Abstract
Triple-stranded RNA was first deduced to form in vitro more than 50 years ago and has since been implicated in RNA catalysis, stability, and small molecule binding. Despite the emerging biological significance of RNA triple helices, it remains unclear how their nucleotide composition contributes to their thermodynamic stability and cellular function. To investigate these properties, we used in vitro RNA electrophoretic mobility shift assays (EMSAs) and in vivo intronless β-globin reporter assays to measure the relative contribution of 20 RNA base triples (N•A–U, N•G–C, N•C–G, N•U–A, and N•G–U) to triple-helical stability. These triples replaced a single internal U•A–U within the known structure of the triple-helical RNA stability element of human metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which contains 10 major-groove base triples. In addition to the canonical C•G–C triple, the noncanonical base triples U•G–C, U•G–U, C•C–G, and U•C–G exhibited at least 30% stability relative to the wild-type U•A–U base triple in both assays. Of these triples, only U•A–U, C•G–C, and U•G–C, when tested as four successive triples, formed stabilizing structures that allowed accumulation of the intronless β-globin reporter. Overall, we find that Hoogsteen-position pyrimidines support triple helix stability and function and that thermodynamic stability, based on EMSA results, is necessary but not sufficient for stabilization activity of the MALAT1 triple helix in cells. These results suggest that additional RNA triple helices containing noncanonical triples likely exist in nature.
Keywords: RNA base triples, RNA triple helix, MALAT1, intronless β-globin reporter assay, RNA stability
INTRODUCTION
RNA triple helices were first deduced to form in vitro more than 50 years ago and have since been implicated as structural elements in eukaryotic, prokaryotic, and viral RNAs (Felsenfeld et al. 1957; Conrad 2014). To date, structural validation of natural RNA triple helices, which we define as three or more stacked major-groove base triples, has been reported for telomerase (Theimer et al. 2005; Cash et al. 2013), bacterial S-adenosylmethionine-II (SAM-II) and class II prequeuosine1 (preQ1) riboswitches (Gilbert et al. 2008; Liberman et al. 2013), group II introns and spliceosomal U2/U6 snRNAs (Toor et al. 2008; Hang et al. 2015), Kaposi's sarcoma-associated herpesvirus (KSHV) polyadenylated nuclear (PAN) RNA (Mitton-Fry et al. 2010), and human metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) (Brown et al. 2014). These RNA triple helices are predominantly composed of canonical U•A–U base triples and typically contain 3–5 consecutive triples. Yet structures with noncanonical triples—that is, triples other than canonical U•A–U and C•G–C—have been identified. For instance, the bacterial SAM-II riboswitch binds its ligand in a U•U•ASAM triple, flanked by two U•A–U major-groove base triples on one side and an A•G–C and G•G–C triple on the other side (Gilbert et al. 2008). In the group II intron from Oceanobacillus iheyensis, a triple helix containing one G•G–U and two C•C–G triples contributes to coordination of the active-site divalent cations required for catalysis (Toor et al. 2008). Finally, the spliceosomal U2/U6 snRNAs from Schizosaccharomyces pombe form a triple helix composed of one A•A–U, one G•G–C, and one U•C–G triple (Hang et al. 2015). These examples of noncanonical base triples suggest that RNA triple helices may vary widely in nucleotide composition.
Recently, RNA triple helices have been implicated in stabilizing vertebrate long noncoding RNAs (lncRNAs), as well as viral and genomic RNAs (for review, see Conrad 2014). For example, a stem–loop structure containing a U-rich internal loop (denoted ENE for expression and nuclear retention element) near the 3′ end of KSHV PAN lncRNA sequesters the downstream poly(A) tail into a triple helix (denoted ENE + A) of five U•A–U major-groove triples and three A-minor triples (Mitton-Fry et al. 2010). The KSHV PAN ENE + A structure acts both in vivo and in vitro to inhibit rapid deadenylation-dependent RNA degradation, enabling PAN RNA to accumulate to high levels in nuclei of lytic-phase KSHV-infected cells (Sun et al. 1996; Conrad and Steitz 2005; Conrad et al. 2006). Similar ENE + A structures have been discovered in the RNAs of diverse classes of viruses through bioinformatics (Tycowski et al. 2012).
More complex ENE + A structures have been identified at the 3′ ends of two human lncRNAs: MALAT1 and multiple endocrine neoplasia β RNA (MENβ) (Brown et al. 2012; Wilusz et al. 2012). The crystal structure of the MALAT1 ENE + A confirmed that the U-rich ENE coalesces with a downstream A-rich tract to form a bipartite triple helix, allowing MALAT1 to accumulate in cells (Brown et al. 2014). Preceded by two A-minor triples, the first triplex segment consists of five U•A–U triples followed by a C•G–C triple, and the second triplex contains four U•A–U triples. Separating these two triple-helical regions is a C–G base pair that resets the helical axis and allows the adjacent formation of these two independent stacks of RNA base triples (see triple helix in Fig. 1A; Brown et al. 2014). The MENβ ENE + A has been proposed to adopt a similar bipartite triple-helical structure based on sequence similarity (Brown et al. 2012).
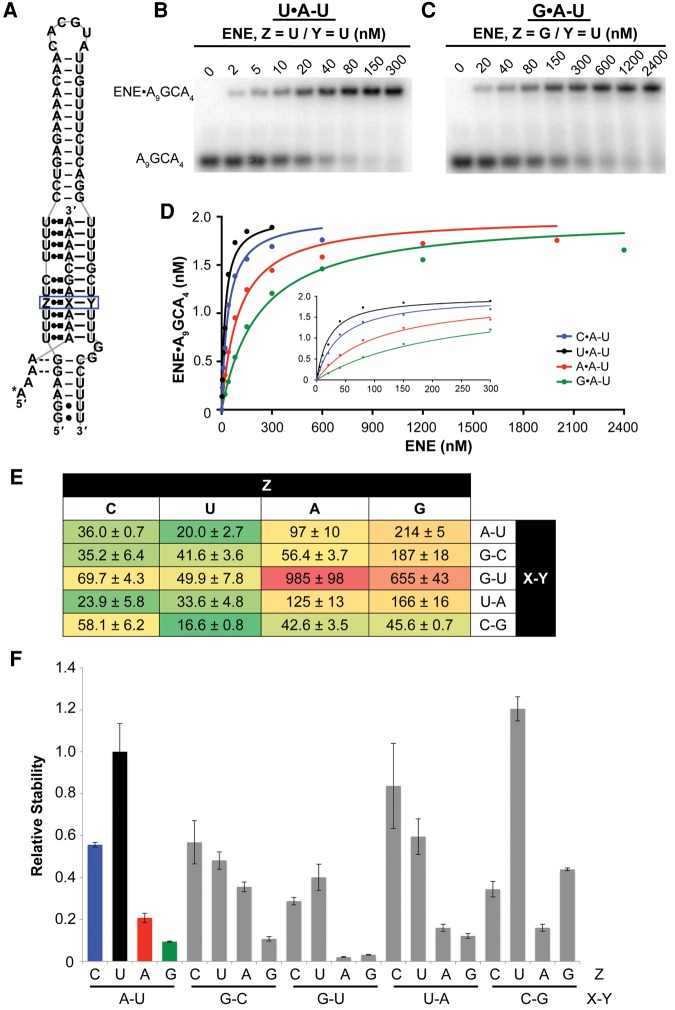
FIGURE 1.
EMSA measuring the thermodynamic stability of RNA base triples within the MALAT1 ENE + A triple helix. (A) Schematic depicting the triple helix formed by the two RNAs used in the EMSA: the MALAT1 ENE construct and the A-rich oligonucleotide. Twenty RNA base triples were tested at the site boxed in blue (Z•X–Y). Watson–Crick base pairs are indicated by dashes, noncanonical base pairs by dots, Hoogsteen base pairs by Leontis–Westhof notation, and A-minor interactions by dashed lines (Leontis and Westhof 2001). An asterisk denotes the 32P-radiolabeled nucleotide. (B,C) Representative gel images for the U•A–U (B) and G•A–U (C) triples. Increasing amounts of the MALAT1 ENE construct were incubated with 2 nM oligonucleotide at pH 7.0, and the ENE-oligonucleotide complex was resolved from free oligonucleotide by native polyacrylamide gel electrophoresis. The fraction of bound oligonucleotide was quantified by autoradiography. (D) Binding curves for the N•A–U triples, in which the C•A–U, U•A–U, A•A–U, and G•A–U triples are shown in blue, black, red, and green, respectively. (E) Heat map of the Kd,app values (in nM) for all base triples. Green shading corresponds to a low Kd,app, yellow shading to an intermediate Kd,app, and red shading to a high Kd,app. (F) The relative stability of base triples was calculated as Kd,app (U•A–U)/Kd,app (Z•X-Y). For both E and F, reported values are the average of at least three replicates ± standard deviation.
Despite the ubiquity and biological importance of RNA triple helices, we understand only poorly how their thermodynamic stability and functional properties are determined by their nucleotide composition. Most RNA triple helices identified to date rely heavily on canonical U•A–U and C•G–C triples (Brown et al. 2014). However, the discovery of noncanonical triples in bacterial riboswitches, the group II intron, and the spliceosome suggests that other nucleotide combinations are possible (Gilbert et al. 2008; Toor et al. 2008; Hang et al. 2015). To probe the effects of altering nucleotide composition within an RNA triple helix, we selected the MALAT1 ENE + A as a model. With nine U•A–U triples and one C•G–C triple, it is the largest of the known naturally occurring major-groove RNA triple helices (Brown et al. 2014). We altered a U•A–U triple at a single position within the MALAT1 ENE + A triple helix and performed electrophoretic mobility shift assays (EMSAs) to determine the relative thermodynamic stability of 20 distinct major-groove RNA base triples (N•A–U, N•G–C, N•C–G, N•U–A, and N•G–U). We subsequently tested the cellular function of these altered MALAT1 triple helices using the intronless β-globin reporter to assess their RNA stabilization activity in cells. We find that Hoogsteen-position pyrimidines favor both MALAT1 triple helix stability and function and that thermodynamic stability is necessary but not sufficient for in vivo stabilization activity. These analyses of noncanonical base triples will assist in the discovery and validation of novel triple-helical RNA structures.
RESULTS AND DISCUSSION
Certain noncanonical RNA base triples with Hoogsteen-position pyrimidines contribute to thermodynamic stability
To measure the relative contribution of distinct base triples to the thermodynamic stability of an RNA triple helix, we chose to vary the nucleotide composition of the fourth U•A–U triple in the MALAT1 ENE + A structure (Fig. 1A, blue box). This position was selected because (i) its internal position in the triple helix might permit us to capture and to analyze triples too weak to form otherwise and (ii) crystallographic data indicated optimal hydrogen bonds for this triple (Brown et al. 2014). At this position, all four standard RNA nucleotides were tested in the Hoogsteen strand (Z in Fig. 1A), with base pairs A–U, G–C, G–U, U–A, and C–G in the Watson (X in Fig. 1A) and Crick (Y in Fig. 1A) strands (strand nomenclature as in Brown et al. 2014).
We performed EMSAs using two RNAs that could associate to form the MALAT1 triple helix: the ENE and a 5′-radiolabeled A-rich tract oligonucleotide mimic (Fig. 1A–D). Apparent equilibrium dissociation constants (Kd,app) were extracted from plots of the concentration of ENE-oligonucleotide complex versus the initial concentration of free ENE (Fig. 1B–D). For the 20 triples tested, Kd,app values varied from 16.6 to 985 nM (Fig. 1E). Relative to the wild-type U•A–U base triple (Kd,app = 20.0 nM), 11 altered triples exhibited thermodynamic stability of at least 30% (Kd,app < 66.7 nM): U•C–G, C•U–A, U•U–A, C•G–C, C•A–U, U•G–C, A•C–G, G•C–G, U•G–U, A•G–C, and C•C–G (Fig. 1F). The results indicate that the primary determinant of RNA base-triple stability in the MALAT1 ENE + A is the identity of the nucleobase in the Hoogsteen position, where pyrimidines are stabilizing. Base-triple stability varied relatively little with different base pairs at the Watson and Crick positions, except for the G–U wobble pair, which formed especially unstable triples, and the U•C–G base triple, which is the most stable triple (Fig. 1F).
Our data can be compared with similar studies of DNA triple helices. The thermodynamic stability of 16 different major-groove DNA base triples at a single site was measured by a quantitative affinity cleavage titration assay, a method for determining association constants (Best and Dervan 1995). Although the differing sequence context and assay conditions in the RNA and DNA base-triple analyses preclude a direct quantitative comparison, the same major conclusions emerge: (i) Hoogsteen-position pyrimidines are more stabilizing than purines, and (ii) the canonical triples U•A–U/T•A–T and C•G–C are among the most stable (Best and Dervan 1995). In the DNA triple helix examined by Best and Dervan, which contained 15 stacked triples, T•A–T and C•G–C were more stable than all other triples tested by at least one order of magnitude, with Kd,app values varying over three orders of magnitude overall (Best and Dervan 1995). In contrast, 10 of the RNA triples tested here exhibited thermodynamic stability comparable to that of U•A–U or C•G–C (Fig. 1E,F), despite their reduced potential to form hydrogen bonds along the Hoogsteen face (e.g., two hydrogen bonds for U•A in U•A–U and no hydrogen bonds for U•C in U•C–G). This overall greater stability of noncanonical RNA base triples compared to noncanonical DNA base triples may be due to (i) differential effects of base stacking (Wang and Kool 1995), which varies with the identity and especially the spatial orientation of the bases involved (Šponer et al. 1996; Brown et al. 2015) and/or (ii) a possible hydrogen bond between the 2′-hydroxyl of the Hoogsteen strand and a phosphate oxygen in the Watson strand (Brown et al. 2014), which has been proposed to explain the greater thermodynamic stability of RNA relative to DNA triple helices (Roberts and Crothers 1992; Holland and Hoffman 1996). Additional studies are needed to establish the exact thermodynamic contributions of hydrogen bonding and base stacking to the stability of DNA and RNA triple helices.
Certain pyrimidine•purine–pyrimidine RNA base triples contribute to the in vivo RNA stabilization activity of the MALAT1 triple helix
We used intronless β-globin reporter (βΔ1,2) assays to determine whether the noncanonical base triples studied in vitro also support the cellular function of the MALAT1 ENE + A as an RNA stability element (Fig. 2A). The 3′-untranslated region of the reporter construct contains nucleotides 8254–8424 from human MALAT1, which includes the ENE, A-rich tract, and tRNA-like structure known as MALAT1-associated small cytoplasmic RNA (mascRNA) (Fig. 2A). Thus, this assay assesses several key steps that potentially contribute to the overall stability of MALAT1 in cells, such as cotranscriptional folding, 3′-end processing by RNase P, assembly of the ENE + A, and the degradation process. This assay has previously suggested that ENE + A structures with weaker triple helices have lower stabilization activity (Conrad and Steitz 2005; Conrad et al. 2006; Brown et al. 2012, 2014). We varied the same base triple in the βΔ1,2-MALAT1 ENE + A + mascRNA construct (Fig. 2A,B) as in the EMSAs (Fig. 1A). Only six particular triples in this position enabled the intronless β-globin transcripts to accumulate to at least 30% of the levels observed for the wild-type U•A–U construct: U•G–C, C•G–C, C•G–U, U•G–U, C•C–G, and U•C–G (Fig. 2C). Consistent with the results of the EMSAs, Hoogsteen-position pyrimidines (Py) exhibited higher stabilization activity than Hoogsteen-position purines (Pu). These cell-based findings, however, revealed a dependence on the nucleotides in the Watson and Crick positions not seen in the EMSAs. Specifically, Watson-position purines and Crick-position pyrimidines exhibit higher stabilization activity, suggesting that a Py•Pu–Py configuration is preferred. Because of this preference for the Py•Pu–Py configuration and the low stabilization activity of N•U–A triples (Fig. 2C), we decided not to test N•U–G triples.
FIGURE 2.
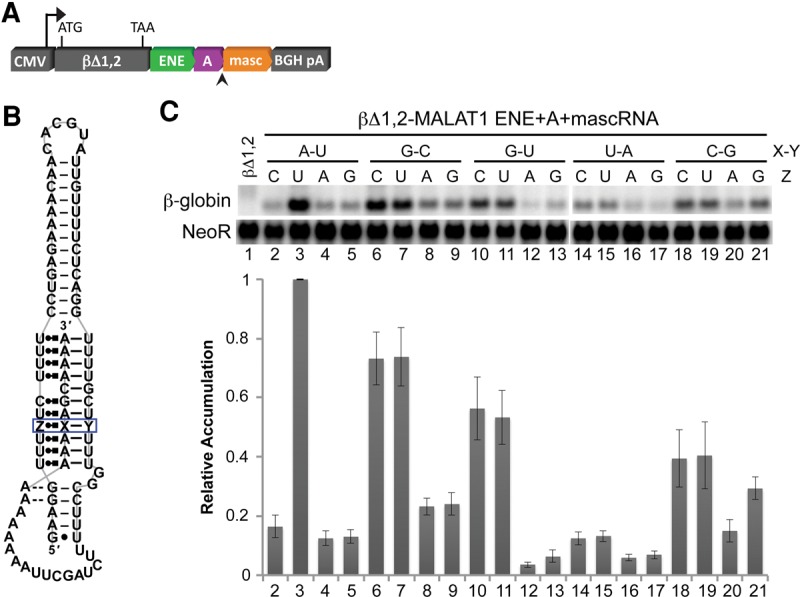
In vivo stabilization activity of MALAT1 derivatives with single base-triple substitutions. (A) The MALAT1 ENE + A + mascRNA segments and derivatives thereof were inserted into the 3′ UTR of cytomegalovirus (CMV) promoter-driven intronless β-globin constructs (βΔ1,2) and were introduced into HEK293T cells. RNase P cleaves the transcript at the site indicated by the arrowhead. (B) The base composition of the MALAT1 triple-helical site boxed in blue was varied to test 20 RNA base triples. Structural notation is as described in Figure 1. (C) Total RNA was isolated and analyzed by Northern blotting for β-globin and NeoR (as a loading and transfection control). The relative accumulation of each mutant β-globin reporter mRNA was normalized to the quantity observed for the MALAT1 construct containing the wild-type U•A–U triple at the boxed site. Measurements are an average of at least three biological replicates ± standard deviation.
We next asked whether those base triples that conferred at least 30% stabilization activity as single substitutions could maintain that activity when inserted at multiple positions within the βΔ1,2-MALAT1 ENE + A + mascRNA reporter system (Fig. 3A,B). Accordingly, we changed all four U•A–U triples in the upper triple helix to C•G–C, U•G–C, C•G–U, U•G–U, C•C–G, or U•C–G triples (Fig. 3B). Of the six variants tested, only the constructs containing C•G–C and U•G–C triples enabled the reporter mRNA to accumulate to at least 30% of the levels observed for the wild-type MALAT1 construct (Fig. 3C). These results suggest that C•G–C and U•G–C base triples have the greatest potential to form a triple helix with stabilization activity in vivo. The low relative accumulation of the reporter containing four consecutive C•G–U, U•G–U, or C•C–G triples implies that these triples can form within an otherwise stable triple helix. Yet they lack the ability to assemble a functional triple helix on their own, at least in the context of the MALAT1 ENE + A structure.
FIGURE 3.
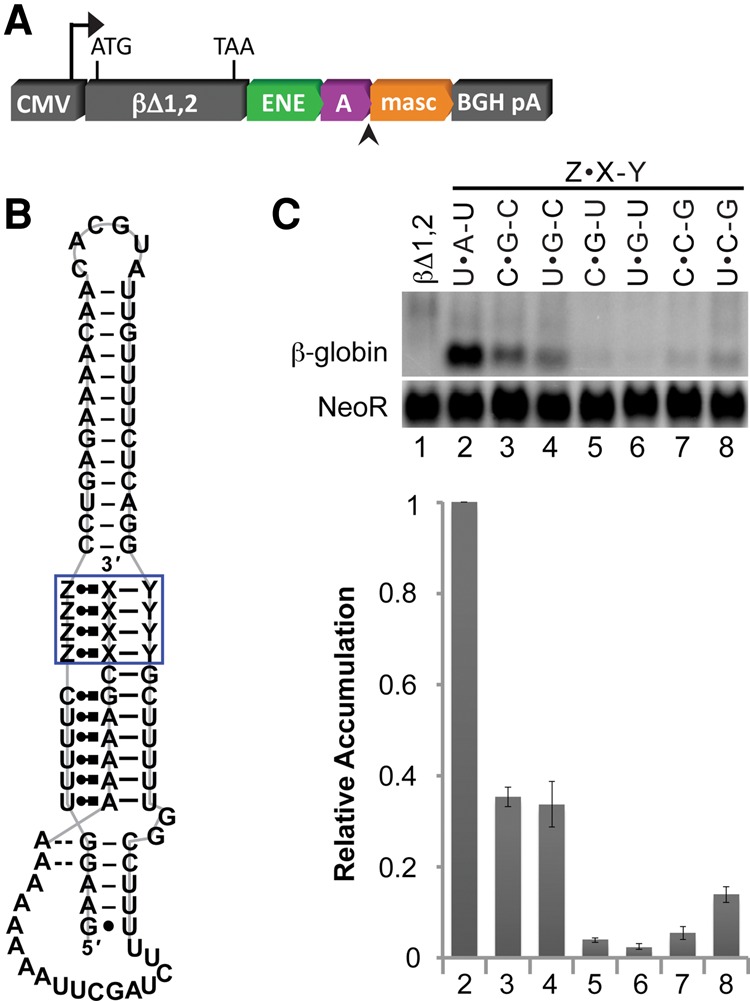
In vivo stabilization activity of MALAT1 derivatives with quadruple base-triple substitutions. (A) Constructs containing the MALAT1 ENE + A + mascRNA and derivatives thereof were prepared and introduced into HEK293T cells as described in Figure 2. (B) The base composition at each of the four sites boxed in blue was varied to test the stabilization activity of selected RNA base triples as a complete, independent triple-helical unit. Structural notation is as described in Figure 1. (C) Total RNA was extracted and analyzed as described in Figure 2. Measurements are an average of at least three biological replicates ± standard deviation.
Thermodynamic stability is necessary but not sufficient for the cellular function of the MALAT1 triple helix
Finally, we compared the relative stability conferred by the noncanonical base triples measured in our two independent assays. Plotting the relative RNA accumulation in the β-globin reporter assay (Fig. 2C) versus the relative thermodynamic stability in native gel-shift assays (Fig. 1F) did not reveal a strong linear correlation (Fig. 4). Considering the complexity of the cellular environment, this result is not surprising. Nonetheless, dividing the plot into quadrants (I–IV in Fig. 4) of high and low thermodynamic stability and cellular RNA accumulation shows that, except for C•G–U (quadrant IV), a base triple exhibits in vivo stabilization activity only if thermodynamically stable (quadrant I). Base triples with low thermodynamic stability almost exclusively exhibit low cellular stabilization activity and have a purine in the Hoogsteen position (quadrant III). Interestingly, not all thermodynamically stable triples possess stabilization activity (quadrant II). We conclude that base triple thermodynamic stability is necessary but not sufficient for in vivo stabilization activity of the MALAT1 ENE + A RNA triple helix.
FIGURE 4.

Relative in vivo RNA stabilization and thermodynamic stability of each base triple. Each of the tested base triples is plotted according to its relative thermodynamic stability (Fig. 1F) and relative cellular RNA stabilization activity (Fig. 2C). Nucleotides at the Watson and Crick positions (X–Y) are indicated by the color of the dots: black for A–U, blue for G–C, red for G–U, green for U–A, and violet for C–G. The nucleotide at the Hoogsteen position (Z) is indicated to the left of each dot. Gray dashed lines at values of 0.3 divide the plot into four quadrants: (I) high thermodynamic stability and high RNA accumulation (six base triples), (II) high thermodynamic stability and low RNA accumulation (six base triples), (III) low thermodynamic stability and low RNA accumulation (seven base triples), and (IV) low thermodynamic stability and high RNA accumulation (one base triple).
There are at least four potential explanations for the discrepancy between triple helix stabilization assessed in vitro and in vivo: (i) pH differences between the in vitro and in vivo assays may affect triple-helical stability; (ii) nucleotide changes could alter the kinetics of triple-helical assembly or the resulting structure (i.e., folded versus unfolded); (iii) nucleotide changes could create a more recognizable substrate for cellular exonucleases or their associated cofactors; and (iv) alternative structures of the MALAT1 ENE + A may alter protein binding, which may subsequently influence the stability of MALAT1 in vivo. For example, the MALAT1 triple helix was previously shown to exhibit a higher third-strand melting temperature at lower pH, suggesting protonation of the Hoogsteen-position cytosine in the C•G–C triple (Brown et al. 2012). Changes in the nucleotide composition may lead to the formation of alternative ENE + A structures, including folds with a more accessible 3′ end. In cells, the rate at which the U-rich internal loop sequesters the 3′ A-rich tract into a triple helix is in competition with the kinetics of 3′-to-5′ RNA decay; therefore, structures with a slow rate of ENE + A assembly may be more susceptible to degradation. In general, 3′-to-5′ exonucleases prefer single-stranded RNA and some preferentially degrade specific nucleotide sequences with large differences in catalytic efficiency (Körner and Wahle 1997; Zuo and Deutscher 2002). The nuclear exosome, in contrast, works with a complex network of RNA-binding proteins, helicases, and a poly(A) polymerase to identify, process, and degrade a variety of RNAs (LaCava et al. 2005; Vaňáčová et al. 2005; Wyers et al. 2005; Houseley et al. 2006; Schmidt et al. 2012; Januszyk and Lima 2014; Thoms et al. 2015). Lastly, another possibility is that a protein specifically binds to the folded or unfolded states of the MALAT1 ENE + A, sterically blocking exonuclease access to the 3′ end of MALAT1 or actively recruiting decay factors to promote degradation of MALAT1. The actual mechanism likely involves contributions from several of the aforementioned factors and requires further investigation.
Although RNA triple helices were first discovered nearly 60 years ago (Felsenfeld et al. 1957), we are only beginning to understand how their stability and biological activity relate to their nucleotide composition. The naturally occurring RNA triple helices identified to date contain only a subset of potential triples, overwhelmingly U•A–U (Brown et al. 2014). Our results imply that nucleotide sequences capable of forming RNA triple helices may be less constrained than initially thought, suggesting a greater diversity in the nucleotide composition of naturally occurring RNA triple helices. Our results will enable more informed searches, which may lead to the discovery and validation of novel major-groove RNA triple helices.
MATERIALS AND METHODS
RNA preparation
A-rich oligonucleotides (5′-A7XAGCA4-3′; GE Dharmacon) were deprotected and 5′-radiolabeled as described previously (Brown et al. 2012). MALAT1 ENE RNAs were transcribed by His-tagged T7 RNA polymerase using PCR-generated templates. Nucleotide substitutions in the MALAT1 ENE were introduced into the DNA template via PCR with chemically synthesized DNA primers (Keck Oligonucleotide Synthesis) containing the appropriate sequence (see Fig. 1A for the sequence). RNAs were gel-purified and processed as described previously (Brown et al. 2012).
EMSA
Increasing concentrations of MALAT1 ENE RNA were combined with 2 nM of the appropriate 5′-radiolabeled oligonucleotide in RNA binding buffer (25 mM sodium cacodylate pH 7.0, 50 mM KCl, 1 mM MgCl2, 0.5 mg/mL E. coli carrier RNA, and 5% v/v glycerol). This mixture was heated at 95°C for 2 min, snap-cooled on ice for 2 min, and incubated at room temperature for 30 min. Samples were immediately loaded onto an 8%–10% native polyacrylamide gel prepared with 1× TB buffer supplemented with 1 mM MgCl2 and electrophoresed at 150 V for ∼2 h. Gels were dried, exposed to a phosphorimager screen, scanned using a Storm 860 (GE Healthcare), and quantitated using ImageQuant software. A plot of ENE-oligonucleotide complex concentration versus the initial concentration of free ENE was fit to Equation 1, E•O = 0.5(Kd,app + E0 + O0) − 0.5[(Kd,app + E0 + O0)2 − 4E0O0]1/2, where E•O denotes the final concentration of the ENE-oligonucleotide complex and E0 and O0 the initial concentrations of ENE and oligonucleotide, respectively. The theoretical binding saturation value (O0) for each of the 20 variants tested was initially quantified by treating both O0 and Kd,app as free parameters and fitting the data to Equation 1. Kd,app values were subsequently extrapolated by fitting the data to Equation 1 with O0 fixed at 2.00 nM, 1.91 nM, and 1.81 nM for variants with >90% saturation, 80%–90% saturation, and <80% saturation, respectively.
Plasmids and mutagenesis
The βΔ1,2 and βΔ1,2-MALAT1 ENE + A + mascRNA plasmids have a pcDNA3 backbone and were described previously (Brown et al. 2012, 2014). Nucleotide substitutions in the βΔ1,2-MALAT1 ENE + A + mascRNA plasmid were created using site-directed mutagenesis.
Intronless β-globin reporter assays
Assays were performed in HEK293T cells as described previously (Brown et al. 2012, 2014).
ACKNOWLEDGMENTS
We thank Seyed Torabi, Kazimierz Tycowski, and Johanna Withers for critical review of the manuscript; Angela Miccinello for editorial work; and all Steitz laboratory members for thoughtful discussions. This work was supported by National Institutes of Health grants K99GM111430 (J.A.B.) and GM026154 and CA193300 (J.A.S.). J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055707.115.
REFERENCES
- Best GC, Dervan PB. 1995. Energetics of formation of sixteen triple helical complexes which vary at a single position within a pyrimidine motif. J Am Chem Soc 117: 1187–1193. [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. 2012. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci 109: 19202–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. 2014. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol 21: 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RF, Andrews CT, Elcock AH. 2015. Stacking free energies of all DNA and RNA nucleoside pairs and dinucleoside-monophosphates computed using recently revised AMBER parameters and compared with experiment. J Chem Theory Comput 11: 2315–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash DD, Cohen-Zontag O, Kim N-K, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. 2013. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci 110: 10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK. 2014. The emerging role of triple helices in RNA biology. Wiley Interdiscip Rev RNA 5: 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Steitz JA. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J 24: 1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. 2006. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell 24: 943–953. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Davies DR, Rich A. 1957. Formation of a three-stranded polynucleotide molecule. J Am Chem Soc 79: 2023–2024. [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182. [DOI] [PubMed] [Google Scholar]
- Hang J, Wan R, Yan C, Shi Y. 2015. Structural basis of pre-mRNA splicing. Science 349: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Holland JA, Hoffman DW. 1996. Structural features and stability of an RNA triple helix in solution. Nucleic Acids Res 24: 2841–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. 2014. The eukaryotic RNA exosome. Curr Opin Struct Biol 24: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner CG, Wahle E. 1997. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 272: 10448–10456. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman JA, Salim M, Krucinska J, Wedekind JE. 2013. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat Chem Biol 9: 353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330: 1244–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Crothers D. 1992. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Xu Z, Mathews DH, Butler JS. 2012. Air proteins control differential TRAMP substrate specificity for nuclear RNA surveillance. RNA 18: 1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šponer J, Leszczyński J, Hobza P. 1996. Nature of nucleic acid–base stacking: nonempirical ab initio and empirical potential characterization of 10 stacked base dimers. Comparison of stacked and H-bonded base pairs. J Phys Chem 100: 5590–5596. [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci 93: 11883–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 17: 671–682. [DOI] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. 2015. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162: 1029–1038. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K, Shu M-D, Borah S, Shi M, Steitz J. 2012. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep 2: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaňáčová Š, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kool ET. 1995. Origins of the large differences in stability of DNA and RNA helixes: C-5 methyl and 2′-hydroxyl effects. Biochemistry 34: 4125–4132. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn C-D, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′-ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. 2002. The physiological role of RNase T can be explained by its unusual substrate specificity. J Biol Chem 277: 29654–29661. [DOI] [PubMed] [Google Scholar]