Summary
Recent research has highlighted a strong correlation between tissue-specific cancer risk and the lifetime number of tissue-specific stem cell divisions. Whether such correlation implies a high unavoidable intrinsic cancer risk has become a key public health debate with dissemination of the ‘bad luck’ hypothesis. Here we provide evidence that intrinsic risk factors contribute only modestly (<10~30%) to cancer development. First, we demonstrate that the correlation between stem-cell division and cancer risk does not distinguish between the effects of intrinsic and extrinsic factors. Next, we show that intrinsic risk is better estimated by the lower bound risk controlling for total stem cell divisions. Finally, we show that the rates of endogenous mutation accumulation by intrinsic processes are not sufficient to account for the observed cancer risks. Collectively, we conclude that cancer risk is heavily influenced by extrinsic factors. These results carry immense consequences for strategizing cancer prevention, research, and public health.
Cancers were once thought to originate from mature tissue cells that underwent de-differentiation in response to cancer progression1. Today, cancers are proposed to originate from the malignant transformation of normal tissue progenitor and stem cells2,3, although this is not uniformly accepted4. Nevertheless, recent research has highlighted a strong correlation of 0.81 between tissue-specific cancer risk and the lifetime population size and cumulative number of cell divisions of tissue-specific stem cells5. However, there has been extensive controversy regarding the conclusion that this correlation implies a very high unavoidable risk for many cancers that are due solely to the intrinsic baseline population size of tissue-specific stem cells6,7. Much discussion has been made to argue against the ‘bad luck’ hypothesis 5–13, yet none offered specific alternatives to quantitatively evaluate the contribution of extrinsic risk factors in cancer development. Applying several distinct modeling approaches, we here provide strong evidence that unavoidable intrinsic risk factors contribute only modestly (<10~30%) to the development of many common cancers.
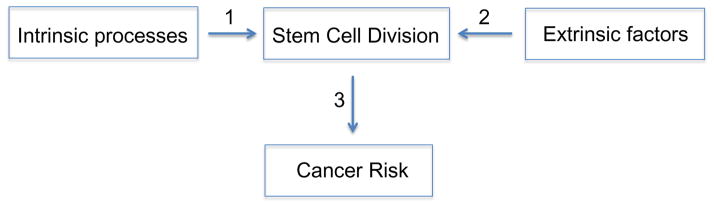
We start by making the conservative and yet conventional assumption that errors occurring during the division of cells, being routes of malignant transformation, can be influenced by both intrinsic processes as well as extrinsic factors (Fig. 1). “Intrinsic processes” include those that result in mutations due to random errors in DNA replication whereas “extrinsic factors” are environmental factors that affect mutagenesis rates (such as UV radiation, ionizing radiation, and carcinogens). For example, radiation can cause DNA damage, which would primarily result in deleterious mutations with functional consequences on cancer development only after cell division. Therefore, extrinsic factors may act through the accumulation of genetic alterations during cell division to increase cancer risk. Accordingly, intrinsic risk would result from those apparently uncontrollable intrinsic processes (Arrow 1, Fig. 1) as well as from those highly modifiable and thus preventable extrinsic factors (Arrow 2, Fig. 1).
Figure 1. A schematic view of how intrinsic processes and extrinsic factors are related to cancer risks through stem-cell division.
This hypothesis maintains the strong role of stem-cell division in imparting cancer risk, but it also illustrates the potential contributions of both intrinsic and extrinsic factors, both operating through stem-cell division. Other effects, e.g. through division of non-stem cells, are not considered here.
Correlation cannot differentiate risks
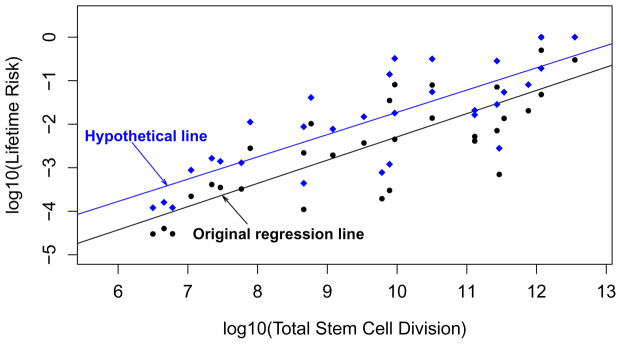
According to the above hypothesis, both intrinsic and extrinsic factors can impart cancer risk through the accumulation of these errors, especially the ‘driver mutations’ (Arrow 3, Fig. 1). As such, a correlational analysis between cancer risk and cell division, for either stem or non-stem cells, is unable to differentiate between the contributions of intrinsic and extrinsic factors. This is best illustrated through a thought experiment where we consider a hypothetical scenario of a sudden emergence of a very potent mutagen globally such as a strong radiation burst from a nuclear fallout that quadruples the lifetime risks for all cancers. In this scenario, it transpires that the proportion of cancer risk explained by intrinsic random errors would be small (at most 1/4 even if we assume all the original risk was due to intrinsic processes). However, if we conduct regression analyses on either the new hypothetical cancer risks or the current cancer risks as reported, against the number of stem-cell divisions5, the correlations from both cases would be 0.81 (Fig. 2). This clearly argues against the implication that ~2/3 of variation could be explained by division-related random intrinsic errors and indicates that correlational analysis cannot distinguish between intrinsic and extrinsic factors.
Figure 2. Correlation analysis of stem-cell division and cancer risk does not distinguish contribution of extrinsic vs. intrinsic factors to cancer risk.
The black dots are data in Fig. 1 (also tabulated in Supplementary Table S1) of the original work by Tomasetti and Vogelstein5. The black line was their original regression line. The blue diamonds represent the hypothesized quadrupled cancer risks due to hypothetical exposure to an extrinsic factor such as radiation. The blue regression line for the hypothetical risk data maintains the same correlation as the original black line, albeit reflecting a much higher contribution of extrinsic factors to cancer risk.
Extrinsic risks by tissue cell turnover
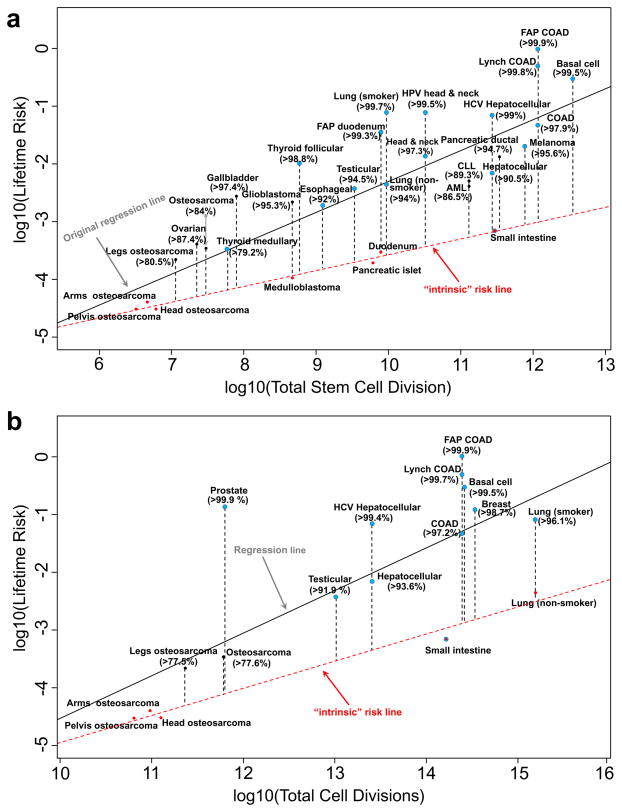
The above conclusion then raises the question of what proportion of total cancer risk is due to extrinsic versus intrinsic factors. In a data-driven approach, we first re-examine the quantitative relationship between the observed lifetime cancer risk and the divisions of the normal tissue stem cells as reported5, with a distinct alternative method. Our rationale is that intrinsic risk, or indeed its upper bound, can be better estimated by the lowest boundary on the plots of cancer risk vs. total tissue stem-cell divisions (red line in Fig. 3a). In other words, intrinsic cancer risk should be determined by the cancer incidence for those cancers with the least risk in the entire group controlling for total stem cell divisions (red dots in Fig. 3a). The argument here is that cancers with the same stem-cell divisions should share the same base of intrinsic cancer risk (if the relationship is causal); if one or more cancers would feature a much higher cancer incidence, for example, lung cancer among smokers vs. non-smokers, then this most likely reflects additional (and probably extrinsic) risk factors (smoking in this case). One could argue that the low-incidence tumor types may have lower incidences because of additional genetic repair mechanisms that restrict evolving malignant cells from accumulating sufficient numbers of genetic alterations required to become fully tumorigenic; however, without more specific data on the operation of repair mechanisms, these could drive the risk up or down, depending on whether they are less or more efficient in any particular tissue. Since, according to our hypothesis, intrinsic risk from stem-cell divisions would define the lowest bound for a given number of stem-cell divisions, we define an “intrinsic” risk line for stem-cell divisions by regressing the smallest cancer risks on any given number of stem-cell divisions (red line, Fig. 3a). The “intrinsic” risk lines themselves are still likely overestimates for the intrinsic risk; however, we should suspect that any cancer risk above that line implies additional biologic determinants, based on which we can compute the percentage of cancer risk not explained by intrinsic “randomness”. As shown in Fig. 3a, most cancer types have very high excess risks relative to the “intrinsic” risk line, indicating large proportions of risks unaccounted by the intrinsic factors, typically larger than 90%. Moreover, these estimated excess risks are very robust – with plausible measurement errors added to the total stem-cell divisions, the resulting excess risks remain essentially intact (Extended Data Table 1).
Figure 3. Estimation of the proportion of lifetime cancer risk that is not due entirely to “bad luck” based on: (a). total tissue stem-cell divisions originally reported in Tomasetti and Vogelstein5, and (b). total tissue cell divisions.
Here red dots are cancers used to compute the “intrinsic” risk linear regression lines (red dashed lines). Blue dots are cancers known to have substantial extrinsic risks from epidemiology studies. The numbers in parentheses are the estimated percentages of cancer risks due to factors other than intrinsic risks.
Although we preformed the initial analysis from a ‘stem-cell theory’ point of view, we wanted to ensure that our results are independent to this specific theory. Furthermore, the lack of reliable data on human tissue stem cell dynamics is a serious concern (see Supplementary Information) rendering the analysis in Fig. 3a less determinate. Thus we separately collected data for the total number of tissue cell divisions that is based on homeostatic tissue cell numbers and their turn-over rates (see Supplementary Information), and analyzed the relationship of cancer risk vs. total tissue cell divisions (Fig. 3b). This approach allows for every dividing cell to be a potential cancer-initiating cell, which would be an application of another cell-of-origin theory of cancer whereby tumors may originate from a hierarchy of cells, from stem cells to committed progenitor cells to differentiated cells4. Mathematically, this can be considered also as an extreme form of stem cell theory where the fraction of stem cells is 1 (this latter formulation then provides an upper bound of the effects of the size of the stem cell population on cancer risk and the role of extrinsic factors). The regression analysis between cancer risk and total tissue cell division shows a high correlation of 0.75, establishing a strong quantitative relationship between cancer risk and total cell division. To dissect the extrinsic vs intrinsic risks, we applied the same rationale and regressed the smallest cancer risks on any given number of cell divisions (red line, Fig. 3b). Although we could only find reliable turn-over data for a subset of tissues, it is remarkable that the conclusion drawn here is nearly identical to that in Fig. 3a, i.e., large proportions of risks that may not be attributable to intrinsic factors, are mostly higher than 90%. It is important to note that here we included breast and prostate cancers – two high-incidence cancers missing in the original stem-cell analysis5. Again, plausible measurement errors have been added to the total cell divisions, and the excess risks remained almost identical (Extended Data Table 1). In summary, irrespective of whether a subpopulation or all dividing cells contribute to cancer, these results indicate that intrinsic factors do not play a major causal role.
Epidemiological evidence
In parallel, numerous epidemiological studies have established strong evidence that many cancers have substantial risk proportions attributed to environmental exposures (Extended Data Table 2). Particularly, for breast and prostate cancers, it has long been observed that large international geographical variations exist in their incidences (5-fold for breast cancer, 25-fold for prostate cancer)14, and immigrants moving from countries with lower cancer incidence to countries with higher cancer rates soon acquire the higher risk of their new country15,16. While several risk factors have been identified for these cancers, no single one can account for their substantial extrinsic risk proportions, suggesting complex mechanisms for their etiologies. Colorectal cancer is another high-incidence cancer that is widely considered to be an environmental disease17, with an estimated 75% or more colorectal cancer risk attributable to diet18. For many other cancers, known environmental risk factors have also been identified. For example, for melanoma, its risk ascribed to sun exposure is around 65–86%19, and for non-melanoma basal and squamous skin cancers, ~90% is attributable to UV20. At least 75% of esophageal cancer, or head and neck cancer are caused by tobacco and alcohol21,22. It is also well known that certain pathogens may dramatically increase the risk of cancers. For instance, HPV may cause ~90% cases in cervical cancer23, ~90% cases in anal cancer24, and ~70% in oropharyngeal cancer25; HBV and HCV may account for ~80% cases of hepatocellular carcinoma26; and H pylori may be responsible for 65–80% of gastric cancer27. These, along with many other reports, provide direct evidence that environmental factors play important roles in cancer incidence and they are modifiable through lifestyle changes and/or vaccinations.
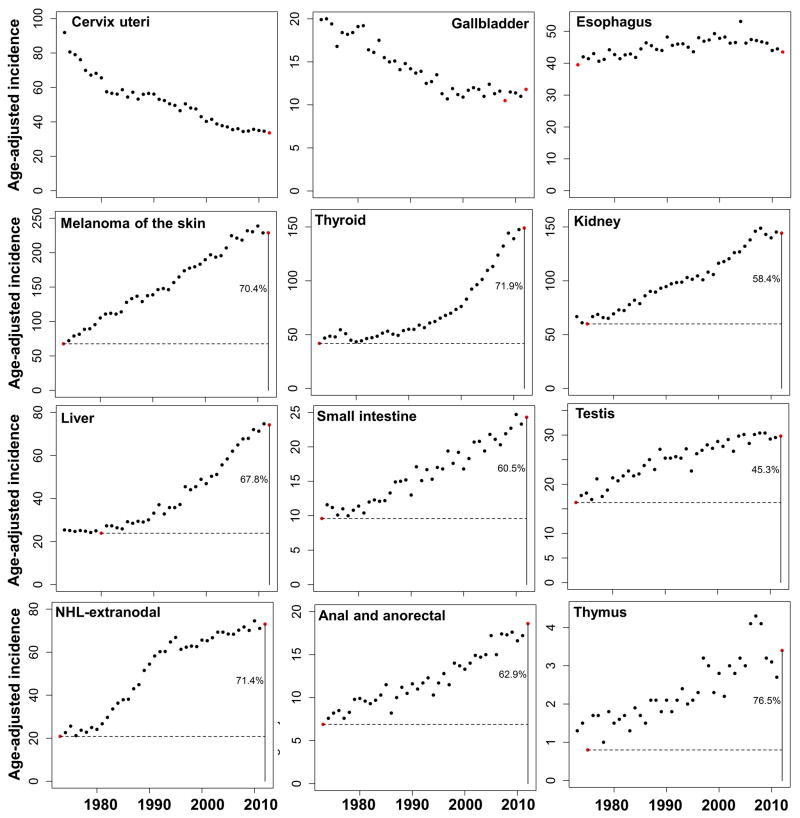
Additionally, analyses of data from the Surveillance, Epidemiology, and End Results Program (SEER) in U.S. between 1973–2012 demonstrate that while many cancers maintain relatively consistent age-adjusted incidence rates, e.g. esophagus cancer, incidences for some cancers, including melanoma, thyroid, kidney, liver, thymus, small intestine, extranodal non-Hodgkin’s lymphoma (NHL), testis, anal and anorectal cancers, have been steadily increasing and their current incidences are substantially higher than their historical minima in the past 40 years28 (Extended Data Fig. 1). Moreover, the mortality trend of lung cancer from 1930–201129, which usually mirrors its incidence trend, shows more than 15-fold increase for lung cancer risk. These significant increases in incidence suggest that substantial risk proportions are attributable to changing environments (e.g. smoking and air pollutants to lung cancers). Collectively, nearly all major cancers have been covered in these epidemiological studies, further supporting the hypothesis of substantial extrinsic risks for most cancers. Remarkably, it should be noted that most of these cancers from the epidemiological and SEER results, except for small intestine (which starts from a very low risk although it is increasing), are located above the red “intrinsic” risk lines in Figs. 3a & 3b (blue points), and accounting for the external factors would move them closer to the proposed ‘intrinsic’ line; thus further supporting the conjecture that the intrinsic line is mainly defined by cancers without compelling known epidemiological risk whereas those above are at higher risks due to extrinsic factors.
Analysis of mutational signatures
Besides epidemiological studies, we evaluated recent studies on mutational signatures in cancer. These are regarded as fingerprints left on cancer genomes by different mutagenic processes30, revealing ~30 distinct signatures among various cancers31. Analysis of these signatures was therefore used to shed light on the proportion of intrinsic versus extrinsic origins of cancer. Two signature mutations, 1A/1B, demonstrated strong positive correlations with age in the majority of cancers, suggesting that they are acquired at a relative constant rate over the lifetime of cancer patients and thus likely result from intrinsic processes; however, all other signature mutations (~30) lack the consistent correlations with age, suggesting they are acquired at different rates in life and thus probably a consequence of extrinsic carcinogen exposures31. Indeed, several mutational signatures have been linked to known factors such as UV and smoking31. We therefore categorized the signatures into intrinsic (type 1A/1B) and extrinsic mutations with known or unknown factors, and summarized their corresponding percentages in the Extended Data Table 3. Significantly, many cancers have substantial extrinsic mutations with known factors. More importantly, cancers known to have substantial environmental risk proportions, e.g. Breast cancer15, Prostate cancer16, Colorectal cancer18, Melanoma19, Head & Neck cancer21, Esophageal cancer22, Cervical cancer23, Liver cancer26, and Stomach cancer27, all harbor large percentages of total extrinsic mutational signatures. This suggests that the percentages of total extrinsic mutational signatures can serve as a good surrogate for extrinsic cancer risks. While a few cancers have relatively large proportions of intrinsic mutations (>50%), the majority of cancers have large proportions of extrinsic mutations, for example, ~100% for Myeloma, Lung and Thyroid cancers and ~80–90% for Bladder, Colorectal and Uterine cancers, indicating substantial contributions of carcinogen exposures in the development of most cancers.
Modeling theoretical intrinsic risk
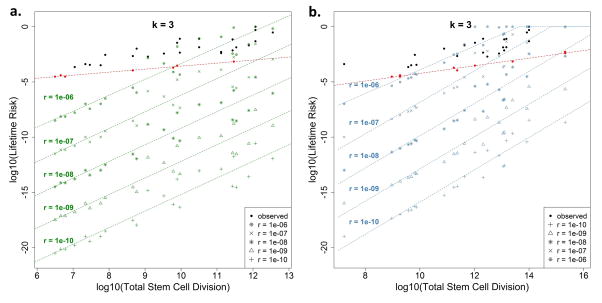
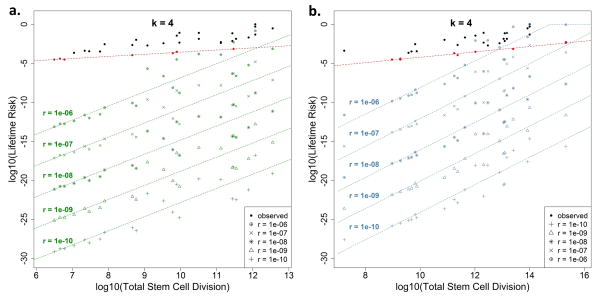
Lastly, in another independent, model-driven approach to dissecting the risk contribution of the intrinsic processes, we modeled the potential lifetime cancer risk due to intrinsic stem-cell mutation errors by varying the number of hits (i.e. driver gene mutations), denoted by k, required for cancer onset. We derived the probability distribution of the propagation of driver gene mutations from one generation to the next, and subsequently established the theoretical relation between cell divisions and the degree of lifetime cancer risk due to intrinsic cell mutation errors alone, which we refer to as the theoretical lifetime intrinsic risk (tLIR). To overcome the limitation of inaccurate estimation in the reported stem cell numbers5, we calculated tLIR using both the reported stem cell number (tLIRsc) and the total tissue cell number (tLIRtt). The latter is equivalent to assuming all homeostatic tissue cells to be stem cells, representing an extreme overestimation of tissue stem cells, which consequently leads to a conservative estimation of the upper bounds in tLIR. The somatic mutation rate in tumors is estimated to be 5 × 10−10 per nucleotide site per cell division32–34. Based on this, in our initial calculation, we used an intrinsic mutation rate (r) of 1 × 10−8 per cell division, which is equivalent to approximately 20 mutable nucleotide sites for each driver gene where the driver will mutate if at least one site mutates. As shown in Figs. 4a and 4b, if only one hit (that is, mutation of one designated driver gene) is required to develop cancer, i.e. k = 1, the lifetime risk for almost all cancers is close to 100%. This confirms that one mutation is not enough for cancer onset (otherwise everyone would theoretically acquire each type of cancer). If two driver gene mutations are needed, k = 2, the modeled intrinsic risk becomes small for cancers with small total number of stem-cell divisions; however it is still very large for those with higher stem-cell divisions and even unreasonably large for some cancers by surpassing the corresponding observed total lifetime cancer risks (Adjusted Basal, COAD, Adjusted Melanoma, Small Intestine, AML and Duodenum). Therefore, it is unlikely that, at least in these cancers, two hits will suffice to induce cancer. Now, if we consider the more reasonable case where three mutations are required35, k = 3, almost all modeled intrinsic risks (both tLIRsc and tLIRtt) drops well below our earlier “intrinsic” risk lines estimated conservatively from the observed data alone (red dashed lines estimated based on observed data following the same mechanism as Fig. 3a). The lifetime risk drops even further for k = 4 and beyond. The extrinsic risks based on the tLIRsc and tLIRtt have been summarized in the Extended Data Table 4. Therefore, this modeling approach demonstrates that cancer risk due to intrinsic stem-cell mutation errors alone is low for almost all cancers that require over 2 mutations, indeed it is lower than the relatively conservative estimate based on data alone (red lines). Since the driver-gene mutation rate in stem-cell division is a key parameter, we further conducted sensitivity analyses with different rates (r = 1 × 10−10 to 1 × 10−6) to examine how this may impact the tLIR (Extended Data Fig. 2 and 3). The results show that for k = 3, when r < 1 × 10−7 (~200 sites for each driver-gene hit), almost all modeled intrinsic risks are below the observed “intrinsic” risk line (red lines); when r = 1 × 10−6 (~2000 sites for each driver-gene hit), the majority of modeled intrinsic risks are still well below the observed “intrinsic” risk lines, particularly those with small total number of divisions (Extended Data Fig. 2). For k = 4, when r < 1 × 10−6, almost all modeled intrinsic risks are below the observed “intrinsic” risk lines estimated through the data-driven approach (Extended Data Fig. 3). These sensitivity analyses demonstrate that our conclusions are highly robust, and that the attribution of intrinsic mutations to lifetime cancer risk through stem-cell divisions, particularly for those cancers with low risk, is rather small, even using widely different intrinsic mutation rates.
Figure 4. Theoretical lifetime intrinsic risks (tLIR) for cancers based on different number of hits (k) required for cancer onset.
The green (a) and blue (b) dashed lines are the “intrinsic” risk lines estimated based on total reported stem cell numbers and total homeostatic tissue cells, respectively. The intrinsic stem cell mutation rate (r) is assumed to be 1 × 10−8 per cell division. The red dashed lines are the “intrinsic” risk lines estimated based on the observed data using the same mechanism as Fig. 3a. Adj. Basal and Adj. Melanoma represent cancer risks after adjusting for the effect of sun exposure and UV.
In summary, we find that a simple regression analysis cannot distinguish between intrinsic and extrinsic factors. We have provided a new framework to quantify the lifetime cancer risks from both intrinsic and extrinsic factors based on four independent approaches that are data-driven and model-driven, with and without using the stem-cell estimations. Importantly, these four approaches provide a consistent estimate of contribution of extrinsic factors of >70–90% in most common cancer types. This concordance lends significant credibility to the overall conclusion on the role of extrinsic factors in cancer development.
Methods
Derivation of the probability of possessing k hits after n cell divisions for one cell
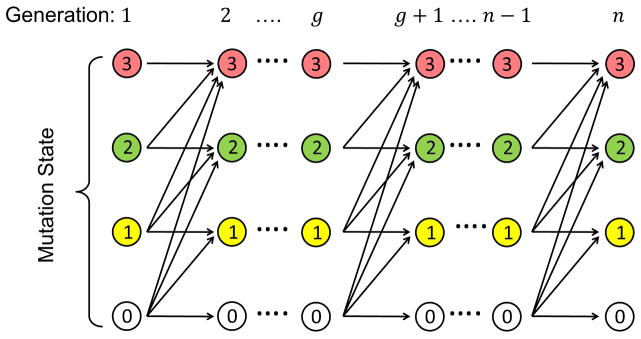
Based on the theory of the clonal stem-cell origin of cancer, in a given tissue, the stem cell would first go through m rounds of symmetric divisions (for each division, each stem cell would divide into two daughter stem cells) to reach a total of S stem cells (S = 2m) at the steady state. Subsequently, these S stem cells would go through a rounds of asymmetric divisions (for each division, each stem cell would yield only one daughter stem cell) throughout the lifetime of the tissue. This means the total number of lifetime stem cell divisions/generations is: n = m + a. Information on the total number of symmetric and asymmetric divisions as well as the total number of stem cells in steady state for various tissues discussed in this work has been extracted from Table S1 of the supplementary materials in Tomasetti and Vogelstein5. With k hits (mutations of k predetermined driver genes) on a stem cell required for cancer onset, the number of possible cell state at a given (stem cell) generation would be k +1, including a zero state with no hit. If we assume that once a hit occurs, it cannot be reversed and therefore be carried to all progeny cells, then a cell state may only transit from lower to higher or equal levels from generation to generation. In the Extended Data Fig. 4, we demonstrate with k = 3 the state transitions of accumulating driver gene mutations. Let Xg denote the number of driver mutations accumulated at generation g and r be the intrinsic driver gene mutation rate due to random errors during DNA replication, the transition probabilities from generation g to g + 1 for all possible states (0 ≤ i ≤ k) are derived as follows:
In particular, for the emission state i = 0:
For the absorbing state i = k:
Based on these, the computing algorithm is derived as follows:
- Set the initial cell state at generation 0:
-
For g = 1, …, n, and 0 ≤ i ≤ k, we compute the following probabilities iteratively:
where n is the total number of divisions that one stem cell may experience during its life time.
Derivation of the theoretical lifetime intrinsic risk (tLIR) of cancer for a given tissue
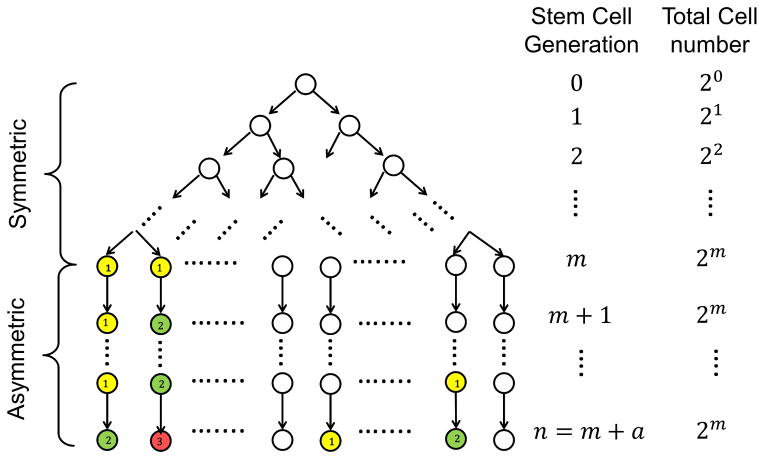
As mentioned afore, for stem cells in a specific tissue, we assume they undergo two phases of divisions (Extended Data Fig. 5): (1) a total of m symmetric divisions before full tissue development, and (2) a total of a asymmetric divisions for normal tissue turnovers. So in a fully developed tissue, there are a total of S = 2m stem cells. For each stem cell, its probability of possessing all k hits for cancer onset after n = m + a divisions is P(Xn = k), which can be calculated from the previous part. Therefore, the theoretical lifetime intrinsic risk (tLIR) of developing cancer, i.e., the probability of at least one stem cell containing k hits during its life time, can be expressed as:
Estimating cancer risk for different tissues
The numbers of symmetric and asymmetric divisions for different tissues were adopted from Table S1 in the supplementary materials of Tomasetti and Vogelstein5. In particular, the number of symmetric divisions, m, is equal to the integer part of log2 S where S is the number of normal stem cells in tissue of origin in the Table S15, and the number of asymmetric divisions, a was the column labeled d in Table S15. Sensitivity analyses have been conducted for scenarios with a broad range of mutation rates, from 1 × 10−10 to 1 × 10−6, and several required hits (k = 1, 2, 3, 4).
Lower-bound estimates of extrinsic risks with the SEER data
As a program of the National Cancer Institute (NCI), SEER (Surveillance, Epidemiology, and End Results Program) is a source of information on cancer incidence and survival in the United States (http://seer.cancer.gov/). The age-adjusted cancer incidences were extracted from the database “SEER 9 Regs Research Data, Nov 2014 Sub (1973–2012) <Katrina/Rita Population Adjustment>”, by using the SEER*Stat 8.2.128. For several cancers, it has been observed that their incidence rates have increased dramatically during the past 40 years (Extended Data Fig. 1). For these cancers, it is reasonable to conjecture that anything above the historical minimum incidence should be attributed to some environmental/extrinsic factors. Therefore, we can establish the following inequality:
Correspondingly, the lower bounds of contributions by extrinsic factors for these cancers can be calculated. As shown in Extended Data Fig. 1, some cancers show substantial contributions from extrinsic factors.
Data and Statistical Analysis
The observed life time cancer risks and the cumulative number of divisions (n) of all stem cells per lifetime are adopted from Table S1 of the supplementary materials by Tomasetti and Vogelstein5. The total tissue cell divisions are from our evaluation of the data (Supplementary Information). For the robustness analysis of Fig. 3 as tabulated in Extended Data Table 1, error terms following the normal distribution with mean 0 and standard deviations of 1 or 0.4 were added to the log10(total stem-cell division) or log10(total cell division). These allows the number of total stem-cell and cell divisions to vary approximately within a range of (1/100 ~ 100) or (1/5 ~ 5) fold(s), respectively. Based on the new data set with measurement errors, the excess risks for each cancer were quantified. This process is repeated for 1,000 times, based on which the mean, the 2.5% and the 97.5% percentiles (namely the 95% confidence intervals) of the excess risk for each cancer are tabulated. In computing the percent of intrinsic versus extrinsic mutations based on mutational signatures from cancer genome, we define the intrinsic mutation as those with signatures 1A/1B, and extrinsic mutation as all other mutational signatures (2–21, R1–R3, U1 and U2). The corresponding data were obtained from the supplemental figures 59–88 in Alexandrov et al31. All statistical analyses and mathematical calculations were performed using R version 3.1.2.
Extended Data
Extended Data Figure 1. Examples of increased cancer incidence trends from 1973 – 2012.
The cancer types include cervix uteri cancer, gallbladder cancer, esophagus cancer, melanoma, thyroid cancer, kidney cancer, liver cancer, small intestine cancer, thymus cancer, anal and anorectal cancer. The horizontal dashed line indicates the historical minimal incidence. The vertical solid line indicates the most recent year. The number represents the minimal percentage of extrinsic risk. The incidence rate is based on per 100,000 people.
Extended Data Figure 2. Sensitivity analysis of different mutation rates on tLIR when the number of hits (k) required is 3.
Theoretical intrinsic lifetime risks (tLIR) for cancers have been calculated, based on five different mutation rates (r = 1 × 10−10, 1 × 10−9, 1 × 10−8, 1 × 10−7, 1 × 10−6). The red dashed lines are the “intrinsic” risk lines based on the observed data following the same estimation mechanism as the intrinsic risk line in Fig. 3a. The green (a) and blue (b) dashed lines are the “intrinsic” risk lines estimated based on total reported stem cell numbers and total homeostatic tissue cells, respectively.
Extended Data Figure 3. Sensitivity analysis of different mutation rates on tLIR when the number of hits (k) required is 4.
Theoretical intrinsic lifetime risks (tLIR) for cancers have been calculated, based on five different mutation rates (r = 1 × 10−10, 1 × 10−9, 1 × 10−8, 1 × 10−7, 1 × 10−6). The red dashed lines are the “intrinsic” risk lines based on the observed data following the same estimation mechanism as the intrinsic risk line in Fig. 3a. The green (a) and blue (b) dashed lines are the “intrinsic” risk lines estimated based on total reported stem cell numbers and total homeostatic tissue cells, respectively.
Extended Data Figure 4.
Intrinsic cancer risk modeling, Part 1/2: Propagation diagram of driver gene mutation states between generations in one stem cell based on which the stem cell mutation transition probabilities from one generation to the next are computed.
Extended Data Figure 5.
Intrinsic cancer risk modeling, Part 2/2: Schema of stem-cell divisions and driver gene mutations based on which the theoretical lifetime intrinsic risks (tLIR) for cancer due to k driver gene mutations are computed.
Here every colored circle represents the mutation of a new driver gene in the given stem cell (yellow: first mutation; green: second mutation; red: third mutation). If the mutation of 3 designated driver genes would induce a cancerous stem cell (k = 3), then this diagram shows a cancer occurrence as the second stem cell in the last generation (generation n) has accumulated all 3 driver gene mutations.
Extended Data Table 1. Robustness analysis on total stem-cell divisions and cell divisions estimates in Fig. 3.
Measurement errors were added to log10(divisions) and 1000 simulations were carried out to calculated the mean and 95% Confidence Interval (CI) of the excess risks. See Methods for details.
| Total stem-cell divisions (Fig. 3A) | Total cell divisions (Fig. 3B) | ||||||
|---|---|---|---|---|---|---|---|
| Name | Observed Risk | Log10 (divisions) | Excess risk | Excess risk 95% CI1 | Log10 (divisions) | Excess risk | Excess risk 95% CI1 |
| AML | 0.0041 | 11.11 | >0.871 | [0.623, 0.962] | NA | NA | NA |
| Basal cell | 0.3 | 12.55 | >0.996 | [0.985, 0.999] | 14.42 | >0.995 | [0.99, 0.998] |
| Breast | 0.123 | NA | NA | NA | 14.54 | >0.987 | [0.974, 0.994] |
| CLL | 0.0052 | 11.11 | >0.899 | [0.701, 0.973] | NA | NA | NA |
| COAD | 0.048 | 12.07 | >0.980 | [0.934, 0.995] | 14.40 | >0.971 | [0.943, 0.986] |
| FAP COAD | 1 | 12.07 | >0.999 | [0.997, 1.000] | 14.40 | >0.999 | [0.997, 0.999] |
| Lynch COAD | 0.5 | 12.07 | >0.998 | [0.994, 1.000] | 14.40 | >0.997 | [0.994, 0.999] |
| Duodenum2 | 3.00E-04 | 9.89 | - | - | NA | NA | NA |
| FAP Duodenum | 0.035 | 9.89 | >0.993 | [0.980, 0.998] | NA | NA | NA |
| Esophageal | 0.00194 | 9.08 | >0.906 | [0.748, 0.975] | NA | NA | NA |
| Gallbladder | 0.0028 | 7.89 | >0.967 | [0.922, 0.991] | NA | NA | NA |
| Glioblastoma | 0.00219 | 8.43 | >0.943 | [0.868, 0.984] | NA | NA | NA |
| Head & neck | 0.0138 | 10.50 | >0.973 | [0.921, 0.992] | NA | NA | NA |
| HPV Head & neck | 0.07935 | 10.50 | >0.995 | [0.985, 0.999] | NA | NA | NA |
| Hepatocellular | 0.0071 | 11.43 | >0.906 | [0.720, 0.975] | 13.41 | >0.932 | [0.872, 0.969] |
| HCV Hepatocellular | 0.071 | 11.43 | >0.991 | [0.969, 0.998] | 13.41 | >0.993 | [0.986, 0.997] |
| Lung (nonsmoker)3 | 0.0045 | 9.97 | >0.938 | [0.835, 0.982] | 15.2 | - | - |
| Lung (smoker) | 0.081 | 9.97 | >0.997 | [0.990, 0.999] | 15.20 | >0.958 | [0.905, 0.982] |
| Medulloblastoma2 | 0.00011 | 8.43 | - | - | NA | NA | NA |
| Melanoma | 0.0203 | 11.88 | >0.960 | [0.872, 0.990] | NA | NA | NA |
| Osteosarcoma | 0.00035 | 7.47 | >0.790 | [0.459, 0.947] | 11.79 | >0.762 | [0.568, 0.887] |
| Arms osteosarcoma2,3 | 4.00E-05 | 6.66 | - | - | 10.99 | - | - |
| Head osteosarcoma2,3 | 3.02E-05 | 6.78 | - | - | 11.1 | - | - |
| Legs osteosarcoma | 0.00022 | 7.05 | >0.727 | [0.306, 0.930] | 11.37 | >0.761 | [0.537, 0.889] |
| Pelvis osteosarcoma2,3 | 3.00E-05 | 6.50 | NA | NA | 10.81 | - | - |
| Ovarian germ cell | 0.000411 | 7.34 | >0.832 | [0.573, 0.958] | NA | NA | NA |
| Pancreatic ductal | 0.013589 | 11.54 | >0.948 | [0.805, 0.987] | NA | NA | NA |
| Pancreatic islet2 | 0.000194 | 9.78 | - | - | NA | NA | NA |
| Prostate | 0.14 | NA | NA | NA | 11.81 | >0.999 | [0.999, 1] |
| Small intestine2,3 | 7.00E-04 | 11.47 | - | - | 14.22 | - | - |
| Testicular | 0.0037 | 9.53 | >0.942 | [0.843, 0.984] | 13.02 | >0.914 | [0.835, 0.959] |
| Thyroid follicular | 0.01026 | 8.77 | >0.986 | [0.964, 0.996] | NA | NA | NA |
| Thyroid medullary | 0.000324 | 7.77 | >0.731 | [0.308, 0.928] | NA | NA | NA |
Confidence Interval.
Cancers used to compute the “intrinsic” risk line based on total stem-cell divisions.
Cancers used to compute the “intrinsic” risk line based on total cell divisions. NA: data not available.
Extended Data Table 2.
Epidemiological studies on the extrinsic risks of various cancers.
| Cancer Types | Extrinsic risk | Examples of potential extrinsic risk factors1 |
|---|---|---|
| Breast | substantial | Oral contraceptive, hormone replacement therapy, lifestyle (diet, smoking, alcohol, weight) |
| Prostate | substantial | Diet, obesity, smoking |
| Lung | >90% | Smoking; air pollutant |
| Colorectal | >75% | Diet, smoking, alcohol, obesity |
| Melanoma | 65–86% | Sun exposure |
| Basal cell | ~90% | UV |
| Hepatocellular | ~80% | HBV, HCV |
| Gastirc | 65–80% | H. pylori |
| Cervical | ~90% | HPV |
| Head & Neck | ~75% | Tobacco, alcohol |
| Esophageal | >75% | Smoking, alcohol, obesity, diet |
| Oropharyngeal | ~70% | HPV |
| Thyroid | >72% | Diet low in iodine, radiation |
| Kidney | >58% | Smoking, obesity, workplace exposures |
| Thymus | >77% | Largely unclear |
| Small intestine | >61% | Diet, smoking, alcohol |
| Extranodal non-Hodgkin’s lymphoma (NHL) | >71% | Chemicals, radiation, immune system deficiency |
| Testis | >45% | Largely unclear |
| Anal and anorectal cancers | >63% | HPV, smoking |
Extended Data Table 3. Percentages of intrinsic vs. extrinsic mutational signatures (MS) with known and unknown causes in different cancer types.
Intrinsic MS includes signatures 1A/B, and extrinsic MS includes signatures 2–21, R1–R3, U1 and U2, excluding signature 11 for Temozolomide, an alkylating agent used for chemotherapy. The blue, yellow and red colors highlight cancers that are have substantial extrinsic risk proportions based on epidemiological, MS with known causes and, MS with unknown causes, respectively. (Data from the supplemental figures 59–88 in Alexandrov et al31)
| Intrinsic MS | Extrinsic MS - Known | Extrinsic MS - Unknown | Extrinsic MS - Total | |
|---|---|---|---|---|
| ALL | 65.8 | 34.2 | 0 | 34.2 |
| AML | 100 | 0 | 0 | 0 |
| Bladder | 14.2 | 71.2 | 14.6 | 85.8 |
| Breast | 35.5 | 60.1 | 4.4 | 64.5 |
| Cervical | 25.3 | 74.7 | 0 | 74.7 |
| CLL | 76.7 | 23.3 | 0 | 23.3 |
| Colorectal | 17.1 | 66 | 16.9 | 82.9 |
| Esophageal | 48 | 25.3 | 26.7 | 52 |
| Glioblastoma | 53.8 | 0 | 46.2 | 46.2 |
| Glioma-Low Grade | 9.2 | 2.8 | 88 | 90.8 |
| Head & Neck | 24.9 | 75.1 | 0 | 75.1 |
| Kidney Chromophobe | 17.4 | 37.5 | 45.1 | 82.6 |
| Kidney Clear Cell | 66.5 | 4.1 | 29.4 | 33.5 |
| Kidney Papillary | 0 | 15.7 | 84.3 | 100 |
| Liver | 10.9 | 21.3 | 67.8 | 89.1 |
| Lung Adenocarcinoma | 9.1 | 73.8 | 17.1 | 90.9 |
| Lung - Small Cell | 0 | 92.8 | 7.2 | 100 |
| Lung-Squamous | 0 | 47 | 53 | 100 |
| Lymphoma B-cell | 46.3 | 33.4 | 20.3 | 53.7 |
| Medulloblastoma | 48.4 | 0 | 51.6 | 51.6 |
| Melanoma | 7.2 | 90.9 | 1.9 | 92.8 |
| Myeloma | 0 | 19.9 | 80.1 | 100 |
| Neuroblastoma | 53.2 | 0 | 46.8 | 46.8 |
| Ovarian | 36.6 | 63.4 | 0 | 63.4 |
| Pancreatic | 49.9 | 50.1 | 0 | 50.1 |
| Pilocytic Astrocytoma | 82.5 | 0 | 17.5 | 17.5 |
| Prostate | 32.2 | 10.2 | 57.6 | 67.8 |
| Stomach | 22.3 | 6.1 | 71.6 | 77.7 |
| Thyroid | 0 | 39.7 | 60.3 | 100 |
| Uterine | 10.7 | 65.5 | 23.8 | 89.3 |
Extended Data Table 4. Percentages of extrinsic risks based on the reported stem-cell estimates and total homeostatic tissue cells, as shown in Fig. 4.
Extrinsic risk = 1 − (tLIRsc or tLIRtt)/observed risk. H.T.O.: Higher than the observed.
| Extrinsic Risks | Based on stem cell estimates | Based on total homeostatic tissue cells | ||||||
|---|---|---|---|---|---|---|---|---|
| Cancer Type | k=1 | k=2 | k=3 | k=4 | k=1 | k=2 | k=3 | k=4 |
| AML | H.T.O. | H.T.O. | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.465 | 1.000 |
| Basal cell | H.T.O. | 0.462 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| CLL | H.T.O. | H.T.O. | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.578 | 1.000 |
| COAD | H.T.O. | H.T.O. | 0.999 | 1.000 | H.T.O. | H.T.O. | 0.928 | 1.000 |
| FAP COAD | H.T.O. | 0.630 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.997 | 1.000 |
| Lynch COAD | H.T.O. | 0.260 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.993 | 1.000 |
| Duodenum | H.T.O. | H.T.O. | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.986 | 1.000 |
| FAP Duodenum | H.T.O. | 0.977 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Esophageal | H.T.O. | 0.946 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.997 | 1.000 |
| Gallbladder | H.T.O. | 1.000 | 1.000 | 1.000 | H.T.O. | 0.974 | 1.000 | 1.000 |
| Glioblastoma | H.T.O. | 0.995 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Head & neck | H.T.O. | 0.631 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.997 | 1.000 |
| HPV Head & neck | H.T.O. | 0.936 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.999 | 1.000 |
| Hepatocellular | H.T.O. | 0.572 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| HCV Hepatocellular | H.T.O. | 0.957 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Lung (nonsmoker) | H.T.O. | 0.971 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Lung (smoker) | H.T.O. | 0.998 | 1.000 | 1.000 | H.T.O. | 0.388 | 1.000 | 1.000 |
| Medulloblastoma | H.T.O. | 0.904 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Melanoma | H.T.O. | 0.444 | 1.000 | 1.000 | H.T.O. | 0.444 | 1.000 | 1.000 |
| Osteosarcoma | H.T.O. | 1.000 | 1.000 | 1.000 | H.T.O. | 0.624 | 1.000 | 1.000 |
| Arms osteosarcoma | H.T.O. | 0.999 | 1.000 | 1.000 | H.T.O. | 0.269 | 1.000 | 1.000 |
| Head osteosarcoma | H.T.O. | 0.999 | 1.000 | 1.000 | H.T.O. | 0.032 | 1.000 | 1.000 |
| Legs osteosarcoma | H.T.O. | 1.000 | 1.000 | 1.000 | H.T.O. | 0.718 | 1.000 | 1.000 |
| Pelvis osteosarcoma | H.T.O. | 1.000 | 1.000 | 1.000 | H.T.O. | 0.542 | 1.000 | 1.000 |
| Ovarian germ cell | H.T.O. | 0.999 | 1.000 | 1.000 | H.T.O. | 0.999 | 1.000 | 1.000 |
| Pancreatic ductal | H.T.O. | 0.806 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Pancreatic islet | H.T.O. | 0.611 | 1.000 | 1.000 | H.T.O. | H.T.O. | 1.000 | 1.000 |
| Small intestine | H.T.O. | H.T.O. | 0.998 | 1.000 | H.T.O. | H.T.O. | 0.684 | 1.000 |
| Testicular | H.T.O. | 0.973 | 1.000 | 1.000 | H.T.O. | H.T.O. | 0.999 | 1.000 |
| Thyroid follicular | H.T.O. | 1.000 | 1.000 | 1.000 | H.T.O. | 0.866 | 1.000 | 1.000 |
| Thyroid medullary | H.T.O. | 0.999 | 1.000 | 1.000 | H.T.O. | 0.785 | 1.000 | 1.000 |
Supplementary Material
Acknowledgments
We thank Dr. Lina Obeid and four anonymous reviewers for their constructive comments. This work was supported in part by NCI grants 97132 and 168409.
Footnotes
Author Contributions: Y.H. formulated the hypothesis. S.W. and Y.H. designed the research. S.W. and W.Z. provided mathematical and statistical analysis. S.W., S.P., W.Z. and Y.H. performed research. S.W., S.P., W.Z. and Y.H. wrote the paper.
The authors declare no competing financial interests.
References
- 1.Sell S. Stem cell origin of cancer and differentiation therapy. Critical reviews in oncology/hematology. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 5.Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashford NA, et al. Cancer risk: Role of environment. Science. 2015;347:727–727. doi: 10.1126/science.aaa6246. [DOI] [PubMed] [Google Scholar]
- 7.Wild C, et al. Cancer risk: Role of chance overstated. Science. 2015;347:728–728. doi: 10.1126/science.aaa6799. [DOI] [PubMed] [Google Scholar]
- 8.Potter JD, Prentice RL. Cancer risk: Tumors excluded. Science. 2015;347:727–727. doi: 10.1126/science.aaa6507. [DOI] [PubMed] [Google Scholar]
- 9.Gotay C, Dummer T, Spinelli J. Cancer risk: Prevention is crucial. Science. 2015;347:728–728. doi: 10.1126/science.aaa6462. [DOI] [PubMed] [Google Scholar]
- 10.Song MY, Giovannucci EL. Cancer risk: Many factors contribute. Science. 2015;347:728–729. doi: 10.1126/science.aaa6094. [DOI] [PubMed] [Google Scholar]
- 11.O’Callaghan M. Cancer risk: Accuracy of literature. Science. 2015;347:729–729. doi: 10.1126/science.aaa6212. [DOI] [PubMed] [Google Scholar]
- 12.Tomasetti C, Vogelstein B. Cancer risk: Accuracy of literature Response. Science. 2015;347:729–731. doi: 10.1126/science.aaa6592. [DOI] [PubMed] [Google Scholar]
- 13.Altenberg L. Statistical Problems in a Paper on Variation In Cancer Risk Among Tissues, and New Discoveries. 2015 http://arxiv.org/pdf/1501.04605v1.pdf.
- 14.Torre LA, et al. Global Cancer Statistics, 2012. Ca-Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 15.Gray J, Evans N, Taylor B, Rizzo J, Walker M. State of the Evidence The Connection Between Breast Cancer and the Environment. Int J Occup Env Heal. 2009;15:43–78. doi: 10.1179/107735209799449761. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu H, et al. Cancers of the Prostate and Breast among Japanese and White Immigrants in Los-Angeles-County. British journal of cancer. 1991;63:963–966. doi: 10.1038/Bjc.1991.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clinics in colon and rectal surgery. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson T, Lund EK. Review article: nutrition, obesity and colorectal cancer. Aliment Pharm Ther. 2007;26:161–181. doi: 10.1111/j.1365-2036.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- 19.Parkin DM, Mesher D, Sasieni P. 13. Cancers attributable to solar (ultraviolet) radiation exposure in the UK in 2010. British journal of cancer. 2011;105(Suppl 2):S66–69. doi: 10.1038/bjc.2011.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh HK, Geller AC, Miller DR, Grossbart TA, Lew RA. Prevention and early detection strategies for melanoma and skin cancer. Current status. Archives of dermatology. 1996;132:436–443. [PubMed] [Google Scholar]
- 21.Blot WJ, et al. Smoking and Drinking in Relation to Oral and Pharyngeal Cancer. Cancer research. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 22.Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterology clinics of North America. 2009;38:27–57. vii. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch FX, et al. Prevalence of Human Papillomavirus in Cervical-Cancer - a Worldwide Perspective. Journal of the National Cancer Institute. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 24.Frisch M, et al. Sexually transmitted infection as a cause of anal cancer. New Engl J Med. 1997;337:1350–1358. doi: 10.1056/Nejm199711063371904. [DOI] [PubMed] [Google Scholar]
- 25.Chaturvedi AK, et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. Journal of Clinical Oncology. 2011;29:4294–4301. doi: 10.1200/Jco.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Serag HB. Epidemiology of Viral Hepatitis and Hepatocellular Carcinoma. Gastroenterology. 2012;142:1264-+. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb PM, et al. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut. 2001;49:347–353. doi: 10.1136/gut.49.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.N.C.I. Surveillance, Epidemiology, and End Results (SEER) Program. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; 2015. SEER*Stat Database: Incidence - SEER 9 Regs Research Data, Nov 2014 Sub (1973–2012) <Katrina/Rita Population Adjustment> - Linked To County Attributes - Total U.S., 1969–2013 Counties. www.seer.cancer.gov. [Google Scholar]
- 29.Dela Cruz CS, Tanoue LT, Matthay RA. Lung Cancer: Epidemiology, Etiology, and Prevention. Clin Chest Med. 2011;32:605-+. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415-+. doi: 10.1038/Nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozic I, Nowak MA. Unwanted Evolution. Science. 2013;342:938–939. doi: 10.1126/science.1247887. [DOI] [PubMed] [Google Scholar]
- 35.Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:118–123. doi: 10.1073/pnas.1421839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.