Abstract
Evolutionary biochemists define enzyme promiscuity as the ability to catalyze secondary reactions that are physiologically irrelevant, either because they are too inefficient to affect fitness or because the enzyme never encounters the substrate. Promiscuous activities are common because evolution of a perfectly specific active site is both difficult and unnecessary; natural selection ceases when the performance of a protein is “good enough” that it no longer affects fitness. Although promiscuous functions are accidental and physiologically irrelevant, they are of great importance because they provide opportunities for evolution of new functions in nature and in the laboratory, as well as targets for therapeutic drugs and tools for a wide range of technological applications.
Keywords: Promiscuity, substrate ambiguity, enzyme, molecular evolution
Once upon a time biology was (thought to be) simple. A gene encoded a single protein, which performed a single function with high specificity. But biology is not simple. Genes can encode multiple proteins, proteins can perform multiple functions, and promiscuous functions abound. Our “linear” thinking about biology is being supplanted by the recognition that understanding the complexity of living systems can best be accomplished by considering the function of the system as a whole in addition to the functions of the individual parts. From this point of view, the origins and implications of promiscuous functions are particularly challenging because we cannot predict every potential promiscuous function – and there may be many thousands - within the context of a particular proteome. This Opinion article will discuss the molecular- and system-level considerations necessary for an integrated perspective on the role of promiscuity in the function, evolution and manipulation of biological systems.
“Promiscuity” means different things to different people
The term “promiscuous” is used to describe enzymes that catalyze more than one reaction. Enzymes commonly display “substrate promiscuity” (a.k.a. substrate ambiguity), which is the ability to carry out a comparable chemical transformation using different substrates. Enzymes that are “catalytically promiscuous” catalyze a secondary reaction that results in a chemical transformation different from that catalyzed with its canonical substrate. The term “promiscuity” is used in this article to encompass both catalytic and substrate promiscuity.
Protein biochemists and molecular biologists often use the term “promiscuous” to describe broad-specificity proteins that bind to multiple interaction partners, which may be ligands, substrates or other macromolecules [1–3]. While this is a reasonable use of the term, evolutionary biochemists prefer to reserve the term “promiscuous” to refer to interactions that are not physiologically relevant [4]. This is a useful distinction, as it communicates not only the property of interest, but also its relevance to physiology and the degree to which it is under selective pressure.
Broad substrate specificity is typical of detoxification enzymes such as glutathione S-transferases [2, 5] and cytochrome P450s [6] and is critical to their ability to protect organisms from the numerous and unpredictable toxins to which they are exposed. Broad-specificity enzymes such as esterases, amidases and phosphatases can allow microbes to initiate degradation of diverse compounds, and can release products such as phosphate and ammonia that are useful to the organism even if the compound cannot be fully degraded. Cases in which broad specificity is important for fitness are clearly different from those in which enzymes catalyze reactions of non-physiological substrates or of physiological substrates with extremely low efficiency.
When an evolutionary biochemist says that a particular enzyme is promiscuous, it is an operational definition meaning that, to the best of our knowledge, the promiscuous activity is physiologically irrelevant. This might be true because the substrate for the promiscuous activity is never encountered by the enzyme, or at least not in concentrations high enough to cause trouble. For example, many chemicals synthesized by humans for industrial or medicinal purposes have never before been present in the biosphere. Even the elaborate natural products synthesized by many organisms to communicate with or to kill other organisms may not be encountered outside of the particular environmental niche in which they are produced.
A promiscuous activity may also be irrelevant because it is too inefficient to influence metabolism and therefore fitness. For instance, gamma-glutamyl phosphate reductase (ProA) from Escherichia coli has a low-level ability to reduce N-acetyl glutamyl phosphate, the substrate for ArgC [7, 8]. However, ProA is unable to substitute for ArgC. The inefficiency of the promiscuous activity, which is several orders of magnitude below that of ArgC, likely precludes sufficient reduction of N-acetyl glutamyl phosphate to support arginine biosynthesis.
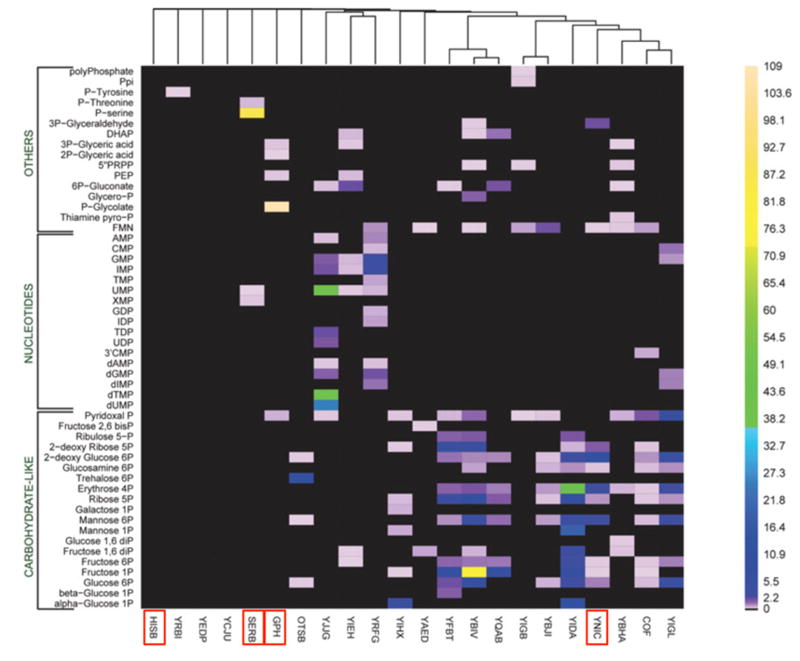
The last few decades have seen the assignment of genes to functions in metabolic pathways that degrade myriad organic compounds, as well as synthesize the standard building blocks of macromolecules and a wide variety of secondary metabolites. However, every genome encodes a number of enzymes that do not participate in known metabolic pathways. Figure 1, which summarizes the activities of 23 haloacid dehalogenase-like phosphatases from E. coli with 80 physiological substrates, exemplifies the conundrum presented by such enzymes [9]. Only a few of the tested enzymes are highly specific. For example, two of the enzymes (HisB and SerB) are involved in amino acid biosynthesis. The substrate for HisB was not available, but, as expected, the enzyme did not have detectable activity with any of the tested substrates. SerB was quite specific for phosphoserine, its physiological substrate. Gph[10] and YniC [9] are involved in detoxification of 2-phosphogycolate and 2-deoxyglucose, respectively. But what can we conclude about the remaining enzymes, many of which have weak activity with a number of substrates? One possibility is that these enzymes participate in as-yet unidentified metabolic pathways, and have high activity with a substrate that was not tested. If so, these lower-level activities would be promiscuous activities. Another possibility is that the broad activity profiles of these enzymes reflect a physiologically important function. For example, YbiV has high activity with fructose 1-phosphate and modest activity with ribose 5-phosphate and glucose 6-phosphate. Each of these sugar phosphates plays a role in primary metabolism, and it seems wasteful to hydrolyze a high-energy phosphoester bond. However, this enzyme might play an important role under phosphate-limited conditions by making phosphate available for more critical processes. This situation may be more common than we appreciate. Not all enzymes need to fit neatly into metabolic pathways to contribute to fitness. The role of some enzymes may be to improve the function of the overall metabolic network, and this role may require broad-substrate specificity.
Figure 1.
A heat map summarizing the activities of 23 haloacid dehalogenase-like proteins from E. coli with 80 physiological substrates. Activities are given in units of μmol/min/mg of protein. Red boxes indicate proteins that are discussed in the text. Reproduced from J. Biol. Chem. 281, 36149–36161 (2006).
Additional difficulties in assessing the physiological relevance of a suspected promiscuous activity arise because of differences in the metabolic networks of organisms. For example, Palmer et al. identified an enzyme in Amycolatopsis that catalyzed racemization of N-acyl amino acids [11]. The racemase activity was later found to be a promiscuous activity of o-succinylbenzoate synthase, an enzyme involved in menaquinone synthesis [12]. By analogy, an enzyme with N-acyl amino acid racemase activity from Geobacillus kaustophilus might be assumed to also be an o-succinylbenzoate synthase. However, clever sleuthing by Sakai et al. [13] revealed that the operon encoding N-acyl amino acid racemase also encodes a succinyl-CoA:D-amino acid N-succinyltransferase and an N-succinyl-L-amino acid hydrolase. These three enzymes participate in a previously unknown pathway for conversion of D-amino acids to L-amino acids via N-acyl amino acid intermediates.
Why does promiscuity exist?
The existence of promiscuous enzymes introduces an element of sloppiness into metabolic networks that we would, overly optimistically, like to think of as perfectly controlled, with no dead-ends, leaks or serendipitous pathways [14] connecting different parts of the network in non-canonical ways. This lack of perfection raises the question: why do promiscuous activities exist? One possible explanation is that “perfect” specificity is unachievable. Active sites of enzymes are loaded with functional groups that position the substrate and/or act as acids, bases, or nucleophiles in a cavity that provides favorable ionic and hydrophobic interactions with the substrate, transition states and intermediates in the reaction. Active sites can often exclude substrates that are too large to fit or that contain charges in places that cause repulsion by active site residues. However, small substrates may fit into a capacious active site, although they may lose some of the binding affinity available to the optimal substrate. Even larger substrates may be able to bind if part of the molecule can protrude from the active site into the solvent. Thus, promiscuity may exist simply because it is impossible to exclude all potential substrates. The existence of secondary “proofreading” active sites in acyl-amino acid tRNA synthetases, which increases the fidelity of loading amino acids onto the 3′-hydroxyl of tRNAs, is a testament to what it can take to prevent the formation of products from inappropriate substrates.
Another explanation for the existence of promiscuous activities is that they are relics of past activities in ancestral generalist enzymes that catalyzed more than one reaction. Given the limited coding capacity of the last universal common ancestor, it is likely that at least some of its enzymes were generalists capable of catalyzing the same chemical reaction using several different substrates [15, 16]. Duplication and divergence of genes encoding generalist enzymes has given rise to extended superfamilies of specialist enzymes that retain the ancestral structural fold and some of its catalytic strategies, but have accumulated mutations that allow higher substrate specificity and sometimes additional catalytic steps [17–19]. Indeed, enzymes in superfamilies often have promiscuous activities that correspond to the physiological functions of other families within the superfamily [20].
Promiscuous activities are important because they provide a repertoire of catalytic activities from which enzymes can be recruited when the environment changes and a new activity becomes importance for fitness. If the selective pressure continues, gene duplication and divergence can lead to the evolution of a new, efficient, and specific enzyme. It is important to recognize that promiscuous activities do not exist because they provide catalytic activities that may become useful in the future. Natural selection can act only upon functions that either impair or improve fitness. Evolution cannot predict what might become useful in the future.
Promiscuous activities are not always inefficient
The enormous catalytic power of enzymes is achieved by a variety of mechanisms that position a substrate in the proper orientation relative to catalytic groups in the active site, remove interfering water molecules, and channel reactive intermediates toward only one of multiple potential fates. Promiscuous activities are usually inefficient due to a less than perfect alignment of the promiscuous substrate with catalytic groups in the active site. For example, the promiscuous activities of HisB, Gph and YtjC with phosphoserine have kcat/KM values of 1.6, 7.6 and 0.8 M−1s−1, respectively, whereas the kcat/KM of SerB, the physiological phosphoserine phosphatase, is 8.7 × 104 M−1s−1 [21]. However, this is not always true; promiscuous activities can be quite efficient.
E. coli homoserine kinase (ThrB) catalyzes the phosphorylation of homoserine, a precursor of threonine, with a kcat/KM of 3.8 × 105 M−1s−1 [14]. The physiological role of ThrB is clearly related to threonine biosynthesis, as thrB is found in an operon with other threonine biosynthesis genes whose expression is controlled in response to cytoplasmic levels of threonine and isoleucine [22]. (Isoleucine is synthesized from threonine.) However, ThrB also catalyzes phosphorylation of 4-hydroxythreonine with a fairly robust kcat/KM of 4.8 × 103 M−1s−1 [14]. Similarly, a highly efficient arylsulfatase from Pseudomonas aeruginosa catalyzes hydrolysis of p-nitrophenyl sulfate with a kcat/KM of 4.9 × 107 M−1s−1,.. The physiological role of the enzyme is clearly hydrolysis of a sulfate ester, as its expression is increased during sulfate starvation and decreased when sulfate is available, and it is encoded in an operon that also encodes a sulfate ester transport system. This enzyme hydrolyzes the alternative and non-physiological substrate bis(4-nitrophenyl) phosphate with a kcat/KM of 2.5 × 105 M−1s−1 [23]. Values of kcat/KM in the range of 103 – 105 M−1s−1 are not uncommon in metabolic enzymes. For example, E. coli acetolactate synthase has a kcat/KM of 6.1 × 103 M−1s−1 [24], and Zymomonas mobilis isocitrate dehydrogenase has a kcat/KM of 3.2 × 103 M−1s−1 [25]. Thus, the rate constants for these apparently promiscuous reactions are within the range of physiologically relevant reactions. In both cases, natural selection has not led to a more specific enzyme, suggesting that the promiscuous activity does not interfere with the physiological activity. This may be the case because the promiscuous substrate is never encountered in vivo, or because occasional conversion of a promiscuous substrate neither generates a toxic product nor impairs flux through a metabolic pathway.
Promiscuity is not just for enzymes
Adventitious interactions with non-canonical molecules are not limited to enzymes. Every macromolecule in cells has the potential for promiscuous interactions with small molecules or other macromolecules in the crowded cytoplasm of cells. A simulation of the E. coli cytoplasm that includes the 50 most abundant macromolecules suggests that proteins have about 25 neighbors at any moment, and encounter over 100 different molecules within 15 μsec [26]. Just as the specificity of enzyme active sites has evolved to be only as specific as it has to be to avoid detrimental effects on fitness, other adventitious interactions in the cell can be tolerated as long as they do not interfere with fitness.
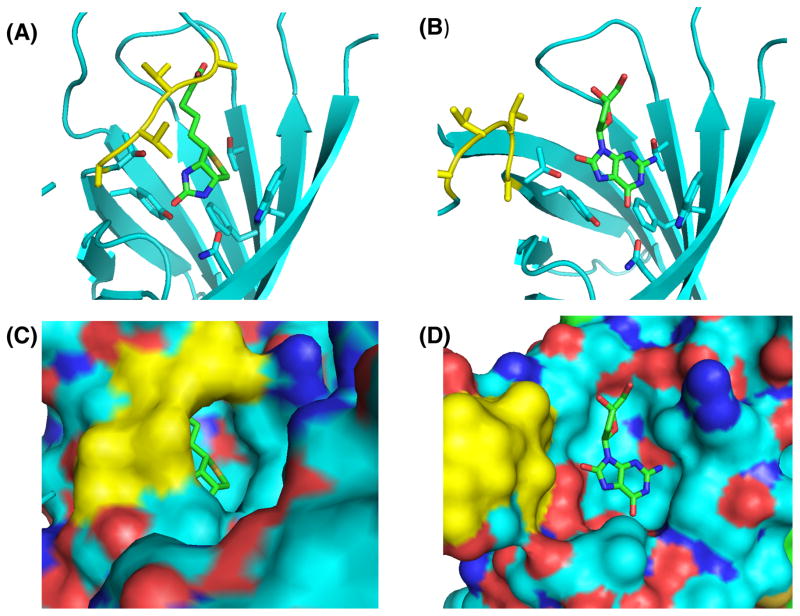
Even avidin, which binds enormously strongly to biotin (KD ≈ 10−15 M) [27], displays promiscuous binding. Avidin is found in the egg white of birds, reptiles and amphibians. It is believed to sequester biotin and consequently prevent bacterial growth [28]. Avidin binds a number of other ligands, including lipoic acid (KD = 0.8 μM) [29], indomethacin (KD = 5.3 μM) [29], 8-oxodeoxyadenosine (KD = 24 μM), 8-oxodeoxyguanosine (KD = 117 μM) and the oligonucleotide CTCGTCT (KD = 40 μM) [30]. Avidin is unlikely to encounter these alternative ligands in its physiological context. Further, the affinities of avidin for these ligands are so much lower than that for biotin that they would not interfere with the important function of avidin even if they were present. The molecular basis for the promiscuous binding is revealed by the crystal structures shown in Figure 2. Binding of biotin induces a conformational change that closes a flexible loop over the binding site. 8-Oxodeoxyguanosine fits nicely into the binding pocket of avidin, but fails to induce the conformational change. The same is true for complexes of avidin with 8-oxodeoxyadenosine (PDB 2A5C) [30] and deoxyguanosine (PDB 2A8G).
Figure 2.
A comparison of the binding of the physiological ligand (biotin) and a non-physiological ligand (8-oxodeoxyguanosine) to avidin. Biotin (A,C) (PDB 2AVI) and 8-oxodeoxyguanosine (B,D) (PDB 2A5B) are shown in the binding pocket of avidin. The flexible loop that closes over the ligand in the biotin-avidin complex is shown in yellow. Cyan, carbon; blue, nitrogen; red, oxygen. C) and d) depict the surface versions of the images shown in a) and b), respectively. Figure was prepared using MacPyMol.
Promiscuity in transcriptional regulators is particularly intriguing, as it can lead to evolution of new regulatory capabilities. Transcriptional regulators bind to specific sites in DNA, and often contain binding sites for small molecule ligands that allow transcription to be tuned according to environmental conditions. The spectrum of sites that can be bound by a transcriptional regulator can be evaluated in vitro using protein-binding microarrays, DNase-Seq, SELEX (Systematic evolution of ligands by exponential enrichment), electrophoretic mobility shift assays, or direct binding assays [31] and in vivo by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) [32]. The in vitro approaches reveal the intrinsic binding properties of the transcription factor, while the in vivo approach reveals what actually happens inside cells when there is competition between proteins for specific binding sites, and competition for binding of proteins among sites with various affinities [33, 34].
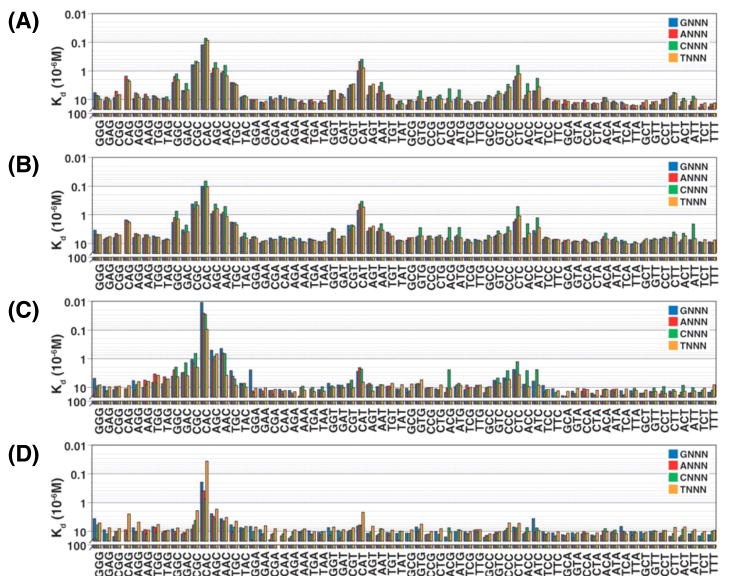
Figure 3 shows the affinities of four related eukaryotic transcription factors for 256 permutations of the binding sequence 5′-CNNNNTG-3′ (where N is any nucleotide) [31]. Three of the four proteins have multiple binding sites with varying affinities. This is clearly not due to an intrinsic limitation on the specificity that can be achieved in this family of transcriptional regulators, as Cbf1 (panel D) is very specific. Values of KD for most “non-target” sites are > 10 μM, whereas the KD for the highest affinity site is < 0.1 μM. Low affinity binding sites may be physiologically irrelevant, but this is certainly not always the case. For example, LuxR, the master regulator of quorum sensing gene expression, directly regulates 105 genes in Vibrio harveyi, some of which have low-affinity binding sites [35]. The variable affinities of different promoter binding sites are believed to allow temporal control of gene expression as concentrations of LuxR rise in response to high population density [36].
Figure 3.
Some transcription factors are more specific than others. Comparison of the affinities of four C-terminally tagged eukaryotic transcription factors belonging to the basic helix-loop-helix (bHLH) family to target DNAs containing all sequence permutations of the four-nucleotide binding site. A) human MAX isoform A; B) human MAX isoform B; C) yeast Pho4p; and D) yeast Cbf1p. The x-axis indicates the final three nucleotides in the sequence. The first nucleotide is indicated by the color of the bar. Reproduced with permission from Science 315, 233–237 (2007).
The up-side and down-side of promiscuous functions
Promiscuity has played an important role in the evolution of superfamilies of enzymes, transcriptional regulators and receptors. A promiscuous function can become physiologically relevant due to a change in environment, such as the introduction of a toxin or a new carbon source. Alternatively, a mutation may increase the opportunity for interactions between two molecules, or increase the consequences of that interaction. For example, a mutation affecting a transporter might lead to higher cytoplasmic levels of a small molecule that can interact with promiscuous proteins, or a mutation affecting a protein can increase its ability to bind a new target, either a small molecule or a macromolecule. A newly important function can be improved by gene duplication and divergence of one copy, ultimately leading to a protein with a new specialized function [37, 38]. The vertebrate receptors for progestagens, glucocorticoids and mineralocorticoids are believed to have evolved in this way from promiscuous binding functions of an ancestral steroid hormone receptor [39].
Promiscuous functions offer a wide range of opportunities for biotechnology and drug development. Promiscuous enzyme activities have been used as a starting point for directed evolution of new biocatalysts for reactions that are difficult or environmentally damaging to achieve via chemical catalysis. Evolved enzymes have been used directly for biocatalysis to make intermediates for pharmaceutical agents [40], and are being developed for detoxification of nerve agents [41, 42]. Evolved enzymes can also be integrated into multi-step pathways in bacteria for “green” production of biofuels and commodity chemicals [43].
Promiscuous enzymes have been exploited for incorporation of unnatural amino acids into proteins [44] and for synthesis of DNA labeled with fluorophores or chemically reactive groups for cross-linking to other molecules [45]. The promiscuous ability of avidin to bind 8-oxodeoxyguanosine [46] has been exploited by commercial ELISA kits that detect oxidatively damaged DNA.
The pharmaceutical industry is based largely on exploitation of promiscuous binding abilities. Many pharmaceutical agents are, by design, molecules that have not previously been encountered and that bind to a particular molecular target in a way that perturbs its function. However, promiscuous binding can also result in toxicity. Off-target promiscuous binding of pharmaceutical agents can cause adverse drug effects. Promiscuous binding of pollutants to hormone receptors can cause endocrine disruption in wildlife [47] and possibly adverse health effects in humans [48]. This phenomenon is not limited to anthropogenic compounds. For example, cone snails inject venom containing hundreds of small disulfide-bridged peptides into their prey. Many of these peptides inhibit ion channels, leading to paralysis within seconds [49, 50]. From the perspective of the prey, the ion channels are engaging in an unfortunate promiscuous binding activity that has lethal consequences.
Concluding remarks
Promiscuity was not generally recognized as important or interesting 20–30 years ago, a time in which protein scientists were making great progress in assigning the physiological roles of proteins and revealing the structure/function relationships that enabled proteins to perform their roles as catalysts, receptors or transcriptional regulators. Low-level binding or catalytic abilities were often identified during the characterization of new proteins, but typically received little attention. Times have changed. The evolutionary and medical implications of promiscuous functions are well recognized, and promiscuous functions have enabled a dazzling array of biotechnological developments; an impressive impact for functions that are physiologically irrelevant and often inefficient.
Highlights.
Promiscuous enzymes catalyze physiologically irrelevant secondary reactions.
Promiscuous activities are not always inefficient.
The concept of promiscuity can be easily extended to non-catalytic proteins.
Promiscuous functions can be the starting point for evolution of new functions.
Promiscuous functions are used for a wide range of biotechnological applications.
Acknowledgments
This work was supported by NIH RO1 GM083285 to S.D.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhat V, et al. Biophysical basis of the promiscuous binding of B-cell lymphoma protein 2 apoptotic repressor to BH3 ligands. J Mol Recognit. 2013;26:501–13. doi: 10.1002/jmr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honaker MT, et al. Enzymatic detoxication, conformational selection, and the role of molten globule active sites. J Biol Chem. 2013;288:18599–611. doi: 10.1074/jbc.M112.445767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayou C, et al. A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science. 2014;343:645–8. doi: 10.1126/science.1248229. [DOI] [PubMed] [Google Scholar]
- 4.Copley SD. Enzymes with extra talents: Moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol. 2003;7:265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 5.Hayes JD, et al. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 6.Iyanagi T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: implications for detoxification. Int Rev Cytol. 2007;260:35–112. doi: 10.1016/S0074-7696(06)60002-8. [DOI] [PubMed] [Google Scholar]
- 7.Yu McLoughlin S, Copley SD. A compromise required by gene sharing enables survival: Implications for evolution of new enzyme activities. Proc Natl Acad Sci U S A. 2008;105:13497–502. doi: 10.1073/pnas.0804804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanal A, et al. Differential Effects of a Mutation on the Normal and Promiscuous Activities of Orthologs: Implications for Natural and Directed Evolution. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuznetsova E, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281:36149–61. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 10.Teresa Pellicer M, et al. Role of 2-phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J Bacteriol. 2003;185:5815–21. doi: 10.1128/JB.185.19.5815-5821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer DR, et al. Unexpected divergence of enzyme function and sequence: “N-acylamino acid racemase” is o-succinylbenzoate synthase. Biochemistry. 1999;38:4252–4258. doi: 10.1021/bi990140p. [DOI] [PubMed] [Google Scholar]
- 12.Thoden JB, et al. Evolution of enzymatic activity in the enolase superfamily: structural studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry. 2004;43:5716–27. doi: 10.1021/bi0497897. [DOI] [PubMed] [Google Scholar]
- 13.Sakai A, et al. Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway for the irreversible conversion of D- to L-amino acids. Biochemistry. 2006;45:4455–62. doi: 10.1021/bi060230b. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, et al. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5′-phosphate synthesis. Mol Syst Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen RA. Enzyme recruitment in evolution of new function. Ann Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 16.Ycas M. On earlier states of the biochemical system. J Theor Biol. 1974;44:145–60. doi: 10.1016/s0022-5193(74)80035-4. [DOI] [PubMed] [Google Scholar]
- 17.Gerlt JA, et al. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 18.Lahiri SD, et al. Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage. Bioorg Chem. 2006;34:394–409. doi: 10.1016/j.bioorg.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–91. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 20.Roodveldt C, Tawfik DS. Shared promiscuous activities and evolutionary features in various members of the amidohydrolase superfamily. Biochemistry. 2005;44:12728–36. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- 21.Yip SH, Matsumura I. Substrate ambiguous enzymes within the Escherichia coli proteome offer different evolutionary solutions to the same problem. Mol Biol Evol. 2013;30:2001–12. doi: 10.1093/molbev/mst105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner JF. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979;76:1706–10. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babtie AC, et al. Efficient catalytic promiscuity for chemically distinct reactions. Angew Chem Int Ed Engl. 2009;48:3692–4. doi: 10.1002/anie.200805843. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz A, et al. Valine 375 and phenylalanine 109 confer affinity and specificity for pyruvate as donor substrate in acetohydroxy acid synthase isozyme II from Escherichia coli. Biochemistry. 2010;49:5188–99. doi: 10.1021/bi100555q. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, et al. Biochemical and molecular characterization of NAD(+)-dependent isocitrate dehydrogenase from the ethanologenic bacterium Zymomonas mobilis. FEMS Microbiol Lett. 2012;327:134–41. doi: 10.1111/j.1574-6968.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- 26.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green NM. Avidin. 1 The Use of (14-C)Biotin for Kinetic Studies and for Assay. Biochem J. 1963;89:585–91. doi: 10.1042/bj0890585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosano C, et al. The X-ray three-dimensional structure of avidin. Biomol Eng. 1999;16:5–12. doi: 10.1016/s1050-3862(99)00047-9. [DOI] [PubMed] [Google Scholar]
- 29.Zsila F. Aromatic side-chain cluster of biotin binding site of avidin allows circular dichroism spectroscopic investigation of its ligand binding properties. J Mol Recognit. 2011;24:995–1006. doi: 10.1002/jmr.1147. [DOI] [PubMed] [Google Scholar]
- 30.Conners R, et al. Recognition of oxidatively modified bases within the biotin-binding site of avidin. J Mol Biol. 2006;357:263–74. doi: 10.1016/j.jmb.2005.12.054. [DOI] [PubMed] [Google Scholar]
- 31.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 32.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 33.Karreth FA, et al. Target competition: transcription factors enter the limelight. Genome Biol. 2014;15:114. doi: 10.1186/gb4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brewster RC, et al. The transcription factor titration effect dictates level of gene expression. Cell. 2014;156:1312–23. doi: 10.1016/j.cell.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kessel JC, et al. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. MBio. 2013;4 doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–67. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 38.Bergthorsson U, et al. Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007;104:17004–9. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eick GN, et al. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012;8:e1003072. doi: 10.1371/journal.pgen.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–94. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 41.Tsai PC, et al. Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents. Biochemistry. 2012;51:6463–75. doi: 10.1021/bi300811t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldsmith M, et al. Evolved stereoselective hydrolases for broad-spectrum G-type nerve agent detoxification. Chem Biol. 2012;19:456–66. doi: 10.1016/j.chembiol.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Abatemarco J, et al. Expanding the metabolic engineering toolbox with directed evolution. Biotechnol J. 2013;8:1397–410. doi: 10.1002/biot.201300021. [DOI] [PubMed] [Google Scholar]
- 44.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 45.Hocek M. Synthesis of Base-Modified 2′-Deoxyribonucleoside Triphosphates and Their Use in Enzymatic Synthesis of Modified DNA for Applications in Bioanalysis and Chemical Biology. J Org Chem. 2014 doi: 10.1021/jo5020799. [DOI] [PubMed] [Google Scholar]
- 46.Struthers L, et al. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal Biochem. 1998;255:20–31. doi: 10.1006/abio.1997.2354. [DOI] [PubMed] [Google Scholar]
- 47.Vos JG, et al. Health effects of endocrine-disrupting chemicals on wildlife, with special reference to the European situation. Crit Rev Toxicol. 2000;30:71–133. doi: 10.1080/10408440091159176. [DOI] [PubMed] [Google Scholar]
- 48.Diamanti-Kandarakis E, et al. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–98. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 50.Lewis RJ, et al. Conus venom peptide pharmacology. Pharmacol Rev. 2012;64:259–98. doi: 10.1124/pr.111.005322. [DOI] [PubMed] [Google Scholar]