ABSTRACT
P2 is the original member of a highly successful family of temperate phages that are frequently found in the genomes of gram-negative bacteria. This article focuses on the organization of the P2 genome and reviews current knowledge about the function of each open reading frame.
KEYWORDS: capsid, DNA replication, lysis, lysogenic conversion, lysogeny, regulation, tail
Introduction
Temperate enterobacteriophage P2 is the best-studied example of a class of temperate phages that are commonly found in γ-proteobacteria. In 1951 Giuseppe Bertani isolated P21 from the Escherichia coli strain in which lysogeny was discovered by Bordet and Reneaux.2 Interest in the P2-like phages was increased by Erich Six's discovery of helper-dependent or “satellite” phage P4, which depends upon P2 late genes for its propagation.3 Since P2-like prophages are so widespread, we have written this article to assist investigators in identifying the functions of P2-like prophages found in the sequence analysis of bacterial genomes. This article summarizes briefly what is known about the P2 genome and gene function, pointing out salient differences with the related and also well-studied phage 186 as well as other P2-related phages of interest. P2 is a member of the subfamily Peduovirinae, which has been further divided into the P2-like viruses and the more distantly related (based on sequence similarity) HP1-like viruses.4 In this review, members of these 2 genera will be referred to as P2-like and HP1-like, and the group collectively as P2-related. A more extensive review by Nilsson and Haggård-Ljungquist can be found in The Bacteriophages.5
Virion and genome structure
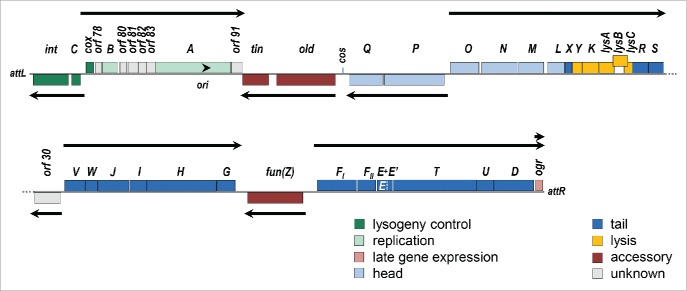
P2 belongs to the family Myoviridae, with an icosahedral capsid 60 nm in diameter and a 135 nm contractile tail (Fig. 1). The P2 genome (NC_001895) consists of a linear dsDNA molecule of 33574 bp, with 19 base, 5′ phosphoryl-terminal, cohesive ends.6,7 There are 42 open reading frames, organized into 10 transcription units. The physical genetic map of P2, depicted in the prophage orientation beginning at the left attachment site (attL), is illustrated in Figure 2. It should be noted, however, that nucleotide numbering of the genome, as reported in the reference sequence in Genbank, is from the left cohesive end rather than from attL. Table 1 summarizes the properties of the genes and their products.
Figure 1.
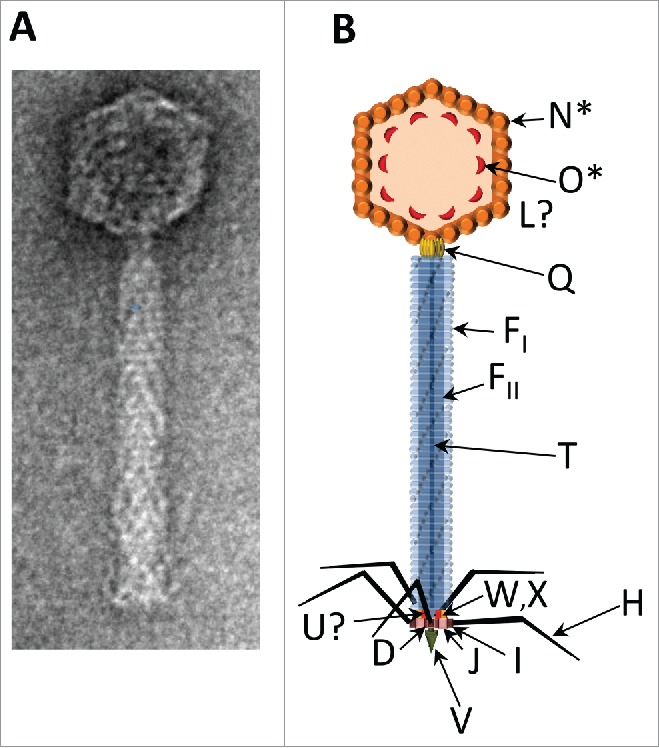
The P2 virion. (A) Electron micrograph of P2, negatively stained with uranyl acetate. This image was generously provided by Dr. Terje Dokland. (B) Schematic illustration of the P2 virion. Arrows indicate the known locations of virion proteins. GpN* is the cleaved form of gpN that constitutes the major capsid protein; gpO* is the fragment of scaffold that remains in the capsid after cleavage. The location in the capsid of the head completion protein gpL is unknown. The dodecameric connector or portal protein, gpQ, lies at the head/tail junction. The tail tube gpFI and tail sheath gpFII are polymerized around the tape measure protein gpT. Arrangement of baseplate components gpW, gpX, gpI, gpJ, gpU, gpD is based on EM studies, known protein-protein interactions, and the location of homologous proteins in the baseplate of bacteriophage T4; assignment for the location of gpU is still tentative. The tail spike is a trimer of gpV, and gpH makes up the tail fibers.
Figure 2.
P2 genetic map. The P2 genome is displayed in prophage orientation, beginning at the leftward attachment site (attL). Genes are color coded by function, as indicated, and genes above the line are read rightwards while those below the line are read leftwards. Arrows indicate the direction and extent of each transcription unit. The arrowhead indicates the location of the replication origin (ori) and direction of replication. The lysB gene is offset to illustrate the overlap with lysC; the dotted white line indicates the end of gene E within the longer E+E’ coding sequence generated by a-1 frameshift.
Table 1.
| Gene | Temporal expression | Size of gene product (aa) | Accession # | Function |
|---|---|---|---|---|
| int | early | 337 | NP_046786 | integrase |
| C | early | 99 | NP_046787 | immunity repressor |
| cox | early | 91 | NP_046788 | repressor of C expression; directionality factor required for excision |
| orf78 | early | 56 | NP_046789 | unknown; highly conserved |
| B | early | 166 | NP_046790 | DNA replication; lagging strand synthesis |
| orf80 | early | 74 | NP_046791 | unknown; highly conserved, DUF2732 |
| orf81 | early | 100 | NP_046792 | unknown; highly conserved |
| orf82 | early | 74 | NP_046793 | unknown; C4 type zinc finger protein, DksA/TraR family |
| orf83 | early | 91 | NP_046794 | unknown; highly conserved |
| A | early | 761 | NP_046795 | DNA replication; site-specific nick at ori |
| orf91 | early | 109 | NP_046796 | unknown, conserved ASCH superfamily domain - possible RNA binding protein |
| tin | constitutive | 253 | NP_046797 | blocks growth of T-even phages |
| old | constitutive | 586 | NP_046798 | nuclease; blocks growth of phage λ |
| Q | late | 344 | NP_046757 | portal |
| P | late | 590 | NP_046758 | terminase lg subunit; DNA-dependent ATPase |
| O | late | 284 | NP_046759 | capsid scaffold; prohead protease |
| N | late | 357 | NP_046760 | major capsid precursor |
| M | late | 247 | NP_046761 | terminase sm subunit |
| L | late | 169 | NP_046762 | capsid completion protein |
| X | late | 67 | NP_046763 | baseplate |
| Y | late | 93 | NP_046764 | holin |
| K | late | 165 | NP_046765 | endolysin |
| lysA | late | 141 | NP_046766 | antiholin |
| lysB | late | 141 | NP_046767 | lysis control, Rz-like spanin component |
| lysC | late | 96 | NP_757382 | lysis control, Rz1-like spanin component |
| R | late | 155 | NP_046768 | tail completion - sheath terminator? |
| S | late | 150 | NP_046769 | tail completion - tube terminator? |
| orf30 | constitutive | 261 | NP_046770 | unknown; highly conserved |
| V | late | 211 | NP_046771 | tail spike |
| W | late | 115 | NP_046772 | baseplate |
| J | late | 302 | NP_046773 | baseplate |
| I | late | 176 | NP_046774 | baseplate |
| H | late | 669 | NP_046775 | tail fiber |
| G | late | 175 | NP_046776 | tail fiber assembly |
| fun(Z) | constitutive | 528 | NP_046777 | FudR sensitivity; blocks phage T5 |
| FI | late | 396 | NP_046778 | tail sheath |
| FII | late | 172 | NP_046779 | tail tube |
| E | late | 91 | NP_046781 | tail assembly chaperone |
| E+E' | late | 142 | NP_046780 | tail assembly chaperone; −1 frameshift extension of gpE |
| T | late | 815 | NP_046782 | tail tape measure |
| U | late | 159 | NP_046783 | tail - tube initiator? |
| D | late | 387 | NP_046784 | baseplate hub |
| ogr | middle/late | 72 | NP_046785 | activator of late transcription; C4 Zn-finger protein |
The lysogeny region
The genes necessary for phage integration/excision and the lysogeny decision lie at the left end of the prophage genome. P2 integrase is a member of the tyrosine recombinase family and promotes the site-specific recombination between the phage and host att sites required for integration of the phage genome.8 The P2 regulatory region contains 2 face-to-face promoters, Pe and Pc, and encodes 2 transcriptional regulators (Fig. 3). The P2 immunity repressor, C, is a small, slightly basic protein of only 99 amino acids that contains an N-terminal helix-turn-helix DNA binding domain.9 Dimers of C protein bind 2 direct repeats, separated by 2 helical turns, which span the −10 region of the early promoter Pe to repress transcription of cox and the replication genes.10 The Cox protein binds to and represses the Pc promoter, which is needed for transcription of the C and int genes.11,12 Cox is a winged HTH protein that oligomerizes as a helical filament and wraps DNA around its outside.13 As is the general case for temperate phages, C also stimulates its own transcription from Pc while Cox represses it. Thus, expression of C leads to lysogenization, whereas Cox expression leads to the lytic cycle. In addition to its role in regulation of lysogenization, Cox also serves as the directionality factor that allows the Int protein to cause prophage excision.14 Many P2-like phages have this arrangement of genes and sites to control the lysis-lysogeny switch.15 However, some P2-like phages and all HP1-like phages characterized thus far have the more complicated arrangement found in the P2-like phage 186.16-18 In this 186-type regulatory switch, there is an additional promoter (Pe) and an additional regulatory protein (CII) involved in expression of the repressor during the establishment of lysogeny19 (Fig. 3). CII is a potent transcription activator that positively regulates transcription from Pe, binding as a dimer to 2 inverted repeat 7-mer half-sites.20 Pe expression is subsequently turned down by the binding of the immunity repressor.
Figure 3.
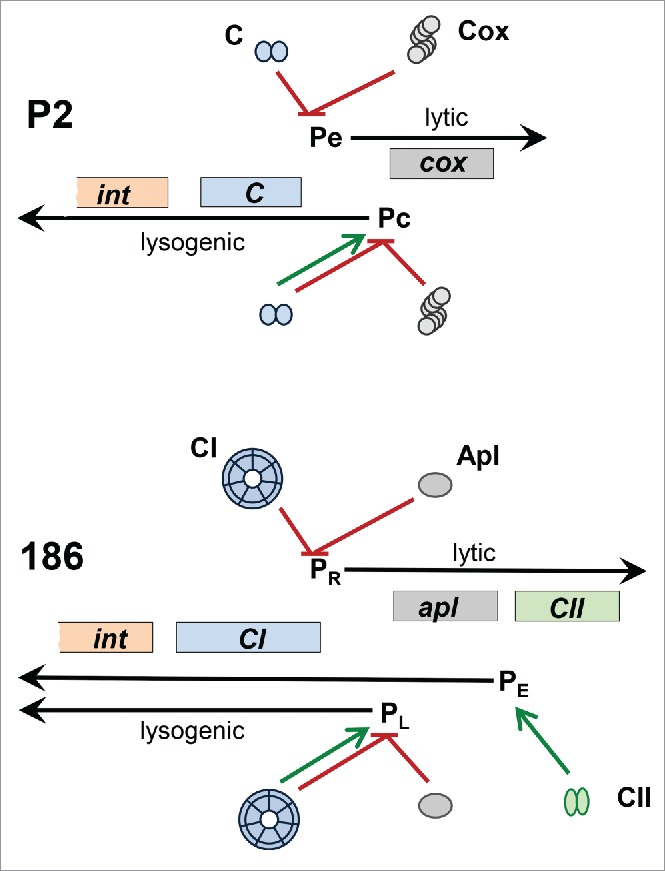
Comparison of the P2 and 186 lysogeny control regions. Black arrows designate the divergent lytic and lysogenic transcripts from the indicated promoters. Genes in the two phages encoding regulatory proteins with equivalent functions are colored the same, as are their corresponding gene products. The promoter targeted by each protein is indicated with a red line if the protein inhibits expression and with a green arrow if the protein activates expression.
The immunity repressor in phages with the 186-type regulatory region is quite different from that in phages with the P2-type region. The 186 repressor, at 192 amino acids, is nearly twice as large as the P2 repressor. It forms an unusual wheel-like heptamer of dimers that provides extended cooperative binding to both adjacent and distant operators.18,21 Like P2 Cox, 186 Apl functions as both a repressor and a directionality factor for excision.17 However, the Cox and Apl proteins from P2-related phages cluster into 2 phylogenetically distinct groups that appear to have co-evolved with the P2-type or 186-type repressor and integrase, respectively.15
The difference in structure between the P2-type and 186-type immunity repressors affects the prophage response to SOS induction. Neither family of repressors contains the LexA cleavage motif commonly found in the repressors of temperate phages. However, while P2 is noninducible,1 186 is induced during the SOS response.22,23 A small accessory gene in 186, tum, is expressed from a LexA-regulated promoter and functions as an antirepressor to effect SOS induction of 186.22-24 The difference in repressor structure also affects the interaction of these 2 phages with the satellite element P4. P4 encodes a protein, Epsilon, which can bind to the P2 C protein to derepress a P2 prophage and allow it to serve as a helper for P4 growth.25,26 In contrast, P4 cannot derepress a 186 prophage and therefore cannot propagate on a 186 lysogen.27
The DNA replication module
The P2 replication genes, A and B, lie in an operon with the cox gene and are flanked by several small open reading frames of unknown function. Following the entry of linear P2 DNA into host cells, the 19 base pair cohesive ends allow the genome to circularize. P2 replicates as circular monomers, unidirectionally from a defined origin.28 The product of gene A nicks the circular DNA at the replication origin, which lies within the A gene.29 GpA binds to the 5′-phosphate, and allows the 3′-hydroxyl to serve as a primer for the leading strand.30 The product of gene B loads the E. coli dnaB helicase onto the replicating fork.31 At the end of a round of replication, the P2 A protein causes the formation of closed circular, daughter DNA molecules.30
The capsid genes
The capsid genes lie in 2 divergently transcribed gene clusters. Gene Q encodes the dodecameric portal protein, which is thought to act as the initiator for capsid assembly, and provides the docking site for the tail proteins and the entry and exit portal for the DNA.32-34 Genes P and M encode the large and small subunits, respectively, of the terminase complex, required for DNA packaging.35 In contrast to the arrangement found in many dsDNA phages, these 2 genes are not adjacent in the P2 genome. Another unusual feature of P2 DNA packaging is that the packaging substrate is closed circular monomeric P2 DNA.36 The M protein is thought to carry the endonuclease activity responsible for converting circular P2 DNA to the linear form with cohesive ends in the presence of empty P2 procapsids and ATP, 37 and the 55 base pair cohesive end site (cos) is also required for this process.38 Gene O encodes a bifunctional 284 amino acid protein. The N-terminal half of gpO functions as a serine protease that cleaves gpO, and a 15.5 kDa N-terminal cleavage product of gpO, O*, remains in the finished capsids.39 The carboxyl terminal 90 amino acids of gpO contain the scaffolding function that causes gpN to form the T =7 prohead.39 The gpO protease also cleaves off the N-terminal 31 amino acids of the major capsid protein, gpN, to yield N*, the protein of the mature capsid.40,41 The product of gene L is not needed for DNA maturation or packaging.42 Instead, gpL is needed to make the finished capsid, and it is found in the capsid.
The lysis genes
The lysis module includes 2 essential genes Y, and K, and 3 accessory genes – lysA, lysB and lysC - which are dispensable under certain conditions of growth. Gene Y encodes a holin, which allows the P2 lysozyme, the product of gene K, to access the peptidoglycan.43,44 The lysA gene product functions as an antiholin, needed to delay the action of gpY until the optimal lysis time.43,44 Amber mutants in lysB exhibit delayed lysis in non-permissive cells.43 The lysC gene, which overlaps the lysB gene, was originally identified by a mutation that overcomes the P2 growth defect conferred by a temperature-sensitive mutation in the β’ subunit of RNA polymerase.45 LysB and LysC are functional homologues of lambda Rz and Rz1, which make up the spanin complex that fuses the inner and outer membranes to complete host cell lysis.46
The tail genes
There are 16 essential P2 tail genes, found within 3 transcription units. Genes X, R and S are at the end of the operon that begins with gene O, and flank the lysis module. Gene X encodes a tail function, according to in vitro complementation studies,43 and gpX has recently been localized to the top of the baseplate.47 Nonsense mutants in gene R make abnormally long tails lacking the head-tail connector, and giant naked tail tubes.48 There is some similarity between gpR and part of T4 phage gp15, which is the connector required for T4 tails to bind to T4 capsids.49 Mutants in gene S produce predominantly normal-appearing but inactive tails, as well as a small number of extended, empty tail sheaths.48 Therefore, gpR and gpS have been proposed to play a role in tail completion and head joining. Tail genes VWJIHG are expressed in a single transcription unit.50-52 In phage 186, however, these genes are cotranscribed with the upstream capsid-lysis-tail gene operon.53 The product of gene V makes up the small spike at the tip of the tail; it is a trimeric iron-binding protein involved in membrane penetration.54-57 The W gene product is homologous to the T4 phage gp25, which is part of the T4 baseplate; it has been localized to the top of the baseplate, 47 and copurifies with gpV.57 The J gene product lies at the edge of the baseplate; 57 gpJ and gpI are believed to make up the baseplate wedges.58 Gene H encodes the tail fiber protein, and gene G is required for tail fiber assembly.59 The tail genes FI, FII, E, T, U and D comprise a single transcription unit.50,52 Gene FI encodes the tail sheath, and FII encodes the tail tube.60 Gene E contains a programmed translational frameshift, allowing it to make a shorter (E) and a longer (E+E’) protein, both of which are essential for phage growth.61 These proteins likely function as chaperones for tail assembly, analogous to the G and G-T proteins of phage lambda.62,63 Because of its exceptional length, gene T is presumed to determine tail length.61 Gene U is likely to encode the tail tube initiator and gene D is believed to encode the central baseplate hub.58 GpD belongs to the same protein domain family (pfam05954: Phage_GPD) as Mu gp44, which forms a trimer with an overall structure like that of the T4 gp27 trimer that forms the central hub of the T4 baseplate.64,65
Control of late gene expression
P2 late promoters share a conserved DNA sequence that is centered 55 base pairs upstream of the transcription start 66,67 and required for late gene expression.68,69 These sequences are recognized and bound by the 72 amino acid P2 Ogr protein, a zinc-binding transcription factor 70 which interacts with the C-terminal domain of the α subunit of E. coli RNA polymerase to stimulate late transcription.71,72 The ogr gene is expressed at middle times after infection from its own promoter and also cotranscribed with the FETUD gene cluster late in infection.73 Unlike ogr, the homologous B gene in 186 is under direct control of the phage immunity repressor.74
Accessory genes (“morons”)
Just to the left of the cos site are 2 genes that are expressed from the prophage and interfere with the growth of 2 other classes of phages. The P2 old gene product interferes with the growth of λ phage and kills E. coli cells that lack the recBCD nuclease-helicase.75 Induction of λ prophage in a P2 old +-lysogenic strain results in partial degradation of many tRNA molecules.76 Purified Old protein has been reported to have 5′ to 3′ exonuclease activity on DNA, as well as an uncharacterized ribonuclease activity.77 The P2 tin gene product inhibits the growth of T-even bacteriophages.78 Tin blocks the action of their single-stranded binding proteins (gp32), thus interfering with T-even phage DNA replication.79
The equivalent region of phage 186 encodes 2 different genes – orf 97, a conserved gene of unknown function, and the tum gene discussed above, required for SOS induction of a 186 prophage.22
The Z/fun gene lies between genes G and FI. In the prophage state this gene causes sensitivity to 5-fluorouracil80 and resistance to bacteriophage T5.81 Mutations in this gene can cause cell killing, eliminating the possibility of lysogenization.82 The Z/fun gene is located at a site that appears to be a hotspot for site-specific insertion of foreign DNA into the genomes of P2-like phages. Ten unrelated sequences were found in this region in P2-like prophages in the E. coli reference collection (ECOR), inserted between a pair of highly conserved long inverted repeats.83 The P2-related Salmonella phage sopEϕ encodes, at this same location, the type III effector SopE, a virulence factor injected into eukaryotic cells via the Salmonella enterica type III secretion system.84
Orf30 lies between genes S and V. It is nonessential, and transcribed constitutively from its own promoter at a low level.49 Homologues of the predicted orf30 gene product are found in many E. coli genomes, but its function remains unknown. Some P2-like phages, such as 186, lack an accessory gene at this location, and the 2 flanking late gene operons are expressed as a single transcription unit.53
The wild type alleles of the old, tin and Z/fun genes provide a selective advantage to bacteria that are lysogenic for P2 by protecting them from infection with other unrelated bacteriophages. Known morons found in other P2-related phages play additional roles, contributing to bacterial virulence or SOS prophage induction. P2-like prophages found in bacterial genome sequences carry a wide variety of accessory genes; the advantages conferred by these genes to the phage or bacterial host and the role of P2-like prophages in the horizontal transfer of accessory genes is an attractive area for further investigation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This review is dedicated to the memory of Giuseppe Bertani and Erich Six. We thank Terje Dokland for the EM image used in Figure 1.
References
- [1].Bertani G. Studies on lysogenesis. I. the mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 1951; 62:293-300; PMID:14888646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bordet J, Reneaux E. L'autolyse microbienne transmissible ou le bacteriophage. Ann Inst Pasteur 1928; 42:1284-335. [Google Scholar]
- [3].Six EW, Klug CA. Bacteriophage P4: A satellite virus depending on a helper such as prophage P2. Virology 1973; 51:327-44; PMID:4571379; http://dx.doi.org/ 10.1016/0042-6822(73)90432-7 [DOI] [PubMed] [Google Scholar]
- [4].Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. Classification of myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 2009; 9:224; PMID:19857251; http://dx.doi.org/ 10.1186/1471-2180-9-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nilsson AS, Haggard Ljungquist E. The P2-like bacteriophages In: RL Calendar, ed. The Bacteriophages. New York: Oxford University Press, 2005:365-90. [Google Scholar]
- [6].Murray K, Murray NE. Terminal nucleotide sequences of DNA from temperate coliphages. Nat New Biol 1973; 243:134-9; PMID:4515740; http://dx.doi.org/ 10.1038/newbio243134a0 [DOI] [PubMed] [Google Scholar]
- [7].Padmanabhan R, Wu R, Calendar R. Complete nucleotide sequence of the cohesive ends of bacteriophage P2 deoxyribonucleic acid. J Biol Chem 1974; 249:6197-6207; PMID:4371195 [PubMed] [Google Scholar]
- [8].Yu A, Bertani LE, Haggard-Ljungquist E. Control of prophage integration and excision in bacteriophage P2: Nucleotide sequences of the int gene and att sites. Gene 1989; 80:1-11; PMID:2676729; http://dx.doi.org/ 10.1016/0378-1119(89)90244-8 [DOI] [PubMed] [Google Scholar]
- [9].Ljungquist E, Kockum K, Bertani LE. DNA sequences of the repressor gene and operator region of bacteriophage P2. Proc Natl Acad Sci U S A 1984; 81:3988-92; PMID:6330728; http://dx.doi.org/ 10.1073/pnas.81.13.3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Massad T, Skaar K, Nilsson H, Damberg P, Henriksson-Peltola P, Haggard-Ljungquist E, Hogbom M, Stenmark P. Crystal structure of the P2 C-repressor: A binder of non-palindromic direct DNA repeats. Nucleic Acids Res 2010; 38:7778-90; PMID:20639540; http://dx.doi.org/ 10.1093/nar/gkq626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saha S, Haggard-Ljungquist E, Nordstrom K. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J 1987; 6:3191-9; PMID:2826134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Saha S, Haggard-Ljungquist E, Nordstrom K. Activation of prophage P4 by the P2 cox protein and the sites of action of the cox protein on the two phage genomes. Proc Natl Acad Sci U S A 1989; 86:3973-7; PMID:2657731; http://dx.doi.org/ 10.1073/pnas.86.11.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Berntsson RP, Odegrip R, Sehlen W, Skaar K, Svensson LM, Massad T, Hogbom M, Haggard-Ljungquist E, Stenmark P. Structural insight into DNA binding and oligomerization of the multifunctional cox protein of bacteriophage P2. Nucleic Acids Res 2014; 42:2725-35; PMID:24259428; http://dx.doi.org/ 10.1093/nar/gkt1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lindahl G, Sunshine M. Excision-deficient mutants of bacteriophage P2. Virology 1972; 49:180-7; PMID:4556923; http://dx.doi.org/ 10.1016/S0042-6822(72)80019-9 [DOI] [PubMed] [Google Scholar]
- [15].Nilsson H, Cardoso-Palacios C, Haggard-Ljungquist E, Nilsson AS. Phylogenetic structure and evolution of regulatory genes and integrases of P2-like phages. Bacteriophage 2011; 1:207-18; PMID:23050214; http://dx.doi.org/ 10.4161/bact.1.4.18470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dodd IB, Kalionis B, Egan JB. Control of gene expression in the temperate coliphage 186. VIII. control of lysis and lysogeny by a transcriptional switch involving face-to-face promoters. J Mol Biol 1990; 214:27-37; PMID:2370665; http://dx.doi.org/ 10.1016/0022-2836(90)90144-B [DOI] [PubMed] [Google Scholar]
- [17].Reed MR, Shearwin KE, Pell LM, Egan JB. The dual role of apl in prophage induction of coliphage 186. Mol Microbiol 1997; 23:669-81; PMID:9157239; http://dx.doi.org/ 10.1046/j.1365-2958.1997.2521620.x [DOI] [PubMed] [Google Scholar]
- [18].Pinkett HW, Shearwin KE, Stayrook S, Dodd IB, Burr T, Hochschild A, Egan JB, Lewis M. The structural basis of cooperative regulation at an alternate genetic switch. Mol Cell 2006; 21:605-15; PMID:16507359; http://dx.doi.org/ 10.1016/j.molcel.2006.01.019 [DOI] [PubMed] [Google Scholar]
- [19].Neufing PJ, Shearwin KE, Camerotto J, Egan JB. The CII protein of bacteriophage 186 establishes lysogeny by activating a promoter upstream of the lysogenic promoter. Mol Microbiol 1996; 21:751-61; PMID:8878038; http://dx.doi.org/ 10.1046/j.1365-2958.1996.351394.x [DOI] [PubMed] [Google Scholar]
- [20].Murchland I, Ahlgren-Berg A, Priest DG, Dodd IB, Shearwin KE. Promoter activation by CII, a potent transcriptional activator from bacteriophage 186. J Biol Chem 2014; 289:32094-108; PMID:25294872; http://dx.doi.org/ 10.1074/jbc.M114.608026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang H, Dodd IB, Dunlap DD, Shearwin KE, Finzi L. Single molecule analysis of DNA wrapping and looping by a circular 14mer wheel of the bacteriophage 186 CI repressor. Nucleic Acids Res 2013; 41:5746-56; PMID:23620280; http://dx.doi.org/ 10.1093/nar/gkt298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lamont I, Brumby AM, Egan JB. UV induction of coliphage 186: Prophage induction as an SOS function. Proc Natl Acad Sci U S A 1989; 86:5492-6; PMID:2664785; http://dx.doi.org/ 10.1073/pnas.86.14.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brumby AM, Lamont I, Dodd IB, Egan JB. Defining the SOS operon of coliphage 186. Virology 1996; 219:105-14; PMID:8623519; http://dx.doi.org/ 10.1006/viro.1996.0227 [DOI] [PubMed] [Google Scholar]
- [24].Shearwin KE, Brumby AM, Egan JB. The tum protein of coliphage 186 is an antirepressor. J Biol Chem 1998; 273:5708-15; PMID:9488703; http://dx.doi.org/ 10.1074/jbc.273.10.5708 [DOI] [PubMed] [Google Scholar]
- [25].Liu T, Renberg SK, Haggard-Ljungquist E. Derepression of prophage P2 by satellite phage P4: Cloning of the P4 epsilon gene and identification of its product. J Virol 1997; 71:4502-8; PMID:9151842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu T, Renberg SK, Haggard-Ljungquist E. The E protein of satellite phage P4 acts as an anti-repressor by binding to the C protein of helper phage P2. Mol Microbiol 1998; 30:1041-50; PMID:9988480; http://dx.doi.org/ 10.1046/j.1365-2958.1998.01132.x [DOI] [PubMed] [Google Scholar]
- [27].Sauer B, Calendar R, Ljungquist E, Six E, Sunshine MG. Interaction of satellite phage P4 with phage 186 helper. Virology 1982; 116:523-34; PMID:6278725; http://dx.doi.org/ 10.1016/0042-6822(82)90145-3 [DOI] [PubMed] [Google Scholar]
- [28].Schnos M, Inman RB. Starting point and direction of replication in P2 DNA. J Mol Biol 1971; 55:31-8; PMID:4926974; http://dx.doi.org/ 10.1016/0022-2836(71)90278-6 [DOI] [PubMed] [Google Scholar]
- [29].Liu Y, Haggard-Ljungquist E. Studies of bacteriophage P2 DNA replication: Localization of the cleavage site of the A protein. Nucleic Acids Res 1994; 22:5204-10; PMID:7816607; http://dx.doi.org/ 10.1093/nar/22.24.5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Odegrip R, Haggard-Ljungquist E. The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage P2. J Mol Biol 2001; 308:147-63; PMID:11327759; http://dx.doi.org/ 10.1006/jmbi.2001.4607 [DOI] [PubMed] [Google Scholar]
- [31].Odegrip R, Schoen S, Haggard-Ljungquist E, Park K, Chattoraj DK. The interaction of bacteriophage P2 B protein with Escherichia coli DnaB helicase. J Virol 2000; 74:4057-63; PMID:10756017; http://dx.doi.org/ 10.1128/JVI.74.9.4057-4063.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rishovd S, Marvik OJ, Jacobsen E, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: Identification and characterization of the portal protein. Virology 1994; 200:744-51; PMID:8178458; http://dx.doi.org/ 10.1006/viro.1994.1238 [DOI] [PubMed] [Google Scholar]
- [33].Rishovd S, Holzenburg A, Johansen BV, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: Structure and function of the connector. Virology 1998; 245:11-7; PMID:9614863; http://dx.doi.org/ 10.1006/viro.1998.9153 [DOI] [PubMed] [Google Scholar]
- [34].Doan DNP, Dokland T. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J Struct Biol 2007; 157:432-6; PMID:17049269; http://dx.doi.org/ 10.1016/j.jsb.2006.08.009 [DOI] [PubMed] [Google Scholar]
- [35].Bowden DW, Calendar R. Maturation of bacteriophage P2 DNA in vitro: A complex, site-specific system for DNA cleavage. J Mol Biol 1979; 129:1-18; PMID:448732; http://dx.doi.org/ 10.1016/0022-2836(79)90055-X [DOI] [PubMed] [Google Scholar]
- [36].Pruss GJ, Wang JC, Calendar R. In vitro packaging of covalently closed circular monomers of bacteriophage DNA. J Mol Biol 1975; 98:465-78; PMID:1104873; http://dx.doi.org/ 10.1016/S0022-2836(75)80080-5 [DOI] [PubMed] [Google Scholar]
- [37].Bowden DW, Modrich P. In vitro maturation of circular bacteriophage P2 DNA. purification of ter components and characterization of the reaction. J Biol Chem 1985; 260:6999-7007; PMID:2987239 [PubMed] [Google Scholar]
- [38].Ziermann R, Calendar R. Characterization of the cos sites of bacteriophages P2 and P4. Gene 1990; 96:9-15; PMID:2265763; http://dx.doi.org/ 10.1016/0378-1119(90)90334-N [DOI] [PubMed] [Google Scholar]
- [39].Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology 2008; 370:352-61; PMID:17931675; http://dx.doi.org/ 10.1016/j.virol.2007.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rishovd S, Lindqvist B. Bacteriophage P2 and P4 morphogenesis: Protein processing and capsid size determination. Virology 1992; 187:548-54; PMID:1546453; http://dx.doi.org/ 10.1016/0042-6822(92)90457-Z [DOI] [PubMed] [Google Scholar]
- [41].Chang JR, Spilman MS, Rodenburg CM, Dokland T. Functional domains of the bacteriophage P2 scaffolding protein: Identification of residues involved in assembly and protease activity. Virology 2009; 384:144-50; PMID:19064277; http://dx.doi.org/ 10.1016/j.virol.2008.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pruss GJ, Calendar R. Maturation of bacteriophage P2 DNA. Virology 1978; 86:454-67; PMID:664241; http://dx.doi.org/ 10.1016/0042-6822(78)90085-5 [DOI] [PubMed] [Google Scholar]
- [43].Ziermann R, Bartlett B, Calendar R, Christie GE. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J Bacteriol 1994; 176:4974-84; PMID:8051010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].To KH, Dewey J, Weaver J, Park T, Young R. Functional analysis of a class I holin, P2 Y. J Bacteriol 2013; 195:1346-55; PMID:23335412; http://dx.doi.org/ 10.1128/JB.01986-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Markov D, Christie GE, Sauer B, Calendar R, Park T, Young R, Severinov K. P2 growth restriction on an rpoC mutant is suppressed by alleles of the Rz1 homolog lysC. J Bacteriol 2004; 186:4628-37; PMID:15231796; http://dx.doi.org/ 10.1128/JB.186.14.4628-4637.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Berry J, Summer EJ, Struck DK, Young R. The final step in the phage infection cycle: The Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol 2008; 70:341-51; PMID:18713319; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06408.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Maxwell KL, Fatehi Hassanabad M, Chang T, Pirani N, Bona D, Edwards AM, Davidson AR. Structural and functional studies of gpX of Escherichia coli phage P2 reveal a widespread role for LysM domains in the baseplates of contractile-tailed phages. J Bacteriol 2013; 195:5461-8; PMID:24097944; http://dx.doi.org/ 10.1128/JB.00805-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lengyel JA, Goldstein RN, Marsh M, Calendar R. Structure of the bacteriophage P2 tail. Virology 1974; 62:161-74; PMID:4607804; http://dx.doi.org/ 10.1016/0042-6822(74)90312-2 [DOI] [PubMed] [Google Scholar]
- [49].Linderoth NA, Julien B, Flick KE, Calendar R, Christie GE. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology 1994; 200:347-59; PMID:8178426; http://dx.doi.org/ 10.1006/viro.1994.1199 [DOI] [PubMed] [Google Scholar]
- [50].Lindahl G. On the control of transcription in bacteriophage P2. Virology 1971; 46:620-33; PMID:4944857; http://dx.doi.org/ 10.1016/0042-6822(71)90065-1 [DOI] [PubMed] [Google Scholar]
- [51].Sunshine M, Six E. Relief of P2 bacteriophage amber mutant polarity by the satellite bacteriophage P4. J Mol Biol 1976; 106:673-82; PMID:789895; http://dx.doi.org/ 10.1016/0022-2836(76)90258-8 [DOI] [PubMed] [Google Scholar]
- [52].Sunshine MG, Thorn M, Gibbs W, Calendar R, Kelly B. P2 phage amber mutants: Characterization by use of a polarity suppressor. Virology 1971; 46:691-702; PMID:4944860; http://dx.doi.org/ 10.1016/0042-6822(71)90071-7 [DOI] [PubMed] [Google Scholar]
- [53].Portelli R, Dodd IB, Xue Q, Egan JB. The late-expressed region of the temperate coliphage 186 genome. Virology 1998; 248:117-30; PMID:9705261; http://dx.doi.org/ 10.1006/viro.1998.9263 [DOI] [PubMed] [Google Scholar]
- [54].Yamashita E, Nakagawa A, Takahashi J, Tsunoda K, Yamada S, Takeda S. The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure. Acta Crystallogr Sect F Struct Biol Cryst Commun 2011; 67:837-41; PMID:21821878; http://dx.doi.org/ 10.1107/S1744309111005999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kageyama Y, Murayama M, Onodera T, Yamada S, Fukada H, Kudou M, Tsumoto K, Toyama Y, Kado S, Kubota K, et al.. Observation of the membrane binding activity and domain structure of gpV, which comprises the tail spike of bacteriophage P2. Biochemistry 2009; 48:10129-35; PMID:19780551; http://dx.doi.org/ 10.1021/bi900928n [DOI] [PubMed] [Google Scholar]
- [56].Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. Phage pierces the host cell membrane with the iron-loaded spike. Structure 2012; 20:326-39; PMID:22325780; http://dx.doi.org/ 10.1016/j.str.2011.12.009 [DOI] [PubMed] [Google Scholar]
- [57].Haggard-Ljungquist E, Jacobsen E, Rishovd S, Six EW, Nilssen O, Sunshine MG, Lindqvist BH, Kim KJ, Barreiro V, Koonin EV. Bacteriophage P2: Genes involved in baseplate assembly. Virology 1995; 213:109-21; PMID:7483254; http://dx.doi.org/ 10.1006/viro.1995.1551 [DOI] [PubMed] [Google Scholar]
- [58].Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol 2012; 726:93-114; PMID:22297511; http://dx.doi.org/ 10.1007/978-1-4614-0980-9_5 [DOI] [PubMed] [Google Scholar]
- [59].Haggard-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol 1992; 174:1462-77; PMID:1531648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Temple LM, Forsburg SL, Calendar R, Christie GE. Nucleotide sequence of the genes encoding the major tail sheath and tail tube proteins of bacteriophage P2. Virology 1991; 181:353-8; PMID:1825255; http://dx.doi.org/ 10.1016/0042-6822(91)90502-3 [DOI] [PubMed] [Google Scholar]
- [61].Christie GE, Temple LM, Bartlett BA, Goodwin TS. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J Bacteriol 2002; 184:6522-31; PMID:12426340; http://dx.doi.org/ 10.1128/JB.184.23.6522-6531.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu J, Hendrix RW, Duda RL. A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. J Mol Biol 2013; 425:3476-87; PMID:23851014; http://dx.doi.org/ 10.1016/j.jmb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Xu J, Hendrix RW, Duda RL. Chaperone–protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J Mol Biol 2014; 426:1004-18; PMID:23911548; http://dx.doi.org/ 10.1016/j.jmb.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kondou Y, Kitazawa D, Takeda S, Tsuchiya Y, Yamashita E, Mizuguchi M, Kawano K, Tsukihara T. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J Mol Biol 2005; 352:976-85; PMID:16125724; http://dx.doi.org/ 10.1016/j.jmb.2005.07.044 [DOI] [PubMed] [Google Scholar]
- [65].Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature 2002; 415:553-7; PMID:11823865; http://dx.doi.org/ 10.1038/415553a [DOI] [PubMed] [Google Scholar]
- [66].Christie GE, Calendar R. Bacteriophage P2 late promoters. transcription initiation sites for two late mRNAs. J Mol Biol 1983; 167:773-90; PMID:6308267; http://dx.doi.org/ 10.1016/S0022-2836(83)80110-7 [DOI] [PubMed] [Google Scholar]
- [67].Christie GE, Calendar R. Bacteriophage P2 late promoters. II. comparison of the four late promoter sequences. J Mol Biol 1985; 181:373-82; PMID:3981640; http://dx.doi.org/ 10.1016/0022-2836(85)90226-8 [DOI] [PubMed] [Google Scholar]
- [68].Christie GE, Anders DL, McAlister V, Goodwin TS, Julien B, Calendar R. Identification of upstream sequences essential for activation of a bacteriophage P2 late promoter. J Bacteriol 2003; 185:4609-14; PMID:12867472; http://dx.doi.org/ 10.1128/JB.185.15.4609-4614.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Grambow NJ, Birkeland NK, Anders DL, Christie GE. Deletion analysis of a bacteriophage P2 late promoter. Gene 1990; 95:9-15; PMID:2129530; http://dx.doi.org/ 10.1016/0378-1119(90)90407-I [DOI] [PubMed] [Google Scholar]
- [70].Lee TC, Christie GE. Purification and properties of the bacteriophage P2 ogr gene product. A prokaryotic zinc-binding transcriptional activator. J Biol Chem 1990; 265:7472-7; PMID:2185249 [PubMed] [Google Scholar]
- [71].Ayers DJ, Sunshine MG, Six EW, Christie GE. Mutations affecting two adjacent amino acid residues in the alpha subunit of RNA polymerase block transcriptional activation by the bacteriophage P2 ogr protein. J Bacteriol 1994; 176:7430-8; PMID:8002564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wood LF, Tszine NY, Christie GE. Activation of P2 late transcription by P2 ogr protein requires a discrete contact site on the C terminus of the alpha subunit of Escherichia coli RNA polymerase. J Mol Biol 1997; 274:1-7; PMID:9398509; http://dx.doi.org/ 10.1006/jmbi.1997.1390 [DOI] [PubMed] [Google Scholar]
- [73].Birkeland NK, Lindqvist BH, Christie GE. Control of bacteriophage P2 gene expression: Analysis of transcription of the ogr gene. J Bacteriol 1991; 173:6927-34; PMID:1938896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dibbens JA, Gregory SL, Egan JB. Control of gene expression in the temperate coliphage 186. X. the cI repressor directly represses transcription of the late control gene B. Mol Microbiol 1992; 6:2643-50; PMID:1447973; http://dx.doi.org/ 10.1111/j.1365-2958.1992.tb01441.x [DOI] [PubMed] [Google Scholar]
- [75].Lindahl G, Sironi G, Bialy H, Calendar R. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc Natl Acad Sci U S A 1970; 66:587-94; PMID:4913204; http://dx.doi.org/ 10.1073/pnas.66.3.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bregegere F. Bacteriophage P2-lambda interference: Inhibition of protein synthesis involves transfer RNA inactivation. J Mol Biol 1974; 90:459-67; PMID:4615170; http://dx.doi.org/ 10.1016/0022-2836(74)90228-9 [DOI] [PubMed] [Google Scholar]
- [77].Myung H, Calendar R. The old exonuclease of bacteriophage P2. J Bacteriol 1995; 177:497-501; PMID:7836278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Smith HS, Pizer LI, Pylkas L, Lederberg S. Abortive infection of Shigella dysenteriae P2 by T2 bacteriophage. J Virol 1969; 4:162-8; PMID: 4896823; PMID:4896823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mosig G, Yu S, Myung H, Haggard-Ljungquist E, Davenport L, Carlson K, Calendar R. A novel mechanism of virus-virus interactions: Bacteriophage P2 tin protein inhibits phage T4 DNA synthesis by poisoning the T4 single- stranded DNA binding protein, gp32. Virology 1997; 230:72-81; PMID:9126263; http://dx.doi.org/ 10.1006/viro.1997.8464 [DOI] [PubMed] [Google Scholar]
- [80].Bertani LE. Lysogenic conversion by bacteriophage P2 resulting in an increased sensitivity of Escherichia coli to 5-fluorodeoxyuridine. Biochim Biophys Acta 1964; 87:631-40; PMID:14220693 [DOI] [PubMed] [Google Scholar]
- [81].Calendar R, Yu S, Myung H, Barreiro V, Odegrip R, Carlson K, Davenport L, Mosig G, Christie GE, Haggård-Ljungquist E. The lysogenic conversion genes of coliphage P2 have unusually high AT content In: Syvanen M., Cado K., ed. Horizontal Gene Transfer. London: Chapman and Hall, 1998:241-252. [Google Scholar]
- [82].Bertani LE. Characterization of clear mutants belonging to the Z gene of bacteriophage P2. Virology 1976; 71:85-96; PMID:775766; http://dx.doi.org/ 10.1016/0042-6822(76)90096-9 [DOI] [PubMed] [Google Scholar]
- [83].Nilsson AS, Karlsson JL, Haggard-Ljungquist E. Site-specific recombination links the evolution of P2-like coliphages and pathogenic enterobacteria. Mol Biol Evol 2004; 21:1-13; PMID:12949155; http://dx.doi.org/ 10.1093/molbev/msg223 [DOI] [PubMed] [Google Scholar]
- [84].Mirold S, Rabsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt WD. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc Natl Acad Sci U S A 1999; 96:9845-50; PMID:10449782; http://dx.doi.org/ 10.1073/pnas.96.17.9845 [DOI] [PMC free article] [PubMed] [Google Scholar]