Abstract
We developed sugarcane plants with improved resistance to the sugarcane borer, Diatraea saccharalis (F). An expression vector pGcry1Ac0229, harboring the cry1Ac gene and the selectable marker gene, bar, was constructed. This construct was introduced into the sugarcane cultivar FN15 by particle bombardment. Transformed plantlets were identified after selection with Phosphinothricin (PPT) and Basta. Plantlets were then screened by PCR based on the presence of cry1Ac and 14 cry1Ac positive plantlets were identified. Real-time quantitative PCR (RT-qPCR) revealed that the copy number of cry1Ac gene in the transgenic lines varied from 1 to 148. ELISA analysis showed that Cry1Ac protein levels in 7 transgenic lines ranged from 0.85 μg/FWg to 70.92 μg/FWg in leaves and 0.04 μg/FWg to 7.22 μg/FWg in stems, and negatively correlated to the rate of insect damage that ranged from 36.67% to 13.33%, respectively. Agronomic traits of six transgenic sugarcane lines with medium copy numbers were similar to the non-transgenic parental line. However, phenotype was poor in lines with high or low copy numbers. Compared to the non-transgenic control plants, all transgenic lines with medium copy numbers had relatively equal or lower sucrose yield and significantly improved sugarcane borer resistance, which lowered susceptibility to damage by insects. This suggests that the transgenic sugarcane lines harboring medium copy numbers of the cry1Ac gene may have significantly higher resistance to sugarcane borer but the sugarcane yield in these lines is similar to the non-transgenic control thus making them superior to the control lines.
Introduction
Sugarcane (Saccharum officinarum L.) is an important sugar crop that is widely cultivated in the tropical and subtropical regions. It provides about 80% of the world sugar [1] and more than 92% of sugar in China [2]. In addition, sugarcane is also a major raw material for ethanol production in countries such as USA and Brazil, and accounts for nearly 90% of the feedstock used in ethanol production [3]. Equally important is the sugarcane borer, Diatraea saccharalis (F.), which is one of the most important lepidopteran pests attacking sugarcane plants and causing more than 10% loss in sugarcane yield worldwide [4]. Damage occurs during the entire crop season and in different tissues resulting in a decreased emergence rate, increased dead heart rate of seedlings, increased stem wind-breakage rate in the adult-plant stage and a reduced sucrose level in the harvest stage. Increasing plant resistance to this insect pest is an effective method to reduce damage by the sugarcane borer. This strategy is also economical and has minimal environmental impact [5,6]. Sugarcane cultivars are complex polyploids with more than 120 chromosomes but without effective insect resistance genes in the sugarcane gene pool [7,8]. This presents a challenge in creating insect-resistant sugarcane cultivars by conventional cross-breeding.
Cry1Ac gene, one of the cry1 genes isolated from Bacillus thuringiensis (Bt), codes for an insecticidal crystal protein, which kills lepidopterans upon entry into the alimentary tract [9]. The first successful insect-resistant transgenic tobacco contained the cry1A(b) gene introduced through the Agrobacterium-mediated transformation [10]. These transgenic tobacco plants were resistant to Manduca sexta. Since then, a number of transgenic plants resistant to lepidopterans have been created including tomato [11], cotton [12], potato [13], corn [14], and rice [15]. Transgenic sugarcane was created by firstly introducing the cry1Ab gene [7] followed by introducing multiple insecticidal genes such as cry1A(b) [16,17], Galanthus nivalis agglutinin (GNA) [18,19], soybean proteinase inhibitors [20], cry1Aa3 [21] and cry1Ac [22,23]. Transgenic sugarcane, resistant to the sugarcane borer, was also generated by transferring the cry1Ab gene driven by the CaMV35S promoter into sugarcane [7]. These transgenic plants resisted insect damage although cry1A(b) expression was low. Sugarcane cultivars ROC16 and YT79-177 were created by particle bombardment of a modified cry1Ac gene, and about 62% of the transgenic plants were resistant to damage by the stem borer in both greenhouse and field trials [23].
In this study, our goal was to improve sugarcane borer resistance in FN15, a newly released sugarcane cultivar with high sucrose content. We investigated the correlation between resistance to sugarcane borers and the copy number of the cry1Ac gene. We determined the level of Cry1Ac protein and investigated how it affected the yield traits and sucrose content. To achieve these goals, the plant expression vector, pGcry1Ac0229 was constructed and introduced into sugarcane by particle bombardment followed by screening and analysis of the transgenic sugarcane lines. It is anticipated that the findings of this study will allow breeding of sugarcanes that are resistant to stem borers.
Materials and Methods
Materials
The cassette containing the cry1Ac gene, 35s promoter and nos terminator in the cry1AcPRD vector was provided by Prof. Illimar Altosaar, University of Ottawa, Canada. The plant expression vector, pGreenⅡ0229, was obtained from the John Innes Centre in England. The sugarcane cultivar, FN15 used for transformation was provided by the Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture, Fujian Agriculture and Forestry University, China. All chemicals used were analytical grade.
Vector construction
The cassette containing the 35s promoter, cry1Ac gene and nos terminator was digested from the cry1AcPRD vector using restriction enzymes, EcoR I and Hind III. The expression vector, pGreenⅡ0229 was also digested with EcoR I and Hind III and linked to the cassette with T4-DNA ligase to create a new plant expression vector termed as pGcry1Ac0229 containing the target gene, cry1Ac (S1 Fig).
Transformation and screening
Transformation was performed based on the PDS 1000/He particle gun operating manual (Bio-Rad, Calif., USA). The embryonic calli of the sugarcane cultivar FN15 were derived from transverse segments of young leaf roll region with the apical meristem. They were cut into 1–2 mm thick discs and cultured in MS medium [24] containing 3.0 mg/L dichlorophenoxyacetic acid (2,4-D) in dark for 2–4 weeks, and used for transformation via bombardment. For each bombardment, 15–20 pieces of embryonic calli with a diameter of 2–3 mm were placed in the centre of a culture dish with MS-based induction medium containing 0.2 mol/L sorbitol and 0.2 mol/L mannitol. Then, they were cultured in dark at 28°C for 4–8 h prior to bombardment. Several culture plates were used to bombard the cry1Ac genes while only two plates were used as controls. One plate served as the control for bombardment with tungsten particles without plasmid DNA, and the other plate was the no bombardment control. The plasmid, pGcry1Ac0229 was coated on tungsten particles (Bio-Rad, 0.7) before bombardment and approximately 1.0 μg of DNA was used for each bombardment. After the procedure, the calli were spread over the surface of the medium and cultured for 1 d at 28°C in dark. Then, the calli were transferred to the MS induction medium containing 2.0 mg/L 2,4-D and 0.75 mg/L phosphinothricin (PPT) and cultured in dark for 2 weeks. For regeneration culture, the calli were transferred to a MS regeneration medium containing 1.0 mg/L 6-Benzyladenine (6-BA), 1.0 mg/L Kinetin (KT) and 0.75 mg/L PPT, and cultured for several cycles of alternating light and dark conditions for 2 weeks at 28°C until sugarcane plantlets regenerated. The plantlets were then transferred to a ½MS rooting medium containing 3.0 mg/L 1-naphthlcetic acid (NAA) until roots developed. Viable putative transformants were transferred to soil in the greenhouse. When the putative transformants grew 5–8 cm in height, they were sprayed with a 3‰ (V/V) Basta (active ingredient phosphinothricin /glufosinate ammonium) solution for preliminary screening.
DNA extraction and primer design
Total genomic DNA was extracted from fresh young leaves of the Basta-resistant plantlets using a CTAB method described previously [25]. The negative control was the FN15 non-transgenic sugarcane without bombardment. All DNA samples were stored at -20°C after measuring the DNA concentration by a UV absorption method. DNA purity was analyzed by the A260/A280 ratio, while the integrity was evaluated by electrophoresis. Primer pairs for polymerase chain reaction (PCR) and real-time quantitative PCR (RT-qPCR) were designed by Primer Premier 5 software. Based on the sequence of the cry1Ac gene, a pair of primers was designed for the amplification of a 211 bp fragment (forward: TATCGTTGCTCAACTAGGTCAGGGTGTC, and reverse: CATTGTTGTTCTGTGGTGGGATTTCGTC) for PCR, and another pair to amplify a 74 bp fragment by RT-qPCR as follows: 1ACQF: 5’-ACCGGTTACACTCCCATCGA-3’, 1ACQR: 5’-CCAGCACCTGGCACGAA-3’, 1ACProbe: 5’- (FAM) TCTCCTTGTCCTTGACACAGTTTCTGCTCA-3’(TAMRA).
PCR analysis
PCR was performed using Eppendorf 5331 (Eppendorf Company, Hamburg, Germany). Each reaction volume was 25 μL containing 2.5 μL PCR buffer (10×), 2.0 μL dNTP mix, 1.0 μL (100 ng μL/L) template DNA, 1.0 μL (10 μmol/L) each of the forward and reverse primers, 0.125 U of Taq DNA polymerase (Takara, Dalian, China), and ddH2O. Following the initial polymerase activation at 95°C for 5 min, 30 cycles were performed at 95°C for 30 s, 56°C for 30 s and 72°C for 30 s with a final extension at 72°C for 10 min. The amplified products were detected on a 1.5% (w/v) agarose gel.
Copy number calculation in transgenic sugarcane lines by RT-qPCR analysis
RT-qPCR was performed using an ABI PRISM 7500 Sequence Detection System (Foster, USA). The reaction volume was 25 μL and contained 12.5 μL of Fast Start Universal Probe Master Mix, 1.0 μL of diluted genomic DNA (25 ng), 1.0 μL (10 μmol/L) each of the forward and reverse primers and sterile ddH2O. The following reaction conditions were used; 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, 60°C for 1 min; 1 cycle of 95°C for 15 s, 60°C for 15 s, 95°Cfor 15 s. Each sample had three replications. In parallel, the pGcry1Ac0229 plasmid DNA was serially diluted to 108, 107, 106, 105, 104, 103, 102, and 101 copies/μL and subjected to RT-qPCR with each reaction having three replications. The plasmid copy number was obtained using the following formula: plasmid copy number (copies/μL) = 6.02 × 1023 (copies/mol) × plasmid concentration (g/μL)/ plasmid molecular weight (g/mol)/660 [26]. Each copy number corresponding to the Ct value was obtained after the reaction. Then, using the lg copies in the X axis, and the Ct value in the Y axis, the standard curve was established to obtain the linear equation Y = kX+b. The total copy number (10Xt) was calculated by relating the Ct value (Yt) and linear equation for the cry1Ac transgenic lines in each reaction. Then, the single cell copy number of each sample was calculated using the following formula: Copies/genome = 10Xt/[25 n g×10−9×6.02×1023/ (10,000 (M bp)×106×660)] [27].
Southern blot analysis
The PCR-positive transgenic sugarcane lines were analyzed by Southern blotting. Primers (forward: TATCGTTGCTCAACTAGGTCAGGGTGTC and reverse: CATTGTTGTTCTGTGGTGGGATTTCGTC) were used to amplify the cry1Ac fragment to prepare the labeled probe using the PCR DIG Probe Synthesis Kit (Roche, Switzerland). About 40 μg of genomic DNA was extracted from fresh young leaves of every transgenic sugarcane line and non-transgenic control, and was completely digested with the restriction enzyme Hind III, which is present once in pGcry1Ac0229. Southern blotting was performed based on the DIG DNA Labeling and Detection Kit manual.
ELISA analysis
ELISA was used to detect Cry1Ac protein expression in transgenic sugarcane lines according to the manufacturer’s instructions in the Qualiplat Kit. Total protein from fresh mature sugarcane leaves and stems was used in ELISA. The cry1Ac crystal protein standard was purchased from Envirologix (Portland, USA) and serially diluted to 5.0 ppb, 2.5 ppb, 1.25 ppb, 0.625 ppb, 0.3125 ppb and 0.15625 ppb with 1×phosphate buffer saline (pH 7.4). The non-transgenic sugarcane line without particle bombardment was used as the negative control. Sterile ddH2O served as the blank control. Each sample had three replications. OD450 value of the standard was obtained after the reaction followed by plotting the concentration of standard in the X axis and the OD450 value in the Y axis to establish the standard curve and form the linear equation y = kx + b. Cry1Ac protein expression (Xt) in the transgenic sugarcane lines was calculated by relating the OD450 value (Yt) to the standard curve.
Trial design and investigation of phenotype traits in sugarcane lines
In isolated field trials, 14 different transgenic cry1Ac sugarcane lines and the non-transgenic controls were evaluated during 2012 to 2013 using a randomized complete block design with three replications. Each block had three 8 m-long rows with 1.3 m space between rows, covering an area of 31.2 m2. The sugarcane crops were harvested in January the following year. Field management was slightly better than commercial cultivation. The fertilizers applied were: nitrogen (N) at 345 kg/ha, phosphorus (P2O5) at 240 kg/ha and potassium (K2O) at 360 kg/ha. At mature stage, yield estimates were obtained from the stalks in the middle rows. The estimates included height, stem diameter, and brix in ten consecutive principal stalks from 10 plants. The number of millable stalks in 5 m long rows was also counted in each block. The cane yield per unit area was calculated based on the area, and number and weight of stalks. Sucrose yield was calculated based on the average sucrose content and cane yield using equations (1): Sucrose content (%) = brix × 1.0825–7.703; and (2): Sucrose yield (t/ha) = Cane yield (t/ha) × Sucrose content (%)
Resistance of transgenic sugarcane to sugarcane borer
Stem segments from transgenic and non-transgenic lines were pre-germinated in an incubator. Upon reaching the 4–5 leaf stage, two 8 d old larvae were placed on each seedling. A total of 8–10 seedlings were infested for each line and maintained in isolation using fly netting. The assay was repeated 3 times. In the field trials, the percentage of stalks damaged by the sugarcane borer and damage symptoms in the attacked leaves and internodes were recorded along with agronomic traits.
Results
Construction of pGcry1Ac0229
Successful construction of the recombinant plasmid, pGcry1Ac0229, was first verified by a single digestion with Hind III and a double digestion with Hind III and EcoR I, which yielded the expected 7,238 bp, and 4,436 bp and 2,802 bp bands, respectively (S2 Fig). Further sequencing demonstrated that this recombinant plasmid had the expected expression cassette inframe (results not shown).
Particle bombardment and resistance screening
The pGcry1Ac0229 plasmid DNA was used to prepare microprojectiles for bombardment transformation, and the embryonic calli of the sugarcane cultivar FN15 were used as receptors. Based on previously determined screening concentrations of PPT (0.75 mg/L), cultures were screened and the putative transformants that survived the stress of PPT were obtained. Wild type cells without PPT resistance gradually turned brown and then died (Fig 1). Regenerated plantlets were transferred to rooting medium without PPT and the total number of survivors was 189. These plantlets were planted in soil in 72-cave trays and the second round of screening in vitro was conducted by spraying a solution containing 3.0 ‰ Basta on the plantlets. After 15–20 d, most of them gradually became yellow, wilted, and dried (S3 Fig). The final total of surviving plants was 26.
Fig 1. Partially regenerated plantlets with PPT resistance in differentiation culture.

PCR identification of resistant sugarcane plants
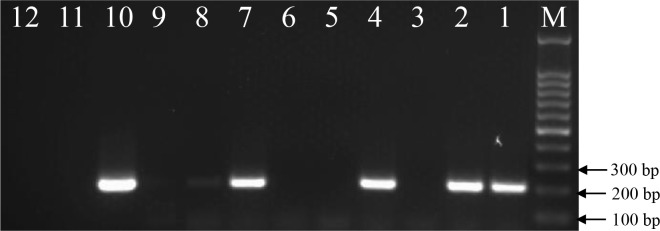
PCR validation was performed on the Basta resistant regenerated plants. A total of 14 independent plants amplified the expected size DNA consistent with the positive control (pGcry1Ac0229), while the negative non-transgenic line and the blank control did not amplify any band (Fig 2). Further sequencing showed that this band was precisely the 211 bp partial sequence of cry1Ac, suggesting that the cry1Ac gene was successfully inserted into the sugarcane genome. A total of 14 among the 26 Basta-resistant regenerated plants tested positive by PCR.
Fig 2. Electrophoresis of PCR amplified products of the candidate cry1Ac transgenic sugarcane plants.
M, DNA Marker; 1–9, Basta-resistant plants; 10, Positive control (plasmid pGcry1Ac0229); 11, Negative control (non-transgenic sugarcane without bombardment); 12, Blank control.
Copy number calculation in transgenic sugarcane plants by RT-qPCR
Fourteen Basta-resistant and cry1Ac gene positive transgenic sugarcane plants were further tested using RT-qPCR to determine cry1Ac gene copy number. A standard curve for quantitative detection of cry1Ac gene based on RT-qPCR was constructed (S4 Fig). The slope was Y = -3.140X+43.622, R2 = 0.998, where Y represents Ct value and X represents the log of starting template copy number. The Ct (18~40) value correlated well with the copy number of the starting template (101~108) (R2 = 0.998). According to the linear equation above, the total copy number (10Xt) of the target gene in samples was calculated by relating the Ct value (Yt). The copy number of cry1Ac gene inserted into a single cell was calculated according to the formula (Table 1):
Table 1. Copy number estimation in different transgenic sugarcane lines with a cry1Ac gene.
| Sugarcane lines | C t Ⅰ | C t Ⅱ | C t Ⅲ | C t mean | Copy number |
|---|---|---|---|---|---|
| T-1 | 30.82 | 30.95 | 30.76 | 30.85±0.10 | 5.13 |
| T-2 | 27.75 | 27.61 | 27.87 | 27.74±0.13 | 49.92 |
| T-3 | 27.35 | 27.35 | 27.64 | 27.45±0.17 | 62.08 |
| T-4 | 32.10 | 32.15 | 31.91 | 32.06±0.13 | 2.11 |
| T-5 | 27.81 | 27.83 | 28.03 | 27.89±0.12 | 44.78 |
| T-6 | 33.12 | 32.73 | 33.02 | 32.96±0.20 | 1.09 |
| T-7 | 31.74 | 32.10 | 32.06 | 31.96±0.20 | 2.26 |
| T-8 | 28.82 | 28.82 | 28.70 | 28.78±0.07 | 23.29 |
| T-9 | 26.62 | 26.51 | 26.66 | 26.60±0.08 | 115.57 |
| T-10 | 26.93 | 27.06 | 27.12 | 27.04±0.10 | 83.68 |
| T-11 | 28.80 | 29.13 | 29.52 | 29.15±0.36 | 17.78 |
| T-12 | 26.72 | 26.66 | 26.83 | 26.73±0.09 | 104.56 |
| T-13 | 26.12 | 26.37 | 26.29 | 26.26±0.13 | 147.92 |
| T-14 | 27.29 | 27.38 | 27.68 | 27.45±0.21 | 61.86 |
| CK FN15 | 34.84 | 34.65 | 34.89 | 34.79±0.13 | 0.28 |
The Ct values of the three replicates had small standard deviations in the different transgenic lines (Table 1). The copy number in the 14 transgenic sugarcane plants ranged from 1 to 148, and they were divided into three groups: high copy number (>80), medium copy number (10~80) and low copy number (<10).
Copy number of transgenic sugarcane lines detected by Southern blot analysis
Based on the copy number determined by RT-qPCR analysis, six independent transgenic lines, including two high (T-9, T-12), two medium (T-2, T-11) and two low (T-1, T-4) copy number lines, were selected for copy number detection by Southern blot analysis. There was no hybridization signal in the two transgenic lines with low copy number and in the non-transgenic control; 3–4 copies were observed in the two medium copy number lines; and 5–6 copies were observed in the two high copy number lines.
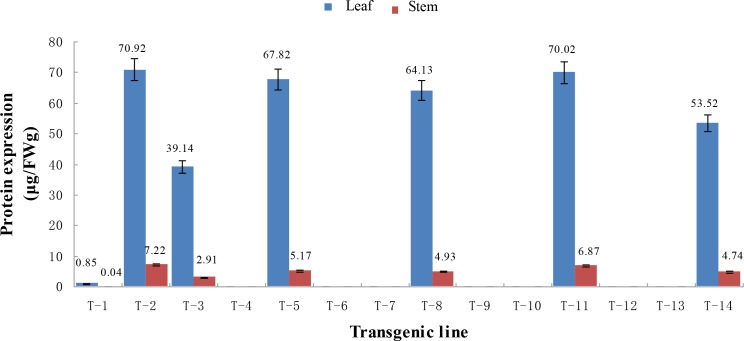
ELISA analysis of Cry1Ac protein content in transgenic sugarcane lines
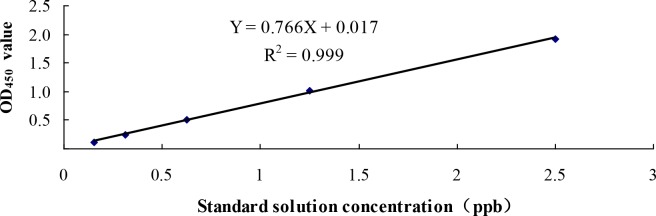
Double-antibody sandwich ELISA was performed to determine the Cry1Ac protein content in mature stage leaves and stems of 14 transgenic sugarcane lines. Standard Bt samples were used to construct a standard curve for quantitative protein detection (Fig 3). The slope was Y = 0.766X+0.017, R2 = 0.999, where Y is absorbance at OD450, and X is the concentration of standard Bt Cry1Ac protein samples. Significant correlation was found between the absorbance and the concentration of the standard protein (R2 = 0.999). Thus, the protein expression of the tested samples was measured based on the linear equation (Fig 4). The results showed that Cry1Ac protein expression was only detected in leaves and stems of seven transgenic lines. Expression in leaves was variable and ranged from a minimum of 0.85μg/FWg in T-1 to a maximum of 70.92 μg/FWg in T-2. In stems, the expression was also variable and ranged from a minimum of 0.04 μg/FWg in T-1 to a maximum of 7.22 μg/FWg in T-2. It should be noted that although there was no Cry1Ac protein expression in the remaining seven transgenic lines, the expression of cry1Ac gene was detected in all these lines.
Fig 3. The standard curve of Cry1Ac protein constructed by ELISA.
Fig 4. Cry1Ac protein expression in the leaves and stems of 14 different transgenic sugarcane lines detected by ELISA.
Agronomic and industrial traits in transgenic sugarcane lines
Agronomic and industrial traits of 14 transgenic lines and the control sugarcane cultivar FN15 were investigated in mature stage plants. As shown in Table 2, univariate statistical analysis indicated that stalk height in transgenic lines T-2, T-3, T-5, T-8, T-11, and T-14 was similar to the control line and no significant difference was observed. Although in some cases slightly higher or lower heights were observed, eight lines had significantly lower heights than the control. Stalk diameter of transgenic lines T-2 and T-3 was not significantly different from the control while in the remaining 12 lines it was significantly less than the control. For brix, only line T-4 was not significantly different compared to the control but the remaining 13 lines were significantly different. Lines T-1, T-6, T-7, T-9, T-10, T-12, and T-13 had significantly higher brix and lines T-2, T-3, T-5, T-8, T-11, and T-14 significantly lower brix than the control. With respect to millable stalks in a block, T-2, T-3, T-5, T-8, T-11, and T-14 had higher and T-1 had lower numbers than the control although there was no statistical significance among these values when compared to the control. The theoretical sucrose yield was calculated based on the height, stalk diameter, brix, and millable stalks in a block. These values were marginally lower in lines T-3, T-8, and T-11 but not significantly different from the control, while the remaining lines had significantly lower values than the control.
Table 2. Agronomic characteristics, industrial traits and the stalks borer damage percentage in transgenic sugarcane lines and the non-transgenic control.
| Transgenic line | H(cm) | D (cm) | Brix (%) | SNB | TSY (kg/block) | TSY (t/ha) | SDR (%) |
|---|---|---|---|---|---|---|---|
| T-1 | 170.67±5.21 b | 2.50±0.07 ef | 22.73±0.06 ab | 184.62±4.62 bc | 26.21±2.91 d | 8.40±0.93 d | 36.67±5.77 bcd |
| T-2 | 207.42±2.16 a | 3.05±0.06 abc | 19.43±0.15 f | 227.69±2.66 a | 46.1±3.45 bc | 14.78±1.11 bc | 13.33±5.77 e |
| T-3 | 213.50±3.88 a | 3.20±0.12 ab | 19.61±0.30 ef | 221.54±7.99 a | 51.65±5.19 ab | 16.56±1.66 ab | 21.67±2.89 cde |
| T-4 | 172.89±0.51 b | 2.41±0.06 f | 21.11±0.14 c | 152.31±9.23 de | 18.1±0.54 de | 5.80±0.17 de | 43.33±5.77 bc |
| T-5 | 207.92±4.42 a | 2.87±0.07 cd | 19.49±0.22 ef | 224.62±7.05 a | 40.52±3.41 c | 12.99±1.09 c | 15.00±5.00 de |
| T-6 | 181.11±2.01 b | 2.37±0.03 f | 22.34±0.16 ab | 166.15±4.62 cd | 21.82±0.37 de | 7.00±0.12 de | 46.67±15.28 b |
| T-7 | 172.89±1.26 b | 2.56±0.04 ef | 22.82±0.25 a | 156.92±9.23 de | 23.74±2.16 de | 7.61±0.69 de | 56.67±5.77 ab |
| T-8 | 203.92±1.66 a | 3.16±0.04 ab | 19.92±0.24 def | 226.15±4.62 a | 50.11±2.63 ab | 16.06±0.84 ab | 16.67±5.77 de |
| T-9 | 150.00±2.19 c | 2.65±0.04 e | 22.33±0.21 ab | 133.85±12.21 ef | 18.2±1.72 de | 5.83±0.55 de | 43.33±5.77 bc |
| T-10 | 135.67±4.58 d | 2.50±0.01 ef | 22.70±0.18 ab | 141.54±9.61 ef | 15.88±1.21 e | 5.09±0.39 e | 53.33±11.55 ab |
| T-11 | 203.50±4.27 a | 3.00±0.10 bc | 20.24±0.10 d | 229.23±7.05 a | 46.68±2.2 abc | 14.96±0.71 abc | 14.33±1.15 e |
| T-12 | 145.00±1.20 cd | 2.66±0.05 de | 22.37±0.23 ab | 153.85±9.61 de | 20.42±1.35 de | 6.54±0.43 de | 53.33±5.77 ab |
| T-13 | 143.67±3.48 cd | 2.54±0.06 ef | 22.19±0.10 b | 124.62±9.23 f | 14.86±1.45 e | 4.76±0.46 e | 46.67±11.55 b |
| T-14 | 211.25±10.13 a | 2.92±0.08 c | 20.09±0.29 de | 220.00±2.66 a | 43.67±1.7 bc | 14.00±0.55 bc | 18.33±5.77 de |
| CK FN15 | 209.92±4.14 a | 3.25±0.14 a | 21.47±0.25 c | 206.15±9.61 ab | 55.86±7.64 a | 17.91±2.45 a | 72.67±2.52 a |
Notes: (1) H: stalk height; D: stalk diameter; SNB: stalk number per block; TSY: theoretical sucrose yield; SDP: stalks borer damage percentage. (2) Data followed by different letters indicate significant difference at 0.05 level (Duncan Test); (3) The area of sub domain is 31.2 m2
Transgenic sugarcane lines are resistant to sugarcane borer
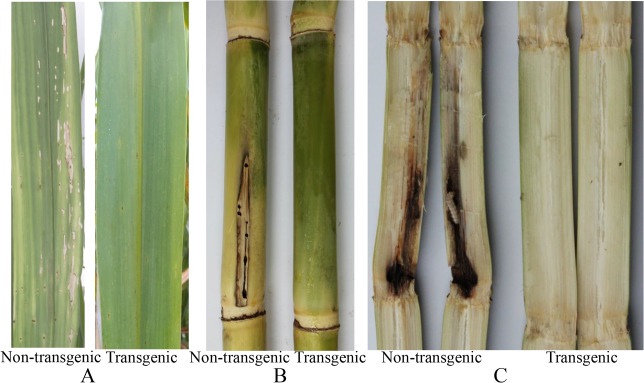
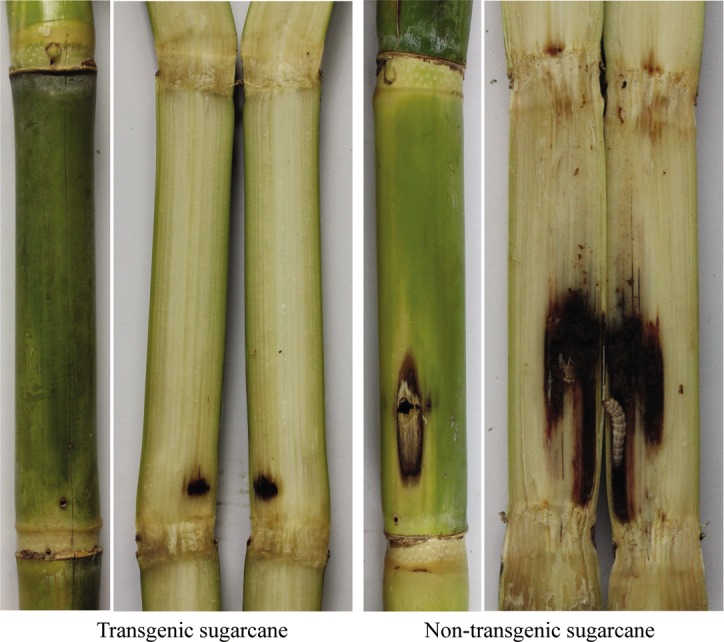
The transgenic sugarcane lines were generally more resistant to sugarcane borer compared to the non-transgenic lines. In the seedling stage, 7 d after inoculation, the typical effects of borer attack leading to dead heart were seen on the non-transgenic plants. The same effects were observed after 10 d in several transgenic lines. Plantlets of non-transgenic sugarcane withered slowly 10 d after inoculation. At 15 d, only transgenic lines continued to grow normally (Fig 5B), while non-transgenics withered or died (Fig 5A). In the transgenic lines, only 10%-20% of borers survived but these appeared weak and were small (Fig 5D). In contrast, the borers in non-transgenic control plants had significantly higher survivorship and were larger (Fig 5C). In the field trials, the transgenic lines were also resistant to the sugarcane borer damage to leaves and stems compared to the non-transgenic control (Fig 6). The borer damage rate in all transgenic lines was lower than the control and differences were significant except for lines T-7, T-10, and T-12. There was no significant difference between the lines T-2, T-3, T-5, T-8, T-11, and T-14, all of which had higher protein expression and a medium cry1Ac copy number (Table 2). Although the transgenic sugarcane lines were still attacked by stem borers, the degree of damage was clearly less than the damage done to the non-transgenic control line (Fig 7).
Fig 5. Bioassay in the seedling stage.

A: Symptoms of non-transgenic sugarcane plantlets challenged with sugarcane borers; B: Symptoms of transgenic sugarcane plantlets challenged with sugarcane borers; C: Sugarcane borer feeding with non-transgenic sugarcane plantlets; D: Sugarcane borer feeding with transgenic sugarcane plantlets.
Fig 6. Typical symptoms caused by natural stem borer infestation in the field.
A: Leaf symptoms; B: Stem symptoms (aspect); C: Stem symptoms (inside stalk).
Fig 7. Damage comparison between transgenic and non-transgenic sugarcane stalks.
Discussion
We constructed the plant expression vector pGcry1Ac0229 containing two expression cassettes, d35s-AMV-cry1Ac-nos, and Nos-bar-nos. The target gene cry1Ac was placed under the control of the constitutively expressed double promoter 35s and the enhancer AMV. These two regulatory sequences significantly increase the expression of exogenous genes in transgenic plants [28]. Consistent with this, the Cry1Ac protein content in the leaves of transgenic sugarcane lines ranged from 0 μg/FWg to 70.92 μg/FWg. A previous study reported that the expression of cry1A(b) regulated by CaMV35S yielded only a maximum of 27.23 ng/mg (i.e. 27.23 μg/FWg) cry1A(b) protein in transgenic sugarcane leaves [7]. This suggests that the double promoter 35s and the enhancer AMV can significantly enhance the expression of target genes.
The Bar gene is a high efficiency selectable marker in plant genetic transformation. It has been widely used in many plant species [29,30] including sugarcane [17,31,32] due to its advantage in screening putative transformants. Sugarcane is more sensitive to PPT than G418 and hygromycin (Hyg) [33]. The screening concentrations of PPT for the sugarcane cultivar FN81-745 and Badila in tissue culture were 0.75–1.0 mg/L PPT, 30 mg/L G418 and 30 mg/L hygromycin [33]. The screening concentration of PPT in the callus differentiation stage of the sugarcane cultivar FN95-1702 (i.e. FN15) was 0.5 mg/L [34]. Putative transformants surviving in tissue culture could be re-screened by spray treatment with Basta herbicide at the seedling stage after soil transplantation. This procedure allows rapid and cost effective identification of transformants because the leaves of transgenic plants will remain green but leaves from non-transgenic leaves turn yellow, wither, and die. In this study, we used 0.75 mg/L PPT to screen the sugarcane cultivar FN15 in callus subcultures and differentiation culture stages. Screening was not performed at the rooting stage. This protocol allowed rapid and normal rooting of putative transformants. It also provided a relatively relaxed selection procedure due to fast growth thus shortening the screening time by 4–6 weeks at the tissue culture stage. A more stringent selection was performed in plantlets on seedling growth stage using 3‰ (V/V) Basta, which resulted in efficient identification of cry1Ac transgenics. Additionally, the bar gene, coding the bialaphos/phosphinothricin resistance protein, is resistant to PPT, the active ingredient in Basta or Glufosinate and Bialaphos in Herbiace [35]. Transgenic plants containing the bar gene are resistant to Basta or Glufosinate and Herbiace. This is also an important agronomic character for sugarcane with respect to weed management due to the slow early stage growth of sugarcane. Thus, transgenic sugarcanes with the bar gene have great potential for commercial use.
It is important to determine the copy number of transgenes in transgenic lines because copy number can affect genetic stability and expression level. The traditional method to estimate copy number of exogenous genes in transgenic plants is by Southern blot analysis. Recently, RT-qPCR technology has been widely used to determine the copy number of exogenous genes [36,37] but the results can be inconsistent with Southern blot analysis. Using quantitative fluorescence PCR, Weng et al. [38] observed that the copy number of gus and npt II genes in 7 out of 10 transgenic events in Brassica napus was consistent with Southern blot analysis. Cheng et al. [39] tested the exogenous gene, gfp in tetraploid upland cotton transgenic lines and showed that the copy number detected by quantitative fluorescence PCR was greater than that observed by Southern blot analysis. Yang et al. [40] analyzed the copy number of npt II in transgenic cotton and found that the copy number detected by Southern blot analysis was less than or equal to that estimated by qPCR analysis. In this study, we established a standard curve to estimate the copy number of cry1Ac gene by SYBR Green quantitative fluorescence PCR. The slope of the reaction was -3.140 and correlation coefficient was 0.998 suggesting a strong correlation between the Ct value and the copy number of the starting template. We then determined the copy number in transgenic lines and found that they ranged from 1 to 148. Because the chromosome ploidy of modern sugarcane is complex and remains undefined [41], the actual ploidy in modern sugarcane varieties including the test cultivar FN15 used here is unclear. Transgenic sugarcane lines were obtained by particle bombardment, which normally introduces multiple copies of exogenous gene expression cassettes and integrates them into the transgenic plant genome [42]. To verify the copy number of the transgenic sugarcane lines, six different transgenic lines with high (>80), medium (10~80) or low (<10) copy numbers of the cry1Ac gene were selected for Southern blotting. The copy numbers estimated by RT-qPCR were higher than those estimated by Southern blotting, which was consistent with previous studies on transgenic rice [43] and cotton [39]. However, the trend in copy number estimation by the two methods was consistent. Inconsistency in copy number estimation by the two methods may result from the following two factors. First, multi-copy tandem integrations in transgenic lines, where the estimated copy number is based on southern blot analysis, are typically lower than the actual number [40,43]. Second, southern blots may lead to poor resolution of the blotting bands and allow detection of only a few of the total bands. When RT-qPCR is used to estimate exogenous gene copy number, the results can be influenced by the slope and correlation coefficient of the standard curve, as well as the concentration of the starting template. In this study, we established a good standard curve with a slope of -3.140 and a correlation coefficient of 0.998 that supports the accurate estimation of the cry1Ac gene copy number.
Protein expression level of exogenous genes can directly affect the target trait and its practical value [17,23]. Transgenic sugarcane lines containing the truncated insecticidal gene m-cry1Ac or the partially modified s-cry1Ac, were previously evaluated to determine integration sites, transgene expression pattern, and level of resistance to insects [23]. Among the 55 PCR positive plants, 17 were determined to be positive using protein analysis and contained a range of 2.2 ng/mg to 50 ng/mg Cry1Ac protein in leaves and 6 to 8 copies of the cry1Ac gene estimated by Southern blot analysis. In our study, there were 14 PCR positive transgenic sugarcane lines but ELISA detected Cry1Ac protein in only 7 lines. Interestingly, RT-qPCR estimation of copy number and ELISA analysis of Cry1Ac protein revealed only one line with low copy number (5 copies) and low Cry1Ac protein content (0.85 μg/FWg in leaf or 0.04 μg/FWg in stem) while the remaining 6 lines had a medium copy number (17~62 copies) and high protein expression (39.14 μg/FWg to 70.92 μg/FWg in leaves or 2.91 μg/FWg to 7.22 μg/FWg in stems). Meanwhile, linear relationship was not observed between the Cry1Ac protein expression level and the copy number of cry1Ac gene in the 6 transgenic lines, T-2, T-3, T-5, T-8, T-11, and T-14 with higher protein expression, which is consistent with previous studies [44,45]. This also indicated that expression of the exogenous gene was likely affected by position effects. It should be noted that Cry1Ac protein was not detected in 7 cry1Ac transgenic sugarcane lines including 3 lines with low copy number (1–2 copies) and 4 lines with high copy number (83–148 copies). Similarly, protein expression was not detected in lines with low copy number (1–2 copies), which may due to a large sugarcane genome [46]. In contrast, a high copy number of an exogenous gene in transgenic plants could result in co-suppression due to multi-copy integration, thus resulting in transgenic silencing [47,48]. The phenomenon of transgenic silencing has been documented in sugarcane [49,50].
Bt protein expression can improve pest resistance in transgenic plants [17,23,51]. The cry1Ab gene has been previously introduced into sugarcane cultivars, Co 86032 and CoJ 64 [17]. Cry1Ab protein in different transgenic events ranged from 0.007% to 1.73% of the total soluble protein in leaf. Transgenic plants had significantly less dead heart at the seedling stage and there was a negative correlation between protein expression and the dead heart rate. Weng et al. [23] analyzed pest resistance of cry1Ac transgenic sugarcane, and found resistance only in lines expressing the Cry1Ac protein more than 9 ng/mg. A positive correlation between the Cry1Ac content and pest resistance was also observed. Lines with low Cry1Ac protein content (1.8 ng/mg) were more susceptible to pests. In the present study, the percentage of damaged stalks ranged from 13.33% to 36.67% in 7 transgenic sugarcane lines expressing Cry1Ac protein, while in the untransformed control line the damage was as high as 76.67%. These results demonstrated that higher Cry1Ac protein expression results in reduced damage. T-2 line, with the highest protein expression in leaves (70.92 μg/FWg) and stems (7.22 μg/FWg) had the lowest percentage of stalks damaged (13.33%), which was consistent with a previous report [23]. Nevertheless, T-1 line with lower expression of Cry1Ac in leaves (0.85 μg/FWg) and stems (0.04 μg/FWg) was also resistant to borer damage (36.67%) which was less than the control (76.67%) and differed from the results reported by Weng et al. [23]. There were limitations in the worm damage evaluation methods used in this study. Assessment was based on the number of wormholes and not the degree of damage. Nevertheless, there was still 13.33% damage in the T-2 line but the damage was confined to the outside of the stalks. In non-transgenic lines, the borers tunneled inside the stalk where they cause relatively more damage (Fig 7).
Weng et al. [23] observed that the agronomic traits of stalk growth and juice quantity were severely affected in transgenic lines compared to the control, but the industrial traits of sucrose content, brix, and purity were not significantly different between the control and transgenic lines. In the present study, we found that the transgenic plant lines had variable height, stalk diameter, millable stalks, and brix. Some transgenic plants did not significantly differ from the control while others were significantly different, which is not in agreement with Weng et al. [23]. Agronomic characteristics and yield, e.g. stalk length, millable stalks and theoretical sucrose yield, in the medium copy number transgenic sugarcane lines were similar to the non-transgenic control. The theoretical sucrose yields of all 14 transgenic plants were lower than the non-transgenic controls suggesting that Bt protein expression may have influenced the normal sugarcane growth while the theoretical sucrose yield in lines T-3 (51.65 kg), T-8 (50.11 kg) and T-11 (46.68 kg) was not significantly different from the non-transgenic control (55.86 kg).
In conclusion, our results suggest that a medium copy number of cry1Ac gene in transgenic sugarcane may be more desirable than too high or too low a number since this appears to compromise gene expression. Higher Cry1Ac protein expression may not be optimum because high protein expression consumes more plant energy and negatively affects agronomic traits. All transgenic lines with medium copy number expressing Cry1Ac protein had relatively equivalent or lower theoretical sucrose yield, compared to controls, and showed significantly improved sugarcane borer resistance. These lines can be potentially used for commercial purposes.
Supporting Information
(TIF)
M: DL15,000+2,000 DNA Ladder; 1: The products of pGcry1Ac0229 digested with Hind III and EcoR I; 2: The products of pGcry1Ac0229 digested with Hind III.
(TIF)
(TIF)
(TIF)
Acknowledgments
This work was supported by the National Nature Science Foundation of China (31271782), the earmarked fund for the Modern Agriculture Technology of China (CARS-20), Scientific Research Fund of Fujian Provincial Education Department (JA12124) and Science and Technology Major Project of Fujian Province (2015NZ0002-2). The authors especially thank Dr. Illimar Altosaar in the University of Ottawa, Canada, for providing the cloning vector, cry1AcPRD.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Nature Science Foundation of China (31271782), the earmarked fund for the Modern Agriculture Technology of China (CARS-20) and Scientific Research Fund of Fujian Provincial Education Department (JA12124) and Science and Technology Major Project of Fujian Province (2015NZ0002-2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Raza G, Ali K, Mukhtar Z, Mansoor M, Arshad M, Asad S (2010) The response of sugarcane (Saccharum officinarum L.) genotypes to callus induction, regeneration and different concentrations of the selective agent (geneticin-418). Afr J Biotechnol 9(51): 8739–8747 [Google Scholar]
- 2.Guo JL, Xu LP, Su YC, Wang HB, Gao SW, Xu JS, Que YX (2013) ScMT2-1-3, a metallothionein gene of sugarcane, plays an important role in the regulation of heavy metal tolerance/accumulation. Biomed Res Int 1:86–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher G, Teixeira E, Hizsnyik ET, Velthuizen HV (2008) Land use dynamics and sugarcane production In Sugarcane Ethanol: Contributions to Climate Change Mitigation and the Environment. Edited by Zuurbier P, Vooren JVD. Wageningen Academic Publishers; 29–62 [Google Scholar]
- 4.Ricaud C, Egan BT, Gillaspie AG, Hughes CG (1989) Diseases of sugarcane: major diseases Elsevier Science Publishers B.V., Netherlands [Google Scholar]
- 5.White WH, Miller JD, Milligan SB, Burner DM, Legendre BL (2001) Inheritance of sugarcane borer resistance in sugarcane derived from two measures of insect damage. Crop Sci 41:1706–1710 [Google Scholar]
- 6.Milligan SB, Balzarini M, White WH (2003) Broad-sense heritabilities, genetic correlations, and selection indices for sugarcane borer resistance and their relation to yield loss. Crop Sci 43:1729–1735 [Google Scholar]
- 7.Arencibia A, Vázquez RI, Prieto D, Téllez P, Carmona ER, CoegoA HL, De la Riva GA, Selman-Housein G (1997) Transgenic sugarcane plants resistant to stem borer attack. Mol Breed 3:247–255 [Google Scholar]
- 8.Zhou DG, Guo JL, Xu LP, Gao SW, Lin QL, Wu QB, Wu LG, Que YX (2014) Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of cry1Ac transgenic sugarcane. Sci Rep 4:4912 10.1038/srep04912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles BH, Ellar DJ (1986) Characterization and partial purification of a plasma membrane receptor for Bacillus thuringiensis var. kurstaki lepidopteran-specific δ-endotoxin. J Cell Sci 83: 89–101 [DOI] [PubMed] [Google Scholar]
- 10.Vaeck M, Reynaerts A, Höfte H, Jansens S, De Beuckeleer M, Dean C, Zabeau M, Van Montagu M, Leemans J (1987) Transgenic plants protected from insect attack. Nature 328: 33–37 [Google Scholar]
- 11.Delannay X, LaVallee BJ, Proksch RK, Fuchs RL, Sims SR, Greenplate JT, Marrone PG, Dodson RB, Augustine JJ, Layton JG, Fishhoff DA (1989) Field performance of transgenic tomato plants expressing the Bacillus thuringinesis var. kurstaki insect control plant. Bio Technol 7:1265–1269 [Google Scholar]
- 12.Perlak FJ, Deaton RW, Armstron TA, Fuchs RL, Sims SR, Greenplate JT, Fishhoff DA (1990) Insect resistant cotton plants. Bio Technol 8: 939–943 [DOI] [PubMed] [Google Scholar]
- 13.Perlak FJ, Stone TB, Muskopf YM, Peterson LJ, Parker GB, Mcpherson SA, Wyman J, Love S, Reed G, Biever D, Fischhoff DA (1993) Genetically improved potatoes: protection from damage by colorado potato beetles. Plant Mol Biol 22: 313–321 [DOI] [PubMed] [Google Scholar]
- 14.Koziel MG, Beland GL, Bowman C, Carozzi NB, Crenshaw R, Crossland L, Dawson L, Desai N, Hill M, Kadwell S, Launis K, Lewis K, Maddox D, McPherson K, Meghji MR, Merlin E, Rhodes R, Warren GW, Wright M, Evola SV (1993) Field performance of elite transgenic maize plants expressing an insecticidal protein derived from Bacillus thuringiensis. Bio Technol 11:194–200 [Google Scholar]
- 15.Fujimoto H, Itoh K, Yamamoto M, Kyozuka J, Shimamoto K (1993) Insect resistant rice generated by introduction of a modified δ-endotoxin gene of Bacillus thuringensis. Bio technol 11:1151–1155 [DOI] [PubMed] [Google Scholar]
- 16.Arencibia AD, Carmona ER, Cornide MT, Castiglione S, O'Relly J, Chinea A, Oramas P, Sala F (1999) Somaclonal variation in insect-resistant transgenic sugarcane (Saccharum hybrid) plants produced by cell electroporation. Transgenic Res 8(5): 349–360 [Google Scholar]
- 17.Arvinth S, Arun S, Selvakesavan RK, Srikanth J, Mukunthan N, Ananda Kumar P, Premachandran MN, Subramonian N (2010) Genetic transformation and pyramiding of aprotinin-expressing sugarcane with cry1Ab for shoot borer (Chilo infuscatellus) resistance. Plant Cell Rep 29(4):383–395 10.1007/s00299-010-0829-5 [DOI] [PubMed] [Google Scholar]
- 18.Sétamou M, Bernal JS, Legaspi JC, Mirkov TE, Legaspi BC (2002) Evaluation of lectin-expressing transgenic sugarcane against stalkborers (Lepidoptera: Pyralidae): effects on life history parameters. J Econ Entomol 95:469–477. [DOI] [PubMed] [Google Scholar]
- 19.Zhangsun DT, Luo SL, Chen RK, Tang KX (2007) Improved agrobacterium-mediated genetic transformation of GNA transgenic sugarcane. Biologia (Bratislava) 62(4):386–393 [Google Scholar]
- 20.Falco MC, Silva-Filho MC (2003) Expression of soybean proteinase inhibitors in transgenic sugarcane plants: effects on natural defense against Diatraea saccharalis. Plant Phys Biochem 41:761–766 [Google Scholar]
- 21.Kalunke RM, Kolge AM, Babu KH, Prasad DT (2009) Agrobacterium mediated transformation of sugarcane for borer resistance using cry1Aa3 gene and one-step regeneration of transgenic plants. Sugar Tech 11(4):355–359 [Google Scholar]
- 22.Weng LX, Deng HH, Xu JL, Li Q, Wang LH, Jiang ZD, Zhang HB, Li QW, Zhang LH (2006) Regeneration of sugarcane elite breeding lines and engineering of strong stem borer resistance. Pest Manage Sci 62:178–187 [DOI] [PubMed] [Google Scholar]
- 23.Weng LX, Deng HH, Xu JL, Li Q, Zhang YQ, Jiang ZD, Li QW, Chen JW, Zhang LH (2011) Transgenic sugarcane plants expressing high levels of modified cry1Ac provide effective control against stem borers in field trials. Transgenic Res 20(4):759–772 10.1007/s11248-010-9456-8 [DOI] [PubMed] [Google Scholar]
- 24.Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Phys 15: 473–492 [Google Scholar]
- 25.Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Bio Rep 11(2): 122–127 [Google Scholar]
- 26.Li HF, Li L, Zhang LJ, Xu YL, Li WF (2010) Standard curve generation of PepT1 gene for absolute quantification using real-time PCR. J Shanxi agr univ 30(4): 332–335 [Google Scholar]
- 27.Xue BT, Guo JL, Que YX, Fu ZW, Wu LG, Xu LP (2014) Selection of suitable endogenous reference genes for relative copy number detection in sugarcane. Int J Mol Sci 15:8846–8862 10.3390/ijms15058846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suo GL, Chen B, Zhang JY, Duan ZY, He ZQ, Yao W, Yue CY, Dai JW (2006) Effects of codon modification on human BMP2 gene expression in tobacco plants. Plant Cell Rep 25: 689–697 [DOI] [PubMed] [Google Scholar]
- 29.Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O'Brien JV, Chambers SA, Adams WR Jr, Willetts NG, Rice TB, Mackey CJ, Krueger RW, Kausch AP, Lemaux PG (1990) Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2(7):603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasil V, Castillo AM, Fromm ME, Vasil IK (1992) Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Bio Technol 10:667–674 [Google Scholar]
- 31.Gallo-Meagher M, Irvine JE (1996) Herbicide resistant transgenic sugarcane plants containing the bar gene. Crop Sci 36:1367–1374 [Google Scholar]
- 32.Manickavasagam M, Ganapathi A, Anbazhagan VR, Sudhakar B, Selvaraj N, Vasudevan A (2004) Agrobacterium mediated genetic transformation and development of herbicide-resistant sugarcane (Saccharum species hybrids) using axillary buds. Plant Cell Rep 23(3):134–143 [DOI] [PubMed] [Google Scholar]
- 33.Luo SL, Lin JY, Zhangsun DT (2003) Selective test of antibiotics and PPT in different stages of sugarcane tissue culture. J Hainan Univ Nat Sci 21 (3):259–265 [Google Scholar]
- 34.Gao SW (2008) Study on genetic transformation of sugarcane with cry gene. Dissertation, Fujian Agriculture and Forestry University
- 35.Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J (1987) Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J 6:2519–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song P, Cai CQ, Skokut M, Kosegi BD, Petolino JF (2002) Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERS™-derived transgenic maize. Plant Cell Rep 20:948–954 [Google Scholar]
- 37.Ji ZG, Gao XJ, Ao JX, Zhang MH, Huo N (2011) Establishment of SYBR Green-base quantitative real-time PCR assay for determining transgene copy number in transgenic soybean. J Northeast Agric Univ 42(10):11–15 [Google Scholar]
- 38.Weng HB, Pan AH, Yang LT, Zhang CM, Liu ZL, Zhang DB (2004) Estimating number of transgene copies in transgenic rapeseed by real-time PCR assay with HMG I/Y as an endogenous reference gene. Plant Mol Bio Rep 22:289–300 [Google Scholar]
- 39.Cheng XY, Zhang J, Chan KM, Liu XK, Hong Y(2008), Quantitative real-time PCR assay to detect transgene copy number in cotton (Gossypium hirsutum). Analytical Biochemistry, 375:150–152 [DOI] [PubMed] [Google Scholar]
- 40.Yang XJ, Liu CL, Zhang CJ, Wu ZX, Zhang XY, Liu K, Fang WP, Li FG (2011) Comparative analysis of exogenous gene copy numbers in transgenic cotton transformed by different methods. J Agr Biotech 19(2):221–229 [Google Scholar]
- 41.D’Hont A, Glaszmann JC (2001) Sugarcane genome analysis with molecular markers: a first decade of research. Proc Int Soc Sugar Cane Technol 24: 556–559 [Google Scholar]
- 42.Zhao Y, Qian Q, Wang HZ, Huang DN (2007) Hereditary behavior of bar gene cassette is complex in rice mediated by particle bombardment. J Genet Genomics 34(9):824–835 [DOI] [PubMed] [Google Scholar]
- 43.Yang LT, Ding JY, Zhang CM, Jia JW, Weng HB, Liu WX, Zhang DB (2005) Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep 23:759–763 [DOI] [PubMed] [Google Scholar]
- 44.Domínguez A, Guerri J, Cambra M, Navarro L, Moreno P, Peña L (2000) Efficient production of transgenic citrus plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep 19(4):427–433 [DOI] [PubMed] [Google Scholar]
- 45.Yang XJ, Liu CL, Zhang XY, Li FG (2010) Molecule characterization and expression of T-DNA integration in transformed plants. Genom Appl Biol 29 (1):125–130 [Google Scholar]
- 46.Tomkins JP, Yu Y, Miller-Smith H, Frisch DA, Woo SS, Wing RA (1999) A bacterial artificial chromosome library for sugarcane. Theor Appl Genet 99:419–424 10.1007/s001220051252 [DOI] [PubMed] [Google Scholar]
- 47.De Bolle MFC, Butaye KMJ, Goderis IJWM, Wouters PFJ, Jacobs A, Delauré SL, Depicker A, Cammue BPA (2007) The influence of matrix attachment regions on transgene expression in Arabidopsis thaliana wild type and gene silencing mutants. Plant Mol Biol 63:533–543 [DOI] [PubMed] [Google Scholar]
- 48.De Paepe A, De Buck S, Hoorelbeke K, Nolf J, Peck I, Depicker A (2009) High frequency of single-copy T-DNA transformants produced by floral dip in CRE-expressing Arabidopsis plants. Plant J 59(4): 517–527 10.1111/j.1365-313X.2009.03889.x [DOI] [PubMed] [Google Scholar]
- 49.Hansom S, Bower R, Zhang L, Potier B, Elliott A, Basnayake S, Cordeiro G, Hogarth DM, Cox M, Berding N, Birch RG (1999) Regulation of transgene expression in sugarcane. In: Proc Int Soc Sug Cane Technol Congr 2:278–290 [Google Scholar]
- 50.Wei HR, Wang ML, Moore PH, Albert HH (2003) Comparative expression analysis of two sugarcane polyubiquitin promoters and flanking sequences in transgenic plants. J Plant Phys 160:1241–1251 [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Tang W, Xu CG, Li XH, Lin YJ, Zhang QF (2005) Transgenic indica rice plants harboring a synthetic cry2A gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor Appl Genet 111:1330–1337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
M: DL15,000+2,000 DNA Ladder; 1: The products of pGcry1Ac0229 digested with Hind III and EcoR I; 2: The products of pGcry1Ac0229 digested with Hind III.
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.