Abstract
Assembly of HIV-1 particles is initiated by the trafficking of viral Gag polyproteins from the cytoplasm to the plasma membrane (PM), where they co-localize and bud to form immature particles. Membrane targeting is mediated by the N-terminally myristoylated matrix (MA) domain of Gag and is dependent on the PM marker phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]. Recent studies revealed that PI(4,5)P2 molecules containing truncated acyl chains [tr-PI(4,5)P2] are capable of binding MA in an “extended lipid” conformation and promoting myristoyl exposure. Here we report that tr-PI(4,5)P2 molecules also readily bind to non-membrane proteins, including HIV-1 capsid (CA), which prompted us to re-examine MA-PI(4,5)P2 interactions using native lipids and membrane mimetic liposomes and bicelles. Liposome binding trends observed using a recently developed NMR approach paralleled results of flotation assays, although the affinities measured under the equilibrium conditions of NMR experiments were significantly higher. Native PI(4,5)P2 enhanced MA binding to liposomes designed to mimic non-raft-like regions of the membrane, supporting proposals that Gag binding may nucleate raft formation. Studies with bicelles revealed a subset of surface and myr-associated MA residues that are sensitive to native PI(4,5)P2, but cleft residues that interact with the 2′-acyl chains of tr-PI(4,5)P2 molecules in aqueous solution were insensitive to native PI(4,5)P2 in bicelles. Our findings call to question extended-lipid MA:membrane binding models, and instead support a model put forward from coarse-grained simulations indicating that binding is mediated predominantly by dynamic, electrostatic interactions between conserved basic residues of MA and multiple PI(4,5)P2 and phosphatidylserine molecules.
Graphical Abstract
Introduction
Assembly of human immunodeficiency virus (HIV) particles is initiated by the cytoplasmic trafficking of viral genomes and virally encoded Gag proteins to assembly sites on the plasma membrane (PM). Genomes are initially anchored to the PM by a small number of Gag proteins (~12 copies or fewer) [1–4], where they diffuse laterally and stochastically, then collide to form dimers [5] and become fixed at sites where additional Gag proteins bind [1–6]. Proteins of the cellular protein sorting machinery are also recruited to assembly sites to promote budding [3,7,8]. Trafficking to the PM is mediated by Gag’s N-terminal matrix domain (MA), which contains a bipartite targeting signal comprising an N-terminal myristoyl group and a highly basic region (HBR). Mutations that either block myristoylation or lead to acylation by unsaturated fatty acids can disrupt trafficking [9], and mutation of conserved HBR residues can cause retargeting of Gag to intracellular compartments [10–12]. Both types of modification inhibit virus production [9–12].
Membrane targeting is also dependent on phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] [10], a member of a family of differentially phosphorylated phosphoinositidyl phosphates (PIPs) that normally function in the recruitment of peripheral membrane proteins to specific organelles [13]. PI(4,5)P2 is considered a major landmark for the inner leaflet of the PM [13]. Induction of enzymes that reduce PM PI(4,5)P2 levels can inhibit Gag binding to the PM and attenuate virus production, whereas induction of enzymes that enhance PI(4,5)P2 levels in endosomal membranes can retarget virus assembly to endosomes [10]. PI(4,5)P2-dependent membrane targeting and/or virus assembly has also been observed for other retroviruses, including the evolutionarily distant Moloney murine leukemia virus and Mason-Pfizer monkey virus (which assembles in the cytoplasm prior to PM binding) [14–17], suggesting that the ability of retroviruses to exploit the PI(4,5)P2 signaling system is evolutionarily conserved.
The inner leaflet of the PM contains other lipid constituents that are enriched in the HIV-1 envelope relative to the PM of the producer cells [including cholesterol, PI(4,5)P2, phosphatidylserine (PS), sphingomyelins, plasmalogen-phosphatidylethanolamine (pl-PE), and saturated fatty acids], indicating that the virus either assembles at sites enriched in these constituents or concentrates them during virus assembly [18–21] (reviewed in:[20,22–25]). These lipid constituents, as well as several specific cellular proteins, are enriched in PM puncta and have been historically referred to as “rafts” or “raft-like microdomains”. By definition, rafts comprise a heterogeneous collection of sterol- and sphingolipid-enriched domains that differ in protein and lipid composition, as well as in temporal stability [26,27]. Studies indicate that the envelope of HIV-1 exists in a liquid-ordered-like state [28], an historical hallmark of rafts [26,27,29,30]. Although raft-like domains are typically enriched with acidic lipids, assembly of HIV-1 Gag proteins at rafts is not due strictly to electrostatic interactions, since cholesterol depletion significantly attenuates virus particle production [31,32]. Some studies indicate that PI(4,5)P2 associates preferentially with lipid rafts [33], but others suggest that PI(4,5)P2 is homogeneously dispersed within the PM of quiescent cells and only co-localizes with lipid rafts upon cellular stimulation [34]. Thus, Gag may either assemble at pre-existing raft-like domains (i.e. domains enriched in cholesterol and acidic lipids), or enrich PI(4,5)P2, cholesterol, and other membrane constituents as it assembles at the PM.
A number of in vitro biophysical methods have been employed in attempts to identify the mechanism utilized by retroviruses to target the PM and acquire a raft-like bilayer coat. Flotation experiments have shown that retroviral Gag constructs bind preferentially to liposomes that contain PI(4,5)P2 and are enriched in both cholesterol and acidic phospholipids [35–38]. NMR and analytical ultracentrifugation studies revealed that HIV-1 MA exists in solution in an equilibrium mixture of myristoyl-sequestered and -exposed states, and that soluble PI(4,5)P2 analogs containing truncated acyl chains [tr-PI(4,5)P2] are capable of binding MA with relatively weak affinities (Kd = 0.08 – 0.15 mM) and promoting myristoyl exposure [39]. NMR studies further showed that tr-PI(4,5)P2 molecules bind in an “extended lipid” conformation, in which the 2′-acyl chain binds to a hydrophobic cleft on MA and the 1′-acyl chain exists in a solvated, unstructured state that could potentially interact with a membrane [39]. Protein foot printing experiments conducted with MA and tr-PI(4,5)P2 are consistent with the NMR-derived structures [40]. The extended lipid-binding model (with one acyl chain embedded in the membrane and the other sequestered by the protein) was originally proposed to explain phospholipid-cytochrome c binding [41–43], and computational studies suggested that this binding mode might be preferred under conditions of negative membrane curvature [41,43,44]. Two other lipid analogs that contain truncated acyl chains were also found to bind MA in solution in an extended lipid conformation, and a “trio engagement” model involving MA interactions with PI(4,5)P2, PS, and PE in extended lipid states was proposed [45]. Recent coarse grained (CG) simulations indicated that extrusion of the seven buried methylenes from the hydrophobic pocket of MA for membrane binding is energetically feasible, costing ~ 8 kCal/mol [46]. However, extrusion of the 20-carbon 2′-acyl chain of PI(4,5)P2 from the membrane is predicted to be significantly more costly (~ 24 kCal/mol [47,48]), and during the entire course of the CG simulations, both acyl chains of PI(4,5)P2 remained buried within the membrane and did not interact with MA [46].
Since the previously determined MA:lipid NMR structures were determined from data obtained in the absence of membranes and using water-soluble PI(4,5)P2 analogs with truncated acyl chains [14,39,40,45,49], the biological relevance of the findings warrants further investigation. In addition, most previous studies of MA interactions with membranes were conducted using flotation assays that required the use of liposomes with high anionic charge and non-equilibrium centrifugation approaches to distinguish membrane-free and -bound MA proteins [35–38,50]. In this study, we employed NMR-based approaches to monitor the influence of PI(4,5)P2 on MA binding to liposomes with biological-like raft and non-raft compositions. We also re-examined the extended lipid hypothesis by characterizing MA interactions with bicelles that contain PI(4,5)P2 molecules with native, full-length acyl chains.
Results
tr-PI(4,5)P2 binds to non-membrane proteins
Previous studies have shown that amphipathic dodecyl-containing detergents can readily interact with hydrophobic surface patches on soluble proteins [51]. We were therefore concerned that truncated analogs of PI(4,5)P2 might also bind to non-membrane associated proteins with hydrophobic surface patches. 2D 1H-15N HSQC spectra were obtained upon titration of diC8-PI(4,5)P2 with three autonomously folded Gag domains: the native N- and C-terminal subdomains of CA (CANTD, and CACTD, respectively), and the nucleocapsid (NC) domain, none of which are believed to interact directly with membrane surfaces during replication[11,50,52,53]. As shown in Supplementary Fig. S1, diC8-PI(4,5)P2 interacts with all three domains. Notably, the affinity of diC8-PI(4,5)P2 for sites on CANTD (Kd as low as 0.25 ± 03 mM) are comparable to the affinity of diC4-PI(4,5)P2 for MA (Kd = 0.15 ± 0.03 mM). Similar results were obtained upon titration of CANTD with 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), a lipid analog that contains 6-carbon acyl chains (Fig. S1d). Thus, the binding mode and affinity of tr-PI(4,5)P2 molecules for MA, and the proposed “extended lipid” binding models proposed to explain PM and raft targeting by the MA domain of Gag [14,39,45,54], may be a consequence of the general affinity of moderately water soluble amphipathic molecules, with hydrophobic acyl chains, for protein surface patches [51].
MA binding to liposomes is enhanced by native PI(4,5)P2
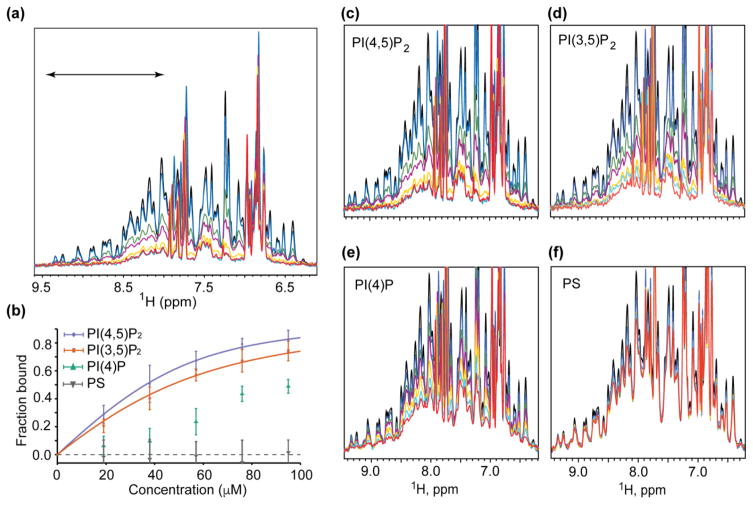
We employed a recently developed NMR approach [55,56] to monitor protein binding to liposomes. This approach allows measurement of the amount of protein that remains free in solution in the presence of liposomes under equilibrium conditions, and is based on the fact that NMR signals are readily detectable for proteins the size of HIV-1 MA (~15kDa) but generally undetectable (or “dark” [55]) for proteins and protein complexes as large as liposomes (>60MDa). Provided that on/off membrane exchange is intermediate-to-slow on the NMR chemical shift time scale, binding to liposomes is expected to render all but the highly flexible MA residues NMR-invisible [55,56]. As shown in Fig. 1a, small signal losses (13%) were observed upon titration of MA with liposomes containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, or here-in abbreviated PC). Titration of MA with PC-liposomes containing increasing amounts of PI(4,5)P2 resulted in increased signal attenuation, indicating that PI(4,5)P2 enhances the affinity of MA for liposomes. Binding isotherms were analyzed by applying non-linear least squares fitting to a single-site binding model, which afforded an apparent MA:PI(4,5)P2 dissociation constant [Kd(apparent)] of 10.2 ± 2.1 μM (Fig. 1b). By comparison, no signal losses were observed upon titration of HIV-1 CA and NC with liposomes, including those prepared with native PI(4,5)P2 (Fig. S2).
Figure 1.
Influence of lipid constituents on MA binding to liposomes. (a) HIV-1 MA binds with increasing affinity to PC liposomes containing increasing molar percentages of PI(4,5)P2: black, free MA in solution; blue, MA + PC liposomes; green, MA + PC-1%PI(4,5)P2; purple, MA + PC-2%PI(4,5)P2; yellow, MA + PC-3%PI(4,5)P2; cyan, MA + PC-4%PI(4,5)P2; red, MA + PC-5%PI(4,5)P2. (b) The spectral region between 9.5 and 8.0 ppm, corresponding to amide signals of structured residues and indicated in (a), was integrated and used to calculate Kd(apparent) binding affinities. (c–f) 1H NMR spectra of MA free in solution (black) and in the presence of PC liposomes in the absence (blue) and presence of increasing amounts of lipid constituents (1 mol%, green; 2 mol%, purple; 3 mol%, yellow; 4 mol%, cyan; and 5 mol%, red).
NMR titrations were conducted with liposomes containing increasing mole percentages of phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] (enriched in late endosomes) and phosphatidylinositol 4-phosphate [PI(4)P] (enriched in Golgi membranes) (Fig. 1b,c,d). PI(3,5)P2 promoted liposome binding about half as well as PI(4,5)P2 [Kd(apparent) = 22.0 ± 1.4 μM], whereas PI(4)P exhibited a more significantly reduced ability to promote MA binding to liposomes [too weak for reliable quantitation], Fig. 1b,e. At similar concentrations, PS was even less effective at promoting liposome binding than the diphosphoinositides, Fig. 1b,f.
Mutations and conditions that modulate myristoyl exposure affect liposome binding
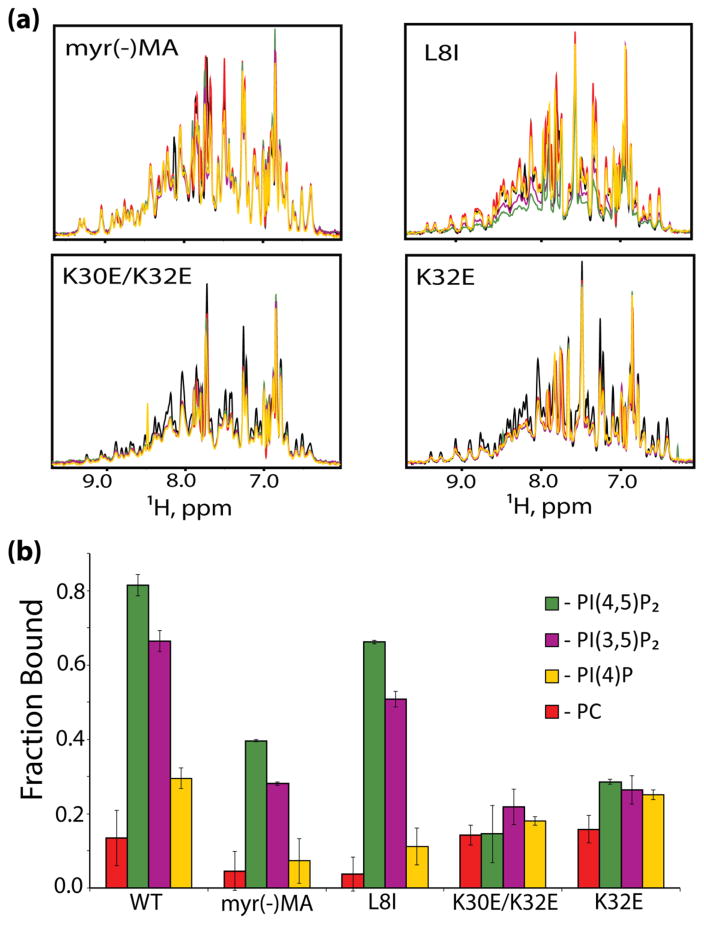
To determine the influence of the N-terminal myristoyl group on liposome binding, we prepared and analyzed MA proteins that either lacked the myristoyl group [myr(−)MA] or contained a mutation (leucine-8 to isoleucine, MAL8I) known to inhibit both myristoyl exposure in vitro and PM targeting in vivo [11,57–59]. As shown in Fig. 2, signal losses upon titration of myr(−)MA with PC liposomes were very small (nearly imperceptible), indicating that ~5% of the proteins were bound to liposomes under the conditions employed. However, myr(−)MA exhibited significant affinity for liposomes containing PI(4,5)P2 (2.5 mol%) or PI(3,5)P2 (2.5 mol%) (40% and 28% myr(−)MA bound, respectively) (Fig. 2b). Similarly, MAL8I bound poorly to PC liposomes, but binding was significantly restored when the liposomes contained PI(4,5)P2 or PI(3,5)P2 (67% and 50% bound, respectively) (Fig. 2b). These results suggest that the myristoyl group contributes to initial membrane association, but is not required for specific recognition and discrimination of phosphoinositidyl phosphates.
Figure 2.
HIV-1 MA mutants do not efficiently bind PI presenting liposomes. (a) 1H NMR spectra of HIV-1 MA mutants in the presence of liposomes containing 2.5 mol% of the different PIs. Colors represent MA alone, black; PC-liposomes, red; PI(4)P, yellow; PI(3,5)P2, purple; PI(4,5)P2, green. (b) Fraction of bound MA mutants in the presence of liposomes with various PIs.
Previous NMR studies showed that myristoyl exposure is enhanced under acidic conditions [60], presumably due to the stabilization of a salt bridge between the protonated side chain of H89 and the acidic side chain of E12 [61,62]. Other studies have indicated that MA binds preferentially to membranes containing lipids with unsaturated acyl chains, which is also compatible with the extended lipid binding models [37]. To test both of these hypotheses, we measured MA binding to liposomes comprising either PC or the fully saturated lipid, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), at pH 5.5 and pH 7.4 (T = 35 °C). We observe that at physiological pH MA bound weakly to both liposomes. However, the affinity of MA for both liposomes was significantly greater (~3-fold) at the lower pH (Fig. S3), consistent with an equilibrium shift towards the myristoyl-exposed species.
Influence of conserved basic residues on liposome binding and virus infectivity
Substitution of conserved HBR residues K30 and K32 by glutamate can result in the retargeting of Gag to intracellular compartments [12,63]. To test the hypothesis that retargeting is due to a gain of affinity for PI(3,5)P2 or PI(4)P, which are the most abundant PIs in late endosomal and Golgi membranes, respectively, we prepared the myristoylated double mutant MA protein (MAK30E/K32E) and measured liposome binding in the presence of physiologically relevant levels of PIs (Fig. 2). As observed for wild-type MA, a small reduction in signal intensity was observed upon titration with PC liposomes, but no additional significant losses were observed for liposomes containing PIs (Fig. 2a). The native, myristoylated MA protein exhibits concentration-dependent myristoyl exposure, presumably stabilized by intermolecular myristoyl-myristoyl interactions [61]. The HBR mutant MA proteins also exhibited concentration-dependent shifts, indicating that the binding defect is not due to a defect in myristoyl exposure. K32 is conserved in over 99.68% of the 4,376 sequenced strains of HIV, whereas K30 is conserved in 27.2% (with 44.9% Arg) (http://www.hiv.lanl.gov/). We therefore constructed a single K32E mutant (MAK32E) and examined its ability to bind liposomes. As shown in Fig. 2, MAK32E exhibits significantly reduced affinity for liposomes and does not discriminate between PIs under the conditions tested, indicating that K32 is a key contributor to both PI(4,5)P2 discrimination and membrane binding.
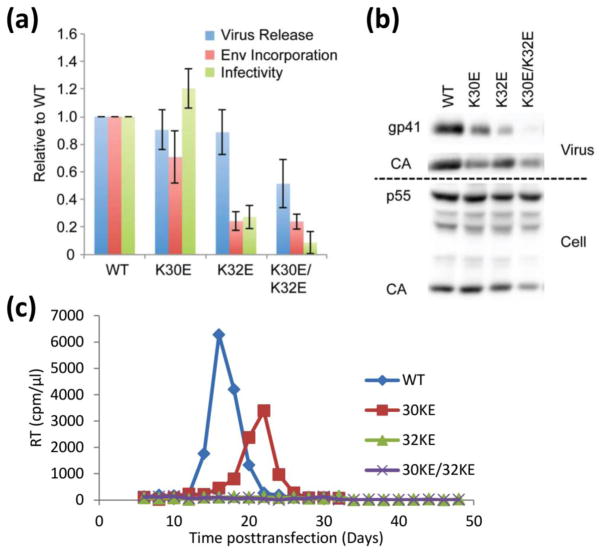
The influence of the HBR mutations on virus production, Env incorporation, virus infectivity, and virus replication was also measured. Viruses containing the Gag-K30E/K32E, Gag-K32E, and Gag-K30E mutants were transfected into HeLa cells, and the efficiency of virus release and Env incorporation was determined by quantitative western blotting (Fig. 3a, b). The infectivity of the released virus particles was also measured in the single-cycle TZM-bl indicator cell assay. Finally, the replication kinetics of the mutants were determined in the Jurkat T-cell line. The K30E mutant exhibited near wild-type viral release, Env incorporation and infectivity, and only modestly delayed replication kinetics (Fig. 3c). In contrast, the K32E mutant displayed severely attenuated Env incorporation and viral infectivity, and even though virus release was only moderately reduced, no replication was discernable. The Gag-K30E/K32E double mutant showed severely abrogated Env incorporation and infectivity, significantly reduced virus released efficiency, and a complete loss of replication, consistent with earlier findings [64]. These results demonstrate that both K30 and K32 are required for PI(4,5)P2 binding and virus release, with K32 apparently playing a more significant role.
Figure 3.
Mutation of MA residues 30 and 32 disrupts HIV-1 particle production, Env glycoprotein incorporation, and particle infectivity. (a, b) HeLa cells were transfected with the indicated molecular clones. Virus and cell lysates were harvested at 48 h, subjected to SDS-PAGE and western blotting with antibodies specific for gp41 and Gag. Single-cycle infectivity was determined on the TZM-bl indicator cell line. Data are presented as luciferase activity normalized for viral input (luciferase signal/RT activity). Densitometries were determined by quantitative western blotting, and virus release calculated as virus CA/(cell p55 + cell CA + virus CA). Env incorporation was determined by calculating the ratio of virion-associated gp41 to CA. (c) The Jurkat T-cell line was transfected with the indicated molecular clones. Aliquots of supernatant were harvested every 2–3 days for RT analysis and the cells were diluted 1/3 in fresh medium.
Influence of other lipid constituents on liposome binding
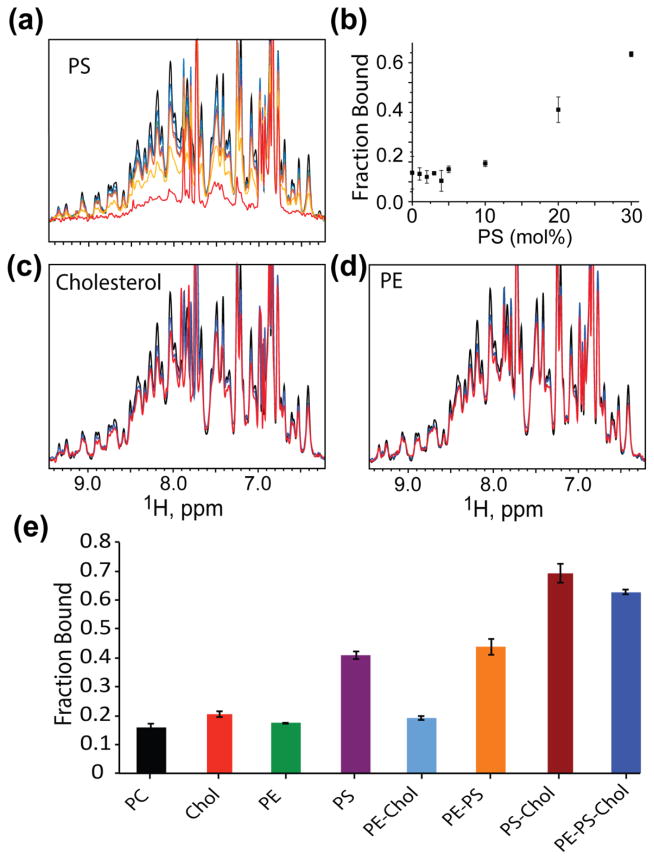
The HIV-1 envelope is enriched in lipids commonly associated with raft-like microdomains, and there is considerable evidence that HIV-1 Gag proteins co-localize and assemble at rafts [31,65,66]. To determine the potential influences of other lipid constituents on membrane binding, we evaluated MA interactions with liposomes containing cholesterol, PE, PS, and PI(4,5)P2. Although PS was relatively ineffective at promoting binding to liposomes at low concentrations (~ 5 mol%; Fig. 1f), the affinity of MA for liposomes increased from ~15% (100% PC liposomes) to ~40% for liposomes that contained 16 mol% PS, Fig. 4. Neither cholesterol nor PE had significant effects on liposome binding when present either individually or together at physiological molar concentrations (up to 44 mol%, and 8 mol%, respectively) (Fig. 4e), and PE did not appear to influence binding when included in liposomes in combination with other lipid constituents (Fig. 4e). However, liposomes that contained both cholesterol and PS bound MA with significantly greater affinity than liposomes containing only PS or cholesterol (chol) (70% bound for PC:Chol:PS = 40:44:16 mol%, Fig. 4e). These findings support proposals that cholesterol may indirectly enhance the ability of PS to bind MA [37]. Since PS contains a polar head group and cholesterol does not (and is generally believed to be buried within the bilayer), enhanced binding could be due to an increase in surface charge density resulting from an increase in the PS/PC ratio.
Figure 4.
Titration of other luminal PM lipids, at physiologically relevant concentrations. (a) Titration of PS at and above physiological concentrations. Colors correspond to MA alone, black; PC-liposomes, blue; 1% PS, green; 5%, purple; 10%, orange; 20% yellow; and 30%, red. (b) Fraction bound of MA in the presence of increasing mol% of PS. (c) and (d) Liposomes containing cholesterol (44 mol%), red; PE (8.36 mol%), red, respectively. Black and blue represent MA alone and PC-liposomes. (e) Bar graphs showing the fraction of MA bound in the presence of liposomes containing different lipid combinations. Percent bound represent liposomes (3.8 mM total lipid concentration) with mixtures of 100% PC, 56.0% PC: 44.0%Chol, 91.6% PC: 8.4% PE, 83.6% PC: 16.4% PS, 47.6% PC: 44.0% Chol: 8.4% PE, 75.2% PC: 8.4% PE: 16.4% PS, 39.6% PC: 44.0% Chol: 16.4% PS, 31.2% PC: 44.0% Chol: 8.4% PE: 16.4% PS.
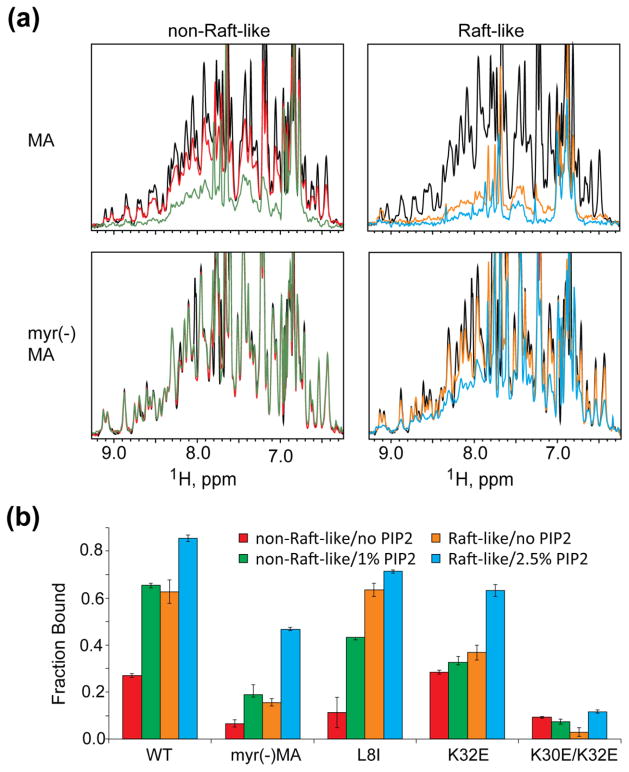
In addition to examining the role of individual lipid constituents on MA binding to PC liposomes, we measured binding to liposomes that contained combinations of lipid constituents at ratios approaching raft-like and non-raft-like regions of the PM inner leaflet (raft-like liposomes = 44 mol% cholesterol, 8 mol% PE, ~3 mol% PI(4,5)P2, 16 mol% PS, 29% PC; non-raft-like liposomes = 17% cholesterol, 6% PE, 1% PI(4,5)P2, 6% PS, 70% PC) [18,19]. (Note: in this work, cholesterol is included as a lipid when calculating mol percentages; see Table S1 for details). A caveat is that the liposomes lack pl-PE, an inner-leaflet PM constituent (~32 mol% abundance), as this lipid is known to induce liposome curvature [67] and is not generally employed in model membrane systems [68]. Thus, the liposomes employed in the present studies contained phosphatidylcholine (PC) as a substitute for pl-PE. Titrations were conducted in the absence and presence of PI(4,5)P2. Unexpectedly, MA exhibited significant affinity for raft-like liposomes, even in the absence of PI(4,5)P2 (Fig. 5). Although MA exhibited very low affinity for non-raft liposomes that lacked PI(4,5)P2 (as expected; see Fig. S3), significant binding was observed to non-raft-like liposomes that contained PI(4,5)P2 at levels expected for non-raft regions of the PM inner leaflet (Fig. 5). These findings suggest that, although HIV-1 virions contain raft-like lipid bilayers, Gag could bind initially to non-raft regions of the PM and either laterally associate with rafts or induce formation of local raft-like structures.
Figure 5.
MA binds with higher affinity to raft-like liposomes. (a) 1H NMR Spectra of HIV-1 MA and myr(−)MA in the presence of non-raft-like and raft-like liposomes. Colors correspond to protein alone (black); non-raft-like liposomes with no PI(4,5)P2 (red) or with 1mol% PI(4,5)P2 (green), and raft-like liposomes with no PI(4,5)P2 (orange) or 2.5mol% PI(4,5)P2 (cyan). (b) Fraction of MA mutants bound to liposomes containing different membrane compositions.
We also determined the relative abilities of HBR and switch-defective mutant MA proteins to bind raft-like and non-raft membrane mimetic liposomes. As observed for wild type MA, MAL8I bound tightly to raft-like liposomes in the absence of PI(4,5)P2, (Fig. 5b). However, this mutant did not exhibit significant affinity for non-raft liposomes, even upon addition of PI(4,5)P2 (Fig. 5b). Myr(−)MA did not significantly bind raft-like liposomes in the absence of PI(4,5)P2, however, PI(4,5)P2 presenting raft-like liposomes were able to significantly enhance liposome binding (Fig. 5b). In contrast, myr(−)MA did not exhibit significant affinity for non-raft liposomes, either in the absence or presence of PI(4,5)P2 (Fig. 5b). The MAK30E/K32E double mutant did not exhibit significant affinity for any of the liposome compositions tested (Fig. 5b). The HBR mutant MAK32E retained an ability to bind raft-like membranes (36%) and experienced a 1.7-fold increase in affinity in the presence of PI(4,5)P2. Interestingly, MAK32E bound with wild type affinity to non-raft like liposomes, and addition of PI(4,5)P2 to liposomes that otherwise have non-raft like compositions did not lead to a significant increase in protein binding (Fig. 5b). These results collectively indicate that (i) the ability to expose the myristoyl group is essential for association with either raft-like or non-raft like membranes, and (ii) K32 plays a significant role in PI(4,5)P2-dependent enhancement of membrane binding.
MA interactions with bicelles
To gain insight into the intermolecular interactions and potential structural changes in MA that occur upon membrane binding, we initiated NMR studies with lipid bicelles. Bicelles have been employed to study a variety of membrane-associated proteins in their active forms [69–72]. Vlach and Saad showed that titration of HIV-1 MA with 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) micelles and DMPC/DHPC bicelles caused NMR chemical shift perturbations consistent with membrane-promoted exposure of the myristoyl group [61]. Unfortunately, global signal broadening precluded further analysis of the MA:bicelle complex. Those studies were conducted with bicelles with long chain (DMPC)/short chain (DHPC) lipid a ratio (q) of 0.5 [45]. We conducted titration experiments a higher DHPC-to-DMPC ratio, q = 0.3 bicelles, which has afforded high quality NMR spectra for other membrane-protein systems [73,74]. In addition, since previous studies showed that HIV-1 MA myristoyl exposure is favored at lower pH [61,62], the bicelle titration studies in the present study were conducted at pH 5.5 (which also enhanced MA binding to liposomes, Fig. S3).
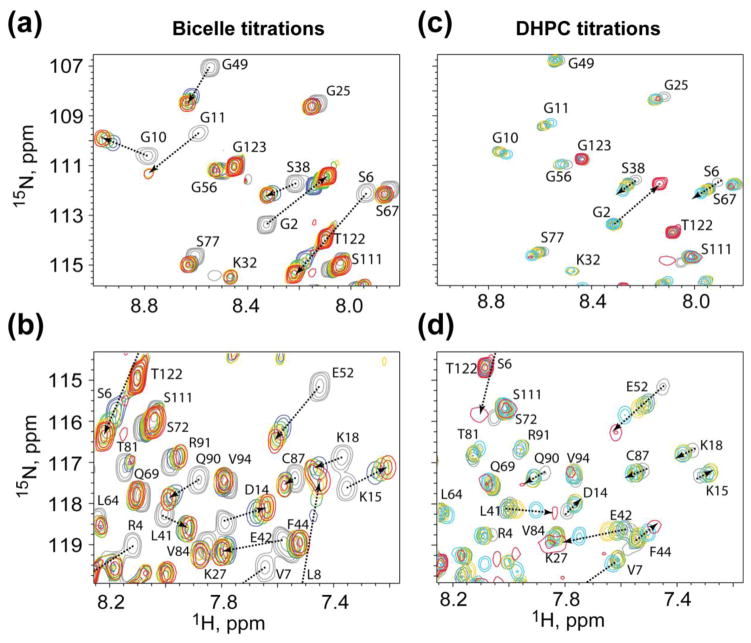
Titration of pre-formed bicelles into MA at lipid concentrations above the critical micelle concentration (CMC) of DHPC resulted in the shifting of amide 1H and 15N NMR signals of N-terminal residues (G2–G11) to values similar to those of the myristoyl-exposed protein conformer [61], indicating that MA binds to the bicelles under the conditions employed, and that binding occurs via the myristoyl-exposed state, Fig. 6a [61]. Significant chemical shift perturbations were also observed for additional residues that are on the surface of MA that were not previously observed to be sensitive to the position of the myristoyl-exposed/sequestered equilibrium [61], Fig. 6b. To determine if these additional perturbations are due to interactions with the short-chain lipid of the DMPC/DHPC bicelles, titrations were conducted with DHPC alone. Interestingly, at concentrations below the DHPC CMC (15 mM) DHPC did not perturb the chemical shifts of the N-terminal residues, indicating that monomeric DHPC does not trigger myristoyl exposure, Fig. 6c. Chemical shift changes indicative of myristoyl exposure were only observed after DHPC concentrations exceeded the CMC (Fig. 6c, red signals). In contrast, the amide signals for the surface residues that are insensitive to the myristoyl switch but exhibited sensitivity to titration with bicelles exhibited significant chemical shift changes at DHPC concentrations well below the CMC, Fig. 6d. Analysis of concentration-dependent chemical shifts afforded a DHPC dissociation constant of 3.8 ± 2.9 mM. Thus, the perturbations observed for these residues in the bicelle titration experiments are likely due to weak interactions with soluble DHPC molecules and do not result from the binding of MA to bicelles or DHPC micelles.
Figure 6.
Titrations of bicelles or micelles promote myristoyl exposure, whereas titrations of monomeric DHPC lipids at sub-CMC concentrations do not. Representative 1H-15N HSQC data showing titration of (a and b) bicelles, or (c and d) soluble DHPC, into MA. Colors correspond to bicelles:MA 0:1, grey; 1:1, blue; 2:1, green; 4:1, yellow; 8:1, red; DHPC:MA 0:1, grey; 8:1, blue; 16:1, green; 32:1, yellow; 64:1, cyan; 256:1, red.
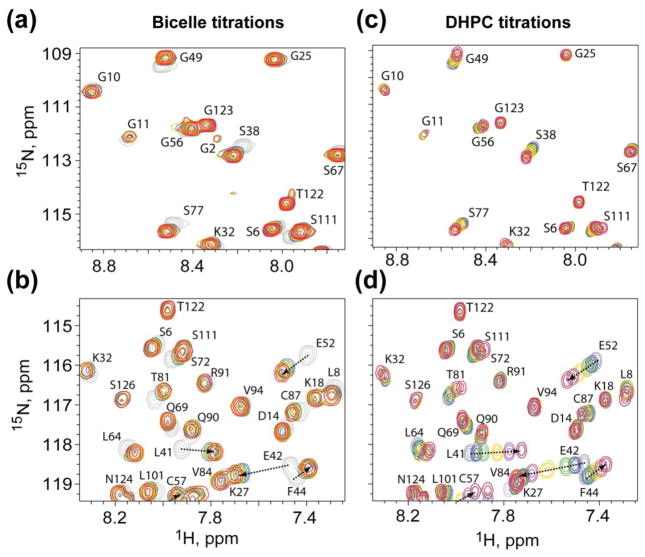
Bicelle and DHPC titration experiments were also conducted with the unmyristoylated matrix protein to determine the influence of the myristoyl moiety on bicelle and DHPC binding. Titration with bicelles did not lead to significant perturbations of the myristoyl switch signals, but did lead to large shifts of the same surface residue signals that were perturbed upon titration of the MA protein with bicelles, Fig. 7a,b. In addition, titration of myr(−)MA with DHPC led to progressive amide chemical shift changes similar to those observed for the MA protein (Fig 7c,d), except that only minor signal broadening was observed at DHPC concentrations that approached or exceeded the CMC (Fig 7c,d, red signals). These findings confirmed that DHPC molecules bind weakly to the surface of myr(−)MA (Kd = 6.4 ± 2.5 mM), and suggest that the protein does not bind with significant affinity to DHPC micelles.
Figure 7.
Titration of bicelles and DHPC do not perturb N-terminal residues for myr(−)MA. Representative 1H-15N HSQC data showing titration of (a and b) bicelles, or (c and d) soluble DHPC, into myr(−)MA. Colors correspond to bicelles:myr(−)MA 0:1, grey; 1:1, blue; 2:1, green; 4:1, yellow; 8:1, red; DHPC:myr(−)MA 0:1, grey; 8:1, blue; 16:1, green; 32:1, yellow; 128:1, purple; 256:1, red.
Finally, we conducted 15N NMR relaxation experiments (600.13 MHz 1H frequency) to determine if the signal broadening observed in the bicelle titration experiments was actually due to binding to bicelles (rather than intermediate-exchange interactions with soluble DHPC). The 15N R1 and R2 values, and the ratio of the average R2/R1 values, for the structured residues of myr(−)MA (residues 20–100; 1.52 s−1, 9.06 s−1, and 5.96, respectively) are consistent with expectations for a 15 kDa protein [75–77], Fig. S4. The average R2/R1 ratio exhibited a small increase in the presence of bicelles (6.96, myr(−)MA:bicelle = 1:2), providing further evidence that myr(−)MA does not bind with significant affinity to bicelles under the conditions employed, and that the chemical shift changes observed upon bicelle titration are due to weak interactions with soluble DHPC. Previous NMR and sedimentation studies revealed that MA weakly self-associates into trimers and higher-order species at concentrations above 0.2 mM [61]. 15N NMR R1 and R2 measurements for MA were therefore made at relatively low protein concentrations (0.05 mM), in which the protein is predominantly monomeric and the measured R1 and R2 rates, and the average R2/R1 ratio (1.48 s−1, 9.70 s−1, and 6.56, respectively) are similar to those observed for the myr(−)MA protein. Unlike observations for the myr(−)MA protein, addition of bicelles (to a MA:bicelle ratio of 1:2) resulted in significant increases in R1 and R2 and to a 200% increase in the average R2/R1 ratio (1.28 s−1, 17.11 s−1, and 13.38, respectively), Fig. S4. These data provide further evidence that the NMR chemical shift changes described above for the MA protein are at least partly due to binding to bicelles.
Influence of native phosphoinositides on MA binding to bicelles
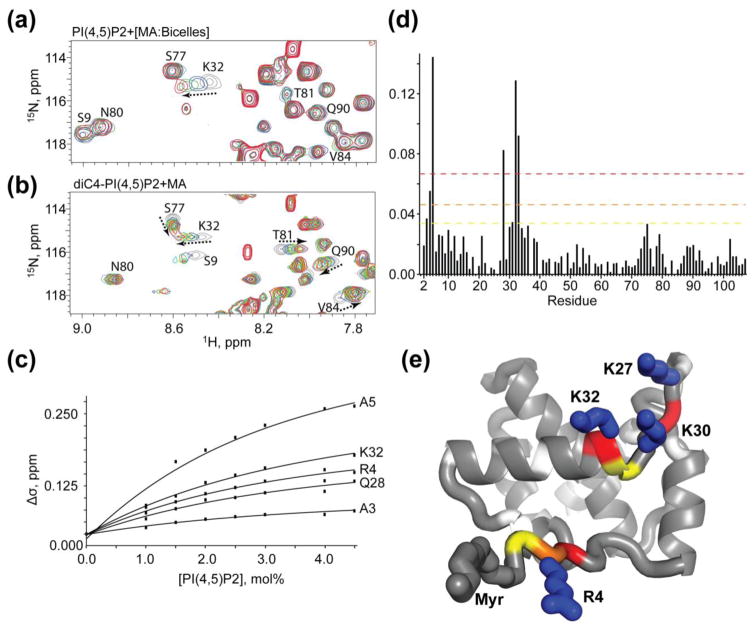
To determine the binding properties of native PI(4,5)P2 in a membrane-like environment, and experimentally test the extended lipid hypothesis, we titrated physiologically relevant molar percentages of native PI(4,5)P2 into a pre-formed MA:bicelle complex. Titration of PI(4,5)P2 resulted in distinct changes in a subset of NMR resonances, Fig. 8a,c,d. The chemical shift changes appear to saturate at PI(4,5)P2/lipid molar ratios of ~5% (Fig 8c), which is similar to estimates of inner-leaflet PI(4,5)P2 concentrations in HIV-1 virions [19]. The resonances that experience the largest changes with addition of PI(4,5)P2 (A3, R4, A5, Q28, and K32) are localized to the N-terminal region of the protein and a loop between helices I and II, Fig. 8e. Importantly, titration of PI(4,5)P2-containing bicelles into MA did not perturb residues between Helices II and IV, including T81, V84, and Q90, that were previously shown to interact with the 2′-acyl chain of tr-PI(4,5)P2 molecules (Fig. 8b) [39,59]. The 2′-acyl chain of native PI(4,5)P2 is highly unsaturated, and significant chemical shift perturbations would have been expected for these residues (due to the proximity of double bond pi electrons) if native PI(4,5)P2 bound to MA in the same extended lipid mode observed for the tr-PI(4,5)P2-MA complex in solution. Although it is conceivable that DHPC interactions somehow inhibited binding of the PI(4,5)P2 2′-acyl chain to the hydrophobic cleft, this seems unlikely since (i) residues of the hydrophobic cleft were unperturbed by DHPC (as observed previously [45]), and (ii) the affinity of DHPC for MA is low (3.75 mM; see above).
Figure 8.
Titration of PI(4,5)P2 into MA:bicelles does not support an extended lipid model. Overlay of 2D 1H-15N HSQC spectra of MA upon titration with (a) bicelle-associated native PI(4,5)P2 and (b) soluble tr-PI(4,5)P2 molecules. Colors correspond to tr-PI(4,5)P2:MA 0:1, grey; 2:1, blue; 4:1, green; 8:1, yellow; 16:1, red; native PI(4,5)P2 0 mol%, grey; 1 mol%, blue; 2 mol%, green; 3 mol%, red. (c) 15N NMR chemical-shift titration data, which fit to 1:1 binding isotherms (Kdapparent = 2.6 ± 0.4 mol%). (d) Histogram displaying the chemical shift perturbation of amide signals caused by addition of 3 mol% PI(4,5)P2 in bicelles to 15N labeled MA. Colors correspond to 1 (yellow), 2 (orange), and 3 (red) standard deviations from the mean noise (below 0.038 ppm). (e) Ribbon representation of MA highlighting the residues corresponding to panel d thresholds that experience to largest perturbation upon titration of PI(4,5)P2 in bicelles (R4, A5, Q28 (adjacent to K27), K30, and K32). Side chains of basic residues that contribute to electrostatic interactions with the acidic phosphates on the inositol head group are labeled.
We also tested for the ability of other physiologically relevant phosphatidylinositides to interact with MA in bicelles. PI(3,5)P2 is a constituent of late endosomal membranes, and titration of MA:bicelles with this membrane marker induced concentration-dependent chemical shift changes similar to those observed for PI(4,5)P2, Fig. S5. Although these findings suggest that MA could potentially target Gag to membranes that contain sufficiently high quantities of PI(3,5)P2, the cellular levels of PI(3,5)P2 are approximately 100-fold lower than those measured for PI(4,5)P2 [78,79], and it is therefore unlikely that PI(3,5)P2 plays an important role in HIV biology. The cellular levels of Golgi constituent PI(4)P are 1–3% (comparable to PI(4,5)P2 levels) [78]. However, titration of PI(4)P into MA-containing bicelles did not lead to detectable NMR spectral changes, Fig. S5.
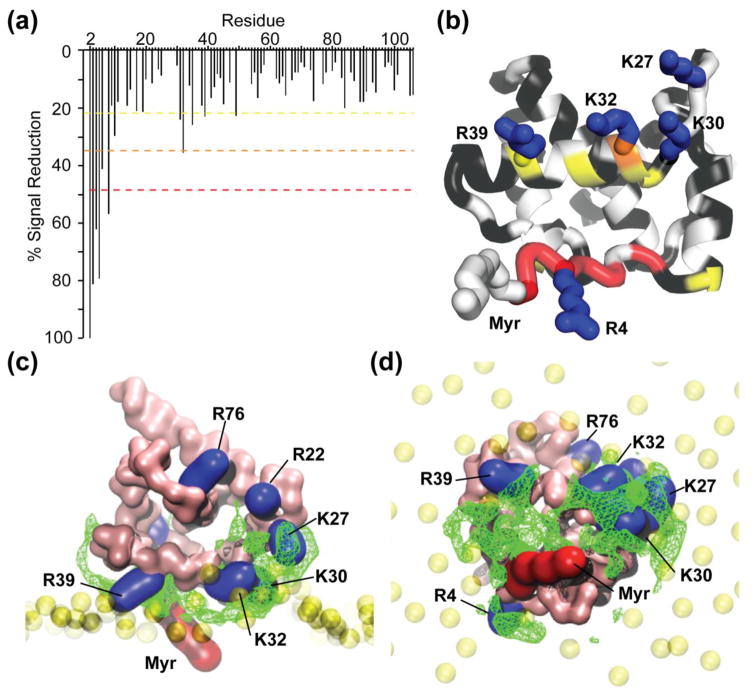
Paramagnetic relaxation enhancement studies of MA bound to bicelles
To gain structural insight into the nature of the MA:bicelle interface, we conducted paramagnetic relaxation enhancement (PRE) experiments with the bicelles containing both PI(4,5)P2 (4.5 mol%) and the paramagnetically labeled lipid, 5-doxyl-phosphatidylcholine (5-DOXYL-PC). 5-DOXYL-PC harbors a nitroxyl radical at position 5 of the sn-2 stearoyl side chain that induces line broadening of NMR resonances for nuclei that are in close proximity to the surface of the bilayer [80]. Significant reductions in amide signal intensity were observed for G2, A3, R4, A5, S6, and L8, indicating that these residues likely reside at or near the surface of the bicelle and closest to the 5-DOXYL-PC nitroxyl moiety, Fig 9a,b. The largest reductions are observed for G2, inferring that the myristoyl group is embedded in the bilayer. Smaller but significant reductions in intensity were also observed for residues that reside on a helix (helix II) located directly adjacent to the exposed myristoyl group. These findings are consistent with empirical models derived from mutagenesis studies [81,82], and with computationally derived models [46,83], that place conserved and essential basic residues of helix II in close proximity to the surface of the membrane.
Figure 9.
NMR and MD simulation data support a revised membrane-binding model. (a) Histogram displaying the loss of amide signal intensities in the HSQC spectra caused by addition of 5-DOXYL-PC to 15N labeled MA in the presence of PI(4,5)P2 containing bicelles. Colors correspond to 1 (yellow), 2 (orange), and 3 (red) standard deviations from the mean noise (below 20%). (b) Ribbon representation of MA highlighting the residues that exhibit significant 5-DOXYL-PC-dependent signal reduction (colors correspond to the dashed lines in panel a; black denotes no signal loss; white denotes not quantitatively measured due to signal overlap. (c and d) Side (c) and bottom (d) views of a representative MA:bilayer structure generated during the course of the CG simulations. The green mesh represents the most statistically occupied regions over time by the PI(4,5)P2 along the MD simulation (>70% of the total simulation duration). Basic residues that exhibit either 5-DOXYL-PC-dependent signal reduction or perturbation upon titration of PI(4,5)P2 in bicelles are represented in blue.
Comparisons with coarse-grained simulations of MA binding to PI(4,5)P2 in bilayers
We compared the PRE and PI(4,5)P2 titration NMR data obtained here with results of coarse-grained simulations employed recently to model MA-membrane interactions [46], Fig. 9c,d. Although the myristoyl moiety migrated from a protein cavity into the membrane during the simulations, the 2′-acyl chain of native PI(4,5)P2 remained within the membrane and did not migrate into the hydrophobic cleft of MA [46] that had previously been identified as a potential 2′-acyl chain binding site [39]. Upon revisiting the population dynamics, we found that PI(4,5)P2 interacts with statistically significant residence times at multiple protein sites during the simulations. In addition, multiple PI(4,5)P2 molecules were often found to be interacting with different regions of the protein simultaneously. Although the predominant site involves the basic residues of the HBR, consistent with the NMR structures of the tr-PI(4,5)P2:MA complexes in solution, other states in the simulations place the PI(4,5)P2 head group in close proximity to R4 and the myristoyl group, and to K27 (Fig 9c,d). All of these residues and/or their adjacent residues exhibit chemical shift sensitivity to the presence of PI(4,5)P2 in bicelles (Fig. 8e). Notably, residues R4 and K27 are separated by ~24 Å, which is consistent with the multi-site binding models suggested by the simulations. Earlier structural and electrostatic modeling suggested that membrane binding by MA may occur in the absence of a unique, high-affinity PI(4,5)P2 binding site [83]. Thus, the PRE and PI(4,5)P2 titration NMR findings presented here are fully compatible with the multi-structure, dynamical binding models suggested by the electrostatic modeling [83] and coarse-grained simulations [46].
Discussion
Previous NMR studies have shown that soluble PI(4,5)P2 analogs containing truncated acyl chains bind MA in an “extended lipid” conformation in aqueous solution, with the 2′-acyl chain inserted within the hydrophobic cleft on the surface of the protein and the 1′-acyl chain exposed to solvent to potentially interact with a membrane [39]. PS, PC, and PE were proposed to promote membrane binding in a similar manner, based on solution-state NMR studies with lipid analogs containing truncated acyl chains [45]. These lipid-binding models are not without precedent, as an extended lipid model was originally proposed to explain how cytochrome-c may be anchored to membranes [41–43]. PI(4,5)P2 molecules containing their native, unsaturated (rather than fully saturated) 2′-acyl chains have been shown to be more efficient at promoting MA binding to vesicle membranes [84], and we observed similar behavior in our NMR-detected liposome binding experiments (data not shown). These findings are compatible with an extended lipid-binding model. However, it is also possible that the modified 2′-acyl chain alters the structure or accessibility of PI(4,5)P2 in the membrane, or alters the membrane itself, in a manner that inhibits MA binding [37,84]. Our finding that PI(4,5)P2 molecules with truncated acyl chains can bind without discrimination to Gag domains that do not associate with membranes, coupled with previous observations that lipids with truncated acyl chains readily bind to hydrophobic surface patches on soluble proteins [51], raises questions about the biological relevance of the extended lipid:MA binding models.
In the present studies, we probed for MA interactions with native PI(4,5)P2 and other membrane constituents using a recently developed NMR-detected liposome binding assay [56]. In the absence of anionic phospholipids, MA exhibited low but detectable affinity for PC liposomes. This finding differs considerably from results obtained using flotation assays [35–38], where MA binding was “absolutely dependent” on the presence of acidic phospholipids [85] and only weak binding was observed for liposomes containing high levels of PS (PC:PS = 2:1; Kd ~ 1 μM) [38,85]. We suspect that the differences in observed affinity may be due to a Le Châtelier’s effect during the flotation experiments, in which the membrane “floats” on one end of the centrifugation tube and free MA is mechanically concentrated at the other end. We found that centrifugation of buffered solutions of MA or myr(−)MA for 45 min in the absence of sucrose gradients (50 μM; 90,000 RPM) resulted in the concentration of ~85% of the protein at the bottom third of the centrifuge tube, leaving only ~5% of the protein in the top third of the tube where the membrane would accumulate (Supplementary Figure S6). Variations in centrifugation conditions, or protein loss/degradation resulting from the multi-step flotation analytical process [86], might at least partially explain well-documented disparities among HIV Gag-membrane binding studies [85].
Because the NMR experiments were conducted under equilibrium conditions that do not require the use of highly anionic lipid compositions, we were able to quantitatively compare the relative influences of different lipid constituents on MA binding to otherwise neutral PC liposomes. Of the lipids tested, PI(4,5)P2 and PI(3,5)P2 exerted the strongest influence on liposome binding, as might be expected based on their high negative charges. Although PI(3,5)P2 is enriched in endosomal membranes relative to the other phosphoinositides, its abundance in cells is significantly lower than that of PI(4,5)P2 (~100-fold lower [13]). It is therefore likely that PM targeting results from the high relative concentration of PI(4,5)P2 rather than differences in affinity of MA for these phosphoinositides. PI(4)P (−2 charge) was also significantly better at promoting liposome binding than PS (−1 charge), as expected on the basis of charge. Interestingly, although cholesterol and PS alone did not significantly promote MA binding to liposomes, even at higher concentrations normally associated with the inner leaflet of the PM (44.0 mol% and 16.4 mol%, respectively; see Table S1), the presence of cholesterol and PS together strongly promoted MA binding to liposomes under the conditions employed. PM cholesterol is critical for virus assembly [18,31,32,65,87–92], and virus-associated cholesterol, which is enriched in the HIV-1 envelope [93], is important for infectivity [94–96]. Although the role of cholesterol in HIV assembly remains unclear [97], one explanation is that it indirectly enhances the ability of anionic lipids to interact with MA, possibly by altering the structure or dynamical properties of the membrane and its constituents. Although this could explain our finding that MA binds more readily to PS-containing liposomes that also contain cholesterol, we cannot rule out the possibility that enhanced binding is due to an increase in surface charge density associated with the increase in PS/PC ratios employed in our assays.
There is also considerable evidence that HIV assembles at cholesterol-rich raft-like microdomains [18,27,30,65,66,93,98], and recent studies have shown that the HIV-1 envelope contains a raft-like lipid composition [18,32,66,95]. However, it is not currently known whether Gag (i) binds initially to non-raft regions of the PM and recruits raft-like membrane constituents, (ii) binds to non-raft regions and then laterally migrates to pre-existing rafts, or (iii) is targeted initially to pre-formed raft-like domains [19,39]. Although our studies show that MA binds with highest affinity to liposomes with raft-like compositions, significant affinity was also observed for liposomes designed to mimic non-raft-like regions of the PM inner leaflet. Our finding that HBR mutations known to cause aberrant membrane targeting in vivo have only a modest influence on binding to raft-like liposomes, but considerably attenuate MA binding to non-raft-like liposomes, appears to support a mechanism in which the initial step of virus assembly involves Gag interactions with non-raft-like regions of the PM.
The HBR residues of MA are critical for virus assembly, and studies have shown that mutation of K30 and/or K32 cause a mistargeting of Gag to late endosomal/multivesicular body compartments [12,63,64,99]. This retargeting phenotype is essentially identical to that induced by depletion of PI(4,5)P2 from the PM [10], suggesting that MA residues K30 and K32 might promote Gag targeting to the PM by interacting directly with PI(4,5)P2. The data presented here are consistent with this hypothesis. We had speculated that mutation of MA residues K30 and K32 might retarget Gag to late endosomes by shifting the phosphoinositide preference from PI(4,5)P2, which is concentrated on the PM, to PI(3,5)P2, which is enriched in late endosomes. This seems to not be the case, as the binding of the K30E/K32E to liposomes is not significantly enhanced by either PI(4,5)P2 or PI(3,5)P2 (Fig. 2b). The mechanisms leading to Gag’s diversion from the PM to late endosomes upon disruption of PI(4,5)P2 binding (via either PI(4,5)P2 depletion or mutation of key PI(4,5)P2-binding residues in MA) remains to be determined.
The above studies enabled quantitative comparison of the relative affinities of MA (and MA mutants) for bilayers with various lipid compositions under equilibrium conditions. However, it was not possible to probe the structural determinants of binding because residues of the globular portion of the liposome-bound protein are NMR-dark. We therefore extended studies to bicelles, which have rotational diffusion properties appropriate for solution-state NMR methods. As observed previously by Saad and co-workers, we found that MA binds to DMPC/DHPC bicelles in a myristoyl-exposed conformation [45]. Our studies further showed that chemical shift perturbations observed for several residues upon titration with bicelles are probably due to weak interactions with free DHPC, and that these interactions do not contribute to the anchoring of the protein to the bicelle. The paramagnetic relaxation enhancement and 15N NMR relaxation experiments indicated that MA is anchored to the bicelles, the myristoyl group is inserted into the bilayer of the bicelle, and the protein is oriented on the surface of the bicelle in a manner predicted by crystallographic [82] and computational [46,83] studies. Results of NMR chemical shift perturbation experiments were also consistent with this binding model. However, we did not observe PI(4,5)P2-dependent NMR chemical shift changes that would be expected for the extended lipid binding mechanism. Specifically, chemical shifts and linewidths of residues of the hydrophobic cleft that were proposed to sequester the unsaturated 2′-acyl chain of PI(4,5)P2 (on the basis of solution-state studies with truncated PI(4,5)P2 analogs [39]) were either unperturbed or exhibited negligible changes upon PI(4,5)P2 titration. Signals for these residues would have been significantly perturbed by the π-bonds of the unsaturated 2′-acyl chain if the extended lipid binding structure had been adopted. Our findings are more compatible with computationally-derived models of the MA:membrane interface [46,83], in which multiple surfaces on MA interact transiently and dynamically with one or more PI(4,5)P2 head groups, and both acyl chains of PI(4,5)P2 remain embedded in the membrane.
In summary, we have obtained quantitative information on the relative contributions of lipid constituents on MA binding to membrane-mimetic liposomes and detergent bicelles under equilibrium conditions. Our findings support proposals that PI(4,5)P2 functions in PM targeting by interacting directly with the MA domain of Gag, but do not support proposals that PI(4,5)P2 adopts an extended lipid conformation. Our findings are fully compatible with computational models in which both acyl chains of PI(4,5)P2 remain anchored in the PM bilayer, and one or more PI(4,5)P2 head groups interact electrostatically and dynamically at multiple sites on the surface of the protein. Data obtained for HBR mutants support an assembly model in which Gag initially associates with non-raft-like regions of the PM. Gag would then either laterally migrate to pre-formed raft-like assembly sites, or recruit raft-like lipid constituents as the Gag proteins assembled. The NMR and computational approaches employed here may now be used to study MA interactions with other viral and cellular constituents that were not amenable to traditional solution-state methods, including MA interactions with the intra-viral domain of Env and tRNAs (underway).
Materials and Methods
Protein expression and purification
HIV-1 MA constructs containing amino acid changes L8I, K30E/K32E, and K32E were made by using QuikChange II site-directed mutagenesis kit (Agilent, Santa Clara, CA) and sequenced at Genewiz (South Plainfield, NJ). Nitrogen-15 labeled HIV-1 MA for bicelle studies was prepared from recombinant E. coli cells as follows: Cells were first grown in 4 L of LB medium at 37 °C [for myristoylated proteins, when the OD600 reached ~ 0.25–0.3 the media was supplemented with myristic acid (10 mg/L; Sigma)] and allowed to grow until the OD600 reached ~ 0.6–0.7. Next, cells were spun down and washed with 1 X M9 salt before transfer to 1 L of M9 minimal medium containing 15NH4Cl as the sole nitrogen source and then growth for 1 h before induction with isopropyl-β-D-1-thiogalactopyranoside. Cells expressing 15N labeled samples were grown at 37 °C for ~ 12–14 h and lysed using a microfluidizer and the proteins were purified by cobalt affinity chromatography (Clontech, Mountain View, CA) and hydrophobicity column chromatography (Butyl column; GE Healthcare) and ion exchange column chromatography (SP column; GE Healthcare) [59,61,81]. Unlabeled MA protein was expressed and purified as described above except that expression was carried out in 2 L of LB for 4 h.
Molecular weights and efficiency of myristoylation were confirmed by electrospray ionization mass spectrometry [Mmeas/Mcalc = 15,746.31±104: 15744.66, 17532.08±143: 15534.31, 15740.22±401: 15746.54, 12746.32±13.98: 15746.54, 15745.39±13.35: 15745.39, for MA and myr(−)MA, MAL8I, MAK30E/K32E, and MAK32E, respectively]. Uniformly 15N-enriched and unlabeled P24-WT, P24-AA, CA-NTD, CA-CTD, and NC were expressed as previously described [61,100–102] and the integrity of the samples were confirmed by mass spectrometry.
Liposome preparation
Liposomes were prepared from stock solutions of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC; Avanti), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine] (POPS; Avanti), L-α-phosphatidylethanolamine (PE; Avanti), L-α-phosphatidylinositol-4,5-bisphosphate ([PI(4,5)P2]; Avanti), 1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,5′-bisphosphate) ([PI(3,5)P2]; Avanti), and L-α-phosphatidylinositol-4-phosphate ([PI(4)P]; Avanti) lipids dissolved in chloroform and used without further purification. Lipids were dried to a thin film using a centrifugal evaporator. The dried lipids were then hydrated with liposome buffer, generating 5.76 mg/mL, (100 mM NaCl, 50 mM sodium phosphate [pH 7.4], and 5 mM beta-mercaptoethanol) and vortexed until the lipid film was completely dissolved. To prepare large unilamellar vesicles by Extrusion (LUVETs), the rehydrated lipid solution was subject to extrusion using a mini-extruder block (Avanti) 28 times through a polycarbonate membrane with a pore size of 100 nm. Liposome samples were used within 24 h of preparation. Molar percentages of heterogeneous liposomes were prepared by decreasing the amount of PC relative to the amount of lipids added.
Bicelle preparation
Bicelle lipids 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) were obtained commercially (Avanti) and used without further purification. DMPC was initially dissolved in benzene:ethanol (95:5) vacuum dried and re-dissolved in NMR buffer (50 mM sodium phosphate [pH 5.5] and 5 mM DTT) containing DHPC to make bicelles with a q values of 0.3, q=[DMPC]/[DHPC].
NMR spectroscopy
NMR data were collected at 35 °C on 600 MHz Bruker Avance III spectrometers equipped with cryogenic probes. Unlabeled protein samples contained 0.05 mM protein. For solution state liposome binding assays, a fraction of the LUV liposome suspensions (total, 866.4 μg lipids) was mixed with a protein sample (~150 μg) with 10% D2O and incubated at ambient temperature for a minimum of 5 minutes before NMR data acquisition in 3 mm NMR tubes. 1H-1D spectra were acquired with 512 transients, 1 sec relaxation delay, 32768 time domain points, and a spectral window of 12019 Hz. Studies with increasing molar percentages of phosphoinositides and other lipid constituents were repeated in triplicate and duplicate, respectively, for estimation of errors (shown as mean ± standard deviation). Experiments with bicelles were conducted with HIV-1 myrMA at concentrations of 0.050–0.450 mM. NMR data was processed using NMRpipe suite [103] and analyzed using NMRViewJ [104,105]. The 1H-15N HSQC NMR spectra of 0.45 mM uniformly 15N-labeled MA was recorded upon titration with 0.9 mM PC-bicelles containing varying molar percentages of PIs (relative to the moles of DMPC lipid). Binding isotherms from 1H-15N NMR HSQC titration experiments were calculated with the NMRViewJ titration analysis tool.
For studies with paramagnetic probes, 1H-15N TROSY-HSQC NMR spectra were obtained for samples containing uniformly 15N-labeled MA (0.45 mM ) PI(4,5)P2 (1.55 mM) (Avanti), and 0.9 mM bicelles. The paramagnetic spin-label probe 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine (5-DOXYL-PC) (Avanti), was dissolved in chloroform, dried by vacuum, then dissolved in an NMR sample containing the MA:bicelle:PI(4,5)P2 complex. The ratio of spin-label to bicelle was 1:4. 1H-15N TROSY-HSQC spectra were collected before and after addition of 5-DOXYL-PC, and intensities of the backbone amide resonances of the protein were monitored and compared. 5-DOXYL-PC did not alter the domain structure as evidenced by the lack of chemical shift perturbations in the NMR spectrum of the protein.
Lipid binding analyses
NMR data were processed using Topspin 3.2 (Bruker, Karlsrube, Germany) and analyzed with dataChord Spectrum Analyst (onemoonscientific.com), where the integrals of each spectrum were obtained using the region from 9.5 to 8.0 ppm, to exclude resonances of the lipids and unstructured regions of the protein. The fraction of protein bound was calculated as a function of the missing signal intensity for each lipid/protein molar ratio, discussed further [56]. The Gnuplot graphing utility (http://www.gnuplot.info) was used for graphing and non-linear least squares fitting of binding isotherms. The following equation was used to obtain relative dissociation constants, Kd [56]:
Where CP is represents the total protein concentration (50 μM for these analyses), CL is the total available lipid concentration (independent variable), n is the number of phosphoinositidyl phosphate lipid molecules bound per protein molecule and y = 1 − I/I0 where I is the integral of the signal for a given CL, and I0 is the integral of the signals where CL = 0. For our analyses, CL was halved to represent the equal distribution of PI(4,5)P2 in the external (exposed for MA binding) and luminal (unavailable for MA binding) leaflets of the PC liposomes, and n is fixed at 1.
15N NMR relaxation measurements and analysis
15N relaxation data for MA were obtained for the labeled protein at 35 °C using the same sample conditions as for the bicelle experiments. Spin-lattice relaxation rate constants (R1) and spin-spin relaxation rate constants (R2) were measured for backbone amide relaxation by two-dimensional inverse-detected methods using conventional methodologies [106–108]. Eight different values for the relaxation times were recorded in an interleaved manner for T1 (=1/R1; T1 delays = 10, 125, 500, 750, 1000, 1500, 2000, and 2500 ms) and T2 (=1/R2; T2 = 16.96, 33.92. 67.84, 118.72, 186.56, 271.36, 373.12, and 508.80 ms) relaxation experiments.
Coarse-grained simulations
The HIV-1 Matrix protein coordinates have been built from the first frame of the PDB entry 2H3I [61]. The HIV-1 matrix protein was mapped into a Coarse-Grained (CG) representation as previously described [46] with the MARTINI force field [109,110]. An elastic network was included between residues less than 0.9 nm distant with a constant force of 500 kJ.mol−1.nm−2, excluding the first nine N-terminal residues (Gly2 to Gly10). The membrane has also been built as previously described [46]. The membrane is a symmetric bilayer that contains 512 lipids on each leaflet with the following molar composition 55% PC, 5% PI(4,5)P2, 15% POPS and 25% POPE. Water molecules and counter-ions (203 Na+) were added in order to neutralize the global charge, so that the final system was further minimized and equilibrated during a first simulation of 1.5 μs. All the energy-minimizations and molecular dynamics simulations have been performed with the Gromacs v4.5.5 software [111]. The steepest descent algorithm has been used for the energy-minimizations. The default parameters for MARTINI MD simulations have been used, the switching function being applied from 0 to 1.2 nm for Coulomb interactions and from 0.9 to 1.2 nm for Lennard-Jones interactions. To establish the PI(4,5)P2 lipid density, a 24 μs simulation was employed [46]. Analyses of lipid densities were performed on the PO4 atoms using tools available in Gromacs.
Replication, release and infectivity of HIV-1 MA mutants
HIV-1 replication was determined in the Jurkat CD4+ T-cell line. Cells were transfected with the NL4-3-derived mutants using DEAE-dextran as described previously [11]. Virus replication was monitored by measuring reverse transcriptase activity in the culture, as described previously [112], from samples collected every two days. Virus release, Env incorporation and infectivity were determined using HeLa cells. Cells were transfected with NL4-3-derived mutants of HIV-1 using PolyJet (Signagen) following the manufacturer’s instructions. After 48 h, 100 μl of supernatant was collected to determine infectivity while the remaining supernatant and cell lysate was harvested to determine virus release and Env incorporation.
To measure virus infectivity, 10 μl of the supernatant was used to infect TZM-bl cells, a HeLa-derived cell line that expresses the receptors necessary to permit HIV infection and that expresses firefly luciferase in response to HIV infection; after 48 h the luciferase signal was measured with Britelite (Perkin Elmer) following the manufacturer’s instructions. Infectivity was calculated as the luciferase signal divided by the reverse transcriptase activity.
To measure virus release and Env incorporation, supernatant was collected, passed through a 0.45 μm filter and centrifuged at 76 000 × g to pellet virus particles. Both the pelleted virions and the remaining cells were lysed by the addition of 2 x Laemmli buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.02% bromophenol blue, 120 mM Tris-Cl pH 6.8). Both lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes for western blotting. Cell lysates were probed with anti-HIV Ig; the virions were probed first with an anti-gp41 Env antibody (2F5) then subsequently with anti-HIV Ig. In all cases, bands were revealed by the use of horseradish peroxidase-conjugated anti-human secondary antibodies, enhanced chemiluminescence and a Chemidoc XRS+ (Biorad). Band intensities were measured using Image Lab software (Biorad). Virus release was calculated as the percentage of CA in supernatant relative to the total Gag (released CA/[released CA+cellular CA+cellular p55]x100). Env incorporation was calculated as the amount of p41 in the supernatant relative to the amount of CA (released gp41/released CA).
Supplementary Material
Highlights.
The influences of lipid constituents on membrane binding by HIV-1 MA were determined.
NMR results differed from those obtained by non-equilibrium flotation assays.
The structural basis for PI(4,5)P2-dependent membrane targeting was re-examined.
Native PI(4,5)P2 does not bind MA in a predicted “extended lipid” conformation.
MA may bind non-raft regions and recruit raft-like constituents during assembly.
Acknowledgments
Support from NIH grants AI30917 and GM103297 (to MFS) and GM106672 (to CS) is gratefully acknowledged. PYM was supported by an NIGMS Initiative for Minority Student Development grant, R25-GM55036.
Footnotes
Note Added in Proof
After this paper was submitted and reviewed, a preprint appeared on-line describing studies of MA binding to synthetic solid-supported bilayers by surface plasmon resonance spectroscopy [113].
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–9. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jouvenet N, Simon SM, Bieniasz PD. Visualizing HIV-1 assembly. J Mol Biol. 2011;410:501–11. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nature cell biology. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Rahman SA, Nikolaitchik OA, Grunwald D, Sardo L, Burdick RC, Plisov S, Liang E, Tai S, Pathak VK, Hu WS. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc Natl Acad Sci U S A. 2016;113:E201–8. doi: 10.1073/pnas.1518572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13:484–96. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita E, Sundquist WI. Retrovirus Budding. Annual Review of Cell and Develpomental Biology. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 8.Ganser-Pomillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindwasser OW, Resh MD. Myristoylation as a target for inhibiting HIV assembly: Unsaturated fatty acids block viral budding. Proc Natl Acad Sci USA. 2002;99:13037–13042. doi: 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single Amino Acid Changes in the Human Immunodeficiency Virus Type 1 Matrix Protein Block Virus Particle Production. Journal of Virology. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono A, Orenstein JM, Freed EO. Role of the Gag Matrix Domain in Targeting Human Immunodeficiency Virus Type 1 assembly. Journal of Virology. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 14.Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Bachtiar I, Piszezek G, Bouamr F, Carter C, Tjandra N. Solution NMR characterizations of oligomerization and dynamics of Equine Infectious Anemia Virus matrix protein and its interactions with PIP2. Biochemistry. 2008;47:1928–1937. doi: 10.1021/bi701984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prchal J, Kroupa T, Ruml T, Hrabal R. Interaction of Mason-Pfizer monkey virus matrix protein with plasma membrane. Front Microbiol. 2013;4:423. doi: 10.3389/fmicb.2013.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamard-Peron E, Julliard F, Saad JS, Roy C, Roingeard P, Summers MF, Darlix J-L, Picart C, Muriax D. Targeting of Murine Leukemia Virus Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the Matrix. J Virol. 2010;84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–6. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. Retroviruses Human Immunodeficiency Virus and Murine Leukemia Virus are enriched in phosphoinositides. J Virol. 2008;82:11228–38. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalonde MS, Sundquist WI. How HIV finds the door. Proc Natl Acad Sci U S A. 2012;109:18631–18632. doi: 10.1073/pnas.1215940109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorizate M, Sachsenheimer T, Glass B, Habermann A, Gerl MJ, Krausslich HG, Brugger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- 22.Campbell SM, Crowe SM, Mak J. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. Journal of Clinical Virology. 2001;22:217–227. doi: 10.1016/s1386-6532(01)00193-7. [DOI] [PubMed] [Google Scholar]
- 23.Chazal N, Gerlier D. Virus Entry, Assembly, Budding, and Membrane Rafts. Microbiology and molecular biology reviews. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 25.Kerviel A, Thomas A, Chaloin L, Favard C, Muriaux D. Virus assembly and plasma membrane domains: which came first? Virus Res. 2013;171:332–40. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–8. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Pike LJ. The challenge of lipid rafts. J Lipid Res. 2009;50(Suppl):S323–8. doi: 10.1194/jlr.R800040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorizate M, Brugger B, Akiyama H, Glass B, Muller B, Anderluh G, Wieland FT, Krausslich HG. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J Biol Chem. 2009;284:22238–47. doi: 10.1074/jbc.M109.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 30.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–99. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 31.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 34.Golub T, Caroni P. PI(4,5)P2-dependent microdomain assemblies capture microtubules to promote and control leading edge motility. J Cell Biol. 2005;169:151–162. doi: 10.1083/jcb.200407058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009;83:12196–203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A. 2010;107:1600–5. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A. 2012;109:18761–6. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chukkapalli V, Hogue IB, Boyko V, Hu W-S, Ono A. Interaction between HIV-1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag-membrane binding. J Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shkriabai N, Datta SK, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 41.Kinnunen PKJ, Koiv A, Lehtonen JYA, Rytomaa M, Mustonen P. Lipid dynamics and peripheral interactions of proteins with membrane surfaces. Chem Phys Lipids. 1994;73:181–207. doi: 10.1016/0009-3084(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 42.Rytomaa M, Kinnunen PKJ. Reversibility of the binding of cytochrome c to liposomes. J Biol Chem. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 43.Touminen EKJ, Wallace CJA, Kinnunen PKJ. Phospholipid-cytochrome c interaction. J Biol Chem. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 44.Kinnunen PKJ. On the molecular-level mechanisms of peripheral protein-membrane interactions induced by lipids forming inverted non-lamellar phases. Chem Phys Lipids. 1996;81:151–166. [Google Scholar]
- 45.Vlach J, Saad JS. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc Natl Acad Sci U S A. 2013;110:3525–30. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlier L, Louet M, Chaloin L, Fuchs P, Martinez J, Muriaux D, Favard C, Floquet N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: revisiting its assembly on membrane domains. Biophys J. 2014;106:577–85. doi: 10.1016/j.bpj.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanford C. Interfacial free energy and the hydrophobic effect. Proc Natl Acad Sci U S A. 1979;76:4175–6. doi: 10.1073/pnas.76.9.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Israelachvili JN, Mitchell DJ, Ninham BW. Theory of self-assembly of lipid bilayers and vesicles. Biochim Biophys Acta. 1977;470:185–201. doi: 10.1016/0005-2736(77)90099-2. [DOI] [PubMed] [Google Scholar]
- 49.Saad JS, Kim A, Ghanam RH, Dalton AK, Vogt MV, Wu Z, Lu W, Summers MF. Mutations that mimic phosphorylation of the HIV-1 matrix protein: Implications for trafficking. Protein Science. 2007;16:1793–1797. doi: 10.1110/ps.072987607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a Membrane-Binding Domain Within the Amino-Terminal Region of Human Immunodeficiency Virus Type 1 Gag Protein Which Interacts with Acidic Phospholipids. Journal of Virology. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kragh-Hansen U, Hellec F, de Foresta B, le Maire M, Moller JV. Detergents as probes of hydrophobic binding cavities in serum albumin and other water-soluble proteins. Biophys J. 2001;80:2898–911. doi: 10.1016/S0006-3495(01)76255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee PP, Linial ML. Efficient Particle Formation Can Occur if the Matrix Domain of Human Immunodeficiency Virus Type 1 Gag Is Substituted by a Myristoylation Signal. Journal of Virology. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Yu X, Lee TH, Essex M. Mutations in the N-Terminal Region of Human Immunodeficiency Virus Type I Matrix Protein Block Intracellular Transport of the Gag Precursor. Journal of Virology. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saad JS. Titrations of myrMA with phosphatidylinositide-(4,5)-bisphosphates containing di-C8 or longer fatty acids (which form micelles) resulted in aggregation and significant NMR signal broadening, which precluded NMR-based structural studies 2005 [Google Scholar]
- 55.Bodner CR, Dobson CM, Bax A. Multiple Tight Phospholipid-Binding Modes of alpha-Synuclein Revealed by Solution NMR Spectroscopy. J Mol Biol. 2009;390:775–90. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceccon A, D’Onofrio M, Zanzoni S, Longo DL, Aime S, Molinari H, Assfalg M. NMR investigation of the equilibrium partitioning of a water-soluble bile salt protein carrier to phospholipid vesicles. Proteins. 2013;81:1776–91. doi: 10.1002/prot.24329. [DOI] [PubMed] [Google Scholar]
- 57.Ono A, Freed EO. Binding of Human Immunodeficiency Virus Type 1 gag to membrane: Role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ono A, Huang M, Freed EO. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makutonina A, Voss TG, Plymale DR, Fermin CD, Norris CH, Vigh S, Garry RF. Human immunodeficiency virus infection of T-lymphoblastoid cells reduces intracellular pH. Journal of virology. 1996;70:7049–55. doi: 10.1128/jvi.70.10.7049-7055.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fledderman EL, Fujii K, Ghanam RH, Waki K, Prevelige PE, Freed EO, Saad JS. Myristate exposure in the human immunodeficiency virus type 1 matrix protein is modulated by pH. Biochemistry. 2010;49:9551–62. doi: 10.1021/bi101245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ono A, Freed EO. Cell-Type-Dependent Tageting of Human Immunodeficiency Virus Type 1 Assembly to the Plasma Membrane and the Multivesicular body. Journal of Virology. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J Virol. 2009;83:5375–87. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–76. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwamoto K, Hayakawa T, Murate M, Makino A, Ito K, Fujisawa T, Kobayashi T. Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys J. 2007;93:1608–19. doi: 10.1529/biophysj.106.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys J. 2010;99:3309–18. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481:45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrison EA, Henzler-Wildman KA. Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile. Biochim Biophys Acta. 2012;1818:814–20. doi: 10.1016/j.bbamem.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–71. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song Y, Mittendorf KF, Lu Z, Sanders CR. Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J Am Chem Soc. 2014;136:4093–6. doi: 10.1021/ja4114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf*GTP. Nat Struct Mol Biol. 2010;17:876–81. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durr UH, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem Rev. 2012;112:6054–74. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohori Y, Okazaki H, Watanabe S, Tochio N, Arai M, Kigawa T, Nishimura C. Flexible and rigid structures in HIV-1 p17 matrix protein monitored by relaxation and amide proton exchange with NMR. Biochim Biophys Acta. 2014;1844:520–6. doi: 10.1016/j.bbapap.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 76.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 77.Campos-Olivas R, Summers MF. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry. 1999;38:10262–10271. doi: 10.1021/bi990991x. [DOI] [PubMed] [Google Scholar]
- 78.Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell. 2006;5:723–31. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shisheva A. Regulating Glut4 vesicle dynamics by phosphoinositide kinases and phosphoinositide phosphatases. Front Biosci. 2003;8:s945–6. doi: 10.2741/1101. [DOI] [PubMed] [Google Scholar]
- 80.Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 2005;44:16064–71. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 82.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal Structures of the Trimeric HIV-1 Matrix Protein: Implications for Membrane Association. Proceedings of the National Academy of Sciences USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murray PS, Li Z, Wang J, Tang CL, Honig B, Murray D. Retroviral matrix domains share electrostatic homology: Models for membrane binding function throughout the viral life cycle. Structure. 2005;13:1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 84.Olety B, Veatch SL, Ono A. Phosphatidylinositol-(4,5)-Bisphosphate Acyl Chains Differentiate Membrane Binding of HIV-1 Gag from That of the Phospholipase Cdelta1 Pleckstrin Homology Domain. J Virol. 2015;89:7861–73. doi: 10.1128/JVI.00794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt MV. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Temmerman K, Nickel W. A novel flow cytometric assay to quantify interactions between proteins and membrane lipids. J Lipid Res. 2009;50:1245–54. doi: 10.1194/jlr.D800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Z, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JE. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retroviruses. 2001;17:1009–19. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- 88.Waheed AA, Ablan SD, Mankowski MK, Cummins JE, Ptak RG, Schaffner CP, Freed EO. Inhibition of HIV-1 replication by amphotericin B methyl ester: selection for resistant variants. J Biol Chem. 2006;281:28699–711. doi: 10.1074/jbc.M603609200. [DOI] [PubMed] [Google Scholar]
- 89.Waheed AA, Ablan SD, Roser JD, Sowder RC, Schaffner CP, Chertova E, Freed EO. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc Natl Acad Sci U S A. 2007;104:8467–71. doi: 10.1073/pnas.0701443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waheed AA, Ablan SD, Soheilian F, Nagashima K, Ono A, Schaffner CP, Freed EO. Inhibition of human immunodeficiency virus type 1 assembly and release by the cholesterol-binding compound amphotericin B methyl ester: evidence for Vpu dependence. J Virol. 2008;82:9776–81. doi: 10.1128/JVI.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waheed AA, Ablan SD, Sowder RC, Roser JD, Schaffner CP, Chertova E, Freed EO. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J Virol. 2010;84:3121–6. doi: 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Til NP, Heutinck KM, van der Rijt R, Paulusma CC, van Wijland M, Markusic DM, Elferink RP, Seppen J. Alteration of viral lipid composition by expression of the phospholipid floppase ABCB4 reduces HIV vector infectivity. Retrovirology. 2008;5:14. doi: 10.1186/1742-4690-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Campbell SM, Crowe SM, Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS. 2002;16:2253–61. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- 95.Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol. 2003;77:8237–48. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao Z, Graham DR, Hildreth JE. Lipid rafts and HIV pathogenesis: virion-associated cholesterol is required for fusion and infection of susceptible cells. AIDS Res Hum Retroviruses. 2003;19:675–87. doi: 10.1089/088922203322280900. [DOI] [PubMed] [Google Scholar]
- 97.Hawkes D, Jones KL, Smyth RP, Pereira CF, Bittman R, Jaworowski A, Mak J. Properties of HIV-1 associated cholesterol in addition to raft formation are important for virus infection. Virus Res. 2015;210:18–21. doi: 10.1016/j.virusres.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 98.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–88. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 99.Freed EO, Martin MA. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 101.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nature Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 102.Zhang H, Curreli F, Waheed AA, Mercredi PY, Mehta M, Bhargava P, Scacalossi D, Tong X, Lee S, Cooper A, Summers MF, Freed EO, Debnath AK. Dual-acting stapled peptides target both HIV-1 entry and assembly. Retrovirology. 2013;10:136. doi: 10.1186/1742-4690-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 104.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 105.Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 106.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–6. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 107.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 108.Kay LE. Protein dynamics from NMR. Nat Struct Biol. 1998 Jul;(Supplement):513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]
- 109.Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- 110.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–24. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 111.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 112.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–47. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barros M, Heinrich F, Datta SA, Rein A, Karageorgos I, Nanda H, Losche M. Membrane Binding of HIV-1 MA Protein: Dependence on Bilayer Composition and Protein Lipidation. J Virol. 2016 doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pike LJ, Han X, Chung KN, Gross RW. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 2002;41:2075–88. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.