Abstract
During development, sensory neurons must choose identities that allow them to detect specific signals and connect with appropriate target neurons. Ultimately, these sensory neurons will successfully integrate into appropriate neural circuits to generate defined motor outputs, or behavior. This integration requires a developmental coordination between the identity of the neuron and the identity of the circuit. The mechanisms that underlie this coordination are currently unknown. Here, we describe two modes of regulation that coordinate the sensory identities of Drosophila melanogaster olfactory receptor neurons (ORNs) involved in sex-specific behaviors with the sex-specific behavioral circuit identity marker fruitless (fru). The first mode involves a developmental program that coordinately restricts to appropriate ORNs the expression of fru and two olfactory receptors (Or47b and Ir84a) involved in sex-specific behaviors. This regulation requires the chromatin modulatory protein Alhambra (Alh). The second mode relies on the signaling from the olfactory receptors through CamK and histone acetyl transferase p300/CBP to maintain ORN-specific fru expression. Our results highlight two feed-forward regulatory mechanisms with both developmentally hardwired and olfactory receptor activity-dependent components that establish and maintain fru expression in ORNs. Such a dual mechanism of fru regulation in ORNs might be a trait of neurons driving plastic aspects of sex-specific behaviors.
The fruitless (fru) gene regulates sexual behavior in flies, but what regulates fru? This study shows that the chromatin modulator Alhambra restricts fru expression to specific neurons in the developing Drosophila olfactory system. In adults, olfactory receptor signaling and the histone acetyltransferase p300 are required to maintain fru expression.
Author Summary
How do individual neurons know what type of a circuit they must integrate into? To correctly assemble neural circuits during development, the identities of neurons must be coordinated with the identities of the circuits into which they will be integrated. How is this process regulated? We have used the olfactory circuits that regulate sex-specific behaviors in Drosophila to answer this question. In Drosophila, the fruitless (fru) gene, which encodes a transcription factor, acts as a molecular marker that labels circuits regulating sex-specific behaviors. Fru is both a necessary and sufficient regulator of sex-specific behaviors like courtship and aggression and is expressed in only about 2,000 interconnected neurons in the fly nervous system. Even though fru expression and function is so critical to sex-specific behavior, the mechanisms regulating its expression in each of the neurons within the circuit are not known. Here, we revealed two different modes of transcriptional regulation of fru during the development of the olfactory receptor neurons that are involved in sex-specific behaviors. In the first mode, the putative chromatin modulator Alhambra (Alh) coregulates both olfactory receptors and fru expression in two olfactory receptor neuron classes during development. In adult flies, the second mode maintains fru expression through the activity of olfactory receptors, calcium signaling, and chromatin modulatory proteins in these olfactory receptor neurons. These two genetic programs separate the developmentally hardwired and the olfactory receptor activity-mediated regulation of fru expression in olfactory circuits, and might represent the molecular mechanisms that mediate the innate and adaptable aspects of odor-guided social behaviors.
Introduction
The assembly of neural circuits dedicated to specific behaviors must be tightly regulated during development, where neurons need not only define their identity as individual neurons but also molecularly and developmentally link themselves with the specific neural circuitry they will be integrated into. Such developmental programs can, for example, establish a connection between sensory circuits tuned to a particular stimulus and the motor pathways that execute the output behavior in response to that stimulus. An elegant example of this is seen in the sex-specific behavioral circuitry of Drosophila melanogaster. This circuitry is regulated by the single transcription factor Fruitless (Fru) [1]. Sex-specific alternative splicing of fru generates the protein product FruM in males only [2–5]. Studies in which the sex-specific splicing of fru was manipulated in both males and females have shown that FruM function is necessary and sufficient for male-specific behaviors [6]. Despite the dramatic nature of these mutant phenotypes, fru is only expressed in a small fraction of the D. melanogaster nervous system. Only about 2,000 interconnected neurons express fru, and the specific activation of fru-positive neurons is sufficient to trigger male-specific behaviors [1,2,6–8]. Thus, fru expression identifies the neural circuitry that controls sex-specific behaviors. Support for this idea comes from the recently identified fru-positive neuronal circuitry that drives the sexually dimorphic response to the pheromone cis-vaccenyl acetate (cVA) [9]. fru is expressed throughout the entire cVA circuitry, from the olfactory receptor neurons (ORNs) that detect the pheromone to the motor neurons that trigger courtship behaviors [10,11]. Despite a large volume of research, it is largely unknown how Fru regulates the development or function of this circuitry and how fru expression is developmentally coordinated with the identity programs for each of the neurons within the circuitry.
Olfaction is a key component of sex-specific behaviors. Flies detect volatile pheromones and other odors important for the initiation of courtship and aggression via olfactory receptors (ORs) expressed in ORNs [12]. Clusters of 1–4 ORNs are housed in sensory hairs called sensilla that cover the surface of the antennae and maxillary palps. ORNs are classified and named by the single OR gene that they express, and each ORN class targets a specific glomerulus in the antennal lobe. FruM function was shown to be specifically required in ORNs for normal courtship behavior [2], and three ORN classes express fru in adult D. melanogaster antennae [12]. Or67d ORNs, the best studied of the fru-positive ORNs, are housed in the at1 sensilla. These neurons detect the male-specific pheromone cVA, which acts as a suppressant of both male—male and male—female courtship [9]. Ir84a ORNs, housed in the ac4 sensilla, also express fru and can detect the availability of food sources and coordinate reproductive behaviors accordingly [13]. Recent studies show that the third fru-positive ORN class, Or47b, detects methyl laurate, a cuticular pheromone, and is necessary for successful copulation [14–17].
Neurons expressing Or67d, Ir84a, and Or47b all express fru and are specifically receptive to olfactory cues required for courtship, but the developmental programs regulating the expression of these three Or genes and coordinating them with fru expression is unknown. During development, combinations of transcription factors diversify ORN precursor cell identities, thus restricting the ORN classes that can be generated in each precursor lineage [18,19]. This “cellular memory” of possible fates is retained through asymmetric divisions as Notch signaling further segregates each possible sensory identity into individual ORNs within the same sensillum [12,20,21]. The retention of cellular identity through multiple cell divisions suggests that changes in chromatin states might also contribute to these programs [22,23]. We speculate that existing OR regulation can be co-opted for coregulation of fru expression in ORNs with sex-specific behavioral functions.
Here, we describe a molecular circuitry with both developmentally hardwired and olfactory-receptor-activity-dependent components that refine and maintain fru expression in Or47b and Ir84a ORNs. In a genetic screen for ORN development, we identified a putative chromatin modulator, Alhambra/AF10 (Alh), which, when mutated, expands both fru/Or47b and fru/Ir84a expression to fru-negative ORNs independently of axon guidance decisions during development. Alh is expressed dynamically in developing ORNs in pupal stages, but during the onset of OR expression it is expressed in ORNs that do not express fru. Once the correct pattern of OR and fru expression is established, the maintenance of fru expression requires Or47b and Ir84a activity in adult flies. This mode of fru regulation in adult ORNs is disrupted in CamKI and histone acetyl transferase p300/CREB Binding Protein (CBP) mutants. Our results suggest that Alh and OR-dependent signaling represent two different modes of fru transcriptional regulation that coordinate, establish, and maintain the fru-positive identity alongside the sensory identities of ORNs.
Results
fru-Positive OR Expression Is Expanded to fru-Negative ORNs in p 1353 Mutants
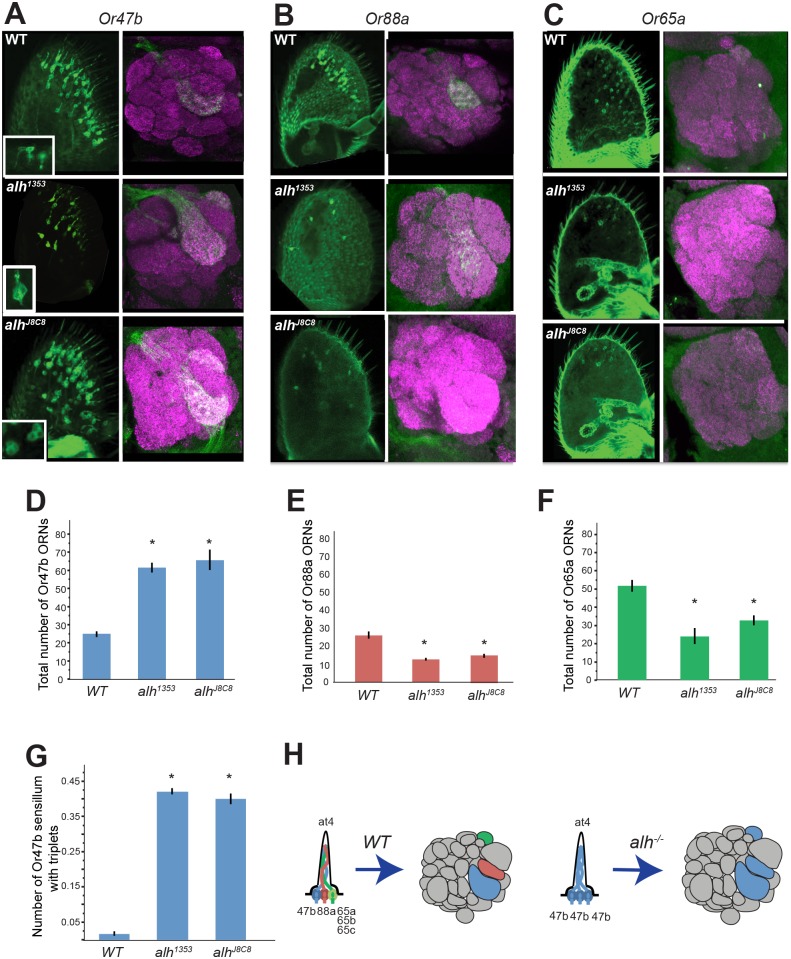
We previously carried out a forward genetic screen for regulators of class-specific ORN development in the olfactory system [24]. This was a histology-based mutagenesis screen where the glomerular targeting patterns of three different classes of ORNs (Or47a, Or47b, and Gr21a) were analyzed in antennal mutant clones in an otherwise heterozygous animal [25]. We specifically looked for mutants with defective ORN projection patterns in the antennal lobe. In this screen, we isolated a mutation (p 1353) that modified the projection pattern of Or47b ORNs in the antennal lobe (Fig 1). We mapped this mutation to the D. melanogaster orthologue of the chromatin modulatory protein AF10, Alhambra (alh). We refer to this mutation as alh 1353 in the rest of the paper. In both alh 1353 and alh J8C8 mutants, Or47b ORN axons project to their normal target, the VA1v glomerulus, as well as the DL3 glomerulus (Fig 1A and 1H). In addition, the VA1v glomerulus expands dorsally, appearing to almost engulf the VA1d glomerulus, leading to a loss of VA1d glomerulus in some of the mutant antennal lobes [26]. The DL3 and VA1d glomeruli are normally innervated by other at4 sensilla ORNs that express Or65a and Or88a, respectively, which develop from the same sensory organ precursor as Or47b ORNs [20].
Fig 1. fru-positive OR expression in at4 sensilla expands to developmentally related fru-negative ORNs in alh mutants.
A) Adult antennae and brains labeled with Or47bGal4 UAS-CD8GFP (green) in wild type and alh mutant clones. Magenta staining in brains is against N-cadherin, a neuropil marker. B) Adult antennae and brains labeled with Or88aGal4 UAS-CD8GFP in wild-type and alh mutant clones. C) Adult antennae and brains labeled with Or65aGal4 UAS-CD8GFP in wild-type and alh mutant clones. D–G) Quantification of cell bodies observed in the adult antennae of WT and alh mutant clones. For all graphs, asterisks indicate significant (p < .05) differences from wild type. Error bars represent standard error of the mean (SEM). An ANOVA was performed for each cell type and followed with Tukey’s Honest Significant Difference (HSD)—see Materials and Methods. D) Total Or47b-positive cells. Wild type flies were significantly different from both alh conditions (p < .0001). n = 10–40. All count data may be found in the Supporting Information as S1 Data. E) Total Or88a-positive cells. Both alh conditions were significantly different from wild-type males (p < .0001). n = 27 − 57. F) Total Or65a-positive cells. Both alh conditions were significantly different from wild-type males (p < .05). n = 9–27. G) Total or47b-positive clusters, normalized by total Or47b-positive cells. Wild type flies were significantly different from all alh conditions (p < .0001). n = 10–40. H) Model: in alh mutants, the Or47b odorant receptor expression is expanded to the other ORNs in the at4 sensilla, at the expense of their native OR expression, but the axons of these ORNs continue to target their original locations in the antennal lobe.
GENOTYPES:
A) eyflp; Or47bGal4/UAS-CD8GFP; FRT82/FRT82Gal80E2F,
eyflp; Or47bGal4/UAS-CD8GFP; FRT82alh 1353/FRT82Gal80E2F,
eyflp; Or47bGal4/UAS-CD8GFP; FRT82alh j8c8/FRT82Gal80E2F
B) eyflp; Or88aGal4/UAS-CD8GFP; FRT82/FRT82Gal80E2F,
eyflp; Or88aGal4/UAS-CD8GFP; FRT82alh 1353/FRT82Gal80E2F,
eyflp; Or88aGal4/UAS-CD8GFP; FRT82alh j8c8/FRT82Gal80E2F
C) eyflp; Or65aGal4/UAS-CD8GFP; FRT82/FRT82Gal80E2F,
eyflp; Or65aGal4/UAS-CD8GFP; FRT82alh 1353/FRT82Gal80E2F,
eyflp; Or65aGal4/UAS-CD8GFP; FRT82alh j8c8/FRT82Gal80E2F
In D. melanogaster, ORNs housed in the same sensillum arise through asymmetric divisions of a single multipotent precursor cells. During these divisions, the axons from the ORNs sort themselves and navigate to different future glomerular regions in the antennal lobe. Unlike mammals, ORs in D. melanogaster are not required for ORN axon guidance [27,28]. In fact, axon sorting decisions that guide ORNs from the same sensillum to distinct glomeruli are made prior to OR expression, suggesting that independent programs regulate the sensory identities and guidance of ORN axons [29]. The defects in the glomerular pattern of Or47b ORN projections in alh mutants could be due to a disruption of developmental programs regulating Or47b ORN axon guidance. Alternatively, the same defects might arise due to a conversion of Or65a and Or88a ORNs to the Or47b sensory identity without affecting the glomerular position of the at4 ORNs. In order to differentiate between these two hypotheses, we analyzed Or47b expression in the antenna. In wild type antennal MARCM clones [30], one Or47b ORN is found in each at4 sensillum. However, in alh 1353 mutant clones, Or47b expression is expanded within the same sensillum, leading to 2–3 Or47b ORNs per sensillum, all of which extend their dendrites into the same sensory bristle (Fig 1A). This was verified by counting the total number of cells in each mutant antenna, a somewhat variable number due to differences in the sizes of the clones produced by MARCM (Fig 1D). To reduce this variability, we also quantified the number of clusters of cells (defined as cell bodies with dendrites that project to the same sensillum) and divided this number by the total number of cells to normalize by the size of the clonal population (Fig 1G). Examination of Or88a and Or65a expression showed a concomitant decrease in the number of Or88a and Or65a ORNs, suggesting a conversion of Or65a and Or88a to Or47b sensory identity (Fig 1B–1F). These changes in Or47b, Or88a, and Or65a expression were also confirmed by qRT-PCR of wild type and alh mutant antennae (S1 Fig). In addition, double-labeling experiments show that the few remaining Or88a ORNs in alh mutants are never found in “clusters” in the antennae, and their axons intermingle with the converted Or47b axons in the VA1d glomerulus (S4A Fig).
We also overexpressed different isoforms of Alh. We hypothesized that if Alh is required to suppress Or88a and activate Or47b fate, overexpression of Alh should result in the decrease of Or47b ORNs and expansion of Or88a fate. Overexpression of the long isoform of alh, alh-L, using elav-GAL4, did not result in any Or88a or Or47b phenotype (S5A Fig). Elav-GAL4-induced expression of alh-S was lethal, suggesting this is the functionally important isoform. We then used heatshock-induced expression of GAL4 to pulse alh-S expression through third instar-10 h after pupanium formation (APF), 10–30 h APF, 24–34 h APF, and 48–58 h APF old pupae. None of these experiments resulted in an expanded Or88a or lost Or47b ORN population (S5 Fig). These results suggest that OR expression in at4 ORNs is more sensitive to loss of alh function rather than overexpression. One possible explanation for this can be that overexpression of alh might not be strong enough to overcome lineage-specific chromatin states around at4 OR genes that are already in place during at4 ORN development at the times of our heatshock. In addition, it is still possible that, due to the highly dynamic expression pattern of alh, there is a very narrow time window during pupal stages that was not captured in our heatshock experiments. Thus, we base our interpretations on the results from the mutant analysis, which suggests a function for Alh in appropriate segregation of ORs identities of ORNs independent of their guidance decisions to specific glomerular zones, uncoupling the genetic programs for sensory receptor selection and axon guidance.
The majority of the other ORN classes representing approximately ten different sensilla, including Or67d ORNs in at1 sensilla (Fig 2D–2F) examined, were not affected by similar sensory conversions (S2 Fig). In contrast, we detected a similar identity conversion in the sensilla housing Ir84a ORNs (Fig 2A–2C). We detected a general decrease in the total number of ac4 sensilla based on cell counts in alh 1353 mutants (Fig 2B). This possibly is due to an earlier function of Alh on precursor patterning based on its expression in a ring of cells on the antennal disc known to give rise to some of the coeloconic and trichoid sensilla fates [31] (S6 Fig), which in alh mutants lead to decreased cell survival. Despite the effects on the total number of sensilla, within the formed sensilla, multiple Ir84a-positive ORNs were observed (Fig 2A–2C).
Fig 2. fru-positive OR expression in ac4 sensilla expands to developmentally related fru-negative ORNs in alh mutants.
A) Adult antennae and brains labeled with Ir84aGal4 UAS-CD8GFP (green) in wild type and alh mutant clones. Magenta staining in brains is against N-cadherin, a neuropil marker. B) Total Ir84a-positive cells. Asterisks indicate significant (p < .05) differences from wild type. Error bars represent SEM. ANOVAs were performed and followed with Tukey’s HSD—see Materials and Methods. Wild type flies were significantly different from all alh conditions (p < .0001). n = 24–50. All count data may be found in the Supporting Information as S1 Data. C) Model: In alh mutants, the Ir84a odorant receptor identity is expanded to other coeloconic ORNs as observed through glomerular innervation. Ir84a expression is expanded to ir75a and ir76a ORNs. D) Adult antennae and brains labeled with Or67dGal4 UAS-CD8GFP (green) in wild type and alh mutant clones in Drosophila. Magenta staining in brains is against N-cadherin, a neuropil marker. E) Total Or67d-positive cells. An ANOVA for this data was not significant. n = 20–30. All count data may be found in the Supporting Information as S1 Data. F) Model: In alh mutants, the expression and axonal targeting patterns of or67d-positive ORNs are unchanged.
GENOTYPES:
A) eyflp; Ir84aGal4/UAS-CD8GFP; FRT82/FRT82Gal80E2F,
eyflp; Ir84aGal4/UAS-CD8GFP; FRT82alh 1353/FRT82Gal80E2F,
eyflp; Ir84aGal4/UAS-CD8GFP; FRT82alh j8c8/FRT82Gal80E2F
D) eyflp; Or67dGal4/UAS-CD8GFP; FRT82/FRT82Gal80E2F,
eyflp; Or67dGal4/UAS-CD8GFP; FRT82alh 1353/FRT82Gal80E2F
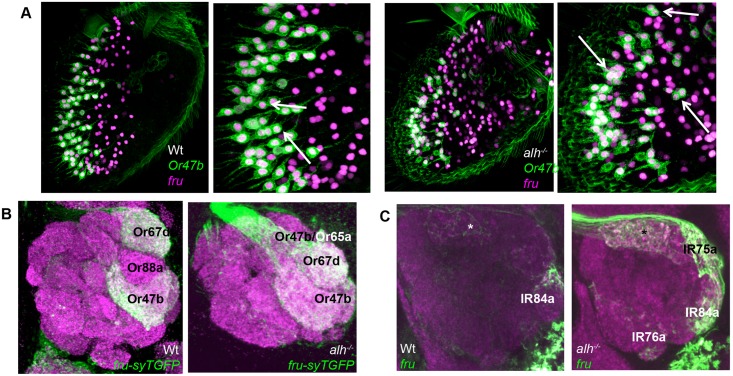
Each Ir84a ORN is normally housed in the ac4 sensilla with its sibling ORNs that express Ir75d and Ir76a (Fig 2C) [31]. Examination of the antennal lobes of alh 1353 mutants showed that in addition to the VL2a glomerulus normally innervated by Ir84a ORNs, Ir84a-positive ORNs were also present in the VM4 glomerulus of Ir76a ORNs (Fig 2A and 2C) [32]. We found that Ir76a expression was decreased in alh mutants (S4C Fig). We also observed Ir84a–positive ORN terminals innervating the DP1l glomerulus housing Ir75a ORNs, which normally reside in ac2 and ac3 sensilla, suggesting that ectopic expression of Ir84a is not restricted to the ac4 ORNs but also expands to other coeloconic sensilla ORNs. From our previous studies, we know that ORN organization and development in coeloconic sensilla is not identical to ORNs in basiconic and trichoid sensilla [18,31,32]. For example, ac2, ac3, and ac4 sensilla all share the same neurons, Ir76b, Or75d, Ir75a and Or76b, in different combinations, and thus they are more developmentally intertwined than other sensilla types, which does not happen in basiconic and trichoid sensilla. It is possible that the differences of Or47b and the Ir84a expression phenotypes in alh 1353 mutants might reflect differences in the developmental programs of trichoid (at4) versus coeloconic (ac4) sensilla. In summary, our results suggest that in alh 1353 mutants, two fru-positive ORs (Or47b and Ir84a) expand to developmentally related ORNs without changing the projection of their axons to appropriate glomerular areas in the antennal lobe.
Fru Expression Accompanies Sensory Conversion in alh 1353 Mutants
Our results demonstrate that Alh regulates some aspects of ORN identity, such as Or expression, but not others, such as axon sorting and guidance. We then asked what other aspects of ORN identity might be regulated by alh 1353. We were intrigued that the two ORNs expanded by the alh 1353 mutation, Or47b and Ir84a, are two of the three ORN classes that express fru. To answer whether other aspects of neuronal identity are also altered in alh 1353 mutants, we examined fru expression.
We observed an expansion of fru expression in alh 1353 mutant antennae (Fig 3). When we examined fru expression closely in alh 1353 mutant ORNs, we found clusters of 2–3 fru-positive ORNs within the same sensillum, mimicking Or47b expression in the alh 1353 mutants (Fig 3B). Indeed, colabeling for Or47b and fru revealed that both were coexpressed in alh mutant ORN clusters (Fig 3B). The expansion of fru expression to multiple at4 and ac4 ORNs observed in the antenna was confirmed when we examined fru-positive axon guidance in the antennal lobes of alh 1353 mutants, which show a striking similarity to the expression patterns of Or47b and Ir84a in alh 1353 mutants. (Fig 3C and 3D). Additional double-labeling experiments show that the innervation of the Va1d glomerulus by fru-positive axons in alh mutants is a result of the expansion of Or47b- and fru-positive axons to this location as previously found. The remaining nonclonal wild type Or88a ORNs in these mutants do not express fruitless (S4B Fig). These results suggest that fru expression accompanies Or47b and Ir84a expansion, and alh 1353 disrupts a program that is normally required to corepress both Or47b/Ir84a and fru expression in inappropriate, yet developmentally related ORNs.
Fig 3. fru expression expands together with Or47b expression in alh mutants.
(A/B) Antennae labeled with fru GAL4 UAS-RedStinger (magenta), and Or47bCD8GFP (green) in wild type and alh mutant clones. Right panels represent higher magnification images. Arrows label Or47b/fru-positive nuclei in wild type images. In alh mutants, arrows point to sensilla with 2–3 Or47b ORNs that are also fru-positive. (C) Antennal lobes labeled with fruGal4 UAS-sytGFP (Z-stack, anterior sections of antennal lobe). (D) Antennal lobes labeled with fruGal4 UAS-CD8GFP (Z-stack, posterior sections of antennal lobe). Asterisks denote fru-labeled glomeruli thought to be innervated by neurons from the antennal sacculus.
GENOTYPES:
(A) wild type: eyFLP/+;Or47bCD8GFP/UAS-RedStinger; FRT82 fru GAL4 /FRT82Gal80E2F
(B) alh mutant: eyFLP/+;Or47bCD8GFP/UAS-RedStinger; FRT82 alh 1353 fru GAL4 /FRT82Gal80E2F
(C) wild type: eyFLP/+; UAS-syTGFP/+; FRT82 fru GAL4 /FRT82Gal80E2F
alh mutant: eyFLP/+; UAS-syTGFP/+; FRT82 alh 1353 fru GAL4 /FRT82Gal80E2F
(D) wild type: eyFLP/+; UAS-CD8GFP/+; FRT82 fru GAL4 /FRT82Gal80E2F
alh mutant: eyFLP/+; UAS-CD8GFP/+; FRT82 alh 1353 fru GAL4 /FRT82Gal80E2F
alh-Dependent Regulation of Or and fru Expression Occurs Early in ORN Development
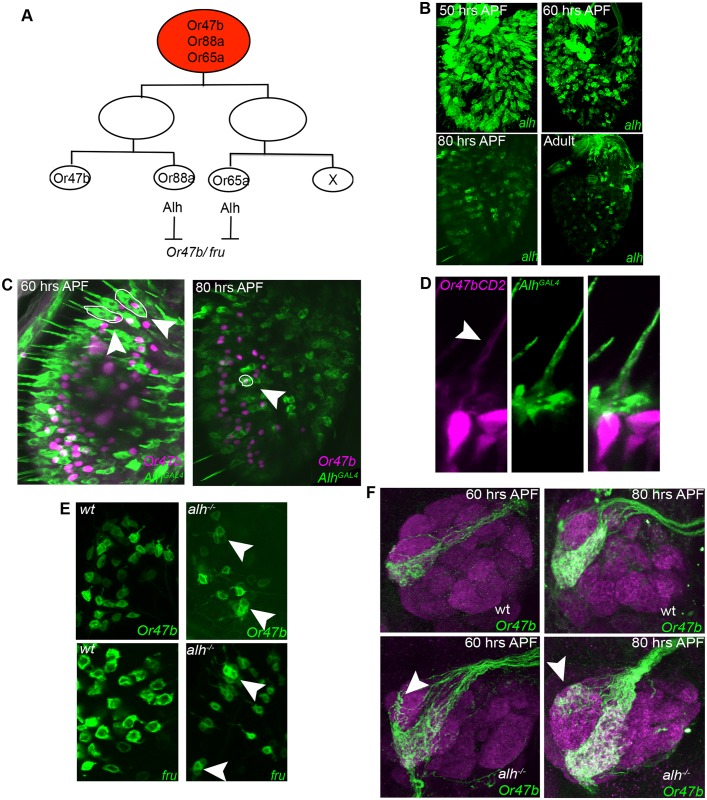
We hypothesized that in wild-type flies Alh, either directly or indirectly, coordinates repression of fru and Or expression in fru-negative ORNs (Fig 4A). If this is the case, then alh should be expressed in fru-negative at4 ORNs and excluded from their fru-positive sibling ORNs at the onset of or expression. To test this, we used three GAL4 enhancer trap lines. Two of the lines have P-element insertions upstream of the first exon of short alh transcripts. The third line has an insertion within the first intron of the short isoform at a similar site to that of alh j8c8 (S3 Fig). All insertions are located within the fifth intron of the long isoforms (S3 Fig).
Fig 4. Alh represses Or47b and fru in developmentally related ORNs in the same sensillum during development.
(A) Asymmetric divisions of neuronal precursors give rise to ORNs in at4 sensilla. From the mutant phenotype, we predict that Alh represses Or47b and fru expression in Or88a and Or65a ORNs. (B) Alh GAL4-driven UAS-CD8GFP expression in developing pupal antennae. (C) Double labeling of Or47b (magenta) and alh (green) around 60–70 h APF (left panel) and 80 h APF. Or47b expression is excluded from alh expressing cells (arrows) but clusters with them (circled in white). (D) High magnification of a sensillum double labeled with Or47b-CD2 (magenta) and alh (green). Arrow points to the dendrite of Or47b ORN innervating the sensory hair together with alh-positive fibers. (E) Enlarged portions of wild-type (left panels) and alh mutant (right panels) antennae at 60–70 h APF, expressing Or47b-Gal4 UASCD8GFP (top panels), or fru-gal4 UASCD8GFP (bottom panels). (F) Wild-type (top panels) and alh mutant (bottom panels) antennal lobes expressing Or47bGal4UASCD8GFP (green) and stained for ncadherin (magenta).
GENOTYPES:
(B) 50hrs: AlhGal4 NP7010/UAS-CD8GFP; 60hrs-adult: AlhGal4 NP7441/UAS-CD8GFP
(C) Or47b-lexA lexOp-tomato:nls AlhGal4 NP6628/UAS-CD8GFP
(D) Or47B-CD2/+; AlhGal4 NP6628/UAS-CD8GFP
(E) eyFLP/+; Or47b-GAL4 UAS-CD8GFP/+; FRT82/FRT82Gal80E2F
eyFLP/+; Or47b-GAL4 UAS-CD8GFP/+; FRT82 alh1353 /FRT82Gal80E2F
or eyFLP/+; UAS-CD8GFP/+; FRT82 fru GAL4 /FRT82Gal80E2F
eyFLP/+; UAS-CD8GFP/+; FRT82 alh1353 fruGAL4/FRT82Gal80E2F
(F) eyFLP/+; Or47b-GAL4 UAS-CD8GFP/+; FRT82 /FRT82Gal80E2F
or eyFLP/+; Or47b-GAL4 UAS-CD8GFP/+; FRT82 alh 1353 /FRT82Gal80E2F
Developmental analysis of the expression of these alh GAL4-driven UAS-CD8GFP reporters showed a highly dynamic but reproducible expression pattern of alh during development (Fig 4B). In the antennal imaginal discs at the third larval instar, Alh is expressed in the central ring, which houses ORN precursors (S6 Fig). Later, 40–50 h after puparium formation (APF), alh is expressed in most ORNs (Fig 4B). At this time, alh-positive ORNs can be seen in sensilla clustered in the lateral regions of the antenna where the at4 and ac4 sensilla are located in adults, as well as neurons in the sacculus. The expression pattern becomes more restricted as pupal development proceeds and is almost absent in the adult.
Our observations suggest that Alh refines and coordinates Or and fru expression among the ORNs in at4 and ac4 sensilla. Developmental analysis presented later in this paper shows that Or47b expression begins at 40 h APF. Colabeling alh and Or47b showed that after 50 h, alh expression is excluded from Or47b ORNs while still present in other at4 ORNs (Fig 4C and 4D). The alh mutant phenotype can be observed at this same time, where clusters of Or47b and fru-positive 2–3 Or47b ORNs are observed in single sensilla (Fig 4E). In addition, the glomerular positioning defects of Or47b ORNs in the antennal lobe are apparent by 60 h APF (Fig 4F). These results suggest that fruitless expression is developmentally coregulated with fru-positive OR expression, and that Alh acts during development to repress the fru-positive OR identity in fru-negative ORNs prior to selection of their appropriate receptors.
Fruitless Expression in ORNs Requires OR Function
In alh mutants, fru expression in the antenna is expanded alongside Or47b and Ir84a expression (Fig 3). The synchronous, expanded expression of both fru and fru-positive ORs in alh mutants suggests a tight relationship between OR and fru expression, regulated by Alh. We posited three possible ways to explain this relationship: 1) Alh represses fru, which is required to activate Or expression; 2) Alh represses Or expression, which can activate fru expression; and 3) Alh represses both fru and Or expression either directly or indirectly, by repressing an activator of both genes. Given the established role of Fru in the regulation of gene expression and courtship [33], our initial prediction was that Fru functions to regulate the expression of Or genes involved in courtship behaviors.
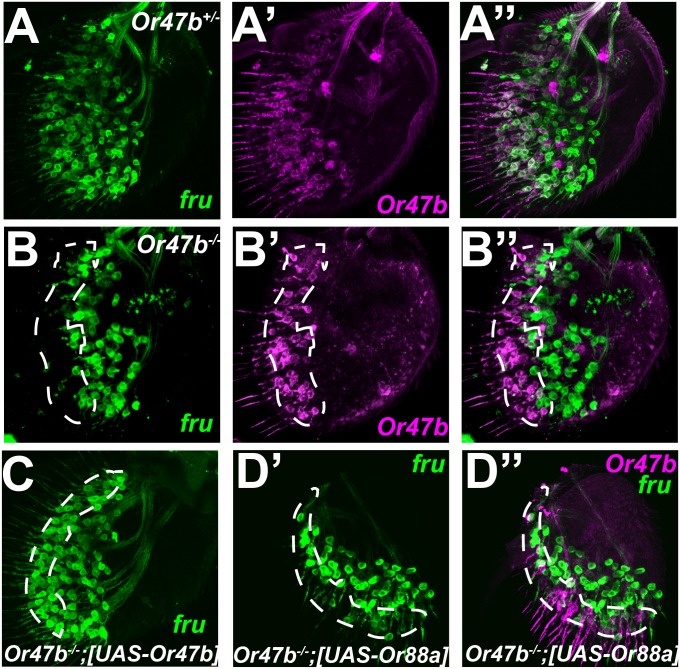
To test this hypothesis, we first asked whether Fru regulates Or47b expression in Or47b ORNs. However, we found that Or47b expression is unaffected in fru mutants, suggesting that Fru does not regulate Or47b expression (S7 Fig). We then examined fru expression in the absence of Or47b function. Surprisingly, we observed that fru expression in Or47b ORNs is abolished in Or47b mutants, (Fig 5B). This finding was confirmed by qRT-PCR (S8 Fig). This phenotype could be rescued by the expression of a UAS-Or47b transgene driven with fru GAL4 (Fig 5C). The expression of UAS-Or88a, a closely related receptor that detects similar ligands to Or47b [17] was partially able to rescue fru expression. This indicates a requirement for the function of ORs activated by the same ligand in fru regulation (Fig 5D and 5D’ and S11 Fig). Expression of Or67d, the third fru-expressing Or gene, which detects cVA, was not able to rescue fru expression in Or47b ORNs (S9D Fig).
Fig 5. fru expression in adult Or47b ORNs requires Or47b function.
(A) Heterozygous Or47b mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP (A) and OR47b-CD2 (A’). (A”) shows the merge of two images. (B) Homozygous Or47b mutant antennae (3–5 d old). (C) Overexpression of UAS-Or47b under the control of fruGal4 in Or47b mutants (14 d old). (D) Overexpression of UAS-Or88a under the control of fruGal4 in Or47b mutants (14 d old).
GENOTYPES:
A–A”: Or47b-CD2 Or47b 2 /+;fru GAL4 UAS-40XCD8GFP/+
B–B”: Or47b-CD2 Or47b 2 /or47b 2;fru GAL4 UAS-40XCD8GFP/+
C: Or47b 2 /Or47b 2;fru GAL4 UAS-40XCD8GFP/UAS-Or47b
D: Or47b-CD2 Or47b 2 /Or47b 2;fru GAL4 UAS-40XCD8GFP/UAS-Or88a
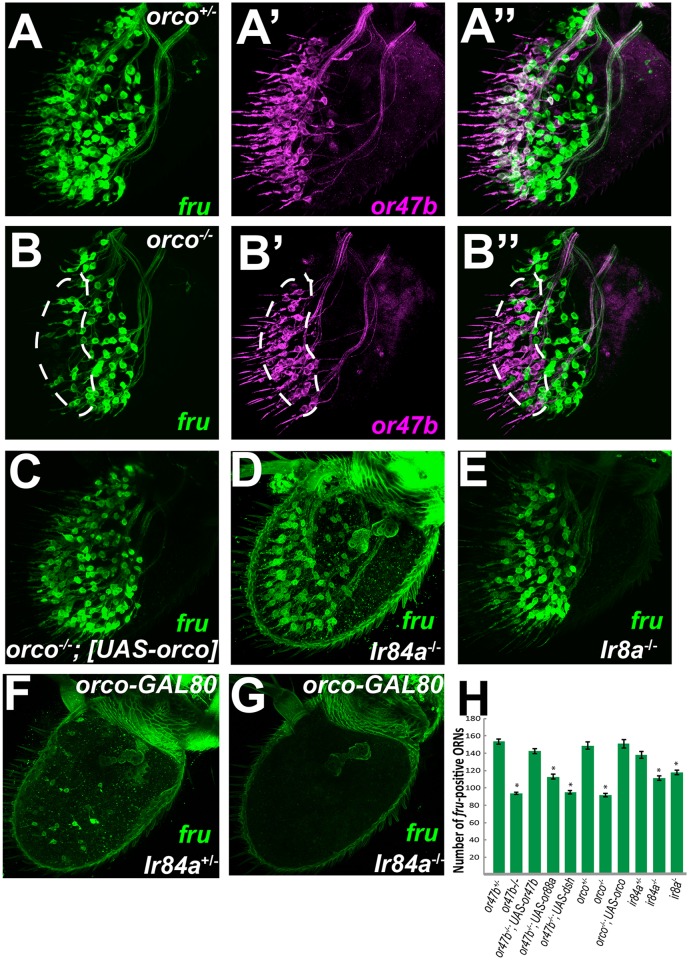
We also abolished Or47b function using mutants for the general Or coreceptor orco [28]. A loss of fru expression, similar to that seen in Or47b mutants, was observed in orco mutants (Figs 6A, 6B and 6H, 8E and S11 Fig) and was confirmed by qRT-PCR (S8 Fig). This phenotype could be rescued by expression of a UAS-orco transgene (Figs 6C and 6H, 8E and S11 Fig).
Fig 6. Or and Ir function is required to regulate fru expression in the adult olfactory system.
(A) Heterozygous orco mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP (A) and OR47b-CD2 (A’). (A”) shows the merge of two images. (B) Homozygous orco mutant antennae (3–5 d old). (C) Overexpression of UAS-orco under the control of fruGal4 in orco mutants (14 d old). (D) Homozygous Ir84a mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP. (E) Homozygous Ir8a mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP. (F) Heterozygous Ir84a mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP and orco-Gal80. (G) Heterozygous Ir8a mutant antennae (3–5 d old) expressing fruGal4 40XUASCD8GFP and orco-Gal80. (H) Quantification and statistical analysis of fru-positive ORN cell bodies observed in adult antennae of the indicated genotypes. n = 5–27. For all graphs, asterisks indicate significant (p < .005) differences from or47b heterozygotes. Error bars represent SEM. A one-way ANOVA was performed and followed with Tukey’s HSD—see Materials and Methods. All count data may be found in the Supporting Information as S1 Data.
GENOTYPES:
A–A”: Or47b-CD2 /+;orco 1 fru GAL4 UAS-40XCD8GFP /+
B–B”: Or47b-CD2 /+;orco 1 fru GAL4 UAS-40XCD8GFP /orco 1
C: +/ UAS-orco; orco 1 fru GAL4 UAS-40XCD8GFP /orco 1
D: Ir84a MI00501 fru GAL4 UAS-40XCD8GFP / Ir84a MI00501
E: Ir8a 1 /Y; fru GAL4 UAS-40XCD8GFP
F: orco-GAL80/+; Ir84a MI00501 fru GAL4 UAS-40XCD8GFP / +
G: orco-GAL80/+; Ir84a MI00501 fru GAL4 UAS-40XCD8GFP / Ir84a MI00501
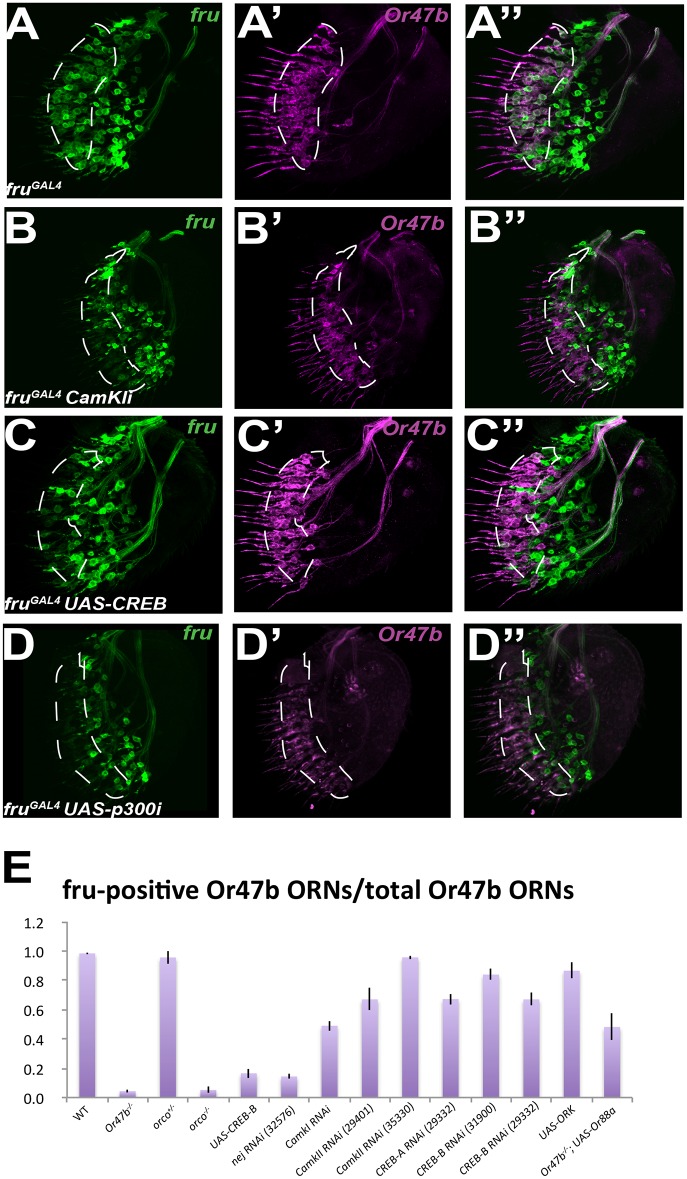
Fig 8. Maintenance of fru expression in adult ORNs requires CamK signaling and p300/CBP.
(A) Wild-type antennae expressing fruGal4 UAS-40XUASGFP (green) and Or47b-CD2 (magenta). (B–D) Antennae expressing fruGal4 UAS-40XUASGFP (green) and Or47b-CD2 (magenta) as well as fruGal4 CamKI RNAi (B), UAS-creb (C), and UAS-p300 RNAi (D). (E) Quantification of antennal fru-positive ORN cell counts for experiments in Figs 5, 6 and 8. Data shown represents the fraction of Or47b-positive cells that are also fru-positive. For all graphs, asterisks indicate significant (p < .01) differences from fru Gal4. Error bars represent SEM. A one-way ANOVA was performed and followed with Tukey’s HSD—see Materials and Methods. Cell count data also graphed in S10 Fig. All raw count data may be found in the Supporting Information as S1 Data.
GENOTYPES:
(A) Or47b-CD2 /+; fru GAL4 UAS-40XCD8GFP
(B) UAS-CamKI RNAi/+; Or47b-CD2/+; fruGAL4 UAS-40XCD8GFP
(C) UAS-CREB/+; Or47b-CD2/+; fruGAL4 UAS-40XCD8GFP
(D) UAS-p300RNAi/+; Or47b-CD2/+; fruGAL4 UAS-40XCD8GFP
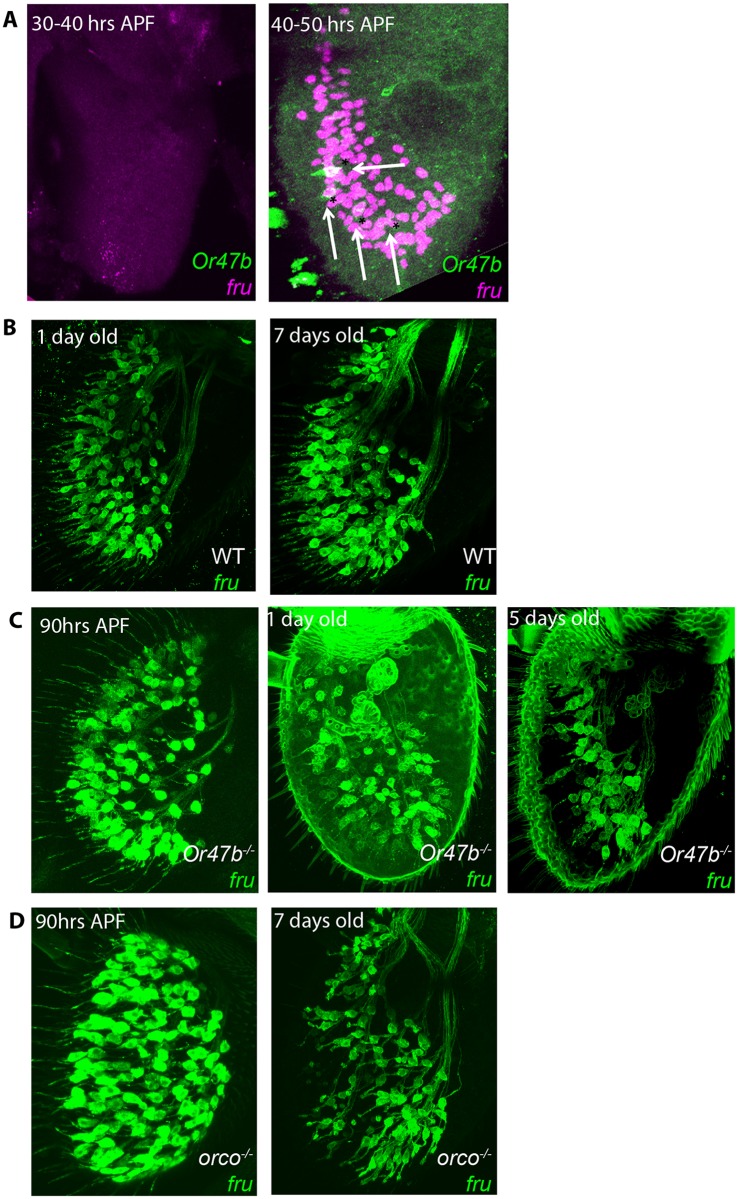
Time-course analysis of antennae showed that Or47b mutant ORNs start showing signs of degeneration by 14 d (S9 Fig). A degeneration phenotype has been reported for orco mutants as well [34]. In order to show that loss of fru expression is not due to degeneration or neuronal death, we colabeled Or47b and fru–positive ORNs. Double-labeling experiments confirmed that in both Or47b and orco mutants, fru expression was dramatically decreased specifically in Or47b ORNs, and that this loss of fru occurred prior to neuronal degeneration (Figs 5B’, 6B’, 8E and S9, S11 Figs). Perturbing Wingless signaling has been shown to suppress orco-dependent neuronal degeneration [34]. fru GAL4-dependent overexpression of disheveled (dsh), a downstream effector of the Wingless pathway, rescues the degeneration defect but not loss of fru expression in Or47b ORNs (S9C Fig). Expression of Or88a was sufficient to rescue Or47b ORN degeneration, and was also partially able to rescue loss of fru expression (Figs 5D, 8E, S9A and S9B and S11 Figs).
We next tested whether the other fru-positive receptors Ir84a and Or67d also regulate fru expression in adult ORNs. Unlike Or47b ORNs, Or67d and orco mutants did not affect fru expression in Or67d ORNs (S9D Fig). On the other hand, analysis of fru expression in Ir84a mutants and mutants of Ir8a (a coreceptor expressed in Ir84a ORNs [35]) showed that the number of fru-positive cell bodies decreases by approximately 20 cells (Fig 6D, 6E and 6H). To confirm that the decrease observed in fru expression in these mutants is restricted to Ir84a ORNs, we used orco-GAL80 to suppress fru GAL4-driven GFP expression in basiconic and trichoid ORNs. This allowed us to observe fru expression only in the Ir84a ORN population of wild type flies and Ir84a mutants. Wild type antennae had approximately 20 fru-positive cell bodies, which represent the Ir84a ORN class (Fig 6F and 6G). On the other hand, we observed a few to no fru-positive cell bodies in Ir84a mutants (Fig 6F and 6G). These findings were confirmed by qRT-PCR (S8 Fig). These results suggest that in addition to Or47b, Ir84a function is also required for fru expression in adult Ir84a ORNs.
OR Function Is Required for Maintenance of fru Expression in Adult ORNs but Not during Development
We predicted that both fru and Or expression must be established in pupal stages as ORNs adopt their final fates (Fig 4A). In wild type flies, the onset of fru expression coincides with the onset of Or47b expression, starting around 40 h APF and absent at earlier stages (Fig 7A). Given the temporal coordination of Or47b expression with fru during pupal development, we wanted to test whether Or47b function is required for the onset of fru expression. To do this, we analyzed fru expression during pupal development in Or47b and orco mutants (Fig 7). In these mutants, we found that fru expression in Or47b ORNs was still detectable by 90 h APF, suggesting that OR function is not required to initiate fru expression and that other factors, such as Alh, establish the correct expression and coupling of fru/Or during ORN development (Fig 7C and 7D). However, fru expression was lost within a few days after eclosion in both Or47b and orco mutants (Fig 7C and 7D). These results indicate that Or47b function is dispensable during the onset of fru expression in pupal stages, but once the flies eclose from their pupal cases, Or47b activity is required to maintain fru expression.
Fig 7. Onset of fru in developing ORNs overlaps with Or47b but is independent of Or47b function.
(A) Wild-type antennae expressing fruGal4 UAS-Redstinger (magenta) and Or47b-CD8GFP (green). (B) Wild-type antennae expressing fruGal4 40XUASCD8GFP. (C) Or47b mutant antennae expressing fruGal4 40XUASCD8GFP. (D) orco mutant antennae expressing fruGal4 40XUASCD8GFP.
GENOTYPES:
(A) Or47b-CD8GFP/+; fruGAL4 UAS-RedStinger/+
(B) +/+; fruGAL4 UAS-40XCD8GFP
(C) Or47b2/Or47b2; fruGAL4 UAS-40XCD8GFP
(D) orco2/orco2; fruGAL4 UAS-40XCD8GFP
CamK1 and p300/CBP Are Involved in Olfactory-Receptor-Dependent Maintenance of fru Expression
In D. melanogaster, ORs are necessary to generate both spontaneous and evoked patterns of ORN activity [9,13,28,36,37]. One possible explanation for the Or-dependent maintenance of fru expression in adult ORNs is that Or-mediated neuronal activity is required. To test the role of neuronal excitability in the regulation of fru expression, we electrically silenced fru-positive ORNs using the potassium channel UAS-dORK. We observed a statistically significant decrease in the number of fru-positive ORN cell bodies in the antenna when these ORNs were electrically silenced (S10A, S11 Figs and Fig 8E). However, double-labeling showed that this decrease was due to a smaller total number of Or47b ORNs. Unlike the OR mutants, the remaining Or47b ORNs continued to express fru. These results suggest that maintenance of fru expression in a subpopulation of ORNs requires OR function independent of neuronal activity.
Next, we investigated the mechanisms that relay Or/Ir activity to the transcriptional machinery in the nucleus. Unlike their mammalian counterparts, odorant receptors in D. melanogaster are not G-protein coupled but instead encode ligand-gated cation channels that conduct both calcium and sodium [38–41]. Given the role of calcium in signaling- and activity-dependent regulation of gene expression, we predicted that calcium signaling could maintain fru transcription in adult ORNs. To test this hypothesis, we screened RNAi knock-down lines of CamKI and CamKII using double-labeling of Or47b and fru expression. We found that disrupting CamKI function using different UAS-CamkI RNAi lines resulted in a decrease in fru expression in the Or47b ORN zone (Fig 8B and 8E, S11 Fig). Loss of CamKII had little yet significant effect on fru expression in Or47b ORNs as well (S10 Fig, S11 Fig). These results suggest that calcium signaling through CamKs contribute to maintenance of fru expression in a proportion of fru-positive ORNs.
CamKI encodes the D. melanogaster orthologue of the vertebrate CamKI/IV, which has been shown to phosphorylate the histone acetyl transferase CBP/p300 and CREB, both of which are activated by receptor signaling and can function to maintain gene expression [42–44]. We first tested the candidate gene Creb-B, which functions in signal-dependent regulation of gene expression in many contexts [45,46]. Overexpression of a dominant negative form of Creb-B, which was previously shown to abolish Creb function [47], did not result a change in fru expression (S10B Fig). RNAi-mediated knock down of either Creb-A or Creb-B had a mild yet significant decrease in fru expression in Or47b ORNs, similar to what was observed for CamKII RNAi knock down (Fig 8E, S11 Fig). In contrast, overexpression of Creb-B resulted in a substantial decrease in fru expression in Or47b ORNs (Fig 8C and 8E and S11 Fig).
The finding that Creb-B overexpression, but Creb-B mutants, shows a dramatic reduction of fru expression is contradictory to the known function of Creb-B as a transcriptional activator. These results suggested that the effect of Creb-B overexpression on fru regulation could be indirect. Thus, we tested the possibility that Creb-B might be titrating a fru regulator that is found in limiting amounts in the ORNs. Creb proteins are known to interact with CBP/p300, which also interacts with many other transcription factors to maintain gene expression [45]. In addition, titrations effects on gene expression due to competition for limiting amounts of CBP/p300 was previously reported [48–52]. Since CBP/p300 is activated through phosphorylation by CamKI [43], we tested the hypothesis that p300 is required to maintain fru expression in Or47b and Ir84a ORNs. Similar to the CamKI mutants, and Creb-B overexpression experiments, an RNAi line targeting p300 showed a strong decrease in fru-positive Or47b ORNs (Fig 8D and 8E, S11 Fig). These results suggest that OR as well as CamK signaling, and CBP/p300 histone acetyl transferase function contribute to the maintenance of fru expression in Or47b ORNs (Fig 9).
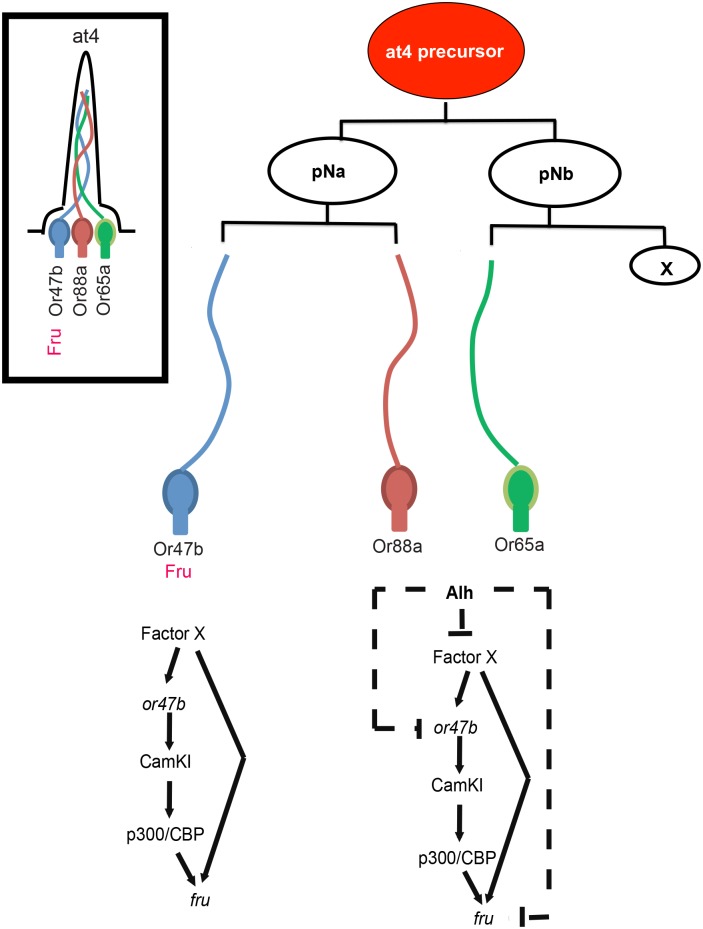
Fig 9. Regulatory feed forward loops establish and maintain fru expression in the olfactory system.
A multipotent precursor cell divides asymmetrically to generate at4 ORN cell types. In Or47b ORNs, a factor X is required to coactivate both Or47b and fru expression during development. In Or65a and Or88a ORNs, Alh, either directly or indirectly through repression of X, is required to repress both Or47b and fru. Once Or47b and fru expression is established in Or47b ORNs, OR function maintains fru expression through p300/CBP.
Discussion
Fru As a Molecular Identity Marker for Sex-Specific Behavioral Circuits
In D. melanogaster, sex-specific behaviors are largely regulated by a single gene, fru, whose expression is restricted to a circuit of approximately 2,000 interconnected neurons [3]. Functional studies of fru-positive neurons support the hypothesis that fru labels the neuronal circuits that drive sex-specific behaviors, but the mechanisms that restrict its expression in the nervous system are not known. If fru is necessary for appropriate circuit formation during development as well as circuit function, its expression in appropriate neurons must be tightly controlled using hardwired developmental programs for neuronal fate such as OR expression. However, it is also known that sex-specific behaviors are adaptable, suggesting a need for a plastic molecular mechanism to modify fru expression and the structure of fru-positive circuits. In this paper, we describe two different modes of transcriptional regulation of fru in ORNs mediating sex-specific behaviors: (1) establishment of fru expression through an Alh-mediated chromatin modulatory pathway that coordinates and refines fru, Or47b, and Ir84a expression in developing ORNs; and (2) maintenance of fru expression through ORs in adult ORNs by calcium signaling and histone acetyl transferase, p300/CBP. To our knowledge, our results provide the first example of a regulatory pathway that coordinates the sensory identity of neurons with the identity of the neural circuit they must integrate into. Our results suggest that this is a distinct step in the developmental program of a neural circuit, one that is independent of axon guidance decisions by the neurons as the circuit develops. These findings also highlight the role of feed-forward regulatory loops that establish and maintain these identities with hardwired and environmentally sensitive components, respectively (Fig 9).
Separate Programs Control Odorant Receptor Choice and Axonal Guidance during ORN development
In mammals, OR genes are critical for appropriate axon guidance through an elegant mechanism that links the amount of guidance molecules produced by the axon to the unique, ligand-independent G-protein coupled receptor (GPCR) activity signature of each Or [53–55]. These guidance molecules interact with gradients of other guidance molecules to sort ORN axons in a class-specific manner as they enter the olfactory bulb [53–55]. In contrast, OR function does not regulate ORN targeting in D. melanogaster. Many groups have shown that there is no contribution of Or genes to the guidance of ORN axons to specific glomerular zones in D. melanogaster [9,27,28,36,37]. The fact that axon guidance is largely completed before OR expression begins [29] also supports the hypothesis that programs of sensory receptor choice and axon guidance are independently regulated.
Nevertheless, receptor identity must be coupled with an appropriate target location to establish the one-neuron/one-receptor/one-glomerulus rule for odortopic mapping in the brain. Since the identity of the ORN precursor cell determines both the receptor expression and guidance programs for all the ORNs it will generate, the expression and axon guidance instructions for each ORN must be segregated together during asymmetric divisions of these precursors. Notch signaling, which acts in a context-independent manner during these divisions, is required to ensure segregation of both sensory and glomerular targeting identities of ORNs [20,22]. The sorting of Or expression and guidance fates throughout precursor division is accomplished through the generation of Notch “ON” and “OFF” cellular states due to asymmetries in Notch signaling components [20,56]. Perturbing Notch function results in the duplication of both sensory fates and glomerular postion of one ORN class at the expense of the other within a sensillum [20]. Segregation of glomerular positioning programs for ORNs in the same sensillum during asymmetric precursor divisions requires Notch-dependent differences in the repulsive cell surface molecule sema-2b [57]. Notch was also shown to affect postmitotic neuronal targeting in the D. melanogaster central nervous system independent of neuronal fate decisions [58]. However, very little is known about the mechanisms that segregate sensory identity of ORNs downstream of Notch. alhambra is the first gene identified that, when mutated, results in a complete conversion of the sensory identity of one ORN in the same sensillum to another while remaining independent of glomerular positioning decisions. This indicates that Alh specifically regulates the proper segregation of individual Or choice programs among ORNs in the same sensillum after glomerular positioning decisions are properly segregated and established. It will be interesting to see whether Or and fru expression decisions driven by Alh are induced downstream of Notch signaling.
The Putative Chromatin Modulator alhambra Coregulates OR and fru Expression
Alh encodes the D. melanogaster orthologue of the leukemia fusion gene AF10, which has been previously shown to interact with chromatin modulators such as proteins in the SWI/SNF remodeling complex and the histone methyl transferase hDOT1 [59,60]. More recently, it was shown that the Caenorhabditis elegans orthologue of alhambra, ZFP-1, interacts with Dot1 and affects cell signaling by regulating RNA polymerase pausing on selected genes [61]. Recent work suggests that polymerase pausing may have many effects on transcriptional regulation, and one such effect appears to be an establishment of permissive chromatin around the paused polymerase [62].
A key feature of the alh mutant phenotype is the repressive effect it has on the expression pattern of both fru and fru-positive OR genes during development. The loss of ORN diversity within the at4 and ac4 sensilla suggests that Alh might modify chromatin in response to signals such as Notch, which operate during asymmetric cell divisions to diversify ORN fates. The phenotypic specificity of alh mutants to at4 and ac4 fru-positive sensilla suggests that Alh functions during the development of these sensilla to ensure fru expression is restricted to only certain neurons that are tuned to specific odors. Alh may function to repress the expression of terminal differentiation genes that work together to define the odor tuning properties of a given ORN (Ors) and the behavioral circuits it will be integrated into (Fru), independently of axon guidance programs (Fig 9).
It is likely that a factor X (Fig 9) is expressed in Or47b ORNs that coordinately activates fru and Or47b. The restriction of the sensory conversion phenotype in alh mutants to fru-positive ORs suggests that Alh might suppress the fru-positive ORs (e.g., Or47b) and promote alternate OR expression (e.g., Or88a), either indirectly, through suppression of factor X, or directly prior to the selection of an alternate receptor by the ORNs in the same sensillum (Fig 9). For example, the fru-positive Or47b identity might be the “default,” since the expression of either Or65a or Or88a seems to require the suppression of Or47b, and Or47b is the first receptor to be expressed in at4 ORNs. Once Or47b expression begins in one of the at4 ORNs, Notch signaling may ensure that Or47b is turned off in the sibling ORNs or precursors by changes in chromatin induced by proteins such as Alh. It is interesting to note that a “default” identity from which other identities diversify is also a phenomenon seen in the diversification of ORN precursor identities, pointing to a modularity in the cell fate programs regulating ORN diversity [18]. The phenotype is slightly more complex for Ir84a, where fru-positive Ir84a expression not only expands to another ORN in the same sensilla subtype but also to an ORN in another coeloconic sensilla. The reason for this discrepancy might be due the differences in developmental programs of coeloconic sensilla ORNs expressing Irs. Several Irs are in fact expressed in multiple neurons across several different coeloconic sensilla subtypes, a phenomenon not seen in canonical ORs expression [32,40]. The third fru-positive OR, Or67d, is not affected in alh mutants. This might be due to the fact that this is the only ORN in the at1 sensillum.
The expansion of OR expression to other sensilla ORNs without affecting guidance was previously reported in atrophin mutants [63]. However, there are multiple differences between the atrophin and alh mutant phenotypes. First, in alh mutants, in both ac4 and at4 sensilla, there is a within-sensilla sensory identity conversion among developmentally related ORNs. In atrophin mutants, expansion does not happen among the ORNs within the same sensillum. Secondly, in atrophin mutants, there is a coexpression of both the endogenous receptor and the ectopically expressed receptor in the affected ORNs. In contrast, in alh mutants, the expansion of one receptor to ectopic ORNs is accompanied by the loss of the endogenous receptor normally expressed in that ORN. Thus, the mutant ORNs express a single receptor rather than two. Loss of alh function converts the sensory identity to one at the expense of another, whereas in atrophin mutants, ORNs maintain a dual sensory identity.
OR Signaling in the Maintenance of fru Expression in Adult Flies
Here, we report for the first time that once the terminal differentiation of Or47b and Ir84a ORNs is complete, OR signaling is required for the maintenance of fru expression. fru expression is lost in an ORN class-specific manner in Or47b and Ir84a mutant adults. This process also requires CamK signaling and the histone acetyl transferase CBP/p300, as mutants in both are associated with a loss of fru expression in adult ORNs. Interestingly, fru expression does not require OR function in Or67d ORNs. This raises the possibility that fru expression in Or67d ORNs is somehow hardwired and not under the control of OR activity.
What, then, is special about Or47b and Ir84a ORNs? fru expression in Or67d ORNs appears unaffected in orco and Or67d mutants. Given the well-established role of Or67d in detection of the male specific pheromone cVA and regulation of courtship, it is possible that fru expression is under a different and more robust regulatory mechanism in these ORNs, one that is independent of OR signaling. Previous studies on the patterns of projection neurons (PNs), which synapse with the fru-positive ORNs have also shown that the organization of Or67d circuit is somehow different from Or47b and Ir84a circuits deeper in the brain. In the lateral horn, a downstream processing center, the axons of the PNs that synapse with Or47b and Ir84a ORNs closely overlap. In fact, they overlap more than any of the other classes of PNs getting input from the 44 ORN classes, while remaining excluded from the Or67d PNs terminals [64]. Aside from these anatomical differences which may indicate possible differences in function, ligand activation of Or and calcium signaling may underlie the Or47b- and Ir84a-dependent maintenance of fru expression. In orco and Or47b mutants, fru expression is lost soon after flies eclose from the pupal case, which might indicate odor-dependent activation of ORs in this process.
Our findings that the expression of Or88a, but not Or67d, in Or47b mutant ORNs can partially rescue the loss of fru expression in Or47b mutants also support the idea Ors are not entirely interchangeable, and that detection of specific fly odors function to maintain fru expression. Recent work has at long last identified the ligands that activate Or47b and Or88a ORNs [17]. While both ORNs have long been known to respond to the complex mixture of compounds found on both male and female cuticles [64], this work establishes that both Or47b and Or88a ORNs respond robustly to a single compound, methyl laurate. On the other hand, Or67d ORNs are activated by cVA, but not by methyl laurate. These responses suggest that the similarity of the ligands detected by Or47b and Or88a ORNs may underlie Or88a’s ability to partially rescue the maintenance of fru expression.
Despite the innate aspects of courtship behaviors and the role of FruM in regulating structure and function of courtship circuits, there is an adaptable aspect to courtship. A particularly interesting example of adaptive courtship behavior is a recently described type of experience-dependent courtship [65]. Males lacking FruM function that have been housed in isolation display very little courtship towards female or male targets. However, when fru mutant males are housed in groups, they acquire the ability to court a wide variety of courtship objects. This learned form of courtship appears to require olfactory input, as no experience-dependent courtship is observed in fru-orco double mutants. It is intriguing to speculate that Or47b, through detection of methyl laurate, may contribute to olfactory experience-dependent modulation of courtship behavior. The receptor-dependent plasticity of fru expression we describe in these ORNs may also be connected to the experience-dependent plasticity of courtship behavior. Recent identification of fru-dependent transcriptional targets showed that fru regulates the expression of many genes that affect different processes in neuronal development in addition to neuronal function [33]. The interconnectedness of fru-positive neuronal circuits [8] and the behavioral function of fru raises the possibility that fru might regulate the flow of information from one fru-positive neuron to another within the circuitry. Thus, loss of fru expression in Or47b or Ir84a mutants might interfere with the integration and communication of Or47b and Ir84a ORNs with the command centers regulating courtship behaviors.
Structural Evolution of ORN Circuits Underlying Sex-Specific Behaviors
Finally, it is tempting to speculate that the processes we describe may operate across Drosophilid and even insect species. Or47b gene sequence and the ligands that activate Or47b and Or88a ORNs are well preserved across Drosophila species [17]. This might also explain the olfactory-experience-dependent courtship learning that occurs in fru M mutants and allows courtship with species other than D. melanogaster [65]. It is plausible to speculate that fru M expression is less robust in ORNs, which have more modulatory effects on courtship, like Or47b and Ir84a. In more innate pathways, driven by the Or67d-dependent detection of cVA, fru expression is independent of OR activity, more robust, and only regulated during development.
It is also important to note the anatomical similarities between the enlarged, fru-positive, dorsolateral portion of the antennal lobe in alh mutants and the macroglomerular complex previously described in Manduca antennal lobes [66]. This enlarged region of the moth antennal lobes is also located in the dorsolateral region and is the region responsive to bombykal, the female sex pheromone [66]. It is possible that in alh mutants, this structure is converted to a more ancient configuration similar to a macroglomerular complex, and that addition of Alh to the gene regulatory networks required for the assembly of the D. melanogaster ORN circuits to further diversify ORN fates from an ancestral state. The anatomical similarities across insect species as well as the conservation of fly-produced mating signals, invite speculation that odor-evoked plasticity is generated by the same mechanisms across insect species, and future studies are needed to investigate whether these mechanisms underlie plasticity of courtship behaviors.
Materials and Methods
Fly Genetics
Fly stocks were maintained on conventional cornmeal-agar-molasses medium at 25°C. The rationale for the genetic screen, from which the alh 1353 mutant was recovered, is described in [24]. Or47b mutants, UAS-Or67d, and UAS-Or88a were kindly provided by Leslie Vosshall, Dean Smith, and John Carlson, respectively. fru GAL4 alleles were obtained from Barry Dickson and Bruce Baker. Other mutant alleles and transgenic lines used in MARCM analyses were obtained from Bloomington and Kyoto Stock Centers. Or47b-LexA transgene was generated by cloning in a 3-kb fragment of Or47b upstream regulatory regions into a LexA plasmid and were injected to obtain transgenics. The expression pattern of the Or47b-LexA transgene was confirmed to match the Or47b-GAL4 line.
Mapping p1353 Mutation
The p 1353 mutation is homozygous lethal. Complementation tests with the deficiency kit on the right arm of third chromosome yielded four deficiencies (Df(3R)Antp17, Df(3R)Antp6, Df (3R)Antp1 and Df(3R)Scx4) that failed to complement the lethality of the p 1353 allele, narrowing the cytological position to a region between 84B–D. Further complementation tests using single gene mutants in the region identified alh j8c8 as allelic to the p 1353 mutation. The alh j8c8 allele is a LacZ enhancer trap line inserted in the first intron of the second transcriptional start site of alh. Phenotypic characterization of Or47b expression in alh j8c8 MARCM clones [30] showed that alh j8c8 phenocopies the p 1353 mutation (Figs 1 and 2). The results from the complementation tests and phenotypic analysis suggest that alh (AF10) is the gene disrupted in p 1353 mutants. alh encodes the D. melanogaster orthologue of the leukemia fusion gene AF10, an epigenetic factor involved in heterochromatin-mediated gene silencing [67–69]. It has two major transcriptional start sites and multiple splice isoforms, which are either “long” isoforms containing the PHD domain, or short isoforms lacking this domain (S3 Fig). We refer to the p 1353 allele as alh 1353 in the paper.
Immunohistochemistry
Staining of adult and pupal brains was performed as previously described [18,24]. Primary antibodies were used in the following dilutions: rabbit α-GFP 1:1,000 (Invitrogen), rat α-Ncad 1:20, rat α-elav 1:100, mouse α-Bruchpilot 1:20 (Developmental Studies Hybridoma Bank), mouse α-CD2 1:1,000 (Serotec). The following secondary antibodies were used: goat α-rabbit-FITC 1:1000, goat α-mouse-Cy3 1:100, goat α-rat-Cy3 1:200, goat α-guinea pig-Cy3 1:500, goat α-rat-Cy5 1:200, goat α-rabbit-Cy5 1:500 (Jackson ImmunoResearch), Alexa 568 goat α-mouse IgG highly cross-adsorbed 1:300 (Invitrogen). Confocal images were taken using an Olympus Fluoview FV1000.
Statistical Analysis
Data was analyzed using JMP.
Total cell counts (Figs 1D–1F and 2B and 2F)
Cells were counted by an experimenter blinded to all genotypes.
A one-way ANOVA was performed for each genotype. Statistical significance was accepted at the p < .01 level.
or47b, F(2,89) = 79.7037, p < .0001.
or88a, F(2,131) = 23.8642, p < .0001.
or65a, F(2,49) = 15.1466, p < .0001.
ir84a, F(2,95) = 31.0864, p < .0001.
or67d, F(1,51) = 0.0786, p = .7804
All simple pairwise comparisons were made using the Tukey’s HSD test. Statistical significance was accepted at the p < .005 level for pairwise comparisons.
Cell cluster ratios (Fig 1G)
Cells were counted by an experimenter blinded to all genotypes.
A one-way ANOVA was performed. Statistical significance was accepted at the p < .01 level.
or47b, F(2,85) = 716.4228, p < .0001.
All simple pairwise comparisons were made using the Tukey’s HSD test. Statistical significance was accepted at the p < .005 level for pairwise comparisons.
Fru-positive cell counts (Fig 6H)
A one-way ANOVA was performed with all fru data: F(10,145) = 68.5683, p < .0001, followed by Tukey-Kramer HSD posthoc tests. Statistical significance was accepted at the p < .005 level for pairwise comparisons.
Fru-positive cell counts (Fig 8D)
A one-way ANOVA was performed with all fru data: F(13,172) = 43.3096, p < .0001, followed by Tukey-Kramer HSD posthoc tests. Statistical significance was accepted at the p < .005 level for pairwise comparisons.
RT-PCR
Antennae from approximately 25 flies were dissected for each genotype, and at least two biological replicates were analyzed for each genotype. RNA was extracted with an RNeasy kit (Qiagen), treated with on-column DNase digestion (Qiagen), and then reverse transcribed into cDNA using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). qPCR was performed using the FastStart Universal SYBR Green Master Mix (Roche) using standard protocol. Expression for each gene was analyzed in triplicate using primers below (Table 1). Ct values were normalized to the expression of actin for each genotype, and these normalized values for experimental and control genotypes were then compared using the delta-delta Ct method. Significance was determined using paired, two-tailed t tests comparing Wt and mutant delta Ct values.
Table 1. qPCR primers.
| Primer Name | Sequence |
|---|---|
| OR47b-qPCR-F | CAAATCTCAGCCTTCTGCGG |
| OR47b-qPCR-R | GATACTGGCACAGCAAACTCA |
| IR84a-qPCR-F | CAGTTGGTCAGGTGTGATGG |
| IR84a-qPCR-R | AAAGTGGATGTTCTGGGTGTG |
| OR65a-qPCR-F | TTGGGATCGATTGTTTGGACC |
| OR65a-qPCR-R | AACCTAGGGCTTTCAACTGGT |
| OR88a-qPCR-F | GGCGGTACCGGAAGTTCTAT |
| OR88a-qPCR-R | GCTGCATTATTTCAGTGAAGTGC |
| fruM-qPCR-F | CCCGCATCCCCTAGGTACAA |
| fruM-qPCR-R | GACTGTTTCGCCCTCGCAGG |
| fruC-qPCR-F | CAAATTTGACCGGCGTGCTAACCT |
| fruC-qPCR-R | AGTCGGAGCGGTAGTTCAGATTGT |
| orco-qPCR-F | GCCTAGATGATTGCTGCATTACT |
| orco-qPCR-R | CGAGGTTGTCATCCTTGCTATT |
| ACT5C-qPCR-F | GGCGCAGAGCAAGCGTGGTA |
| ACT5C-qPCR-R | GGGTGCCACACGCAGCTCAT |
Generating Or67d Mutants Using CRISPR
A targeted deletion of the Or67d coding region was generated using the CRISPR Cas9 system. Guide RNA sequences that flank the Or67d coding region were:
Or67d 5’ chiRNA sense oligo- 5’ CTTCGTGCTTTCGATTATTTTTCC 3’
Or67d 5’ chiRNA antisense oligo- 5’ AAACGGAAAAATAATCGAAAGCAC 3’
Or67d 3’ chiRNA sense oligo- 5’ CTTCGAAGGCCAAGATGGTTGCTG 3’
Or67d 3’ chiRNA antisense oligo- 5’ AAACCAGCAACCATCTTGGCCTTC 3’
The sense and antisense oligo for each chiRNA were annealed and then cloned into the BbsI site of the pU6-BbsI-chiRNA plasmid (Addgene). The Or67d locus was replaced with a 3xP3-DsRed selectable marker and attP phage recombination site by homology-directed repair. Homology arms of approximately 1 kb upstream and downstream of the Cas9 target sites were amplified from genomic DNA from the genotype into which they would be injected, y1 M vas-Cas9.GFP-ZH-2A w1118, using the following primers:
5’ homology arm forward- 5’ TTCCCACCTGCAAATTCGCTTACCCAAAAAGGGCGGCTG 3’
5’ homology arm reverse- 5’ CACACACCTGCCCCCCTACAAAATAATCGAAAGCGCCAC 3’
3’ homology arm forward- 5’ AGGCCTCTGAGGGGTGTTGGGAGGTC 3’
3’ homology arm reverse- 5’ GTGATTCTGCAGCTGCCAACGGGAAGCAATCT 3’
The homology arms were cloned into the 5’ and 3’ multiple cloning sites of the pHD-DsRed-attP plasmid (Addgene) with AarI (5’ arm), StuI (3’ arm) and PstI (3’ arm).
The embryo injection mixture contained the two pU6-BbsI-chiRNA plasmids at 100 ng/mL each and the pHD-DsRed-attP donor plasmid at 500 ng/mL. F1 progeny of the injected embryos were screened for DsRed expression in the adult eye and correct targeting of the Or67d locus was confirmed by PCR and DNA sequencing of the locus.
Supporting Information
(XLSX)
Data shown represents the fold change (normalized by the ΔΔCt method) in the expression of selected genes in the antenna as compared to MARCM control flies. A value of 1 indicates no change from control. All fold change data may be found in the Supporting Information as S1 Data.
(TIF)
Connectivity of ORNs in wild type (top panels) and alh 1353 mutant (bottom panels) antennal lobes.
(TIF)
(A) alh splice isoforms (letters on the right denote the name of the isoform). The Alh j8c8 allele and the alh NP lines used in expression analysis are inserted in the first intron of the short isoforms, and the fifth intron of the long isoforms. (B) Major protein domains found in long and short Alh isoforms.
(TIF)
A) Double-labeling of Or47b and Or88a in WT and alh mutant antennae and antennal lobes. Or88a expression is not expanded to other ORNs in alh mutant antennae. B) Double-labeling of Or88a and fru in WT and alh mutant antennae and antennal lobes. Or88a expression does not overlap with fru expression in wild type or alh mutant antennae. C) Ir76a expression is decreased in alh mutant antennae and antennal lobes.
(TIF)
A) Expression of the long isoform of alh (alh-L) under the control of elav-GAL4 does not change the expression of either Or47b and or Or88a as assayed with direct fusion reporters in adult male antennae. (Expression of the short isoform of alh (alh-S) under the control of elav-GAL4 is lethal). B) Overexpression of the short isoform does not affect Or47b or Or88a expression. Flies were heat-shocked at 37°C for one hour during larval and pupal development at the indicated ages.
(TIF)
AlhGal4 expression in 3L larvae shows a spatial restriction to the center of the eye-antennal disc.
(TIF)
(TIF)
Data shown represents the fold change (normalized by the ΔΔCt method) in the expression of selected genes in the antenna as compared to w1118 control flies. A value of 1 indicates no change from control. Asterisks indicate p < .05 as measured by two-tailed t tests comparing Ct values of each genotype to controls. All fold change data may be found in the Supporting Information as S1 Data.
(TIF)
(A) Degeneration defects in Or47b mutants are rescued by UAS-dsh and UAS-Or88a overexpression using fru GAL4. (B) Expression of Or88a in Or47b mutants (bottom) partially rescues the loss of fru-positive ORN axon terminals (fru-syTGFP) in Or47b target glomerulus (top). (C) Loss of fru expression in or47b mutants is independent of neuronal degeneration. Degenerating Or47b ORN cell bodies are apparent by 14 d (left), which are rescued by overexpression of UAS-dsh (middle). However, despite the rescue of neuronal death, fru is still not expressed in or47b mutant antennae (right). (D) Or67d expression is not able to rescue the degeneration and fru expression defects in Or47b mutants (left panels). Or67d mutants do not have defects in fru expression and they do not degenerate by 14 d (right panels).
(TIF)
Fru expression is not affected by neuronal silencing using fru GAL4 driven UAS-ORK expression (A), or loss of Creb2a (B) and CamkII (C) function using fru GAL4-driven UAS-RNAi expression. Inhibition of PKA (D) and Creb (F) function by fru GAL4-driven UAS-PKA inhibitor and UAS-CREB DN expression, respectively, also does not affect fru expression. Constitutive activation of CamKII also does not have an effect (E).
(TIF)
See Fig 8 in main text for significance. All raw count data may be found in the Supporting Information as S1 Data.
(TIF)
Acknowledgments
We would like to thank Leslie Vosshall, Dean Smith, John Goodwin, Bruce Baker, Thomas Hummel, and Barry Dickson for providing stocks, and Anne West, Rebecca Yang, Hiro Matsunami, Dave McClay, Dan Tracey and members of the Volkan lab for comments on the manuscript.
Abbreviations
- APF
after pupanium formation
- CBP
CREB Binding Protein
- cVA
cis-vaccenyl acetate
- GPCR
G-protein coupled receptor
- HS
Honest Significant Difference
- OR
olfactory receptor
- ORN
olfactory receptor neuron
- PN
projection neuron
- SEM
standard error of the mean
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
PCV is supported by funds from Duke University and the Whitehall Foundation (2013-05-12). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2013;14: 681–92. 10.1038/nrn3567 [DOI] [PubMed] [Google Scholar]
- 2. Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436 10.1038/nature03859 [DOI] [PubMed] [Google Scholar]
- 3. Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121: 785–794. 10.1016/j.cell.2005.04.027 [DOI] [PubMed] [Google Scholar]
- 4. Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, et al. Aberrant Splicing and Altered Spatial Expression Patterns in fruitless Mutants of Drosophila melanogaster. Genetics. 2000;154: 725–745. http://www.genetics.org/content/154/2/725.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anand A, Villella A, Ryner LC, Carlo T, Goodwin SF, Song HJ, et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158: 1569–95. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1461753&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural Circuitry that Governs Drosophila Male Courtship Behavior. Cell. 2005;121: 795–807. 10.1016/j.cell.2005.04.026 [DOI] [PubMed] [Google Scholar]
- 7. Kimura K-I, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438: 229–33. 10.1038/nature04229 [DOI] [PubMed] [Google Scholar]
- 8. Yu JY, Kanai MI, Demir E, Jefferis GSXE, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20: 1602–14. 10.1016/j.cub.2010.08.025 [DOI] [PubMed] [Google Scholar]
- 9. Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446: 542–546. 10.1038/nature05672 [DOI] [PubMed] [Google Scholar]
- 10. Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452: 473–477. 10.1038/nature06808 [DOI] [PubMed] [Google Scholar]
- 11. Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. Nature Publishing Group; 2010;468: 686–690. 10.1038/nature09554 [DOI] [PubMed] [Google Scholar]
- 12. Rodrigues V, Hummel T. Development of the Drosophila olfactory system. Adv Exp Med Biol. 2008;628: 82–101. 10.1007/978-0-387-78261-4_6 [DOI] [PubMed] [Google Scholar]
- 13. Grosjean Y, Rytz R, Farine J-P, Abuin L, Cortot J, Jefferis GSXE, et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478: 236–240. 10.1038/nature10428 [DOI] [PubMed] [Google Scholar]
- 14. Root CM, Masuyama K, Green DS, Enell LE, Nässel DR, Lee C-H, et al. A Presynaptic Gain Control Mechanism Fine-Tunes Olfactory Behavior. Neuron. 2008;59: 311–321. 10.1016/j.neuron.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang L, Han X, Mehren J, Hiroi M, Billeter J-C, Miyamoto T, et al. Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci. 2011;14: 757–762. 10.1038/nn.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakurai A, Koganezawa M, Yasunaga K, Emoto K, Yamamoto D. Select interneuron clusters determine female sexual receptivity in Drosophila. Nat Commun. Nature Publishing Group; 2013;4: 1825 10.1038/ncomms2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dweck HKM, Ebrahim SAM, Thoma M, Mohamed AAM, Keesey IW, Trona F, et al. Pheromones mediating copulation and attraction in Drosophila. Proc Natl Acad Sci U S A. 2015;112: E2829–2835. 10.1073/pnas.1504527112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q, Ha TS, Okuwa S, Wang Y, Wang Q, Millard SS, et al. Combinatorial rules of precursor specification underlying olfactory neuron diversity. Curr Biol. Elsevier Ltd; 2013;23: 2481–2490. 10.1016/j.cub.2013.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, Alenius M. Combinatorial Activation and Repression by Seven Transcription Factors Specify Drosophila Odorant Receptor Expression. PLoS Biol. 2012;10: e1001280 10.1371/journal.pbio.1001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat Neurosci. 2007;10: 153–160. 10.1038/nn1832 [DOI] [PubMed] [Google Scholar]
- 21. Ray A, van Der Goes Van Naters W, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53: 353–69. 10.1016/j.neuron.2006.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Endo K, Karim MR, Taniguchi H, Krejci A, Kinameri E, Siebert M, et al. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat Neurosci. Nature Publishing Group; 2011;15: 224–233. [DOI] [PubMed] [Google Scholar]
- 23. Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A. Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 2012;26: 2483–2498. 10.1101/gad.201665.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GSB, Gunning D, et al. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319: 1256–60. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37: 221–31. 10.1016/S0896-6273(02)01183-2 [DOI] [PubMed] [Google Scholar]
- 26. Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15: 1535–47. 10.1016/j.cub.2005.07.034 [DOI] [PubMed] [Google Scholar]
- 27. Dobritsa A a., Van Der Goes Van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37: 827–841. [DOI] [PubMed] [Google Scholar]
- 28. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43: 703–14. 10.1016/j.neuron.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 29. Jefferis GSXE, Hummel T. Wiring specificity in the olfactory system. Semin Cell Dev Biol. 2006;17: 50–65. 10.1016/j.semcdb.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 30. Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24: 251–4. nprot.2006.320 [pii]\r [DOI] [PubMed] [Google Scholar]
- 31. Li Q, Barish S, Okuwa S, Maciejewski A, Brandt AT, Reinhold D, et al. A Functionally Conserved Gene Regulatory Network Module Governing Olfactory Neuron Diversity. PLoS Genet. 2016;12: e1005780 10.1371/journal.pgen.1005780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, et al. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. J Neurosci. 2011;31: 13357–13375. 10.1523/JNEUROSCI.2360-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neville MC, Nojima T, Ashley E, Parker DJ, Walker J, Southall T, et al. Male-Specific Fruitless Isoforms Target Neurodevelopmental Genes to Specify a Sexually Dimorphic Nervous System. Curr Biol. The Authors; 2014;24: 229–241. 10.1016/j.cub.2013.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chiang A, Priya R, Ramaswami M, Vijayraghavan K, Rodrigues V. Neuronal activity and Wnt signaling act through Gsk3-beta to regulate axonal integrity in mature Drosophila olfactory sensory neurons. Development. 2009;136: 1273–1282. 10.1242/dev.031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional Architecture of Olfactory Ionotropic Glutamate Receptors. Neuron. 2011;69: 44–60. 10.1016/j.neuron.2010.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones WD, Cayirlioglu P, Grunwald Kadow I, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445: 86–90. 10.1038/nature05466 [DOI] [PubMed] [Google Scholar]
- 37. Elmore T, Smith DP. Putative drosophila odor receptor OR43b localizes to dendrites of olfactory neurons. Insect Biochem Mol Biol. 2001;31: 791–798. 10.1016/S0965-1748(00)00184-3 [DOI] [PubMed] [Google Scholar]
- 38. Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452: 1007–U10. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- 39. Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452: 1002–1006. 10.1038/nature06850 [DOI] [PubMed] [Google Scholar]
- 40. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell. Elsevier Inc.; 2009;136: 149–162. 10.1016/j.cell.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4: 240–257. 10.1371/journal.pbio.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wayman G a., Tokumitsu H, Davare M a., Soderling TR. Analysis of CaM-kinase signaling in cells. Cell Calcium. Elsevier Ltd; 2011;50: 1–8. 10.1016/j.ceca.2011.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, et al. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34: 235–244. [DOI] [PubMed] [Google Scholar]
- 44. Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, et al. γCaMKII Shuttles Ca2+/CaM to the Nucleus to Trigger CREB Phosphorylation and Gene Expression. Cell. Elsevier; 2014;159: 281–294. 10.1016/j.cell.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nonaka M, Kim R, Sharry S, Matsushima A, Takemoto-Kimura S, Bito H. Towards a better understanding of cognitive behaviors regulated by gene expression downstream of activity-dependent transcription factors. Neurobiol Learn Mem. Elsevier Inc.; 2014;115: 21–29. 10.1016/j.nlm.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 46. Bito H, Takemoto-Kimura S. Ca(2+)/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium. 2003;34: 425–430. 10.1016/S0143-4160(03)00140-4 [DOI] [PubMed] [Google Scholar]
- 47. Eresh S, Riese J, Jackson DB, Bohmann D, Bienz M. A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J. 1997;16: 2014–22. 10.1093/emboj/16.8.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parry GC, Mackman N. Role of cyclic AMP response element-binding protein in cyclic AMP inhibition of NF-kappaB-mediated transcription. J Immunol. 1997;159: 5450–6. http://www.ncbi.nlm.nih.gov/pubmed/9548485 [PubMed] [Google Scholar]
- 49. Ollivier V, Parry GCN, Cobb RR, de Prost D, Mackman N. Elevated Cyclic AMP Inhibits NF- B-mediated Transcription in Human Monocytic Cells and Endothelial Cells. J Biol Chem. 1996;271: 20828–20835. 10.1074/jbc.271.34.20828 [DOI] [PubMed] [Google Scholar]
- 50. Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, et al. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci U S A. 1997;94: 1074–9. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=19746&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, et al. A CBP Integrator Complex Mediates Transcriptional Activation and AP-1 Inhibition by Nuclear Receptors. Cell. 1996;85: 403–414. 10.1016/S0092-8674(00)81118-6 [DOI] [PubMed] [Google Scholar]
- 52. Topper JN, DiChiara MR, Brown JD, Williams AJ, Falb D, Collins T, et al. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc Natl Acad Sci U S A. 1998;95: 9506–11. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=21368&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science (80-). 2006;314: 657–61. 10.1126/science.1131794 [DOI] [PubMed] [Google Scholar]
- 54. Imai T, Sakano H. Axon-axon interactions in neuronal circuit assembly: Lessons from olfactory map formation. Eur J Neurosci. 2011;34: 1647–1654. 10.1111/j.1460-9568.2011.07817.x [DOI] [PubMed] [Google Scholar]
- 55. Nakashima A, Takeuchi H, Imai T, Saito H, Kiyonari H, Abe T, et al. Agonist-Independent GPCR Activity Regulates Anterior-Posterior Targeting of Olfactory Sensory Neurons. Cell. Elsevier Inc.; 2013;154: 1314–1325. 10.1016/j.cell.2013.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131: 965–973. 10.1242/dev.01074 [DOI] [PubMed] [Google Scholar]
- 57. Joo W, Sweeney L, Liang L, Luo L. Linking cell fate, trajectory choice, and target selection: Genetic analysis of sema-2b in olfactory axon targeting. Neuron. 2013;78: 673–686. 10.1016/j.neuron.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Langen M, Koch M, Yan J, De Geest N, Erfurth M-L, Pfeiffer BD, et al. Mutual inhibition among postmitotic neurons regulates robustness of brain wiring in Drosophila . Elife. 2013;2: 1–21. 10.7554/eLife.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, De Bruijn DRH, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99: 275–281. 10.1182/blood.V99.1.275 [DOI] [PubMed] [Google Scholar]
- 60. Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L Links Histone Methylation to Leukemogenesis. Cell. 2005;121: 167–178. 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- 61. Cecere G, Hoersch S, Jensen MB, Dixit S, Grishok A. The ZFP-1(AF10)/DOT-1 Complex Opposes H2B Ubiquitination to Reduce Pol II Transcription. Mol Cell. Elsevier Inc.; 2013;50: 894–907. 10.1016/j.molcel.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. Nature Publishing Group; 2012;13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alkhori L, Öst A, Alenius M. The corepressor Atrophin specifies odorant receptor expression in Drosophila. FASEB J. 2014;28: 1355–64. 10.1096/fj.13-240325 [DOI] [PubMed] [Google Scholar]
- 64. van der Goes van Naters W, Carlson JR. Receptors and Neurons for Fly Odors in Drosophila. Curr Biol. 2007;17: 606–612. 10.1016/j.cub.2007.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pan Y, Baker BS. Genetic Identification and Separation of Innate and Experience-Dependent Courtship Behaviors in Drosophila. Cell. Elsevier Inc.; 2014;156: 236–248. 10.1016/j.cell.2013.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hansson BS, Christensen TA, Hildebrand JG. Functionally distinct subdivisions of the macroglomerular complex in the antennal lobe of the male spinx moth Manduca sexta. J Comp Neurol. 1991;312: 264–278. [DOI] [PubMed] [Google Scholar]
- 67. Bahri SM, Chia W, Yang X. The Drosophila homolog of human AF10/AF17 leukemia fusion genes (Dalf) encodes a zinc finger/leucine zipper nuclear protein required in the nervous system for maintaining EVE expression and normal growth. Mech Dev. 2001;100: 291–301. 10.1016/S0925-4773(00)00539-6 [DOI] [PubMed] [Google Scholar]
- 68. Perrin L, Bloyer S, Ferraz C, Agrawal N, Sinha P, Dura JM. The leucine zipper motif of the Drosophila AF10 homologue can inhibit PRE-mediated repression: implications for leukemogenic activity of human MLL-AF10 fusions. Mol Cell Biol. 2003;23: 119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Perrin L, Dura JM. Molecular genetics of the Alhambra (Drosophila AF10) complex locus of Drosophila. Mol Genet Genomics. 2004;272: 156–161. 10.1007/s00438-004-1042-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data shown represents the fold change (normalized by the ΔΔCt method) in the expression of selected genes in the antenna as compared to MARCM control flies. A value of 1 indicates no change from control. All fold change data may be found in the Supporting Information as S1 Data.
(TIF)
Connectivity of ORNs in wild type (top panels) and alh 1353 mutant (bottom panels) antennal lobes.
(TIF)
(A) alh splice isoforms (letters on the right denote the name of the isoform). The Alh j8c8 allele and the alh NP lines used in expression analysis are inserted in the first intron of the short isoforms, and the fifth intron of the long isoforms. (B) Major protein domains found in long and short Alh isoforms.
(TIF)
A) Double-labeling of Or47b and Or88a in WT and alh mutant antennae and antennal lobes. Or88a expression is not expanded to other ORNs in alh mutant antennae. B) Double-labeling of Or88a and fru in WT and alh mutant antennae and antennal lobes. Or88a expression does not overlap with fru expression in wild type or alh mutant antennae. C) Ir76a expression is decreased in alh mutant antennae and antennal lobes.
(TIF)
A) Expression of the long isoform of alh (alh-L) under the control of elav-GAL4 does not change the expression of either Or47b and or Or88a as assayed with direct fusion reporters in adult male antennae. (Expression of the short isoform of alh (alh-S) under the control of elav-GAL4 is lethal). B) Overexpression of the short isoform does not affect Or47b or Or88a expression. Flies were heat-shocked at 37°C for one hour during larval and pupal development at the indicated ages.
(TIF)
AlhGal4 expression in 3L larvae shows a spatial restriction to the center of the eye-antennal disc.
(TIF)
(TIF)
Data shown represents the fold change (normalized by the ΔΔCt method) in the expression of selected genes in the antenna as compared to w1118 control flies. A value of 1 indicates no change from control. Asterisks indicate p < .05 as measured by two-tailed t tests comparing Ct values of each genotype to controls. All fold change data may be found in the Supporting Information as S1 Data.
(TIF)
(A) Degeneration defects in Or47b mutants are rescued by UAS-dsh and UAS-Or88a overexpression using fru GAL4. (B) Expression of Or88a in Or47b mutants (bottom) partially rescues the loss of fru-positive ORN axon terminals (fru-syTGFP) in Or47b target glomerulus (top). (C) Loss of fru expression in or47b mutants is independent of neuronal degeneration. Degenerating Or47b ORN cell bodies are apparent by 14 d (left), which are rescued by overexpression of UAS-dsh (middle). However, despite the rescue of neuronal death, fru is still not expressed in or47b mutant antennae (right). (D) Or67d expression is not able to rescue the degeneration and fru expression defects in Or47b mutants (left panels). Or67d mutants do not have defects in fru expression and they do not degenerate by 14 d (right panels).
(TIF)
Fru expression is not affected by neuronal silencing using fru GAL4 driven UAS-ORK expression (A), or loss of Creb2a (B) and CamkII (C) function using fru GAL4-driven UAS-RNAi expression. Inhibition of PKA (D) and Creb (F) function by fru GAL4-driven UAS-PKA inhibitor and UAS-CREB DN expression, respectively, also does not affect fru expression. Constitutive activation of CamKII also does not have an effect (E).
(TIF)
See Fig 8 in main text for significance. All raw count data may be found in the Supporting Information as S1 Data.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.