ABSTRACT
Purpose
The aim of this study was to identify β-lactamase-producing oral anaerobic bacteria and screen them for the presence of cfxA and BlaTEM genes that are responsible for β-lactamase production and resistance to β-lactam antibiotics.
Material and Methods
Periodontal pocket debris samples were collected from 48 patients with chronic periodontitis and anaerobically cultured on blood agar plates with and without β-lactam antibiotics. Presumptive β-lactamase-producing isolates were evaluated for definite β-lactamase production using the nitrocefin slide method and identified using the API Rapid 32A system. Antimicrobial susceptibility was performed using disc diffusion and microbroth dilution tests as described by CLSI Methods. Isolates were screened for the presence of the β-lactamase-TEM (BlaTEM) and β-lactamase-cfxA genes using Polymerase Chain Reaction (PCR). Amplified PCR products were sequenced and the cfxA gene was characterized using Genbank databases.
Results
Seventy five percent of patients carried two species of β-lactamase-producing anaerobic bacteria that comprised 9.4% of the total number of cultivable bacteria. Fifty one percent of β-lactamase-producing strains mainly Prevotella, Porphyromonas, and Bacteroides carried the cfxA gene, whereas none of them carried blaTEM. Further characterization of the cfxA gene showed that 76.7% of these strains carried the cfxA2 gene, 14% carried cfxA3, and 9.3% carried cfxA6. The cfxA6 gene was present in three Prevotella spp. and in one Porphyromonas spp. Strains containing cfxA genes (56%) were resistant to the β-lactam antibiotics.
Conclusion
This study indicates that there is a high prevalence of the cfxA gene in β-lactamase-producing anaerobic oral bacteria, which may lead to drug resistance and treatment failure.
Keywords: Anaerobic bacteria, β-lactamases, Prevotella, Porphyromonas, β-lactams
INTRODUCTION
The human mouth harbours a complex microbial community containing aerobic and anaerobic bacteria. These bacteria cause polymicrobial opportunistic oral and extra oral infections. Anaerobic bacteria such as Porphyromonas gingivalis, Treponema denticola, Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Prevotella nigrescens, Parvimonas micra, and Eubacterium nodatum cause periodontal diseases, odontogenic abscesses, orofacial infections, and have been implicated in brain abscesses 26 , 31 . Some of these bacteria are also isolated from sputum samples collected from patients with cystic fibrosis and ICU patients with aspiration pneumonia 17 , 21 . β-lactam antibiotics are often prescribed to treat these infections. However, studies have shown that many oral anaerobic bacteria have developed resistance to β-lactam antibiotics because of the production of β-lactamases 12 .
Bacterial resistance to β-lactam antibiotics has been attributed to resistance genes, present on chromosomal or plasmid DNA that can be transferred between commensal and pathogenic bacteria 30 . Studies have shown that there is a high prevalence of cfxA genes responsible for the β-lactamase production in Prevotella and Capnocytophaga species isolated from periodontal pockets 6 , 13 . In addition, cfxA and cfxA2 genes have been isolated from oral infection sites as well as from the causative organisms isolated from these infection sites 7 , 16 . It has been suggested that this cfxA/cfxA2 partition could be partly related to the genus and partly to the geographical origin of the enzyme-producing strains 9 . CfxA2 and cfxA3 genes have been detected in anaerobic bacteria isolated from patients in France, United Kingdom, Norway, Argentina, and the United States of America 11 , 20 , 25 , whereas cfxA6 have only been detected in anaerobic bacteria isolated from patients in Argentina 5 . No data are available from Africa on the production of β-lactamases by oral bacteria and the genes responsible for the production of these enzymes in patients with chronic periodontitis. This study was therefore conducted to isolate and identify β-lactamase-producing oral anaerobic bacteria from the periodontal pockets of patients with chronic periodontitis and to detect the genes responsible for the production of these enzymes.
MATERIAL AND METHODS
Study population
This study was conducted in the Oral Health Centre, Johannesburg in 2012. Forty-eight patients diagnosed with severe to moderate forms of chronic periodontitis using classification provided by the American Association of Periodontology 1 and with pocket depths of five millimeters and more were invited to participate in the study. Ethics clearance was obtained from the Human Research Ethics Committee (certificate no.: M 110112) and written consent was obtained from all participants. Patients with a history of previous periodontal treatment, necrotizing ulcerative gingivitis, diabetes, or those who had taken systemic antimicrobials or anti-inflammatory drugs four weeks prior to the study were excluded.
After careful removal of supragingival plaque, a sterile paper point was inserted into the two deepest pockets and left in place for ten seconds. Paper points were pooled in 1 mL of reduced transport fluid and processed within an hour of sampling to ensure the viability of anaerobic bacteria.
Isolation and identification of β-lactamase producing bacteria
The samples were vortexed for 30 s, serially diluted in phosphate buffered saline, and 0.1 mL of 1/10 to 1/10000 dilutions were plated on 5% blood agar plates supplemented with 5 mg/L of haemin (Sigma-Aldrich, Johannesburg, Gauteng, South Africa) and 1 mg/L of menadione (Sigma-Aldrich, Johannesburg, Gauteng, South Africa) for determining the total anaerobic bacterial count. The proportion of the microbiota resistant to amoxicillin was determined by plating samples on blood agar plates supplemented with 3 µg/mL of amoxicillin with and without 0.75 µg/mL of clavulanic acid 15 . The plates were anaerobically incubated for one week at 37oC and the number of colony forming units (cfu) was counted. The bacteria that grew on amoxicillin but not on the amoxicillin-clavulanic acid plate were identified using the API 32A system (BioMerieux, Midrand, Gauteng, South Africa) and evaluated for β-lactamase production using the Nitrocefin Paper Disc Spot test. Although other techniques can be used, the API 32A system was the only one available at the time.
Detection of β-lactamase genes
DNA was extracted from β-lactamase producing isolates using a technique described by Handal, et al. 13 (2005). A loopful of culture was inoculated into a sterile eppendorf tube containing 10 µL of 10× PCR buffer, 15 mM MgCl2 (Qiagen, Whitehead Scientific products, Johannesburg, South Africa), and 90 µL of sterile distilled water. The inoculated buffer was boiled at 95°C for 10 min, cooled on ice, and centrifuged at 5000 rpm for 10 min. The supernatant was harvested and stored at -70°C until required.
DNA was amplified using the Polymerase Chain Reaction (PCR) technique. A 25 µL reaction mixture containing 12.5 µL of 2× PCR Master Mix, ٢.٥ µL of sterile nuclease-free water, 5 µL of 5 µM primer (For blaTEM GTATGGATCCTCAACATTTCCGTGTCG and ACCAAAGCTTAATCAGTGAGGCA, and for blaCfxA GCAAGTGCAGTTTAAGATT and GCTTTAGTTTGCATTTTCATC) was added to 5 µL of template DNA. Samples were amplified in an iCycler thermal cycler (BIO-RAD, USA) using the PCR conditions described by Handal, et al. 12 (2005) Escherichia coli ATCC 25746 (University of Copenhagen) was used as a positive control for amplification of bla TEM. The P. intermedia isolate containing the bla cfxA gene was used as the positive control for the amplification of that gene. For both PCR reactions, the negative control consisted of sterile water. Amplified PCR products were isolated using gel electrophoresis, sequenced, and further characterized by the BLAST option of the nucleotide GenBank│European Molecular Biology Laboratory - databases.
Antimicrobial susceptibility
All the 85 β-lactamase producing isolates were subjected to antimicrobial susceptibility using a disk diffusion test and the results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) performance standards for antimicrobial disk susceptibility tests 4 . Minimum inhibitory concentrations (MICs) were performed using the microbroth dilution method according to CLSI Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria 3 . Only 17 strains were successfully revived and included in this assay.
The data were analyzed using the STATA statistical package (College Station, Texas, USA).
RESULTS
Demography and the prevalence of β-lactamase-producing anaerobic oral bacteria
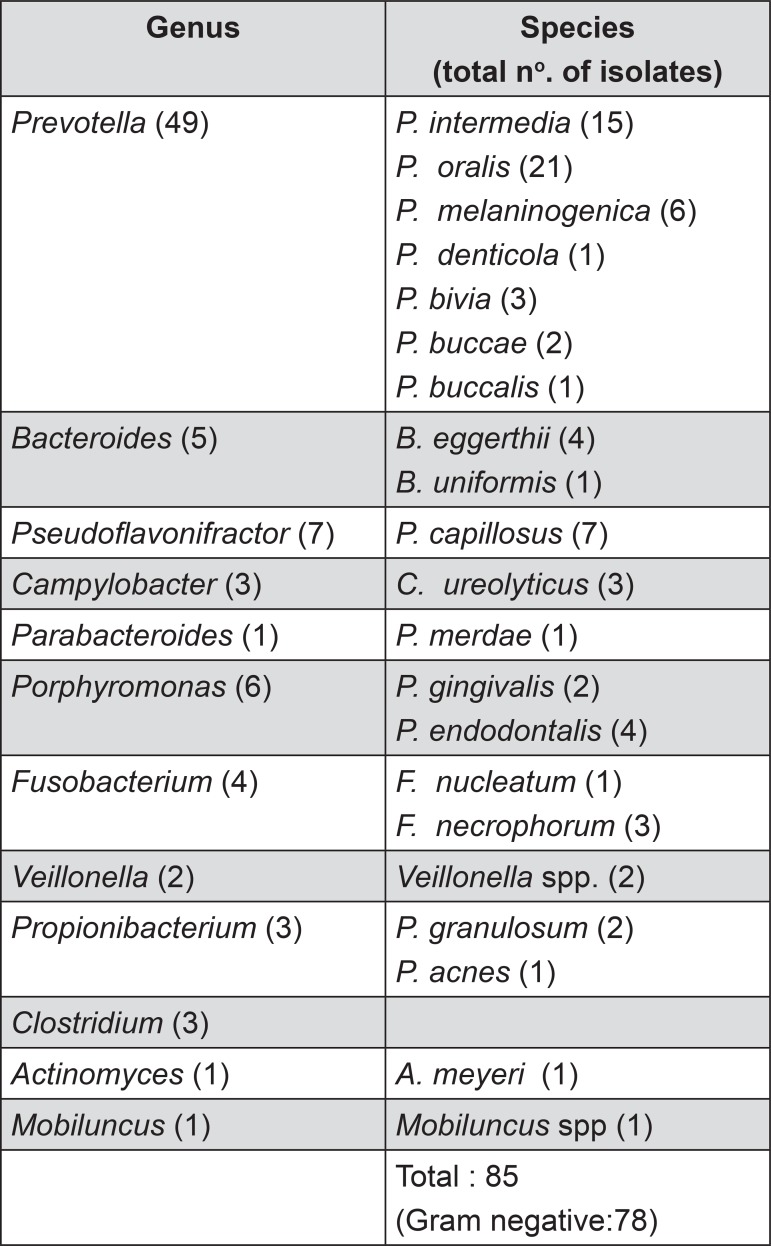
The mean age of the patients was 52 with a range from 32 to 83 years of age. Fifty-eight percent of the patients were female. The average pocket depth of the sampled pockets was 7 mm with a range of 5 mm to 13 mm. Mean total anaerobic bacterial count on control plate was 1.8x106 cfu/mL. Mean total anaerobic bacterial count on the amoxicillin plate was 1.9x105 cfu/mL. Mean total anaerobic bacterial count on the augmentin plate was 5.9x104 cfu/mL. Seventy-five percent of patients carried on average two species of β-lactamase-producing anaerobic oral bacteria, which constitute 9.4% of the total cultivable number of bacteria. All the isolates that grew on the blood agar plate containing amoxicillin tested positive for β-lactamase with the nitrocefin test. Seventy-eight of the 85 isolates of β-lactamase-producing bacteria were mainly gram negative black pigmented anaerobes (Figure 1).
Figure 1. β-lactamase-producing oral anaerobic bacteria.
Detection of β-lactamase genes
Fifty-one percent of β-lactamase-producing isolates carried the bla CfxA gene, whereas none carried bla TEM (Table 1). Further characterization of the cfxA gene showed that 76.7% of these isolates carried the cfxA2 gene (showing 100% similarity to cfxA2 GenBank accession no. AM940016 of Bacteroides ovatus), 14% carried the cfxA3gene (Genbank accession no. AY860640 of Capnocytophaga ochracea plasmid Pcap Mob A), and 9.3% carried cfxA6 (Genbank accession no. FN376426 100% similarity with Prevotella intermedia partial cfxA6 gene, strain PI51). Most of these isolates were from the black pigmented anaerobic bacteria such as Prevotella spp. and Bacteroides spp. The Minimal Inhibitory Concentration (MIC) for penicillin and amoxicillin was performed on the 17 isolates that carried cfxA. The results showed that up to 56% of these isolates were resistant to β-lactam antibiotics (Table 2).
Table 1. The prevalence of cfxA genes in the β-lactamase-producing oral anaerobic bacteria.
| β-lactamase gene (n=85) | Genus | Total no. of | No. of isolates with cfxA genes (%) | ||
|---|---|---|---|---|---|
| positive isolates (%) | cfxA2 | cfxA3 | cfxA6 | ||
| cfxA | Prevotella spp | 21 | 14 | 4 | 3 |
| Porphyromonas spp | 4 | 3 | - | 1 | |
| Bacteroides spp | 12 | 10 | 2 | - | |
| Fusobacterium spp | 2 | 2 | - | - | |
| Clostridium spp | 2 | 2 | - | - | |
| Propionobacterium spp | 2 | 2 | - | - | |
| Total | 43 (50.6) | 33 (76.7) | 6 (14) | 4 (9.3) | |
| BlaTEM | As above | 0 | - | - | - |
Table 2. Antimicrobial susceptibility (Disc diffusion test) of β-lactamase-producing anaerobic bacteria against β-lactam antibiotics.
| No of isolates resistant to β-lactam antibiotics (%) | ||||
|---|---|---|---|---|
| aDisc diffusion test (n=85) | ||||
| cfxA gene | Ampicillin | Penicillin | ||
| R | S | R | S | |
| Present (n=43) | 18 (41.86) | 25 (58.14) | 24 (55.81) | 19 (44.19) |
| Absent (n=42) | 14 (33.33) | 28 (66.67) | 8 (19.05) | 34 (80.95) |
aDisc diffusion test (disk content 10 µg): R: Resistant ≤28 mm), S: Susceptible ≥29 mm)
Antimicrobial susceptibility
All 85 β-lactamase-producing isolates were subjected to the disc diffusion test for ampicillin and penicillin. The results showed that 42 to 56% of the isolates containing resistance genes were resistant to β-lactam antibiotics (Table 2). MIC test, which is more accurate, showed that 53 to 59% of isolates that carried the resistance gene were resistant and another 6% had intermediate susceptibility to β-lactam antibiotics (Table 3).
Table 3. Antimicrobial susceptibility (Broth dilution test) of β-lactamase-producing anaerobic bacteria against β-lactam antibiotics.
| cfxA gene | No of isolates resistant to β-lactam antibiotics (%) | |||||
|---|---|---|---|---|---|---|
| aBroth dilution test (n=17) | ||||||
| Amoxicillin | Penicillin | |||||
| R | I | S | R | I | S | |
| Present | 9 (52.94) | 0 | 8 (47.06) | 10 (58.82) | 1 (5.89) | 6 (35.29) |
aBroth dilution test: R: Resistant (≥2 µg/ml), I: Intermediate susceptibility (1 µg/ml), S: Susceptible (≤0.5 µg/ml)
DISCUSSION
In this study, Prevotella spp. and Bacteroides spp. were the most prevalent β-lactamase-producing bacteria and 43% and 75% of these bacteria, respectively, carried the cfxA gene. These findings are similar to the results obtained in the North American, French, and Norwegian population 9 , 13 . The cfxA genes are known to be present in Pseudomonas aeruginosa, gut flora including bacteroides and oral bacteria such as Prevotella and Capnocytophaga 2 , 8 , 13 . These genes are responsible for the resistance to penicillins and cephalosporins. The presence of these genes in oral commensals is always a cause for concern because this resistance can spread to serious pathogens and cause resistance to extended-spectrum cephalosporins as well. In addition, these normal commensals do cause extraoral infections which require antibiotic treatment. For example, Capnocytophaga spp containing the cfxA3-β-lactamase-producing gene have been isolated from patients with leukemia and neutropenia 22 , 23 . Similarly, cfxA gene-containing resistant Prevotella spp have been isolated from patients with cystic fibrosis and intra-abdominal infections 28 , 29 . The presence of these resistance genes in the oral microbiota could result in the commensals serving as reservoirs of antibiotic resistance 30 .
This study also showed that 77% of β-lactamase producing strains carried cfxA2 genes. In the Prevotella spp, there was a predominance of the cfxA gene in North America, cfxA2 in France 20 , 25 , and both genes in the United Kingdom 16 . Horizontal gene transfer might explain the spread of closely related gene sequences among these periodontal species 6 . Although the cfxA3 gene has been found mainly in Capnocytophaga spp. 11 , 23 , in our study it was closely associated with Prevotella and Bacteroides spp. Interestingly, although cfxA6 was initially detected in Prevotella spp. (Genbank accession no. FN376426) and also found in the present study, no other studies have detected this gene. In addition, the presence of cfxA6 in Porphyromonas spp. has not been previously reported.
In this study, 10% of the periodontal pocket microbiota in 75% of the patients produced β-lactamase enzymes; and there were on average two species per patient. Of these β-lactamase producers, only 50.6% carried cfxA genes. However, oral bacteria that live in a biofilm environment, which is highly stressful and competitive, are known to adapt to genetic transfer. Metagenomic and bioinformatic studies have confirmed that oral bacteria play a major role in horizontal gene transfer 18 because it improves their chance of survival, increases virulence, and alters their metabolism and drug resistance. Both plasmid and chromosomal-borne transfer of antibiotic resistance has been shown in oral bacteria 10 . Transformation through eDNA present in the plaque 14 and membrane vesicles 24 has also been described. In addition, a highly mobile Tn916, like as transposon that facilitates conjugation, has been found in many oral bacteria such as Streptococci species, F. nucleatum, Eubacterium species, Veillonella species, and Actinobacillus species. Therefore, a potential reservoir may transfer resistance genes to other drug sensitive bacteria from these 10% of drug resistant bacteria.
Some β-lactamase-producing anaerobes which carried β-lactamase genes were sensitive to β-lactam antibiotics (Table 3) with high MIC values of 0.125 µg/mL. This phenomenon has also been reported by other studies 30 , 32 . This is possibly because β-lactamases are ubiquitous in bacteria, and when produced in small quantities they may contribute little to antibiotic resistance, but could play a physiological role in peptidoglycan metabolism 19 . Garcia, et al. 8 (2008) has also shown that cfxA gene containing Bacteroides spp. can show varying quantities of β-lactamase enzyme activity. In addition, some β-lactamases are constitutive whereas others require induction, which suggests that the mere presence of these genes may not contribute to the β-lactam antibiotics as shown by more reliable broth dilution techniques (53% and 59% resistance to amoxicillin and penicillin, respectively) 27 . Nevertheless, in nonresponsive patients antibiotics other than β-lactam should be considered. Our results also showed that many β-lactamase-producing bacteria were resistant to β-lactam antibiotics, but did not carry the cfxA or Bla TEM gene, which suggests that there may be other genes responsible for the production of this enzyme (Table 2). This study only studied cfxA or Bla TEM because these genes responsible for the production of β-lactamases are known to be present in oral bacteria. Other genes such as CepA/CblA are only occasionally identified in oral bacteria 9 .
One of the limitations of our study was the identification scheme. APi is an old technique based on biochemical reactions and was the only technique available in our laboratory at the time of this study. The identification percentage similarities of 80 and above were accepted as the final identification.
In conclusion, seventy-five percent of patients carried β-lactamase-producing anaerobic bacteria. CfxA2, cfxA3, and cfxA6 genes were detected in 51% of these organisms, which comprised 10% of the total cultivable oral microbiota in our patients with chronic periodontitis. The cfxA6 gene was found in Prevotella and Porphyromonas spp, which has epidemiological implications. Fifty-six percent of the isolates that carried cfxA genes were resistant to β-lactam antibiotics, which suggest that in nonresponsive patient antibiotics other than β-lactam should be considered.
ACKNOWLEDGMENTS
We thank Arshnee Moodley from The Department of Veterinary Disease Biology, Faculty of Health and Medical Sciences, University of Copenhagen for the E. coli ATCC 25746 culture, and the University of the Witwatersrand for the financial support through FRC grant.
REFERENCES
- 1.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 2.Celesk R, Robillard NJ. Factors influencing the accumulation of ciprofloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33:1921–1926. doi: 10.1128/aac.33.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute . Antimicrobial Susceptibility Testing of anaerobic bacteria; approved standard - sixth edition. Clinical and Laboratory Standards Institute document M11-A6. Wayne: CLSI; 2004. 39 [Google Scholar]
- 4.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial disk susceptibility tests; approved standard – ninth edition. Clinical and Laboratory Standards institute document M2-A9. Wayne: CLSI; 2006. 99 [Google Scholar]
- 5.Fernández-Canigia L, Cejas D, Gutkind G, Radice M. Detection and genetic characterization of β-lactamases in Prevotella intermedia and Prevotella nigrescens isolated from oral cavity infections and peritonsillar abscesses. Anaerobe. 2015;33:8–13. doi: 10.1016/j.anaerobe.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira LQ, Avelar KE, Vieira JM, Paula GR, Ap Colombo, Domingues RM, et al. Association between the cfxA gene and transposon Tn4555 in Bacteroides distasonis strains and other Bacteroides species. Curr Microbiol. 2007;54:348–353. doi: 10.1007/s00284-006-0411-0. [DOI] [PubMed] [Google Scholar]
- 7.Fosse T, Madinier I, Hannoun L, Giraud-Morin C, Hitzig C, Charbit Y, et al. High prevalence of cfxA β-lactamase in aminopenicillin-resistant Prevotella strains isolated from periodontal pockets. Oral Microbiol Immunol. 2002;17:85–88. doi: 10.1046/j.0902-0055.2001.00096.x. [DOI] [PubMed] [Google Scholar]
- 8.García N, Gutiérrez G, Lorenzo M, García JE, Píriz S, Quesada A. Genetic determinants for cfxA expression in Bacteroides strains isolated from human infections. J Antimicrob Chemother. 2008;62:942–947. doi: 10.1093/jac/dkn347. [DOI] [PubMed] [Google Scholar]
- 9.Giraud-Morin C, Madinier I, Fosse T. Sequence analysis of cfxA2-like beta-lactamases in Prevotella species. J Antimicrob Chemother. 2003;51:1293–1296. doi: 10.1093/jac/dkg221. [DOI] [PubMed] [Google Scholar]
- 10.Guiney DG, Bouic K. Detection of conjugal transfer systems in oral black-pigmented Bacteroides spp. J Bacteriol. 1990;172:495–497. doi: 10.1128/jb.172.1.495-497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handal T, Giraud-Morin C, Caugant DA, Madinier I, Olsen I, Fosse T. Chromosome- and plasmid-encoded β-lactamases in Capnocytophaga spp. Antimicrob Agents Chemother. 2005;49:3940–3943. doi: 10.1128/AAC.49.9.3940-3943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handal T, Olsen I. Antimicrobial resistance with focus on oral beta-lactamases. Eur J Oral Sci. 2000;108:163–174. doi: 10.1034/j.1600-0722.2000.108003163.x. [DOI] [PubMed] [Google Scholar]
- 13.Handal T, Olsen I, Walker CB, Caugant DA. Detection and characterization of β-lactamase genes in subgingival bacteria from patients with refractory periodontitis. FEMS Microbiol Lett. 2005;242:319–324. doi: 10.1016/j.femsle.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol. 2010;59:345–349. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 15.Herrera D, van Winkelhoff AJ, Dellemijn-Kippuw N, Winkel EG, Sanz M. Βeta-lactamase producing bacteria in the subgingival microflora of adult patients with periodontitis. A comparison between Spain and The Netherlands. J Clin Periodontol. 2000;27:520–525. doi: 10.1034/j.1600-051x.2000.027007520.x. [DOI] [PubMed] [Google Scholar]
- 16.Iwahara K, Kuriyama T, Shimura S, Williams DW, Yanagisawa M, Nakagawa K, et al. Detection of cfxA and cfxA2, the beta-lactamase genes of Prevotella spp., in clinical samples from dentoalveolar infection by real-time PCR. J Clin Microbiol. 2006;44:172–176. doi: 10.1128/JCM.44.1.172-176.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauterbach E, Voss F, Gerigk R, Lauterbach. M. Bacteriology of aspiration pneumonia in patients with acute coma. Intern Emerg Med. 2014;9:879–885. doi: 10.1007/s11739-014-1120-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Chen X, Skogerbø G, Zhang P, Chen R, He S, et al. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics. 2012;100:265–270. doi: 10.1016/j.ygeno.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Livermore DM. Determinants of the activity of beta-lactamase inhibitor combinations. J Antimicrob Chemother. 1993;31(Suppl A):9–21. doi: 10.1093/jac/31.suppl_a.9. [DOI] [PubMed] [Google Scholar]
- 20.Madinier I, Fosse T, Giudicelli J, Labia R. Cloning and biochemical characterization of a class A β-lactamase from Prevotella intermedia. Antimicrob Agents Chemother. 2001;45:2386–2389. doi: 10.1128/AAC.45.8.2386-2389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam E. Emerging cystic fibrosis pathogens and the microbiome. Paediatr Respir Rev. 2014;15(Suppl 1):13–15. doi: 10.1016/j.prrv.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Martino R, Rámila E, Capdevila JA, Planes A, Rovira M, Ortega MD, et al. Bacteremia caused by Capnocytophaga species in patients with neutropenia and cancer: results of a multicenter study. Clin Infect Dis. 2001;33:E20–E22. doi: 10.1086/322649. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto T, Matsubara M, Oana K, Kasuga E, Suzuki T, Hidaka E, et al. First case of bacteremia due to chromosome-encoded CfxA3-beta-lactamase-producing Capnocytophaga sputigena in a pediatric patient with acute erythroblastic leukemia. Eur J Med Res. 2008;13:133–135. [PubMed] [Google Scholar]
- 24.Olsen I, Tribble GD, Fiehn NE, Wang BY. Bacterial sex in dental plaque. 10.3402/jom.v5i0.20736.J Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker AC, Smith CJ. Genetic and biochemical analysis of a novel Amber class A beta lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrobial Agents Chemother. 1993;37:1028–1036. doi: 10.1128/aac.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samaranayake LP. Essential microbiology for Dentistry. Edinburgh: Churchill Livingstone; 2002. pp. 233–238. [Google Scholar]
- 27.Schuetz AN. Antimicrobial resistance and susceptibility testing of anaerobic bacteria. Clin Infect Dis. 2014;59:698–705. doi: 10.1093/cid/ciu395. [DOI] [PubMed] [Google Scholar]
- 28.Sherrard LJ, Schaible B, Graham KA, McGrath SJ, McIlreavey L, Hatch J, et al. Mechanisms of reduced susceptibility and genotypic prediction of antibiotic resistance in Prevotella isolated from cystic fibrosis (CF) and non-CF patients. J Antimicrob Chemother. 2014;69:2690–2698. doi: 10.1093/jac/dku192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran CM, Tanaka K, Watanabe K. PCR-based detection of resistance genes in anaerobic bacteria isolated from intra-abdominal infections. J Infect Chemother. 2013;19:279–290. doi: 10.1007/s10156-012-0532-2. [DOI] [PubMed] [Google Scholar]
- 30.Wilke MS, Lovering AL, Strynadka NCJ. β-lactam antibiotic resistance: a current structural perspective. Cur Opin Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Wu PC, Tu MS, Lin PH, Chen YS, Tsai HC. Prevotella brain abscesses and stroke following dental extraction in a young patients: a case report and review of the literature. Intern Med. 2014;53:1881–1887. doi: 10.2169/internalmedicine.53.1299. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, Chen J, He J, Miao X, Xu M, Wu X, et al. Antimicrobial resistance and prevalence of resistance genes of obligate anaerobes isolated from periodontal abscesses. J Periodontol. 2014;85:327–334. doi: 10.1902/jop.2013.130081. [DOI] [PubMed] [Google Scholar]