Abstract
The transcription factor cAMP-response element-binding protein (CREB) is involved in neuronal plasticity. Phosphorylation activates CREB and an increased level of phosphorylated CREB is regarded as an indicator of CREB-dependent transcriptional activation. In honeybees (Apis mellifera) we recently demonstrated a particular high abundance of the phosphorylated honeybee CREB homolog (pAmCREB) in the central brain and in a subpopulation of mushroom body neurons. We hypothesize that these high pAmCREB levels are related to learning and memory formation. Here, we tested this hypothesis by analyzing brain pAmCREB levels in classically conditioned bees and bees experiencing unpaired presentations of conditioned stimulus (CS) and unconditioned stimulus (US). We demonstrate that both behavioral protocols display differences in memory formation but do not alter the level of pAmCREB in bee brains directly after training. Nevertheless, we report that bees responding to the CS during unpaired stimulus presentations exhibit higher levels of pAmCREB than nonresponding bees. In addition, Trichostatin A, a histone deacetylase inhibitor that is thought to enhance histone acetylation by CREB-binding protein, increases the bees’ CS responsiveness. We conclude that pAmCREB is involved in gating a bee's behavioral response driven by an external stimulus.
The transcription factor cAMP-response element-binding protein (CREB) is widespread in the animal kingdom. It was cloned in various vertebrate and invertebrate animals, including honeybees (Apis mellifera), where it is termed Apis mellifera CREB (AmCREB) (Gonzalez et al. 1989; Eisenhardt et al. 2003, 2006; Sadamoto et al. 2004; Song et al. 2009; van den Berg et al. 2010). CREB is activated following post-translational modifications such as phosphorylation by protein kinase A (PKA). This enables the binding of co-factors such as the CREB-binding protein (CBP) thereby inducing CREB-mediated transcription (Yamamoto et al. 1990; Chrivia et al. 1993). CREB is involved in different processes of the nervous system. It is activated following learning thereby inducing transcription underlying long-term memory (LTM) formation (Alberini 2009). Furthermore, CREB is regulating the basic intrinsic excitability of neurons, a mechanism that might be important for the allocation of the memory trace (Benito and Barco 2010; Barco and Marie 2011; Yiu et al. 2014). Honeybee workers display a division of labor, which is age-dependent (Beshers and Fewell 2001; Zayed and Robinson 2012). They perform in-hive duties such as brood care and food processing for the first 2–3 wk after hatching and start to forage for nectar and pollen outside the hive thereafter (Seeley 1982). Interestingly, AmCREB was identified as a potential regulator of age-dependent behavior in honeybees (Chandrasekaran et al. 2011; Ament et al. 2012; Khamis et al. 2015). In line, we recently demonstrated an age-dependent increase in the abundance of phosphorylated AmCREB (pAmCREB) in the central bee brain and a subpopulation of the mushroom body neurons, the inner compact cells (IC) (Gehring et al. 2016). This increase parallels the worker bees’ switch from working solely inside the hive to flying out of the hive for orientation flights and foraging (Seeley 1982; Capaldi et al. 2000; Degen et al. 2015) suggesting an involvement of AmCREB-dependent processes in these behaviors. Given that the ability to learn and to form memories is a precondition for successful explorative behavior and foraging, we hypothesize that pAmCREB in the central brain is linked to learning and memory formation.
We tested this hypothesis in individual bees trained in an appetitive Pavlovian conditioning paradigm that is based on olfactory conditioning of the proboscis extension response (PER) (Takeda 1961; Bitterman et al. 1983; Giurfa and Sandoz 2012; Matsumoto et al. 2012). In this paradigm, honeybees learn the association between an initially neutral olfactory stimulus, the conditioned stimulus (CS), and a sucrose stimulus, the unconditioned stimulus (US). Once the association between the olfactory CS and the rewarding US has been learned, the CS alone elicits the PER, resembling the conditioned response.
Following conditioning, memories of different stability are formed, defined by the time point of their retrieval and their underlying molecular mechanisms. Four different memories are distinguished: short-term memory (STM), mid-term memory (MTM), early long-term memory (eLTM), and late long-term memory (lLTM) (Müller 2013; Eisenhardt 2014). Their formation depends on the number of conditioning trials and the intertrial interval (Menzel 1990; Gerber et al. 1998; Menzel et al. 2001; Friedrich et al. 2004; Felsenberg et al. 2012). Moreover, the sequence of stimulus presentations and the temporal relationship between CS and US are critical for memory stability and the underlying molecular mechanisms (Felsenberg et al. 2015).
In this study, we characterize differences in memory stability and sensitivity to a transcriptional inhibitor between classically conditioned bees and bees that experienced the unpaired presentation of CS and US. We exploit the differences we found in memory formation between those two protocols in order to compare the level of bee brain pAmCREB following paired and unpaired presentations of CS and US. Our experiments did not reveal a learning-dependent alteration of pAmCREB. Nevertheless, we report that bees responding to the CS during unpaired CS–US presentation exhibit higher levels of pAmCREB than bees that do not respond. Furthermore, we demonstrate that the histone deacetylase (HDAC) inhibitor Trichostatin A (TSA) enhances the bees’ odor responsiveness before and during unpaired training. Since CBP binds to phosphorylated CREB and acts as histone acetyltransferase (HAT), we conclude that pAmCREB is involved in regulating the threshold for a behavioral response driven by an external stimulus in honeybees.
Results
Transcription-dependent memory formation following paired training
In the present study, we explored the role of the high abundance of pAmCREB in the mushroom bodies of the honeybee brain. Previous studies in vertebrates demonstrated an involvement of phosphorylated CREB in the formation of transcription-dependent LTMs (for review, see Alberini et al. 2009). Therefore, we hypothesized that pAmCREB is involved in the formation of a transcription-dependent LTM in honeybees and proposed that an altered pAmCREB level would be observed after training that leads to a transcription-dependent LTM but not after training that does not. In order to test this hypothesis, we had to identify two training protocols in the beginning of our study: One that leads to a transcription-dependent LTM and one that does not result in a transcription-dependent LTM.
In honeybees transcription-dependent late LTM (lLTM) can be retrieved three and four days after classical conditioning in an appetitive olfactory conditioning paradigm (Wüstenberg et al. 1998; Menzel et al. 2001; Friedrich et al. 2004; Lefer et al. 2012). Here we used a classical conditioning protocol with three CS–US stimulations presented with an intertrial interval of 10 min (Fig. 1A). From here on we term this protocol paired training protocol. When we started our study it remained unknown whether this protocol leads to a transcription-dependent lLTM. Therefore, we first had to examine the transcription-dependence of 24 and 72 h memories following training with this protocol.
Figure 1.
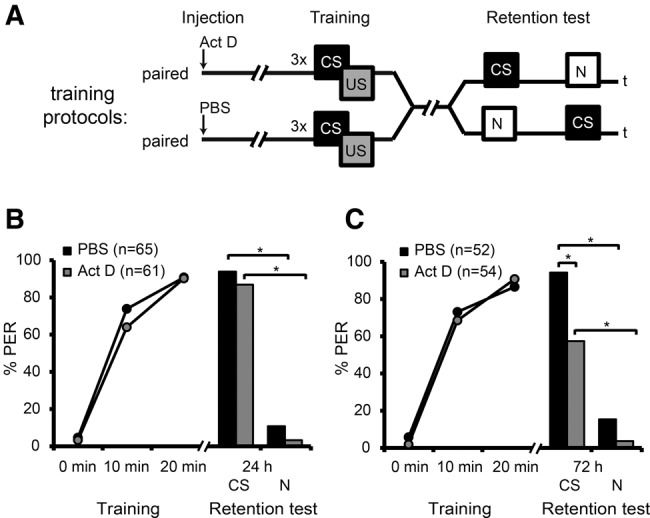
Transcription-dependent late long-term memory formation following paired training. (A) Schematic representation of the behavioral experiment. Bees were injected with actinomycin D (Act D) or phosphate buffered saline (PBS) 90 min prior to a paired training and memory retention was tested 24 h and 72 h after the training with the conditioned stimulus (CS) and a novel odor (N). (B,C) Percentage of bees responding to the presented odor during paired training and memory retention test 24 h (B) and 72 h (C) after training. During the memory retention test, half of the bees of each group received the CS first and 10 min later the novel odor. For the other half of the bees, the sequence was reversed. Because the subgroups did not significantly differ in their CS responses during training or memory retrieval, their results were pooled (training: rmANOVA factor odorAct D 24 h: F(1,59) = 0.23; factor odorPBS 24 h: F(1,63) = 0.15, factor odorAct D 72 h: F(1,52) = 0.002, for all P > 0.05; PBS-injected bees of the 72 h group could not be tested with rmANOVA due to exact equal means in the subgroups, retention test: Fisher's exact tests between the subgroups “first odor CS” and “first odor novel,” all P > 0.05). (*) Significant differences: P < 0.0125 (Bonferroni-adjusted) detected with Fisher's exact and McNemar tests. (US) unconditioned stimulus, (PER) proboscis extension response.
Bees received a systemic injection of the transcriptional inhibitor actinomycin D (Act D, 1.5 mM) or the solvent phosphate buffered saline (PBS) 90 min before paired training and were tested with the CS and a novel odor 24 or 72 h later (Fig. 1A).
During paired training, the CS response levels of the Act D-injected group and the PBS-injected group increased from the first to the third trial and did not differ between the two groups (rmANOVA factor trial 24 h: F(2,248) = 287.56 and factor trial72 h: F(2,208) = 235.15, for both P < 0.001; factor injection24 h: F(1,124) = 0.90 and factor trial × injection24 h: F(2,248) = 0.95, for both P > 0.05; factor injection72 h: F(1,104) = 0.10 and factor trial × injection72h: F(2,208) = 0.70, for both P > 0.05) (Fig. 1B,C). Memory retrieval 24 h later revealed a significantly higher responsiveness to the CS compared with the novel odor indicating odor-specific memory formation (McNemar tests with Bonferroni adjustment, P < 0.0125). No effect of the Act D- and PBS-treatment was observed (Fisher's exact tests with Bonferroni adjustment, P > 0.0125 for CS and novel odor) (Fig. 1B). After 72 h, both groups showed a significantly higher responsiveness to the CS compared with the novel odor (McNemar tests with Bonferroni adjustment, P < 0.0125) (Fig. 1C). A significant difference in responsiveness to the CS, but not to the novel odor, was observed between the Act D-treated group and the PBS-treated group (Fisher's exact test with Bonferroni adjustment, P < 0.0125 for clove, P > 0.0125 for 1-hexanol) (Fig 1C). Accordingly, we demonstrated that the paired protocol leads to an odor-specific, transcription-dependent lLTM.
During unpaired training sensitization takes place
As second training protocol we wanted to use one that does not lead to transcription-dependent lLTM although the same number of CS–US trials is presented as in the paired protocol. We chose an unpaired training protocol during which the bees received alternating CS- and US-presentations with an inter-stimulus interval of 5 min. First, we examined learning and memory formation in bees trained with the unpaired protocol as it was unclear what is learned during unpaired training and whether unpaired training leads to memory formation in honeybees.
In a first experiment, we analyzed two groups that had either been exposed to one unpaired CS- and US-presentation with an intertrial interval of 5 min or to one CS-only trial. After stimulus exposure, both groups were subdivided into two groups each. One group was tested first with the CS and 7 min later with the novel odor or vice versa (Fig. 2A). In the group that received one unpaired training trial, significantly more bees responded to the CS and to the novel odor than in the group that received the CS-only presentation (Fisher's exact tests with Bonferroni adjustment, P < 0.0125 for CS unpaired versus CS CS-only and Nunpaired versus NCS-only) (Fig. 2B). Both groups, the unpaired group and the CS-only group, did not significantly differ in their response levels to the CS and to the novel odor (McNemar tests with Bonferroni adjustment, P > 0.0125 for CSunpaired versus Nunpaired and CSCS-only versus NCS-only) (Fig. 2B). These results demonstrate that the odor responsiveness is enhanced in bees that received unpaired training compared with bees that receive a CS-only presentation. Because the unpaired group did not differ in their responses to the CS and the novel odor, we conclude that the enhancement of odor responses during unpaired training are US-dependent and therefore are based on sensitization.
Figure 2.
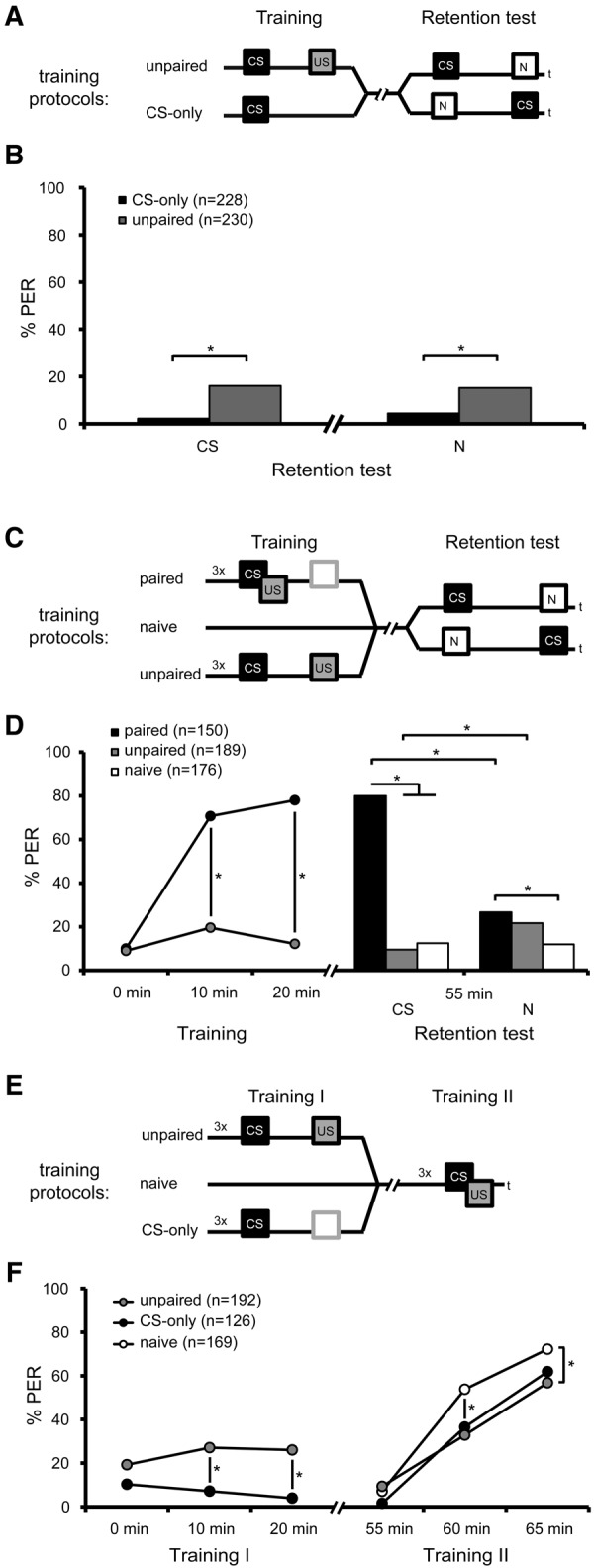
Learning and memory formation during unpaired CS- and US-presentation. (A) Schematic representation of the behavioral experiment. Bees were trained with one unpaired presentation of CS and US or were exposed to the CS alone (CS-only). After stimulus exposure, both groups were subdivided into two groups each. One group was tested first with the CS and then with the novel odor (N) or vice versa. (B) Percentage of bees responding to the presented odor (CS or novel odor) during the memory retention test after the presentation of the US. The sequence of odor presentation did not significantly alter the bees’ responsiveness to the odors and therefore the results were pooled. (Fisher's exact tests of CS responses and novel odor responses between the subgroups “first odor CS” and “first odor novel,” all P > 0.05). (C) Schematic representation of the behavioral experiment. Naive, paired, or unpaired trained bees were tested 55 min after training with the odor used during the training phase (CS) and a novel odor (N). (D) Percentage of bees responding to the presented odor during training and memory retention tests 55 min after training. During memory retention the sequence of the tested odors did not significantly alter the bees’ odor responsiveness for each odor (Training: rmANOVA factor odorpaired: F(1,148) = 0.01, factor odorunpaired F(1,187) = 2.08, P > 0.05, Retention test: Fisher's exact tests between the subgroups “first odor CS” and “first odor novel,” all P > 0.05). Thus we pooled the results of each odor. (E) Schematic representation of the retardation of acquisition assay. Unpaired trained bees, bees exposed to CS-only trials and naive bees (Training I) underwent paired training 55 min later (Training II). (F) Percentage of bees responding to the presented odor during Training I and Training II of the retardation of acquisition assay. (*) Significant differences: P < 0.0056 (Bonferroni-adjusted) detected with Fisher's exact and McNemar tests (B), P < 0.05 detected with Tukey HSD post hoc tests after rmANOVA (D), P < 0.016 (Bonferroni-adjusted) detected with Fisher's exact and McNemar tests (F)
In the second experiment, we asked about memory formation following unpaired training this time with three CS- and US-presentations. We compared acquisition and 1 h memory retention of the unpaired group, the paired group, and a naive group. All three groups were tested with the CS and a novel odor (Fig. 2C).
During the training phase, the CS responses of the unpaired group differed significantly from the paired group at the second and third trial (rmANOVA: factor trial: F(2,674) = 142.00, P < 0.001; factor training: F(1,337) = 190.25, P < 0.001; trial × training: F(2,674) = 97.00, P < 0.001, post hoc Tukey HSD test paired versus unpaired: trial 1, P = 0.99; trial 2, P = 0.00; trial 3, P = 0.00; paired: trial 1 versus trail 2, P = 0.01, trial 1 versus trial 3, P = 0.93; paired: trial 1 versus trial 2, P = 0.00, trial 1 versus trial 3, P = 0.00) (Fig. 2D). During the retention test, significantly more animals of the unpaired group responded to the novel odor than to the CS, whereas significantly more animals in the paired group responded to the CS than to the novel odor (McNemar test with Bonferroni adjustment, P < 0.0056 for both tests) (Fig. 2D). In addition, the responses of the unpaired group to both odors were comparable to the responses of the naive group (Fisher's exact test with Bonferroni adjustment, P > 0.0056 for CS and novel odor). Responses of the paired group to both the CS and the novel odor differed significantly from the responses of the naive group (Fisher's exact test with Bonferroni adjustment, P < 0.0056 for CS and novel odor).
These results demonstrate that an odor-specific 1 h memory was not formed in the unpaired group. The probability of a CS response in the unpaired group was similar to that in the naive group, indicating that bees from the unpaired group were not sensitized by the unpaired training 55 min later.
Formation of a memory for the inhibitory properties of the CS
In the previous experiment, the unpaired group responded less to the CS than to the novel odor, suggesting that the CS might have acquired inhibitory properties during the unpaired CS–US presentations. This was the case in studies where honeybees received repeated unreinforced CS presentations or multiple unpaired CS and US presentations (Bitterman et al. 1983; Chandra et al. 2010). Therefore, in the next experiment, we asked whether unpaired trained bees form a memory about inhibitory properties of the CS, which can be demonstrated by using a retardation of acquisition assay. Because it is known that mere exposure to the CS without reinforcement retards subsequent acquisition in honeybees (latent inhibition, Chandra et al. 2010), we used a group of bees that had been exposed only to the CS (CS-only group) as a control group.
We analyzed three groups of bees. In the first training phase (Training I), one group received unpaired training as indicated above (unpaired group), the second group received only CS presentations (CS-only group), and the third group remained naive (naive group). Subsequently, all groups received additional paired training (Training II) to analyze whether the unpaired group and the CS-only group shows a retardation of acquisition compared to the naive group (Fig. 2E). During the first training phase, the response level of the unpaired group differed significantly from that of the CS-only group (rmANOVA: factor trial: F(2,636) = 0.75, P > 0.05; factor training: F(1,318) = 29.37, P < 0.001; factor trial × training: F(2,636) = 3.82, P < 0.05, post hoc Tukey HSD test unpaired versus CS-only: trial 1, P = 0.26; trial 2, P = 0.00; trial 3, P = 0.00) (Fig. 2F), indicating again that unpaired CS- and US-presentations result in an enhanced responsiveness to the CS compared with presentation of the CS alone.
During the second training phase, all groups showed an increase in CS responsiveness from the first to the third training trial, thus indicating learning (rmANOVA factor trial: F(2,968) = 294.84, P < 0.001). The unpaired and the CS-only group differed in their CS responsiveness from the naive group (rmANOVA factor training: F(2,484) = 8.25, P < 0.001). The CS-only group differed in their CS response from the naive group at the second trial, the unpaired group and the naive group differed at the second and third trial (rmANOVA factor trial × training: F(4,968) = 5.15, P < 0.001, post hoc Tukey HSD test unpaired versus naive: trial 1, P = 0.99; trial 2, P = 0.00; trial 3, P = 0.01; CS-only versus naive: trial 1, P = 0.97; trial 2, P = 0.01; trial 3, P = 0.48) (Fig. 2F).
Taken together, the unpaired training and the CS-only presentation retard the acquisition during a subsequent paired training phase to a similar extent, indicating that bees of the unpaired group and the CS-only group formed a memory about inhibitory properties of the CS.
No transcription-dependent memory about inhibitory properties of the CS following unpaired training
Next, we examined the formation of transcription-dependent memories about the inhibitory effect of the CS acquired during unpaired training.
We analyzed two groups, the naive group and the unpaired group, and injected the transcriptional inhibitor Act D or the solvent PBS 90 min before the unpaired training (Training I) started. All four groups received paired training (Training II) 3, 24, or 72 h after unpaired training (Training I) (Fig. 3A).
Figure 3.
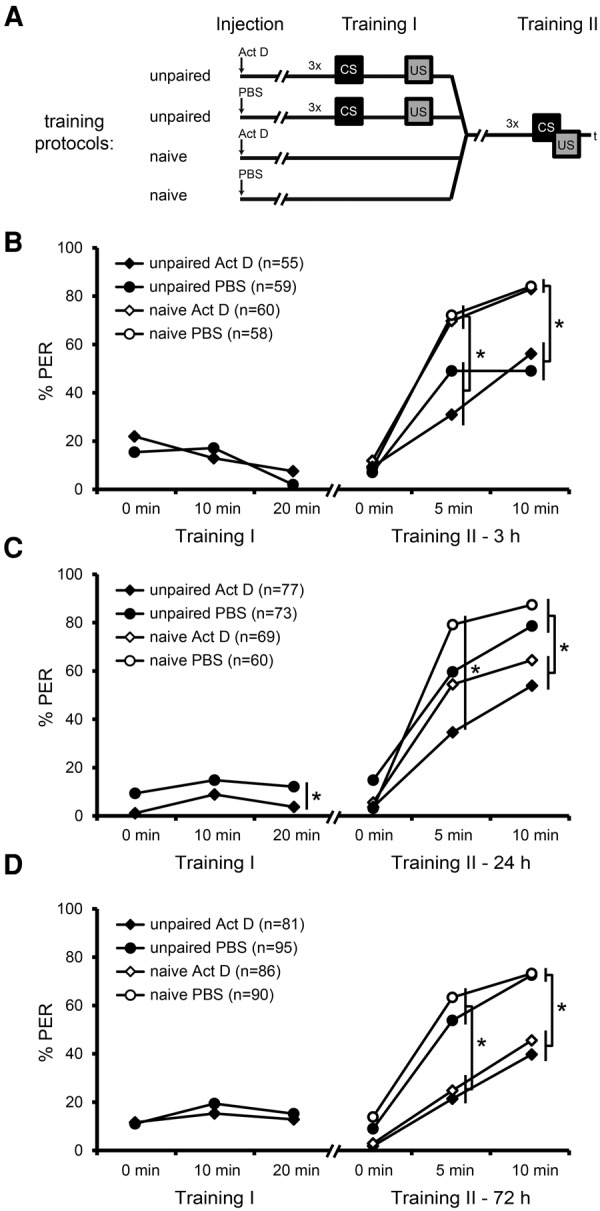
Unpaired training leads to the formation of a 3 and a 24 h, but not a 72 h memory about inhibitory properties of the CS. (A) Schematic representation of the retardation of acquisition assay. Bees were injected with Act D or PBS 90 min prior to unpaired training (Training I) and received paired training (Training II) 3, 24, and 72 h following unpaired training. (B–D) Percentage of bees responding to the CS during Training I and Training II after 3 h (B), 24 h (C), and 72 h (D). (*) Significant differences: P < 0.05 detected with Tukey HSD post-hoc tests after rmANOVA.
In two of the three experiments, the Act D-injected bees did not differ from the PBS-injected bees in their responsiveness toward the CS during the unpaired CS–US presentations (Training I) (rmANOVA factor injection3 h: F(1,112) = 0.36, factor injection72 h: F(1,174) = 0.27, for both P > 0.05, but see factor injection24 h: F(1,148) = 6.15 P < 0.05) (Fig. 3B–D). In one of the three experiments, the unpaired trained bees showed a decrease in CS responsiveness from the first to the third trial (rmANOVA factor trial3 h: F(2,224) = 7.55, P < 0.001, but see factor trial24 h: F(2,296) = 2.7 and factor trial72 h: F(2,348) = 1.85, for both P > 0.05) (Fig. 3B–D).
During paired training 3 h later (Training II) (Fig. 3B), the unpaired and the naive group showed an increase in CS responsiveness from the first to the third trial (rmANOVA factor trial: F(2,456) = 179.4, P < 0.001), indicating successful learning. A difference in responsiveness to the CS was observed between the unpaired and the naive group (rmANOVA factor training: F(1,228) = 32.45, P < 0.001). The unpaired trained bees showed a lower CS response at the second and third training trial than the naive bees and, accordingly, a retardation of acquisition (rmANOVA factor trial × training: F(2,456) = 12.86, P < 0.001, post hoc Tukey HSD test unpaired versus naive: trial 1, P = 0.99; trial 2, P = 0.00; trial 3, P = 0.00). No effect of Act D was observed (rmANOVA factor injection: F(1,228) = 0.16, P > 0.05) (Fig. 3B). We conclude that 3 h after unpaired training a memory has been formed for the inhibitory properties of the CS. This memory is not transcription-dependent.
During paired training 24 h later (Training II) (Fig. 3C), the unpaired and the naive group showed an increase in CS responsiveness from the first to the third trial (rmANOVA factor trial: F(2,550) = 292.69, P < 0.001), indicating successful learning. A difference in responses to the CS was observed between the unpaired and the naive group (rmANOVA factor training: F(1,275) = 5.38, P < 0.05). The unpaired trained bees showed a lower CS response than the naive bees at the second training trial and thus a retardation of acquisition (rmANOVA factor trial × training: F(2,550) = 9.78, P < 0.001, post hoc Tukey HSD test unpaired versus naive: trial 1, P = 0.927; trial 2, P = 0.000; trial 3, P = 0.381) (Fig. 3C). An effect of Act D on the bees’ CS responses was observed (rmANOVA factor injection: F(1,275) = 25.11, P < 0.001). Act D-injected bees showed a significantly lower CS responsiveness than PBS-injected bees at the second and third training trial, demonstrating an effect of Act D on learning during paired training (Training II) (rmANOVA factor trial × injection: F(2,550) = 8.75, P < 0.001, post hoc Tukey HSD test PBS-injected versus Act D-injected animals: trial 1, P = 0.90; trial 2, P = 0.00; trial 3, P = 0.00) (Fig. 3C). However, Act D did not impair the CS responsiveness of a particular group, i.e., unpaired or naive bees, during paired training (Training II) (rmANOVA factor training × injection: F(1,275) = 0.54, P > 0.05), indicating that, independently of each other, both factors influence the CS responsiveness of the different groups. Thus, 24 h after unpaired training, bees formed a memory about the inhibitory properties of the CS, but this memory is not transcription-dependent. However, inhibition of transcription 24 h before the retardation of acquisition assay inhibits acquisition per se.
During paired training 72 h later (Training II) (Fig. 3D), the unpaired and the naive group showed an increase in CS responsiveness from the first to the third trial (rmANOVA factor trial: F(2,696) = 215.66, P < 0.001). No difference in responsiveness to the CS was observed between the unpaired and the naive group (rmANOVA factor training: F(1,348) = 1.77, P > 0.05; factor trial × training: F(2,696) = 0.31, P > 0.05) (Fig. 3D). Significantly fewer bees of the Act D-injected group showed a response compared to the PBS-injected group (rmANOVA factor injection: F(1,348) = 61.52, P < 0.001). Act D-injected bees of the unpaired and the naive group also showed a significantly lower response than the PBS-injected bees at the second and third trial, demonstrating an effect of Act D on learning during paired training (Training II) (rmANOVA factor trial × injection: F(2,696) = 15.77, P < 0.001, post hoc Tukey HSD test PBS-injected versus Act D-injected animals: trial 1, P = 0.29; trial 2, P = 0.00; trial 3, P = 0.00) (Fig. 3D). These results demonstrate that no 72 h memory about the CS’ inhibitory properties has been formed. However, inhibition of transcription 72 h before the retardation of acquisition inhibits the bees’ CS response during paired training.
Taken together, our findings demonstrate that bees form a 3 h memory (MTM) and a 24 h memory (eLTM) but no 72 h memory (lLTM) about the CS’ inhibitory properties after unpaired training. The MTM and the eLTM are not susceptible to the transcriptional inhibitor Act D, indicating that memory formation does not depend on transcription. Accordingly, we do not expect an involvement of transcription factors in memory formation following unpaired training. Therefore, the unpaired group is an appropriate control group for subsequent experiments examining pAmCREB following paired training.
The level of pAmCREB in the central brain is not altered immediately after olfactory classical conditioning
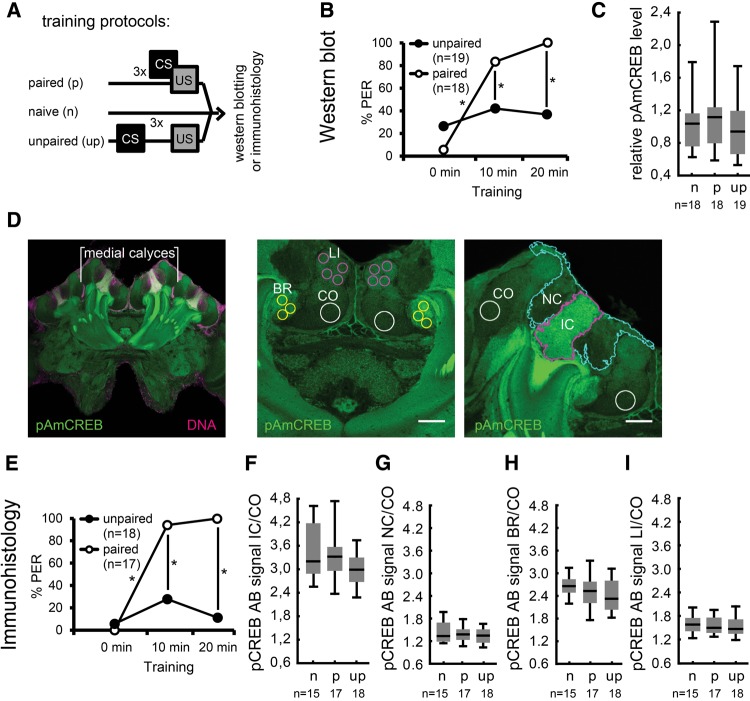
Next, we compared the pAmCREB level of bees that were subjected to the paired training protocol (paired group), the unpaired training protocol (unpaired group) and a naive group, which was left untrained, in Western blot analysis and immunohistological experiments (Fig. 4A). To exclude age-dependent variation in the level of pAmCREB (Gehring et al. 2016), age-matched bees were used.
Figure 4.
The level of pAmCREB does not differ between paired, unpaired trained and naive bees. (A) Schematic representation of the behavioral experiments that preceded the quantification of pAmCREB levels using Western blot analysis (B,C) or immunohistochemistry (D–I). Brains of naive, paired, or unpaired trained bees were analyzed. (B) Behavior of bees subsequently undergoing Western blot analysis. Shown are the percentages of bees responding to the CS during training. (C) Relative levels of pAmCREB in the central brain detected on Western blots. Naive (n), paired (p), and unpaired (up) trained groups did not differ in the level of pAmCREB in the central brain directly after the last US presentation. (D) Learning-dependent changes in the intensity of fluorescence-labeled pAmCREB in subpopulations of mushroom body neurons. (Left) Overview of the central honeybee brain stained with the pCREB antibody (green) and a DNA stain (magenta). The medial calyces were measured. (Center, right) Magnification of medial calyces showing the regions of interest (ROIs) measured during pCREB antibody signal intensity quantification. (Center) ROIs were placed in lip (magenta line), collar (white line), and basal ring (yellow line). (Right) ROIs comprised the inner compact cells (magenta line), and the noncompact cells (blue line) somata regions. (E) Behavior of bees subsequently undergoing immunohistochemistry. Shown are the percentages of bees responding to the CS during training. (F–I) Relative pCREB antibody signal intensities measured in different regions of the mushroom bodies of naive (n), paired (p), or unpaired (up) trained bees: inner compact cell somata (F), noncompact cell somata (G), basal ring (H), and lip (I). Naive (n), paired (p), and unpaired (up) trained groups did not differ in signal intensities directly after training. Box blots show median, 25% and 75% quartiles and value range (min–max). (*) Significant differences: P < 0.05 detected with Tukey HSD post hoc tests after rmANOVA. (IC) inner compact cell somata, (NC) noncompact cell somata.
During training, the paired group differed from the unpaired group in its responsiveness toward the CS at the second and third trial, and a significant increase in the responses to the CS from the first to the second trial was observed in the paired group but not in the unpaired group (rmANOVA factor trial: F(2,70) = 29.51, P < 0.001; factor training: F(1,35) = 8.47, P < 0.01; factor trial × training: F(2,70) = 16.85, P < 0.001, post hoc Tukey HSD test paired trial 1 versus unpaired trial 1, P = 0.60; paired trial 2 versus unpaired trial 2, P = 0.02; paired trial 3 versus unpaired trial 3, P = 0.00; paired trial 1 versus paired trial 2, P = 0.00; unpaired trial 1 versus unpaired trial 2, P = 0.66) (Fig. 4B). Immediately after the last US presentation, bees of the paired and unpaired group and, at similar time points, naive bees were sacrificed and central bee brains were dissected. Western blot analyses were performed to quantify the relative level of pAmCREB. The analysis revealed no significant difference in pAmCREB levels between bees that received paired training, unpaired training and naive animals (Kruskal–Wallis ANOVA over all groups: H(2,N = 55) =0.40, P > 0.05) (Fig. 4C). Thus, neither the paired training shown to induce lLTM (see above) nor the unpaired training altered the level of pAmCREB in the central bee brain immediately following training.
The level of pAmCREB in IC is not altered immediately after olfactory classical conditioning
Next, we measured pAmCREB levels in subpopulations of mushroom body neurons: in the inner compact cells (IC) that display a high abundance of pAmCREB, and in the noncompact cells (NC), where the level of pAmCREB is rather low (Gehring et al. 2016). The same experiment as described above was performed, but instead of processing the bee brains for Western blotting, the brains were fixed and pAmCREB was immunolabeled. The pCREB antibody signal in defined brain regions of bees experiencing paired training, unpaired training and of naive bees was measured (Fig. 4D). Similar to the results of the Western blot analysis (Fig. 4B), the paired group differed from the unpaired group in its responsiveness toward the CS at the second and third trial during training, and a significant increase in the responses to the CS from the first to the second trial was observed in the paired group but not in the unpaired group (rmANOVA factor trial: F(2,66) = 41.91, P < 0.001; factor training: F(1,33) = 72.67, P < 0.001; factor trial × training: F(2,66) = 25.27, P < 0.001, post-hoc Tukey HSD test paired trial 1 versus unpaired trial 1, P = 0.99; paired trial 2 versus unpaired trial 2, P = 0.00; paired trial 3 versus unpaired trial 3, P = 0.00; paired trial 1 versus paired trial 2, P = 0.00; unpaired trial 1 versus unpaired trial 2, P = 0.19) (Fig. 4E). Immediately after the last US presentation, brains from the paired and unpaired group and, at similar time points, from naive bees were fixed and immunostainings were performed. The pCREB antibody signal intensity was measured in regions of interest (ROIs) in the IC, the NC, and their dendritic input regions, the basal ring and the lip region (Fig. 4D). Consistent with the observed results of the Western blot analysis of central bee brains (Fig. 4C), no significant differences in relative pAmCREB levels were found in the region of IC somata, NC somata, lip, basal ring, and basal ring subregions between paired trained, unpaired trained, and naive bees (Kruskal–Wallis ANOVA over all groups, IC: H(2,N = 50) = 4.05, NC: H(2,N = 50) = 0.57, basal ring: H(2,N = 50) = 4.37, lip: H(2,N = 50) = 1.23, P > 0.05 for all tests) (Fig. 4F–I). In summary, these results demonstrate that neither paired training, i.e., olfactory classical conditioning, nor unpaired training, i.e., unpaired presentations of the CS and the US, altered the pAmCREB levels in the central brain or in the investigated regions of the mushroom bodies immediately following training.
The level of pAmCREB correlates with the CS responsiveness of a bee during unpaired CS and US presentations
Above, we demonstrated that the CS responsiveness is reduced during paired training when bees are injected with a transcription inhibitor 24 h beforehand (Fig. 3C,D). Accordingly, we hypothesize that the bee's responsiveness to the CS is depending on transcription taking place before acquisition and therefore related to the abundance of activated transcription factors. Thus, we next examined whether the level of pAmCREB might be correlated with a bee's responsiveness to the CS. We tested this hypothesis by analyzing bees of the previous experiment according to their response to the CS. In contrast to the paired group, the unpaired group consisted of bees that never responded to the CS (nonresponder bees) and bees that did respond to the CS at any of the CS presentations during unpaired training (responder bees) (Fig. 5A). Therefore, we compared the pAmCREB levels in nonresponder and responder bees of the unpaired group. The level of pAmCREB detected in the central brains of responder bees was significantly higher than that found in the central brains of nonresponder bees (Mann–Whitney U-test, P < 0.05) (Fig. 5B). The same correlation was observed in the IC somata (Mann–Whitney U-test, P < 0.05) (Fig. 5C). However, the NC somata, the basal ring and the lip did not differ in the intensity of the pCREB antibody signal between responder and nonresponder bees (Mann–Whitney U-test, P > 0.05 for all tests) (Fig. 5D–F). Thus, in the unpaired group, the behavioral response of individual bees toward the CS correlated with the level of pAmCREB in the central bee brain or, more specifically, in the IC. These results suggest that the bees’ CS responsiveness during unpaired training is linked to the altered level of pAmCREB observed in IC somata.
Figure 5.
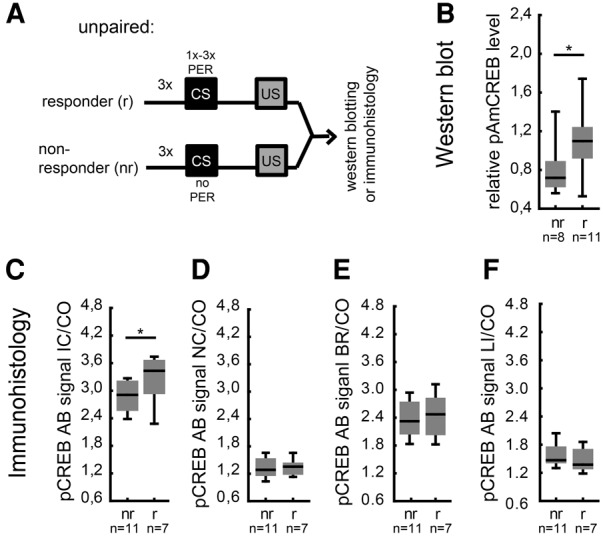
The level of pAmCREB correlates with a bee's CS responsiveness during unpaired training. (A) Schematic representation of the behavioral criterion used to identify responder and nonresponder bees from the unpaired trained group shown in Figure 4. (B) Relative levels of pAmCREB in the central brain of responder and nonresponder bees detected on Western blots. Bees that responded to at least one CS presentation during the unpaired training (responders) differed in the level of pAmCREB compared with bees that did not respond to the CS (nonresponders). (C–F) Relative pCREB antibody signal intensities measured using immunohistochemistry in different mushroom body regions of responder and nonresponder bees of the unpaired group: inner compact cell somata (C), noncompact cell somata (D), basal ring (E), and lip (F). Different pCREB antibody signal intensities detected between responders and nonresponders in the inner compact cell somata but not in the noncompact cell somata, basal ring, or lip. Box blots show median, 25% and 75% quartiles and value range (min–max). (*) Significant differences: P < 0.05 detected with Mann–Whitney U-tests. (IC) inner compact cell somata, (NC) noncompact cell somata, (BR) basal ring, (LI) lip, (CO) collar.
The CS responsiveness of a bee during unpaired CS and US presentations depends on protein acetylation
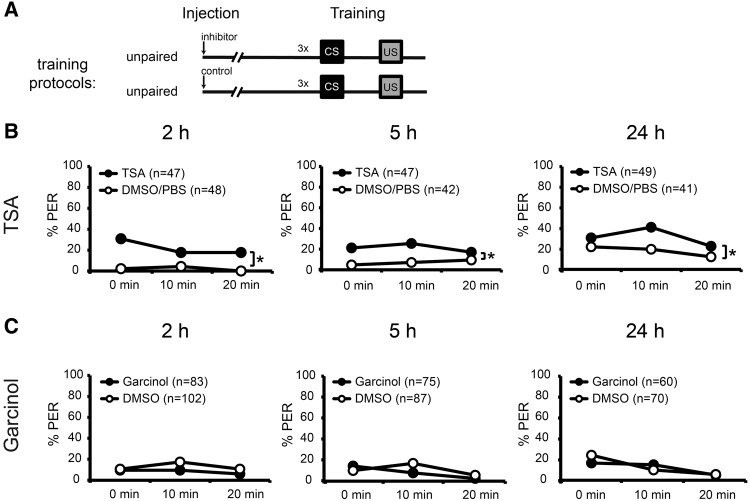
In the last experiment, we examined whether interference with the consequences of CREB phosphorylation alters the honeybees responsiveness during unpaired training. In vertebrate animals, CREB phosphorylation at S133 promotes recruitment of the co-activator protein CBP and its paralog p300 thereby leading to CREB-dependent transcription (Yamamoto et al. 1990; Chrivia et al. 1993; Montminy 1997; Du et al. 2000). CBP has a histone acetyltransferase activity (HAT) and acetylates histones and other nuclear proteins including CREB itself (Bannister and Kouzarides 1996; Parker et al. 1996; Lu et al. 2003; Valor et al. 2013). Especially histone H3K18 is acetylated with high specificity by CBP and acetylated H3K18 is associated with transcriptional activation (Wang et al. 2008; Barber et al. 2012; Henry et al. 2013; Valor et al. 2013). In the honeybee genome, a histone acetyltransferase p300-like protein is encoded (NCBI reference sequence: XP_006568897), that is homolog to Mus musculus CBP (NCBI reference sequences: NP_001020603.1) and p300 (NCBI reference sequences: NP_808489.4) indicating that CBP/p300 can be found in honeybees. Based on these homologies, we propose that, in the honeybee, pAmCREB recruits CBP/p300, which then acetylates H3K18. We concluded from our results above that a high level of pAmCREB is linked to an enhanced odor responsiveness of honeybees during unpaired training. If this conclusion holds true, an enhancement of histone acetylation at H3K18 should enhance the odor responsiveness as well. In order to test this hypothesis, we interfered with acetylation of H3K18 before unpaired training. In honeybee brains, acetylation of H3K18 is enhanced 2 h after systemic injection of TSA, a histone deacetylase (HDAC) inhibitor, and reduced after injection of Garcinol, a HAT inhibitor (Merschbaecher et al. 2012). Therefore, we utilized both inhibitors to test our hypothesis.
We carried out two experiments where we injected half of the bees with either TSA or Garcinol and the other half with the corresponding vehicles. The injected bees were then further divided into groups receiving unpaired training 2, 5, or 24 h after injection (Fig. 6A).
Figure 6.
Inhibition of histone deacetylases enhances CS responsiveness. (A) Schematic representation of the behavioral experiments. Bees were injected with the histone deacetylase inhibitor Trichostatin A (TSA), the histone acetyl transferase inhibitor, Garcinol or the respective vehicles 2, 5, or 24 h prior to unpaired training (B) Percentage of bees responding to the CS during unpaired training after injection of Trichostatin A (B) or Garcinol (C). (*) Significant differences: P < 0.05 detected with Tukey HSD post hoc tests after rmANOVA.
Significantly more bees injected with the HDAC inhibitor TSA (1.65 mM) responded to the CS during unpaired training 2, 5, and 24 h after injection than bees from the control group (rmANOVA factor injection2 h: F(1,93) = 19.44, P < 0.001, factor injection5 h: F(1,87) = 6.95, P < 0.01, factor injection24 h: F(1,88) = 5.57, p < 0.05) (Fig. 6B). The injection of Garcinol (6 mM) did not significantly affect the bees’ CS responses (rmANOVA factor injectionGarcinol 2 h: F(1,183) = 2.19, P > 0.05, rmANOVA factor injectionGarcinol 5 h: F(1,160) = 0.67, P > 0.05, rmANOVA factor injectionGarcinol 24 h: F(1,128) = 0.08, P > 0.05) (Fig. 6C).
Taken together, these results demonstrate that an inhibition of HDACs, that has been shown to result in an enhanced histone acetylation, leads to an enhanced CS responsiveness in honeybees, whereas an inhibition of HATs in general does not affect the bees’ response to the CS during unpaired training. We conclude from these findings that an increasing level of protein acetylation enhances the CS responsiveness. This finding is in line with our conclusion of an involvement of AmCREB-dependent processes in regulating the bee's odor response during unpaired training
Discussion
Here, we shed light on the role of pAmCREB in the central brain of honeybees. We investigated whether changes in the abundance of pAmCREB might be related to learning and memory formation. We did not observe an altered pAmCREB level directly after learning. In contrast, our results indicate individual differences of pAmCREB before learning and suggest a role for pAmCREB in the central brain of honeybees in gating the responsiveness to external stimuli.
The CS acquires mixed properties during unpaired training
We examined differences in learning and memory formation following the learning protocols used for the experimental (paired group) and the control group (unpaired group). We demonstrate that unpaired training gives rise to two learning processes, sensitization and the acquisition of CS inhibitory properties. Similar results were obtained in rats, where the CS acquires excitatory and inhibitory properties during unpaired training (Droungas and LoLordo 1994). A CS with “mixed” properties has also been reported during backward conditioning in honeybee and vertebrates (Williams and Overmier 1988; Felsenberg et al. 2013) indicating that two opposing properties of a CS are learned whenever the CS’ meaning is ambiguous during training.
Early actinomycin D application inhibits acquisition
In line with several other studies in bees, we report the formation of an lLTM about excitatory properties of the CS after paired training, which is sensitive to the transcription inhibitor Act D (Wüstenberg et al. 1998; Menzel et al. 2001; Friedrich et al. 2004; Lefer et al. 2012). We were not able to retrieve lLTM after unpaired training. Thus, memories about the inhibitory properties formed after unpaired training are less stable than memories formed after forward conditioning with the same trial number. This result resembles findings from backward conditioning in honeybees (Felsenberg et al. 2015) and fruit flies Drosophila melanogaster (Diegelmann et al. 2013) where memories about the inhibitory properties of the CS are less stable than memories about the excitatory properties of the same CS formed after paired training.
Unexpectedly, our experiments revealed the inhibition of the acquisition if Act D was applied one day and three days before paired training. Act D treatment of honeybees reduces transcription by 60%–65% between 40 min and up to 2–2.5 h after systemic injection (Wüstenberg et al. 1998; Menzel et al. 2001). Act D intercalates in the DNA, preferentially blocking transcription in transcription bubbles that have already been unwound by DNA helicases (Sobell 1985; Paramanathan et al. 2012). Thus, Act D transiently blocks ongoing gene transcription in naive bees. We exclude a role for these genes in sucrose perception because all analyzed bees responded to the US during each US presentation. An effect on odor perception one day after Act D treatment seems unlikely as well because Act D did not inhibit the CS response during memory retention 24 h after paired training (see Fig. 1). Thus, Act D might down-regulate constitutively expressed proteins important for the bees’ ability to learn.
The level of pAmCREB is not altered directly after classical conditioning
We did not detect any change in the pAmCREB level in the central honeybee brain or in the somata of intrinsic mushroom body neurons (IC or NC) directly after paired training compared with unpaired trained or naive bees. This is surprising because CREB plays an important role during LTM formation in various species and LTM-inducing training protocols give rise to an increase of neuronal pCREB levels in certain brain areas (Stanciu et al. 2001; Mizuno et al. 2002; Ribeiro et al. 2003; Trifilieff et al., 2006; Liu et al. 2008; Porte et al. 2008). Furthermore, lLTM formation in honeybees results in transcription-dependent structural changes in the lip of the mushroom body calyx (Hourcade et al. 2010), suggesting a learning-specific activation of transcription factors in this neuropil.
Several explanations for our finding can be envisaged: First, in mice, a biphasic pCREB peak can be observed 15 min and 8 h following water maze learning in the CA1 region of the hippocampus (Trifilieff et al., 2006) and a particular high pCREB level was reported 3–6 h after context-dependent fear conditioning in hippocampus, parietal cortex and amygdaloid nuclei (Stanciu et al. 2001). Thus, it might well be that in our experiment the pAmCREB level is enhanced in the central brain of the paired group compared with the unpaired and the naive group but at a later time point rather than immediately after training.
Second, in the fruit fly D. melanogaster, the formation of a hunger-dependent appetitive LTM depends on the interaction of CREB with the cAMP-regulated transcriptional coactivator (CRTC), but not on the cofactor CBP (Hirano et al. 2013). CRTC binds to CREB at the C-terminal basic leucine zipper domain and not the phosphorylated kinase-inducible domain (Luo et al. 2012). Thus, the formation of a hunger-dependent appetitive LTM might be independent of CREB phosphorylation at the PKA consensus site. This might also apply to honeybee lLTM formation, which depends on the bees’ feeding state (Friedrich et al. 2004). Third, research in the fruit fly suggests that the rate limiting step for the D. melanogaster dCREB2 activity is not the phosphorylation of the S133 homologous serine residue but the nuclear entry and DNA binding of dCREB2, which is regulated by phosphorylation of residues other than the S133 homologous one (Horiuchi et al. 2004). Moreover, the nuclear entry rate of pCREB2 correlates with memory formation (Fropf et al. 2013). Thus, it might well be that learning-dependent activation of AmCREB is correlated with other mechanisms such as the nuclear entry and DNA binding instead of phosphorylation at the S133 homologous serine residue.
The level of pAmCREB correlates with a bee's responsiveness toward the CS
We report that, immediately after training, bees responding to the CS during unpaired training showed an increased level of pAmCREB compared with bees that did not respond at all, suggesting a correlation between the amount of pAmCREB and the bees’ CS responsiveness. There are two possible explanations for this finding: First, some bees might have higher pAmCREB levels per se, causing the difference in behavior toward the CS. Second, the level of pAmCREB was changed due to learning during the unpaired training, resulting in the formation of a transcription-dependent lLTM.
The latter seems very unlikely because no difference in the level of pAmCREB was observed between bees experiencing unpaired training and naive bees. Furthermore, the formation of a LTM about the CS excitatory properties was not observed although the US enhanced the response to the CS during unpaired training. In addition, we demonstrate that the LTM about the CS inhibitory properties is not transcription-dependent. Therefore, we propose that individual bees differ in the basal pAmCREB level in the central brain and that these differences play a role in setting a threshold for the CS responsiveness: a low level of pAmCREB is linked to a low CS responsiveness of bees, whereas a high level of pAmCREB is linked to a high CS responsiveness.
We found that TSA, which enhances histone acetylation in honeybees (Merschbaecher et al. 2012), enhances the bees’ CS responsiveness before and during unpaired training. Because an enhanced phosphorylation of CREB at S133 results in an enhanced binding of CBP (Du et al. 2000) and CBP is a transcriptional coactivator with acetyltransferase activity (for review, see Valor et al. 2013), these results are in line with the notion of a causal relationship between CREB phosphorylation and the bee's stimulus responsiveness. Further support for the idea that the level of phosphorylated CREB is important for gating the response to an external stimulus comes from studies in vertebrates where CREB, CBP and histone acetylation play a role in behavioral sensitization (Kumar et al. 2005; Levine et al. 2005; Renthal et al. 2007; Fasano et al. 2009; Sanchis-Segura et al. 2009a,b; Malvaez et al. 2011; Bilbao et al. 2014). Behavioral sensitization is a process in which repeated exposure to addictive drugs leads to a long-lasting enhancement of drug-induced behavior (Robinson and Berridge 1993; Stewart et al. 1993; Vanderschuren et al. 2001). Interestingly, the extent of behavioral responses following behavioral sensitization is correlated with the level of pCREB in certain brain areas (Nona et al. 2013).
Phosphorylated AmCREB and nonassociative learning
In our experiments with honeybees, it remains unclear whether the pAmCREB level in the IC or the central bee brain is linked to the bees’ ability to respond to an external stimulus per se or whether it is related to sensitization observed during unpaired training, and thus to the ability to learn nonassociatively. To our knowledge, a link between the level of basal pCREB and the ability to learn nonassociatively has not yet been demonstrated and learning and STM formation is thought to be independent of new gene expression and CREB activity. Nevertheless, Gruart et al. (2012) demonstrated that an up-regulation of CREB activity in the mouse hippocampus enhances acquisition in an associative learning paradigm. The authors discussed this finding to be a result of the enhanced expression of CREB target genes. In line, Suzuki et al. (2011) reported that the up-regulation of CREB activity before learning results in an enhanced CS responsiveness during STM retention as well as in the expression of the brain-derived neurotropic factor (BDNF) and demonstrate that BDNF improves STM retention.
Taken together, these findings suggest that up-regulation of CREB activity leads to an altered CREB-dependent gene expression contributing to an altered acquisition and STM retention. In the broader sense, this notion is supported by our finding that the inhibition of transcription with Act D hours before associative learning leads to an inhibition of the CS responsiveness during acquisition (see above). In sum, it seems to be quite clear that the level of activated CREB before learning affects the CS responsiveness during learning and memory retention, but it remains unclear whether it is attributed to an enhanced ability to learn or to enhanced stimulus responsiveness per se.
A role of pAmCREB-associated stimulus responsiveness in foraging?
Previously, we demonstrated a high abundance of pAmCREB in the IC of the honeybee mushroom bodies (Gehring et al. 2016) and, in this study, found a correlation between the level of pAmCREB in these neurons and the bees’ CS responsiveness. Little is known about the function of the IC, but they have been hypothesized to be involved in foraging behavior (Kiya et al. 2007). Interestingly, studies in ants suggest that individuals differing in their response thresholds enable a colony to respond flexibly to varying intrinsic and extrinsic demands (Robinson et al. 2009, 2012). Therefore, one might speculate that AmCREB-dependent processes in the IC linked to the CS responsiveness influence the foraging decision of individual honeybees. Support for a role of CREB in regulating behavior of social insects comes from bioinformatics analyses of genomic and transcriptomic data (Simola et al. 2013a,b). The honeybee and several social ant species can be discriminated from solitary species based on the abundance of CREB-binding sites (Simola et al. 2013a). In addition, in the ant Camponotus floridanus, a differential recruitment of CBP to enhancer sequences in different castes (female minors, female majors, and males) has been reported (Simola et al. 2013b). Furthermore, histone acetylation has been shown to be involved in regulating task-specific behavior in these ants (Simola et al. 2016). These results suggest a prominent role for CBP- and, accordingly, CREB-dependent gene activation in processes related to task-specific behavior of these castes. Based on our findings we therefore hypothesize that CREB-dependent processes might be important to regulate task-specific behavior of social insects in general and that the IC could be a subpopulation of mushroom body neurons that underlies the regulation of this behavior in honeybees.
Material and Methods
Animals and cohort experiments
Honeybees (Apis mellifera) leaving their hives located at the Freie Universität Berlin were caught, harnessed, and fed in the evening as described in Felsenberg et al. (2011).
For experiments exploring the learning-dependency of pAmCREB levels in the honeybee brain, we utilized age-matched cohorts that were prepared as described in Gehring et al. (2016). The bees were collected at the age of 19 or 20 d from inside the hive between 10 a.m. and 2 p.m. and were kept in a cage inside an incubator for 48 h (34°C, >75% humidity, water and 0.88 M sucrose solution, i.e., white refined household sugar dissolved in tap water, ad libitum). Next, they were restrained in plastic tubes and treated similarly as the other bees for behavioral experiments.
Training with different behavioral protocols started the following morning. Before training, the bees were allowed to adapt to their surroundings near the conditioning place for >30 min. Bees that were tested for memory retention and retardation of acquisition 24 or 72 h after training were fed on subsequent evenings after the training phase with no more than 16 µL (4 times 4 µL) sucrose solution (0.88 M) and were kept in a dark, humid box at room temperature.
From January to March 2012 and 2013, behavioral experiments were performed with bees collected from a caged colony in a glasshouse in order to examine the 1 h memory about the excitatory and inhibitory properties of the odor following paired and unpaired training (Fig. 2). Experiments examining the transcription-dependency of memory formation after paired and unpaired training (Figs. 1, 3) were conducted from July to October 2014 with bees caught from a hive in the garden. Experiments analyzing the role of protein acetylation were conducted from June to September 2015 (Fig. 6). Cohort experiments examining the effect of learning on the pAmCREB level in honeybee brains were conducted from July to September 2011 and from May to September 2012 (Figs. 4–6).
Behavioral experiments
Injection protocol
To explore the effect of transcription-, HAT-, and HDAC-inhibitors on the behavior of bees during and after training, drugs or their solvents were systemically injection into the flight muscle (Felsenberg et al. 2011). For the investigation of the transcriptional dependence of memories, bees were injected 90 min prior to the paired or unpaired training with 1.5 mM actinomycin D (Act D, Sigma-Aldrich) or the solvent PBS (137 mM NaCl; 2,7 mM KCl; 10,1 mM; Na2HPO4; 1,8 mM KH2PO4, pH 7.2). To examine the role of histone acetylation, bees were injected 2, 5, or 24 h prior to the unpaired training with 6 mM Garcinol (Abcam; dissolved in 100% DMSO), 1.65 mM Trichostatin A (TSA, Abcam; dissolved in 20% DMSO in PBS) or the appropriate solvent as control.
Behavioral protocols
Bees received paired presentations of conditioned stimulus (CS) and unconditioned stimulus (US) (paired training), unpaired presentations of CS and US (unpaired training), three CS presentations (CS-only training) or no stimulus presentation at all (naive). For simplicity, the abbreviations CS and US were used as equivalent to the odor clove oil (Bombastus-Werke) and the sucrose solution (1.25 M, sucrose dissolved in tap water), respectively, in every protocol.
Paired trained bees received three overlapping CS–US presentations (training trials) with an intertrial interval of 10 min. One trial consisted of placing the bee for 10 sec without any stimulus presentation in front of an exhaust (placing) followed by the presentation of the CS for 5 sec and the US for 4 sec, with an inter-stimulus interval of 3 sec (2 sec overlap). After an additional 10 sec of placing, bees were put back into the rack (Felsenberg et al. 2011). In experiments examining the 1 h memory about the excitatory properties of the CS, 5 min after each training trial, bees were placed in the training context without stimulation for 30 sec (the duration of a paired training trial) to balance the time spent in front of the exhaust (blank trial). During the training phase, a control group (naive) receiving no stimulation was placed near the conditioning place (together with the other bees). In the unpaired group, the bees received unpaired presentations of the CS and the US in alternating sequence with an interval of 5 min between the onset of each stimulus. The CS was presented as the first stimulus. One CS presentation consisted of 10 sec placing, 5 sec presentation of the CS, and 10 sec placing. One US presentation consisted of 10 sec placing, 4 sec presentation of the US, and 10 sec placing. After each stimulus presentation, the bees were put back into the rack. The CS-only group was treated like the unpaired group but with a blank trial instead of the US presentation.
Bees that did not respond to any of the US presentations during training and to the US presented after the last retention test (see below) were excluded from the analysis. The occurrence of a proboscis extension response (PER) during each odor presentation was recorded and the percentage of responses (% PER) was calculated.
Memory retention
Bees underwent memory retention tests ∼5 min (Fig. 2F), 1 h (55 min) (Fig. 2B), 24 h (Fig. 1B), or 72 h (Fig. 1C) after training onset. The different training groups (paired, unpaired, and naive) were divided into subgroups that were either exposed to the CS or to the novel odor 1-hexanol. Ten minutes later, animals that previously experienced the CS were exposed to 1-hexanol or vice versa. After the retention tests, the bees’ ability to extend their proboscis was tested by eliciting the PER with the US (sugar water test).
Retardation of acquisition assay
Memory retention for the inhibitory properties of the CS was tested in a retardation of acquisition assay (Figs. 2D, 3A–D). This assay is based on the assumption that inhibitory and excitatory properties of a CS add up during acquisition (Hammond 1968; Rescorla 1969; Papini and Bitterman 1993). Accordingly, classical conditioning with a CS that has previously acquired inhibitory properties results in a retarded acquisition. In the retardation of acquisition assay, after the initial training (Training I, training protocols see above), all groups received paired training with three paired CS–US trials with an intertrial interval of 5 min (Training II). One trial of the paired training consisted of 10 sec placing in front of an exhaust followed by presentation of the CS for 5 sec and the US for 4 sec, with an inter-stimulus interval of 3 sec (2 sec overlap). After an additional 10 sec of placing, bees were put back into the rack.
Treatment of bees for pAmCREB quantification
For the pAmCREB quantification experiments, bees were anesthetized directly after the last US presentation (in the paired and the unpaired group), their head capsule was opened and brains were further processed for Western blotting or immunohistochemistry.
Western blotting
Directly after the last stimulation during training, bees were anesthetized on ice, a window was cut into the head capsule, and trachea and glands were removed. Dissected and homogenized central bee brains were subjected to sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described in Gehring et al. (2016).
As primary antibodies, we used a monoclonal rabbit pCREB antibody directed against a synthetic phosphopeptide corresponding to residues surrounding S133 of human CREB (#9198, Cell Signaling Technology 1:1500) (see also Gehring et al. 2016) and, as standard, a monoclonal mouse α-tubulin antibody (#CP06, Calbiochem, Sandhausen, Germany, 1:3000). The primary antibodies were detected by secondary antibodies coupled to horseradish peroxidase directed against mouse IgG (for tubulin) or rabbit IgG (for pCREB detection) (1:10,000 for anti-mouse IgG and 1:5000 for anti-rabbit IgG, Sigma-Aldrich). Chamiluminescence signals were captured with a LAS1000 camera and the software Image Reader LAS-1000 2.60 (Fujifilm Corporation). Signal intensities were quantified with the program MultiGauge version 3.0 (Fujifilm). The acquired value of each 35-kDa band detected with the pCREB antibody was normalized to the mean value of all 35-kDa bands on the same blot. The value of each band detected with the α-tubulin antibody was normalized to the mean value of all α-tubulin bands. Finally, the ratio between the normalized pCREB and α-tubulin values was calculated for each sample. One sample represented one bee brain.
Immunohistochemistry
Immunohistochemistry was done as described in Gehring et al. (2016).
Brain slices were incubated with the pCREB antibody (1:75, see above) to label pAmCREB. To reliably identify the localization of lip, collar, basal ring, IC, and NC in the bee brain, slices were counterstained with Alexa Fluor 546 Phalloidin (0.3 units; Invitrogen) to label neuronal f-actin (Frambach et al. 2004; Groh et al. 2004) and SytoxGreen (1:2000, Invitrogen) to label cell nuclei. As secondary antibody we used a Cy5-conjugated goat anti-rabbit antibody (1:200, Jackson ImmunoResearch Laboratories).
Microscopy, image processing, and data acquisition
Labeled brain slices were scanned with a confocal laser-scanning microscope (TCS SP2, Leica Microsystems) equipped with an argon laser (488 nm), a helium-neon laser (633 nm), and a green helium-neon laser (543 nm). Optical sections were taken at a resolution of 1024×1024 pixels using a 40× or a 63× oil-immersion objective (HCX PL APO, Leica Microsystems). The channels of the triple-labeled preparations were pseudocolored and merged using the Zeiss LSM Image Browser (Version 3.2.0.115, Carl Zeiss Microscopy GmbH).To measure the intensity of pCREB antibody signals, one slice per bee was chosen at a depth of ∼250 µm where central complex and medial lobes of the mushroom bodies were present ensuring scanning of similar regions in each bee brain. Optical sections were taken in one optical plane from both medial mushroom body calyces with the 40× objective, and microscope settings were kept identical for comparable slices of other bees. Digital images were further processed and pCREB antibody signal intensities were measured as described in Gehring et al. (2016). Briefly, we measured the mean gray value of several regions of interest (ROIs) in the scanned images. ROIs of variable pixel sizes contained the entire area either of the IC or of the NC somata, and ROIs with a fixed size (radius of 50 pixels) were placed in the basal ring, the region containing IC dendrites, and the lip (see Fig. 4D; Gehring et al. 2016). ROIs with a radius of 100 pixels located in the dense region of the collar, a region displaying only background staining, served as control for all measurements. The synaptic regions were identified by f-actin counterstaining, whereas the DNA counterstaining helped to distinguish between IC and NC somata. The measured mean gray value of each ROI was normalized to the averaged gray values of the control regions in the collar. To obtain one single value per region and per bee, the mean of the normalized data of each region was calculated.
Statistical analysis
The values from quantitative Western blot analyses and from the quantification of pCREB antibody signal intensities were in most cases nonnormally distributed. Therefore groups were tested with the Kruskal–Wallis test and the two-tailed Mann–Whitney U-test. The αlevel was set at 0.05.
In behavioral experiments, differences in CS responsiveness during training between groups were tested using repeated measures analysis of variance (rmANOVA). The Tukey HSD test was used as post hoc test. To reveal within-group and between-group effects during retention tests with the CS and the novel odor, either Fisher's exact test (between-group comparisons) or McNemar χ2 test (within-group comparisons) was used. α-levels were Bonferroni adjusted. All statistical analyses were performed with Statistica 12 (StatSoft).
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research (BMBF), Grant number: 01GQ0941 (to D.E. within the Bernstein Focus Neuronal Basis of Learning) and by the Deutsche Forschungsgemeinschaft (DFG), Grant numbers: EI 512/2–1 (to D.E. as part of the joint project FOR 1363 Biogenic amines in insects: coordination of physiological processes and behavior).
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.040964.115.
References
- Alberini CM. 2009. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 89: 121–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament SA, Blatti CA, Alaux C, Wheeler MM, Toth AL, Le Conte Y, Hunt GJ, Guzmán-Novoa E, DeGrandi-Hoffman G, Uribe-Rubio JL, et al. 2012. New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. Proc Natl Acad Sci 109: E1801–E1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384: 641–643. [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. 2012. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Marie H. 2011. Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol Neurobiol 44: 330–349. [DOI] [PubMed] [Google Scholar]
- Benito E, Barco A. 2010. CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci 33: 230–240. [DOI] [PubMed] [Google Scholar]
- Beshers SN, Fewell JH. 2001. Models of division of labor in social insects. Annu Rev Entomol 46: 413–440. [DOI] [PubMed] [Google Scholar]
- Bilbao A, Rieker C, Cannella N, Parlato R, Golda S, Piechota M, Korostynski M, Engblom D, Przewlocki R, Schutz G, et al. 2014. CREB activity in dopamine D1 receptor expressing neurons regulates cocaine-induced behavioral effects. Front Behav Neurosci 8: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman ME, Menzel R, Fietz A, Schäfer S. 1983. Classical conditioning of proboscis extension in honeybees (Apis mellifera). J Comp Physiol 97: 107–119. [PubMed] [Google Scholar]
- Capaldi EA, Smith AD, Osborne JL, Fahrbach SE, Farris SM, Reynolds DR, Edwards AS, Martin A, Robinson GE, Poppy GM, et al. 2000. Ontogeny of orientation flight in the honeybee revealed by harmonic radar. Nature 403: 537–540. [DOI] [PubMed] [Google Scholar]
- Chandra SBC, Wright GA, Smith BH. 2010. Latent inhibition in the honey bee, Apis mellifera: is it a unitary phenomenon? Anim Cogn 13: 805–815. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, Price ND, Robinson GE. 2011. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci 108: 18020–18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859. [DOI] [PubMed] [Google Scholar]
- Degen J, Kirbach A, Reiter L, Lehmann K, Norton P, Storms M, Koblofsky M, Winter S, Georgieva PB, Nguyen H, et al. 2015. Exploratory behaviour of honeybees during orientation flights. Anim Behav 102: 45–57. [DOI] [PubMed] [Google Scholar]
- Diegelmann S, Klagges B, Michels B, Schleyer M, Gerber B. 2013. Maggot learning and Synapsin function. J Exp Biol 216: 939–951. [DOI] [PubMed] [Google Scholar]
- Droungas A, LoLordo VM. 1994. Evidence for simultaneous excitatory and inhibitory associations in the explicitly unpaired procedure. Learn Motiv 25: 1–25. [Google Scholar]
- Du K, Asahara H, Jhala US, Wagner BL, Montminy M. 2000. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol Cell Biol 20: 4320–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt D. 2014. Molecular mechanisms underlying formation of long-term reward memories and extinction memories in the honeybee (Apis mellifera). Learn Mem 12: 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt D, Friedrich A, Stollhoff N, Müller U, Kress H, Menzel R. 2003. The AmCREB gene is an ortholog of the mammalian CREB/CREM family of transcription factors and encodes several splice variants in the honeybee brain. Insect Mol Biol 12: 373–382. [DOI] [PubMed] [Google Scholar]
- Eisenhardt D, Kuhn C, Leboulle G. 2006. The PKA-CREB system encoded by the honeybee genome. Insect Mol Biol 15: 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano S, Pittenger C, Brambilla R. 2009. Inhibition of CREB activity in the dorsal portion of the striatum potentiates behavioral responses to drugs of abuse. Front Behav Neurosci 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J, Gehring KB, Antemann V, Eisenhardt D. 2011. Behavioural pharmacology in classical conditioning of the proboscis extension response in honeybees (Apis mellifera). Jove 47: 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J, Dombrowski V, Eisenhardt D. 2012. A role of protein degradation in memory consolidation after initial learning and extinction learning in the honeybee (Apis mellifera). Learn Mem 19: 470–477. [DOI] [PubMed] [Google Scholar]
- Felsenberg J, Plath JA, Lorang S, Morgenstern L, Eisenhardt D. 2013. Short- and long-term memories formed upon backward conditioning in honeybees (Apis mellifera). Learn Mem 21: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J, Dyck Y, Feige J, Ludwig J, Plath JA, Froese A, Karrenbrock M, Nolle A, Heufelder K, Eisenhardt D. 2015. Differences in long-term memory stability and AmCREB level between forward and backward conditioned honeybees (Apis mellifera). Front Behav Neurosci 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frambach I, Rössler W, Winkler M, Schürmann F-W. 2004. F-actin at identified synapses in the mushroom body neuropil of the insect brain. J Comp Neurol 475: 303–314. [DOI] [PubMed] [Google Scholar]
- Friedrich A, Thomas U, Muller U. 2004. Learning at different satiation levels reveals parallel functions for the cAMP-protein kinase A cascade in formation of long-term memory. J Neurosci 24: 4460–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fropf R, Tubon TC Jr, Yin JC. 2013. Nuclear gating of a Drosophila dCREB2 activator is involved in memory formation. Neurobiol Learn Mem 106: 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring KB, Heufelder K, Kersting I, Eisenhardt D. 2016. The abundance of phosphorylated Apis mellifera CREB in the honeybee's mushroom body inner compact cells varies with age. J Comp Neurol 524: 1165–1180. [DOI] [PubMed] [Google Scholar]
- Gerber B, Wustenberg D, Schutz A, Menzel R. 1998. Temporal determinants of olfactory long-term retention in honeybee classical conditioning: Nonmonotonous effects of the training trial interval. Neurobiol Learn Mem 69: 71–78. [DOI] [PubMed] [Google Scholar]
- Giurfa M, Sandoz JC. 2012. Invertebrate learning and memory: fifty years of olfactory conditioning of the proboscis extension response in honeybees. Learn Mem 19: 54–66. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs W III, Vale WW, Montminy MR. 1989. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature 337: 749–752. [DOI] [PubMed] [Google Scholar]
- Groh C, Tautz J, Rössler W. 2004. Synaptic organization in the adult honey bee brain is influenced by brood-temperature control during pupal development. Proc Natl Acad Sci 101: 4268–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruart A, Benito E, Delgado-García JM, Barco A. 2012. Enhanced cAMP response element-binding protein activity increases neuronal excitability, hippocampal long-term potentiation, and classical eyeblink conditioning in alert. J Neurosci 32: 17431–17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond LJ. 1968. Retardation of fear acquisition by a previously inhibitory CS. J Comp Physiol Psychol 66: 756–759. [DOI] [PubMed] [Google Scholar]
- Henry RA, Kuo Y-M, Andrews AJ. 2013. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochem 52: 5746–5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Masuda T, Naganos S, Matsuno M, Ueno K, Miyashita T, Horiuchi J, Saitoe M. 2013. Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science 339: 443–446. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Jiang W, Zhou H, Wu P, Yin JC. 2004. Phosphorylation of conserved casein kinase sites regulates cAMP-response element-binding protein DNA binding in Drosophila. J Biol Chem 279: 12117–12125. [DOI] [PubMed] [Google Scholar]
- Hourcade B, Muenz TS, Sandoz JC, Rossler W, Devaud JM. 2010. Long-term memory leads to synaptic reorganization in the mushroom bodies: a memory trace in the insect brain? J Neurosci 30: 6461–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis AM, Hamilton AR, Medvedeva YA, Alam T, Alam I, Essack M, Umylny B, Jankovic BR, Naeger NL, Suzuki M, et al. 2015. Insights into the transcriptional architecture of behavioral plasticity in the honey bee Apis mellifera. Sci Rep 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiya T, Kunieda T, Kubo T. 2007. Increased neural activity of a mushroom body neuron subtype in the brains of forager honeybees. PLoS One 2: e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. 2005. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48: 303–314. [DOI] [PubMed] [Google Scholar]
- Lefer D, Perisse E, Hourcade B, Sandoz J, Devaud JM. 2012. Two waves of transcription are required for long-term memory in the honeybee. Learn Mem 20: 29–33. [DOI] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. 2005. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci 102: 19186–19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RY, Fioravante D, Shah S, Byrne JH. 2008. cAMP response element-binding protein 1 feedback loop is necessary for consolidation of long-term synaptic facilitation in Aplysia. J Neurosci 28: 1970–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hutchins AE, Doyle CM, Lundblad JR, Kwok RP. 2003. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J Biol Chem 278: 15727–15734. [DOI] [PubMed] [Google Scholar]
- Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai WW, Talai A, Mayo KE, Montminy M, Radhakrishnan I. 2012. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc Natl Acad Sci 109: 20865–20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA. 2011. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci 31: 16941–16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Menzel R, Sandoz JC, Giurfa M. 2012. Revisiting olfactory classical conditioning of the proboscis extension response in honey bees: a step toward standardized procedures. J Neurosci Methods 211: 159–167. [DOI] [PubMed] [Google Scholar]
- Menzel R. 1990. Learning, memory and “cognition” in honey bees. In Neurobiology of comparative Cognition (ed. Kesner RP, Olton DS), pp. 237–292. Erlbaum Inc, Hillsdale, N.J. [Google Scholar]
- Menzel R, Manz G, Menzel R, Greggers U. 2001. Massed and spaced learning in honeybees: the role of CS, US, the intertrial interval, and the test interval. Learn Mem 8: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merschbaecher K, Haettig J, Mueller U. 2012. Acetylation-mediated suppression of transcription- independent memory: bidirectional modulation of memory by acetylation. PLoS One 7: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Yamada K, Maekawa N, Saito K, Seishima M, Nabeshima T. 2002. CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav Brain Res 133: 135–141. [DOI] [PubMed] [Google Scholar]
- Montminy M. 1997. Transcriptional regulation by cyclic AMP. Annu Rev Biochem 66: 807–822. [DOI] [PubMed] [Google Scholar]
- Müller U. 2013. Memory phases and signaling cascades in honeybees. In Invertebrate learning and memory (ed. Randolf Menzel PRB), pp. 433–441. Elsevier, London. [Google Scholar]
- Nona CN, Guirguis S, Nobrega JN. 2013. Susceptibility to ethanol sensitization is differentially associated with changes in pCREB, trkB and BDNF mRNA expression in the mouse brain. Behav Brain Res 242: 25–33. [DOI] [PubMed] [Google Scholar]
- Papini MR, Bitterman M. 1993. The two-test strategy in the study of inhibitory conditioning. J Exp Psychol Anim Behav Process 19: 342–352. [DOI] [PubMed] [Google Scholar]
- Paramanathan T, Vladescu I, McCauley MJ, Rouzina I, Williams MC. 2012. Force spectroscopy reveals the DNA structural dynamics that govern the slow binding of Actinomycin D. Nucleic Acids Res 40: 4925–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. 1996. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol 16: 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte Y, Buhot MC, Mons NE. 2008. Spatial memory in the Morris water maze and activation of cyclic AMP response element-binding (CREB) protein within the mouse hippocampus. Learn Mem 15: 885–894. [DOI] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE III, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. 2007. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56: 517–529. [DOI] [PubMed] [Google Scholar]
- Rescorla RA. 1969. Pavlovian conditioned inhibition. Psychol Bull 72: 77–94. [Google Scholar]
- Ribeiro MJ, Serfozo Z, Papp A, Kemenes I, O'Shea M, Yin JC, Benjamin PR, Kemenes G. 2003. Cyclic AMP response element-binding (CREB)-like proteins in a molluscan brain: cellular localization and learning-induced phosphorylation. Eur J Neurosci 18: 1223–1234. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. 1993. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res Rev 18: 247–291. [DOI] [PubMed] [Google Scholar]
- Robinson EJ, Feinerman O, Franks NR. 2009. Flexible task allocation and the organization of work in ants. Proc Biol Sci 276: 4373–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EJ, Feinerman O, Franks NR. 2012. Experience, corpulence and decision making in ant foraging. J Exp Biol 215: 2653–2659. [DOI] [PubMed] [Google Scholar]
- Sadamoto H, Sato H, Kobayashi S, Murakami J, Aonuma H, Ando H, Fujito Y, Hamano K, Awaji M, Lukowiak K, et al. 2004. CREB in the pond snail Lymnaea stagnalis: cloning, gene expression, and function in identifiable neurons of the central nervous system. J Neurobiol 58: 455–466. [DOI] [PubMed] [Google Scholar]
- Sanchis-Segura C, Jancic D, Jimenez-Minchan M, Barco A. 2009a. Inhibition of cAMP responsive element binding protein in striatal neurons enhances approach and avoidance responses toward morphine—and morphine withdrawal-related cues. Front Behav Neurosci 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. 2009b. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology 34: 2642–2654. [DOI] [PubMed] [Google Scholar]
- Seeley TD. 1982. Adaptive significance of the age polyethism schedule in honeybee colonies. Behav Ecol Sociobiol 11: 287–293. [Google Scholar]
- Simola DF, Wissler L, Donahue G, Waterhouse RM, Helmkampf M, Roux J, Nygaard S, Glastad KM, Hagen DE, Viljakainen L, et al. 2013a. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res 23: 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Ye C, Mutti NS, Dolezal K, Bonasio R, Liebig J, Reinberg D, Berger SL. 2013b. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res 23: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Graham RJ, Brady CM, Enzmann BL, Desplan C, Ray A, Zwiebel LJ, Bonasio R, Reinberg D, Liebig J, et al. 2016. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 351: aac6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell HM. 1985. Actinomycin and DNA transcription. Proc Natl Acad Sci 82: 5328–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Sun Y, Zhang Y, Li M. 2009. Molecular cloning and characterization of Bombyx mori CREB gene. Arch Insect Biochem 71: 31–44. [DOI] [PubMed] [Google Scholar]
- Stanciu M, Radulovic J, Spiess J. 2001. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: relationship to Fos production. Mol Brain Res 94: 15–24. [DOI] [PubMed] [Google Scholar]
- Stewart R, Gatto G, Lumeng L, Li T-K, Murphy J. 1993. Comparison of alcohol-preferring (P) and nonpreferring (NP) rats on tests of anxiety and for the anxiolytic effects of ethanol. Alcohol 10: 1–10. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Fukushima H, Mukawa T, Toyoda H, Wu L-j, Zhao M-g, Xu H, Shang Y, Endoh K, Iwamoto T, et al. 2011. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J Neurosci 31: 8786–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K. 1961. Classical conditioned response in the honey bee. J Insect Physiol 6: 168–179. [Google Scholar]
- Trifilieff P, Herry C, Vanhoutte P, Caboche J, Desmedt A, Riedel G, Mons N, Micheau J. 2006. Foreground contextual fear memory consolidation requires two independent phases of hippocampal ERK/CREB activation. Learn Mem. 13:349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valor LM, Viosca J, Lopez-Atalaya JP, Barco A. 2013. Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr Pharm Des 19: 5051–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg M, Verbaarschot P, Hontelez S, Vet LEM, Dicke M, Smid HM. 2010. CREB expression in the brains of two closely related parasitic wasp species that differ in long-term memory formation. Insect Mol Biol 19: 367–379. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, De Vries TJ, Wardeh G, Hogenboom FA, Schoffelmeer AN. 2001. A single exposure to morphine induces long-lasting behavioural and neurochemical sensitization in rats. Eur J Neurosci 14: 1533–1538. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA, Overmier JB. 1988. Backward inhibitory conditioning with signaled and unsignaled unconditioned stimuli: distribution of trials across days and intertrial interval. J Exp Psychol Anim Behav Process 14: 26–35. [Google Scholar]
- Wüstenberg D, Gerber B, Menzel R. 1998. Long- but not medium-term retention of olfactory memories in honeybees is impaired by actinomycin D and anisomycin. Eur J Neurosci 10: 2742–2745. [DOI] [PubMed] [Google Scholar]
- Yamamoto KK, Gonzalez GA, Menzel P, Rivier J, Montminy MR. 1990. Characterization in transcription of a bipartite activator factor CREB. Cell 60: 611–617. [DOI] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. 2014. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron 83: 722–735. [DOI] [PubMed] [Google Scholar]
- Zayed A, Robinson GE. 2012. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu Rev Genet 46: 591–615. [DOI] [PubMed] [Google Scholar]