Transient groundwater dynamics in a coastal aquifer: the effects of tides, the lunar cycle, and the beach profile. Water Resour Res 49:2473–2488. doi: 10.1002/wrcr.20075. [DOI] [Google Scholar]
- 6.Moore WS. 1999. The subterranean estuary: a reaction zone of ground water and sea water. Mar Chem 65:111–125. doi: 10.1016/S0304-4203(99)00014-6. [DOI] [Google Scholar]
- 7.Bedford KW. 1992. The physical effects of the Great Lakes on tributaries and wetlands. J Great Lakes Res 18:571–589. doi: 10.1016/S0380-1330(92)71323-9. [DOI] [Google Scholar]
- 8.Crowe AS, Meek GA. 2009. Groundwater conditions beneath beaches of Lake Huron, Ontario, Canada. Aquat Ecosyst Health Manag 12:444–455. doi: 10.1080/14634980903354825. [DOI] [Google Scholar]
- 9.Piggot AM, Klaus JS, Johnson S, Phillips MC, Solo-Gabriele HM. 2012. Relationship between enterococcal levels and sediment biofilms at recreational beaches in South Florida. Appl Environ Microbiol 78:5973–5982. doi: 10.1128/AEM.00603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamahara KM, Layton BA, Santoro AE, Boehm AB. 2007. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environ Sci Technol 41:4515–4521. doi: 10.1021/es062822n. [DOI] [PubMed] [Google Scholar]
- 11.Ge Z, Nevers MB, Schwab DJ, Whitman RL. 2010. Coastal loading and transport of Escherichia coli at an embayed beach in Lake Michigan. Environ Sci Technol 44:6731–6737. doi: 10.1021/es100797r. [DOI] [PubMed] [Google Scholar]
- 12.Russell TL, Yamahara KM, Boehm AB. 2012. Mobilization and transport of naturally occurring enterococci in beach sands subject to transient infiltration of seawater. Environ Sci Technol 46:5988–5996. doi: 10.1021/es300408z. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, He X, Yan T. 2015. Differential decay of wastewater bacteria and change of microbial communities in beach sand and seawater microcosms. Environ Sci Technol 49:8531–8540. doi: 10.1021/acs.est.5b01879. [DOI] [PubMed] [Google Scholar]
- 14.Shah AH, Abdelzaher AM, Phillips M, Hernandez R, Solo-Gabriele HM, Kish J, Scorzetti G, Fell JW, Diaz MR, Scott TM, Lukasik J, Harwood VJ, McQuaig S, Sinigalliano CD, Gidley ML, Wanless D, Ager A, Lui J, Stewart JR, Plano LRW, Fleming LE. 2011. Indicator microbes correlate with pathogenic bacteria, yeasts and helminthes in sand at a subtropical recreational beach site. J Appl Microbiol 110:1571–1583. doi: 10.1111/j.1365-2672.2011.05013.x. [DOI] [PubMed] [Google Scholar]
- 15.Ishii S, Hansen DL, Hicks RE, Sadowsky MJ. 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ Sci Technol 41:2203–2209. doi: 10.1021/es0623156. [DOI] [PubMed] [Google Scholar]
- 16.Yamahara KM, Sassoubre LM, Goodwin KD, Boehm AB. 2012. Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Appl Environ Microbiol 78:1733–1745. doi: 10.1128/AEM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloutier DD, Alm EW, McLellan SL. 2015. Influence of land use, nutrients, and geography on microbial communities and fecal indicator abundance at Lake Michigan beaches. Appl Environ Microbiol 81:4904–4913. doi: 10.1128/AEM.00233-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solo-Gabriele HM, Harwood VJ, Kay D, Fujioka RS, Sadowsky MJ, Whitman RL, Wither A, Caniça M, Carvalho da Fonseca R, Duarte A, Edge TA, Gargaté MJ, Gunde-Cimerman N, Hagen F, McLellan SL, Nogueira da Silva A, Novak Babič M, Prada S, Rodrigues R, Romão D, Sabino R, Samson RA, Segal E, Staley C, Taylor HD, Veríssimo C, Viegas C, Barroso H, Brandão JC. 2016. Beach sand and the potential for infectious disease transmission: observations and recommendations. J Mar Biol Assoc UK 96:101–120. doi: 10.1017/S0025315415000843. [DOI] [Google Scholar]
- 19.United States Environmental Protection Agency. 2012. Recreational water quality criteria. Report 820-F-12-058. United States Environmental Protection Agency, Washington, DC. [Google Scholar]
- 20.Cabelli VJ, Dufour AP, McCabe LJ, Levin MA. 1982. Swimming-associated gastroenteritis and water quality. Am J Epidemiol 115:606–616. [DOI] [PubMed] [Google Scholar]
- 21.Byappanahalli MN, Whitman RL, Shively DA, Ting WT, Tseng CC, Nevers MB. 2006. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J Water Health 4:313–320. [DOI] [PubMed] [Google Scholar]
- 22.Halliday E, Gast RJ. 2011. Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environ Sci Technol 45:370–379. doi: 10.1021/es102747s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staley C, Dunny GM, Sadowsky MJ. 2014. Environmental and animal-associated enterococci. Adv Appl Microbiol 87:147–186. doi: 10.1016/B978-0-12-800261-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 24.Ran QH, Badgley BD, Dillon N, Dunny GM, Sadowsky MJ. 2013. Occurrence, genetic diversity, and persistence of enterococci in a Lake Superior watershed. Appl Environ Microbiol 79:3067–3075. doi: 10.1128/AEM.03908-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2015. Species sorting and seasonal dynamics primarily shape bacterial communities in the Upper Mississippi River. Sci Total Environ 505:435–445. doi: 10.1016/j.scitotenv.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed W, Staley C, Sadowsky MJ, Gyawali P, Sidhu JPS, Palmer A, Beale DJ, Toze S. 2015. Toolbox approaches using molecular markers and 16S rRNA gene amplicon data sets for identification of fecal pollution in surface water. Appl Environ Microbiol 81:7067–7077. doi: 10.1128/AEM.02032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Human Microbiome Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaidos E, Rusch A, Ilardo M. 2011. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol 13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- 31.Böer SI, Hedtkamp SIC, van Beusekom JEE, Fuhrman JA, Boetius A, Ramette A. 2009. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J 3:780–791. doi: 10.1038/ismej.2009.29. [DOI] [PubMed] [Google Scholar]
- 32.Cui H, Yang K, Pagaling E, Yan T. 2013. Spatial and temporal variation in enterococcal abundance and its relationship to the microbial community in Hawaii beach sand and water. Appl Environ Microbiol 79:3601–3609. doi: 10.1128/AEM.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halliday E, McLellan SL, Amaral-Zettler LA, Sogin ML, Gast RJ. 2014. Comparison of bacterial communities in sands and water at beaches with bacterial water quality violations. PLoS One 9:e90815. doi: 10.1371/journal.pone.0090815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton RJ, Huse SM, Morrison HG, Peake CS, Sogin ML, McLellan SL. 2013. Shifts in the microbial community composition of Gulf Coast beaches following beach oiling. PLoS One 8:e74265. doi: 10.1371/journal.pone.0074265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. 2009. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75:5227–5236. doi: 10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2015. Evaluation of water sampling methodologies for amplicon-based characterization of bacterial community structure. J Microbiol Methods 114:43–50. doi: 10.1016/j.mimet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Claesson MJ, Wang QO, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinformatics J 7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 41.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- 45.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 47.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 48.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acar EF, Sun L. 2013. A generalized Kruskal-Wallis test incorporating group uncertainty with application to genetic association studies. Biometrics 69:427–435. doi: 10.1111/biom.12006. [DOI] [PubMed] [Google Scholar]
- 50.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert JA, Steele JA, Caporaso JG, Steinbrueck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han SK. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155. doi: 10.3354/ame028141. [DOI] [Google Scholar]
- 53.Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. 2013. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci U S A 110:4651–4655. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ. 2013. Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115:1147–1158. doi: 10.1111/jam.12323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.