Abstract
Although the source of drinking water (DW) used in hospitals is commonly disinfected, biofilms forming on water pipelines are a refuge for bacteria, including possible pathogens that survive different disinfection strategies. These biofilm communities are only beginning to be explored by culture-independent techniques that circumvent the limitations of conventional monitoring efforts. Hence, theories regarding the frequency of opportunistic pathogens in DW biofilms and how biofilm members withstand high doses of disinfectants and/or chlorine residuals in the water supply remain speculative. The aim of this study was to characterize the composition of microbial communities growing on five hospital shower hoses using both 16S rRNA gene sequencing of bacterial isolates and whole-genome shotgun metagenome sequencing. The resulting data revealed a Mycobacterium-like population, closely related to Mycobacterium rhodesiae and Mycobacterium tusciae, to be the predominant taxon in all five samples, and its nearly complete draft genome sequence was recovered. In contrast, the fraction recovered by culture was mostly affiliated with Proteobacteria, including members of the genera Sphingomonas, Blastomonas, and Porphyrobacter. The biofilm community harbored genes related to disinfectant tolerance (2.34% of the total annotated proteins) and a lower abundance of virulence determinants related to colonization and evasion of the host immune system. Additionally, genes potentially conferring resistance to β-lactam, aminoglycoside, amphenicol, and quinolone antibiotics were detected. Collectively, our results underscore the need to understand the microbiome of DW biofilms using metagenomic approaches. This information might lead to more robust management practices that minimize the risks associated with exposure to opportunistic pathogens in hospitals.
INTRODUCTION
Despite the use of disinfectants in drinking water distribution systems (DWDS), bacteria are able to colonize different parts of DWDS, such as building plumbing systems and fixtures (e.g., sinks, showerheads, and faucets) (1–3). Previous studies have shown that several organisms associated with DWDS can tolerate the effects of disinfectant compounds because of their ability to form biofilms (4, 5). Unlike planktonic forms, bacteria in biofilms are more resistant to sterilization procedures and antimicrobial exposure, showing in some cases a MIC up to 1,000-fold higher than that of their planktonic counterparts (6). Hence, biofilm formation in response to disinfectant treatment can increase the resistance to common cleaning protocols and promote the transfer of antibiotic resistance genes among the biofilm members, producing multidrug-resistant bacteria (7, 8).
Although the frequency of nosocomial infections caused by bacteria located in hospital water supplies is traditionally thought to be low (9), this infection route has regained attention due to the increase in hospital-acquired infections in recent years and the presence of opportunistic pathogens in biofilms located in hospital premise plumbing and medical devices (10–12). Microorganisms forming a biofilm can detach and be transferred to surfaces, medical equipment, and human individuals (13). Biofilms located on hospital showerheads can as such be an important reservoir for nosocomial infections (1, 14, 15). Previous studies of the microbial community composition of showerhead biofilms have identified nontuberculous mycobacteria (NTM), some of which are considered opportunistic pathogens that are commonly found in natural environments (i.e., soil and water), as well as in the built environment, including hospitals (10, 16, 17). Some NTM species have been linked to hypersensitivity pneumonitis, cervical lymphadenitis, allergies, and respiratory problems, mainly in immunocompromised individuals (18, 19). NTM growing in biofilms have been identified in drinking water systems, on polyvinyl chloride (PVC) surfaces, and on showerheads from hospitals, houses, and workplaces (20–22). Their frequent occurrence in such habitats may be explained by their ability to survive stressors commonly found in distribution systems, such as oligotrophic conditions, chlorination, and hot temperatures (23, 24). However, most previous surveys reporting the occurrence and prevalence of mycobacteria in DW have been restricted to 16S rRNA gene fragment analysis, lacking resolution at the species level (25), and to culture-based techniques (17, 26, 27), which often provide a biased representation of the sample due to the selective lab media, culture conditions, and volume of the sample processed.
The gene functions that underlie the ecological success of most DW bacteria in DWDS remain poorly described, in part due to the lack of genetic information on microbial groups commonly inhabiting DWDS. Hence, metagenome sequencing (i.e., random sequencing of total community DNA extracts) has recently been used to examine the functional network of complex microbial communities (28, 29). In spite of the rise of infections by opportunistic premise plumbing pathogens, relatively few studies have assessed the diversity of biofilms growing in DWDS at the metagenome level, especially in health care units (30–33). Most previous reports are based on 16S rRNA gene amplicon surveys that are limited in scope as far as accurately predicting exposure risks. Therefore, in this study, we characterized the biofilm microbial communities of shower hoses in a hospital using shotgun metagenome sequencing and evaluated the genetic diversity and relative abundance of antibiotic and disinfectant resistance present. We also compared the metagenomics findings to those obtained by a substantial collection of genome sequences of isolates (n = 94) recovered from the same samples and those of previous studies from other hospitals and the built environment.
MATERIALS AND METHODS
Sample collection.
The samples used in this study were collected during four consecutive days in 2012 from 40 showerheads located in different rooms within an Ohio hospital. Drinking water in this building normally contains a free chlorine residual of 0.8 mg/liter, and the average water temperature and pH are 20°C and 8.4, respectively. In addition, the concentrations of several metals (Cr, Cu, Fe, Ni, Sr, Sn, and Pb) were measured using an inductively coupled plasma-mass spectrometry (ICP-MS) device, according to U.S. EPA Method 200.8 (34), and were found to be below regulatory thresholds (e.g., Al, 52 to 65 μg/liter; P, 155 to 170 μg/liter; S, 20 to 22 mg/liter; K, 0.5 to 2.4 mg/liter), with limited variation from room to room.
To minimize collection time, the entire showerheads were removed with the shower hoses, water was discarded, and the showerheads were transferred to sterile plastic bags, which were then placed in coolers containing ice packs. Hoses were removed and split open with a sterile knife to expose the inner luminal surfaces. Biofilms from the shower hoses were collected by scraping the inner surfaces with sterile spatulas. The biomass was then transferred to sterile conical tubes and resuspended in phosphate buffer. Five of the samples were randomly selected for metagenomic studies, while all samples were used for conventional microbiological culture. Samples were processed within 4 h of collection time.
Culturing and identification of isolates.
For isolation, an aliquot (1 ml) of the resuspended biomass was used to grow heterotrophic bacteria. Biofilm samples were diluted, processed in duplicate, and spotted onto R2A agar plates (35), which were then incubated at 25°C for 5 to 7 days. Colonies were restreaked onto R2A agar plates to obtain single isolated colonies. Using sterilized toothpicks, >2,000 pure colonies were carefully scraped from the R2A agar plates and resuspended in 30 μl of sterile molecular-grade water. Resuspended cells (2 μl) were used to partially amplify the 16S rRNA gene using universal primers 8F and 787F. The amplification conditions and sequencing analysis conducted were the same as those described elsewhere (36).
High-throughput sequencing.
The five samples used for the metagenomic analyses were filtered onto polycarbonate membranes and stored at −20°C until further processing. Total DNA was extracted from these filters using an UltraClean soil DNA kit (MoBio Laboratories), as previously described (25).
A subset of all strains isolated in this study were subjected to whole-genome sequencing. This subset represented strains with the most common colony morphotypes and included strains from the samples used in metagenome sequencing.
Total DNA extracted from polycarbonate filters and from selected isolates was normalized to 5 ng/μl, and libraries were constructed using the Illumina TruSeq preparation protocol and sequenced on an Illumina HiSeq 2000 using a 100-bp paired-end read approach, according to the instructions of the manufacturer (Illumina, San Diego, CA).
Read trimming and de novo assembly.
Raw reads from the metagenomes and isolate genomes were trimmed using SolexaQA, with a Q 20 Phred score cutoff (37); sequences of <50 bp after trimming and/or with Illumina adaptors at the 3′ end were discarded. Assembly of the metagenomes was performed using the previously described hybrid protocol (38), which combines Velvet (39), SOAPdenovo (40), and Newbler 2.0 (41) assemblers, using k-mer values from 31 to 63. Table S1 in the supplemental material shows the statistics of the shower hose metagenomes. For the isolate genomes, trimmed reads were assembled using SPAdes assembler with “–sc –careful” and error correction options (42).
Taxonomic classification of the biofilm microbial communities.
Taxonomic classification of assembled metagenomic contigs was carried out using MyTaxa, with default parameters (likelihood score, ≥0.5) (43). In addition, the taxonomic affiliation of 16S rRNA gene fragments recovered from metagenomes and isolate genome reads was determined using the Ribosomal Database Project (RDP) classifier (44), with the RDP 16S rRNA database release 11.3 (45) at a 97% nucleotide sequence identity level.
Metagenomic functional gene assignment and abundance analysis.
Protein-coding genes in assembled contigs >5 kbp were identified by MetaGeneMark using default parameters (46). Functional annotation was based on BLASTp (47) searches of the predicted amino acid sequences against the UniProt/Swiss-Prot database (48), using a cutoff for a match of at least 30% identity and 50% of the length of the query protein sequence covered in the alignment. The abundance of protein functions in each data set was calculated as the number of (assembled) protein sequences assigned to the function above the cutoff divided by the total number of annotated proteins predicted in the respective sample.
Predicted proteins associated with antibiotic resistance mechanisms were identified by BLASTp searches against the antibiotic resistance database (ARDB) (49) composed of 23,137 antibiotic resistance genes (ARG), with a threshold E value of 1e−10 and at least 70% of the query sequence covered by the BLAST alignment (higher stringency than that described above in order to reduce the frequency of false-positive matches, as previously suggested [50]).
Genome equivalents in the metagenomic data sets were calculated as follows: Hidden Markov Model (HMM) searches of 101 universally conserved single-copy genes (51) against the individual unassembled metagenomic reads were performed using HMMER3 version 3.1 (http://hmmer.janelia.org/) (52), with default settings. Ten models, which represented more than one family or extremely conserved families at the sequence level (rpoC, rpoC1, pheT-bacteria, pheT-archaea, proS-bacteria, proS-archaea, glyS, alpha-glyS, era, and the tRNA synthase class I gene), were excluded from further analysis. The median sequencing depth (number of reads/bp) of the remaining 91 HMM models was determined and taken as a proxy of 1 genome equivalent (i.e., the corresponding proteins should be encoded by every genome in the sample). The number of copies per cell of a target gene was estimated as the sequencing depth of that gene (number of reads/bp) divided by the normalizing factor, i.e., the median number of reads/bp of the 91 universal genes.
Open reading frame (ORF) prediction and functional annotation of protein-coding genes in the isolate or population (bin) genomes (see below) were performed as described above for metagenomes. Proteins were assigned to the functional categories using Gene Ontology terms (53). In addition, genome completeness was estimated by the recovery of the 91 universal single-copy genes based on HMM searches. The contamination rate was defined as the percentage of the universal genes found in multiple copies in an isolate or the population genome.
Recovery of genomes from metagenomes (binning).
Assembled contigs for each data set were clustered using MaxBin (54), an expectation maximization-based algorithm that combines differential coverage and tetranucleotide compositional information to bin contigs into population genomes. Additionally, population genomes (bins) were visually inspected for uniform coverage across the genome sequence and a consistent phylogenetic signal of universal genes, which was confirmed using CONCOCT (55). Taxonomic affiliation of bins was based on MyTaxa analysis, and the results were further validated by inspecting the results of BLASTp searches of universal genes predicted in the bins against the NCBI RefSeq database using the lowest common ancestor (LCA) algorithm of MEGAN (56), essentially as previously performed (57).
Potential virulence factors (VFs) in the Mycobacterium bin were identified by BLASTp searches of its predicted proteins against the Virulence Factors of Pathogenic Bacteria (58) and PATRIC databases (59), using a cutoff E value of 1e−10 and at least 70% of the query aligned sequence.
Accession numbers.
All raw sequence data sets were deposited in the Sequence Read Archive database at the NCBI under BioProject accession number PRJNA299404 and accession number SRP065069, and binned genome sequences are available at http://enve-omics.ce.gatech.edu/data/showerheads.
RESULTS
Composition of the microbial community of shower hose biofilms.
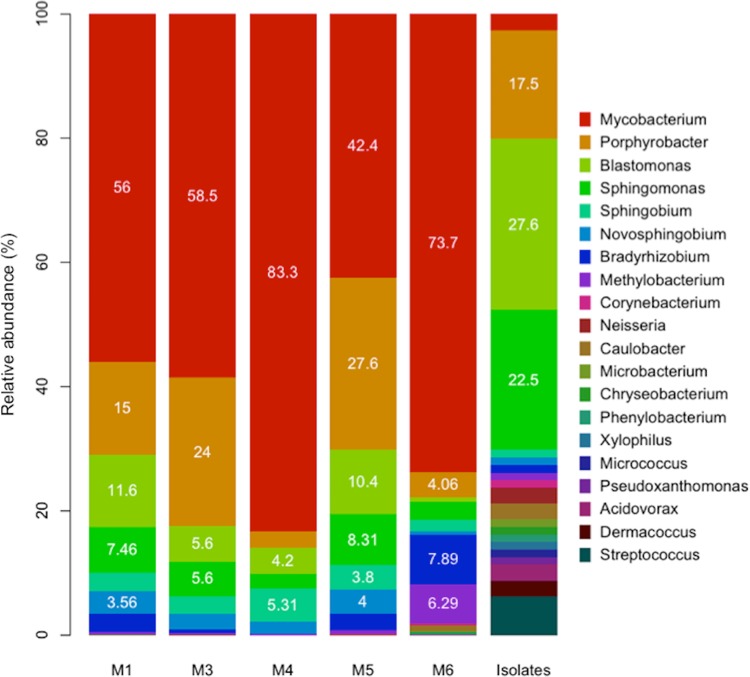
The taxonomic assignment based on 16S rRNA gene-containing metagenomic reads showed that shower hose biofilms contained actinobacteria closely related to the genus Mycobacterium (average ± standard deviation [SD] relative abundance, 42.2% ± 13% of the total; data are from the results with 5 samples), Proteobacteria closely related to the genera Erythrobacter (average ± SD, 9.4% ± 3%), Sphingomonas (average ± SD, 6.6% ± 2.6%), Novosphingobium (average ± SD, 4.2% ± 1.4%), and Bradyrhizobium (average ± SD, 5.2% ± 3.2%), and other less-abundant bacterial genera affiliated with the phyla Bacteroidetes (average ± SD, 4.1% ± 3%) and Firmicutes (average ± SD, 1.2% ± 1%) (Fig. 1; see also Table S1 in the supplemental material). Similar results were obtained based on best match analysis of predicted protein sequences recovered in the assembled metagenomic contigs against complete available genome sequences (see Table S1 in the supplemental material).
FIG 1.
Taxonomic composition of the shower hose biofilms based on 16S rRNA gene fragments recovered from the metagenomes and isolates. The relative abundances (y axis) of the 16S rRNA gene-containing reads recovered from the metagenomes (normalized by the total number of classified 16S rRNA gene-containing reads in each metagenome) and the cultured fraction (normalized by the number of isolates; last column) for the major genera present in each sample (x axis) are shown.
Overall, in all five shower hose metagenomes, the dominant population corresponded to a previously unclassified Mycobacterium sp. most closely related to Mycobacterium rhodesiae and Mycobacterium tusciae, showing ∼85% genome aggregate average nucleotide identity (ANI) (60). The second most abundant population genome was affiliated with Blastomonas, which shared 77% of its proteins at ∼84% average amino acid identity (AAI) to the closely related Blastomonas sp. strain AAP53 reference genome (see Table S3 in the supplemental material).
Analysis of partial 16S rRNA gene sequences of >1,850 R2A isolates revealed that the vast majority (>74%) belonged to the Proteobacteria phylum (data not shown). Specifically, 23% (22/94) of the isolates whose genomes were fully sequenced as part of this study were affiliated with the genus Blastomonas, followed by Sphingomonas (18%) and Porphyrobacter (14%) (see Table S2 in the supplemental material). Several isolates were assigned to the genera Streptococcus (n = 4), Dermacoccus (n = 2), Acidovorax (n = 4), Neisseria (n = 3), and Mycobacterium (n = 2) (Fig. 1).
A comparison of the recovered Blastomonas species population genome against the Blastomonas isolates showed an ANI of 99.9% (SD, 0.01) sharing approximately 91% of its protein sequences. These results suggest that the Blastomonas isolates are representatives of the population recovered in the metagenomes, presumably representing members of the same population (60). In contrast, the average ANI of the recovered Mycobacterium species population and the two isolates classified as Mycobacterium sp. indicated that they indeed belong to the same genus but represent distinct populations and presumably species (ANI, 82.41%; SD, 0.02), and they are low-abundance members of the biofilm community.
The discrepancy between the taxonomic profiles of the culture-dependent and culture-independent results was presumably attributable to the culture medium and growth conditions used, which favored the recovery of Blastomonas species (61). While many mycobacterial species can grow on R2A, it should be noted that some mycobacteria are slow growers and can take up to 8 weeks (or longer) to grow on media typically used for the propagation of mycobacteria (62). Nonetheless, the genome isolates were useful as reference genomes for evaluating genome coverage and confirming species identification (see Fig. S2 in the supplemental material). Our results also showed that a substantial fraction (≥20%) of drinking water microbial communities growing on the shower hose surfaces can be cultured with the described medium, contrasting with the 1 to 2% or less for several complex natural environments, such as soils (63).
Presence of opportunistic pathogens.
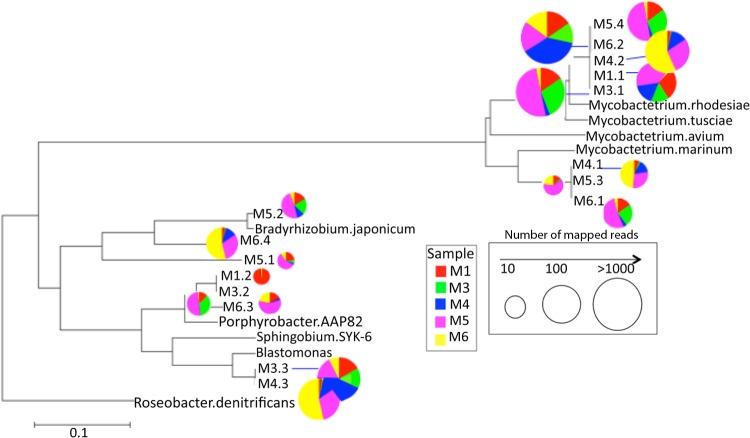
The taxonomic classification of metagenomic sequences and the genomes of isolates (see above) revealed the presence of potential opportunistic pathogens in shower hose biofilms (e.g., members of Sphingomonas, Rhizobium, Mycobacterium mucogenicum, and Neisseria perflava). Notably, the most abundant population recovered in the metagenomes (average ± SD relative abundance among samples, 66.7% ± 8.21% of the total) represented a close relative of M. rhodesiae and M. tusciae. These two mycobacterial species are considered potential opportunistic pathogens, since they have been identified as the causative agent of pulmonary and disseminated infections in immunocompromised individuals (10, 64–66) (Fig. 2).
FIG 2.
Phylogenetic relationships and relative abundance of the populations recovered in the shower hose metagenomes. The tree shows all 30S ribosomal protein S9 sequences assembled from the metagenomes and selected reference sequences from publicly available genomes (denoted by complete species names). The radius of the pie charts indicates the number of reads mapping to the specific protein sequence related to the node, and the colors represent the five different data sets (see figure key). Roseobacter denitrificans was used as an outgroup. The phylogenetic tree was constructed using the neighbor-joining algorithm with 1,000 bootstrap replicates in MEGA5 (91). The scale bar represents the number of substitutions per site.
Phylogenetic analysis showed that the assembled protein sequences of this Mycobacterium-like population genome are linked to a novel species based on relatively low ANI values (∼85%; see Fig. S2 in the supplemental material) to known mycobacterial species (60). Remarkably, the recruitment of metagenomic reads against the recovered mycobacterial genome revealed that this population was the most abundant and distinct from rare (less-abundant) cooccurring relatives in the samples (see Fig. S4 in the supplemental material). Further, reads with >99% nucleotide identity to the reference represented around 62.5% of the total Mycobacterium-like sequences in the metagenomes, and overlapping reads sampling the same part of the genome produced a star-like phylogeny (see Fig. S4 in the supplemental material), suggesting that this is an abundant and homogenous clonal (or nearly clonal) population. Predicted proteins from this population shared 85.6% AAI (77% of the total number of proteins in the population bin) with M. tusciae and 86.2% AAI (76% of the total number of predicted proteins) with M. rhodesiae.
Functional annotation of the recovered Mycobacterium species population genome revealed a number of proteins related to virulence and host colonization previously identified in other NTM species, including M. rhodesiae, M. smegmatis, and M. bovis (see Table S3 in the supplemental material). In particular, our analysis identified several key proteins for (i) biogenesis and central metabolism inside host cells, such as the pantothenate synthetase gene (panC), aspartate-1-decarboxylase gene (panD), and superoxide dismutase gene (sodC), together with genes for (ii) insertion into the host cell via complement-mediated phagocytosis, including fibronectin-binding protein C (fbpC2) and the fibrinogen-binding protein (fbpA), and (iii) genes for protection against oxygen free radicals delivered by host cells, such as catalase-peroxidase (katG) and the sigma factor (sigF) (Table 1).
TABLE 1.
Description of the proteins present in the metagenomes associated with biofilm formation, antibiotic and disinfectant resistance mechanisms, and virulence
| Mechanism | Protein name | Gene(s) | Avg (SD) abundancea |
|---|---|---|---|
| Biocide resistance | |||
| Attachment, invasion, and peroxide resistance | DNA binding protein | dps | 0.27 (0.16) |
| Protective role, oxidative stress defense | Thioredoxin reductase | trxB | 0.51 (0.05) |
| Copper/zinc superoxide dismutase | sodC | 0.07 (0.01) | |
| Putative alcohol dehydrogenase D | adhD | 0.09 (0.08) | |
| Redox-sensitive transcriptional regulator | soxR | 0.04 (0.01) | |
| Alkyl hydroperoxide reductase protein | ahpF | 0.06 (0.02) | |
| Glutathione reductase | gorA | 0.01 (0.008) | |
| Manganese superoxide dismutase | sodA | 0.03 (0.02) | |
| RNA polymerase sigma factor | rpoS | 0.1 (0.07) | |
| Hydrogen peroxide-inducible gene activator | oxyR | 0.14 (0.04) | |
| DNA repair | Exodeoxyribonuclease III | xthA | 0.12 (0.07) |
| Resistance to copper and silver | Cation efflux system protein | cusA | 0.09 (0.03) |
| Multidrug efflux pump systems | RND family | acrB mdtB | 0.02 (0.01) |
| Multidrug resistance protein | emrK | 0.02 (0.01) | |
| ABC transporter ATPase | PGP3 | 0.01 (0.01) | |
| Biofilm formation | |||
| Heat shock protein | groEL1 | 0.27 (0.06) | |
| Biosynthesis | Glutamate synthase | gltB | 0.08 (0.02) |
| Growth | Putative membrane protein | mmpL4 | 0.07 (0.05) |
| Biofilm detachment | Glutathione synthetase | ghsB | 0.08 (0.02) |
| Carbon metabolism | Phosphoenolpyruvate carboxykinase | pckA | 0.05 (0.04) |
| Metabolism | Mycocerosic acid synthase | mas | 0.05 (0.04) |
| Exopolysaccharide biosynthesis and modification | Extracellular polymeric substance | EPS | 0.07 (0.04) |
| Biofilm development | GDP mannose dehydrogenase | algD | 0.09 (0.09) |
| Virulence and antigenic variation | |||
| Possible role in virulence and antigenic variation | Uncharacterized PE-PGRS family proteinb | PE_PGRS33 | 0.22 (0.16) |
| Required for virulence | Cholesterol oxidase | choD | 0.06 (0.04) |
| ABC transporter ATP-binding/permease protein | Rv1747 | 0.11 (0.09) | |
| Serine/threonine-protein kinase | pknF | 0.09 (0.08) | |
| Probable cation-transporting ATPase G | ctpG | 0.04 (0.04) | |
| Probable copper-exporting P-type ATPase V | ctpV | 0.03 (0.05) | |
| Known virulence factors | |||
| Protection against oxygen free radicals | Peroxidase/catalase | katG | 0.23 (0.04) |
| Increased resistance to reactive oxygen intermediates | Sigma factor | sigF | 0.11 (0.08) |
| Secreted protein and virulence determinant factor | Glutamine synthase | glnA1 | 0.07 (0.04) |
| Facilitate the adhesion of bacteria to the mucosal surface | Fibronectin binding proteins | fbpC2 and fbpA | 0.02 (0.01) |
| Essential for ESX-1 secretion system and DNA conjugation | Extracellular mycosin protease | mycP1 | 0.05 (0.03) |
| Transposition | Insertion element IS6110 | MRA_0012 | 0.06 (0.04) |
| Transposase for insertion sequence element IS1081 | YIA3_RHISP | 0.09 (0.06) | |
| Uncharacterized protein encoded by y4hP | NGR_a03340 | 0.18 (0.13) | |
| Transposase for insertion sequence element IS6120 | PUV_09480 | 0.11 (0.07) | |
| Uncharacterized protein encoded by y4jA-y4nE-y4sE | NGR_a03150 | 0.07 (0.04) | |
| Insertion element ISR1 | YIA3_RHISP | 0.06 (0.04) |
Standard deviation (4th column) represents the variation observed among the five metagenomes. Relative abundance was based on the number of predicted proteins assigned to a particular function divided by the total number of annotated metagenomic proteins, previously mentioned in Materials and Methods.
PE-PGRS, Pro-Glu polymorphic GC-rich sequence.
We also identified members of the Sphingomonas genus in the shower hose biofilms. Members of this genus were previously isolated from hospital water sources and associated with urinary tract infections and peritonitis (12, 67). Notably, cases of bacteremia have been reported, including one in a hospital in Taiwan (68) and another in a cardiovascular intensive care unit (ICU) in a hospital in Turkey (69). Since these reports are based on nonsequencing methods (e.g., pulsed-field gel electrophoresis and blood cultures), it was not possible to perform a more detailed comparison to the isolates and populations recovered in the present study. 16S rRNA gene sequence analysis showed that the closest relative for several of our isolates was Sphingomonas koreensis (99% nucleotide identity), which has been identified as the causative agent of meningitis in at least one previous study (70). Taken together, it is likely that the Sphingomonas isolates recovered here represent opportunistic pathogens.
Disinfectant resistance mechanisms.
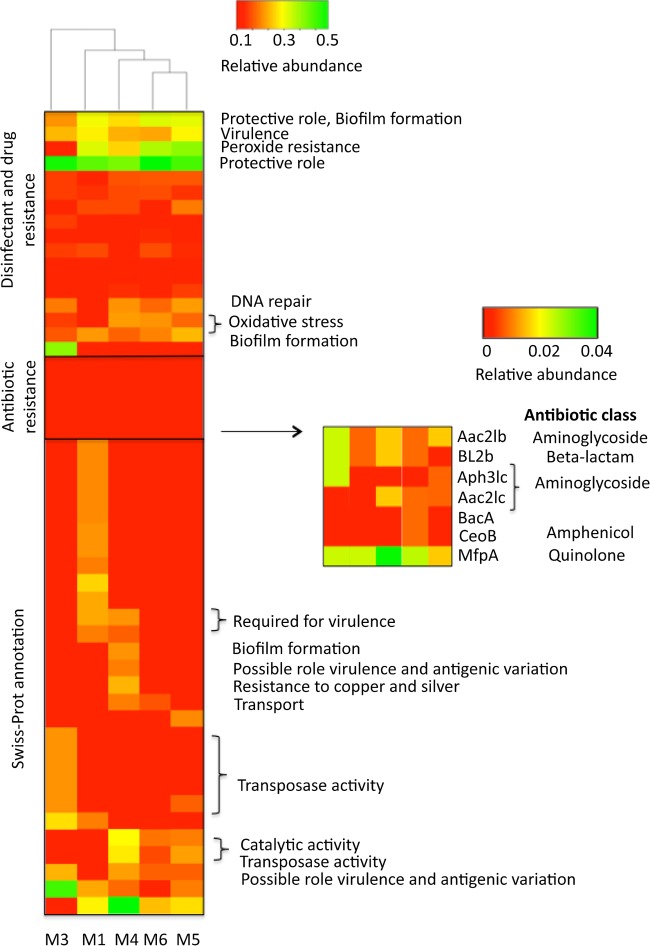
Several genes associated with resistance to disinfectants applied to municipal water treatment were recovered in both metagenome and isolate genomes. For instance, we recovered genes encoding proteins with participation in SoxR, OxyR, and SOS systems that have been experimentally identified as conferring protection against oxidative stress (71, 72). These functions were at least 10 times more frequent (i.e., number of distinct gene alleles detected) in the metagenomes relative to all completed bacterial genomes with similar genome sizes available in NCBI, as of January 2016 (number of genomes used, 442; genome size, 2 to 4 Mb; average P value, 0.00065, t test), indicating that the shower hose environment selects for the functions. In addition, we identified multidrug efflux pump genes, including those encoding the ABC, small multidrug resistance (SMR), and resistance-nodulation-division (RND) systems, which can confer resistance to disinfectants and antibiotics (for the antibiotic mechanisms, see below) in Gram-negative biofilm members affiliated with Sphingomonas, Porphyrobacter, and Blastomonas. All corresponding protein sequences showed high amino acid identity (>40%) and conservation of functional domains with their experimentally verified homologs (Fig. 3; see also Table S4 in the supplemental material).
FIG 3.
Relative abundance of functional genes in the shower hose metagenomes. (Top to bottom) The heat map on the left is composed of 7 proteins involved in antibiotic resistance, 12 proteins involved in disinfectant resistance mechanisms and EPS production, and the 30 most abundant proteins annotated with UniProt DB (rows) for each sample (columns). The small heat map on the right represents a magnification of the main heat map, focusing on the antibiotic resistance genes (note the difference in scale). Relative abundance of a gene/function was defined as the fraction of total annotated proteins with the particular function. The antibiotic class denotes the classification of the antibiotics based on WHO ATC code J01 (WHO Collaborating Centre for Drug Statistics Methodology [http://www.whocc.no/atc_ddd_index/]). The cladogram was constructed using complete linkage hierarchical clustering with Euclidean distance, as implemented on gplot package in R (92). A detailed description of all the proteins plotted in the heat map is in Table S4 in the supplemental material.
Antibiotic resistance mechanisms.
A BLAST analysis of the metagenomic proteins against the Antibiotic Resistance Genes Database (ARDB) revealed that the sampled organisms likely have proteins that underlie resistance to at least four distinct antibiotic classes: β-lactamases, quinolones, aminoglycosides, and amphenicols. Overall, the M3 metagenome presented the highest percentage of cells carrying ARG, specifically the β-lactamase gene (bl2B) (23.1% of total), mycobacterial fluoroquinolone resistance protein A gene (mfpA) (28.3%), which is involved in DNA mimicry mechanisms (73), and the aminoglycoside 2′-N-acetyltransferase gene (aac2Ib) that acetylates aminoglycoside antibiotics, preventing their binding to the bacterial ribosome (74) (Table 2). Thus, the dominance of Mycobacterium in sample M3 (53%) was also reflected in the antibiotic resistance profile of this sample, since 66.6% of the contigs containing ARG were phylogenetically affiliated with this genus.
TABLE 2.
Abundance of antibiotic resistance genes recovered from the shower hose metagenomes
| ARG symbol | Function | Antibiotic resistance | Genome equivalents (%)a |
||||
|---|---|---|---|---|---|---|---|
| M1 | M3 | M4 | M5 | M6 | |||
| BL2b | β-Lactamase | Penicillin, cephalosporin | 10.9 | 23.1 | 6.2 | 0 | 2.5 |
| MfpA | Mycobacterium fluoroquinolone resistance protein A | Fluoroquinolone, ciprofloxacin, sparfloxacin | 4.2 | 17.6 | 4.3 | 16.7 | 5.5 |
| Aac2Ib | Aminoglycoside 2′-N-acetyltransferase | Netilmicin, tobramycin, dibekacin, gentamicin | 1.3 | 38.3 | 2.7 | 0 | 2.8 |
| Aph3Ic | Aminoglycoside O-phosphotransferase | Paromomycin, neomycin, kanamycin, lividomycin, ribostamycin, gentamicin B | 0 | 1.9 | 0 | 0.2 | 0 |
| Aac2Ic | Aminoglycoside N-acetyltransferase | Gentamicin, netilmicin, tobramycin, dibekacin | 0 | 0 | 0 | 18.7 | 31.6 |
| CeoB | Resistance-nodulation-division transporter system; multidrug resistance efflux pump | Chloramphenicol | 0 | 0 | 0 | 0 | 0.5 |
| BacA | Undecaprenyl pyrophosphate phosphatase | Bacitracin | 0 | 0 | 0 | 0 | 0.4 |
The values represent the genome equivalents of each gene calculated using its sequencing depth divided by the normalizing factor of the corresponding data set, outlined in Materials and Methods.
The second most abundant population genome recovered from the metagenomes, which was also well represented among the isolates (unlike the abundant mycobacterial population), was assigned to Blastomonas and contained genes likely conferring resistance to aminoglycoside, macrolide, and bacitracin antibiotics. Indeed, a comparison between the 16S RNA gene sequences obtained from the shower hose Blastomonas isolates in this study and those obtained from Blastomonas strains isolated from a tap water sample in Portugal (GenBank accession no. HF930725.1) (64) revealed high sequence identity (>97%); therefore, these two isolates likely represent the same or highly related species. The Portuguese tap water isolate was highly resistant to antibiotics, based on an ATB PSE EU (bioMérieux) susceptibility test, mostly to the aminoglycoside antibiotic class, including gentamicin and tobramycin. This finding was consistent with the gene content predicted in the Blastomonas isolates of our study. In addition, other genes conferring resistance to penicillin, cephalosporin, paromomycin, neomycin, lividomycin, ribostamycin, and chloramphenicol were detected in the metagenomes, albeit at much lower abundances (present in <5% of the genome equivalents).
Comparisons to other similar environments.
We compared 16S rRNA gene fragments recovered from the shower hose metagenomes against 16S rRNA gene sequences available from DWDS pipes located in Florida (75) and a surface in the intensive care unit (ICU) of a hospital ward in Spain (76). This analysis revealed distinct taxonomic profiles between these and our shower hose metagenomes (see Fig. S1 in the supplemental material). Most notably, the shower hose data sets presented higher abundances of sequences related to Mycobacteriaceae (an average of 38% in shower hose versus 0.03% in the ICU surface and 6% in pipes), followed by Sphingomonadaceae (18% in shower hose versus 5% in the ICU surface and 0.03% in pipes), and Erythrobacteraceae (13% in shower hose versus 0.09% in the ICU surface and 0% in pipes). Distinctively, members of the Methylococcaceae order dominated the DWDS pipe sample (83% of the total) but were essentially absent in the other two data sets. In addition, Staphylococcaceae and Enterobacteriaceae dominated the ICU ward surface of the hospital (22% and 20% of the total, respectively) but were in low abundance in the other data sets.
A comparison of the shower hose metagenomes with available metagenomes from diverse natural water ecosystems in similar temperate geographic regions indicated that the shower hose metagenomes were enriched in virulence factors (3.64% of total metagenomic reads) and antibiotic resistance functions (0.032%) compared to metagenomes from the Pearl River (China) (0.072% and 0.011% of total reads annotated as virulence factors and antibiotic resistance genes, respectively), and wintertime (0.071% and 0.010%, respectively) and summertime (0.340% and 0.072%, respectively) samples from Lake Lanier (GA, USA). Compared to a drinking water treatment plant located in the Pearl River Delta in China (0.008%), the showerhead metagenomes were enriched in these two functions (0.17%) (see Fig. S3 in the supplemental material).
DISCUSSION
This study analyzed biofilms of shower hoses in a hospital and found that most metagenomic sequences were associated with members of the genera Mycobacterium, Erythrobacter, Sphingomonas, and Novosphingobium. These findings were consistent with those from previous studies showing that mycobacterial populations are frequently abundant in DWDS because of their high resistance to chlorine, monochloramine, and other disinfectant compounds in water systems (1, 22, 77). The high abundance of mycobacterial populations in the shower hose biofilms contrasted with their low abundance or absence in microbial communities on the surface of the ICU in a hospital in Spain, which consisted predominantly of Staphylococcaceae and Enterobacteriaceae (see Fig. S1 in the supplemental material). The abundance of these two bacterial groups in the Spanish hospital might be the result of these organisms being continuously shed by incoming patients and hospital staff and therefore may not be waterborne in nature.
Another possible explanation for the dominance of Mycobacterium-like sequences is related to the particular physicochemical features of the shower hose, such as pipe material that is either galvanized (zinc coated) or made of copper, and the disinfectants and low organic carbon content of the water, which selectively favor the growth of some mycobacterial populations (78, 79). Because of the identification of several pathogenicity factors and antibiotic resistance genes (Fig. 3), as well as its high relatedness to characterized NTM (i.e., in terms of both gene content and amino acid similarity), the recovered Mycobacterium species population might represent an opportunistic pathogen. Therefore, our findings revealed that microbial biofilms in hospital shower hoses are characterized by a distinct composition, including previously nondescribed species, which require more attention due to their potential implications for health (see also below). Nonetheless, it should be noted that this Mycobacterium species genome encoded, in general, fewer virulence factors than those of its close relatives M. tusciae (66.6% of total virulence factors [VFs] of M. tusciae were present in the Mycobacterium species population) and M. rhodesiae (80.9% of total VFs shared), indicating that this population might represent a member of NTM with comparatively fewer public health implications.
In addition, some of the isolates were affiliated with disease-causing bacteria. The isolates CCH10-H12 and CCH6-A12 were most closely affiliated with Neisseria perflava (98% and 100% 16S rRNA gene identity, respectively). This bacterium is a common oral commensal of the human upper respiratory tract but occasionally can cause endocarditis, peritonitis, and complicated bacteremia, mainly in individuals with immune suppression (80). Further, two Mycobacterium isolates were below the detection limit of our metagenomic effort (rare members of the biofilm) and most closely assigned to M. mucogenicum (100% 16S rRNA gene identity). Compared to other mycobacterial species, this is a fast-growing organism and is commonly involved in catheter-related infections and nosocomial outbreaks caused by contaminated hospital equipment and water sources (81, 82). The divergence between the Mycobacterium species recovered by culture-dependent and -independent methods was probably due to the fact that incubation time and culture media were not suitable for isolating the most abundant Mycobacterium sp., which was recovered with the metagenomic approach used here (close relative to M. rhodesiae and M. tusciae). Therefore, even though the frequency with which the aforementioned organisms cause infections is probably lower than that of some of the other most commonly encountered opportunistic pathogens, such as members of the Burkholderia and Ralstonia genera, it is quite likely that they represent a health risk, especially for immunocompromised patients. Collectively, these findings suggest that more attention needs to be given to biofilms growing on shower hoses and other surfaces in clinical settings due to their potential to represent a health risk. Current and future studies held by the Hospital Microbiome and the Indoor Environment Projects (33, 83), analyzing hundreds of samples and from various hospital settings, would add to the picture of the microbial communities presented here and the assessment of the associated risk for public health.
In addition to mycobacteria, members of other abundant genera present in the shower hose biofilms, namely, Porphyrobacter, Blastomonas, and Sphingomonas, have also been frequently found in water-related environments, such as swimming pools, bulk water, and faucets, presumably because of their ability to survive disinfection regimes (3, 84). In particular, these bacterial groups are considered to play an important role in the formation and dynamics of biofilms because of their high production potential for exopolysaccharide (EPS) and ability to colonize surfaces (85). Members of these genera also have the ability to coaggregate with other community members, contributing to effective colonization and expansion of biofilms (84). In view of the frequent occurrence of Sphingomonadaceae in hospital tap water and their high survival in the air of the indoor environment, this group has been identified as a frequent contaminant of medical devices (67, 69, 76). Although these organisms were less abundant than mycobacteria in shower hose biofilms (see Fig. S2 in the supplemental material), their occurrence in these environments may be linked to resistance to cleaning and disinfection due to known adaptive mechanisms and biofilm-forming ability.
Biocide agents have a strong influence on the bacterial community structure and may increase the frequency of antibiotic-resistant bacteria (86). Exposure to chlorine can stimulate the expression of efflux pumps and drug resistance operons, as well as induce mutations in some genes leading to increased antibiotic resistance (87). Some of the antibiotic resistance signatures observed in the shower hose metagenomes have been reported to be triggered by biocide exposure; these include the chloramphenicol, kanamycin, and penicillin resistance genes (87, 88). Further, previous studies have observed that several Mycobacterium species can modify the cell membrane fatty acid composition in response to stress conditions, producing an altered permeability to biocide and antibiotic compounds (89, 90). Several of the known proteins that underlie this altered permeability, such as those involved in lipid metabolism and mycolic acid biosynthesis, e.g., long-chain fatty acid ligase (Facl), membrane protein (MmpL3), mycolic acid methyltransferase (MmaA), and GroEL, were encoded in the shower hose metagenomes. Accordingly, the acquisition of the antibiotic resistance profile identified in the biofilm community may, to a certain extent, be directly influenced by chlorine exposure. However, the direct testing of this hypothesis and quantification of the effect of chlorine exposure would require additional experiments.
The bacterial populations recovered from the metagenomes were validated through analysis of the presence/absence (completeness) and phylogenetic identity (contamination) of single-copy genes. These binned populations represented consistent biological units with limited, if any, contaminating sequences from other populations based on the phylogenetic analysis of single-copy genes (e.g., see Fig. S2 in the supplemental material). Also, the genome sequences of the isolates recovered from the same samples were used to validate several of the bins at almost-complete high-quality draft genome sequences (see Table S3 in the supplemental material). For example, the binned Blastomonas population genome showed high nucleotide identity values (ANI, 99.9%; SD, 0.01) and remarkable synteny with the Blastomonas isolate genomes (see Fig. S5 in the supplemental material). In contrast to Blastomonas, the recovery of an abundant uncultivated Mycobacterium population, without known sequenced representatives and 100% completeness, was achieved using binning approaches. The fact that a number of functional gene sequences were recovered using culture-dependent and culture-independent approaches (i.e., both genome isolates and metagenomes), as well as the high relative abundance in situ (e.g., Mycobacterium species and Blastomonas populations), suggests that many of the bacteria in these biofilms were alive, further highlighting their ability to withstand the harsh conditions within DW systems. Finally, although the variation in the abundance of the dominant populations among the samples was, in general, limited, certain populations, such as the Mycobacterium sp., showed substantial differences in abundance (Fig. 1). These differences were not attributable to the measured physicochemical parameters of the water of the shower hoses, which typically do not vary much among samples, or some characteristics (e.g., floor) of the hospital rooms sampled, and thus are likely due to random sampling events.
Altogether, the results reported here reveal novel metagenomic information relevant to microbial exposure in the built environment. As some of the identified mycobacterial populations are related to previously identified pathogens, they may represent an uncharacterized pool of potential nosocomial pathogens growing in biofilms attached to showerhead surfaces. While further evidence is needed to determine if the abundant Mycobacterium sp. and some other less-abundant biofilm populations represent a high risk to patients and health care workers, the data suggest that they should be carefully examined due to their chlorine-resistant phenotype and the presence of several important antibiotic resistance genes in their genomes. Because of the persistence of several community members across samples, the potential for release from the biofilm and adhesion to medical devices, and the presence of antibiotic resistance genes in the biofilm community, our findings call for more attention to the biofilms growing on showerheads, as they might constitute a public health risk. In conclusion, our findings further highlight the increasing importance of metagenomic surveys to better understand the functional genetic network (or microbiome) in clinical settings and in DW distribution systems (22).
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Rodgers for helping during sample collection and for providing comments on early drafts. We also thank Simoni Triantafyllidou for sharing physicochemical data and related discussions.
This study was supported by the U.S. Environmental Protection Agency. Maria J. Soto-Giron was supported by a COLCIENCIAS-Colombian Government doctoral scholarship. Hodon Ryu was the recipient of a National Research Council Senior Research Fellowship. The U.S. Environmental Protection Agency, through its Office of Research and Development, partially funded, managed, and collaborated in the research described here. This work has been subjected to the agency's administrative review and has been approved for external publication.
Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03529-15.
REFERENCES
- 1.Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A 106:16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poitelon JB, Joyeux M, Welte B, Duguet JP, Prestel E, Lespinet O, DuBow MS. 2009. Assessment of phylogenetic diversity of bacterial microflora in drinking water using serial analysis of ribosomal sequence tags. Water Res 43:4197–4206. doi: 10.1016/j.watres.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Yu Z, Guo H, Liu M, Zhang H, Yang M. 2012. Pyrosequencing analysis of eukaryotic and bacterial communities in faucet biofilms. Sci Total Environ 435–436:124–131. [DOI] [PubMed] [Google Scholar]
- 4.Berry D, Xi C, Raskin L. 2006. Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol 17:297–302. doi: 10.1016/j.copbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Revetta RP, Gomez-Alvarez V, Gerke TL, Curioso C, Santo Domingo JW, Ashbolt NJ. 2013. Establishment and early succession of bacterial communities in monochloramine-treated drinking water biofilms. FEMS Microbiol Ecol 86:404–414. doi: 10.1111/1574-6941.12170. [DOI] [PubMed] [Google Scholar]
- 6.Araújo P, Lemos M, Mergulhão F, Melo L, Simões M. 2011. Antimicrobial resistance to disinfectants in biofilms, p 826–834. In Mendez-Vilas A. (ed), Science against microbial pathogens: communicating current research and technological advances. Formatex, Badajoz, Spain. [Google Scholar]
- 7.Schwartz T, Kohnen W, Jansen B, Obst U. 2003. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol 43:325–335. doi: 10.1111/j.1574-6941.2003.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 8.Falkinham JO III, Pruden A, Edwards M. 2015. Opportunistic premise plumbing pathogens: increasingly important pathogens in drinking water. Pathogens 4:373–386. doi: 10.3390/pathogens4020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams MM, Armbruster CR, Arduino MJ. 2013. Plumbing of hospital premises is a reservoir for opportunistically pathogenic microorganisms: a review. Biofouling 29:147–162. doi: 10.1080/08927014.2012.757308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin JH, Lee EJ, Lee HR, Ryu SM, Kim HR, Chang CL, Kim YJ, Lee JN. 2007. Prevalence of non-tuberculous mycobacteria in a hospital environment. J Hosp Infect 65:143–148. doi: 10.1016/j.jhin.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Szymańska J. 2007. Bacterial contamination of water in dental unit reservoirs. Ann Agric Environ Med 14:137–140. [PubMed] [Google Scholar]
- 12.Kilic A, Senses Z, Kurekci AE, Aydogan H, Sener K, Kismet E, Basustaoglu AC. 2007. Nosocomial outbreak of Sphingomonas paucimobilis bacteremia in a hemato/oncology unit. Jpn J Infect Dis 60:394–396. [PubMed] [Google Scholar]
- 13.Guinto CH, Bottone EJ, Raffalli JT, Montecalvo MA, Wormser GP. 2002. Evaluation of dedicated stethoscopes as a potential source of nosocomial pathogens. Am J Infect Control 30:499–502. doi: 10.1067/mic.2002.126427. [DOI] [PubMed] [Google Scholar]
- 14.Falkinham JO III, Iseman MD, de Haas P, van Soolingen D. 2008. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health 6:209–213. [DOI] [PubMed] [Google Scholar]
- 15.Vornhagen J, Stevens M, McCormick DW, Dowd SE, Eisenberg JN, Boles BR, Rickard AH. 2013. Coaggregation occurs amongst bacteria within and between biofilms in domestic showerheads. Biofouling 29:53–68. doi: 10.1080/08927014.2012.744395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker BC, Ford MA, Gruft H, Falkinham JO III. 1983. Epidemiology of infection by nontuberculous mycobacteria. IV. Preferential aerosolization of Mycobacterium intracellulare from natural waters. Am Rev Respir Dis 128:652–656. [DOI] [PubMed] [Google Scholar]
- 17.Dailloux M, Blech MF. 1992. Do water mycobacteria present any infectious risk in immunocompromised patients? Aggressologie 33(Spec No 2):84–86. (In French.) [PubMed] [Google Scholar]
- 18.Primm TP, Lucero CA, Falkinham JO III. 2004. Health impacts of environmental mycobacteria. Clin Microbiol Rev 17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace RJ Jr, Brown BA, Griffith DE. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu Rev Microbiol 52:453–490. doi: 10.1146/annurev.micro.52.1.453. [DOI] [PubMed] [Google Scholar]
- 20.Carter G, Wu M, Drummond DC, Bermudez LE. 2003. Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J Med Microbiol 52:747–752. doi: 10.1099/jmm.0.05224-0. [DOI] [PubMed] [Google Scholar]
- 21.Nishiuchi Y, Tamura A, Kitada S, Taguri T, Matsumoto S, Tateishi Y, Yoshimura M, Ozeki Y, Matsumura N, Ogura H, Maekura R. 2009. Mycobacterium avium complex organisms predominantly colonize in the bathtub inlets of patients' bathrooms. Jpn J Infect Dis 62:182–186. [PubMed] [Google Scholar]
- 22.Gomez-Alvarez V, Revetta RP, Santo Domingo JW. 2012. Metagenomic analyses of drinking water receiving different disinfection treatments. Appl Environ Microbiol 78:6095–6102. doi: 10.1128/AEM.01018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkinham JO., III 2003. Mycobacterial aerosols and respiratory disease. Emerg Infect Dis 9:763–767. doi: 10.3201/eid0907.020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amoils S. 2009. Microbiology: showering with bacteria. Nature 461:360. doi: 10.1038/461360a. [DOI] [PubMed] [Google Scholar]
- 25.Revetta RP, Matlib RS, Santo Domingo JW. 2011. 16S rRNA gene sequence analysis of drinking water using RNA and DNA extracts as targets for clone library development. Curr Microbiol 63:50–59. doi: 10.1007/s00284-011-9938-9. [DOI] [PubMed] [Google Scholar]
- 26.Hilborn ED, Covert TC, Yakrus MA, Harris SI, Donnelly SF, Rice EW, Toney S, Bailey SA, Stelma GN Jr. 2006. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl Environ Microbiol 72:5864–5869. doi: 10.1128/AEM.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussein Z, Landt O, Wirths B, Wellinghausen N. 2009. Detection of non-tuberculous mycobacteria in hospital water by culture and molecular methods. Int J Med Microbiol 299:281–290. doi: 10.1016/j.ijmm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Luo C, Rodriguez RL, Johnston ER, Wu L, Cheng L, Xue K, Tu Q, Deng Y, He Z, Shi JZ, Yuan MM, Sherry RA, Li D, Luo Y, Schuur EA, Chain P, Tiedje JM, Zhou J, Konstantinidis KT. 2014. Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl Environ Microbiol 80:1777–1786. doi: 10.1128/AEM.03712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez RL, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. 2015. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J 9:1928–1940. doi: 10.1038/ismej.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuhata K, Kato Y, Goto K, Saitou K, Sugiyama J, Hara M, Fukuyama M. 2007. Identification of yellow-pigmented bacteria isolated from hospital tap water in Japan and their chlorine resistance. Biocontrol Sci 12:39–46. doi: 10.4265/bio.12.39. [DOI] [PubMed] [Google Scholar]
- 31.McLean JS, Lombardo MJ, Ziegler MG, Novotny M, Yee-Greenbaum J, Badger JH, Tesler G, Nurk S, Lesin V, Brami D, Hall AP, Edlund A, Allen LZ, Durkin S, Reed S, Torriani F, Nealson KH, Pevzner PA, Friedman R, Venter JC, Lasken RS. 2013. Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res 23:867–877. doi: 10.1101/gr.150433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, Ziegler MG, Chitsaz H, Allen AE, Yooseph S, Tesler G, Pevzner PA, Friedman RM, Nealson KH, Venter JC, Lasken RS. 2013. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci U S A 110:E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lax S, Gilbert JA. 2015. Hospital-associated microbiota and implications for nosocomial infections. Trends Mol Med 21:427–432. doi: 10.1016/j.molmed.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Creed J, Brockhoff C, Martin T. 1994. US-EPA Method 200.8: determination of trace elements in waters and wastes by inductively coupled plasma-mass spectrometry. Environmental Monitoring Systems Laboratory Office of Research and Development, Environmental Protection Agency, Cincinnati, OH: https://www.epa.gov/sites/production/files/2015-08/documents/method_200-8_rev_5-4_1994.pdf. [Google Scholar]
- 35.Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu H, Henson M, Elk M, Toledo-Hernandez C, Griffith J, Blackwood D, Noble R, Gourmelon M, Glassmeyer S, Santo Domingo JW. 2013. Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl Environ Microbiol 79:196–204. doi: 10.1128/AEM.02802-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo C, Tsementzi D, Kyrpides NC, Konstantinidis KT. 2012. Individual genome assembly from complex community short-read metagenomic datasets. ISME J 6:898–901. doi: 10.1038/ismej.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL. 2009. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res 19:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo C, Rodriguez RL, Konstantinidis KT. 2014. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42:e73. doi: 10.1093/nar/gku169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. 2006. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res 34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B, Pop M. 2009. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X, Meng Z, Zhao F, Liu D, Ma J, Qin N, Xiang C, Xiao Y, Li L, Yang H, Wang J, Yang R, Gao GF, Wang J, Zhu B. 2013. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun 4:2151. [DOI] [PubMed] [Google Scholar]
- 51.Haft DH, Selengut JD, Brinkac LM, Zafar N, White O. 2005. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 21:293–306. doi: 10.1093/bioinformatics/bti015. [DOI] [PubMed] [Google Scholar]
- 52.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The Gene Ontology Consortium, Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu YW, Tang YH, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 56.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albertsen HM, Chettier R, Farrington P, Ward K. 2013. Genome-wide association study link novel loci to endometriosis. PLoS One 8:e58257. doi: 10.1371/journal.pone.0058257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Xiong Z, Sun L, Yang J, Jin Q. 2012. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40:D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 61.Narciso-da-Rocha C, Vaz-Moreira I, Manaia CM. 2014. Genotypic diversity and antibiotic resistance in Sphingomonadaceae isolated from hospital tap water. Sci Total Environ 466-467:127–135. [DOI] [PubMed] [Google Scholar]
- 62.Tsukamura M. 1976. Numerical classification of slowly growing mycobacteria. Int J Syst Evol Microbiol 26:409–420. [Google Scholar]
- 63.Torsvik V, Øvreås L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245. doi: 10.1016/S1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 64.Tortoli E, Kroppenstedt RM, Bartoloni A, Caroli G, Jan I, Pawlowski J, Emler S. 1999. Mycobacterium tusciae sp. nov. Int J Syst Bacteriol 1999:1839–1844. [DOI] [PubMed] [Google Scholar]
- 65.Curry E, Yehia M, Roberts S. 2008. CAPD peritonitis caused by Mycobacterium rhodesiae. Perit Dial Int 28: 97–99. [PubMed] [Google Scholar]
- 66.Rahman SA, Singh Y, Kohli S, Ahmad J, Ehtesham NZ, Tyagi AK, Hasnain SE. 2014. Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis. mBio 5(6):e02020-14. doi: 10.1128/mBio.02020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsueh PR, Teng LJ, Yang PC, Chen YC, Pan HJ, Ho SW, Luh KT. 1998. Nosocomial infections caused by Sphingomonas paucimobilis: clinical features and microbiological characteristics. Clin Infect Dis 26:676–681. doi: 10.1086/514595. [DOI] [PubMed] [Google Scholar]
- 68.Lin JN, Lai CH, Chen YH, Lin HL, Huang CK, Chen WF, Wang JL, Chung HC, Liang SH, Lin HH. 2010. Sphingomonas paucimobilis bacteremia in humans: 16 case reports and a literature review. J Microbiol Immunol Infect 43:35–42. doi: 10.1016/S1684-1182(10)60005-9. [DOI] [PubMed] [Google Scholar]
- 69.Meric M, Willke A, Kolayli F, Yavuz S, Vahaboglu H. 2009. Water-borne Sphingomonas paucimobilis epidemic in an intensive care unit. J Infect 58:253–255. doi: 10.1016/j.jinf.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 70.Marbjerg LH, Gaini S, Justesen US. 2015. First report of Sphingomonas koreensis as a human pathogen in a patient with meningitis. J Clin Microbiol 53:1028–1030. doi: 10.1128/JCM.03069-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pagán-Ramos E, Song J, McFalone M, Mudd MH, Deretic V. 1998. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol 180:4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dhandayuthapani S, Mudd M, Deretic V. 1997. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol 179:2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. 2005. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 74.Ainsa JA, Martin C, Gicquel B, Gomez-Lus R. 1996. Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob Agents Chemother 40:2350–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelly JJ, Minalt N, Culotti A, Pryor M, Packman A. 2014. Temporal variations in the abundance and composition of biofilm communities colonizing drinking water distribution pipes. PLoS One 9:e98542. doi: 10.1371/journal.pone.0098542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poza M, Gayoso C, Gómez MJ, Rumbo-Feal S, Tomás M, Aranda J, Fernández A, Bou G. 2012. Exploring bacterial diversity in hospital environments by GS-FLX Titanium pyrosequencing. PLoS One 7:e44105. doi: 10.1371/journal.pone.0044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perkins SD, Mayfield J, Fraser V, Angenent LT. 2009. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl Environ Microbiol 75:5363–5372. doi: 10.1128/AEM.00658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Falkinham JO., III 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev 9:177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, Edwards M, Falkinham JO III, Pruden A. 2012. Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems Appl Environ Microbiol 78:6285–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lavigne JP, Le Bayon A, Michaux-Charachon S, Arich C, Bouziges N, Campello C, Sotto A. 2004. Neisseria subflava subsp. perfiava bacteremia: a case study and literature review. Med Mal Infect 34:331–332. (In French.) doi: 10.1016/S0399-077X(04)00144-1. [DOI] [PubMed] [Google Scholar]
- 81.Adékambi T. 2009. Mycobacterium mucogenicum group infections: a review. Clin Microbiol Infect 15:911–918. doi: 10.1111/j.1469-0691.2009.03028.x. [DOI] [PubMed] [Google Scholar]
- 82.Kline S, Cameron S, Streifel A, Yakrus MA, Kairis F, Peacock K, Besser J, Cooksey RC. 2004. An outbreak of bacteremias associated with Mycobacterium mucogenicum in a hospital water supply. Infect Control Hosp Epidemiol 25:1042–1049. doi: 10.1086/502341. [DOI] [PubMed] [Google Scholar]
- 83.Arnold C. 2014. Rethinking sterile: the hospital microbiome. Environ Health Perspect 122:A182–187. doi: 10.1289/ehp.122-A182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rickard AH, Leach SA, Hall LS, Buswell CM, High NJ, Handley PS. 2002. Phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Appl Environ Microbiol 68:3644–3650. doi: 10.1128/AEM.68.7.3644-3650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bereschenko LA, Stams AJ, Euverink GJ, van Loosdrecht MC. 2010. Biofilm formation on reverse osmosis membranes is initiated and dominated by Sphingomonas spp. Appl Environ Microbiol 76:2623–2632. doi: 10.1128/AEM.01998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJ. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248. doi: 10.1093/jac/dkv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karumathil DP, Yin HB, Kollanoor-Johny A, Venkitanarayanan K. 2014. Effect of chlorine exposure on the survival and antibiotic gene expression of multidrug resistant Acinetobacter baumannii in water. Int J Environ Res Public Health 11:1844–1854. doi: 10.3390/ijerph110201844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang JJ, Hu HY, Tang F, Li Y, Lu SQ, Lu Y. 2011. Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res 45:2775–2781. doi: 10.1016/j.watres.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 89.Steed K. 2003. Effect of growth in biofilms upon antibiotic and chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Master's thesis. Virginia Polytechnic Institute and State University, Blacksburg, VA. [Google Scholar]
- 90.Armstrong JL, Calomiris JJ, Seidler RJ. 1982. Selection of antibiotic-resistant standard plate count bacteria during water treatment. Appl Environ Microbiol 44:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Warnes GR, Bolker B, Bonebakkeri L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2009. gplots: various R programming tools for plotting data. R package version 2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.