Abstract
Endocytosis and postendocytic sorting of epidermal growth factor (EGF) receptor (EGFR) are the major regulators of EGFR signaling. EGFR endocytosis and ubiquitin-dependent lysosomal targeting are also considered to be the prototypic experimental system for studying the molecular mechanisms of stimulus-induced and constitutive endocytic trafficking. Therefore, elucidation of the mechanisms of EGFR endocytosis and its regulation of the signaling network is essential not only for better understanding of the EGFR biology but also for defining general regulatory principles in the endocytosis system. Comprehensive analysis of these mechanisms requires quantitative and physiologically relevant methodological approaches for measuring the rates of EGFR internalization, degradation, and recycling. Basic experimental protocols described in this chapter cover a combination of single-cell microscopy and biochemical methods that are used to follow EGF-induced endocytosis of EGFR in real time, measure the kinetic rate parameters of EGFR internalization and recycling, and analyze EGF-dependent ubiquitination and degradation of EGFR.
INTRODUCTION
Epidermal growth factor (EGF) receptor (EGFR) plays an important role in the regulation of cell proliferation, differentiation, survival, and motility both in development and adulthood (Sibilia et al., 2007). At least six ligands for EGFR in addition to the best characterized EGF have been described (Henriksen, Grandal, Knudsen, van Deurs, & Grøvdal, 2013). Upon ligand binding to EGFR at the cell surface, receptors dimerize, which leads to activation of its intrinsic tyrosine kinase activity, and tyrosine phosphorylation of the cytoplasmic domain of the receptor as well as other cytoplasmic substrates (Lemmon & Schlessinger, 2010). These phosphorylation events trigger several signal transduction cascades ultimately leading to altered gene expression.
At the same time, activated EGFR is rapidly endocytosed through clathrin-dependent and clathrin-independent pathways. It is proposed that clathrin-mediated endocytosis of EGFR has limited capacity and is saturated by the excess of EGF: EGFR complexes at the cell surface (when high EGF concentrations are used) (Sorkin & Goh, 2009). Therefore, measurement of the EGFR internalization rates through clathrin pathway requires the use of low, physiological EGF concentrations. After internalization into early endosomes, EGF-receptor complexes are capable of recycling back to the plasma membrane but are also retained in endosomes and eventually sorted to late endosomes and lysosomes for degradation (Sorkin & Goh, 2009). EGFR ubiquitination by the E3 ligase Cbl is the key mechanism mediating lysosomal targeting of EGFR and many other endocytic cargos (Eden, Huang, Sorkin, & Futter, 2012; Hislop & von Zastrow, 2011; Weinberg & Drubin, 2014). The acceleration of internalization and lysosomal targeting of activated EGFR results in the reduction of EGFR protein levels and downregulation of EGFR-dependent signaling as part of the negative feedback regulation loop (Sorkin & von Zastrow, 2009).
The key role of EGFR trafficking in regulation of signaling processes underscores the importance of understanding the molecular mechanism of this trafficking. However, despite extensive studies for more than three decades, these mechanisms, in particular, those of the internalization step, remain elusive. Therefore, the use of standardized, universally accepted, and quantitative methodologies is vital for studying EGFR endocytosis in diverse experimental model systems. Analysis using a combination of such methodologies should allow careful reinterpretation and reconciliation of numerous contradictory experimental observations and proposed models of EGF endocytosis.
1. OBJECTIVES AND RATIONALE
Internalization rates of EGF-occupied EGFR were traditionally measured by monitoring the uptake of radiolabeled EGF (125I-EGF) in the cell. 125I-EGF is also used to measure the rate of recycling of internalized 125I-EGF:EGFR complexes back to the cell surface, and the rate of 125I-EGF degradation. Because the bulk of endosomal EGF:EGFR complexes remain intact in endosomes, methods involving 125I-EGF indirectly measure EGFR recycling and degradation. While 125I-EGF-based methods remain most sensitive and quantitative, combining these methods with optical microscopy and direct EGFR protein quantification assays is the most desirable approach to conduct the comprehensive analysis of EGFR endocytosis. Availability of various biologically active labeled derivatives of EGF, numerous antibodies, and genetically encoded fluorescent fusion proteins of EGFR makes such analysis to be highly feasible. Importantly, unprecedented increase in the sensitivity of microscopy imaging systems provides an opportunity to follow endocytosis of EGFR activated by low, physiological concentrations of fluorescent EGF in living cells and in real time. Therefore, in this chapter we focus on the description of single-cell microscopy analysis of EGFR endocytosis, 125I-EGF-based methods of measuring internalization and recycling rates, and simple methods of measuring EGF-induced degradation of EGFR and EGFR ubiquitination using immunoprecipitation and western blotting. Additionally, we demonstrate the utility of these methods for examining the effects of two chemical compounds, Dyngo-4a and primaquine (PQ), on internalization and recycling of EGFR, respectively. Dyngo-4a is thought to inhibit the activity of the large GTPase dynamin that is necessary for the scission of vesicles during clathrin-mediated and several types of clathrin-independent endocytosis (Ferguson & De Camilli, 2012; McCluskey et al., 2013; Mettlen, Pucadyil, Ramachandran, & Schmid, 2009). PQ has been shown to inhibit recycling of the transferrin receptor by an unknown mechanism (Raub & Newton, 1991; van Weert, Geuze, Groothuis, & Stoorvogel, 2000).
2. MATERIALS
2.1 REAGENTS
- EGFR-expressing cells:
- UMSCC2 (University of Michigan squamous cell carcinoma of head and neck)
- HeLa cells
- Mouse NIH 3T3 cells stably expressing human EGFR (NIH3T3/EGFR)
Dulbecco minimum essential medium (DMEM)
Fetal bovine serum (FBS)
Normal calf serum
Bovine serum albumin (BSA)
Binding medium (DMEM plus 0.1% (w/v) BSA)
HEPES buffer
Ca2+, Mg2+-free PBS (CMF-PBS)
Tween-20
pH 42.3–4.5 acetate buffer (0.2 M sodium acetate buffer, 0.5 M NaCl)
pH 2.5–2.8 acetate buffer (0.2 M acetic acid, 0.5 M NaCl)
Trichloroacetic acid (TCA)
Phosphotungstic acid (PTA)
TCA/PTA (10%/2% w/v) in water
Phosphate buffer saline (PBS)
Tris buffer saline (TBS)
Sample buffer 1× (62.5 mM Tris pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, bromophenol blue)
TGH buffer (1% (v/v) Triton X-100, 10% (v/v) glycerol, 50 mM HEPES pH 7.4)
Lysis buffer (TGH buffer, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovandate,10 mM N-ethylmaleimide, 10 µg/mL aprotinin, 10 µg/mL leupeptin, and protease inhibitors)
Recombinant human culture-grade EGF; receptor-grade mouse EGF (for labeling); 125I-EGF; EGF-Rhodamine (EGF-Rh); Alexa Fluor® 647-labeled EGF complex (EGF-A647).
Wheat germ agglutinin (WGA) Oregon Green
Paraformaldehyde (16% solution; electron microscopy grade).
Cycloheximide
DMSO
Dyngo-4a
ProLong® Antifade mounting media
Collagen-coated glass bottom 35 mm MatTek dishes
6-, 12-, and 24-well plastic cell culture plates
35 and 60 mm plastic cell culture dishes
Mouse monoclonal antibody 528 to EGFR (ATTC)
Rabbit polyclonal antibody 1005 to EGFR (Santa Cruz)
Mouse monoclonal antibody P4D1 to ubiquitin (Santa Cruz)
2.2 MAJOR INSTRUMENTS
Zeiss Axio Observer Z1 inverted fluorescence microscope equipped with 63× Plan Apo PH NA 1.4 objective, computer-controlled spherical aberration correction unit, Yokogawa CSU-X1 spinning disk, Photometrics Evolve 16-bit EMCCD camera, environmental chamber, piezo stage controller and lasers (405, 445, 488, 515, 561, and 640 nm lines), all controlled by SlideBook software (Intelligent Imaging Innovation, Denver, CO).
Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE)
Cobra Gamma-counter (Packard Instruments)
3. BASIC TECHNIQUES
3.1 CELL CULTURE
Cells expressing EGFR are grown and maintained in DMEM supplemented with serum at 37 °C, humidity and 5% CO2. 10% (v/v) and 5% (v/v) FBS is used to grow HeLa and UMSCC2, respectively. NIH 3T3/EGFR cells are grown in DMEM supplemented with 10% (v/v) NCF. Cells are plated at 40,000–50,000 cells/cm2 and grown to a desired confluency. Prior to any experiment, cells are serum-starved to minimize basal EGFR activity. Starvation is typically performed in binding medium for 6–16 h.
3.2 SDS-PAGE AND WESTERN BLOT
Protein are resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% acrylamide gels that are used to allow optimal resolution in the high-molecular weight range (150 kDa and higher) to clearly detect EGFR (170 KDa) and its slowly migrating, posttranslationally modified forms. Proteins are transferred to the nitrocellulose membrane at 500 mA for 3.5 h. The membrane is then blocked with TBS containing 0.02% (v/v) Tween-20 and 5% (w/v) nonfat milk (blocking solution) either 1 h at room temperature or overnight at 4 °C with constant agitation. Immunoblotting with primary and secondary antibodies is performed in TBS/Tween buffer following manufacturer’s recommendations. The blots are washed with TBS/Tween after incubations with antibodies. Membranes are scanned using Odyssey LI-COR imager, and the data are analyzed using LI-COR software.
3.3 FLUORESCENCE MICROSCOPY
MatTek dishes (living cells) or subject glass with mounted coverslips (fixed cells) are placed onto the microscope stage adaptor. Constant temperature (37 °C), humidity, and 5% CO2 atmosphere are maintained throughout the duration of live-cell imaging. The capture integration time and other imaging parameters (number of optical section in the z-stack, stepsize between sections, time intervals between 3-D images during time-lapse imaging) are set to be identical during image acquisition of experimental variants and to allow nonsaturated fluorescence signals.
4. EXPERIMENTAL STRATEGIES
4.1 IMAGING EGFR ENDOCYTOSIS IN LIVING CELLS BY FLUORESCENCE MICROSCOPY, AND THE EFFECTS OF DYNAMIN INHIBITOR ON THIS PROCESS
4.1.1 Theory
Fluorescently labeled EGFR ligands like EGF conjugated to Rhodamine (EGF-Rh) are used to visualize endocytosis of ligand-occupied EGFR in single cells. This type of experiments was first performed more than three decades ago (Haigler, Ash, Singer, & Cohen, 1978). However, only recently, new microscopy systems, such as spinning disk confocal microscope, that are capable of continuous 3-D time-lapse imaging of endocytosis of labeled EGF at physiological concentrations (1–4 ng/mL) in living cells without significant photobleaching and phototoxicity, have become available. Imaging the entire cell in 3-D, rather than a single confocal or optical plane is necessary to obtain truly quantitative information about the kinetics of EGF endocytosis in addition to the information about the morphology of various EGF-containing compartments in different parts of the cell. Certainly, endocytosis is a highly temperature-dependent process, and therefore, the assay is carried out at 37 °C.
4.1.2 Experimental protocol
Cells are grown in 35 mm MatTek dishes until 50–60% confluency and serum-starved in binding medium.
The dish with 2 mL of medium is placed onto the microscopic stage in the environmental chamber. Time-lapse imaging is started by acquiring a z-stack of 10–20 x–y confocal images (0.2–0.4 µm stepsize) through 561 nm (EGF-Rh) or 640 nm (EGF-A647) channels with the desired time intervals and image capture time of 100 ms or less.
After two to three time points, 200 µL of prewarmed binding medium containing fluorescent EGF and additional HEPES is rapidly added to the dish to ensure the final concentration of labeled EGF and HEPES buffer of 2–10 ng/mL and 10 mM respectively.
3-D imaging is continued during EGF injection and further continued for the desired time, typically, 5–60 min.
4.1.3 Variation: examination of the effect of Dyngo-4a on EGFR endocytosis
Cells are grown in 35 mm MatTek dishes or on 12 mm round coverslips placed in 24-well dishes until ~50–70% confluency and serum-starved.
Cells are washed with DMEM and preincubated with 30 µM of Dyngo-4a or vehicle (DMSO) in DMEM for 30 min at 37 °C.
Cells in MatTek dishes are used for live-cell imaging of fluorescent EGF as in Steps 2–4 (Section 4.1.2) above.
Cells on coverslips are incubated in binding medium containing additional 10 mM HEPES and fluorescent EGF (2–10 ng/mL) in CO2 incubator or water bath.
At desired time points (5–60 min), EGF endocytosis is stopped by placing the plates firmly on ice and washing cells twice with ice-cold PBS.
Coverslips are fixed with freshly prepared paraformaldehyde (4%) in PBS for 30 min at 4 °C and mounted on subject glass.
Optional: After fixation and three 5-min washes with PBS, the cells are incubated with 0.5 µg/mL WGA-Oregon Green in PBS for 10 min at room temperature to label the plasma membrane.
Coverslips are then washed with PBS and mounted.
3-D confocal images through 488 nm (Oregon Green), 561 nm (EGF-Rh), or 640 nm (EGF-A647) channels are captured. Image acquisition times and other parameters of imaging depend on the cell type, EGFR levels and other experimental factors.
4.1.4 Results
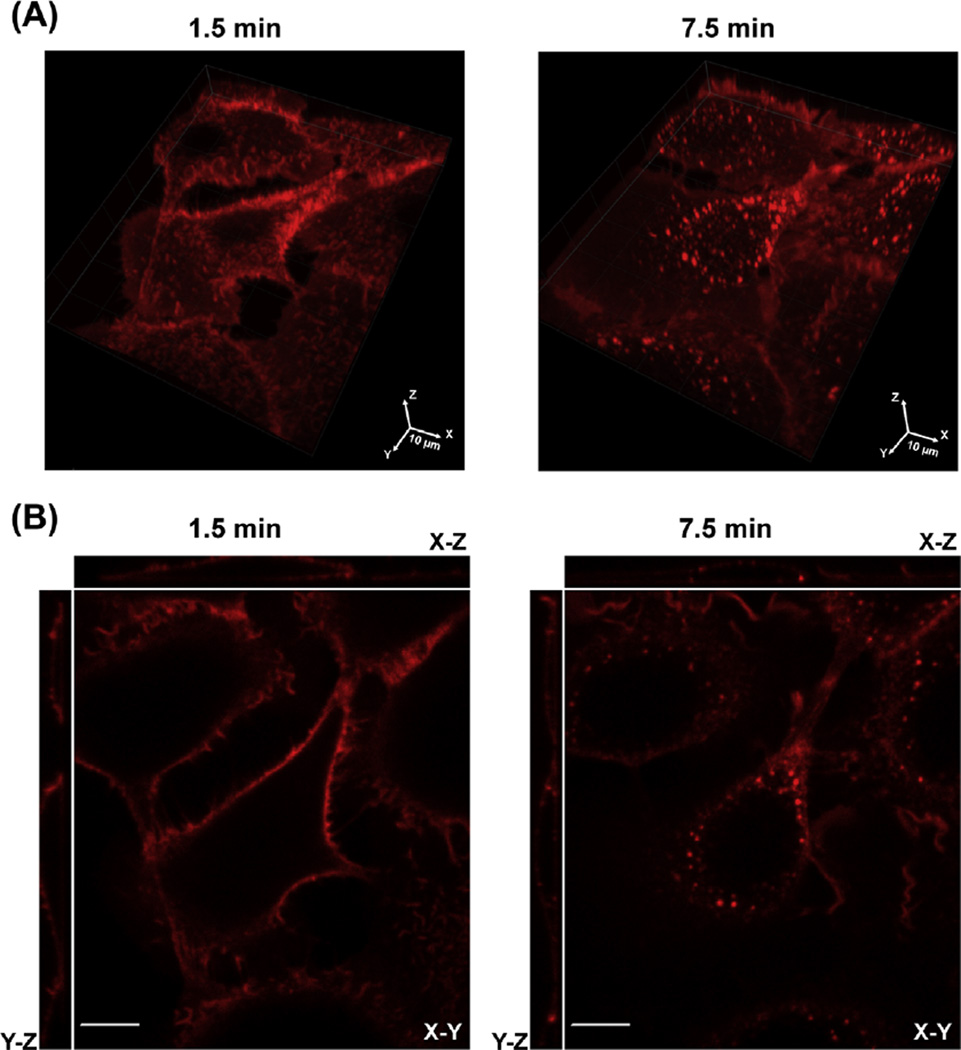
In the example presented in Figure 1, living UMSCC2 cells are imaged by capturing 20 x–y images with 400 nm z-plane stepsize every 90 s for 15 min. Addition of EGF-Rh (4 ng/mL) to the cells resulted in rapid binding of EGF-Rh to surface receptors (1.5 min; Figure 1) and accumulation of EGF-Rh in endosomes (7.5 min; Figure 1). 3-D volume (x–y–z) image (Figure 1(A)) that allows visualization of the entire image stack and the “3-D view” image (Figure 1(B)) that shows single x–y, x–z, and y–z confocal planes are presented. These images demonstrate localization of EGF-Rh mostly at the cell surface at the 1.5-min time point (cell perimeter, filopodia, ruffles). Bright vesicular peripheral and perinuclear compartments containing EGF-Rh observed on the 7.5-min image are indicative of predominantly endosomal localization of EGF-Rh, although some diffusely distributed fluorescence in the cell perimeter (plasma membrane) is also observed.
FIGURE 1. Time-lapse imaging of UMSCC2 cells stimulated with EGF-Rhodamine (EGF-Rh).
Cells were imaged before and after EGF-Rh (4 ng/mL) stimulation for 15 min. Z-stacks of 20 x–y images (400 nm z-stepsize) of Rhodamine fluorescence were acquired every 1.5 min. 1.5 and 7.5 min images are shown. 3-D volume view image (A) and “3-D view” image (single confocal x–y, y–z, and x–z sections) are presented (B). Scale bars, 10 µm. (See color plate)
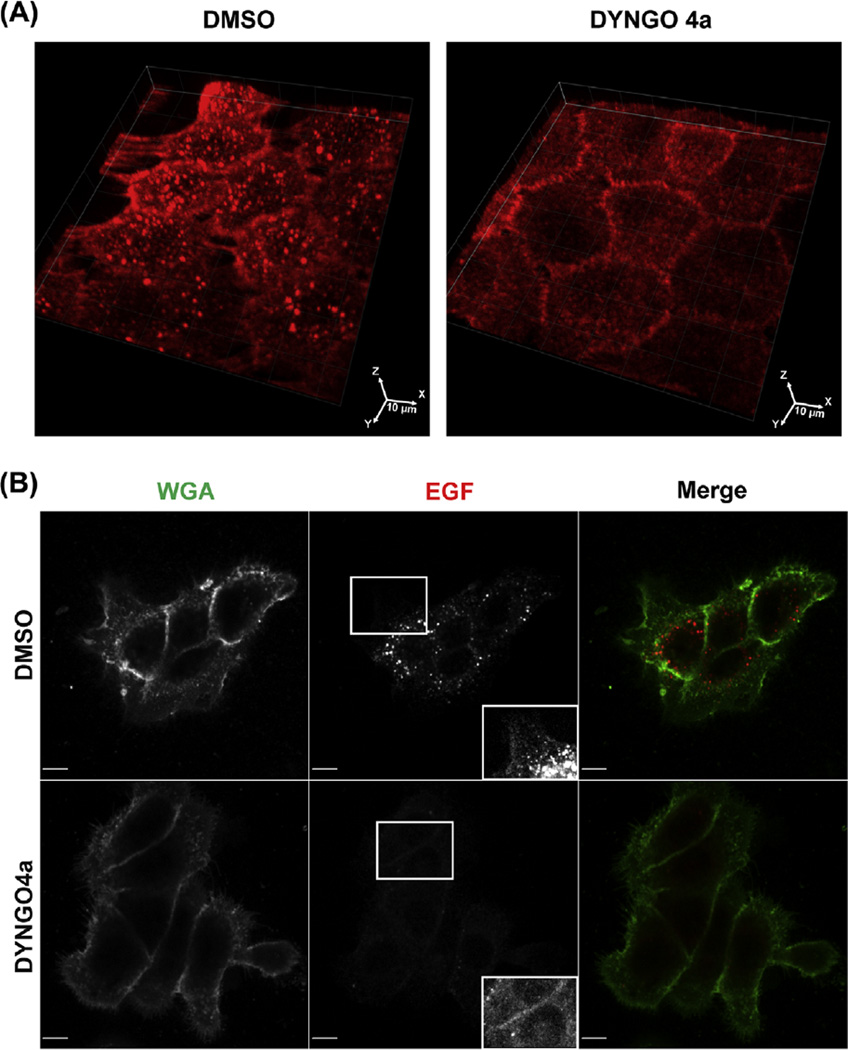
The strong inhibitory effect of Dyngo-4a on EGF-Rh internalization in UMSCC2 cells examined in living cells is demonstrated in Figure 2(A). The same effect is observed in fixed cells in which the cell surface was labeled with WGA-Oregon Green (Figure 2(B)). In these images of cells incubated with EGF-Rh for 15 min, most EGF-Rh containing endosomes do not overlap with WGA-Oregon Green staining in control cells and are practically absent in Dyngo-4a treated cells.
FIGURE 2. Effect of Dyngo-4a on EGF-Rhodamine (EGF-Rh) endocytosis in UMSCC2 cells.
(A) Cells were incubated 30 min with Dyngo-4a 30 µM or vehicle (DMSO) and then imaged before and after EGF-Rh (4 ng/mL) stimulation as in Figure 1. 3-D volume view images of control and Dyngo-4a pretreated cells that were incubated with EGF-Rh for 5 min are shown. (B) Cells preincubated or not with Dyngo-4a were incubated with EGF-Rh for 15 min in the presence of inhibitor or vehicle, fixed and labeled with wheat germ agglutinin (WGA)-Oregon Green as described in the text. Insets show high magnification images of the peripheral cell regions marked by white rectangles. All image acquisition parameters and intensity scales are the same for corresponding images of control and Dyngo-4a-treated cells. Scale bars, 10 µm. EGF, epidermal growth factor. (See color plate)
4.1.5 Considerations
Various preparations of fluorescently labeled EGF are commercially available. Direct conjugates of EGF with fluorochromes are preferable to use as compared to preparations consisting of the complex of biotinylated EGF with streptavidin, such as EGF-A647. Such complexes may be multivalent, and thus leading to receptor cross-linking. Actual concentration of EGF in these preparations could be difficult to determine. Among direct EGF conjugates, conjugates to fluorophores with high brightness and photostability, such as Rhodamine or TexasRed, are preferable compared with less stable FITC.
Endosomes and other compartments move rather rapidly, and therefore can move during an acquisition of a 3-D image. Therefore, shortest image capture times and minimal number of planes are recommended.
The activity of Dyngo-4a is dramatically reduced when albumin or serum is present ((McCluskey et al., 2013) and our observations). Therefore, BSA is omitted during preincubation of cells with this inhibitor.
We noticed that additional incubations of EGF-Rh labeled cells after fixation result in the substantial loss of EGF-Rh bound to the cell surface due to poor fixation of this conjugate by formaldehyde.
4.2 MEASUREMENT OF EGFR INTERNALIZATION RATES USING 125I-EGF
4.2.1 Theory
The internalization assay is performed as a short time-course of 125I-EGF uptake (less than 5–6 min depending on cell type) to minimize the influence of recycling of 125I-EGF:EGFR complexes on the overall uptake. Because endocytosis is highly temperature-dependent, the assay is performed at 37 °C. The key step of this assay is stripping of surface 125I-EGF at low pH to separate the surface and internalized pools of 125I-EGF. Either Na+ acetate or glycine/HCl buffers (pH 2.5–2.8), the latter combined with 3 Murea, are effective in rapid removal of 125I-EGF bound to surface receptors without affecting intracellular 125I-EGF (Haigler, Maxfield, Willingham, & Pastan, 1980; Wiley & Cunningham, 1982).
4.2.2 Experimental protocol
Cells are plated into 12-well dishes and grown for 2–3 days until confluency.
125I-EGF-containing binding medium (0.5 mL per well) is prepared. Unlabeled EGF (50–100 M excess) is added to the half of the medium with 125I-EGF—this is the “nonspecific” medium. It is important to prepare some excess of the 125I-EGF-containing medium to save an aliquot for measuring the specific activity of 125I-EGF.
The dish is placed in the water bath at 37 °C, and two wells are quickly washed with warm (37 °C) binding media (~1 mL). 0.5 mL of prewarmed (37 °C) specific (not containing cold EGF) or nonspecific 125I-EGF containing media are added to two wells simultaneously, preferably, using two pipetteman, one in each hand. These cells correspond to the longest time point (3–6 min).
The same procedure (Step 3) is repeated with other pairs of wells at 30 s or 1-min intervals. Rapid handling of all steps of this procedure is critical because of the short incubations of cells with 125I-EGF.
After the last pair of wells has been incubated with 125I-EGF, the dish is placed on ice to stop endocytosis, and the radioactive media are rapidly aspirated. In a very short time-course experiments (0–3 min), the actual timing of 125I-EGF medium addition to cells are recorded because the fast pace of the assay often does not allow the precisely planned timing of incubations.
The monolayers are rapidly washed three times with ~1 mL/well ice-cold DMEM followed by vacuum aspiration to remove as much unbound 125I-EGF as possible after the last wash. This step is performed as quickly as possible to avoid significant dissociation of 125I-EGF from surface receptors during washes.
All wells are then incubated for ~5 min with the pH 2.8 Na+ acetate buffer (1 mL/well) at 4 °C to strip surface-bound 125I-EGF followed by with another short acetate buffer wash (0.5 mL/well). Both acidic washes are combined and transferred to γ-counter vials to determine the amount of surface-bound 125I-EGF.
The cells are lysed in 1 mL of 1 N NaOH for ~1 h at 37 °C to determine the amount of internalized 125I-EGF.
4.2.3 Variation: effect of inhibitors on 125I-EGF endocytosis
To test whether endocytosis is dynamin-dependent, the cells are preincubated with Dyngo-4a in the medium without BSA as described in Section 4.1.3, and the protocol further follows Steps 2–8 above. Dyngo-4a is also present in the 125I-EGF-containing uptake medium.
4.2.4 Results
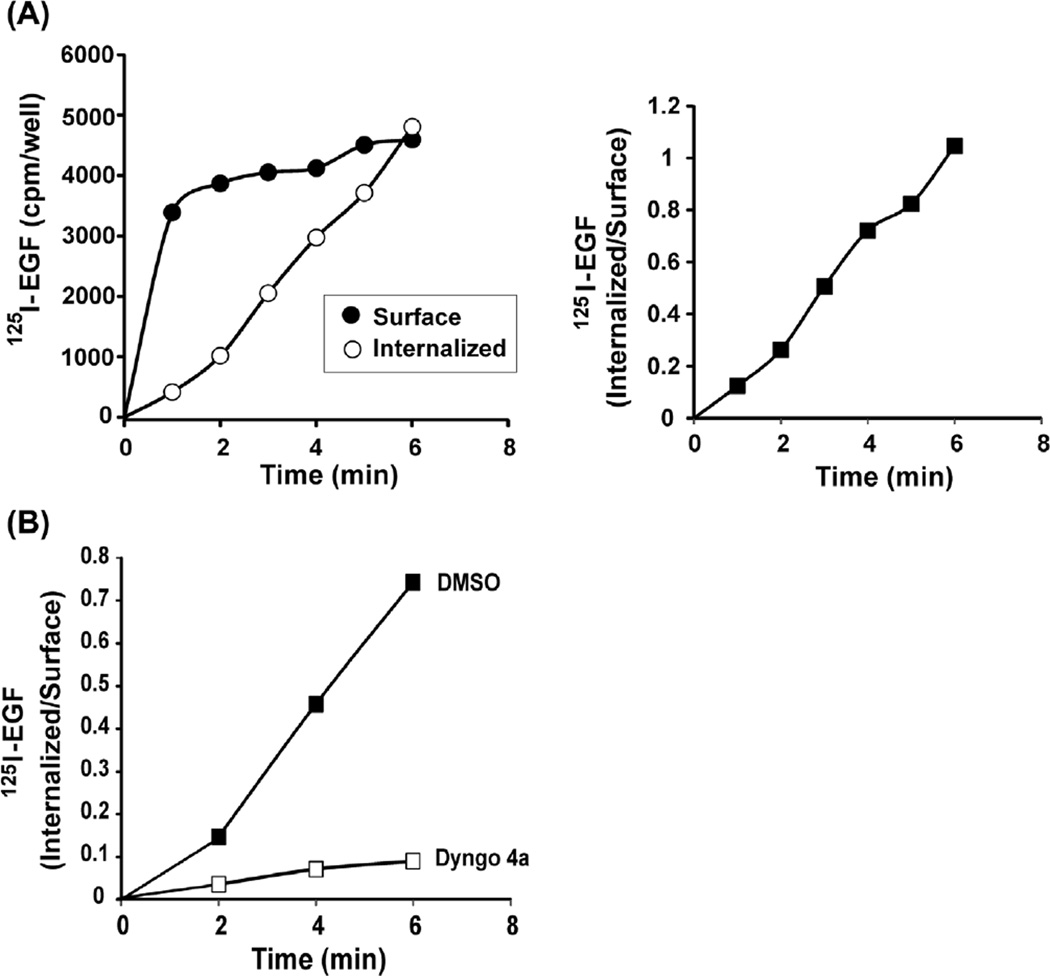
Figure 3(A) shows an example of the data obtained in the 125I-EGF internalization experiment. The amounts of internalized (I) and surface (S) 125I-EGF, and the ratio I/S are calculated as follows. The specific radioactivity of the acid-sensitive (surface) and acid-insensitive (internalized) fractions is obtained by subtracting nonspecific counts from the corresponding counts of “specific” wells. Nonspecific radioactivity is typically not more than 5–10% of the total counts. Internalization is thought to be a first-order kinetics process where the specific rate of internalization depends on the concentration of EGF-receptor complexes at the cell surface. The calculation of the specific internalization rate constant ke can be performed if the 125I-EGF concentration at the cell surface does not significantly change (or remains constant) during the time-course of 125I-EGF uptake. Under these conditions the I/S ratio displays a linear dependence on time, and therefore, ke corresponds to the linear regression coefficient of this dependence (Wiley & Cunningham, 1982). The calculation of the ke value yields 0.18 min−1 (Figure 3(A)). Figure 3(B) demonstrates an example of 125I-EGF internalization experiment in which EGFR endocytosis is inhibited by Dyngo-4a. The ke value in the presence of Dyngo-4a is 0.04 min−1.
FIGURE 3. Internalization of 125I-EGF.
(A) NIH 3T3/EGFR cells were incubated with 1 ng/mL 125I-EGF for 1–6 min at 37 °C, and the amount of surface and internalized radioactivity was determined as described in Section 4.2. (B) UMSCC2 cells were incubated with 30 µM Dyngo-4a in DMEM, and the internalization assay with 1 ng/mL 125I-EGF was performed as in (A).
4.2.5 Considerations
The assay is typically performed when cells are confluent to maximize specific 125I-EGF binding and minimize binding of 125I-EGF to plastic surfaces.
These single 12-well dish experiments must be repeated multiple times to obtain statistically reliable values of ke.
When high concentrations of 125I-EGF are used, significant amount of surface EGFRs is rapidly internalized leading to a substantial decrease of empty EGFR levels at the cell surface, thus resulting in nonlinear I/S time dependence and making it impossible to use the linear regression coefficient for calculation of ke values. In such cases, the I/S ratio plotted against incubation time is considered to be an approximate measure of an apparent internalization rate.
4.3 DETECTION OF EGFR UBIQUITINATION
4.3.1 Theory
The EGFR ubiquitination assay is based on efficient immunoprecipitation of EGFR and detection of ubiquitin immunoreactivity in immunoprecipitated EGFR by western blotting. Typically, EGFR is significantly ubiquitinated only after activation by the ligand. In most cells, the maximal extent of EGFR ubiquitination is observed after 3–5 min of cell stimulation with EGF at 37 °C.
4.3.2 Experimental protocol
Cells are grown in 60 mm dishes until ~80–90% confluency, and serum-starved.
Cells are incubated with or without EGF (2–20 ng/mL) in binding medium for 0–60 min at 37 °C.
The incubation is stopped by placing the dishes on ice and washing three times with ice-cold CMF-PBS.
400 µL of ice-cold lysis buffer containing protease, phosphatase, and deubiquitination inhibitors is added to each dish; the cells are scraped with the rubber cell lifter, and transferred to 1.5 mL tubes.
The tubes are placed on the nutator for 10-min agitation at 4 °C to complete cell lysis in, and the lysates are cleared by centrifugation at 16,000 × g for 10 min at 4 °C. Supernatants are transferred to new tubes.
Aliquots (10%) of cleared lysates are saved, and EGFR monoclonal antibody 528 (10 µg) is added to the rest of the lysate. The lysates are incubated with the antibody on the nutator for 3–4 h of agitation at 4 °C.
75 µL of Protein A Sepharose (20% solution) are added to each tubes, and the incubation on the nutator continues for 1 h at 4 °C.
Samples are centrifuged (16,000 × g; 2 min) to pellet the beads. Aliquots of supernatants (10% of total volume) are saved (for evaluation of the efficiency of immunoprecipitation) and the rest of supernatant is aspirated.
Sepharose beads are washed two times with 700 µL/tube of TGH buffer containing 1 mM sodium orthovandate and 10 mM NEM, and one time with the same buffer supplemented with 0.5 M NaCl.
After as much of the last wash as possible is aspirated using protein gel loading tips, 100 µL of 1× sample buffer is added, tubes are briefly vortexed and heated at 95 °C for 5 min to release from beads and denature proteins.
Aliquots of lysates (Step 6) and supernatants (Step 8) are mixed with an equal volume of the 2× sample buffer, and heated at 95 °C for 5 min.
Immunoprecipitates, lysates, and supernatants are resolved by SDS-PAGE.
Proteins are transferred to nitrocellulose, and western blotting is performed with antiubiquitin antibody followed by secondary antimouse antibodies conjugated to far-red fluorescent dye (IRDye-680) and the detection of the fluorescence signal using the Odyssey LI-COR imager. Subsequently, blot is probed with anti-EGFR 1005 (total EGFR) followed by the secondary antibody (labeled with IRDye-800). A part of the blot corresponding to lanes loaded with lysates is probed with an antibody to a housekeeping protein to control for equal loading.
4.3.3 Results
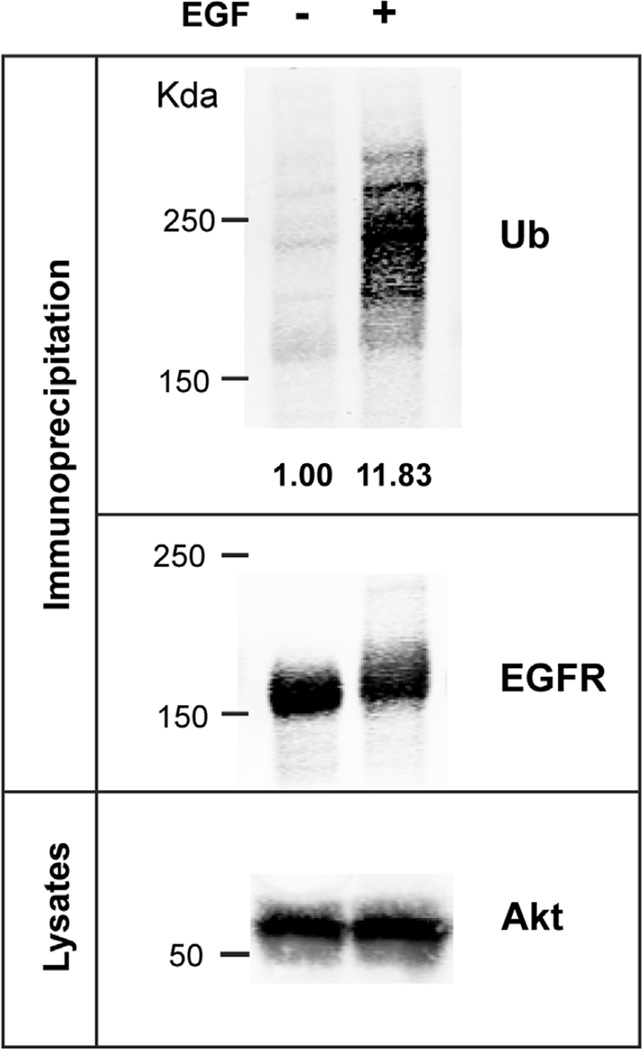
In the experiment presented in Figure 4, UMSCC2 cells were stimulated with 4 ng/mL EGF for 5 min. Very little EGFR ubiquitination is detected in nonstimulated cells whereas a strong signal of high-molecular weight ubiquitin immunoreactivity is detected in EGF-treated cells. Several ubiquitinated EGFR species are resolved which reflects heterogeneity of EGFR ubiquitination by one to three short (mostly diubiquitin) ubiquitin chains. In our experience the appearance of ubiquitinated EGFR immunoreactivity varies from a single smear above 170 kDa to several high-molecular weight bands (as in Figure 4) in different experiments. Similar amounts of EGFR in immunoprecipitates from cells treated or not with EGF, and equal amount of protein in the lysates used for immunoprecipitation is evident based on the intensities of EGFR and Akt (that is used as loading control in this experiment) bands, respectively.
FIGURE 4. Epidermal growth factor receptor (EGFR) ubiquitination.
UMSCC2 cells were incubated with or without 4 ng/mL epidermal growth factor (EGF) for 5 min at 37 °C. Cells were solubilized, and EGFR immunoprecipitated as described in Section 4.3.2. EGFR immunoprecipitates and lysates were resolved by electrophoresis, and western blotting was performed using antibodies to ubiquitin, EGFR, and Akt (loading control). The extent of EGFR ubiquitination is calculated as the amount of ubiquitin normalized to the amount of EGFR is shown under the image of the ubiquitin blot.
4.3.4 Considerations
Lysis buffer must be supplemented with NEM, inhibitor of deubiquitination enzymes to preserve ubiquitination of EGFR.
Antibodies to the EGFR extracellular domain (such as EGFR-528) tend to be more efficient in immunoprecipitating native EGFR than antibodies to the intracellular domain of EGFR.
EGFR ubiquitination can be detected in cells treated with EGF at 4 °C.
Western blot detection of ubiquitinated and total EGFR can be performed simultaneously because different antibody species are used and two corresponding secondary antibodies labeled with different fluorophores can be detected simultaneously by two-channel LI-COR imaging.
4.4 LIGAND-INDUCED EGFR DEGRADATION
4.4.1 Theory
The simplest method to examine degradation of the EGFR protein is using western blot detection of the amount of EGFR in cell lysates. Since under steady-state cell growth conditions degradation of a protein is compensated by its synthesis, measurements of the degradation rates are carried out in cells incubated with cycloheximide, that inhibits protein synthesis. Because EGF stimulation dramatically accelerates EGFR degradation, protein synthesis does not significantly contribute in the total concentration of EGFR in EGF-stimulated cells, and therefore, cycloheximide treatment is not essential for measurement of the rate of EGF-induced EGFR degradation.
4.4.2 Experimental protocol
Cells are grown in 35 or 60 mm dishes, until they reach ~80–90% confluency, and serum-starved.
Cells are treated with cycloheximide (50 µg/mL) for 2 h to block de novo synthesis of EGFR.
2–100 ng/mL EGF is added to several dishes (depending on the desired time-course) whereas other dishes are continued to be incubated in the cycloheximide-containing binding medium without EGF.
At the desired time points (0–24 h) the incubation is stopped by placing the plates firmly on ice and washing three times with ice-cold CMF-PBS. Aspirate as much as CMF-PBS as possible, and store dishes at −80 °C until the completion of the longest incubation.
The cells are lysed in the lysis buffer and the lysates are cleared as described in Section 4.3.2 with the exception that the lysis buffer does not include sodium orthovanadate and NEM.
Protein concentration is measured in each sample using Bradford method.
Aliquots of lysates with equal protein amounts are resolved on SDS-PAGE, proteins transferred to nitrocellulose, and EGFR is detected by western blotting as described in Section 4.3.2 above.
4.4.3 Results
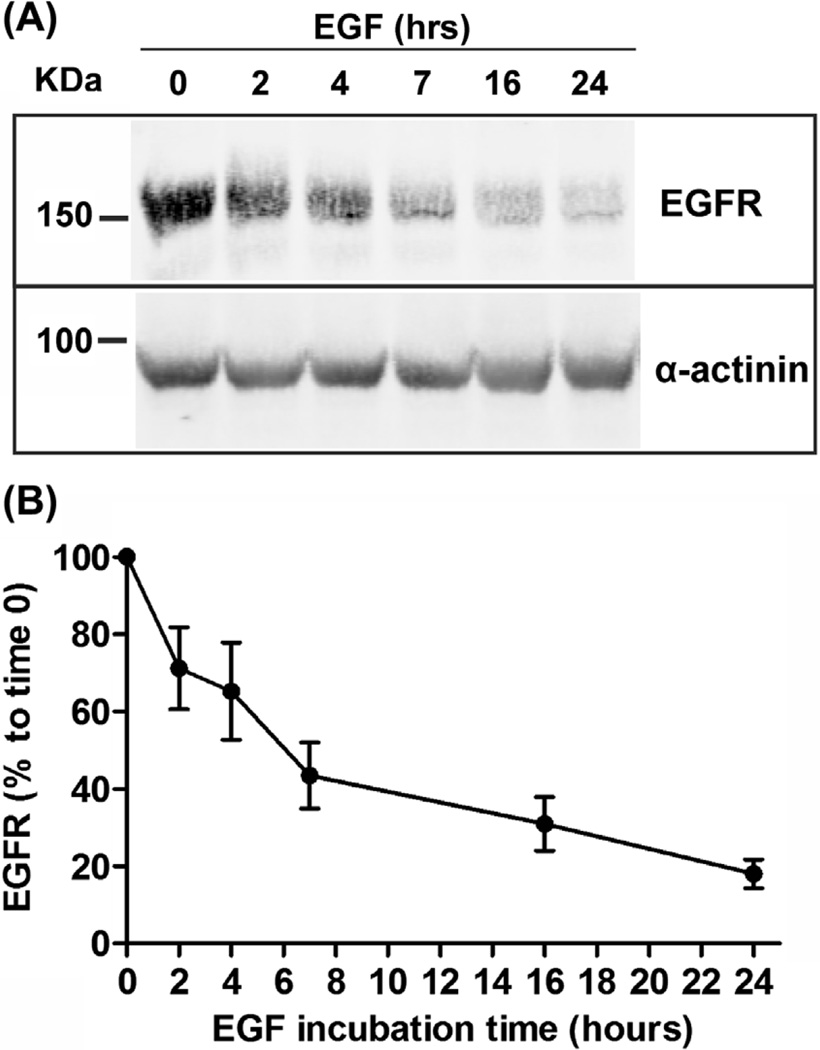
As shown in Figure 5, treatment of UMSCC2 cells with EGF results in rapid decrease in the amount of EGFR. Quantitation of the immunoreactive signal intensities of EGFR and α-actinin (loading control) was performed using LI-COR software. The normalized amounts of EGFR (EGFR/α-actinin signals) are expressed as percent of this amount at time zero (Figure 5(B)). These calculations yielded the half-life of EGF-activated EGFR protein of ~5 h.
FIGURE 5. Epidermal growth factor (EGF)-induced EGF receptor (EGFR) degradation.
UMSCC2 cells were incubated with 80 ng/mL EGF for 0–24 h. (A) EGFR was detected by western blotting in cell lysates using antibodies 1005. α-actinin is as loading control. (B) Quantification of EGFR immunoreactivity from two experiments including the data presented in (A).
4.4.4 Considerations
Because most EGFR antibodies that are used in western blotting target EGFR intracellular domain, these antibodies do not efficiently recognize phosphorylated and ubiquitinated EGFR. Therefore, the lysis buffer used in this protocol does not include sodium orthovandate and NEM, so that EGFR is dephosphorylated and deubiquitinated upon cell lysis. Alternatively, complete lysis buffer can be used, but the immunoblotting should be performed with an antibody to the extracellular EGFR domain, for instance, monoclonal anti-EGFR 05-104 (Millipore).
Time-course should be designed based on the predicted half-life of the EGFR protein. For native, endogenous EGFR, the lesser the amount of the receptor, the shorter its half-life. For instance, in contrast to UMSCC2 cells, HeLa cells expressing low level of EGFR degrade activated EGFR with the half-life of 45–60 min.
4.5 MEASUREMENTS OF THE RATE OF EGF:EGFR COMPLEX RECYCLING USING 125I-EGF, AND THE EFFECT OF PQ ON THIS PROCESS
4.5.1 Theory
Monitoring 125I-EGF recycling as a method to measure recycling of EGF–EGFR complexes is based on the observation that EGF does not significantly dissociate from EGFR in endosomes until it reaches the lysosome (Sorkin, Teslenko, & Nikolsky, 1988). The schematics of this protocol is presented in Figure 6(A). The recycling assay includes loading early endosomes with 125I-EGF-receptor complexes by allowing cells to endocytose 125I-EGF for short times at 37 °C, followed by stripping the nonendocytosed, surface-bound 125I-EGF by the mild acidic buffer treatment at 4 °C. When “125I-EGF-loaded cells” are returned to 37 °C, a pool of 125I-EGF-receptor complexes are recycled back to the cell surface, where 125I-EGF is released from the receptor and incapable of rebinding to cells due to the excess of unlabeled EGF in the medium and full occupancy of surface EGFR with unlabeled EGF. During the 37 °C chase incubation a pool of internalized 125I-EGF-receptor complexes are also sorted from early endosomes to late endosomes and lysosomes where 125I-EGF-receptor complexes are degraded by proteolytic enzymes. This degradation results in the release of mono- and di-125iodotyrosines, which rapidly pass through the membranes and accumulate in the extracellular media. These low-molecular weight products are not precipitable by TCA and can be biochemically separated from intact 125I-EGF. Only negligible amounts of TCA-soluble 125I can be detected inside the cells, and therefore the amount of TCA-soluble 125I in the medium reflects EGF degradation. The sum of intact 125I-EGF (TCA-precipitable) detected in the medium and at the cell surface (acid wash stripped) after the 37 °C chase incubation of 125I-EGF-loaded cells corresponds to 125I-EGF-receptor complexes that were recycled from endosomes to the plasma membrane.
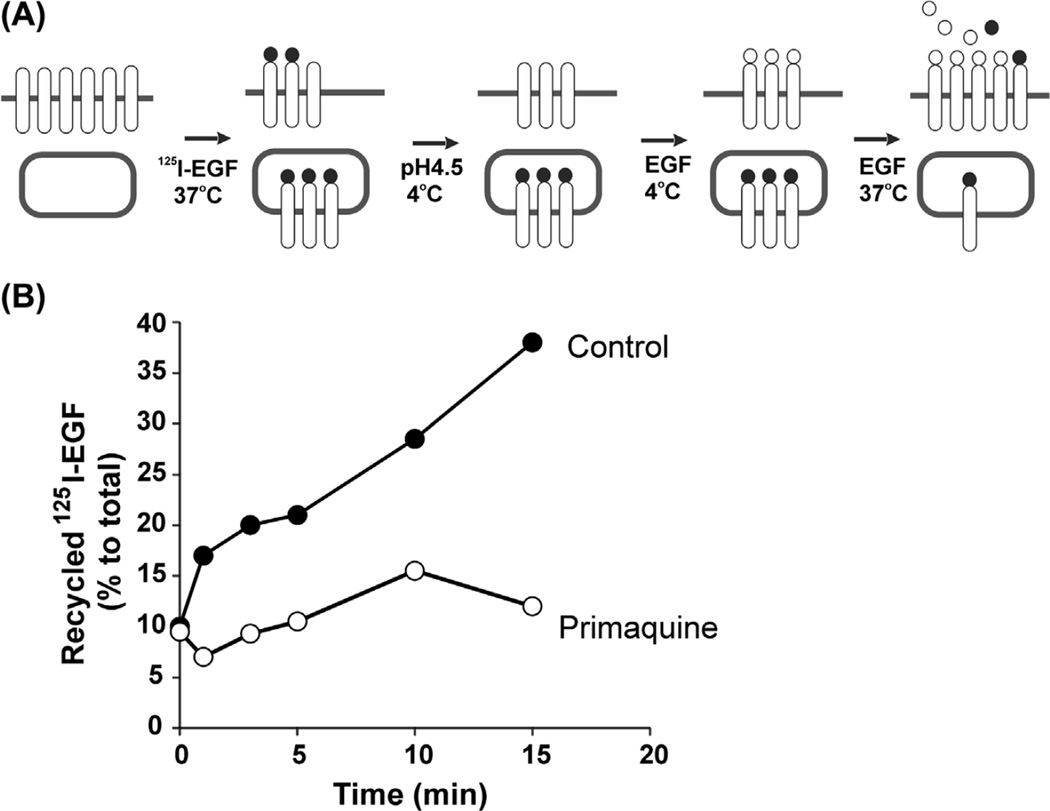
FIGURE 6. Recycling of 125I-EGF.
(A) Incubation steps in the experimental protocol to measure recycling of 125I-EGF. (B) NIH 3T3/EGFR cells were incubated with 5 ng/mL 125I-EGF for 5 min at 37 °C, surface 125I-EGF was removed by the mild acidic buffer wash, and the recycling assay was performed as described in Section 4.5. The chase incubation medium was supplemented or not with 300 µM primaquine.
4.5.2 Experimental protocol
The cells grown in 35 mm culture dishes are incubated with 1–20 ng/mL 125I-EGF in binding medium for 5–10 min at 37 °C to allow for endocytosis of 125I-EGF-receptor complexes into early endosomes. Control dishes are incubated with the same 125I-EGF-containing media supplemented with a 50-fold excess of unlabeled EGF to determine nonspecific radioactivity.
The dishes are placed on ice to stop endocytosis, and then rapidly washed three times with 1 mL of ice-cold media. The cells are incubated with 1 mL of pH 4.5 acetate buffer for 2.5 min and then additionally rinsed with 1 mL of the same buffer to remove cell-surface 125I-EGF. The resulting cells contain 125I-EGF only in endosomes and are referred to as “125I-EGF-loaded cells.”
The 125I-EGF-loaded cells are incubated in binding medium containing 200 ng/mL unlabeled EGF for 40 min at 4 °C to occupy surface EGFR.
-
The media are aspirated, and the cells further incubated with fresh prewarmed (37 °C) media containing 200 ng/mL EGF (1 mL/dish) at 37 °C (in the water bath for short times (less than 15 min) or CO2 incubator for longer times) to allow traffic of endosomal 125I-EGF-receptor complexes. The length of 37 °C chase incubations of EGF-loaded cells is discussed below.
Three dishes (two incubated with “specific” media and one with “nonspecific” media) are kept on ice. These dishes represent a “zero” time point. While other dishes are incubated at 37 °C, the prewarmed media are added to “zero-time point” dishes and immediately collected into γ-counter vials.
At the end of the chase incubations, the cells are placed on ice, and the media are collected into γ-counter vials.
Surface-bound 125I-EGF is stripped by the pH 2.8 acetate buffer wash (as in the internalization assay Section 4.2.2) and collected into γ-counter vials.
The cells are then solubilized in 1N NaOH to measure the amount of intracellular 125I-EGF as described in Section 4.2.2.
The media collected at Step 5 (1 mL) is mixed with 0.3 mL of 10% TCA/2% PTA. The mixture is kept at 4 °C for 1 h or longer and centrifuged at 5000 × g. The supernatants are transferred to new γ-counter tubes to determine the amount of degraded 125I-EGF. The pellets are dissolved in 1 mL of 1 N NaOH to determine the amount of nondegraded, intact 125I-EGF.
4.5.3 Variation: effect of PQ on recycling
After Steps 1–3, Step 4 (chase incubation at 37 °C) is performed using the same media but containing 300 µM PQ.
Follow Steps 5–8 in Protocol Section 4.5.2.
4.5.4 Results
The example of an 125I-EGF recycling experiment in NIH3T3/EGFR cells is presented in Figure 6(B). The recycled 125I-EGF is expressed as percent of total (cells plus medium) 125I-EGF. For calculation, first, specific counts are obtained by subtracting nonspecific radioactivity of all fractions from the counts of the same fractions of “specific” dishes. The total amount of 125I-EGF associated with cells and media is calculated for each dish as the sum of second (pH 2.8) acid wash (surface 125I-EGF, Step 6), intracellular 125I-EGF (Step 7), intact medium 125I-EGF (TCA-precipitated radioactivity, Step 8), and degraded medium 125I-EGF (Step 8). The amount of degraded 125I-EGF was negligible during the first 15-min chase, and therefore, is not plotted in Figure 6(B). The amount of recycled 125I-EGF is calculated by summing the surface 125I-EGF and intact medium 125I-EGF. The percent of each 125I-EGF pool (intracellular, recycled, and degraded) relative to the total cell/medium-associated 125I-EGF is calculated for each time point. Figure 6(B) shows that PQ strongly inhibits recycling of 125I-EGF.
4.5.5 Considerations
Because handling of multiple dishes in this type of experiments is difficult, it is recommended to use not more than two “specific” and one “nonspecific” dishes for each time point of the chase incubation (Step 5 below). The entire time-course experiment is repeated two to three times to ensure the statistical significance of the data.
Confluent cells offer better signal/noise ratio because nonspecific binding of 125I-EGF to plastic is minimized.
Because of its small size, EGF polypeptide is not efficiently precipitated by TCA alone. Therefore, addition of PTA is important for complete precipitation of intact 125I-EGF.
Recycling and degradation are considered to be first-order kinetics processes. This means that their specific rate constants are proportional to the concentration of 125I-EGF in early/sorting endosomes. However, the calculation of specific rate constants as the linear regression coefficients is difficult due to the decrease in the concentration of 125I-EGF in early/sorting endosomes during the 37 °C chase incubation. Also, presence of unlabeled EGF and its contribution to overall EGF concentrations in various compartments makes calculations of specific rate constants challenging. Therefore, it is important to interpret the results of the recycling/degradation assay as providing values of the apparent rates. These apparent rates should be compared between experimental variants under conditions of the same starting concentration of 125I-EGF in endosomes, and preferably, the same total EGFR numbers per cell.
SUMMARY
In this chapter, we describe several basic experimental procedures that are designed to perform quantitative analysis of EGFR internalization, ubiquitination, degradation, and recycling. These assays take advantage of the availability of biologically active radiolabeled EGF and fluorescent conjugates of EGF, and antibodies that efficiently immunoprecipitate native receptor and detect denatured EGFR by immunoblotting. The specific details in the design of these methods are based on the experience of using these assays to study EGFR traffic in the variety of cell types expressing a wide range of EGFR levels including various mutants of EGFR.
REFERENCES
- Eden ER, Huang F, Sorkin A, Futter CE. The role of EGF receptor ubiquitination in regulating its intracellular traffic. Traffic. 2012;13(2):329–337. doi: 10.1111/j.1600-0854.2011.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nature Reviews Molecular Cell Biology. 2012;13(2):75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler H, Ash JF, Singer SJ, Cohen S. Visualization by fluorescence of the binding and internalization of epidermal growth factor in human carcinoma cells A-431. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(7):3317–3321. doi: 10.1073/pnas.75.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler HT, Maxfield FR, Willingham MC, Pastan I. Dansylcadaverine inhibits internalization of 125I-epidermal growth factor in BALB 3T3 cells. The Journal of Biological Chemistry. 1980;255(4):1239–1241. [PubMed] [Google Scholar]
- Henriksen L, Grandal MV, Knudsen SL, van Deurs B, Grøvdal LM. Internalization mechanisms of the epidermal growth factor receptor after activation with different ligands. PLoS One. 2013;8(3):e58148. doi: 10.1371/journal.pone.0058148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop JN, von Zastrow M. Role of ubiquitination in endocytic trafficking of G-protein-coupled receptors. Traffic. 2011;12(2):137–148. doi: 10.1111/j.1600-0854.2010.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey A, Daniel JA, Hadzic G, Chau N, Clayton EL, Mariana A, et al. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013;14(12):1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochemical Society Transactions. 2009;37(Pt 5):1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub TJ, Newton CR. Recycling kinetics and transcytosis of transferrin in primary cultures of bovine brain microvessel endothelial cells. The Journal of Cellular Physiology. 1991;149(1):141–151. doi: 10.1002/jcp.1041490118. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75(9):770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Experimental Cell Research. 2009;315(4):683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Teslenko L, Nikolsky NN. The endocytosis of epidermal growth factor in A431 cells: a pH of microenvironment and the dynamics of receptor complexes dissociation. Experimental Cell Research. 1988;175:192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nature Reviews Molecular Cell Biology. 2009;10(9):609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanWeert AW, Geuze HJ, Groothuis B, Stoorvogel W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. The European Journal of Cell Biology. 2000;79(6):394–399. doi: 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- Weinberg JS, Drubin DG. Regulation of clathrin-mediated endocytosis by dynamic ubiquitination and deubiquitination. Current Biology. 2014;24(9):951–959. doi: 10.1016/j.cub.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS, Cunningham DD. The endocytotic rate constant. A cellular parameter for quantitating receptor-mediated endocytosis. The Journal of Biological Chemistry. 1982;257(8):4222–4229. [PubMed] [Google Scholar]