Abstract
Containing both AP2 domain and B3 domain, RAV (Related to ABI3/VP1) transcription factors are involved in diverse functions in higher plants. A total of eight TsRAV genes were isolated from the genome of Thellungiella salsuginea and could be divided into two groups (A- and B-group) based on their sequence similarity. The mRNA abundance of all Thellungiella salsuginea TsRAVs followed a gradual decline during seed germination. In Thellungiella salsuginea seedling, transcripts of TsRAVs in the group A (A-TsRAVs) were gradually and moderately reduced by salt treatment but rapidly and severely repressed by ABA treatment. In comparison, with a barely detectable constitutive expression, the transcriptional level of TsRAVs in the group B (B-TsRAVs) exhibited a moderate induction in cotyledons when confronted with ABA. We then produced the “gain-of-function” transgenic Arabidopsis plants for each TsRAV gene and found that only 35S:A-TsRAVs showed weak growth retardation including reduced root elongation, suggesting their roles in negatively controlling plant growth. Under normal conditions, the germination process of all TsRAVs overexpressing transgenic seeds was inhibited with a stronger effect observed in 35S:A-TsRAVs seeds than in 35S:B-TsRAVs seeds. With the presence of NaCl, seed germination and seedling root elongation of all plants including wild type and 35S:TsRAVs plants were retarded and a more severe inhibition occurred to the 35S:A-TsRAV transgenic plants. ABA treatment only negatively affected the germination rates of 35S:A-TsRAV transgenic seeds but not those of 35S:B-TsRAV transgenic seeds. All 35S:TsRAVs transgenic plants showed a similar degree of reduction in root growth compared with untreated seedlings in the presence of ABA. Furthermore, the cotyledon greening/expansion was more severely inhibited 35S:A-TsRAVs than in 35S:B-TsRAVs seedlings. Upon water deficiency, with a wider opening of stomata, 35S:A-TsRAVs plants experienced a faster transpirational water loss than wild type and 35S:B-TsRAVs lines. Taken together, our results suggest that two groups of TsRAVs perform distinct regulating roles during plant growth and abiotic defense including drought and salt, and A-TsRAVs are more likely than B-TsRAVs to act as negative regulators in the above-mentioned biological processes.
Introduction
The RAV (Related to ABI3/VP1) family represents a unique group of plant-specific transcription factors since some members within this family contain two DNA-binding domains, e.g. a Basic 3 (B3) domain and an Apetala2 (AP2) domain. First identified in the viviparous1 (vp1) mutant from Zea mays and the ABA insensitive-3 (abi3) mutant from Arabidopsis, the B3 domain has been shown to be involved in the process of seed maturation and ABA regulation of gene expression [1,2,3]. The AP2 domain could be found within many AP2/ERF family members and has been implicated in plant hormone signal transduction and responses to biotic, pathogenic, and environmental cues.
In Arabidopsis, there are thirteen RAV family members [4], six of which contain both B3 domain and AP2 domain [5,6]. So far, only AtRAV1 and AtRAV2 have been functionally investigated in several aspects of plant physiology and development. Transgenic Arabidopsis plants overexpressing AtRAV1 or AtRAV2 were reported to result in growth-retarded phenotypes including reduced lateral root, smaller rosette leaf and slightly late flowering [4,7]. Transgenic tobacco lines constitutively expressing a soybean RAV2 ortholog also exhibited slower plant growth rate, reduced root elongation and delayed flowering, especially under short day conditions [8]. In contrast, rav1 and rav2 single mutants showed slightly promoted growth patterns in the early stage of development [4]. Considering these results, RAV1 and RAV2 have been believed to act as negative regulators during plant growth and floral transition.
RAV genes have also been shown to be induced by various biotic and abiotic environmental stimuli including pathogen infection, salicylic acid, osmotic stress, cold, high salinity, wounding and exogenous hormone application [9,10,11,12,13]. However, contradictory results including the repressed expression of AtRAV1 and RAV2 by drought, salt and ABA treatment have also been reported [4]. Thus, whether or not and how RAV transcription factors are involved in the adaptive strategies of stressed plants still remains unclear. It is well known that both drought and high salinity can trigger the production of ABA, and drought-inducible genes could also be activated by salt-stress and ABA. In literature, Arabidopsis rav1 mutant has been shown to exhibit an ABA insensitivity to root growth inhibition, and conversely overexpression of AtRAV1 or AtRAV2 in transgenic Arabidopsis resulted in an increased sensitivity to ABA inhibition of seed germination and root growth [14]. However, in another work published recently, AtRAV1 overexpressing and T-DNA insertion mutant lines have been described as insensitive and hypersensitive, respectively, to ABA inhibition of root growth [15]. In addition to the above-mentioned discrepancies in gene expression profiles and induction patterns, results of functional analyses were not always consistent either. For example, transgenic cotton (Gossypium hirsutum) expressing AtRAV1 and 2 showed improved resistance to drought and water usage efficiency [16]. Overexpression of a pepper RAV1 was reported to enhance the tolerance to NaCl [10] and transgenic tomato overexpressing its endogenous SlRAV2 also resulted in tolerance to bacterial wilt [17]. In contrast to these promising results, overexpression of GhRAV1 in Arabidopsis resulted in transgenic plants sensitive to high salinity, drought and exogenous ABA instead [18]. AtRAVs were also recently demonstrated to play negative roles in drought and salt tolerance in an ABA-independent manner [4]. So far, there is no clear explanation for these experimental discrepancies. In addition, all functional analyses performed on RAVs to date have been focused on RAV1, RAV2 and their homologues, and little information exists in literature about the expression patterns and function roles of other RAV gene family members.
In order to make a systematic investigation on the responses of RAV family members to salt and ABA, and analyze their respective contribution to salt and drought tolerance, we isolated all eight RAV subfamily members that contain both AP2 and B3 domains from T. salsuginea, a close relative of Arabidopsis with a exceptionally high resistance to cold, drought, salt and oxidative stresses [19,20,21,22]. Compared to A. thaliana, a modest expansion of the RAV family occurred to the genome of T. salsuginea, which might be associated with the adaptive strategies of T. salsuginea to its extreme environment. Interestingly, two groups of ThRAVs separated by their characteristic sequence features also exhibited distinctively different induction patterns when confronted with salt and ABA treatment. When introduced into Arabidopsis, overexpression of A-TsRAVs and B-TsRAVs resulted in various responses towards abiotic stresses including NaCl, water loss and ABA treatment, implying their different regulatory roles during plant growth and development.
Materials and Methods
Plant materials, growing conditions and stress treatments
Seeds of T. salsuginea were obtained from Dr. Yinxin Li at the Institute of Botany of the Chinese Academy of Sciences. To investigate the responses of TsRAVs to NaCl and ABA, seeds of T. salsuginea were germinated on 1/2 MS medium and grown vertically on plates for 15 days. Upon stress treatments, seedlings were first acclimated in 1/2 liquid medium for half an hour before being treated by 200 mM NaCl or 5 μM ABA. Rosette leaves and roots were separately sampled from three biological replicates at 0, 0.5, 1, 3, 6, 12, and 24 h time points.
Sequence isolation and analysis
Sequence alignments of TsRAV and AtRAV proteins were performed with Clustal W [23] using default parameters. A phylogenetic tree was constructed with MEGA 5.0 [24] using the neighbor-joining method and the tree reliability was set to 1000 bootstrap replicates. The identification of B3 and AP2 domains was based on the genome annotation and verified by the InterProScan and Pfam online tools with defaulted parameters [25,26,27]. Motif identification within protein sequences was carried out using the MEME Suite tool version 4.11.0 [28] with the following parameters: optimum width of 5–50 amino acids, any number of repetitions of a motif, and the maximum number of motifs set at 30 with a E-value<4.6e-5 and a P-value<1.1e-5.
Gene isolation, expression profile analyses and construction of transgenic plants
Total RNA of T. salsuginea seedling was extracted using the EasyPure Plant RNA Kit (TransGen, China) and cDNA synthesis was performed using the EasyScript® First-Strand cDNA Synthesis SuperMix (TransGen, China) with the Oligo(dT)18 reverse primer. Amplifications of actin were used as controls for the semi-quantitative PCR analysis. Quantitative Real-Time PCR (qRT-PCR) analysis of cDNA was performed on a PikoReal 96 Real-time Thermal Cycler and PikoReal Software (V2.2) (Thermo Fisher Scientific, Finland) using Real Master Mix SYBR Green I (NEWBIO, China). The following thermal cycle conditions were used: 95°C for 2 min, followed by 45 cycles of 95°C for 20 s and 58°C for 20 s, 72°C for 30 s. All reactions were performed in triplicate from three independent pooled samples (50 plants per sample). Relative quantification of specific mRNA levels was analyzed using the cycle threshold (Ct) 2-ΔΔCt method. Relative expression levels are normalized using the housekeeping gene actin and shown in folds of the expression value in untreated samples. The sequences of all primers used in this study are listed in S1 Table.
To generate Arabidopsis transgenic lines overexpressing TsRAVs, the cDNA fragment of each TsRAV was cloned into the pK2GW7I expression vector under the control of CaMV 35S promoter. The plasmids were introduced into Agrobacterium tumefaciens strain individually to transform Arabidopsis thaliana ecotype Columbia (Col-0) using the floral dip method [29]. The kanamycin-resistant seedlings were transplanted into soil and homozygous T3 seeds were used for the following transgenic analyses.
Stress and hormonal treatments on transgenic plants
For seed germination under abiotic stresses, transgenic T3 seeds were surface-sterilized and germinated on 1/2 MS agar medium containing 100 mM NaCl or 1 μM ABA for 7 days at 22°C with a 16 h-light/8 h-dark cycle. Fifty seeds per replicate were examined with three replicates for each treatment. For measurement of root growth, Arabidopsis seeds were germinated on 1/2 MS agar medium for 5 days and the seedlings were transferred to fresh medium containing 200 mM NaCl or 30 μM ABA, and plates were placed vertically on shelves to facilitate comparison of root growth. To determine the drought tolerance in a quantitative manner, leaves were detached from each plant and placed in Petri dishes that were kept at 40% relative humidity in a growth chamber, and the loss of fresh weight was determined at the indicated time points. Stomatal aperture measurements were performed on epidermis from leaves of 4-week-old plants and values are the means for at least 50 apertures from at least three experiments.
Results and Discussion
Identification and classification of T. salsuginea RAV family
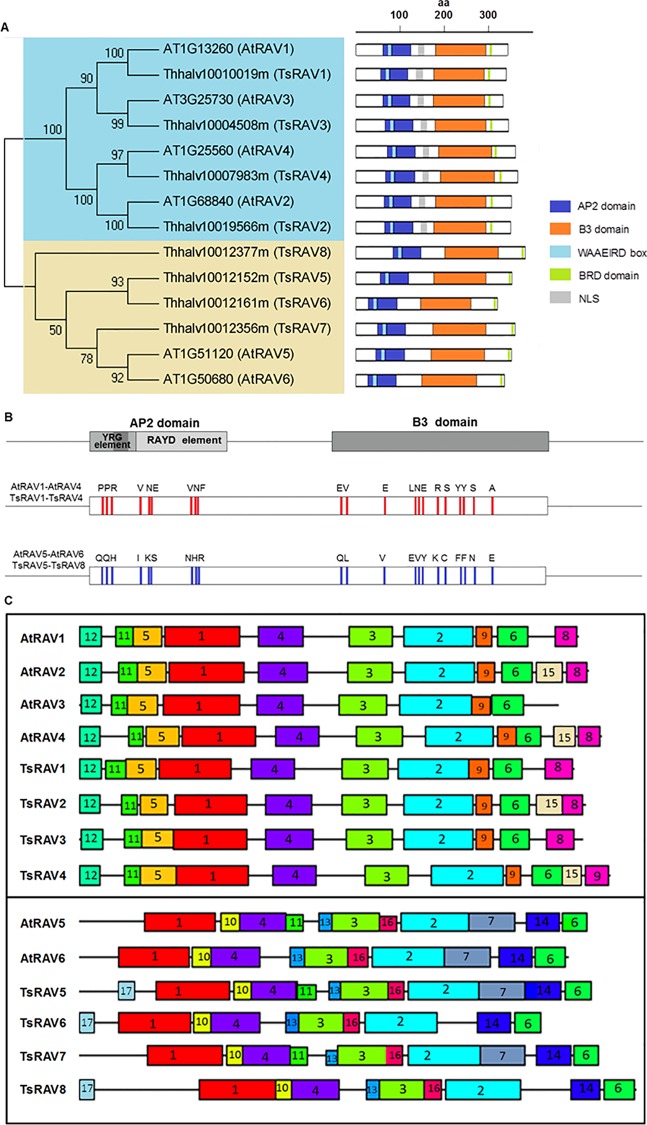
The Arabidopsis genome contains six RAVs that contain both AP2 domain and B3 domain [30] and AtRAVs share 35–79% amino acid identity throughout their full-length sequences. Using RAV sequences from Arabidopsis and other species as query sequences to search the T. salsuginea genome database, a total of 8 TsRAVs were isolated with the predicted full-length proteins ranging from 320 to 384 amino acids (S2 Table). In the phylogenetic tree, eight TsRAVs and six AtRAVs obviously split into two groups (Fig 1A). The group A includes TsRAV1-4 and AtRAV1 (At1g13260), AtRAV2/TEM2 (AT1g68840), AtRAV3/RAV1L (At3g25730) and AtEDF1/TEM1 (AT1g25560), and the group B includes TsRAV5-8 and two AtRAVs (At1g51120 and At1g50680) (Fig 1A). At their N-terminal regions, all eight TsRAVs contain an AP2 DNA-binding domain that recognizes a consensus CAACA sequence [31] and includes the conserved YRG and RAYD elements as well as a conserved 7-aa WAAEIRD box motif [32] (Fig 1A). At the C-terminal regions, all eight TsRAVs contain a B3 domain that recognizes a consensus CACCTG sequence and a 15-aa B3 repression domain (BRD) (GNSKTLRLFGVNMEC) that is responsible for the repressive activity in many B3 super-family members (Fig 1A). A short peptide composed of five amino acid residues, R/KLFGV, has been demonstrated to be crucial to maintain the repressive activity of the BRD domain [33]. In this work, TsRAVs in group A and group B include a RLFGV and M/KLFGV core sequence, respectively, which was also identified in rice RAV homologues [33]. The function of RAVs as transcriptional repressors has been demonstrated in several plant species. For example, TEM1 and GmRAV can repress flowering by binding to and repressing the promoters of FT genes in Arabidopsis and soybean, respectively [34,35]. Notably, each A-TsRAV but not a single B-TsRAV contains a nuclear localization sequence, which might contribute significantly to their roles as transcriptional regulators (Fig 1A). In addition to the BRD domain, a close comparison of the amino acid sequences between A-TsRAVs and B-TsRAVs revealed that they are characterized by distinctly different amino acids at some highly conserved positions and these group-specific residues might in turn affect the secondary structure of RAV proteins as well as their DNA-binding activities (Fig 1B). We also analyzed the motif composition of AtRAVs and TsRAVs using the MEME program. In total, 17 conserved motifs were detected and some of them appeared to be highly divergent between two groups (Fig 1C). For example, motifs 5 and 12 located in front of the AP2 domain, motif 9 within the B3 domain and motif 8 at the C-end of the RAV protein are highly conserved in the group A-TsRAVs, but are absent in the B-group RAVs. On the contrary, motif 10 within the AP2 domain, motifs 13 and 16 within the B3 domain and motif 14 located in front of the BRD domain are present in all group B-TsRAVs but missing in the group A-TsRAVs (Fig 1C).
Fig 1. Sequence characterization of RAV family members of Thellungiella salsuginea and Arabidopsis thaliana.
(A) Phylogenetic tree of the RAV family members in Thellungiella salsuginea and Arabidopsis thaliana. The phylogenetic tree was constructed using full-length protein sequences by the maximum-likelihood method with MEGA 5.0 and a bootstrap value of 1,000. The two major phylogenetic clades are designated as groups A and B. Shown on the right are diagrams of RAV proteins with information on the structure and position of different protein domains. (B) RAV subfamily-specific amino acids and their locations along the RAV full-length sequences. The amino acid sequences in boxes represent the conserved AP2 and B3 DNA-binding domains, which are characteristic of RAV transcription factors. The locations of the conserved YRG and RAYD elements are indicated as well. (C) Schematic illustrations of the types and distributions of motifs for each TsRAV family members with a same group. Motifs were identified using the MEME search tool and numerically marked according to their statistical significance (low E-value) in a descending order.
Transcriptional change of TsRAVs during seed germination and their responses to salt and ABA application
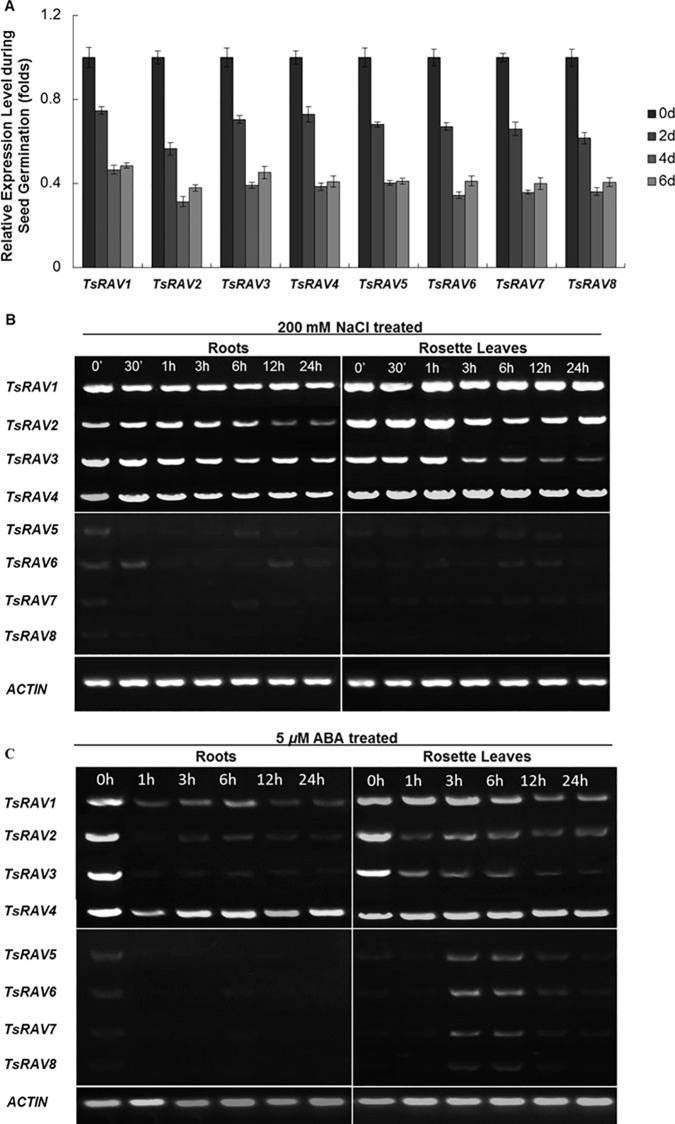
We then determined the transcript level of eight TsRAV genes during Thellungiella salsuginea seed germination. It has been reported that in Arabidopsis, the expression level of AtRAV1 was very low in imbibed seed and was dramatically increased during seed germination, and its down-regulation could slowed down seed germination [15]. However, in this work, within six days after imbibition, the transcripts of all TsRAVs experienced a persistent and significant decrease until the end of the germination process, indicating that TsRAVs are negatively involved in seed germination process (Fig 2A).
Fig 2. Expression of TsRAV genes during seed germination and in response to salt and ABA treatments.
(A) qRT-PCR assay of TsRAVs transcription in Thellungiella salsuginea during seed germination. Analyses were performed on seeds at 0, 2, 4, and 6 DAI (days after imbibition). The transcription levels of genes were quantified relative to that of actin. Each data bar represents the means ± SE (Standard Error) of three replicates. (B) Semi-quantitative RT-PCR assay of TsRAVs transcription in Thellungiella salsuginea seedlings upon 200 mM NaCl treatment. Total RNA was extracted at various time intervals from leaves and roots of 15-d-old seedlings. The cDNA samples are normalized using an actin gene as an internal control. (C) Semi-quantitative RT-PCR assay of TsRAVs transcription in Thellungiella salsuginea seedlings upon 5 μM ABA treatment. Total RNA was extracted at various time intervals from leaves and roots of 15-d-old seedlings. The cDNA samples are normalized using an actin gene as an internal control.
We then analyzed the transcriptional changes of all TsRAVs in response to salt and ABA in seedling rosette leaves and roots. First, when treated by 200 mM NaCl, similar responses were observed in these two types of tissues for all eight TsRAV genes (Fig 2B). In the group A, TsRAV1-4 exhibited a high constitutive expression level under normal conditions. Upon NaCl stress, TsRAV1 and 4 remained a stable expression level within 24 hours, while TsRAV2 and 3 showed a gradual and moderate decrease in their expression level (Fig 2B). In the group B, TsRAV5-8 only accumulated a detectable yet low level of transcripts normally and did not significantly respond to the presence of NaCl (Fig 2B). In literature, the expression level of AtRAV1 and AtRAV2 has been reported to be up-regulated by various external and environmental cues including low temperature, darkness, wounding, drought, salt, and pathogen attack [10,11,36]. RAV1 orthologs from Brassica napus, pepper and cotton have also been reported to be inducible by NaCl [10,13,18]. However, the expression of AtRAV1 and 2 were recently shown to be repressed by drought and salt [4] just as described in this work. A possible explanation for these discrepancies might be the internal developmental regulations of RAV genes [37] and/or related to the time points and tissue types that were collected for analyses. Although TsRAVs were not found to be inducible by salt in this study, we could not rule out the possibility of them to play a role in response to salt stress.
ABA is a plant hormone known for its involvement in various signal transduction pathways and exogenous ABA is capable of inhibiting seed germination and early seedling growth [38,39]. In a previous work, the expression of AtRAV1-3 were shown to be repressed significantly by ABA treatment for 24 hours [4]. In this work, upon the treatment of 5 μM ABA, A-TsRAVs and B-TsRAVs responded to ABA in a distinctly different manner and only A-TsRAVs but not B-TsRAVs were transcriptionally repressed (Fig 2C). TsRAV1 showed a moderate and continuous decline in its expression level and the transcript level of TsRAV4 kept fairly stable, and the responses of both genes in shoot and root were similar (Fig 2C). For TsRAV2 and TsRAV3, their transcript abundances in roots was rapidly repressed by ABA within one hour and then disappeared drastically thereafter; while in rosette leaves, their transcripts followed a gradual decrease (Fig 2C), indicating the differential responses of two tissues to ABA application. For B-TsRAVs, despite that their expression level kept fairly low in root regardless of the presence of ABA, they could be transiently but obviously triggered by ABA in leaves with a transcriptional peak observed at 3 h and 6 h (Fig 2C). Thus, when exposed to NaCl or ABA, the only significant induction observed in the current work happened to B-TsRAVs in the ABA-treated leaves (Fig 2B and 2C). Under normal or salty conditions, B-TsRAVs always maintained a low expression level while A-TsRAVs showed a high level of constitutive expression and were repressed to various extents by salt or ABA application. In summary, although that the elevated expression of RAV1 by ABA has previously been reported in cotton and pepper [10,18], our results were consistent with the transcriptional repression of AtRAV1 by exogenous ABA [15]. Meanwhile, we revealed that upon abiotic stress treatment, TsRAVs could respond in a significantly different manner in various types of plant tissues, which might be one of the factors contributing to the discrepancy of different experimental systems.
Effects of TsRAVs overexpression on Arabidopsis seed germination and early seedling growth with or without NaCl
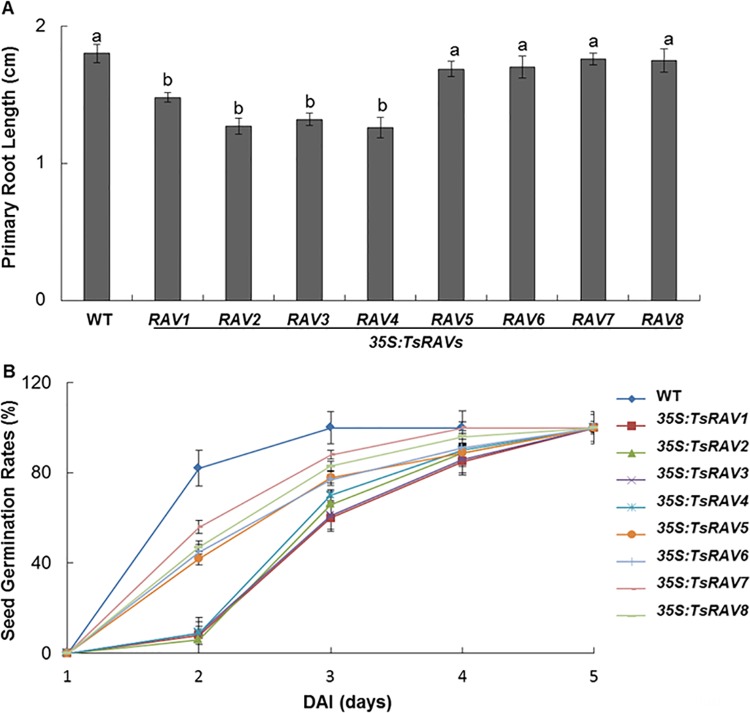
To understand the involvement of TsRAV genes in response to stressed conditions, we generated the “gain-of-function” transgenic lines by introducing each of the eight TsRAV genes individually into Arabidopsis under the control of the CaMV 35S promoter. For each TsRAV transgene, three homozygous overexpressing (OE) lines with the highest transgene expression level were selected for further analysis (S1 Fig). The previously reported inhibitory effects of AtRAV1 and AtRAV2 on plant growth [7,34,40,41] supported their roles as negative regulators during Arabidopsis growth and development [1]. In this work, we observed severe root growth retardation in 35S:A-TsRAVs transgenic lines but not in 35S:B-TsRAVs lines (Fig 3A). The only morphological defect detected in the vegetative and floral tissues was the narrower and longer leaves shown by 35S:TsRAV7 transgenic plants (S2 Fig).
Fig 3. Phenotypic characterization of 35S:TsRAVs transgenic Arabidopsis plants under normal conditions.
(A) Primary root length of 7-d-old 35S:TsRAVs transgenic Arabidopsis seedlings grown on 1/2 MS media. Each data bar represents the means ± SE of three replicates. More than 50 seedlings were measured for each replicates. Different letters indicate significant differences among means (P<0.05 by Tukey’s test). (B) Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds during a 5-day period on normal 1/2 MS media. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicated.
We then compared the germination rates of the TsRAVs-overexpressing transgenic seeds with that of wild type seeds. With wild type seeds basically completed germination within 2 days after imbibition (DAI), transgenic seeds overexpressing TsRAVs were inhibited in germination process to different extents, and the germination of A-TsRAVs transgenic seeds was much more repressed than that of B-TsRAVs transgenic seeds (Fig 3B). This difference was most significant at DAG 2 with the average germination rate of A-TsRAV transgenic lines as 8% compared to that of B-TsRAVs as 47% and that of WT as 82% (Fig 3B). Previously, the RAV1-overexpressing transgenic Arabidopsis have been reported to show normal seed germination pattern compared with wild type in the absence of salt [4], whereas our results indicated that even without any stress, the germination rates of TsRAVs overexpressing lines have already been reduced significantly.
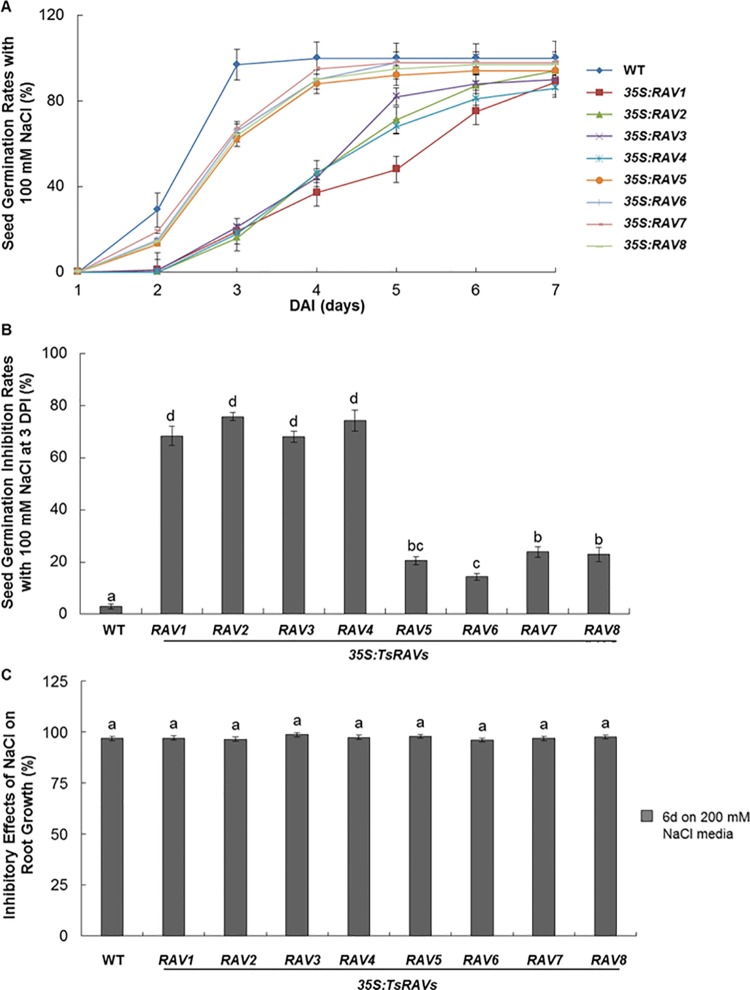
Salts at high concentration could inhibit the germination of Arabidopsis and ABA plays a role in this process [42]. When wild type and transgenic seeds were germinated on media containing 50, 75 or 100 mM NaCl, all types of TsRAVs transgenic seeds experienced a considerable reduction in their germination rates (S3 Fig and Fig 4). However, the inhibitory effect of A-TsRAVs overexpression on seed germination was much more significant than that of B-TsRAVs overexpression (S3 Fig and Fig 4). At DPI 3 when nearly all WT seeds were germinated on the salt-containing media, the germination rates of B-TsRAVs and A-TsRAVs transgenic seeds reached 60% and 20%, respectively (Fig 4A and 4B), indicating that as negative regulators of seed germination process, A-TsRAVs were much more competent than B-TsRAVs. A previous work by Li and co-workers also reported that the inhibition effect of NaCl on the germination of RAV transgenic seeds was greater than on that of wild type seeds, although that they did not detect any significant difference in their germination rates on normal 1/2 MS medium [18]. In summary, our data showed that overexpression of RAV genes did not enhance the salt tolerance of transgenic plants at the seed germination stage, which was contrary to the report that pepper RAV1 improved salt tolerance when overexpressed in transgenic Arabidopsis lines [10].
Fig 4. NaCl sensitivity of 35S:TsRAVs transgenic Arabidopsis plants.
(A) Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 100 mM NaCl. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate. (B) Inhibitory effect of 100 mM NaCl on 35S:TsRAVs transgenic Arabidopsis seed germination rates. Each data bar represents the mean ± SE of three replicates. More than 50 seedlings were measured in each replicate. Different letters indicate significant differences among means (P<0.05 by Tukey’s test). (C) Inhibitory effect of 200 mM NaCl on 35S:TsRAVs transgenic Arabidopsis seedling root elongation. Seedlings were grown on normal media for 5 days before being transferred onto 1/2 MS medium with 200 mM NaCl and grown for other 6 days. Each data bar represents the mean ± SE of three replicates. More than 50 seedlings were measured in each replicate. Different letters indicate significant differences among means (P<0.05 by Tukey’s test)
We then determined whether or not TsRAVs-overexpressing seedlings have improved salt tolerance during early developmental stage by transferring 5-day-old seedlings from normal 1/2 MS media to NaCl-containing media. As described earlier, under normal conditions, 35S:A-TsRAVs lines had shorter root length whereas 35S:B-TsRAVs lines had similar root length as WT. After transferring to medium supplemented with 200 mM NaCl, both WT and TsRAVs OE lines were inhibited by about 96% in their primary root length (S4 Fig and Fig 4C). However, when the inhibitory effect of NaCl on root elongation was calculated against their respective performance on normal media, it could be seen that all TsRAVs transgenic plants responded to NaCl to a similar extent (Fig 4C), indicating overexpression of A-TsRAVs and B-TsRAVs in Arabidopsis did not change the sensitivity of transgenic plants to salt.
Effects on germination and early seedling growth of TsRAVs-transgenic Arabidopsis with or without ABA
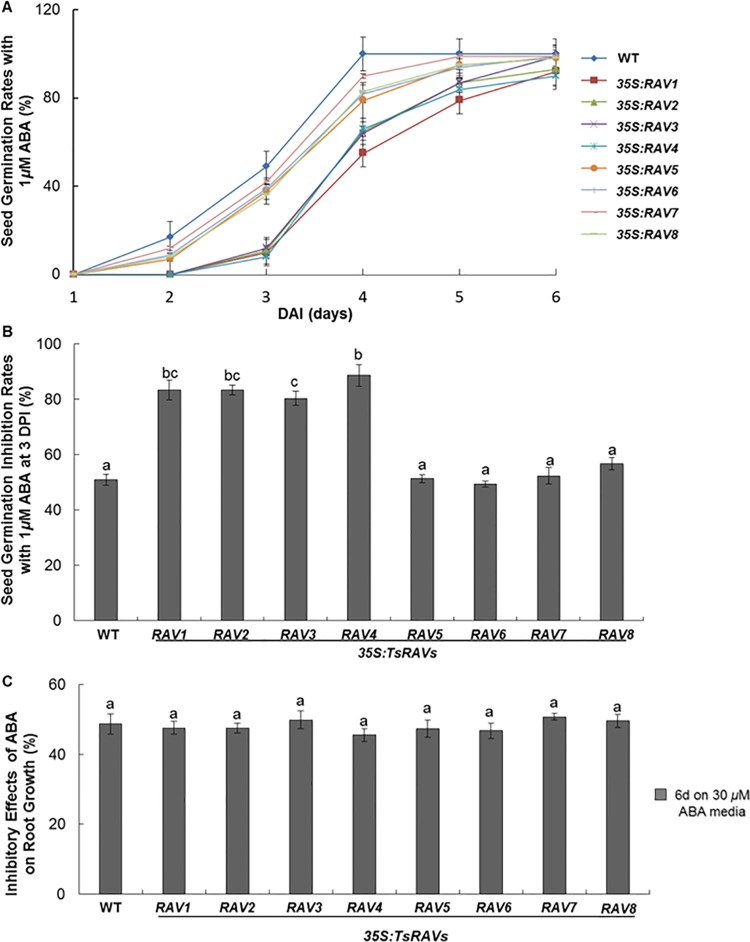
Abscisic acid (ABA) is a key phytohormone regulating many important plant growth and developmental events [39,43]. In the developing embryo, ABA accumulation regulates seed development, storage product accumulation, seed maturation, and seed dormancy [39,44]. The ABA content follows a rapid decline during imbibition process [45,46] and exogenous ABA inhibits seed germination and early seedling growth [38,39]. In the available literature, contrary results existed about the effect of RAV overexpression on the sensitivity of transgenic plant towards ABA inhibition during seed germination [4]. In this work, with the presence of 1 μM ABA, seed germination was significantly inhibited in WT and all transgenic lines (Fig 5A). With the germination rate at DAG 3 of WT seeds changed from 100% to 46%, the average germination rate of A-TsRAVs and B-TsRAVs transgenic seeds dropped from 64% to 10% and from 81% to 38%, respectively (Fig 5A). When the inhibitory effects of ABA were calculated against their normal germination rates, B-TsRAVs transgenic seeds were affected to a same extent as WT seeds, but to a much less extent than A-TsRAVs transgenic seeds (Fig 5B). Our work was consistent with the previous report that exogenous ABA inhibited the germination of both wild type and GhRAV1 overexpression transgenic seeds with a much greater degree of inhibition observed in the transgenic seeds than in wild type [18]. However, results contrary to our data also existed and a recent work revealed that RAV1-overexpressing lines showed strong ABA-insensitive phenotypes during seed germination and early seedling development, whereas RAV1-underexpressing lines were more sensitive to ABA than wild-type plants [15].
Fig 5. ABA sensitivity of 35S:TsRAVs transgenic Arabidopsis plants.
(A) Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 1 μM ABA. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate. (B) Inhibitory effect of 1 μM ABA on 35S:TsRAVs transgenic Arabidopsis seed germination rates. Each data bar represents the mean ± SE of three replicates. More than 50 seedlings were measured in each replicate. Different letters indicate significant differences among means (P<0.05 by Tukey’s test). (C) Inhibitory effect of 30 μM ABA on 35S:TsRAVs transgenic Arabidopsis seedling root elongation. Seedlings were grown on normal media for 5 days before being transferred onto 1/2 MS medium with 30 μM ABA and grown for other 6 days. Each data bar represents the mean ± SE of three replicates. More than 50 seedlings were measured in each replicate. Different letters indicate significant differences among means (P<0.05 by Tukey’s test).
Since ABA inhibits the growth of Arabidopsis seedlings, we also investigated the ABA sensitivity of various transgenic lines in terms of seedling root growth. After transferring onto vertical agar plates supplemented with 30 μM ABA, compared to the root growth of their respective untreated controls, both wild-type and all TsRAVs transgenic lines was similarly inhibited by about 50% (S4 Fig and Fig 5C), implied that RAV transcription factors might not play a direct role in ABA signaling during early seedling development. A previous work showed that 10 μM ABA could strongly inhibit the root growth of CaRAV1 overexpressing transgenic plants, but did not affect that of wild-type plants [10]. However, in another work, 10-day-old seedlings of wild type, rav mutant and RAV2-, RAV3-overexpressing Arabidopsis plants showed a similar reduction in root growth when exposed to 1 μM of ABA [4], which demonstrated a normal responsiveness to ABA in terms of root growth inhibition as described for TsRAVs in our work. Interestingly, in the same work, the root growth of the AtRAV1-overexpressing plants was not inhibited by ABA treatment despite of their shorter initial root length, so the authors hypothesized that only RAV1 may be involved with the regulation of ABA sensitivity in Arabidopsis [4]. In contrast, data in this work indicated that overexpression of all 8 TsRAV genes did not alter the responsiveness of transgenic plants towards ABA and an altered ABA-sensitivity only occurred to the 35S:A-TsRAVs transgenic plants at the seed germination stage.
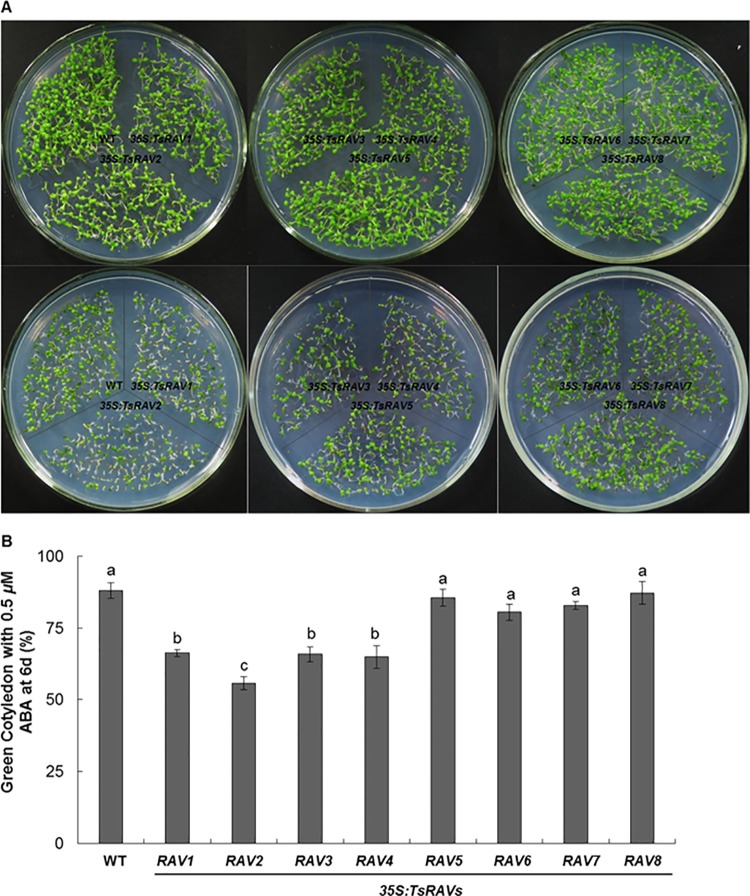
It has been reported that when grown on MS plus ABA, the RAV1-overexpressing plants showed higher cotyledon-greening percentage than wild-type plants, and much lower cotyledon-greening percentages have been reported for RAV RNAi lines and rav1 mutants [15], so we also determined the green cotyledon percentages of both wild type and TsRAVs transgenic lines in the current study. As shown in Fig 6A, there was no significant difference in green cotyledon rate between TsRAVs transgenic lines and wild type on normal 1/2 MS media, just as described previously [15]. In the presence of exogenous ABA, the green seedling rates of both wild type and TsRAVs overexpressing lines were significantly decreased. Compared to WT and B-TsRAVs transgenic lines, A-TsRAVs transgenic lines had significantly fewer open and green leaves after 6 days (Fig 6A). After being treated by 0.5 μM ABA for 6 days, nearly 88% of wild type seedlings stayed green, while only 55–66% and 79–87% green seedlings was recorded for A-TsRAVs and B-TsRAVs transgenic lines, respectively (Fig 6B). Thus, the ABA-induced earlier leaf etiolation and lower chlorophyll content were much more severe in 35S:A-TsRAVs transgenic lines, suggesting that A-TsRAVs but not B-TsRAVs might be involved in the ABA-regulated photosynthesis and leaf senescence.
Fig 6. Cotyledon-greening analysis on 35S:TsRAVs transgenic Arabidopsis seedlings.
(A) Phenotypic comparison of wild-type and 35S:TsRAVs transgenic Arabidopsis seedlings after grown on normal 1/2 MS medium (upper panel) or on 1/2 MS medium with 0.5 μM ABA for 6 days (lower panel). (B) Cotyledon-greening percentages of 35S:TsRAVs transgenic Arabidopsis seedlings after grown on 1/2 MS medium with 0.5 μM ABA for 6 days. Each data bar represents the mean ± SE of three replicates. More than 100 seeds were measured in each replicate. Different letters indicate significant differences among means (P<0.05 by Tukey’s test).
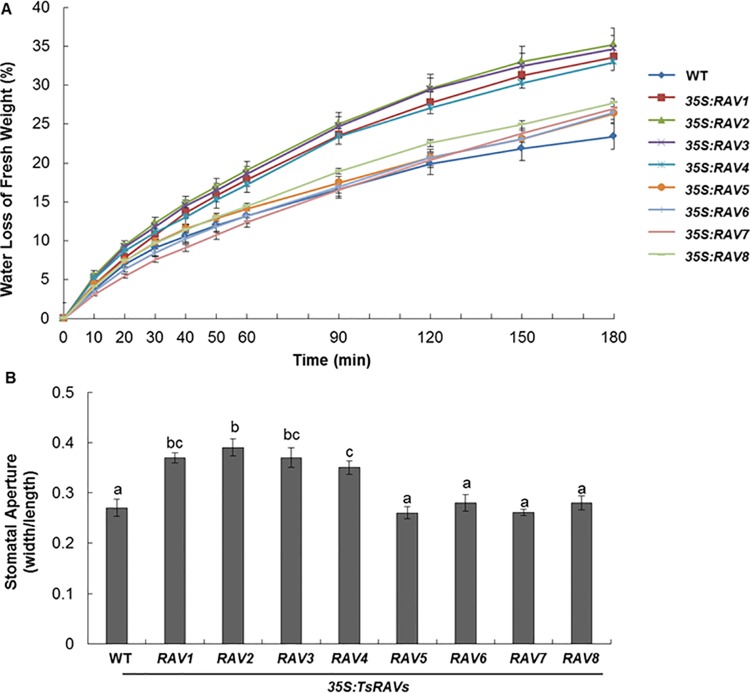
Water deficit tolerance of transgenic Arabidopsis overexpressing TsRAVs
Water stress can induce the synthesis of ABA within plants and ABA acts directly on guard cells to induce stomatal closure and minimize water loss through transpiration [47]. Previously, transgenic plants overexpressing pepper RAV1 have been shown to be more tolerant to water stress [10], whereas AtRAVs-overexpressing transgenic plants have been reported to exhibit higher transpirational water loss than wild type [4]. Thus, we cut leaves from the 4-week soil-grown 35S:TsRAVs transgenic plants, placed them on a filter paper in an ambient temperature and measured the transpirational water loss over a 3-hour period. Our results showed that 35S:A-TsRAVs experienced a significantly faster water loss than both wild type and 35S:B-TsRAVs plants (Fig 7A). Since water loss of leaves is associated with the ABA-regulated stomatal movement, which is one of the appropriate criteria to check the ABA sensitivity, we then analyzed the stomatal opening of TsRAVs-overexpressing transgenic plants in parallel with wild type plants. Under same conditions, a wider opening of stomata was detected in 35S:A-TsRAVs transgenic plants than both wild type and 35S:B-TsRAVs transgenic plants (Fig 7B), indicating that constitutive overexpression of A-TsRAVs, but not B-TsRAVs could result in a wider stomatal opening, an increased transpiration rate, and a comprised drought tolerance, which was consistent with the previous notion that a faster water loss in RAV1-overexpressing plants was caused by the incapacity of stomata of to respond to ABA [4].
Fig 7. Different water loss rates of wild-type and 35S:TsRAVs transgenic Arabidopsis plants.
(A) Weight loss in fresh leaves under water deficit of 4-week-old wild-type and 35S:TsRAVs transgenic Arabidopsis plants. Each data bar represents the mean ± SE of three replicates. (B) Width/length ratio of the rosette leaf stomatal aperture of 4-week-old wild-type and 35S:TsRAVs transgenic Arabidopsis plants. Each data bar represents the mean ± SE of three replicates and different letters indicate significant differences among means (P<0.05 by Tukey’s test).
In summary, based on their distinctively different sequence characters and responses to salt and ABA stresses, RAV transcriptional factors originated from Thellungiella salsuginea could be divided into two major groups with four members within each group. Under normal conditions, 35S:A-TsRAVs lines exhibited a moderately shortened root length, which indicated the roles of TsRAV1-4 as negative growth regulators. Base on the performance of each TsRAVs transgenic line under various ABA-related abiotic stresses, functional analyses revealed the highly consistent results among four members within a same group and distinctively different results between two groups. Compared to 35S:A-TsRAVs plants, 35S:B-TsRAVs plants only exerted a low degree or little inhibitory effect on seed germination and seedling growth, suggesting that ABA signaling pathway might have only been perturbed in 35S:A-TsRAVs plants, but not in 35S:B-TsRAVs plants. Moreover, the salt and ABA hypersensitivity conferred by A-TsRAVs overexpression only occurred to the germination stage, but not to the young seedling stage, indicating their inhibitory effect was modulated developmentally. Under water deficit conditions, 35S:A-TsRAVs plants, but not 35S:B-TsRAVs plants, became more susceptible to transpiration water loss than control plants due to their impaired stomatal aperture regulation. Therefore, despite of the presence of a B3 domain and a BRD repression domain in all eight TsRAVs, the presence or absence of a nuclear localization sequence, the core sequences within the BRD domains as well as some highly conserved group-specific residues might together affect their DNA-binding potentials and activities. Data present hereby suggests that two subsets of TsRAVs perform different physiological roles during plant growth, especially in ABA-mediated signaling pathway, and they might be regulated by different mechanisms and/or in turn modify different sets of downstream target genes.
Supporting Information
Characterization of 35S:TsRAVs transgenic Arabidopsis by semi qRT-PCR. Four independent overexpression lines with similar level of transgene expression were chosen for the following analysis.
(TIF)
(TIF)
Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 50 mM NaCl. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate. Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 75 mM NaCl. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate.
(TIF)
After germination, seedlings were first grown on normal media for 5 days before being transferred onto 1/2 MS medium with 200 mM NaCl or 30 μM ABA and grown for other 6 days.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by the National Transgenic Major Project (2014ZX0800404B) and Central Government Shared Marine Appropriation Expenditure by State Oceanic Administration (HYSYJ-HB-2010-BH-3).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Transgenic Major Project (2014ZX0800404B) and Central Government Shared Marine Appropriation Expenditure by State Oceanic Administration (HYSYJ-HB-2010-BH-3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu M, Kang HK, Son SH, Kim SK, Nam KH (2014) A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol 55: 1892–1904. 10.1093/pcp/pcu118 [DOI] [PubMed] [Google Scholar]
- 5.Magnani E, Sjolander K, Hake S (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16: 2265–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Soltis PS, Wall K, Soltis DE (2006) Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol 23: 107–120. [DOI] [PubMed] [Google Scholar]
- 7.Osnato M, Castillejo C, Matias-Hernandez L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- 8.Zhao L, Hao D, Chen L, Lu Q, Zhang Y, Li Y, et al. (2012) Roles for a soybean RAV-like orthologue in shoot regeneration and photoperiodicity inferred from transgenic plants. J Exp Bot 63: 3257–3270. 10.1093/jxb/ers056 [DOI] [PubMed] [Google Scholar]
- 9.Feng JX, Liu D, Pan Y, Gong W, Ma LG, Luo JC, et al. (2005) An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol 59: 853–868. [DOI] [PubMed] [Google Scholar]
- 10.Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK (2006) Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61: 897–915. [DOI] [PubMed] [Google Scholar]
- 11.Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Wang Z, Wang X, Dong J, Ren J, Gao H (2009) Isolation and characterization of GoRAV, a novel gene encoding a RAV-type protein in Galegae orientalis. Genes Genet Syst 84: 101–109. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang J, Sun CC, Zhou XR, Xiong AS, Zhang J (2011) Isolation and characterization of an AP2/ERF-RAV transcription factor BnaRAV-1-HY15 in Brassica napus L. HuYou15. Mol Biol Rep 38: 3921–3928. 10.1007/s11033-010-0508-1 [DOI] [PubMed] [Google Scholar]
- 14.Mittal A, PhD dissertation Texas Tech University (2012) Overexpression and interactions of Arabidopsis thaliana RAV1 (Related to Abscisic Acid Insensitive 3/ Viviparous 1), RAV2, RAV2-Like and ABI5 in transgenic cotton (Gossypium hirsutum): Effects on drought avoidance and fiber quality: 161.
- 15.Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF (2014) Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J 80: 654–668. 10.1111/tpj.12670 [DOI] [PubMed] [Google Scholar]
- 16.Mittal A, Gampala SS, Ritchie GL, Payton P, Burke JJ, Rock CD (2014) Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J 12: 578–589. 10.1111/pbi.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CW, Su RC, Cheng CP, Sanjaya, You SJ, Hsieh TH, et al. (2011) Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol 156: 213–227. 10.1104/pp.111.174268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XJ, Li M, Zhou Y, Hu S, Hu R, Chen Y, et al. (2015) Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One 10: e0118056 10.1371/journal.pone.0118056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amtmann A (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12. 10.1093/mp/ssn094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, et al. (2004) Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, et al. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–215. 10.1093/nar/gkn785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37: D205–210. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54. [DOI] [PubMed] [Google Scholar]
- 29.Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Kong L, Zhang M, Lv Y, Liu Y, Zou M, et al. (2013) Genome-Wide Identification, Phylogeny, Evolution and Expression Patterns of AP2/ERF Genes and Cytokinin Response Factors in Brassica rapa ssp. pekinensis. PLoS ONE 8: e83444 10.1371/journal.pone.0083444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci U S A 94: 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda M, Ohme-Takagi M (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975. 10.1093/pcp/pcp048 [DOI] [PubMed] [Google Scholar]
- 34.Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343. 10.1016/j.cub.2008.07.075 [DOI] [PubMed] [Google Scholar]
- 35.Lu Q, Zhao L, Li D, Hao D, Zhan Y, Li W (2014) A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS One 9: e89145 10.1371/journal.pone.0089145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee D, Polisensky DH, Braam J (2005) Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytol 165: 429–444. [DOI] [PubMed] [Google Scholar]
- 37.Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, et al. (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957. 10.1093/jxb/erq206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkelstein RR, Lynch TJ (2000) Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol 122: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14 Suppl: S15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu YX, Wang YX, Liu XF, Li JY (2004) Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res 14: 8–15. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Luo Q, Yang C, Han Y, Li W (2008) A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227: 1389–1399. 10.1007/s00425-008-0711-7 [DOI] [PubMed] [Google Scholar]
- 42.Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–355. 10.1016/j.cub.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nambara E, Marion-Poll A (2003) ABA action and interactions in seeds. Trends Plant Sci 8: 213–217. [DOI] [PubMed] [Google Scholar]
- 45.Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, et al. (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488. [DOI] [PubMed] [Google Scholar]
- 46.Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8: 183–187. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard Cell Signal Transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of 35S:TsRAVs transgenic Arabidopsis by semi qRT-PCR. Four independent overexpression lines with similar level of transgene expression were chosen for the following analysis.
(TIF)
(TIF)
Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 50 mM NaCl. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate. Germination rates of 35S:TsRAVs transgenic Arabidopsis seeds on 1/2 MS media with 75 mM NaCl. Each data bar represents the means ± SE of three replicates. More than 100 seeds were measured in each replicate.
(TIF)
After germination, seedlings were first grown on normal media for 5 days before being transferred onto 1/2 MS medium with 200 mM NaCl or 30 μM ABA and grown for other 6 days.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.