Abstract
This study was performed to determine the effects of feeding a fiber-rich fraction of Brassica vegetables on the immune response through changes in enteric bacteria and short-chain fatty acid (SCFA) production in normal mice. The boiled-water-insoluble fraction of Brassica rapa L. (nozawana), which consists mainly of dietary fiber, was chosen as a test material. A total of 31 male C57BL/6J mice were divided into two groups and housed in a specific-pathogen-free facility. The animals were fed either a control diet or the control diet plus the insoluble B. rapa L. fraction for 2 weeks and sacrificed to determine microbiological and SCFA profiles in lower-gut samples and immunological molecules. rRNA-based quantification indicated that the relative population of Bacteroidetes was markedly lower in the colon samples of the insoluble B. rapa L. fraction-fed group than that in the controls. Populations of the Eubacterium rectale group and Faecalibacterium prausnitzii, both of which are representative butyrate-producing bacteria, doubled after 2 weeks of fraction intake, accompanying a marginal increase in the proportion of colonic butyrate. In addition, feeding with the fraction significantly increased levels of the anti-inflammatory cytokine interleukin-10 (IL-10) and tended to increase splenic regulatory T cell numbers but significantly reduced the population of cells expressing activation markers. We demonstrated that inclusion of the boiled-water-insoluble fraction of B. rapa L. can alter the composition of the gut microbiota to decrease the numbers of Bacteroidetes and to increase the numbers of butyrate-producing bacteria, either of which may be involved in the observed shift in the production of splenic IL-10.
INTRODUCTION
Animals coexist with microbial symbionts that act as an integral component of the host's physiology in the gastrointestinal (GI) tract (1). The vast majority of GI bacteria are strict anaerobes that derive energy from fermentation, by which indigestible complex carbohydrates (cellulose, pectin, gums, beta-glucan, and lignin) are converted to short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate, as terminal electron acceptors (2–4). As each member of the GI microbiota has its own preferences for different energy sources (5, 6), the profile of dominant species in the human gut microbiota can potentially be modified by the types of dietary carbohydrate in the diet.
Among the SCFAs, butyrate acts as a primarily effective molecule on physiological regulatory systems of the host gut and as an energy source for the colonic epithelium of the host. There has long been interest in the immunomodulatory and anti-inflammatory effects of butyrate on colonic epithelial cells (7–9). The production of butyrate in the GI tract is supported by specific groups of bacteria, i.e., butyrate producers, which are considered to play important roles in the maintenance of gut health (10, 11). The main human colonic butyrate producers belong to two groups of Gram-positive firmicutes: Faecalibacterium prausnitzii in the Clostridium leptum (or clostridial cluster IV) cluster, and Eubacterium rectale/Roseburia spp. in the Blautia coccoides (formerly Clostridium coccoides, clostridial cluster XIVa) (12–15), whereas there are other butyrate-producing groups that have been detected in humans (16–18).
Vegetables belonging to the Brassica genus (Brassicaceae family) contain a number of nutrients with health-promoting properties, such as anticancer actions (19) and effects on cholesterol metabolism (20). These vegetables (Brassica oleracea L. [kale, cabbage, and broccoli], and Brassica rapa L. [turnip]) are unique in that they are rich sources of fiber, carotenoids, and foliate, as well as polyphenols and sulfur-containing compounds (21). Vegetable dietary fiber is suggested to act as an effective prebiotic by inducing major shifts in gut microbial composition and by affecting the mucosal immune system (22–24). However, experimental studies have yielded inconclusive results to explain how the effective components in Brassica vegetables affect the immune response through bacterial fermentation, especially whether they selectively increase butyrate production. Brassica rapa L. (nozawana), which is a traditionally and regionally planted vegetable in Japan, contains high levels of fiber (33% on a dry-matter basis) (25). We chose this vegetable as an example of a supplement and examined the effects of the insoluble (i.e., fiber) fraction on immunological molecules in response to the intestinal community structure in mice.
MATERIALS AND METHODS
Preparation of B. rapa L.
Fresh B. rapa L. was soaked in pure water and autoclaved at 121°C for 20 min. The autoclaved samples were then homogenized (AM-3, Ace homogenizer; Nissei Co. Ltd., Tokyo, Japan), and the resultant suspensions were centrifuged at 2,215 × g for 10 min to remove large residues. In order to collect a functional fraction of B rapa L. extract, the supernatants were also centrifuged at 20,630 × g for 5 min, and the pellets were lyophilized in a freeze-dryer (FDU-1200; Eyela, Tokyo, Japan) and used as the extracts. One-gram samples of fresh B. rapa L. yielded ca. 0.3 mg of residual extract, which was considered to be composed mainly of highly insoluble fiber (25, 26).
Animals and tissue sampling.
Mice were cared for according to the Guide for the Care and Use of Experimental Animals of Shinshu University (27). Due to limitations in the capacity of the facility and animal management, the data presented in this paper were obtained over the course of three feeding trials with exactly the same design (10 animals for trial 1 and 12 animals for trials 2 and 3). Five-week-old female C57BL/6 mice were housed in a specific-pathogen-free facility. The standard diet (67% carbohydrate, 19% protein, 4% fat, and 4% ash) consisted mainly of casein (190 g/kg of diet), corn starch (300 g/kg), sucrose (330 g/kg), cellulose (47 g/kg), soybean oil (22 g/kg), lard (18 g/kg), vitamins, and minerals. After a 1-week acclimatization period on the control diet, the animals were split into two groups. The two groups were fed the same standard diet ad libitum, and one group received once-daily oral administration of the extract of B. rapa L. (resuspended to 2 mg/ml of water and 20 mg/kg of body weight [BW]/day for the B. rapa L. group), while the other group received water in the same manner (control group). The quantity and the periods of administration of the extract were determined according to our preliminary study (unpublished data). Feed and water were supplied ad libitum. Body weight was measured once daily in this feeding period. Tube feeding lasted 2 weeks, and three animals were excluded from the study because of irregular body weight decreases (one was in the control group during trial 1, and of the other two, one was in the control group and the other in the B. rapa L. group during trial 2). Therefore, 16 animals in the B. rapa L. group and 15 animals in the control group completed the feeding trials. Mice were sacrificed by cervical dislocation, and tissues, including the colon, cecum, spleen, mesenteric lymph nodes (mLNs), and Peyer's patches (PPs), were collected and weighed. After being measured, colonic and cecal contents (∼0.10 g) were subsampled in 1 ml of phosphate-buffered saline (PBS) and mixed thoroughly to equalize the distribution in the buffer. The sampling position of colonic contents from each mouse was unified to the middle part of colon. Thereafter, other organs (small intestine, liver, heart, and stomach) were separated and weighed.
Microbial analyses.
Total RNAs were extracted from the prokaryotic cells in the suspensions using an RNeasy Plus minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Solutions of the extracted RNA were stored at −80°C until use. An RNA-based, sequence-specific rRNA cleavage method was applied to monitor active bacterial populations in the intestinal samples (28). For the detection and quantification of the respective bacterial groups, the following probes were used: Bac303m (Bacteroides and Prevotella); Erec482m (B. coccoides-Eubacterium rectale group); Rfla1269 (Ruminococcus flavefaciens), Rbro730m (Ruminococcus bromii), and Fprau645 (F. prausnitzii); and Lab158m (Lactobacillus-Enterococcus group). These probes were applied separately under the same reaction conditions described in previous studies (29, 30). We employed two additional probes, Rrec584 (E. rectale) (31) and Clept866 (C. leptum subgroup) (32). Probe validation was conducted according to the methods described in our previous report (33), using Roseburia faecis JCM 17581 and Ruminococcus albus JCM 14654T as reference strains for plotting standard digestion curves. By doing so, we determined the reaction conditions (formamide percentage and digestion coefficient) to be 10% and 0.90 for Rrec584 and 5% and 0.86 for Clept866, respectively. Bacterial genomic DNAs were extracted from the prokaryotic cells of the suspensions using a QIAamp DNA stool minikit (Qiagen) according to the manufacturer's instructions. Solutions of the extracted DNA were stored at −80°C until use. Quantitative real-time PCR analysis for total bacterial DNA was performed as described previously (34). Real-time PCR primers and conditions for amplification of the butyryl-coenzyme A (CoA):acetate CoA-transferase gene were published previously (35, 36).
Organic acid measurements.
Cooled cecal and colonic content samples (∼0.05 g) were weighed and dispersed in 1 ml of sterilized water. Suspensions were centrifuged at 10,000 × g at 4°C for 5 min. The supernatants were used to analyze the organic acids with a high-performance liquid chromatography (HPLC) system equipped with an electroconductivity detector (LC-20 model; Shimadzu Corp., Kyoto, Japan) as described previously (28).
Isolation and culture of cells from spleen, mLNs, and PPs.
Spleen, mLNs, and PPs were removed from each mouse in both groups, and single-cell suspensions were passed through 40-μm-pore-size cell strainers (BD Falcon, Franklin Lakes, NJ). To deplete red blood cells, spleen cells were treated with 0.17 M Tris-HCl buffer (pH 7.65) containing 0.83% NH4Cl. For culture of the spleen cells, the cells were resuspended at a concentration of 1 × 107 cells/ml in RPMI 1640 medium containing 10% fetal bovine serum (FBS) plus 10,000 U/ml penicillin G and 10 mg/ml streptomycin. The cells were cultured in 96-well flat-bottomed plates in the presence or absence of 0.1 or 1 μg/ml lipopolysaccharide (LPS) from Escherichia coli 055:B5 (Sigma, St. Louis, MO) for 48 h at 37°C under 5% CO2.
Flow cytometry.
For analysis of cell surface molecules, we used fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (GK1.5), phycoerythrin (PE)-conjugated anti-CD8 (53-6.7), PE-conjugated CD11b (M1/70), allophycocyanin (APC)-conjugated CD69 (H1.2F3), APC-conjugated CD11c (HL3), FITC-conjugated anti-H-2Kb (AF6-88.5), PE-conjugated anti-I-Ab (AF6-120.1), and 7-amino-actinomycin D (7-AAD). These antibodies were purchased from BioLegend (San Diego, CA). Cells from spleens, mLNs, and PPs were stained with fluorescently labeled monoclonal antibodies (MAbs) and 7-AAD. The expression levels were evaluated by flow cytometry (FACSCalibur; Becton Dickinson, San Jose, CA).
Regulatory T (Treg) cell staining.
After being stained with FITC-conjugated anti-CD4 and APC-conjugated anti-CD25 (clone PC61; BioLegend) MAbs, cells were fixed and permeabilized using the FlowX FoxP3 fixation and permeabilization buffer kit (R&D Systems, Minneapolis, MN). Permeabilized cells were stained with PE-conjugated anti-mouse Foxp3 MAb (clone 150D; BioLegend). Stained cells were subsequently analyzed by flow cytometry (FACSCalibur; Becton Dickinson).
ELISA.
The levels of interleukin-10 (IL-10) production in culture supernatants of the spleen cells from each mouse individually were measured using an enzyme-linked immunosorbent assay (ELISA) kit (eBioscience, San Diego, CA), according to the manufacturer's instructions. We used 3-fold-diluted supernatants to detect IL-10 levels by ELISA.
Statistical analyses.
Measurements were analyzed by the unpaired Student t test with Stata 13.1 (Stata Corp., College Station, TX). In all analyses, a P value of <0.05 was taken to indicate statistical significance.
RESULTS
Body and organ weights.
The body weights and organ weights of mice are shown in Table 1. Body weights showed no differences between the two groups at any time point during the feeding period, so mice in each treatment group were considered to grow normally. There were also no differences in cecum weights between the groups. Colon weight was higher in the B. rapa L. group than in the controls, suggesting that the undigested B. rapa L. fraction reached the colon and probably became a substrate for colonic microbial fermentation. No difference was observed with respect to the weights of other organs (small intestine, liver, heart, and stomach) (data not shown).
TABLE 1.
Body weights and organ weights of cecum and colon in control mice orally administered water and mice administered B. rapa L. extracta
| Mouse group | Body wt (g) | Cecum wt (mg) | Colon wt (mg) |
|---|---|---|---|
| Control | 20.5 ± 0.9 | 537.9 ± 131.1 | 218.6 ± 39.6 |
| B. rapa L. | 20.2 ± 0.9 | 492.5 ± 86.4 | 251.5 ± 47.0b |
Data are expressed as means ± standard deviations (SD).
Significant difference between the control and B. rapa L. groups (P < 0.05).
Analysis of colonic samples.
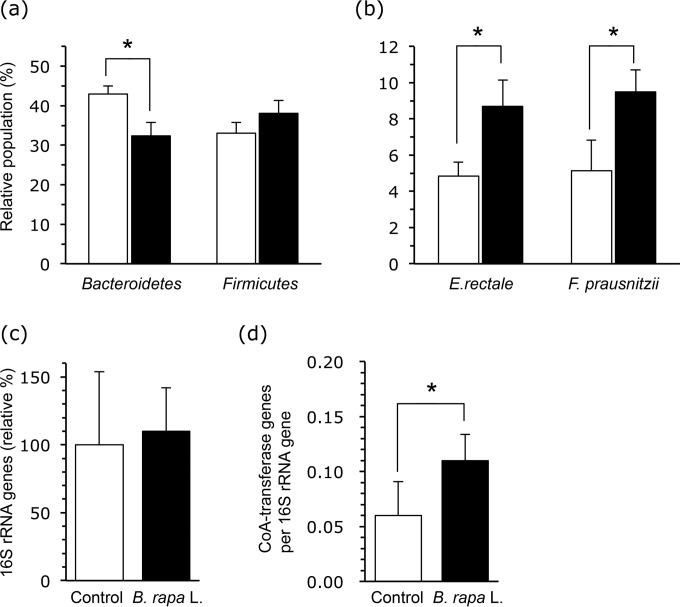
Bacterial profiles in the colonic contents of the mice are shown in Fig. 1. rRNAs of the phylum Bacteroidetes, particularly Bacteroides and Prevotella spp. (determined with the Bac303m probe), and the phylum Firmicutes, particularly the B. coccoides-E. rectale group and C. leptum subgroup (sum of values of Erec482, Clept866, and Lact158), constituted the major fraction of the bacterial community (approximately 75% of the total 16S rRNA). At the phylum level, the Bacteroidetes content was lower, while the Firmicutes content tended to be higher, in B. rapa L.-fed mice than in controls (Fig. 1a). In the lower group level, the relative populations of F. prausnitzii (determined with Fprau645) and E. rectale (determined with Rrec584) were higher in B. rapa L.-fed mice (Fig. 1b) than in the control. The relative populations of R. flavefaciens and R. bromii were 1.4% ± 0.8% and 2.1% ± 0.8%, respectively, and there were no differences between groups (data not shown). The Lactobacillus-Enterococcus group was shown to constitute approximately 1% of the total rRNA of colon samples, and there were no differences between the two groups. We also determined total bacterial numbers and the butyryl-CoA:acetate CoA-transferase gene copy numbers in colonic contents by quantitative PCR (Fig. 1c and d). While total bacterial numbers were not significantly different between treatments, the butyryl-CoA:acetate CoA-transferase gene copy numbers were higher in the B. rapa L. group than in the controls.
FIG 1.
Effects of oral administration of the extract of B. rapa L. on the colonic bacterial community and the level of a gene involved in butyrate generation. (a) Relative bacterial populations at the phylum level (among the Bacteroidetes, Bacteroides and Prevotella; and among the Firmicutes, the B. coccoides-E. rectale group and C. leptum subgroup). (b) Relative bacterial populations in the representative butyrate-producing bacteria (E. rectale and F. prausnitzii). The data shown in panels a and b were obtained using the sequence-specific rRNA cleavage method. (c) Total bacterial 16S rRNA copies in colonic samples. (d) Data are expressed as the relative gene copy numbers per colonic contents, assuming that the average for the control group is 100. The number of butyryl-CoA–CoA transferase genes is relative to the number of 16S rRNA genes. Error bars indicate standard deviations (SD) from the results for all mice used in three independent experiments performed together (control group, n = 15; B. rapa L. group, n = 16). The levels of significance of differences between the control (white bars) and B. rapa L. (black bars) groups were determined by Student's t test (*, P < 0.05).
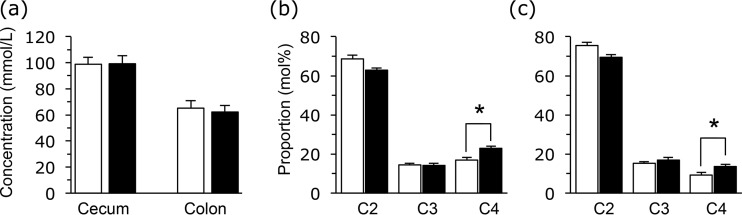
The total organic acids (SCFAs and lactate) of the cecal and colonic contents were not different between the two groups (Fig. 2a). SCFA proportions were also determined, and the butyrate concentrations were higher in the B. rapa L. group than in the controls (Fig. 2b and c). Lactate and valerate were minor constituents (<0.5 mmol/kg of sample) of the samples.
FIG 2.
Effects of oral administration of the extract of B. rapa L. on total SCFA concentrations in cecum and colon samples (a) and relative molar proportions of acetate (C2), propionate (C3), and butyrate (C4) in the cecum sample (b) and colon sample (c). Data are represented in the same manner as described in the legend to Fig. 1.
Immunological analyses.
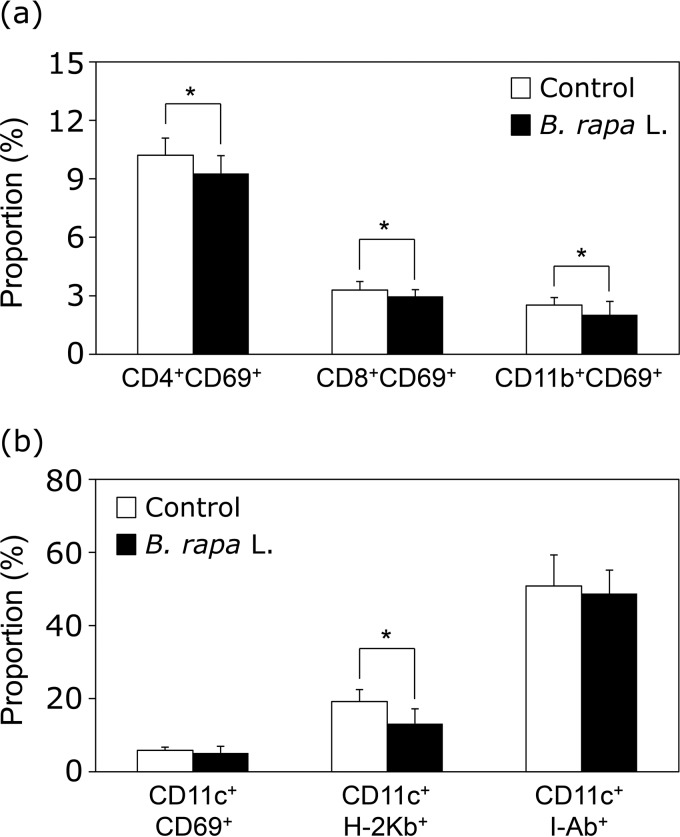
The early activation marker CD69 was examined in spleen cells of mice orally administered the extract of B. rapa L. CD69 expression levels on CD4+, CD8+, or CD11b+ cells in B. rapa L.-administered mice were downregulated compared with expression levels in the same cells in the controls (Fig. 3a). In addition, oral administration of B. rapa L. extract significantly decreased the expression of major histocompatibility complex class I (MHC-I) (H-2Kb) molecules, but not CD69 or MHC-II (I-Ab) molecules, on CD11c+ dendritic cells (Fig. 3b).
FIG 3.
Oral administration of the extract of B. rapa L. downregulates the expression of CD69 in spleen cells. The extract of B. rapa L. (20 mg/kg of BW/day) or water was administered orally to mice for 14 days, and then spleen cells were collected. (a) Spleen cells were stained with anti-CD4, anti-CD8α, anti-CD11c, and anti-CD69 MAbs, and expression levels of the early activation marker CD69 on CD4+ T cells, CD8+ T cells, and CD11b+ cells were evaluated by flow cytometry. (b) Flow cytometry was performed to determine the expression of CD69, H-2Kb, or I-Ab on CD11c+ cells using anti-CD69, anti-H-2Kb, anti-I-Ab, and anti-CD11c MAbs. Data are presented in the same manner as described in the legend to Fig. 1.
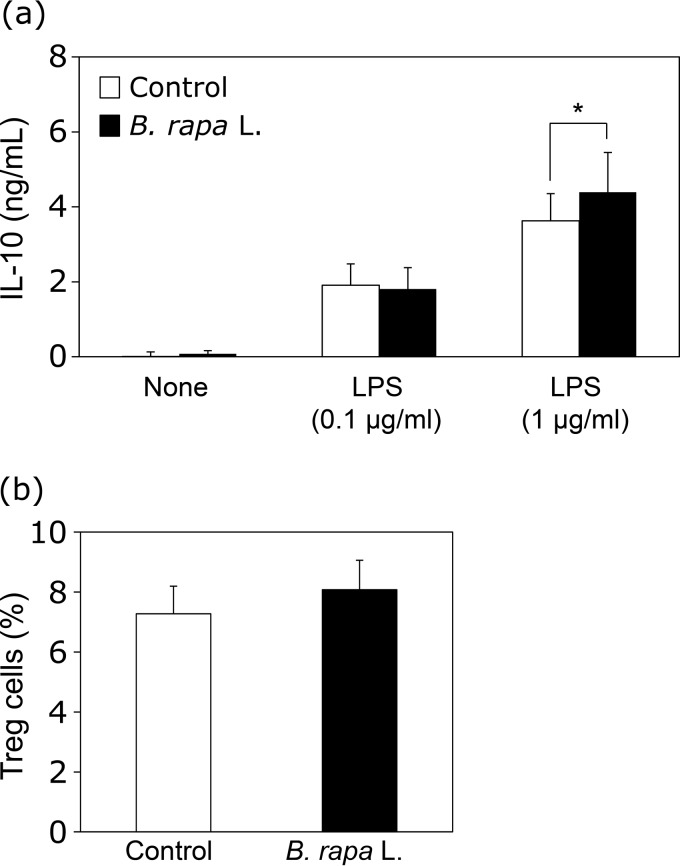
To determine the immunoregulatory effects of B. rapa L., IL-10 production by spleen cells was analyzed. Dietary supplementation with B. rapa L. extract induced IL-10 production by spleen cells stimulated with LPS (Fig. 4a). Furthermore, the proportion of Treg cells tended to increase in mice orally administered B. rapa L. extract (Fig. 4b) (P = 0.06). These findings suggested that oral administration of B. rapa L. extract induces immunoregulatory effects, including decreases in activation markers and increases in IL-10 production and Treg cells.
FIG 4.
Enhancement of IL-10 production and Treg cells in mice orally administered the extract of B. rapa L. Sampling of spleen cells was conducted in the same manner as described in the legend of Fig. 3. (a) Spleen cells (5 × 105 cells/well) from mice were stimulated with LPS (0.1 and 1 μg/ml) for 48 h. IL-10 production levels in the supernatants were measured by ELISA. (b) Spleen cells from mice orally administered the extract of B. rapa L. (20 mg/kg of BW/day) or water were stained with anti-CD4, anti-CD25, and anti-Foxp3 MAbs, and the percentages of Treg cells were evaluated by flow cytometry. Data are presented in the same manner as described in the legend to Fig. 1.
DISCUSSION
Diet is regarded as a major contributing factor directing the bacterial population in the large intestine. Prebiotics are nondigestible food ingredients, such as celluloses, fibers, and other oligosaccharides, such as resistant starch, fructooligosaccharides (FOS), and xylooligosaccharides (XOS), which beneficially affect the host by modulating the intestinal microbiota (4, 37–40). Certain dietary constituents, for example, resistant starch, are known to increase the bacterial production of butyrate in the large intestine, which is generally regarded as having a beneficial effect (41). Controversially, the limited ingestion of fiber in the diet was suggested to be a critical factor in disease onset linked to the gut microbiome (42).
In this experiment, we profiled the changes in active populations of the microbiota instead of determining the 16S rRNA gene amplicons of specific groups. Our results showed that there were significant shifts in the active populations in the colonic microbiota, but this method did not allow us to know the exact metabolisms employed by the active populations. For example, we found two remarkable responses with regard to the composition of the colonic microbiota with the introduction of the insoluble B. rapa L. fraction. First, the Bacteroidetes population was reduced in animals treated with this extract. In our previous study, long-term administration (12 weeks) of a product of kale (Brassica oleracea) to BL6 mice resulted in an elevated proportion of Firmicutes and a reduced population of Bacteroidetes. As a consequence, the Bacteroidetes/Firmicutes ratio in colon samples was lower in the kale ingestion group than in the controls (28). Thus, the administration of Brassica vegetables may yield a common tendency to decrease the population of Bacteroidetes, although the reasons remain unknown. Another response was a 2-fold increase in the number of strains of E. rectale and Faecalibacterium, both of which are representative butyrate-producing bacteria, after 2 weeks of insoluble B. rapa L. fraction intake, and this was probably accompanied by an increase in the molar proportion of butyrate. Although the total colonic SCFA concentration remained unchanged and the increase in the molar proportion of butyrate in the B. rapa L. group was numerically small, total colonic contents increased (Table 1); afterwards, an increase in colonic butyrate production was expected. In other reports, 5% to 10% of the total microbiota species were shown to be related to E. rectale and Roseburia, and 5% to 15% were F. prausnitzii (13, 43, 44), in accordance with our results. Butyrate-producing strains exhibit different growth profiles on various substrates, which include starch, inulin, FOS, and XOS (10, 45, 46). In this study, we could not determine which plant-derived components were effective as substrates for these bacteria. However, it is possible that butyrate-producing bacteria are particularly dependent on the dietary fiber in the fraction to maintain their populations in the colon. The nature and processing of the fiber must be determined to provide sufficient production of butyrate. We also quantified the butyryl-CoA:acetate CoA-transferase gene copy numbers using degenerate primers that recognize multiple phylogenetic groups as another benchmark for determining butyrate-producing bacteria (14, 35, 41). The results of this study suggested the increase in the copy numbers of the gene in the B. rapa L. group to be in good accordance with an increase in the number of butyrate-producing organisms, although we did not measure the expression levels of the butyryl-CoA:acetate CoA-transferase gene. It is regarded that the butyrate-CoA–to–butyrate pathway using extracellular acetate is involved in major intracellular reductive pathways in the gut (47); specifically, there may be a metabolic relay from fiber-fermenting bacteria that produce acetate to stimulate butyrate producers and acetate-utilizing strains (10, 22, 48).
Butyrate is suggested to influence various aspects of gut physiology beyond simply acting as a crude caloric source (49, 50). These effects result in anticancer activity and can also be useful in the treatment of some chronic digestive diseases (51). The contributions of the gut microbiota to the development of the immune system have been extensively characterized (24, 52–56). The microbiota drives the immune system, which allows the host to tolerate the large amount of antigens present in the gut (immunological tolerance) by Treg cells (57, 58). Treg cells contribute to the homeostasis of the immune system by suppressing the immune responses of other cells via IL-10 (59). Some commensal bacteria, including fiber-fermenting species, appear to preferentially drive T-regulatory lymphocyte development (60–63). In addition, butyrate decreases intestinal expression levels of tumor necrosis factor alpha (TNF-α), IL-1β, and IL-6 in patients with Crohn's disease (12, 64, 65). In this context, these microbes may affect host immune function by producing SCFAs. In the present study, the B. rapa L. fraction significantly increased production of the anti-inflammatory cytokine IL-10, and it significantly reduced the induction of activation markers (i.e., CD69 on CD4+, CD8+, and CD11b+ cells). Our results were generally in accordance with previous findings and support the role of the microbiota in the development of the mucosal adaptive immune system. Additionally, LPS, which is released from dead cells of Gram-negative bacteria, such as Bacteroidetes, has been suggested to be correlated with in vivo IL-10 production (66, 67). In relation to this, in another in vitro experiment, we found that the induction of IL-10 production from spleen cells was stimulated with the B. rapa L. extract (unpublished data), suggesting that B. rapa L. may inherently possess components that directly affect a systematic immune response unrelated to enhanced butyrate production. The reason why responses in immune molecules were not as prominent as those of the colonic bacterial community may have been partially due to the use of healthy mice, which did not require strong immunoregulation responses.
Oral administration of B. rapa L. extract significantly decreased the expression of MHC-I molecules, but not CD69 or MHC-II molecules, on dendritic cells. MHC-I molecules are generally used in the presentation of endogenous antigens to CD8+ T cells. In some cases, however, exogenous antigen can enter the MHC-I presentation pathway of dendritic cells (cross-presentation) (68). In addition, IL-10 inhibits MHC-I molecule expression on dendritic cells and converts the immature type into the tolerogenic type (69). In this study, the expression levels of CD69 and MHC-II molecules on dendritic cells were not changed by the oral administration of B. rapa L., but MHC-I molecule expression decreased. Also, IL-10 production from spleen cells stimulated with LPS was higher in mice orally administered the B. rapa L. extract than in controls. Therefore, dietary B. rapa L. might induce the functional or population changes in dendritic cells by the enhancement of IL-10 production.
Taken together, our findings indicate synchronized relationships between changes in the GI bacterial community structure in normal mice fed the insoluble B. rapa L. fraction and the colonic induction of the splenic expression of IL-10. These changes probably occurred concurrently, due to the induction of Treg cells. This study also implies that the increased generation of butyrate derived from a food component rich in dietary fiber may have a suppressive effect on gut immune functions mediated by changes in the microbiota. SCFAs have been reported to act through cell surface signaling receptors, such as G protein-coupled receptors (GPRs), to achieve some of their functions, including immunological responses (7, 70, 71). Determination of the responses of GPRs will be necessary to evaluate the increased butyrate production by B. rapa L. administration acting jointly with the immune responses. Further studies are also required to determine which components in the insoluble fraction of B. rapa L. affect the butyrate producers and other members of the microbiota.
ACKNOWLEDGMENTS
This work was financially supported by Integration Research for Agriculture and Interdisciplinary Fields from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
We thank S. Yonekura and H. Fujii for their suggestions regarding this work and T. Shimosato for his critical comments during the preparation of the manuscript.
Funding Statement
This study was supported by the NARO Bio-oriented Technology Research Advancement Institution (BRAIN): Integration research for agriculture and interdisciplinary fields (to S.T. and Y.U.)
REFERENCES
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrin P, Pierre F, Patry Y, Champ M, Berreur M, Pradal G, Bornet F, Meflah K, Menanteau J. 2001. Only fibres promoting a stable butyrate producing colonic ecosystem decrease the rate of aberrant crypt foci in rats. Gut 48:53–61. doi: 10.1136/gut.48.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Topping DL, Clifton PM. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81:1031–1064. [DOI] [PubMed] [Google Scholar]
- 5.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guamer F, Pederson O, de Vos WM, Brunak S, Dore J, MetaHIT Consortium, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. . 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. 2010. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev 23:366–384. doi: 10.1017/S0954422410000247. [DOI] [PubMed] [Google Scholar]
- 8.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Säemann MD, Böhmig GA, Österreicher CH, Burtscher H, Parolini O, Diakos C, Stöckl J, Hörl WH, Zlabinger GJ. 2000. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J 14:2380–2382. [DOI] [PubMed] [Google Scholar]
- 10.Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 12.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benus RFJ, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJM, Whelan K. 2010. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr 104:693–700. doi: 10.1017/S0007114510001030. [DOI] [PubMed] [Google Scholar]
- 14.Hippe B, Zwielehner J, Liszt K, Lassl C, Unger F, Haslberger AG. 2011. Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age. FEMS Microbiol Lett 316:130–135. doi: 10.1111/j.1574-6968.2010.02197.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Finegold SM, Song Y, Lawson PA. 2008. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58:1896–1902. [DOI] [PubMed] [Google Scholar]
- 16.Vital M, Penton CR, Wang Q, Young VB, Antonopoulos DA, Sogin ML, Morrison HG, Raffals L, Chang EB, Huffnagle GB. 2013. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome 1:8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vital M, Gao J, Rizzo M, Harrison T, Tiedje JM. 2014. Diet is a major factor governing the fecal butyrate-producing community structure across Mammalia, Aves and Reptilia. ISME J 9:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. mBio 5(2):e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manchali S, Chidambara Murthy KN, Patil BS. 2012. Crucial facts about health benefits of popular cruciferous vegetables. J Funct Foods 4:94–106. doi: 10.1016/j.jff.2011.08.004. [DOI] [Google Scholar]
- 20.Aiso I, Inoue H, Seiyama Y, Kuwano T. 2014. Compared with the intake of commercial vegetable juice, the intake of fresh fruit and komatsuna (Brassica rapa L. var. perviridis) juice mixture reduces serum cholesterol in middle-aged men: a randomized controlled pilot study. Lipids Health Dis 13:1–8. doi: 10.1186/1476-511X-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higdon JV, Delage B, Williams DE, Dashwood RH. 2007. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res 55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 23.Hooper LV, Macpherson AJ. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 24.Vieira AT, Teixeira MM, Martins FS. 2013. The role of probiotics and prebiotics in inducing gut immunity. Front Immunol 4:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ministry of Education, Culture, Sports, Science and Technology-Japan. 2005. Nozawana (leaves, raw; item no. 06229), standard tables of food composition in Japan, 5th ed Ministry of Education, Culture, Sports, Science and Technology—Japan, Tokyo, Japan. [Google Scholar]
- 26.Chang MC, Morris WC. 1990. Effect of heat treatments on chemical analysis of dietary fiber. J Food Sci 55:1647–1650. doi: 10.1111/j.1365-2621.1990.tb03591.x. [DOI] [Google Scholar]
- 27.Animal Care and Use Committee, Shinshu University. 2014. Guide for the care and use of experimental animals. Shinshu University, Nagano, Japan. [Google Scholar]
- 28.Uyeno Y, Katayama S, Nakamura S. 2014. Changes in mouse gastrointestinal microbial ecology with ingestion of kale. Benef Microbes 5:345–349. doi: 10.3920/BM2013.0073. [DOI] [PubMed] [Google Scholar]
- 29.Uyeno Y, Sekiguchi Y, Tajima K, Takenaka A, Kurihara M, Kamagata Y. 2010. An rRNA-based analysis for evaluating the effect of heat stress on the rumen microbial composition of Holstein heifers. Anaerobe 16:27–33. doi: 10.1016/j.anaerobe.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Uyeno Y, Kawashima K, Hasunuma T, Wakimoto W, Noda M, Nagashima S, Akiyama K, Tabata M, Kushibiki S. 2013. Effects of cellooligosaccharide or a combination of cellooligosaccharide and live Clostridium butyricum culture on performance and intestinal ecology in Holstein calves fed milk or milk replacer. Livest Sci 153:88–93. doi: 10.1016/j.livsci.2013.02.005. [DOI] [Google Scholar]
- 31.Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lay C, Sutren M, Rochet V, Saunier K, Doré J, Rigottier-Gois L. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol 7:933–946. doi: 10.1111/j.1462-2920.2005.00763.x. [DOI] [PubMed] [Google Scholar]
- 33.Uyeno Y, Sekiguchi Y, Sunaga A, Yoshida H, Kamagata Y. 2004. Sequence-specific cleavage of small-subunit (SSU) rRNA with oligonucleotides and RNase H: a rapid and simple approach to SSU rRNA-based quantitative detection of microorganisms. Appl Environ Microbiol 70:3650–3663. doi: 10.1128/AEM.70.6.3650-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550. [DOI] [PubMed] [Google Scholar]
- 35.Louis P, Flint HJ. 2007. Development of a semiquantitative degenerate real-time PCR-based assay for estimation of numbers of butyryl-coenzyme a (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louis P, Young P, Holtrop G, Flint HJ. 2010. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol 12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 37.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 38.Whelan K, Judd PA, Preedy VR, Simmering R, Jann A, Taylor MA. 2005. Fructooligosaccharides and fiber partially prevent the alterations in fecal microbiota and short-chain fatty acid concentrations caused by standard enteral formula in healthy humans. J Nutr 135:1896–1902. [DOI] [PubMed] [Google Scholar]
- 39.Scott KP, Martin JC, Duncan SH, Flint HJ. 2014. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 40.Benyacoub J, Rochat F, Saudan K-Y, Rochat I, Antille N, Cherbut C, von der Weid T, Schiffrin EJ, Blum S. 2008. Feeding a diet containing a fructooligosaccharide mix can enhance Salmonella vaccine efficacy in mice. J Nutr 138:123–129. [DOI] [PubMed] [Google Scholar]
- 41.Louis P, Scott KP, Duncan SH, Flint HJ. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 42.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aminov RI, Walker AW, Duncan SH, Harmsen HJ, Welling GW, Flint HJ. 2006. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl Environ Microbiol 72:6371–6376. doi: 10.1128/AEM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. 2012. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. 2007. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol 73:6526–6533. doi: 10.1128/AEM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonel AJ, Alvarez-Leite JI. 2012. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care 15:474–479. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 50.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost F, Brummer R-J. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. [DOI] [PubMed] [Google Scholar]
- 51.Neish AS. 2009. Microbes in gastrointestinal health and disease. Gastroenterology 136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maslowski KM, Mackay CR. 2011. Diet, gut microbiota and immune responses. Nat Immunol 12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 54.Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 55.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noverr MC, Huffnagle GB. 2004. Does the microbiota regulate immune responses outside the gut? Trends Microbiol 12:562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T. 2009. Regulatory T cells: how do they suppress immune responses? Int Immunol 21:1105–1111. doi: 10.1093/intimm/dxp095. [DOI] [PubMed] [Google Scholar]
- 60.Watzl B, Girrbach S, Roller M. 2005. Inulin, oligofructose and immunomodulation. Br J Nutr 93:S49–S55. doi: 10.1079/BJN20041357. [DOI] [PubMed] [Google Scholar]
- 61.Seifert S, Watzl B. 2007. Inulin and oligofructose: review of experimental data on immune modulation. J Nutr 137:2563S–2567S. [DOI] [PubMed] [Google Scholar]
- 62.Meijer K, de Vos P, Priebe MG. 2010. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr Opin Clin Nutr Metab Care 13:715–721. doi: 10.1097/MCO.0b013e32833eebe5. [DOI] [PubMed] [Google Scholar]
- 63.Galisteo M, Duarte J, Zarzuelo A. 2008. Effects of dietary fibers on disturbances clustered in the metabolic syndrome. J Nutr Biochem 19:71–84. doi: 10.1016/j.jnutbio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 64.Tedelind S, Westberg F, Kjerrulf M, Vidal A. 2007. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol 13:2826–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Sabatino A, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi F, Tinozzi S, Corazza G. 2005. Oral butyrate for mildly to moderately active Crohn's disease. Aliment Pharmacol Ther 22:789–794. doi: 10.1111/j.1365-2036.2005.02639.x. [DOI] [PubMed] [Google Scholar]
- 66.Abraham C, Medzhitov R. 2011. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLoughlin RM, Mills KH. 2011. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J Allergy Clin Immunol 127:1097–1107. doi: 10.1016/j.jaci.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 68.Heath WR, Carbone FR. 2001. Cross-presentation, dendritic cells, tolerance and immunity. Annu Rev Immunol 19:47–64. doi: 10.1146/annurev.immunol.19.1.47. [DOI] [PubMed] [Google Scholar]
- 69.Steinbrink K, Wölfl M, Jonuleit H, Knop J, Enk AH. 1997. Induction of tolerance by IL-10-treated dendritic cells. J Immunol 159:4772–4780. [PubMed] [Google Scholar]
- 70.Aoyama M, Kotani J, Usami M. 2010. Butyrate and propionate induced activated or non-activated neutrophil apoptosis via HDAC inhibitor activity but without activating GPR-41/GPR-43 pathways. Nutrition 26:653–661. doi: 10.1016/j.nut.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki S, Kuwahara A. 2008. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol 59:251–262. [PubMed] [Google Scholar]