Abstract
Ancient bacteria originated from metal-rich environments. Billions of years of evolution directed these tiny single cell creatures to exploit the versatile properties of metals in catalyzing chemical reactions and biological responses. The result is an entire metallome of proteins that use metal co-factors to facilitate key cellular process that range from the production of energy to the replication of DNA. Two key metals in this regard are iron and zinc, both abundant on Earth but not readily accessible in a human host. Instead, pathogenic bacteria must employ clever ways to acquire these metals. In this review we describe the many elegant ways these bacteria mine, regulate, and craft the use of two key metals (iron and zinc) to build a virulence arsenal that challenges even the most sophisticated immune response.
Keywords: metal, iron, zinc, pathogenesis, heme, transporters, regulation
The resting state of iron and zinc in the host
Bacterial pathogens must procure essential metals when they invade their mammalian hosts, but metal distribution within the host varies due to their respective chemistries and biological functions. Iron and zinc, the 2nd and 27th most abundant metals in the earth's crust, respectively1,2, are essential nutrients for virtually all living organisms3,4. Iron primarily exists as two cations, the oxidized ferric (Fe3+) form and the reduced ferrous (Fe2+) form5. The gain or loss of an electron from these ions is required for multiple important biological functions, such as oxygen carrying by hemoglobin, electron transport chain reactions, and DNA biosynthesis6,7. Zinc exists solely as Zn2+ and as such is unable to perform redox reactions8. Consequentially, organisms take advantage of these functional differences between iron and zinc to use the metals in distinctly different biological processes. Because oxygen is a major component of the air, we live in an oxidative environment, and as such the oxidized form of iron (i.e. Fe3+) is the most stable and dominant form (e.g. rust)5,9. However, this ferric form of iron is insoluble under common aerobic conditions. Thus, the incorporation of iron into biological structures can be challenging. In contrast, the ferrous form of iron (Fe2+) is relatively soluble under aerobic conditions and is found in most natural water sources5,9.
Although iron makes up less than 0.01% of human body weight (2-4 grams)10, it is absolutely necessary for strong bones and oxygen binding to hemoglobin and myoglobin6,11. Zinc similarly comprises 2-3 grams of human body weight, which is distributed primarily in skeletal muscle and bone12. Zinc is found throughout the body and is redistributed from the blood to the liver during pathology, an action that presumably recycles this metal13. Both metals are intertwined with the host's immune system. The status of zinc affects important functions of host immunity, including lymphocyte production and function, monocyte recruitment, and cytokine production14–17. Iron is used to catalyze the formation of reactive oxygen species (ROS) during macrophage-based killing of bacteria18. A dedicated organelle, the phagolysosome, uses the ability of iron to cycle in Fenton reactions and generate ROS, which harms bacterial membranes, proteins, and DNA19. A description of common host and bacterial factors involved in the exploitation of metals is shown in Table 1. Trafficking of iron and zinc inside of the mammalian host remains parallel to each other. Iron is mainly absorbed from the diet in the duodenum and upper jejunum, in the forms of heme (e.g. meat) or non-heme (e.g. plant), both of which are fractionally absorbed (in the case of iron) 20,21. The majority of dietary zinc comes from red meat, poultry, and seafood22. Zinc is absorbed throughout the intestinal tract facilitated by membrane ZnT and Zrt-, Irt-like protein ZIP transporters as well as cysteine rich intestinal protein (CRIP)23,24. DMT1 (divalent metal transporter 1) and HCP1 (heme carrier protein 1) are responsible for iron and heme absorption, respectively, in the duodenum25,26. The majority of iron that pathogens encounter (∼75% of host iron) will be used as a heme cofactor incorporated into hemoglobin (e.g. during erythropoiesis) which coordinates oxygen for its delivery to tissues and cells25,27,28. Zinc, on the other hand, is more broadly used. This metal is incorporated into about 10% of human proteins, of which over 300 enzymes require Zn2+ for metabolic and regulatory functions2,4,29. An invading pathogen will find 90% of host zinc in skeletal muscle and bone, with some present in organs like the spleen, liver, and kidneys12,30. In these tissues and circulating cells, host zinc is present at 100-500 μM concentrations intracellularly, a large portion of which is bound to metallothioneins31,32. Intracellular zinc is further compartmentalized within the cytosol (50%), nucleus (30-40%), and membranes2,33. Like iron, the remaining zinc, about 0.1%, is present in blood serum (1.25μg/ml serum) bound to albumin (73-91%), macroglobin (9-27%), or various serum proteins and amino acids (2-8%)34–36. Iron and zinc are of such critical importance that their loss must quickly be replenished. For example, humans lose iron daily through sweating, shedding of surface cells, and gastrointestinal blood loss, making dietary replenishment of iron a necessary activity37. Too little iron results in anemia and is the most common and widespread nutritional disorder in the world21. The physiological importance of zinc to humans was first described in 1963, and today zinc deficiency is a global health concern – thought to affect prenatal development, childhood growth, and infection susceptibility38–41. Organisms must have ways to regulate metal concentrations however, since excessive levels are toxic. Excess iron can result in iron overload or haemochromatosis25,42, a case of iron toxicity that damages organs because iron catalyzes Fenton reactions which generate damaging and toxic ROS43–46. Haemochromatosis also fosters a more beneficial environment for invasive and opportunistic pathogens47. Unlike iron that has two stable oxidation states (Fe2+ and Fe3+), zinc only has one stable oxidation state (Zn2+), and thus cannot directly induce generation of ROS. However, excess zinc facilitates ROS formation in neuronal cells, an effect caused by mitochondrial zinc transport and subsequent disruption of the mitochondrial membrane48–50. Zinc toxicity can lead to nausea, vomiting, and diarrhea in humans, which is associated with the suppression of copper absorption and alteration of lipoprotein profiles51,52.
Table 1.
Host and bacterial factors involved in iron and zinc exploitation.
| Iron | Protein Type | Localization | Function | Reference |
|---|---|---|---|---|
| Host sources of iron | Heme containing proteins | Cell membranes, Cytoplasm | Transport electrons and oxygen during respiration | 203 |
| Transferrin | Blood, interstitial fluid | Sequester iron in blood and interstitial fluid | 65,203 | |
| Lactoferrin | Secretory fluids | Sequester iron in secretory fluids | 58 | |
| Ferritin | Cytoplasm | Store iron to balance intracellular iron concentrations | 100 | |
| labile iron pool | Cytoplasm | Buffer intracellular iron concentrations | 204 | |
| Iron acquisition systems | Membrane receptors | Cell membranes | Actively transport iron from the environments | 205 |
| ABC transporters | Cell membranes | Actively transport iron from the environments | 206 | |
| Siderophores | Secreted | Chelates iron with high affinity | 207–209 | |
| Utilization of acquired iron | Heme biosynthesis | Cytoplasm | Transport electrons and bind diatomic gases in respiration, defend oxidative stress | 210 |
| Iron-sulfur protein biosynthesis | Cytoplasm | Synthesize dNTPs, produce energy, and defend against oxidative stress | 211 | |
| Zinc | Protein Type | Localization | Function | Reference |
| Host sources of zinc | Metallothioneins | Cytoplasm | Zinc buffering, suppress inflammatory cytokine secretion | 32,212 |
| Zincosomes | Cytoplasm | Zinc storage and buffering | 33 | |
| Metalloproteinases | Cytoplasm, membrane, and secreted | Degrade extracellular matrix, direct cellular differentiation and tissue morphogenesis | 213–215 | |
| Calprotectin | Cytoplasm – secreted by neutrophils | Chelate zinc and manganese at site of infection | 71 | |
| S100 proteins | Cytoplasm – secreted by neutrophils | Regulate cell proliferation and differentiation. Chelate metals at site of infection | 216 | |
| Zinc fingers | Cytoplasm/nucleus | Transcription factors, nucleases, polymerases, ribosomes | 217,218 | |
| Serum albumin | Blood, interstitial fluid | Maintain osmotic pressure, carry metabolites | 219 | |
| α-2-macroglobulin | Blood | Inhibits bacterial proteases via entrapment | 220 | |
| Zinc acquisition systems | ZIP (Zrt-Irt-like protein) | Cell Membrane | Diffusion | 140 |
| Znu (ABC) | Cell Membrane | Active Transport | 13 | |
| Zincophores | Secreted | Putatively bind zinc for transport | 221 | |
| Calprotectin Binding Protein | Secreted | Binds calprotectin for transport | 131 | |
| Utilization of zinc during pathogenesis | Metalloproteases | Secreted | Compromise epithelial and endothelial barriers, interfere with clotting cascade, cleave immune proteins to evade clearance. | 146,149,222 |
The sequestration of iron and zinc by the host
Frustratingly for the pathogen, they cannot directly access the host reserves of iron and zinc, as their availability is very low due to nutritional immunity. Nutritional immunity is the term given to the host's ability to restrict bacterial access to critical nutrients upon an infection, during which metals such as iron and zinc are heavily sequestered by high affinity binding proteins or kept in organelles that are not accessible to bacteria53. In addition, these metals are strongly associated with cellular components (such as iron in hemoglobin and ferritin and zinc bound to proteins, nucleic acids, and membranes) and therefore are not readily available unless the cell is in a diseased state2. Free zinc levels of mammalian hosts have been measured in the picomolar range for cytosol and plasma – while that of iron is 10-24 M in mammalian blood54–56 – although micromolar concentrations of zinc can be present in airway epithelia and mucosal membranes57.
Mining the metals a bacterium needs to replicate, grow, and survive is challenging, and mammals use a variety of tactics to keep iron and zinc away from bacterial pathogens. Some of these mechanisms include the global regulation of metal homeostasis on a systemic basis. This includes the production of the hormone hepcidin, the host master iron balance regulator58,59. Elevated levels of hepcidin leads to degradation of ferroportin, the only known cellular iron exporter in vertebrates that facilitates the release of iron to the circulatory system25,26,60. Hepcidin also induces a decrease in the expression of proteins regulated by the IRE/IRP (IRE: iron response element, IRP: iron response proteins) system, including duodenal iron absorption proteins and HCP125,26,60. Similarly, global regulation of zinc storage is mediated by hormones. Glucagon and epinephrine increase metallothionein expression and zinc storage in liver tissue61. Likewise, detection of bacterial invaders via LPS can induce IL-6 expression which in turn increases metallothionein expression and reduces free zinc concentrations62,63. Conversely, glucocorticoid signaling can induce zinc secretion from pancreatic cells64.
Other mechanisms of regulation use secreted or circulating factors that keep metals sequestered. This includes transferrin and NGAL (neutrophil gelatinase-associated lipocalin). The blood protein transferrin sequesters free iron in the circulatory system such that only the peripheral cells expressing the cognate transferrin-iron receptors can transport the transferrin-bound iron65. There is evidence that bacteria commonly target host transferrin, as it is undergoing rapid evolution to avoid recognition by bacterial pathogens66. However, transferrin is not the only molecule with iron sequestering properties, as NGAL binds ferric-siderophore complexes67,68. Siderophores are small, high affinity ferric iron binding molecules synthesized by bacteria that constitute an important cog of bacterial iron uptake69. Siderophore-NGAL binding further increases proinflammatory cytokine (e.g. IL6) production, likewise increasing stimulation of the host immune responses70. The main host-secreted zinc chelation protein is calprotectin. This protein is secreted by neutrophils at the site of infection and binds zinc and manganese to limit their availability to the pathogen71. Indeed, calprotectin is found in zinc depleted abscesses of S. aureus and can limit other forms of microbial growth in vitro72,73. Overall, the net effect of the above host actions during inflammation is that the amount of free metals in the circulatory system and tissue remains very low, keeping iron and zinc out of the hands of pathogenic bacteria74. Moreover, the alteration of the cellular iron and zinc availability may have other consequences including lymphocyte proliferation and activation75–77.
The bacterial acquisition of metals from the host
Facing the obstacles posed by nutritional immunity, a successful bacterial pathogen must develop efficient strategies to acquire metals from various host resources to facilitate their infection, survival, and replication. Because of the importance of these metals to the host as well, such resources are seemingly plentiful. The challenge, however, is to usurp the multiple ways the host limits access to these resources which include transferrin/lactoferrin, heme, and iron storage proteins (e.g. ferritin) as shown in Figure 1. They also include the zinc storage protein metallothionein and the zinc chelation protein calprotectin, along with zinc-associated proteins like serum albumin, alpha-2 macroglobulin, metalloproteases, and zinc-finger regulatory proteins. Here, we discuss known strategies pathogenic bacteria use to raid host resources by mining these metals.
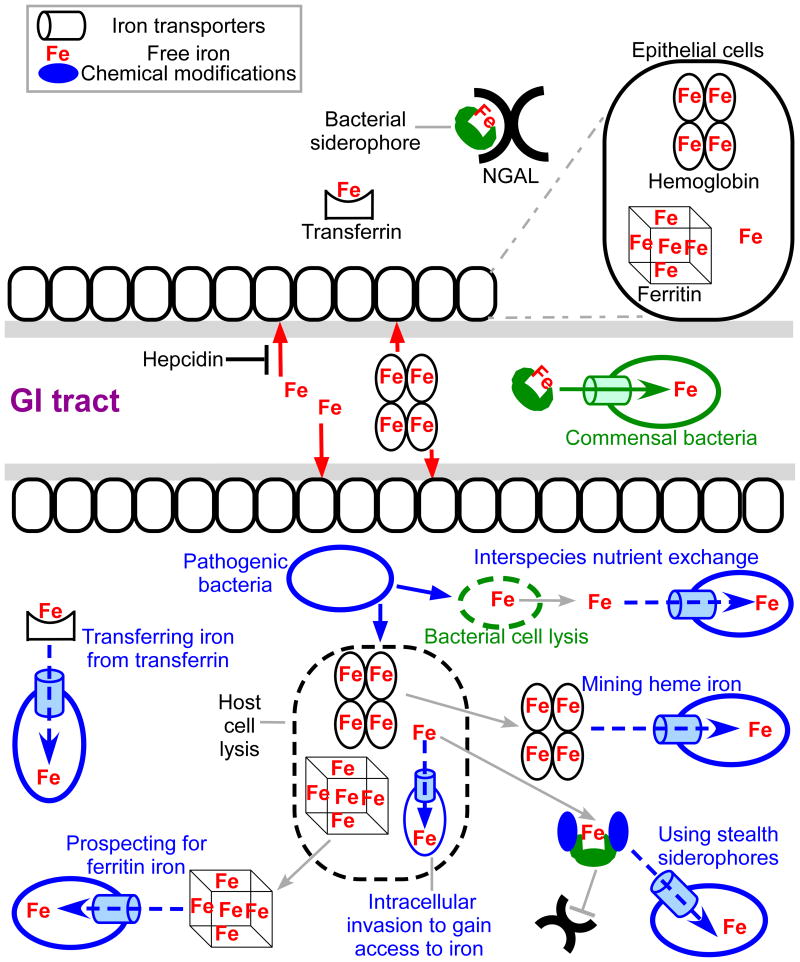
Figure 1. Bacterial iron uptake in the host.
Under normal conditions, commensal bacteria of the GI tract use siderophore-based iron uptake systems to obtain iron. Upon infection, the host uses nutritional immunity to restrict bacterial access to essential nutrients including iron (top panel). Host iron limitation includes hepcidin- mediated reduction of circulatory iron and/or the production of NGAL to prevent siderophores from chelating free iron. Additionally, iron is kept unavailable for bacteria by being bound to heme or proteins such as transferrin or ferritin. Bacterial pathogens employ diverse strategies to counter nutritional immunity (bottom panel), including the utilization of transferrin/lactoferrin, heme/heme-containing proteins, iron storage proteins such as ferritin, blocking the host from recognizing their siderophores, utilizing other species siderophores, and even invading into the cytoplasm of host cells.
Transferring iron from transferrin
Transferrin, the blood plasma glycoprotein that preferentially binds ferric iron, is the primary tool for delivering absorbed iron to cells. Lactoferrins are proteins of the transferrin family and are found in various secretory fluids (e.g. milk and tears)58. In pathogenic Neisseria species, the outer membrane anchored protein TbpB (Transferrin-binding protein B) binds and transfers holo-transferrin to the outer membrane receptor TbpA, where iron is extracted and shuttled across the outer membrane78. Resembling the Fe-Ent transport system in non-pathogenic E. coli79, TbpA-mediated iron uptake requires the TonB-ExbB-ExbD complex to transduce energy and allow a conformational change in the N-terminal plug domain, dislodging it from the channel and allowing iron to pass through where it is picked up by the periplasmic protein FbpA (Ferric binding protein component A)78. Finally, FbpA shuttles the iron to the inner membrane ABC transporter FbpBC that transports iron into the cytoplasm, and this process is influenced by periplasmic anion content78,80. In a similar way, the outer membrane proteins LbpAB (lactoferrin binding protein AB) are involved in ferric-lactoferrin complex uptake in pathogenic Neisseria species81,82. Haemophilus influenzae also contains a homolog of TbpA and is able to remove iron from host transferrin83. For Mycobacterium tuberculosis, the iron from the holo-transferrin can be either extracted by its siderophore carboxymycobactin, subsequently transported in via a mycobactin-dependent or mycobactin-independent pathway, or holo-transferrin itself can be internalized involving GAPDH and other surface proteins84. Thus, although transferrin is a major component of nutritional immunity, and exhibits growth restrictive properties, pathogenic bacteria have also evolved transport systems to target transferrin as an iron source85.
Mining heme iron
Heme and heme-containing proteins account for the most abundant source of iron in the host. Not surprisingly, bacterial pathogens have developed various strategies to mine iron from this resource. There are several mechanisms by which bacteria gain access to host heme86. Free heme is recognized by TonB-dependent outer membrane receptors in gram negative bacteria or cell wall anchored receptors in gram positive bacteria86. Free heme is also recognized and bound by secreted bacterial proteins named hemophores that have high affinity for the heme moiety and are made by both gram positive and gram negative bacteria87. Hemophores also actively extract heme from heme-containing proteins88,89, utilizing specific residues in the heme binding pocket to promote the loss of heme from hemoglobin90. Additionally, the hemophores may stimulate the dissociation of hemoglobin tetramers into dimers and monomers, which have a lower affinity for the heme and increase its loss from the globin91. The hemophores have a higher affinity for heme, which will be bound at equilibrium. Once bound, the heme can be transferred to cognate surface receptors where it is then moved across the cell wall or membrane into the cytoplasm, where heme can be degraded to liberate iron89. For example in Bacillus anthracis, the causative agent of anthrax, the surface anchored proteins IsdC (Isd: iron-regulated surface determinant), Hal (heme-acquisition leucine-rich repeat protein), and possibly BslK (Bacillus surface layer protein K) are involved in scavenging the heme moiety from heme containing proteins92–94. B. anthracis also secretes two hemophores IsdX1 and IsdX2, which extract heme from host heme containing proteins and shuttle them to receptors in the bacterial envelope95. Both the receptors and the hemophores use the NEAT (N-terminal near-iron transporter) domains to interact with the heme moiety through a highly conserved YXXXY motif96. It is interesting to note that, Hbp2 (heme/hemoglobin-binding protein 2), a NEAT-domain containing hemophore in Listeria monocytogenes, can scavenge heme but its activity is dependent on a non-canonical tyrosine residue, suggesting an unprecedented mechanism of heme binding by this protein97. The NEAT domain has been recognized as being very important in gram-positive biology. In addition to important roles in making bacteria more virulent93, they also may serve as recombinant vaccine candidates for pathogens such as Staphylococcus aureus98,99 and B. anthracis (Balderas and Maresso, unpublished data). In gram negatives, HasA (heme acquisition system component A) represents a family of highly conserved hemophores identified in Serratia marcescens, Pseudomonas aeruginosa, Pseudomonas fluorescens, Yersinia pestis, and Yersinia enterocolitica87. HasA is secreted via the type I secretion pathway and may capture heme from hemoglobin. The TonB–dependent outer membrane receptor HasR interacts with HasA to facilitate heme transfer and uptake87.
Prospecting for ferritin iron
Ferritins are tightly regulated storage proteins that deposit and release iron to maintain its safe level within the host100. Normally, ferritins are cytosolic and their extracellular concentrations are very low (<0.01% of the extracellular transferrin)101. In sputum and bronchoalveolar lavage fluid, there are higher levels of ferritin, which increases during diseased states (e.g. cystic fibrosis patients)101. Not surprisingly, lung pathogens possess the ability to take advantage of this iron source. For example, P. aeruginosa secretes extracellular proteases that lyse the ferritin and release its stored ferric iron, which are reduced by secreted bacterial molecules (e.g. pyocyanin) and possibly get transported in via the Feo iron transport system101. Similarly, another lung pathogen, Burkholderia cenocepacia, can use ferritin as an iron source in a protease-dependent manner102. Bacillus cereus also uses ferritin as an iron source. In this pathogen, the surface protein IlsA (iron-regulated leucine rich surface protein type A) recognizes and binds ferritins, leading to the destabilization and subsequently release of ferric iron ions, which are chelated by the bacterial siderophore bacillibactin and transported via the cognate membrane transporter FeuABC (ferric bacillibactin uptake protein components ABC)103. Thus, it appears that when labile iron in circulation is not available, bacteria can prospect into deep host reserves such as ferritin to satisfy their requirement for this metal.
Bacterial countermeasures to overcome host iron sequestration
Some pathogenic bacteria can chemically modify their secreted siderophores to evade recognition by host siderophore-binding proteins like NGAL. For example, Salmonella species, uro- and avian pathogenic E. coli strains, and certain Klebsiella strains (e.g. K. pneumonia) are able to synthesize variations of the catecholate siderophore Ent that is glycosylated104. The glycosylation benefits these bacterial pathogens and contributes to virulence by interfering with NGAL binding through steric hindrance of the added bulky glucose groups104–106. Yersinia species, some E. coli and K. pneumoniae strains are able to synthesize a structurally different siderophore termed yersiniabactin (a mixed ligands siderophore). The uptake of yersiniabactin depends on the TonB-dependent outer membrane receptor FyuA and its importance for bacterial virulence was demonstrated in Y. enterocolitica, E. coli and K. pneumonia but not in Y. pestis107–110. Strains of E. coli, S. flexneri, and K. pneumonia produce the hydroxamate siderophore aerobactin, whose role in pathogenesis is important in some cases but dispensable in others111–114. Another way to fine tune the siderophore based iron uptake system in bacterial pathogens is to “amplify” its iron uptake ability. An example is the asymptomatic bacteriuria caused by E. coli strain 83972. When compared to its commensal counterpart, it has additional abilities to synthesize and transport in salmochelin, aerobactin, and yersiniabactin106. The redundancy of the iron transport systems contributes significantly to its colonization in the urinary tract106. This feature gives the pathogen the versatility to satisfy its iron needs in different environmental niches.
Deep prospecting: iron uptake by intracellular bacteria
Nutrient levels in the extracellular milieu are under tight control by the host. The intracellular environment, however, is very nutrient rich with higher concentrations of several growth-promoting factors. The intracellular environment offers additional benefits for bacteria in that there is a low level of antimicrobial peptides, antibiotics, and humoral antibodies. But entry into host cells comes at great risk for bacteria; eukaryotic cells have intracellular sensors that activate alarms if bacterial components are detected115. In addition, cells contain specialized organelles called phagolysosomes that harness the harmful effects of low pH and/or reactive oxygen species to kill bacteria116. However, some bacteria are ideally adapted to survive and replicate in this environment, which confers a selective advantage by occupying a niche where very few bacteria are capable of thriving. For example, all Shigella subgroups, S. flexneri, S. sonnei, S. dysenteriae, and S. boydii, are able to grow intracellularly in host epithelial cells117. Multiple iron uptake systems in S. flexneri contribute to iron uptake intracellularly, including the Iuc (transporter for the native siderophore aerobactin), Feo, and Sit (transporter for manganese and ferrous iron)111,112. Each of the three iron uptake systems is dispensable when tested in a cell culture model but a triple mutant cannot survive in cells111. Furthermore, monitoring gene expression during intracellular pathogenesis shows activation of the sitA and fhuA promoters, indicating they may have a role in intracellular iron acquisition in S. flexneri112. Francisella tularensis is also capable of replicating intracellularly by escaping the phagosome of macrophages. Once inside of the macrophages, F. tularensis upregulates the host transferrin receptor TfR1118. The increased level of transferrin receptors is believed to benefit F. tularensis intracellular growth due to the increase of the labile iron pool, which represents a freely available iron source for intracellular bacterial pathogens118. Similarly, once inside of the monocytes, N. gonorrhoeae upregulates hepcidin, NGAL, and NRAMP1 (Natural resistance-associated macrophage protein 1, which shuttles iron from the late endosome and phagolysosome to the cytosol to store in ferritins), downregulates labile iron-detoxifying enzyme BDH2 (short chain 3-hydroxybutyrate dehydrogenase), with a net effect being an increase of the labile iron pool to facilitate N. gonorrhoeae survival intracellularly119. Thus, it would seem that some of the same mechanisms used by extracellular bacteria to gain access to and modulate iron levels are also used by intracellular bacteria in the host cytoplasm.
The bacterial acquisition of zinc
Plundering host zinc is also critical for the survival of intracellular pathogens. Many of them require the Zn ABC transporters for replication and full virulence. This is true for Listeria monocytogenes, Salmonella enterica, Brucella abortus, and Yersinia pestis120–122. Under Zn2+ deficient conditions, like those thought to be encountered in the intestine or in blood, bacteria employ ABC transporters homologous to the ZnuABC system in E. coli. Here, the periplasmic binding protein ZnuA binds a single zinc ion with high affinity, and upon contact with the ZnuB permease, the complex actively transports zinc through the inner membrane driven by ATP hydrolysis of the ZnuC ATPase123,124. These ABC transporters are found across Gram positive and Gram negative species125, and are commonly considered virulence factors121,126,127. Importantly, these transporters can serve as antigenic targets for vaccines, and inoculation of mutant strains lacking transporters can confer resistance to wild-type infections128,129. Conversely, host-induced zinc toxicity is likely a problem, as putative zinc efflux pumps are required for M. tuberculosis to survive in macrophages130. Interestingly, N. meningitidis was recently shown to scavenge host zinc from calprotectin, suggesting a mechanism to subvert neutrophil-mediated killing131. Unfortunately, little is known about the ability of other bacterial pathogens to target host zinc-binding proteins for zinc acquisition.
The regulation of bacterial metal uptake
Generally, iron uptake systems are regulated by the bacterial protein Fur (ferric uptake regulator), with evidence that small RNAs are involved as well132–136. When facing iron deficient conditions, such regulation allows bacteria to increase the expression of the genes needed to import iron. The basic principles of bacterial iron transport also hold true for zinc. Similar to bacteria employing Fur to regulate intracellular iron levels, they rely on Zur (zinc uptake regulator), which is a Fur family homolog protein, to regulate Zn2+ uptake mechanisms. Interestingly, E. coli derived Fur binds zinc to form active dimers, but this zinc binding activity is not necessary for Fur mediated regulation in other bacteria137,138. This evidence suggests possible crosstalk between iron and zinc homeostasis mechanisms. Upon binding Zn2+, Zur proteins actively bind to DNA and suppress transcription of downstream genes associated with zinc import, like the ABC transporters139. This negative feedback loop prevents the toxic buildup of intracellular zinc and induces expression of zinc acquisition mechanisms when the metal is limiting. Bacteria might also import zinc into the cytosol with ZIP transporters; however, they are only known to be present in E. coli140,141. While their presence is generally necessary for full virulence, it is unclear whether these transporters alone are sufficient to maintain an infection, or if like iron, some liberation of zinc from host protein and cellular stores is also required. Finally, some non-specific transporters can import both metals. This is true of ZupT (Zinc uptake protein component T), which in addition to transporting zinc can also transport ferrous iron141.
The use of metals to power bacterial virulence
Bacteria use the metals they acquire to drive key cellular processes, some of which were briefly mentioned above. These activities are necessary for growth and replication of the microbe, which in turn sustains and propagates the infection. What sometimes is lost in this consideration is that acquired metals are important catalysts for two broadly conserved and critically important types of bacterial hydrolases that directly interface with the host and/or the host response to infection. Examples include the production of metalloproteases and lactamases that require zinc for their catalytic activity. It is becoming increasingly clear that, much like iron, zinc is essential to the survival of a pathogen during host infection, but perhaps in a different way. Whereas iron serves as a co-factor in processes related to energy transduction through respiration, zinc can be crafted into factors that interact with the host on several levels. In the final part of this review, we consider the importance of metals in the use of bacterial weapons of warfare – the very virulence factors bacteria use to overcome the host barriers to infection.
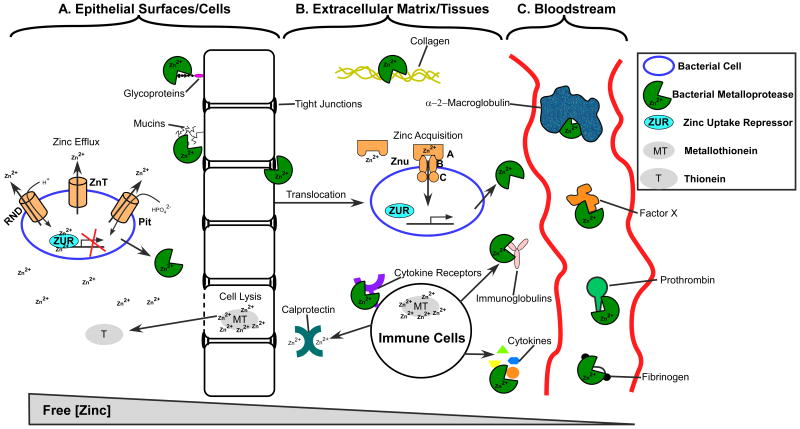
Proteases are enzymes that hydrolyze peptide bonds in proteins or peptides. They can be exoproteases – which cleave at the amino or carboxy terminus of proteins, or endoproteases – which are capable of cleaving at one or multiple sites within a protein. Proteases are categorized by the catalytic residue in their active site. This includes aspartic, threonine, serine, and cysteine proteases, with these residues driving catalysis. For a comprehensive review on the classes and activities of the multitude of known proteases, please see references142–144. A critical feature of many proteases is that one or more metals serve as a co-factor for catalysis, the so-called metalloproteases. Most bacterial metalloproteases are secreted and use zinc as the metal cofactor. Zinc metalloproteases contain variations on the typical HEXXH binding motif, which coordinates a single Zn2+ ion with three amino acids, usually histidine and glutamate, but sometimes aspartate and cysteine residues. The catalytic cleft is composed of a tridentate site with a coordinated water molecule145,146. Mechanistically, zinc metalloproteases cleave peptide bonds via nucleophilic attack on the carbonyl carbon in the peptide – an action performed by the deprotonized water molecule. During the transition state, zinc helps to stabilize the negatively charged intermediate product. The final products exit the catalytic site upon hydrolysis by the water molecule and creation of amine and carboxyl termini on the new peptide fragments147. Metalloproteases typically exhibit broad specificity, as has been described for vEP of Vibrio fulnificus, InhA1 of Bacillus anthracis, and ZmpB of Burkholderia cenocepacia148–150. The broad specificity of bacterial metalloproteases may actually suit the pathogen's needs by facilitating the disruption of physiologically important host processes, including the breakdown of barriers, the destruction of key signaling intermediates, and the release of nutrients such as metals from host metalloproteins. For example, collagen is the main component of skin, tendons, and cartilage. It is a fibrous, structural protein that is present in connective tissues and comprises 25-33% of all proteins in mammals. It is also a common target of zinc metalloproteases, resulting in compromised host barriers that spread infection and delay immune clearance151. Some examples of collagenolytic proteases are B. anthracis Npr599 and InhA1, both of which cleave collagen types I and IV in vitro149, and the Burkholderia cenocepacia metalloproteases ZmpB and ZmpA150,152. Tissue disruption can also occur by cleavage of tight cell junctions. Zona occluden-1 is a tight junctional protein which is cleaved by Pseudomonas aeruginosa pseudolysin, Vibrio cholera hemagglutinin, and B. anthracis InhA1; the latter thought to cause increased blood brain barrier permeability and dissemination of bacilli153–155. Immune components can also be directly cleaved by metalloproteases. This is true for the IgA protease of Streptococcus sanguis and the immunoglobulin protease of S. marcescens156,157. This also includes mirabilysin of Proteus mirabilis and pseudolysin of P. aeruginosa which both cleave IgG158,159. Interestingly, the host is thought to directly target the zinc status of bacteria in infected tissues as a nutritional immunity strategy. Specifically, neutrophils that are recruited to infection sites secrete the metal chelator protein calprotectin, which mainly binds zinc and manganese. As stated above, calprotectin is found in zinc-depleted S. aureus abscesses, and it can reduce other forms of microbial growth in vitro72,160,161. It may be that the chelation of zinc by the host has a direct effect of preventing bacterial metalloproteases from acquiring this critical metal co-factor.
Metalloproteases can also interfere with immune clearance by interfering with signaling cascades. Lethal toxin from B. anthracis induces endothelial disruption by cleaving MAP kinases162. InhA1 can also cleave prothrombin and factor X to induce clotting163. Similarly, fibrinogen is cleaved by Serratia marcescens to interfere with the extracellular matrix and coagulation cascade157. Cytokines or interleukins (IL) are the recruitment signal for neutrophils and macrophages, and they can also be disrupted by pathogenic bacteria to avoid immune clearance. Examples include the cleavage of IL-2 by Legionella pneumophila metalloprotease, and cleavage of the IL-6 receptor by supernatants of S. marcescens and other bacteria164,165. An overview of zinc metalloprotease virulence mechanisms and their host substrates is shown in Figure 2. The broad use of such metals in mechanisms like these further supports the notion that blocking the ways bacteria attain these metals might serve as both an anti-infective and anti-virulence strategy.
Figure 2. The role of zinc in bacterial pathogenesis.
A. Bacterial pathogens encounter higher concentrations of zinc at epithelial surfaces, where lysed cells release metallothioneins that liberate zinc upon oxidative stress. To combat Zn2+ toxicity, bacteria employ efflux transporters like RND, ZnT and Pit. ZUR proteins are bound to bacterial DNA and prevent transcription of zinc uptake mechanisms. Metalloproteases cleave mucins, glycoproteins, and tight cell junctions to allow bacteria to translocate into other tissues. B. At sites of infection and translocation, the host can reduce available zinc by secreting the zinc chelator calprotectin. Once in zinc deficient environments, bacterial ZUR proteins relieve transcriptional repression and zinc uptake mechanisms are expressed, such as the Znu ABC transporter. Here, metalloproteases can cleave collagen, cytokine receptors, cytokines and immunoglobulins to further disrupt tissues and interfere with immune signaling. C. When present in the bloodstream, host α-2-macroglobulin can inactivate metalloproteases via entrapment. However, metalloproteases can cleave fibrinogen, prothrombin, and factor X to disrupt the clotting cascade and permit further dissemination.
An intriguing an understudied aspect of metalloproteases is their potential role in nutrient acquisition. Much work has been done to elucidate the amino acid acquisition systems of intracellular pathogens. These bacteria redirect host autophagy and lysosomal degradation pathways to liberate free amino acids, a concept termed nutritional virulence166,167. Extracellular proteases are known to degrade hemoglobin, transferrin, and other iron and heme containing compounds168–170. Presumably these functions are dedicated to acquiring iron, but their potential role in amino acid acquisition has not yet been defined. However, it was recently discovered that V. cholera employs the metalloprotease VchC to help utilize collagen as its sole nutrient source171, and that B. anthracis metalloprotease InhA1 can degrade hemoglobin as an amino acid source in vitro172. With this information we should consider the possibility that metalloproteases and proteases in general not only interfere with host defense mechanisms, but can also release essential metals and amino acids from a distance for bacteria to scavenge.
Finally, metals like zinc are important in other bacterial processes, including the break-down of life-saving antibiotics. It is widely recognized that modern medicine is on the precipice of a microbial-induced disaster. The rise of strains (and enzymes) that are resistant to (and can inactivate) commonly used and recently developed antibiotics is risking nearly 80 years of progress in successfully treating once life-threatening bacterial infections. Much of this resistance is driven by a large class of enzymes localized to the bacterial surface termed metallo-β-lactamases. These enzymes cleave the β-lactam ring of antibiotics that include the penicillins, carbapenems, cephalosporins, and monobactams173. Similar in mechanism to the metalloproteases, metallo-β-lactamases require zinc cations in the catalytic cleft to exert their full activity. A water molecule performs nucleophilic attack on the carbonyl carbon in the β-lactam ring while zinc stabilizes the negatively charged intermediate. This reaction breaks the β-lactam ring, which can no longer inactivate the bacterial transpeptidase that makes the cell wall174. Metallo-β-lactamases are distributed across dozens of gram-positive and gram-negative species, with the most notorious in recent times being NDM-1 (first discovered in a K. pneumoniae strain isolated from a patient that visited New Delhi)174–176. Since then, NDM-1 has been discovered in clinical isolates in the United Kingdom, Japan, Pakistan, United States, and Canada, and is found in multiple gram-negative genre like Escherichia and Acinetobacter177. Despite the critical importance of such enzymes in undermining the medical miracle of antibiotics, it is not understood the sources of, or mechanism by which host zinc is incorporated into these enzymes.
Future work and perspectives
Not all bacteria are pathogens. A number of bacteria, which are now recognized as the microbiome and commonly found on or in body surfaces such as the gastrointestinal and respiratory tract, skin, and nares, often exert beneficial effects on our health178,179. One commensal bacterium that lives in the human gut is Escherichia coli, and it utilizes several iron-uptake mechanisms to compete not only with the host but also other bacteria occupying the intestinal niche. One such mechanism is to synthesize siderophores, which are secreted into the surrounding environment. Siderophores bind free iron by virtue of their high affinity and are then imported via the cognate membrane transporters180. A second mechanism commensal E. coli uses to attain iron is through the use of two transport systems. The ferric-dicitrate transport system transports in citrate181. Citrate is a common component of our daily diet and can be found in many foods such as green leafy vegetables and fruits and thus found in our intestinal tract182. It also, by virtue of its structure, can weakly chelate iron and often is bound to this metal. The ferrous iron transport system shuttles in free ferrous iron183. Due to the fact that most commensal bacteria live in the lower intestine where anaerobiosis and acidification are common and favors ferrous iron184, having this system may be a benefit in this environment. Finally, bacteria of the intestinal microbiome can utilize xenosiderophores. Xenosiderophores are siderophores that demonstrate cross species and even cross kingdom activity, i.e. synthesized by one species but are able to be utilized by different species.
Bacteroides species are opportunistic pathogens and another representative of the commensal bacteria179. Similar to E. coli, Bacteroides species possess the ferrous iron transport system185. B. fragilis has a putative siderophore mediated iron transport system186, but the siderophore has not yet been identified187. B. fragilis, however, has the ability to utilize heme and hemoglobin as an iron source, a feature that is associated with it being an opportunistic pathogen and distinguishes itself from the discussion of commensals that take up metals such as the nonpathogenic strains of E. coli188,189. Recently, one member of Bacteroidetes phylum demonstrated iron acquisition from transferrin, but the medical significance of this finding is not known190.
In summary, although there are clear examples of commensal bacteria that inhabit the skin or GI tract and utilize a multitude of systems to attain essential metals, the fact that they are utilized for colonization of the host would suggest that they also can be perceived as virulence factors. In this context, they may not directly participate in the pathological consequences of the infection but certainly are needed to maintain a relationship with the host that may “break bad” when the host is immunocompromised.
When a bacterial pathogen infects the host, it also encounters a polymicrobial environment, and must develop ways to compete for essential nutrients such as iron and zinc with the microbiome. One strategy is to take advantage of other microbes to fulfill nutrient requirements via inter- and intraspecies metabolite usage191 S. aureus is an opportunistic pathogen mostly found in the human respiratory tract and on the skin, and represents a good example of the interspecies metabolite usage192. In the presence of S. aureus, P. aeruginosa produces a staphylolytic protease LasA, which targets the glycyl-glycine and glycyl-alanine bonds of the pentaglycine interpeptide bridge in the S. aureus peptidoglycan, leading to S. aureus lysis. The lysed S. aureus serves as the iron pool for P. aeruginosa to support its growth193. H. influenza also benefits from the presence of S. aureus because the hemolysins (α, β, and γ) produced by S. aureus help lyse erythrocytes to release nutrients (e.g. heme) to facilitate H. influenza growth. The mixture of staphylococcal strains deficient in menaquinone biosynthesis with those lacking heme biosynthesis reaches the wild type level of growth in vitro and remains fully virulent when tested in a murine model of osteomyelitis194. The restoration is explained by the ability of the menaquinone biosynthesis mutant to synthesize and supply heme to the population194. As more investigation focuses on the ecosystem of the microbiome and its relation to human disease, the mechanisms by which interspecies nutrient exchange occurs will become more evident.
The importance of metal uptake during infections and the seemingly continuous development of resistance against antibiotics compels consideration of the inhibition of metal uptake for antibacterial drug development195,196. Indeed, an increasing number of studies have evaluated the effectiveness of targeting bacterial iron metabolism as an antibacterial strategy, with efficacy demonstrated in some cases but not others197–201. Additionally, the “Trojan horse” strategy shows promise, where siderophore-like molecules are loaded with toxic drugs196,202. Considering the multiple roles metalloproteases display in virulence, as well as the critical requirement of metals in β-lactamase activity, there exists a need to understand how these important enzymes become loaded with zinc. Future studies should be directed towards testing the clinical validity of these ideas as well as exploring new therapeutic entry points that disrupt bacterial metal homeostasis.
Supplementary Material
Acknowledgments
The authors offer the sincerest of apology for not being able to cite all relevant works (due to page constraints). This work was supported in part by grants AI097167 and AI116497 from the National Institutes of Health. We thank the Maresso laboratory for comments and suggestions.
References
- 1.Wicander R, Monroe J. Essentials of geology. 4th. Cengage Learning; Boston, MA: 2005. pp. 63–64. [Google Scholar]
- 2.Vallee B, Falchuk H. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 3.Cairo G, Bernuzzi F, Recalcati S. Genes Nutr. 2006;1:25–39. doi: 10.1007/BF02829934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 5.Kappler A, Straub KL. Rev Mineral Geochemistry. 2005;59:85–108. [Google Scholar]
- 6.Winter WE, Bazydlo LA, Harris NS. Lab Med. 2014;45:92–102. doi: 10.1309/lmf28s2gimxnwhmm. [DOI] [PubMed] [Google Scholar]
- 7.Huang M, Parker MJ, Stubbe J. J Biol Chem. 2014;289:28104–28111. doi: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kambe T, Tsuji T, Hashimoto A, Itsumura N. Physiol Rev. 2015;95:749–784. doi: 10.1152/physrev.00035.2014. [DOI] [PubMed] [Google Scholar]
- 9.Lu S, Gischkat S, Reiche M, Akob DM, Hallberg KB, Küsel K. Appl Environ Microbiol. 2010;76:8174–8183. doi: 10.1128/AEM.01931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggett P. In: Present Knowledge in Nutrition. 10th. Erdman J, Macdonald I, Zeisel S, editors. Wiley-Blackwell; Washington, DC.: 2012. pp. 506–20. [Google Scholar]
- 11.Harris MM, Houtkooper LB, Stanford VA, Parkhill C, Weber JL, Flint-Wagner H, Weiss L, Going SB, Lohman TG. J Nutr. 2003;133:3598–3602. doi: 10.1093/jn/133.11.3598. [DOI] [PubMed] [Google Scholar]
- 12.Wastney ME, Aamodt RL, Rumble WF, Henkin RI. Am J Physiol. 1986;251:R398–408. doi: 10.1152/ajpregu.1986.251.2.R398. [DOI] [PubMed] [Google Scholar]
- 13.Cerasi M, Ammendola S, Battistoni A. Front Cell Infect Microbiol. 2013;3:108. doi: 10.3389/fcimb.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moulder K, Steward M. Clin Exp Immunol. 1989;77:269–274. [PMC free article] [PubMed] [Google Scholar]
- 15.Gross R, Osdin N, Fong L, Newberne P. Am J Clin Nutr. 1979;32:1260–1265. doi: 10.1093/ajcn/32.6.1260. [DOI] [PubMed] [Google Scholar]
- 16.Weston WL, Huff JC, Humbert J, Hambidge K, Neldner K, Walravens PA. Arch Dermatol. 1977;113:422–5. [PubMed] [Google Scholar]
- 17.Nishida K, Hasegawa A, Nakae S, Oboki K, Saito H, Yamasaki S, Hirano T. J Exp Med. 2009;206:1351–64. doi: 10.1084/jem.20082533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klebanoff SJ, Kettle J, Rosen H, Winterbourn CC, Nauseef WM. J Leukoc Biol. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slauch JM. Mol Microbiol. 2011;80:580–3. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes J, Beton D, Brown DA. Gut. 1968;9:323–324. doi: 10.1136/gut.9.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Food and Agriculture Organization/World Health Organization. Vitamin and mineral requirements in human nutrition : report of a joint FAO/WHO expert consultation. 2nd. 2004. [Google Scholar]
- 22. [May 19, 2015];Natl Institutes Heal. http://ods.od.nih.gov/factsheets/Zinc-HealthProfessional/
- 23.Wang X, Zhou B. IUBMB Life. 2010;62:176–82. doi: 10.1002/iub.291. [DOI] [PubMed] [Google Scholar]
- 24.Hempe JM, Cousins RJ. J Nutr. 1992;122:89–95. doi: 10.1093/jn/122.1.89. [DOI] [PubMed] [Google Scholar]
- 25.Silva B, Faustino P. Biochim Biophys Acta. 2015;1852:1347–1359. doi: 10.1016/j.bbadis.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Przybyszewska J, Żekanowska E. Gastroenterol Rev. 2014;4:208–213. doi: 10.5114/pg.2014.45102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen HL, Peterson CT, Reddy MB, Hanson KB, Swain JH, Sharp RL, Alekel DL. Int J Sport Nutr Exerc Metab. 2006;16:281–295. doi: 10.1123/ijsnem.16.3.281. [DOI] [PubMed] [Google Scholar]
- 28.Chiabrando D, Mercurio S, Tolosano E. Haematologica. 2014;99:973–983. doi: 10.3324/haematol.2013.091991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Tang JW, Ma WQ, Feng J. Biol Trace Elem Res. 2010;133:325–34. doi: 10.1007/s12011-009-8437-3. [DOI] [PubMed] [Google Scholar]
- 31.Plum LM, Rink L, Haase H. Int J Environ Res Public Health. 2010;7:1342–1365. doi: 10.3390/ijerph7041342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukada T, Yamasaki S, Nishida K, Murakami M, Hirano T. J Biol Inorg Chem. 2011;16:1123–34. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eide DJ. Biochim Biophys Acta. 2006;1763:711–22. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Folin M, Contiero E, Vaselli GM. Biometals. 1994;7:75–79. doi: 10.1007/BF00205198. [DOI] [PubMed] [Google Scholar]
- 35.Foote JW, Delves HT. J Clin Pathol. 1984;37:1050–1054. doi: 10.1136/jcp.37.9.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad AS, Oberleas D. J Lab Clin Med. 1970;76:416–25. [PubMed] [Google Scholar]
- 37.Centers for Disease Control and Prevention. Morb Mortal Wkly Rep. 1998;47(RR3):1. [Google Scholar]
- 38.Prasad S. Am J Clin Nutr. 1991;53 doi: 10.1093/ajcn/53.2.403. [DOI] [PubMed] [Google Scholar]
- 39.Shah D, Sachdev HPS. Br J Nutr. 2001;85 doi: 10.1079/bjn2000301. [DOI] [PubMed] [Google Scholar]
- 40.Imdad A, Bhutta ZA. BMC Public Health. 2011;11(Suppl 3):S22. doi: 10.1186/1471-2458-11-S3-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayo-Wilson E, Junior J, Imdad A, Dean S, Chan X, Chan E, Jaswal A, Bhutta Z. Cochrane Database Syst Rev. 2014;5 doi: 10.1002/14651858.CD009384.pub2. [DOI] [PubMed] [Google Scholar]
- 42.Yun S, Vincelette ND. Crit Rev Oncol Hematol. 2015;95:12–25. doi: 10.1016/j.critrevonc.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Sawada H, Hao H, Naito Y, Oboshi M, Hirotani S, Mitsuno M, Miyamoto Y, Hirota S, Masuyama T. Arterioscler Thromb Vasc Biol. 2015;35 doi: 10.1161/ATVBAHA.115.305586. [DOI] [PubMed] [Google Scholar]
- 44.Di Lorenzo F. Neuro Endocrinol Lett. 2015;36 [PubMed] [Google Scholar]
- 45.Haldar M, Kohyama M, So AYL, Wumesh K, Wu X, Briseno CG, Satpathy AT, Kretzer NM, Rajasekaran NS, Wang L, Egawa T, Igarashi K, Baltimore D, Murphy TL, Murphy KM. Biol Trace Elem Res. 2015;6 [Google Scholar]
- 46.Walling C, Goosen A. J Am Chem Soc. 1973;95:2987–91. [Google Scholar]
- 47.Khan Fa, Fisher Ma, Khakoo Ra. Int J Infect Dis. 2007;11:482–487. doi: 10.1016/j.ijid.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Kim EY, Koh JY, Kim YH, Sohn S, Joe E, Gwag BJ. Eur J Neurosci. 1999;11:327–334. doi: 10.1046/j.1460-9568.1999.00437.x. [DOI] [PubMed] [Google Scholar]
- 49.Dineley KE, Richards LL, Votyakova TV, Reynolds IJ. Mitochondrion. 2005;5:55–65. doi: 10.1016/j.mito.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Clausen A, McClanahan T, Ji SG, Weiss JH. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0083347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fosmire G. Am J Clin Nutr. 1990;51:225–227. doi: 10.1093/ajcn/51.2.225. [DOI] [PubMed] [Google Scholar]
- 52.Hooper P, Visconti L, Garry P, Johnson G. J Am Med Assoc. 1980;244:1960–1961. [PubMed] [Google Scholar]
- 53.Hood MI, Skaar EP. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krężel A, Maret W. J Biol Inorg Chem. 2006;11:1049–62. doi: 10.1007/s00775-006-0150-5. [DOI] [PubMed] [Google Scholar]
- 55.Magnesons GR, Puvathingal JM, Ray WJ. J Biol Chem. 1987;262:11140–11148. [PubMed] [Google Scholar]
- 56.Lewis JP. Periodontology. 2010;52:94–116. doi: 10.1111/j.1600-0757.2009.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zalewski PD, Truong-Tran AQ, Grosser D, Jayaram L, Murgia C, Ruffin RE. Pharmacol Ther. 2005;105:127–49. doi: 10.1016/j.pharmthera.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Cassat JE, Skaar EP. Cell Host Microbe. 2014;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shetty PS. Nutrition, Immunity and Infection. CABI; Oxfordshire, UK: 2010. pp. 88–95. [Google Scholar]
- 60.Ganz T. Blood. 2011;117:4425–4433. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cousins R, Dunn M, Leinart A, Yedinak K, DiSilvestro R. Am J Pathol. 1986;251:688–694. doi: 10.1152/ajpendo.1986.251.6.E688. [DOI] [PubMed] [Google Scholar]
- 62.Gaetke LM, Mcclain CJ, Talwalkar RT, Shedlofsky SI. Am J Physiol. 1997;272:E952–6. doi: 10.1152/ajpendo.1997.272.6.E952. [DOI] [PubMed] [Google Scholar]
- 63.Gabay C, Kushner I. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 64.Guo L, Lichten LA, Ryu MS, Liuzzi JP, Wang F, Cousins RJ. Proc Natl Acad Sci U S A. 2010;107:2818–23. doi: 10.1073/pnas.0914941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gkouvatsos K, Papanikolaou G, Pantopoulos K. Biochim Biophys Acta. 2012;1820:188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Barber MF, Elde NC. Science. 2014;12:1362–6. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 68.Steigedal M, Marstad A, Haug M, Damas JK, Strong RK, Roberts PL, Himpsl SD, Stapleton A, Hooton TM, Mobley HLT, Hawn TR, Flo TH. J Immunol. 2014;193:6081–6089. doi: 10.4049/jimmunol.1401528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miethke M, Marahiel MA. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holden VI, Lenio S, Kuick R, Ramakrishnan SK, Shah YM, Bachman MA. Infect Immun. 2014;82:3826–3836. doi: 10.1128/IAI.01849-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kehl-Fie TE, Skaar EP. Curr Opin Chem Biol. 2010;14:218–24. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli M, Nacken W, Chazin WJ, Skaar EP, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 73.Sohnle PG, Collins-lech C, Wiessner J. J Infect Dis. 1991;163:187–192. doi: 10.1093/infdis/163.1.187. [DOI] [PubMed] [Google Scholar]
- 74.Nairz M, Schroll A, Demetz E, Tancevski I, Theurl I, Weiss G. Immunobiology. 2014;220:280–294. doi: 10.1016/j.imbio.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 75.Cherayil BJ. Arch Immunol Ther Exp. 2011;58:407–15. doi: 10.1007/s00005-010-0095-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cherayil B. Immunol Res. 2011;50:1–9. doi: 10.1007/s12026-010-8199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dennis MK, Field AS, Burai R, Ramesh C, Whitney K, Bologa CG, Oprea TI, Yamaguchi Y, Hayashi S, Sklar La, Hathaway HJ, Arterburn JB, Prossnitz ER. Cell. 2014;156:1223–34. [Google Scholar]
- 78.Noinaj N, Buchanan SK, Cornelissen CN. Mol Microbiol. 2013;86:246–257. doi: 10.1111/mmi.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma L, Kaserer W, Annamalai R, Scott DC, Jin B, Jiang X, Xiao Q, Maymani H, Massis LM, Ferreira LCS, Newton SMC, Klebba PE. J Biol Chem. 2007;282:397–406. doi: 10.1074/jbc.M605333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parker Siburt CJ, Mietzner TA, Crumbliss AL. Biochim Biophys Acta - Gen Subj. 2012;1820:379–392. doi: 10.1016/j.bbagen.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Biswas GD, Sparling PF. Infect Immun. 1995;63:2958–2967. doi: 10.1128/iai.63.8.2958-2967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prinz T, Meyer M, Pettersson A, Tommassen J. J Bacteriol. 1999;181:4417–4419. doi: 10.1128/jb.181.14.4417-4419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gray-Owen SD, Schryvers AB. Infect Immun. 1995;63:3809–3815. doi: 10.1128/iai.63.10.3809-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boradia VM, Malhotra H, Thakkar JS, Tillu VA, Vuppala B, Patil P, Sheokand N, Sharma P, Chauhan AS, Raje M, Raje CI. Nat Commun. 2014;5:4730. doi: 10.1038/ncomms5730. [DOI] [PubMed] [Google Scholar]
- 85.Rooijakkers SHM, Rasmussen SL, McGillivray SM, Bartnikas TB, Mason AB, Friedlander AM, Nizet V. J Biol Chem. 2010;285:27609–27613. doi: 10.1074/jbc.M110.154930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Contreras H, Chim N, Credali A, Goulding CW. Curr Opin Chem Biol. 2014;19:34–41. doi: 10.1016/j.cbpa.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cescau S, Cwerman H, Létoffé S, Delepelaire P, Wandersman C, Biville F. Biometals. 2007;20:603–13. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 88.Ghigo J, Létoffé S, Wandersman C. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheldon JR, Heinrichs DE. FEMS Microbiol Rev. 2015;39:592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- 90.Ekworomadu MT, Poor CB, Owens CP, Balderas Ma, Fabian M, Olson JS, Murphy F, Balkabasi E, Honsa ES, He C, Goulding CW, Maresso AW. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spirig T, Malmirchegini GR, Zhang J, Robson SA, Sjodt M, Liu M, Kumar KK, Dickson CF, Gell Da, Lei B, Loo JA, Clubb RT. J Biol Chem. 2013;288:1065–1078. doi: 10.1074/jbc.M112.419119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maresso AW, Chapa TJ, Schneewind O. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balderas MA, Nobles CL, Honsa ES, Maresso AW. J Bacteriol. 2012;194:5513–21. doi: 10.1128/JB.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Honsa ES, Maresso AW. BioMetals. 2011;24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 95.Maresso AW, Garufi G, Schneewind O. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Honsa ES, Maresso AW, Highlander SK. PLoS One. 2014;9:e104794. doi: 10.1371/journal.pone.0104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malmirchegini GR, Sjodt M, Shnitkind S, Sawaya MR, Rosinski J, Newton SM, Klebba PE, Clubb RT. J Biol Chem. 2014;289:34886–34899. doi: 10.1074/jbc.M114.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, Mcneely T, Noble L, Brown MJ, Zorman JK, Wang XM, Pancari G, Fan H, Isett K, Burgess B, Bryan J, Brownlow M, George H, Meinz M, Liddell ME, Kelly R, Schultz L, Montgomery D, Onishi J, Losada M, Martin M, Ebert T, Tan CY, Schofield TL, Nagy E, Meineke A, Joyce JG, Kurtz MB, Caulfield MJ, Jansen KU, Mcclements W, Anderson AS. Infect Immun. 2006;74:2215–2223. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joshi A, Pancari G, Cope L, Bowman EP, Cua D, Proctor RA, McNeely T. Hum Vaccin Immunother. 2012;8:336–346. doi: 10.4161/hv.18946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saito H. Nagoya J Med Sci. 2014;76:235–54. [PMC free article] [PubMed] [Google Scholar]
- 101.Dehner C, Morales-Soto N, Behera R, Shrout J, Theil E, Maurice P, Dubois J. J Biol Inorg Chem. 2013;18:371–81. doi: 10.1007/s00775-013-0981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whitby PW, VanWagoner TM, Springer JM, Morton DJ, Seale TW, Stull TL. J Med Microbiol. 2006;55:661–668. doi: 10.1099/jmm.0.46199-0. [DOI] [PubMed] [Google Scholar]
- 103.Segond D, Abi Khalil E, Buisson C, Daou N, Kallassy M, Lereclus D, Arosio P, Bou-Abdallah F, Nielsen Le Roux C. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Müller SI, Valdebenito M, Hantke K. BioMetals. 2009;22:691–695. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- 105.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Watts RE, Totsika M, Challinor VL, Mabbett AN, Ulett GC, De Voss JJ, Schembri Ma. Infect Immun. 2012;80:333–344. doi: 10.1128/IAI.05594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 108.Brumbaugh AR, Smith SN, Subashchandrabose S, Himpsl SD, Hazen TH, Rasko Da, Mobley HLT. Infect Immun. 2015;83:1443–1450. doi: 10.1128/IAI.02904-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lawlor MS, O'Connor C, Miller VL. Infect Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pradel E, Lemaître N, Merchez M, Ricard I, Reboul A, Dewitte A, Sebbane F. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reeves SA, Gonzales EG, Payne SM. Infect Immun. 2003;71:1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Payne SM, Wyckoff EE, Murphy ER, Oglesby AG, Boulette ML, Davies NML. BioMetals. 2006;19:173–180. doi: 10.1007/s10534-005-4577-x. [DOI] [PubMed] [Google Scholar]
- 113.Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, Gao S, Liu X. BMC Microbiol. 2012;12:143. doi: 10.1186/1471-2180-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nassif X, Sansonetti PJ. Infect Immun. 1986;54:603–608. doi: 10.1128/iai.54.3.603-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, S Akashi-takamura, Miyake K, Zhang J, Lee WP, Forsberg LS, Carlson RW, Dixit VM. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 116.O'Riordan M, Portnoy Da. Trends Microbiol. 2002;10:361–364. doi: 10.1016/s0966-842x(02)02401-0. [DOI] [PubMed] [Google Scholar]
- 117.Payne SM. Mol Microbiol. 1989;3:1301–6. doi: 10.1111/j.1365-2958.1989.tb00281.x. [DOI] [PubMed] [Google Scholar]
- 118.Pan X, Tamilselvam B, Hansen EJ, Daefler S. BMC Microbiol. 2010;10:64. doi: 10.1186/1471-2180-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zughaier SM, Kandler JL, Shafer WM. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Corbett D, Wang J, Schuler S, Lopez-Castejon G, Glenn S, Brough D, Andrew PW, Cavet JS, Roberts IS. Infect Immun. 2012;80:14–21. doi: 10.1128/IAI.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. Infect Immun. 2007;75:5867–76. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Desrosiers DC, Bearden SW, Mier I, Abney J, Paulley JT, Fetherston JD, Salazar JC, Radolf JD, Perry RD. Infect Immun. 2010;78:5163–77. doi: 10.1128/IAI.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patzer S, Hantke K. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 124.Hantke K. Biometals. 2001;14:239–249. doi: 10.1023/a:1012984713391. [DOI] [PubMed] [Google Scholar]
- 125.Ma Z, Jacobsen F, Giedroc D. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabri M, Houle S, Dozois CM. Infect Immun. 2009;77:1155–64. doi: 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weston BF, Brenot A, Caparon MG. Infect Immun. 2009;77:2840–8. doi: 10.1128/IAI.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim S, Watanabe K, Shirahata T, Watarai M. J Vet Med Sci. 2004;66:1059–1063. doi: 10.1292/jvms.66.1059. [DOI] [PubMed] [Google Scholar]
- 129.Yang X, Becker T, Walters N, Pascual DW. Infect Immun. 2006;74:3874–9. doi: 10.1128/IAI.01957-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. Cell Host Microbe. 2011;10:248–59. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stork M, Grijpstra J, Bos MP, C Mañas Torres, Devos N, Poolman JT, Chazin WJ, Tommassen J. PLoS Pathog. 2013;9:e1003733. doi: 10.1371/journal.ppat.1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carpenter BM, Whitmire JM, Merrell DS. Infect Immun. 2009;77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Troxell B, Hassan HM. Front Cell Infect Microbiol. 2013;3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Porcheron G, Habib R, Houle S, Caza M, Lepine F, Daigle F, Masse E, Dozois CM. Infect Immun. 2014;82:5056–5068. doi: 10.1128/IAI.02287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Murphy ER, Payne SM. Infect Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reinhart AA, Powell DA, Nguyen AT, O'Neill M, Djapgne L, Wilks A, Ernst RK, Oglesby-Sherrouse AG. Infect Immun. 2015;83:863–875. doi: 10.1128/IAI.02707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lewin AC, Doughty Pa, Flegg L, Moore GR, Spiro S. Microbiology. 2002;148:2449–2456. doi: 10.1099/00221287-148-8-2449. [DOI] [PubMed] [Google Scholar]
- 138.Pecqueur L, D'Autréaux B, Dupuy J, Nicolet Y, Jacquamet L, Brutscher B, Michaud-Soret I, Bersch B. J Biol Chem. 2006;281:21286–21295. doi: 10.1074/jbc.M601278200. [DOI] [PubMed] [Google Scholar]
- 139.Outten CE, Halloran TVO. Science (80- ) 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 140.Hantke K. Curr Opin Microbiol. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 141.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. J Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gonzales T, Robert-Baudouy J. FEMS Microbiol Rev. 1996;18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 143.Kaman WE, Hays JP, Endtz HP, Bikker FJ. Eur J Clin Microbiol Infect Dis. 2014;33:1081–7. doi: 10.1007/s10096-014-2075-1. [DOI] [PubMed] [Google Scholar]
- 144.Raju RM, Goldberg AL, Rubin EJ. Nat Rev Drug Discov. 2012;11:777–89. doi: 10.1038/nrd3846. [DOI] [PubMed] [Google Scholar]
- 145.Vallee BL, Auld DS. Proc Natl Acad Sci U S A. 1990;87:220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miyoshi S, Shinoda S. Microbes Infect. 2000;2:91–8. doi: 10.1016/s1286-4579(00)00280-x. [DOI] [PubMed] [Google Scholar]
- 147.Wu JW, Chen XL. Appl Microbiol Biotechnol. 2011;92:253–62. doi: 10.1007/s00253-011-3532-8. [DOI] [PubMed] [Google Scholar]
- 148.Chang AK, Kim HY, Park JE, Acharya P, Park I, Yoon SM, You HJ, Hahm K, Park JK, Lee JS. J Bacteriol. 2005;187:6909–6916. doi: 10.1128/JB.187.20.6909-6916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chung M-C, Popova TG, Millis Ba, Mukherjee DV, Zhou W, Liotta La, Petricoin EF, Chandhoke V, Bailey C, Popov SG. J Biol Chem. 2006;281:31408–18. doi: 10.1074/jbc.M605526200. [DOI] [PubMed] [Google Scholar]
- 150.Kooi C, Subsin B, Chen R, Pohorelic B, Sokol Pa. Infect Immun. 2006;74:4083–93. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Harrington DJ. Infect Immun. 1996;64:1885–1891. doi: 10.1128/iai.64.6.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kooi C, Corbett CR, Sokol PA. J Bacteriol. 2005;187:4421–4429. doi: 10.1128/JB.187.13.4421-4429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Azghani A, Gray LD, Johnson AR. Infect Immun. 1993;61:0–5. doi: 10.1128/iai.61.6.2681-2686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu Z, Nybom P, Magnusson K. Cell Microbiol. 2000;2:11–17. doi: 10.1046/j.1462-5822.2000.00025.x. [DOI] [PubMed] [Google Scholar]
- 155.Mukherjee DV, Tonry JH, Kim KS, Ramarao N, Popova TG, Bailey C, Popov S, Chung MC. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Plaut AG, Genco RJ, Tomasi TB. J Immunol. 1974;113 [PubMed] [Google Scholar]
- 157.Molla A, Matsumoto K, Oyamada I, Katsuki T, Maeda H. Infect Immun. 1986;53:522–529. doi: 10.1128/iai.53.3.522-529.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Loomes LM, Kerr MA, Senior BW. J Med Microbiol. 1993;39:225–232. doi: 10.1099/00222615-39-3-225. [DOI] [PubMed] [Google Scholar]
- 159.Brezski RJ, Jordan RE. MAbs. 2010;2:212–20. doi: 10.4161/mabs.2.3.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kehl-Fie T, Chitayat S, Hood MI, Dama S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Cell Host Microbe. 2012;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nisapakultorn K, Ross KF. Infect Immun. 2001;69:4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Warfel JM, Steele AD, Agnillo FD. Am J Pathol. 2005;166:1871–1881. doi: 10.1016/S0002-9440(10)62496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kastrup CJ, Boedicker JQ, Pomerantsev AP, Moayeri M, Pompano RR, Kline TR, Sylvestre P, Shen F, Leppla H, Tang W, Ismagilov RF. Nat Chem Biol. 2008;4:742–750. doi: 10.1038/nchembio.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mintz CS, Miller RD, Gutgsell NS, Malek T. Infect Immun. 1993;61:3416–3421. doi: 10.1128/iai.61.8.3416-3421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vollmer P, Walev I, Rose-john S, Bhakdi S. Infect Immun. 1996;64:3646–3651. doi: 10.1128/iai.64.9.3646-3651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Abu Kwaik Y, Bumann D. Cell Microbiol. 2013;15:882–90. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 167.Price C, Al Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Science (80- ) 2011;334 doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 168.Carlsson J, Hogling JD, Sundqvist G. J Med Microbiol. 1984;0:39–46. doi: 10.1099/00222615-18-1-39. [DOI] [PubMed] [Google Scholar]
- 169.Otto BR, van Dooren SJ, Nuijens JH, Luirink J, Oudega B. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Guan SM, Nagata H, Shizukuishi S, Wu JZ. Anaerobe. 2006;12:279–82. doi: 10.1016/j.anaerobe.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 171.Park BR, Zielke Ra, Wierzbicki IH, Mitchell KC, Withey JH, Sikora AE. J Bacteriol. 2015;197:1051–64. doi: 10.1128/JB.02329-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Terwilliger A, Swick MC, Pflughoeft KJ, Pomerantsev A, Lyons CR, Koehler TM, Maresso A. J Bacteriol. 2015;197:2400–2411. doi: 10.1128/JB.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Walsh TR, Toleman MA, Poirel L, Nordmann P. Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Palzkill T. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tong Y, Guo M. Arch Biochem Biophys. 2009;481:1–15. doi: 10.1016/j.abb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fuursted K, Schøler L, Hansen F, Dam K, Bojer MS, Hammerum AM, Dagnæs-Hansen F, Olsen A, Jasemian Y, Struve C. Microbes Infect. 2012;14:155–8. doi: 10.1016/j.micinf.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 177.Gayathri D, Eramma NK, Devaraja TN. Int J Biol Med Res. 2012;3:1870–1874. [Google Scholar]
- 178.Lopetuso LR, Scaldaferri F, Franceschi F, Gasbarrini A. Best Pr Res Clin Gastroenterol. 2014;28:995–1002. doi: 10.1016/j.bpg.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 179.Martín R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermúdez-Humarán LG. Microb Cell Fact. 2013;12:71. doi: 10.1186/1475-2859-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Neilands JB. Horiz Biochem Biophys. 1978;5:65–98. [PubMed] [Google Scholar]
- 181.Braun V, Mahren S, Ogierman M. Curr Opin Microbiol. 2003;6:173–180. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 182.Eisner BH, Asplin JR, Goldfarb DS, Ahmad A, Stoller ML. J Urol. 2010;183:2419–2423. doi: 10.1016/j.juro.2010.02.2388. [DOI] [PubMed] [Google Scholar]
- 183.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. BioMetals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 184.Flint HJ, Scott KP, Louis P, Duncan SH. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 185.Rocha ER, Smith CJ. In: Iron uptake and homeostasis in microorganisms. Andrews SC, Cornelis P, editors. Caister Academic Press; Norwich, UK.: 2010. pp. 155–165. [Google Scholar]
- 186.Otto BR, Verweij-van Vught AMJJ, van Doorn J, Maclaren DM. Microb Pathog. 1988;4:279–287. doi: 10.1016/0882-4010(88)90088-5. [DOI] [PubMed] [Google Scholar]
- 187.Neilands JB. In: Biochemistry of metal micronutrients in the Rhizosphere. Manthey J, Crowley DE, Luster DG, editors. CRC Press; Boca Raton, FL: 1994. pp. 20–21. [Google Scholar]
- 188.Otto BR, Sparrius M, Verweij-van Vught AMJJ, MacLaren DM. Infect Immun. 1990;58:3954–3958. doi: 10.1128/iai.58.12.3954-3958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Otto BR, Kusters JG, Luirink J, De Graaf FK, Oudega B. Infect Immun. 1996;64:4345–4350. doi: 10.1128/iai.64.10.4345-4350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Manfredi P, Lauber F, Renzi F, Hack K, Hess E, Cornelis GR. Infect Immun. 2015;83:300–310. doi: 10.1128/IAI.02042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Peters BM, Jabra-Rizk MA, O'May Ga, William Costerton J, Shirtliff ME. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nair N, Biswas R, Götz F, Biswas L. Infect Immun. 2014;82:2162–2169. doi: 10.1128/IAI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mashburn LM, Jett AM, Akins DR, Whiteley M. J Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hammer ND, Cassat JE, Noto MJ, Lojek LJ, Chadha AD, Schmitz JE, Creech CB, Skaar EP. Cell Host Microbe. 2014;16:531–537. doi: 10.1016/j.chom.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ballouche M, Cornelis P, Baysse C. Recent Pat Antiinfect Drug Discov. 2009;4:190–205. doi: 10.2174/157489109789318514. [DOI] [PubMed] [Google Scholar]
- 196.Mislin GL, Schalk IJ. Metallomics. 2014;6:408–20. doi: 10.1039/c3mt00359k. [DOI] [PubMed] [Google Scholar]
- 197.Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV. Antimicrob Agents Chemother. 2012;56:5419–5421. doi: 10.1128/AAC.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fernandes SS, Nunes A, Gomes AR, de Castro B, Hider RC, Rangel M, Appelberg R, Gomes MS. Microbes Infect. 2010;12:287–294. doi: 10.1016/j.micinf.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 199.Visca P, Bonchi C, Minandri F, Frangipani E, Imperi F. Antimicrob Agents Chemother. 2013;57:2432–2433. doi: 10.1128/AAC.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gentile V, Frangipani E, Bonchi C, Minandri F, Runci F, Visca P. Pathogens. 2014;3:704–719. doi: 10.3390/pathogens3030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Banin E, Brady KM, Greenberg EP. Appl Environ Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. [Apr 18, 2015];Publichealthwatch. https://publichealthwatch.wordpress.com/2014/09/10/novel-antibiotic-may-offer-new-approach-to-treating-drug-resistant-superbugs/
- 203.Lane DJR, Merlot AM, Huang MLH, Bae DH, Jansson PJ, Sahni S, Kalinowski DS, Richardson DR. Biochim Biophys Acta. 2015;1853:1130–1144. doi: 10.1016/j.bbamcr.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 204.Correnti C, Strong RK. J Biol Chem. 2012;287:13524–31. doi: 10.1074/jbc.R111.311829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miethke M. Metallomics. 2013;5:15–28. doi: 10.1039/c2mt20193c. [DOI] [PubMed] [Google Scholar]
- 206.Schalk IJ, Guillon L. Amino Acids. 2013;44:1267–77. doi: 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 207.Bertrand S. [Aug 11, 2015]; http://bertrandsamuel.free.fr/siderophore_base/siderophores.php.
- 208.Hider R, Kong X. Nat Prod Rep. 2010;27:637–57. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 209.Crumbliss AL, Harrington JM. Adv Inorg Chem. 2009;61:179–250. [Google Scholar]
- 210.Hederstedt L. Biochim Biophys Acta - Bioenerg. 2012;1817:920–927. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 211.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Biochim Biophys Acta. 2013;1827:923–937. doi: 10.1016/j.bbabio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 212.Isani G, Carpenè E. Biomolecules. 2014;4:435–57. doi: 10.3390/biom4020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jackson BC, Nebert DW, Vasiliou V. 2010;4:194–201. doi: 10.1186/1479-7364-4-3-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Edwards DR, Handsley MM, Pennington CJ. Mol Aspects Med. 2008;29:258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sterchi EE, Socker W, Bond JS. Mol Aspects Med. 2010;29:309–328. doi: 10.1016/j.mam.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Curr Mol Med. 2013;13:24–57. [PMC free article] [PubMed] [Google Scholar]
- 217.Laity JH, Lee BM, Wright PE. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 218.Coleman JE. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- 219.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Mol Aspects Med. 2012;33:209–90. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 220.Rehman AA, Ahsan H, Khan FH. J Cell Physiol. 2013;228:1665–75. doi: 10.1002/jcp.24266. [DOI] [PubMed] [Google Scholar]
- 221.Kallifidas D, Pascoe B, Owen Ga, Strain-Damerell CM, Hong H-J, Paget MSB. J Bacteriol. 2010;192:608–11. doi: 10.1128/JB.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hase CC, Finkelstein RA. Microbiol Rev. 1993;57:823–837. doi: 10.1128/mr.57.4.823-837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.