Summary
Carbon catabolite repression (CCR) is a regulatory phenomenon implemented by bacteria to hierarchically organize carbohydrate utilization in order to achieve maximal growth. CCR is likely of great importance to Streptococcus pneumoniae because the human host sites inhabited by this pathogen represent complex carbohydrate environments. In this species, inactivation of the prototypical Gram-positive CCR master regulator, ccpA, attenuates virulence in mice but does not relieve CCR of most metabolic enzymes, suggesting CcpA-independent CCR mechanisms predominate. Here we show the activities of three transcriptional regulators constitute the majority of transcriptional CCR of galactose metabolism operons. We determined seryl-phosphorylated histidine phos-phocarrier protein (HPr-Ser~P)-mediated regulation is a major CCR mechanism and an essential activity in the pneumococcus, as an HPr point mutation abolishing HPrK/P-dependent phosphorylation was not tolerated nor was deletion of hprk/p. The HPr-Ser~P phosphomimetic mutant HPr S46D had reduced phos-photransferase system transport rates and limited induction of CCR-repressed genes. These results support a model of pneumococcal CCR in which HPr-Ser~P directly affects the activity of CcpA while indirectly affecting the activity of pathway-specific transactional regulators. This report describes the first CcpA-independent CCR mechanism identified in the pneumococcus and the first example of lethality from loss of HPr-Ser~P-mediated CCR in any species.
Introduction
Streptococcus pneumoniae (pneumococcus) is a Gram-positive asymptomatic colonizer of the human nasophar-ynx, capable of spreading to disparate sites throughout the body of a susceptible host. Previous work from multiple labs collectively supports a model in which pneumococcus ‘grazes’ on glycosylated host mucosal surfaces through the combined action of multiple surface-associated gly-cosidases, carbohydrate transporters and intracellular catabolic enzymes (King et al., 2004; 2006; Manco et al., 2006; Burnaugh et al., 2008; Yesilkaya et al., 2008; Dalia et al., 2010; King, 2010; Terra et al., 2010; Limoli et al., 2011; Marion et al., 2011a,b; 2012; Bidossi et al., 2012; Buckwalter and King, 2012). Expression of these factors is likely regulated in response to carbohydrate availability. Carbon catabolite repression (CCR) is a regulatory phenomenon used by both Gram-positive and negative bacteria to achieve maximum growth in mixed carbohydrate environments. CCR organizes utilization of available carbohydrates into a hierarchy by repressing transporters and metabolic enzymes for less easily metabolized carbohydrates when a preferred carbohydrate is available (Deutscher et al., 2006). Given the carbohydrate diversity at the sites occupied by the pneumococcus and the large fraction of their genome dedicated to carbohydrate metabolism, we hypothesize that CCR is of utmost importance to this pathogen’s fitness.
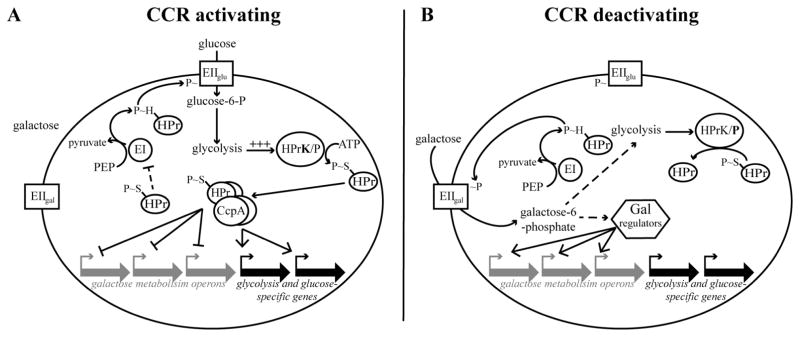
Gram-positive species accomplish CCR by two main activities: (i) inhibition of transport of non-preferred carbohydrates and (ii) transcriptional regulation of metabolic genes by global and local regulators (Deutscher et al., 1994; 2006; Deutscher and Küster, 1995; Darbon et al., 2002; Titgemeyer and Hillen, 2002; Warner and Lolkema, 2003; Schumacher et al., 2004; Lorca et al., 2005; Fujita, 2009; Marciniak et al., 2012). These two branches of CCR are interlocked by their mutual dependence on the histidine-containing phosphocarrier protein (HPr) (Deutscher et al., 2006). The central role of HPr in CCR is intimately tied to its decisive role in phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) transport. As the liaison between the initial energy supplying event (PEP hydrolysis by Enzyme I) and the carbohydrate-specific EII components, HPr governs the decision of which EII complex to power and therefore which carbohydrate to transport (Fig. 1) (Poolman, 1993). Similar to PTS transport, HPr’s involvement in CCR also depends on a pivotal phosphorylation event. Metabolism of a preferred carbohydrate elevates the concentration of intracel-lular glycolytic intermediates, stimulating HPr kinase/ phosphorylase (HPrK/P) to phosphorylate HPr at a conserved serine residue (Fig. 1A) (Deutscher et al., 2006). Because seryl-phosphorylated HPr (HPr-Ser~P) is a poor substrate for EI-dependent phosphorylation, less HPr-His~P is produced, and PTS transport is restricted, ultimately enabling prioritization of glucose transport (see reviews by Dahl, 2002; Deutscher et al., 2006). Restriction of transport also limits the formation of intracellular carbohydrate inducers that would otherwise induce expression of corresponding metabolic genes, by affecting the activity of dedicated transcriptional regulators (Fig. 1A) (Bissett and Anderson, 1974a,b; Abranches et al., 2004; Zeng et al., 2010; 2012). Concomitantly, HPr-Ser~P affects global transcriptional control by serving as a co-repressor for the master CCR transcriptional regulator, catabolite control protein A (CcpA) (Fig. 1A) (Warner and Lolkema, 2003; Schumacher et al., 2004; Deutscher et al., 2006; Deutscher, 2008). This complex represses transcription of non-preferred carbohydrate metabolism genes while activating transcription of central and preferred carbohydrate metabolism genes (Titgemeyer and Hillen, 2002; Schumacher et al., 2004). CCR is relieved when decreased availability of the preferred carbohydrate results in decreased flux through glycolysis, and hence less stimulation of HPrK/P’s kinase activity. Under these circumstances, HPrK/P resorts to its phosphorylase activity and dephosphorylates HPr-Ser~P, replenishing the supply of HPr available to participate in PTS transport. Transport of non-preferred carbohydrates resumes and CcpA-dependent transcriptional repression is relieved (Fig. 1B).
Fig. 1. Model of carbon catabolite repression in Gram-positive species.
A depicts a cell enacting carbon catabolite repression in response to a mixed carbohydrate environment; the cell is in the presence of galactose and glucose. The preferred carbohydrate glucose is efficiently transported via its phosphoenolpyruvate transport system (PTS). Glucose-6-phosphoate is then metabolized by glycolysis, causing a rapid increase in glycolytic intermediates. Glycolytic intermediates stimulate the kinase activity of HPrK/P, a dual kinase/phosphorylase. HPrK/P phosphorylates HPr at a conserved serine residue, resulting in HPr S~P. HPr S~P serves two main functions. The first function involves restricting PTS transport. HPr S~P is a poor substrate for EI-dependent phosphorylation, resulting in decreased HPr H~P in the cell. The galactose PTS is outcompeted by the glucose PTS for the remaining supply of HPr H~P. Glucose transport predominates. The second function of HPr S~P is as a co-regulator of the master CCR transcriptional regulator, CcpA. As part of this complex, CcpA recognizes pseudopalindromic sequences proximal to or within catabolite-regulated genes, resulting in repression of expression of non-preferred carbohydrate metabolism genes and activation of expression of glucose transport and metabolism genes.
B depicts the release of CCR occurring when the glucose supply is diminished. Decreased availability of glucose results in reduced glucose transport, culminating in reduced flux through glycolysis. Hence, the kinase activity of HPrK/P is no longer stimulated and it resorts to its phosphorylase activity and dephosphorylates HPr S~P. Decreased levels of HPr S~P relieve CcpA-dependent regulation. Increased availability of HPr for EI-dependent phosphorylation allows galactose transport by the galactose PTS. Galactose transport results in intracellular galactose 6-phosphate, which activates transcriptional regulators that upregulate galactose metabolism operons.
Of these classic Gram-positive CCR mechanisms, only the role of CcpAin pneumococcal biology has been studied (Giammarinaro and Paton, 2002; Iyer et al., 2005; Kaufman and Yother, 2007; Carvalho et al., 2011). The pneumococcus encodes a CcpA homolog and the genome harbors over 300 putative CcpA binding sites, suggesting CcpA is a master CCR regulator in this organism (van Opijnen et al., 2009). A microarray study of a ccpA deletion strain confirmed that CcpA does indeed have an extensive regulatory influence in the pneumococcus (Carvalho et al., 2011). Although CcpA is required for establishing colonization and invasive disease, inactivation of ccpA does not alleviate CCR of all or even most catabolite-repressed enzymes (Giammarinaro and Paton, 2002; Iyer et al., 2005). These enzymatic assay results suggest that similar to other species, the pneumococcus uses both CcpA-dependent and CcpA-independent methods of CCR.
In this study, we sought to uncover how the pneumo-coccus accomplishes CcpA-independent CCR. Our goal was to determine if pneumococcal HPr-Ser~P can participate in CCR. We found that under CCR conditions, transport of a non-preferred carbohydrate is restricted, and transcriptional repression of the corresponding metabolism genes occurs independently of and in conjunction with CcpA. We identified three examples of predicted carbohydrate-binding transcriptional regulators whose activity profiles are consistent with them responding to intracellular carbohydrate inducers. In order to assess the potential contribution of HPr to these observed phenotypes, we characterized two HPr point mutants classically used to investigate HPr-Ser~P-mediated CCR. Consistent with this hypothesis, HPr S46D, a phosphomimetic mutant of HPr-Ser~P, mimicked a constitutive CCR state evident by reduced PTS transport rates and reduced induction of non-preferred carbohydrate metabolism genes. The HPr S46A mutant, which disables HPr-Ser~P-mediated CCR in related species, however, was lethal in the pneumococ-cus. This result strongly suggests that some or all of HPr-Ser~P’s CCR activities are uniquely essential to the pneumococcus. We found that supplying wild-type (WT) ptsH (HPr) in trans or deleting ptsI (EI) in cis were both able to rescue the lethality of the HPr S46A mutation. We propose an updated model in which CcpA and HPr-Ser~P globally co-regulate metabolism in response to glucose availability, while modulation of carbohydrate transporter activities by HPr-Ser~P allows for exclusion or uptake of non-preferred carbohydrates that are subsequently sensed and responded to by a network of carbohydrate-specific transcriptional regulators. This organization of CCR allows for temporal regulation of carbohydrate metabolism both globally and on a carbohydrate-specific basis.
Results
Pneumococcus inhibits galactose transport in favor of glucose transport and metabolism
For the purpose of investigating CCR mechanisms in pneumococcus, media containing the preferred carbohydrate glucose and the non-preferred carbohydrate galac-tose were used as a model for the induction and alleviation of CCR. The monosaccharide galactose was chosen as it is prevalent in many of the host niches occupied by pneu-mococcus (Buckwalter and King, 2012). Additionally, as pneumococcus encodes both the Leloir and the tagatose 6-phosphate pathways for galactose metabolism, it is likely galactose is transported by both PTS and non-PTS transporters, offering a unique opportunity to investigate a larger scope of possible CCR activities.
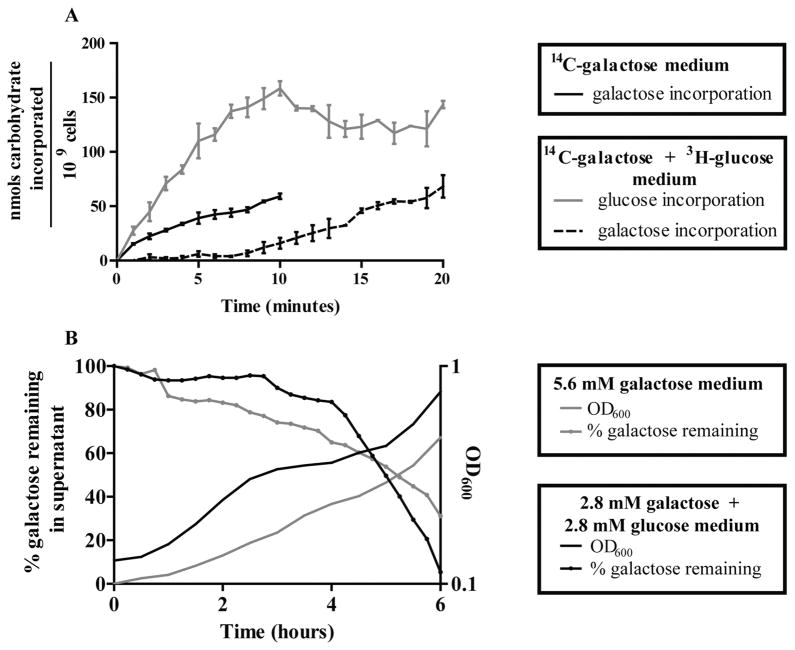
A hallmark of CcpA-independent CCR in Gram-positive species is restriction of non-preferred carbohydrate transport through both PTS and non-PTS transporters when glucose is available. Two approaches were taken to determine if glucose causes inhibition of non-preferred carbohydrate transport in the pneumococcus. First, a radiolabeled carbohydrate incorporation assay was used to measure incorporation rates of galactose in the presence and absence of glucose. Cells preconditioned to galactose-containing medium were assayed over time for uptake of 14C-D-galactose from galactose-only medium or 14C-D-galactose and 3H-D-glucose from medium containing both glucose and galactose. As expected, the former accumulated 14C-D-galactose with an appreciable rate (Fig. 2A). When glucose and 3H-D-glucose were included in the assay medium, however, 14C-D-galactose incorporation was initially reduced to a nearly undetectable level, while 3H-D-glucose incorporation proceeded (Fig. 2A). This result demonstrates that the activity of pre-existing galactose transporters is inhibited when glucose becomes available. 14C-D-galactose incorporation increased from 0.9 ± 0.2 nmol galactose/min/109 cells in the first 7 min of the experiment to 4.7 ± 0.3. This increase at 7 min coincided with the plateauing of 3H-D-glucose accumulation. Based on the measured rate of glucose incorporation, at this point in the assay, the glucose concentration in the medium had diminished, suggesting the rate of galactose accumulation increased because the available glucose concentration had dropped below the threshold for maintaining CCR. Based on the speed at which galactose uptake was first inhibited and latter restored, we conclude that S. pneumoniae uses at least one CCR mechanism that does not require regulation at the level of transcription.
Fig. 2. Galactose transport and metabolism are inhibited in the presence of the preferred carbohydrate glucose.
A. Galactose incorporation was measured for WT in the presence and absence of glucose using a radiolabeled carbohydrate incorporation assay. Cells preconditioned to galactose were washed and exposed to a mixture of unlabeled and C14-labeled galactose or a mixture of C14-labeled galactose, H3-labeled glucose and 100 μM of each unlabeled carbohydrate at 37°C. Accumulation of each radioisotope over time was measured by standard liquid scintillation methods. Black solid line represents galactose incorporation in the absence of glucose. Gray solid line represents glucose incorporation in the presence of galactose. Black dashed line represents galactose incorporation in the presence of glucose. Values are reported as the total nanomoles of carbohydrate accumulated / 109 cells by accounting for the concentration of unlabeled carbohydrate and the colony-forming units in each reaction. Each point is the average of three biological replicates, and error bars are the standard error of the mean.
B. The extent of galactose metabolism by WT in the presence and absence of glucose was determined by measuring the concentration of galactose remaining in culture supernatants. WT cells pre-grown in galactose chemically defined medium (CDM) were washed and inoculated into 5.6 mM galactose or 2.8 mM glucose plus 2.8 mM galactose CDM. OD600 was measured every 30 min (smooth lines). Supernatant aliquots were collected every 15 min and assayed for total galactose using the Amplex Red Galactose/Galactose Oxidase Kit (Molecular Probes) (dotted lines). A galactose standard curve was used to convert absorbance readings to moles of galactose present. Data are shown as the percentage of the original concentration of galactose remaining at the point of collection. A representative pair of experiments is shown.
As a second approach to determine if glucose causes inhibition of non-preferred carbohydrate transport, a galactose oxidase assay was used to monitor galactose concentrations in culture supernatants over time. Cells preconditioned to galactose-containing medium were back-diluted and grown in media containing only galactose, or galactose in the presence of a limiting amount of glucose. Throughout the 6 h of growth in galactose-only medium, the density of the culture increased progressively, while the concentration of galactose remaining in the supernatant decreased steadily (Fig. 2B). In contrast, when a limiting amount of glucose was provided that was still sufficient to sustain appreciable growth, a diauxic growth curve resulted in which two periods of exponential growth were interspersed by a lag in growth occurring roughly between 3 and 4 h (Fig. 2B). The galactose concentration in the supernatant remained constant throughout the first phase of exponential growth indicating galactose was not being transported and metabolized (Fig. 2B, through 3 h). The galactose concentration in the supernatant decreased sharply starting at 4 h, coinciding with the end of the lag phase and the beginning of the second exponential growth phase (Fig. 2B). This concurrence is indicative of galactose being consumed in the second growth phase of the diauxic growth curve. Collectively, the diauxic growth curve and dynamics of galactose consumption are consistent with sequential transport and utilization of carbohydrates, a hallmark of CCR.
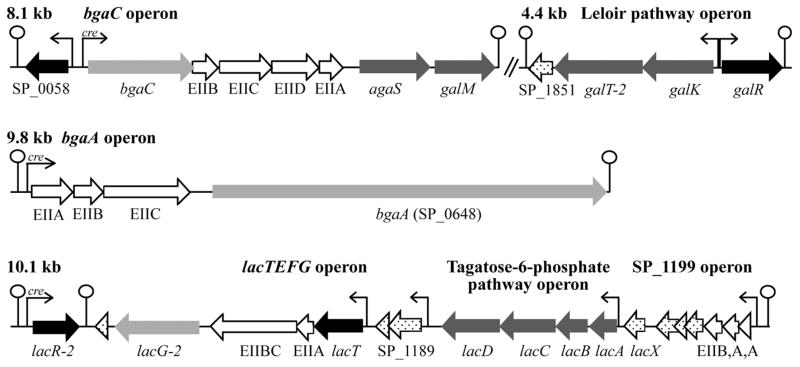
Fig. 3.
Predicted galactose metabolism operons in Streptococcus pneumoniae. Gene color codes are as follows: white: PTS transporter; white with black polka dots: hypothetical or degenerate gene; light gray: β-galactosidase; dark gray: metabolic enzymes of the Leloir and tagatose 6-phosphate pathways; and black: putative transcriptional regulators. Predicted CcpA-binding sites are indicated by ‘cre’ in the promoter regions. Operons are drawn nearly to scale. The regulator genes SP_0058, lacR-2 and galR were individually inactivated by replacement with a spectinomycin resistance cassette.
Transcriptional CCR is largely independent of CcpA activity
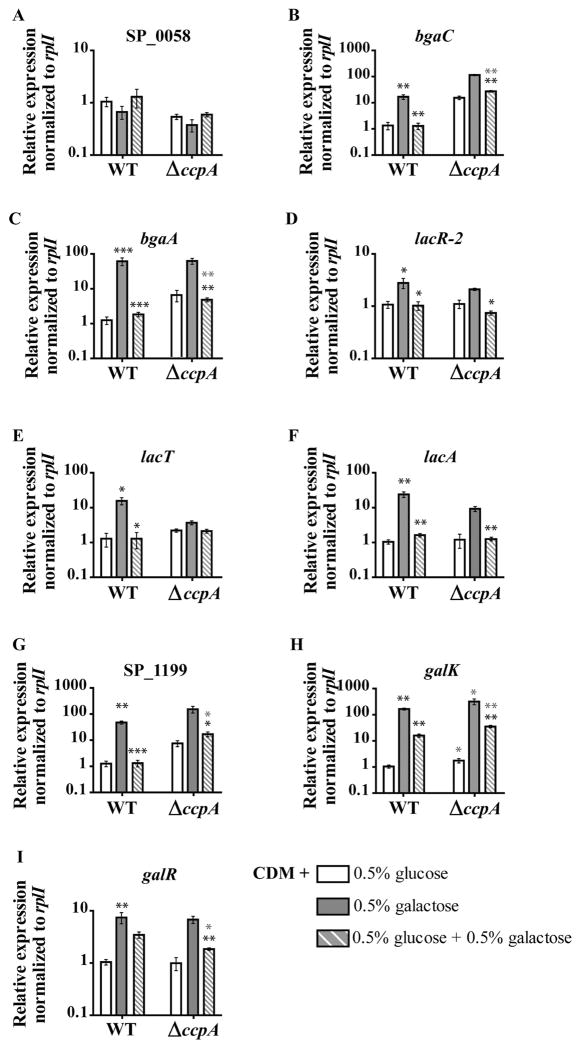
The results presented above are consistent with conservation of HPr-Ser~P’s role in inhibition of non-preferred carbohydrate transport. Next, we wanted to address a possible role of HPr-Ser~P in transcriptional CCR. Because HPr-Ser~P in related species affects transcription not only through co-repression with CcpA, but also through indirectly affecting the activity of dedicated transcriptional regulators by limiting transport of inducers, we first sought to establish a framework for dissecting these two types of transcriptional CCR in the pneumococcus. As such, using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), we determined the expression profiles of all galactose metabolism genes (shown in Fig. 3) during growth in a non-inducing condition (glucose), an inducing condition (galactose) and a CCR-repressing condition (glucose plus galactose). As shown in Fig. 4, in WT cells, all galactose-associated genes (with the exception of SP_0058) show galactose-dependent induction; the expression during growth on galactose is greater than during growth in glucose-only medium. These genes also show glucose-dependent CCR; expression in the CCR condition is lower than that of the galactose-only condition. These levels of repression are statistically significant for all genes except galR. Unlike the rest of the galactose metabolism genes, glucose-dependent CCR of galK and galR is incomplete; expression in the CCR condition is greater than that in the glucose-only condition (Fig. 4H and I respectively). Surprisingly, deletion of ccpA did not fully alleviate the glucose-dependent CCR of any one of these genes, meaning expression during growth on glucose plus galactose is still lower than expression during growth on galactose only (Fig. 4A–I). Deletion of ccpA abolished induction of lacT expression during growth on galactose, making it impossible to address CCR of this gene in this strain. Some genes, such as lacA and galK, showed no sign of CcpA-dependent CCR regulation. The expression of lacA in the glucose plus galactose condition is equivalent to that of WT in the same condition (Fig. 4F). In ΔccpA, galK expression in all conditions was about twofold greater than that of WT cells in the same condition, meaning the magnitude of glucose-dependent CCR was equivalent between these two strains. These results suggest that the pneumococcus relies heavily on a CcpA-independent mechanism to accomplish transcriptional CCR.
Fig. 4. Transcriptional CCR is largely independent of CcpA activity.
A–I. Transcription of representative genes from the galactose metabolism operons (shown in Fig. 3) during mid-exponential growth of WT and ΔccpA was assessed by qRT-PCR. Cells were grown to mid-exponential phase in CDM with one of the following: 0.5% glucose (white bars), galactose (gray bars) or 0.5% glucose and 0.5% galactose (gray and white striped bars), the latter condition representing a carbon catabolite-repressive condition. The average of at least five biological replicates normalized to rplI and relative to the WT 0.5% glucose sample is graphed for each, with error bars representing the standard error of the mean. Statistical significance is indicated as follows. For WT, black asterisks over gray bars indicate genes with a statistically significant difference in expression between the glucose-only and galactose-only conditions. For both strains, black asterisks over striped bars indicate expression in the CCR condition (glucose plus galactose) was different with statistical significance from the galactose-only condition within each strain. Gray asterisk represents expression in a given condition in the mutant strain was different with statistical significance from expression in the WT strain grown in the same condition. For all, one asterisk (*) indicates P value ≤ 0.05, two asterisks (**) indicate P value ≤ 0.01 and three asterisks (***) indicate P value ≤ 0.001.
Identification of inducer-responsive transcriptional regulators
Because CcpA is not solely responsible for transcriptional CCR of the galactose metabolism operons, we reasoned local transcriptional regulators may contribute to transcriptional CCR of these genes. There are four putative transcriptional regulators encoded proximally to these galactose metabolism operons; SP_0058, SP_1182 (lacR-2), SP_1187 (lacT) and SP_1854 (galR). (Fig. 3, shown in black). SP_0058, GalR and LacR-2 all share homology with characterized carbohydrate-binding transcriptional regulators in other species, while LacT is a conserved PTS regulatory domain containing RNA-binding transcriptional antiterminator, making them all likely candidates for responding to carbohydrate inducers or PTS transport activity (Oskouian and Stewart, 1990; van Rooijen et al., 1993; Alpert and Siebers, 1997; Ajdić and Ferretti, 1998; Lindner et al., 2002; Rigali et al., 2002; Aravind and Anantharaman, 2003; Graille et al., 2005; Tomoyasu et al., 2013; Afzal et al., 2014). In order to determine if the three predicted carbohydrate-binding transcriptional regulators contribute to CCR of the galactose metabolism operons, each was deleted individually in the WT and ΔccpA backgrounds. The resulting strains were assessed for relief of CCR of the galactose metabolism operons by qRT-PCR.
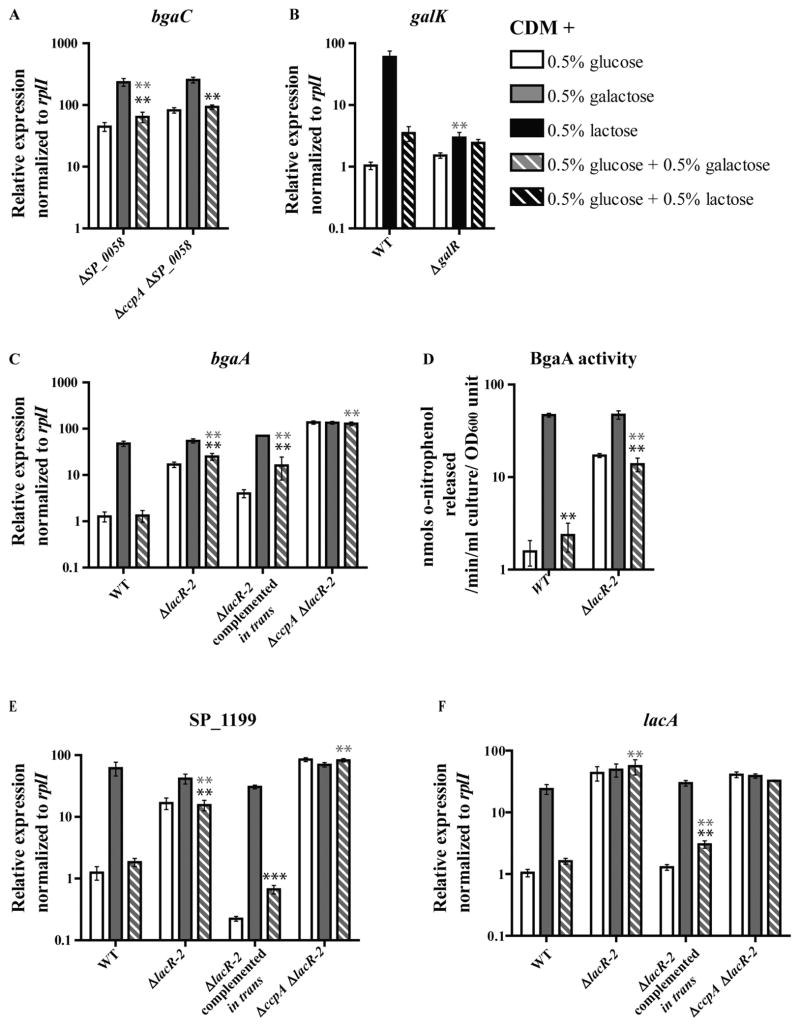
Deletion of SP_0058 resulted in de-repression of the bgaC operon (Fig. 5A, bgaC is shown as a representative of this operon). Similar to ΔccpA, CCR of this operon was not completely abolished in the ΔSP_0058 strain. In order to determine if together CcpA and SP_0058 constitute the sole CCR regulators of this operon, the ΔccpA ΔSP_0058 double mutant was tested. A small but consistent level of transcriptional CCR was still detectable in this strain, suggesting another transcriptional CCR regulator influences this operon. Interestingly, bgaC transcription in ΔccpA ΔSP_0058 was not statistically different from transcription in ΔSP_0058 in the same condition, suggesting deletion of SP_0058 disrupts CcpA-dependent repression of this operon.
Fig. 5.
Characterization of SP_0058, GalR and LacR-2 as transcriptional regulators of galactose operons. Transcription of representative genes from the galactose metabolism operons (shown in Fig. 3) during mid-exponential growth of regulator mutants was assessed by qRT-PCR. Cells were grown to mid-exponential phase in CDM with the following carbohydrates: 0.5% glucose (white bars), galactose (gray bars) or 0.5% glucose and 0.5% galactose (gray and white striped bars), the latter condition representing a carbon catabolite-repressive condition. For analysis of GalR regulation, WT and ΔgalR strains were grown in 0.5% lactose (black bars) in place of galactose, and 0.5% glucose plus 0.5% lactose (black and white striped bars) in place of glucose plus galactose. The average of at least five biological replicates normalized to rplI and relative to the WT 0.5% glucose sample (from Fig. 4) is graphed for each, with error bars representing the standard error of the mean.
A. Relative expression of bgaC is shown as a representation of regulation of the bgaC operon in ΔSP_0058 and ΔccpAΔSP_0058.
B. Relative expression of galK is shown as a representation of regulation of the galK operon in ΔgalR. Relative expressions of bgaA, SP_1199 and lacA are shown as representations of regulation of their respective operons in ΔlacR-2, ΔlacR-2 complemented in trans and ΔccpAΔlacR-2 in C, E and F respectively.
D. The effect of the lacR-2 deletion on BgaA activity was determined by monitoring the cleavage of o-nitrophenyl-galactopyranoside (ONPG) at absorbance 420 nm, in lysates of mid-exponential cultures grown in the standard carbohydrate conditions. Data reported represent the average of six biological replicates, with error bars representing the standard error of the mean. For all panels, statistical significance is indicated as follows. Black asterisk indicates expression in the CCR condition (glucose plus galactose) is different with statistical significance from the galactose-only condition within each strain. Gray asterisk represents expression in a given condition in the mutant strain was different with statistical significance from expression in the WT strain grown in the same condition. For both, one asterisk (*) indicates P value ≤ 0.05, two asterisks (**) indicate P value ≤ 0.01 and three asterisks (***) indicate P value ≤ 0.001.
Deletion of the second putative regulator, GalR, resulted in an inability to grow on galactose as a sole carbohydrate. This result suggests the regulatory activity of GalR is required for growth on galactose (Table 1). The ΔgalR strain is able to grow on lactose. In WT cells, lactose induces this operon to a comparable extent as galactose, making it a relevant substitute for galactose induction (compare Figs 4H and 5B). As such, qRT-PCR analysis of galKT-2 induction in ΔgalR was conducted using lactose as the inducing condition and glucose plus lactose as the CCR condition. Deletion of galR in the pneumococcus resulted in nearly uninduced levels of galKT-2 transcription in all conditions tested (Fig. 5B, galK is shown as a representative of this operon). These results strongly suggest GalR is an activator of the Leloir pathway genes in pneu-mococcus. In a variety of Gram-positive and negative species, GalR is a transcriptional auto-repressor and a repressor of the Leloir pathway genes (galKTE) (Choy and Adhya, 1992; Ajdić and Ferretti, 1998). Interestingly, the genomic architecture of the Leloir pathway operon and galR, and the (pseudo)palindromic GalR operator sequence in between these two promoters are both features conserved in the pneumococcus genome, and yet the function of GalR as a repressor is not conserved (Ajdić et al., 1996; Zeng et al., 2012).
Table 1.
Doubling times under various conditions.
| Strain | Doubling time (min ± SD)
|
|||
|---|---|---|---|---|
| 0.5% glucose | 0.5% galactose | 0.5% lactose | THY | |
| WT | 54 ± 2 | 77 ± 5 | 73 ± 2 | 39 ± 4 |
| ΔccpA | 98 ± 3 | 127 ± 13 | 88 ± 1 | 59 ± 7 |
| HPr S46D | 105 ± 6 | 121 ± 10 | 116 ± 7 | 43 ± 2 |
| ΔSP_0058 | 50 ± 1 | 76 ± 2 | 76 ± 3 | 38 ± 1 |
| ΔccpAΔSP_0058 | 108 ± 7 | 125 ± 14 | 200 ± 16 | 71 ± 17 |
| ΔgalR | 58 ± 1 | No growth | 80 ± 2 | 40 ± 4 |
| ΔlacR-2 | 59 ± 1 | 80 ± 2 | 77 ± 7 | 34 ± 2 |
| ΔccpAΔlacR-2 | 212 ± 18 | 247 ± 14 | 261 ± 40 | 71 ± 3 |
The data are presented as the average and standard deviation of six independent cultures in either chemically defined medium with the indicated carbohydrates, or THY, as detailed in Experimental procedures.
LacR-2 was recently shown to regulate the tagatose 6-phosphate pathway operon in D39 (Afzal et al., 2014). Deletion of lacR-2 relieved transcriptional CCR of many galactose metabolism operons (Fig. 5C–F). In the ΔlacR-2 strain, the unlinked bgaA operon was transcriptionally de-repressed (Fig. 5C, bgaA is shown as a representative of this operon). These results were corroborated by a β-galactosidase assays measuring BgaA activity (Fig. 5D). Compared with WT, ΔlacR-2 had uniformly higher BgaA activity in both conditions containing glucose. The SP_1199 operon was also de-repressed in the ΔlacR-2 strain (Fig. 5E). Similar to deletion of ccpA, deletion of lacR-2 did not entirely disrupt CCR of the bgaA or SP_1199 operons. Deletion of both ccpA and lacR-2, however, abolished all glucose-dependent CCR of these operons, demonstrating that these two regulators constitute the only transcriptional CCR regulators of these operons (Fig. 5C and E respectively). In contrast, transcription of lacA operon was completely de-repressed under all conditions in the ΔlacR-2 strain (Fig. 5F). This result suggests LacR-2 is the sole transcriptional CCR regulator of this operon. Corroborating these results, deletion of ccpA had no additional effect on lacA transcription as a single deletion mutant or in combination with lacR-2 (Figs 3F and 5F). The regulatory consequences of deleting lacR-2 were completely complemented in nearly all conditions by expressing lacR-2 in trans (Fig. 5C, E and F). The only deviation from WT-like expression in the complemented strain was a less than twofold increase in bgaA expression compared with WT in the glucose plus galactose condition.
Identification and validation of LacR-2 consensus operator sequence
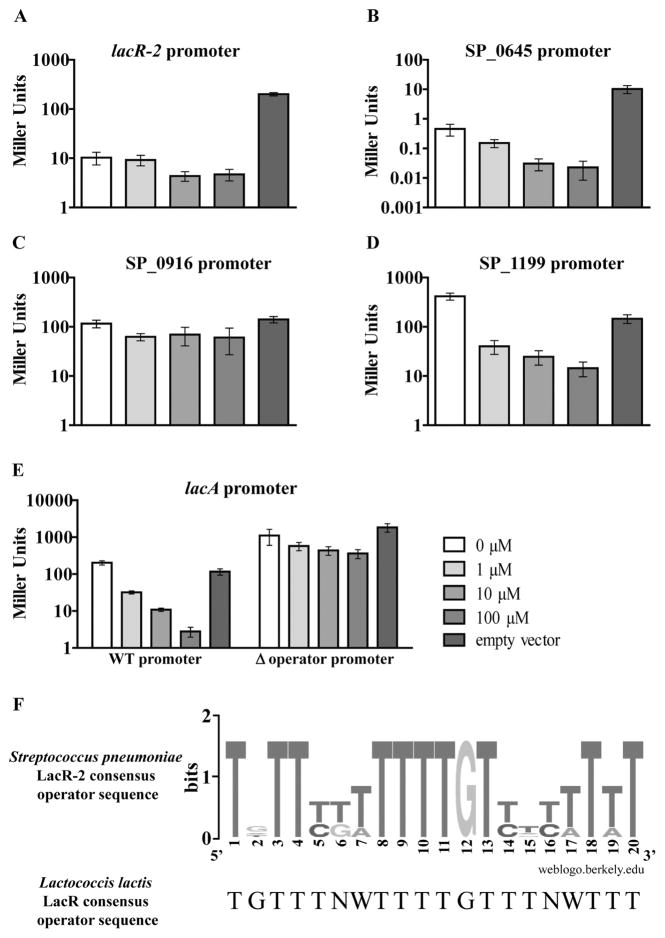
Because LacR-2 was found to affect the regulation of multiple galactose metabolism operons and because it has been identified as a regulator of a key virulence factor in Streptococcus intermedius (Tomoyasu et al., 2013), we wanted to distinguish between the possibility of it being a dedicated transcriptional regulator or a global regulator. In order to do so, we sought to determine LacR-2’s consensus operator sequence to enable determination of direct regulation of these galactose operons and prediction of additional genes or operons under its direct regulation. Pneumococcal LacR-2 has high sequence homology to LacR transcriptional regulators in other species. Using the 20-base pair consensus operator sequence from one such regulator, LacR from Lactococcus lactis (van Rooijen et al., 1993), a computational search was performed to uncover potential LacR-2 operator sites in the TIGR4 genome (Fig. 6). The search yielded eight sites throughout the genome with at least 90% homology to the L. lactis sequence suggesting LacR-2 is a semi-global regulator (Table 2).A number of these sites were just upstream of the putative −35 box of the promoter of galactose utilization operons including the bgaA operon, the lacA operon and lacR-2’s own promoter. In order to test for direct regulation of these predicted operator sites by LacR-2, we developed a LacZ-based operator validation assay in the heterologous host Escherichia coli. This system consisted of two parts: cloning putative LacR-2-regulated promoter regions in place of lacZ’s endogenous transcriptional regulation elements in E. coli MG1655 and IPTG-inducible expression of lacR-2 from a plasmid in these strains. These strains and empty vector controls were grown with varying concentrations of IPTG, and LacZ activity was measured. We tested four promoter regions from those listed in Table 2 (Fig. 6A–D). A strain containing the SP_1199 promoter region controlling lacZ was also tested as it was de-repressed in ΔlacR-2 (Fig. 6E).
Fig. 6. Validation of predicted LacR-2 operator sequences in Escherichia coli MG1655.
A–E. A LacZ-based reporter system in the heterologous host E. coli was used to validate the predicted LacR-2 operator sequences listed in Table 2. This system consisted of two parts: cloning LacR-2 regulated promoter regions in place of lacZ’s endogenous transcriptional regulation elements in E. coli MG1655 and IPTG-inducible expression of lacR-2 from a plasmid in these strains. The Miller assay was used to measure LacZ activity of the resulting strains under increasing concentrations of IPTG (shown by increasing intensity of gray). To determine auto-regulation, the lacR-2 promoter with both predicted operator sequences was tested in A. To determine direct regulation of SP_0645, SP_0916 and SP_1199, their respective promoter fusion strains were tested in part B, C and D respectively. To test for direct regulation of the lacA operon, its promoter fusion strain and the isogenic operator deletion version were tested in E. For each section, bars represent the average of six biological replicates with error bars representing the standard error of the mean. F. These confirmed operator sequences were used to generate a consensus sequence logo. Searching the TIGR4 genome for this revised sequence predicted no additional sites beyond the sites that had already been validated by this assay.
Table 2.
Predicted LacR-2 operator sequences.
| Sequence | Accuracy (%) | Distance from −35 (nts) | Gene immediately downstream |
|---|---|---|---|
| TTTTTGTTTTTGTTTTATTT | 95 | −14 | lacR2 |
| TGTTTTATTTTGTTGTTTTT | 95 | −4 | lacR2 |
| TGTTCTTTTTTGTCTCTTTT | 90 | −8 | SP_0645 (bgaA operon) |
| TGTTTGTTTTTGTTATTTAT | 90 | −7 | lacA |
| TGTTTCATTTGTTTTATTTT | 90 | n/a | SP_0375 (6-phosphogluconate dehydrogenase) |
| TGTTTATTTTGGTTTCTGTT | 90 | −39 | SP_0916 (lysine decarboxylase) |
| TTTTTGATTTTTTTTGATTT | 90 | +32 | SP_0987 (hypothetical) |
| TTTTTTATTTTTTTTTATTT | 90 | +15 | SP_1058 (hypothetical) |
This chart shows the predicted LacR-2 operator sequences in the TIGR4 genome and their similarity (% accuracy) to the Lactococcus lactis consensus sequence used for the homology search. The relative distance of each operator from the nearest predicted promoter and the corresponding gene name are indicated. n/a signifies no promoter could be predicted. Bold indicates genes involved in galactose metabolism.
Of the predicted operators listed in Table 2 that were tested by this assay, all showed decreased LacZ activity with increased IPTG concentrations except for the SP_0916 promoter strain. This strain had comparable LacZ activity in all concentrations of IPTG tested, suggesting LacR-2 does not regulate this promoter under the conditions tested (Fig. 6C). The other predicted operator strains tested, including the SP_1199 promoter strain, all showed dose-dependent decreases in LacZ activity with increased IPTG (Fig. 6A, B, D and E). This result suggests LacR-2 is auto-regulatory and represses the bgaA operon, the lacA operon and the SP_1199 operon directly.
To confirm that the repressive effect of LacR-2 in these reporter strains is due specifically to the predicted operator sequence and not due to non-specific interactions resulting from overexpression of lacR-2, a strain was made in which the lacA operon promoter lacking the predicted operator sequence was cloned upstream of lacZ (Fig. 6E, Δoperator strain). The dose-dependent repressive effect on LacZ activity was lost in the Δoperator strain, suggesting LacR-2 repression of this strain is dependent on the presence of the predicted operator sequence. These confirmed operator sequences were used to generate a consensus sequence logo, which differed slightly from the L. lactis consensus (Fig. 6F). Searching the TIGR4 genome for this revised sequence predicted no additional sites beyond the sites confirmed by our assay, suggesting the remainder of promoter regions listed in Table 2 is not in fact LacR-2-regulated.
As a final test of the role of the validated operator sequences in regulation of galactose metabolism operons, we made a pneumococcal strain bearing a mutated version of the operator sequence preceding the lacA promoter (PlacA*). The PlacA* strain had a three-base pair substitution (TGT→ACA) in this putative LacR-2 operator, designed to disrupt the most highly conserved central region of the pneumococcal LacR-2 operator consensus sequence (Fig. 6F). In all conditions, lacA transcription in this strain was de-repressed to a level equivalent to that of the WT strain during growth on galactose (Fig. 7). That is to say, mutation of the putative LacR-2 operator sequence had a similar effect on lacA transcription as galactose-dependent induction in WT cells. In each condition except the glucose plus galactose condition, lacA transcription in this strain was also equivalent to the levels in ΔlacR-2 under the corresponding condition. In the glucose plus galactose condition, lacA transcription in ΔlacR-2 was consistently about threefold higher than in the PlacA* strain, suggesting LacR-2 maintains some ability to repress expression despite the operator mutation. Taken with the dose-dependent repression of this promoter observed in the E. coli system, this analysis supports the conclusion that LacR-2 is a semi-dedicated regulator that directly represses three galactose metabolism operons and is auto-regulated.
Fig. 7.
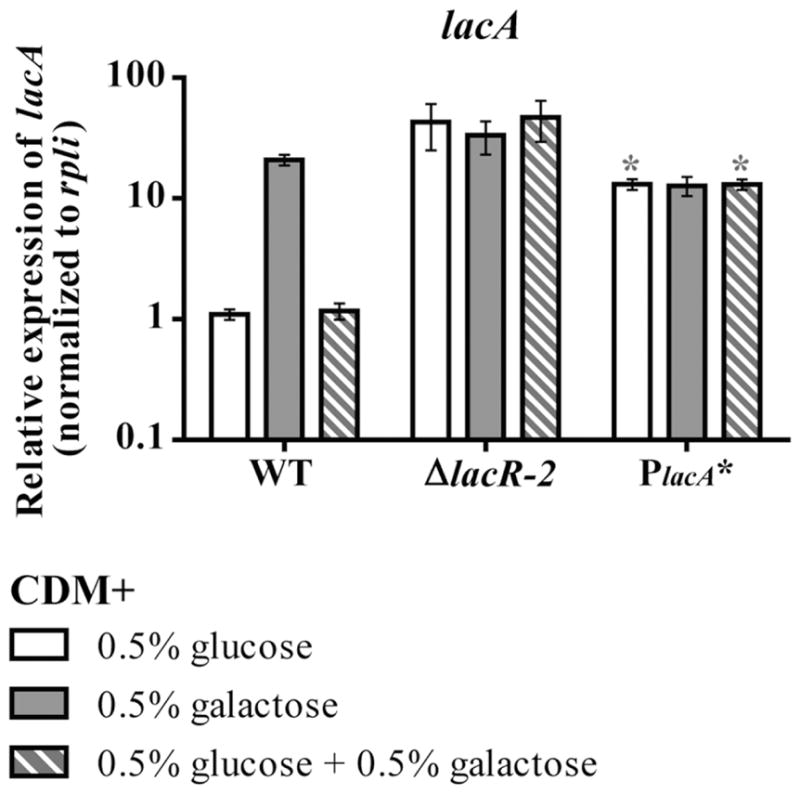
Analysis of lacA transcription in a LacR-2 operator mutant in pneumococcus. lacA transcription during mid-exponential growth of was assessed by qRT-PCR in WT, ΔlacR-2 and the PlacA* strain, which has a three-base pair substitution (TGT→ACA) in this putative LacR-2 operator. Cells were grown to mid-exponential phase in CDM with the following carbohydrates: 0.5% glucose (white bars), galactose (gray bars) or 0.5% glucose and 0.5% galactose (gray and white striped bars), the latter condition representing a carbon catabolite-repressive condition. Asterisk indicates a statistically significant difference in expression compared to the corresponding WT sample (P value ≤ 0.05)
HPr S46D recapitulates the phenotypes of a carbon catabolite-repressed state
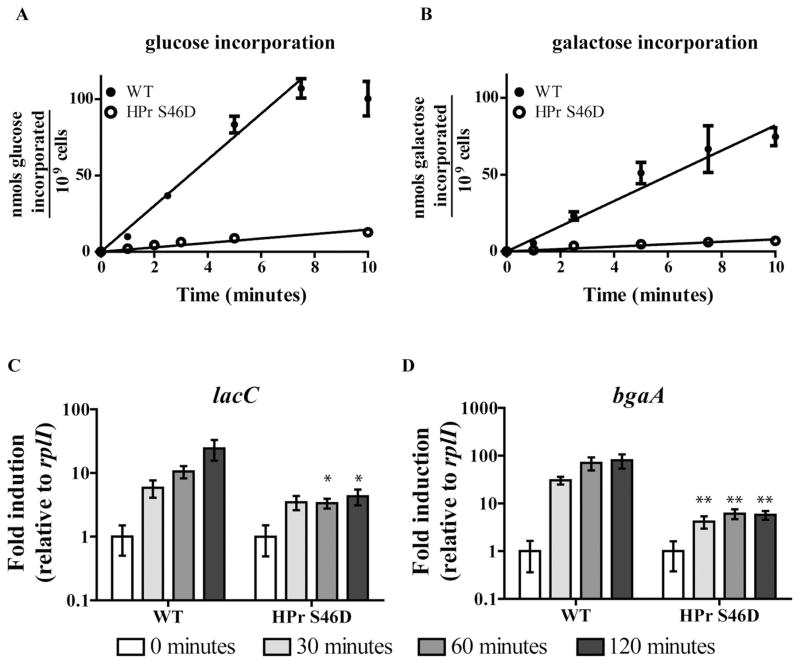
Thus far, we have established that utilization of a non-preferred carbohydrate is inhibited at the level of transport, and transcriptional CCR of the associated metabolism genes is accomplished largely by local transcriptional regulators with predicted carbohydrate-binding motifs. To test the possibility that HPr-Ser~P mediates some or all of these phenotypes, we sought to introduce the two mutations of HPr’s regulatory seryl residue that are classically used to investigate HPr-mediated CCR in Gram-positive species: HPr S46A and HPr S46D (Reizer et al., 1989; Deutscher et al., 1994; 2006). In related species, HPr S46A cannot be phosphorylated by HPrK/P, and hence cannot exclude inducers or co-repress with CcpA. The aspartic acid residue of HPr S46D serves as a phospho-mimetic mutation, giving the opposite phenotype; perpetual exclusion of inducers and co-repression with CcpA. To begin assessment of HPr-Ser~P-mediated CCR in the pneumococcus, the HPr S46D mutant was characterized. As HPr-Ser~P is a poor participant in PTS transport, PTS transport rates of the HPr S46D mutant are expected to be reduced. Compared with the WT incorporation rates of 15.08 ± 0.45 and 8.21 ± 0.46 nmol carbohydrate/min/109 cells for glucose and galactose, respectively, the HPr S46D mutant’s rates were decreased by 10-fold for both carbohydrates (1.45 ± 0.01 and 0.80 ± 0.01), as determined by radiolabeled carbohydrate incorporation assays (Fig. 8A and B). HPr S46D mutants in related species typically exhibit growth defects on PTS transported carbohydrates, presumably in part because of the diminished rates of transport of these substrates (Zeng and Burne, 2010). HPr S46D in the pneumococcus had growth defects when grown in media containing either glucose or galactose as sole sources of carbohydrate (Table 1).
Fig. 8. Characterization of the HPr S46D mutant in pneumococcus.
A. The rate of glucose incorporation was determined for WT (closed circles) and HPr S46D (open circles) using the carbohydrate radiolabeled incorporation assay. Cells preconditioned to unlabeled glucose were washed before exposure to a mixture of unlabeled glucose plus H3-labeled glucose at 37°C, and accumulation of the latter was determined in filtered samples over time according to standard liquid scintillation method. Values are reported as total nanomoles of carbohydrate accumulated / 109 cells by accounting for the concentration of unlabeled carbohydrate and the colony-forming units in each reaction. Each point is the average of at least six biological replicates, and error bars are the standard error of the mean. Rates were determined by the slope of the best fit line.
B. The rate of galactose incorporation was determined for WT (closed circles) and HPr S46D (open circles) exactly as described in A, replacing unlabeled glucose with galactose and H3-labeled glucose with C14-labeled galactose.
C and D. Galactose-dependent induction of lacC (C) and bgaA (D) in WT and HPr S46D was determined as follows. Mid-exponential phase THY cultures were washed twice and resuspended in CDM lacking carbohydrates. One sample was collected at this point (white bars), and additional samples were collected 30, 60 and 120 min after the addition of galactose such that the final concentration was 0.5% (progressively darker gray bars). Fold induction is reported as the average ratio of each time point over the 0 min control, relative to rplI expression within each strain. Five biological replicates of each strain were analyzed from multiple days. Asterisks indicate a statistically significant difference in expression compared to the corresponding WT sample. One asterisk (*) indicates P value ≤ 0.05, and two asterisks (**) indicate P value ≤ 0.01.
As discussed for related Gram-positive species, transport of non-preferred carbohydrates supplies inducers, which induce expression of non-preferred carbohydrate metabolic genes by modulating the activity of dedicated carbohydrate-binding transcriptional regulators. HPr-Ser~P-mediated CCR prevents this transcriptional induction by restricting non-preferred carbohydrate transport and by co-repressing with CcpA. In order to determine if the HPr S46D mutant in pneumococcus recapitulates these two aspects of HPr-Ser~P-mediated transcriptional CCR, a qRT-PCR-based galactose induction experiment was conducted. WT and HPr S46D cells pre-grown to mid-exponential phase in rich medium were washed and then exposed to galactose CDM. Samples were collected periodically and total RNA was isolated for qRT-PCR analysis. Two genes shown to be induced by galactose and CCR repressed by glucose were selected for analysis (Fig. 3). The first gene, lacC, is CCR repressed independently of CcpA(Fig. 4F, lacA is shown as an example of this operon). For the second gene, bgaA, direct repression by CcpA constitutes a portion of the transcriptional CCR occurring at this operon (Fig. 4C). The CcpA-binding site upstream of this operon has been validated in another serotype of pneumococcus and is conserved in TIGR4 (Kaufman and Yother, 2007). Both genes are subject to direct repression by LacR-2 (Figs 6 and 7). By comparing CCR of these two genes, we are able to dissect both CcpA-dependent and CcpA-independent transcriptional effects of the HPr S46D mutation. As expected, upon exposure to galactose, WT exhibited a temporal induction in both lacC and bgaA transcript levels (Fig. 8C and D respectively). The HPr S46D mutant showed decreased induction for both genes. In this strain, lacC transcription by 120 min of galactose exposure was sixfold lower than that of WT. Because of this operon is not subject to CcpA-dependent regulation, this result demonstrates HPr S46D has a repressive effect on transcription independently of CcpA. This effect is likely due to failure to fully deactivate LacR-2. By 120 min, bgaA was induced 80-fold in WT, but only about 5-fold in HPr S46D, representing a 16-fold reduced induction. The difference in magnitude of inhibition caused by the HPr S46D mutation may be due to CcpA repressing bgaA but not lacC. This result is consistent with HPr S46D having a two-pronged effect on bgaA transcription; it maintains CcpA-dependent repression by acting as a co-repressor, while also preventing deactivation of LacR-2 by inhibiting galactose transport. Collectively, these experiments support the conclusion that HPr-Ser~P’s activity restricting non-preferred carbohydrate transport and co-repressing with CcpA both contribute to transcriptional CCR of galac-tose metabolism in the pneumococcus.
HPr-Ser~P-mediated CCR is an essential activity in the pneumococcus
Although the HPr S46D mutant was easily isolatable by standard co-transformation procedures (see the Experimental procedures section for details), we could not recover an HPr S46A mutant, suggesting this mutation is lethal in the pneumococcus. To address this possibility, a copy of WT ptsH-encoding HPr and its predicted promoter region were introduced during the co-transformation process as part of the selectable co-transformation plasmid (pG+host9-HPr, Table 3). Inclusion of a WT copy of ptsH enabled recovery of HPr S46A chromosomal mutant with a similar frequency as the HPr S46D mutant (data not shown). The fact that WT ptsH supplemented in trans was sufficient to restore the co-transformation frequency is consistent with the HPr S46A mutation being lethal.
Table 3.
Strains and plasmids used in this study.
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Streptococcus pneumoniae | ||
| Wild type | TIGR4 | Aaberge et al., 1995 |
| HPr S46D | ptsH 136TCA → 136GAC | This study |
| HPr S46A + pG+host9-HPr | ptsH 136TCA → 136GCT | This study |
| HPr H15A | ptsH 43CAC → 43GCG | This study |
| HPr H15A/S46A + pG+host9-HPr | ptsH 43CAC → 43GCG and 136TCA → 136GCT | This study |
| HPr S46A ΔpstI | ptsH 136TCA → 136GCT pstI::spec | This study |
| ΔccpA | ccpA::cat | Laboratory strain |
| ΔSP_0058 | SP_0058::spec | This study |
| ΔlacR-2 | lacR-2::spec | This study |
| ΔlacR-2 complemented in trans | lacR-2::spec, with lacR-2 promoter and coding region cloned into the SP_1773 locus | This study |
| ΔgalR | galR::spec | This study |
| ΔccpAΔSP_0058 | ccpA::cat, SP_0058::spec | This study |
| ΔccpAΔlacR-2 | ccpA::cat, lacR-2::spec | This study |
| PlacA operator mutant | Mutation of predicted LacR-2 operator preceeding the tagatose 6-phosphate operon promoter, TGTTACATTTACATATTTAT→TGTTTGTTTTACATATTTAT | This study |
| Escherichia coli | ||
| TG1 | K-12 supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5, (rK−mK−) | Laboratory strain |
| PSP_0645 – lacZ | MG1655 Δ (lacI-promoter of lacZ), replaced with SP_0645 promoter region, kanR | This study |
| PSP_0916 – lacZ | MG1655 Δ (lacI-promoter of lacZ), replaced with SP_0916 promoter region, kanR | This study |
| PlacA – lacZ | MG1655 Δ (lacI-promoter of lacZ), replaced with pneumococcus lacA promoter region, kanR | This study |
| PlacR-2 – lacZ | MG1655 Δ (lacI-promoter of lacZ), replaced with pneumococcus lacR-2 promoter region, kanR | This study |
| PlacAΔoperator – lacZ | MG1655 Δ (lacI-promoter of lacZ), replaced with pneumococcus lacA promoter region with 20-base pair deletion of the predicted LacR-2 operator, kanR | This study |
| Plasmids | ||
| pG+host9 | 4.8 kb pAMβ1 derivative conferring erythromycin resistance. Low copy, termosensitive ori | van Opijnen and Camilli, 2012 |
| pG+host9-HPr | pG+host9 with hpr and promoter region cloned in using SmaI and XhoI sites | This study |
| pDL993 | p15A ori, WT pLac promoter, lacI with native promoter, tetR | Courtesy of David Lasinski |
| pDL993_lacR-2 | pDL993 with lacR-2 cloned downstream of the pLac promoter using SmaI and EcoRI sites | This study |
As an additional test of HPr S46A lethality, we conducted a plasmid loss experiment in an attempt to rid HPr S46A of the temperature-sensitive pG+host9-HPr plasmid. If the HPr S46A mutation is lethal, this strain will die upon loss of pG+host9-HPr at a non-permissive temperature and/or display distorted plasmid loss kinetics compared with the WT (pG+host9-HPr) control. These strains were grown in triplicate at permissible temperature (30°C) with erythromycin for selection of plasmid-containing cells. Upon reaching mid-exponential phase, the cultures were washed and back-diluted into fresh medium without selection and to a non-permissive temperature (37°C). The cultures were grown for up to 18 successive generations by back-diluting them into fresh medium every six generations. Samples were plated periodically to determine the proportion of plasmid-bearing cells remaining. As expected for a lethal mutation, the HPr S46A strain showed reduced growth and ultimately death at the non-permissive temperature (Fig. 9A). Of the HPr S46A cells recovered prior to each back dilution, all had maintained the plasmid, whereas the WT cultures exhibited a logarithmic rate of plasmid loss throughout the 18 generations of growth at 37°C, demonstrating the kinetics of pG+host-HPr loss was also aberrant in the mutant (Fig. 9B). These data are strongly indicative of the HPr S46A mutation being lethal in the pneumococcus.
Fig. 9. HPr S46A is a lethal mutation. WT and HPr S46A both carrying the complementing pG+host9-HPr plasmid were grown for successive generations in 0.5% galactose CDM at a non-permissive temperature (37°C), by back-diluting every six generations.
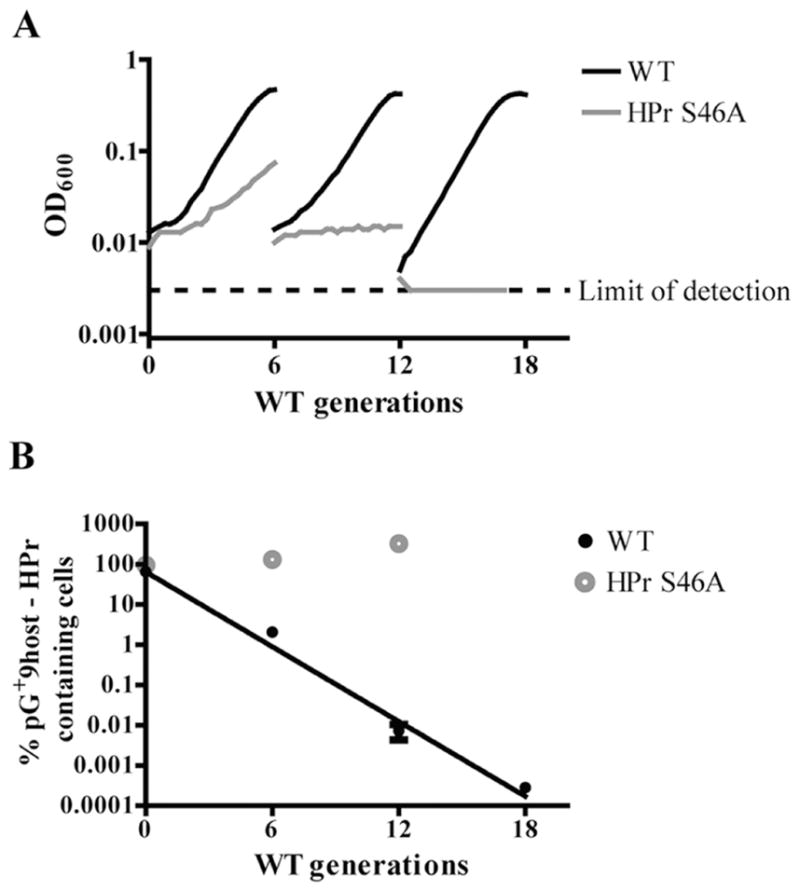
A. Successive growth curves of a representative of each strain are shown (WT: black line; HPr S46A: gray line). Samples were plated every six generations with and without selection at 30°C to determine the proportion of plasmid-bearing cells remaining (WT: closed black circles; HPr S46A: gray open circles).
B. The percent of plasmid-containing cells is reported as the average of three biological replicates with error bars representing the standard error of the mean. A linear regression was fit to the WT data, demonstrating a linear relationship between plasmid loss and log10 number of generations.
HPr S46A lethality is rescued by deletion of ptsI but not by an HPrH15A mutation
By preventing the formation of HPr-Ser~P, the HPr S46A mutation increases the pool of HPr that can be phosphorylated by EI in related species (Vadeboncoeur et al., 1991; Deutscher et al., 1994; 2006). It is possible that when more HPr substrate is available, the pneumococcus is uniquely sensitive to increased expenditure of PEP by EI, or it expends more PEP than other species through faster EI auto-phosphorylation and/or EI-HPr phosphorylation kinetics. In order to determine if deletion of ptsI in cis can rescue the lethality of the HPr S46A mutation, we transformed WT pneumococcus with a single splicing by overlap extension (SOE) PCR construct bearing both the HPr S46A codon change and a spectinomycin cassette replacing the ptsI open reading frame (see Experimental procedures for details). Under standard transformation conditions, we were able to recover transformants with a similar frequency as transformation with the control construct lacking the HPr S46A codon change (data not shown). Colony PCR of the resulting transformants confirmed the majority contained the HPr S46A codon change, showing deletion of ptsI in cis rescues the lethality of the HPr S46A mutation.
According to current understanding of the PTS system, deletion of ptsI abolishes EI-dependent PEP cleavage, and by extension EI-dependent HPr phosphorylation. In order to determine if eliminating EI-dependent HPr phosphorylation is sufficient to rescue the HPr S46A mutation, we sought to create an HPr H15A/S46A double mutant. A single SOE PCR product bearing both codon changes was transformed into WT pneumococcus by standard co-transformation procedures (see Experimental procedures for details). A control transformation using pG+host9-HPr as the selectable marker was done in parallel. We were able to recover the HPr H15A/S46A double point mutant only when WT ptsH was provided in trans (data not show). This result recapitulates our results from trying to recover the HPr S46A mutation. Because both deletion of ptsI and mutation of HPr H15A are anticipated to abolish EI-dependent phosphorylation of HPr, but only the former is capable of rescuing an HPr S46A mutation, these results suggest that specifically EI-dependent cleavage of PEP or another EI-dependent HPr-independent activity contributes to the lethality of HPr S46A.
Discussion
By studying both transport of a non-preferred carbohydrate and transcription of the corresponding metabolic genes, we have demonstrated the fundamental features of HPr-Ser~P-mediated CCR are active and likely essential in the pneumococcus. First, we showed that galactose transport can be inhibited in the presence of glucose. Direct inhibition of pre-existing non-preferred carbohydrate transporters is a CCR activity distinct from all others, which act at a transcriptional level. In Gram-positive bacteria, this activity is most commonly associated with a rise of HPr-Ser~P levels in the cell. Our results resemble the immediate inhibition of non-preferred carbohydrate transport characteristic of inducer exclusion in other organisms, but our assays did not distinguish between inhibition of PTS transporters versus inhibition of non-PTS transporters (Viana et al., 2000). That is to say, the reduction of galactose transport in the presence of glucose could be due to increased competition between PTS systems for limited quantities of HPr-His~P, the direct inhibition of non-PTS transporters by HPr-Ser~P or a combination of multiple mechanisms (Deutscher et al., 2006; Zeng and Burne, 2010). Although there is no known ATP-binding cassette (ABC) transporter or symporter for galactose in the pneumococcus, we showed that the Leloir pathway operon has the unique feature of being subject to incomplete CCR; the inclusion of glucose in inducing conditions does not completely prevent induction of this pathway. This result supports the idea that pneumococcus is able to differentially affect the activity of galactose PTS versus non-PTS transporters.
We identified three carbohydrate-binding transcriptional regulators whose regulation of galactose metabolism genes is fitting with a model in which restriction of galactose transport affects their regulatory activity. We show LacR-2 is a semi-dedicated repressor of galactose metabolism operons while the classic repressor of the Leloir pathway, GalR, in fact is an activator in the pneumococcus. GalR is also an activator of the Leloir pathway in Streptococcus thermophilus, a species that has an uncommon preference for lactose over glucose (Vaughan et al., 2001). It will be interesting to determine why and how the pneumococcus has evolved to allow expression of the Leloir pathway genes during glucose-dependent CCR conditions, and if GalR’s activity as an activator is involved in this phenotype.
Galactose transport restriction in the HPr S46D mutant further supports the idea that the concurrent rise of HPr-Ser~P and fall of HPr-His~P can influence the activity of these regulators by affecting galactose transport. We strengthened this connection by analyzing galactose-dependent transcriptional induction of a CcpA-repressed gene (bgaA) and a gene that is not subject to CcpA-dependent regulation (lacC) in WT and HPr S46D. lacC was not fully induced in HPr S46D, demonstrating HPr S46D has a transcriptional effect independent of CcpA. From our transcriptional regulator characterization studies, we know LacR-2 is the sole transcriptional CCR regulator of lacC. The lack of full induction of lacC in the HPr S46D strain, therefore, is likely due to failure to completely relieve LacR-2-dependent repression due to decreased transport of its galactose-derived inducer. This conclusion is consistent with reports of LacR responding to intracellular concentrations of PTS-derived galactose derivatives in other species (Oskouian and Stewart, 1990; van Rooijen et al., 1992; 1993; Zeng et al., 2012). Induction of bgaA was even more drastically restricted in HPr S46D compared with WT. We demonstrate that LacR-2 and CcpA constitute the only two transcriptional CCR regulators of bgaA. The considerably greater magnitude of inhibition of bgaA transcriptional induction in HPr S46D is consistent with HPr S46D maintaining CcpA-dependent repression in addition to limiting the amount of inducer available to relieve LacR-2 dependent repression of this operon.
An ideal follow-up experiment to these results would be to show that glucose-dependent CCR is compromised in a strain unable to seryl-phosphorylate HPr. However, we found that the classic mutation used to abolish HPrK/P-dependent seryl-phosphorylation of HPr, HPr S46A, is not tolerated in the pneumococcus. To our knowledge, this is the first example of this mutation being lethal in any species. The lethality may be due to the intended effect of preventing HPrK/P-dependent phosphorylation of HPr or to an unintended effect on another aspect of HPr’s function. We could not isolate an hprK/P null mutant, and other studies have identified this gene as being essential as well, further supporting the conclusion that the HPr S46A mutation is lethal from loss of HPr-Ser~P-mediated CCR (van Opijnen et al., 2009; van Opijnen and Camilli, 2012). Two separate genetic manipulations were able to rescue the HPr S46A mutant lethality: expression of WT ptsH in trans and deletion of ptsI. Under the same conditions used to recover the HPrS46A-ΔptsI mutant, we were unable to recover any HPr H15A/S46A double mutants except when WT ptsH was supplied in trans. This result suggests EI but not EI-dependent phosphorylation of HPr contributes to the lethality of the HPr S46A mutant. Ultimately, we believe our results are most suggestive of loss of HPr-Ser~P having multifarious effects in the cell that cumulatively cause the lethality of this mutation. These effects include but are not limited to loss of CcpA-dependent regulation and possibly misregulation of EI activity.
No matter the exact cause of the lethality, the results presented in this study strongly suggest HPr-Ser~P and its associated activities are of fundamental importance to pneumococcal physiology. We are unaware of any other species that shows such complete dependence on HPr-Ser~P-mediated CCR. No CCR activity in the pneumococcus (meaning repression measured transcriptionally or enzymatically) to date has been shown to be solely due to CcpA regulation. That is to say, each gene and metabolic enzyme analyzed is still subject to glucose-dependent CCR in the ccpA null strain (Giammarinaro and Paton, 2002; Iyer et al., 2005; Kaufman and Yother, 2007). This result is in contrast to other organisms that rely more heavily on CcpA and other semi-global CCR DNA-binding regulators (Servant et al., 2005; Deutscher et al., 2006; Abranches et al., 2008; Deutscher, 2008; Fujita, 2009). Extensive exploration of CcpA-independent mechanisms of CCR in Streptococcus mutans has shown this species employs a number of mechanisms centered on HPr-Ser~P activity, yet the HPr S46A mutation is tolerated in this species (Abranches et al., 2003; 2004; 2008; Ajdić and Pham, 2007; Zeng and Burne, 2008; 2010; Zeng et al., 2010).
Together, these observations suggest that the pneumococcus’ requirement of HPr-Ser~P is unique. It is worth pursuing further analysis of HPr-Ser~P activity in order to clarify details of the activities presented in this study, uncover any additional mechanisms of CcpA-independent CCR and to elucidate why the HPr S46A mutation is lethal in this species. Trying to introduce more moderate mutations at position 46 of HPr, such as threonine, will be of value to determining the importance of HPr-Ser~P to the pneumococcus (Reizer et al., 1989; Deutscher et al., 1994; 2006). Relief of CCR can also be studied by making mutations that specifically diminish glucose transport. These types of mutants are unable to use glucose efficiently enough to evoke CCR. The most common mutation used in related Streptococci is deletion of the EIIAB component of the major glucose PTS system (manL of manLMN) (Vadeboncoeur, 1984; Lortie et al., 2000; Zeng and Burne, 2010; Bidossi et al., 2012). Of note, ManL also frequently has its own CCR activities, including powering transport of non-preferred carbohydrate transporters and inactivating non-PTS transporters via a phosphorylation event (Abranches et al., 2003; Zeng and Burne, 2008; 2010). Surprisingly, the activity of this transporter as the major glucose transporter may not be conserved in the pneumococcus and therefore its role in enacting glucose-dependent CCR is also in question (Bidossi et al., 2012). Although this example does not exclude the possibility of other glucose transporters having a role in CCR, it does further illustrate how the CCR strategy in the pneumococcus seems to be uniquely streamlined toward HPr-Ser~P-dependent mechanisms.
Experimental procedures
Bacterial strains and culture conditions
All strains and plasmids used are listed in Table 3. Unless otherwise noted, S. pneumoniae TIGR4 (serotype 4) and isogenic mutants were grown at 37°C and 5% carbon dioxide in Todd Hewitt broth supplemented with 0.5% yeast extract (THY; BD Biosciences), chemically defined medium (CDM) with varying carbohydrates as described in the text (Kloosterman et al., 2006) or on 5% sheep’s blood agar plates (Northeast Laboratory Services). All liquid culture conditions were supplemented with 300 U ml−1 of catalase (Worthington Biochemicals). Antibiotics were used at the following concentrations for selection of pneumococcal mutants: chloramphenicol (4 μg ml−1), spectinomycin (200 μg ml−1) and erythromycin (0.1 μg ml−1). E. coli strains used for cloning and for the LacR-2 operator sequence validation experiments were cultivated at 37°C in Luria–Bertani (LB) with aeration or on LB plates, unless otherwise noted. Antibiotics were used at the following concentrations for selection and propagation of E. coli mutants: kanamycin (50 μg ml−1), tetracycline (2 μg ml−1) and erythromycin (150 μg ml−1).
Generation of pneumococcal mutants
Marked deletions were created by natural transformation using standard methods (Bricker and Camilli, 1999). Point mutations were generated by co-transformation (Maguin et al., 1996; Dalia et al., 2014). SOE PCR was used to generate DNA constructs for these purposes, as described previously (Horton et al., 1990). All primers used for SOE PCR are listed in Supporting Information Table S1. For co-transformation, competent cells were simultaneously given 100 ng of pG+host9 plasmid carrying an erythromycin cassette as the selectable marker and 2.0 μg of the unselected SOE construct containing the desired mutation. At least 20 erythromycin-resistant transformant colonies were screened for incorporation of the unselected point mutation by PCR as described (Cha et al., 1992).
For acquisition of the HPr S46A mutation, WT TIGR4 ptsH was provided in trans on the pG+host9 co-transformation plasmid. The 369-base pair region encompassing ptsH and its predicted promoter region was amplified from genomic DNA using primers designed to incorporate SmaI and XhoI restriction sites at the 5′ and 3′ ends of the product, respectively, allowing for cloning into pG+host9 downstream of the erythromycin cassette, generating pG+host9-HPr. This plasmid was maintained in E. coli TG1.
The LacR-2 trans complemented strain was created by replacing the neutral gene SP_1773 (van Opijnen et al., 2009) with lacR-2 and its predicted promoter region. In order to minimize potential complications inherent to the introduction of constitutive antibiotic resistance cassettes, the CHESHIRE cassette (Weng et al., 2009) was included in the SOE PCR construct upstream of the lacR-2 promoter region. The CHESHIRE cassette encodes ermAB, enabling temporary selection of transformants on erythromycin and subsequent removal of almost the entire cassette via the cassette-encoded fucose-inducible cre recombinase and flanking lox sites (Weng et al., 2009). Selection and curing of the CHESHIRE cassette were performed as described previously (Weng et al., 2009). All mutations and plasmids generated in this study were confirmed by Sanger sequencing by the Tufts University Core Facility or Eton Bioscience (http://www.etonbio.com).
Determination of cis mutations that rescue the lethality of HPr S46A
In order to determine if deletion of ptsI rescues the lethality of the HPr S46A mutation, an SOE PCR construct containing both the HPr S46A codon change and a spectinomycin cassette replacing the entire ptsI open reading frame was introduced into WT by natural transformation. Transformants were selected on spectinomycin plates and screened for incorporation of the codon change as described above.
In order to determine if an HPr H15A mutation in cis rescues the lethality of the HPr S46A mutation, an SOE PCR construct containing both the HPr H15A and the HPr S46A codon changes was co-transformed into WT using either the pG+host9 or the pG+host9-HPr co-transformation plasmids for selection of competent cells. Transformants were screened for incorporation of both codon changes as described above. As a control for the viability of the HPr H15A mutation alone, an SOE PCR construct containing only the HPr H15A codon change was also introduced into the WT background as described.
Growth analysis of pneumococcal strains
Growth at 37°C and ambient CO2 levels were determined in 96-well plates using a BioTek Synergy HT plate reader without aeration (BioTek Instruments). Mid-exponential THY cultures of each strain were washed once and back-diluted to 0.01 in THY or CDM with 0.5% of either glucose, galactose or lactose, with catalase (300 U ml−1). Optical density at 600 nm (OD600) was monitored over the course of 15 h. At least six biological replicates from at least two different days were assayed for each strain.
Radiolabeled carbohydrate uptake assays
Single carbohydrate incorporation assays were performed generally as described previously but with the following modifications (Marion et al., 2011a). Each strain was collected from a blood agar plate, washed and resuspended to an OD600 of 0.2 in CDM with 0.5% glucose or 0.5% galactose. Cell suspensions were incubated at 37°C for 1 h, after which they were collected, washed twice in CDM containing no added carbohydrate and resuspended to the original volume in CDM with no added carbohydrate. Cell suspensions were kept on ice until the start of the experiment. The cells were added to a chilled flask containing an equivalent volume of 200 μM unlabeled glucose or galactose CDM with 1 −Ci of D-glucose [C6-H3] (American Radiochemicals) or D-galactose [C1-C14] (Perkin Elmer). Volumes and concentrations were arranged to ensure optimal mixing of reaction components, such that in a final volume of 6.5 ml, the final OD600 was 0.2 and the final unlabeled carbohydrate concentration was 100 −M. Immediately upon adding the cells, a 1 ml sample was taken (time 0), and the flask was put into a 37°C water bath. Additional 1 ml samples were taken at 1, 2.5, 5, 7.5 and 10 min for WT and 1, 2, 3, 5 and 10 min for HPr S46D after moving the flask to the 37°C water bath. Each 1 ml sample was applied to a 0.45 μm nitrocellulose filter assembled on a vacuum apparatus, and washed twice with 1 ml of PBS (EMD Millipore). Filters were dried and thoroughly mixed with 10 ml of Ecoscint H scintillation fluid (National Diagnostics), before measuring disintegrations per minute (DMPs) on a Beckman LS 6500 Scintillation System (Beckman). The 0 min value was used as a correction for non-specific association of radioactivity to the cells. DMPs were converted to nanomoles of total carbohydrate incorporated per 109 cells using DMP to μCi standard curves generated for D-glucose [C6-H3] or D-galactose [C1-C14], and normalizing to the number of cells based on dilution plating of cell solutions. The graphed data represent the average value of at least six biological replicates tested on multiple days, with error bars representing the standard error of the mean.
For the dual carbohydrate incorporation assay, cells were preconditioned to 0.5% galactose, washed and resuspended as described above. Each cell suspension was split into two unequal parts. The first part was assayed for galactose incorporation, exactly as described above except that the cells were pre-acclimated to 37°C prior to the addition of carbohydrate, and 500 μl samples were collected each minute for 10 min starting at 1 min after the addition of galactose. The second fraction of the cell suspension was used to assay dual incorporation of glucose and galactose simultaneously. The total reaction volume was increased to 10 ml to accommodate collecting 500 μl samples every minute for 20 min beginning at 1 min after the addition of carbohydrate. Final unlabeled carbohydrate concentrations were 100 μM and 1 μCi of each radioactive carbohydrate. The pre-acclimation step was added because it helped synchronize the two parts of this experiment. Data collection and processing were identical except for including a C14 bleed-through correction for the D-glucose [C6-H3] DMP measurements. Measurement of the D-galactose [C1-C14] standard curve samples using the wide tritium channel showed that at most, there was 10% bleed-through. As a ‘flat rate’ correction, 10% of the D-galactose [C1-C14] DMP measurement was subtracted from each D-glucose [C6-H3] measurement in the dual uptake experiments. If anything, this is an over-correction and artificially decreases the perceived glucose transport value. This is not of major concern to the experimental design as we are most concerned with accuracy of D-galactose [C1-C14] measurement.
Galactose oxidase assay for determination of galactose concentration in culture supernatants
A galactose oxidase assay was used to measure galactose concentrations in culture supernatants as previously described (Taniai et al., 2008). Mid-exponential WT cultures grown in galactose CDM were washed and inoculated at an OD600 of 0.05 in either 0.1% galactose (5.6 mM) or 0.05% glucose plus 0.05% galactose CDM (2.8 mM each) with catalase. OD600 was measured every 30 min, and 200 μl samples were collected every 15 min by centrifuging at room temperature, 4000 × g for 2 min. The resulting supernatant was incubated at 80°C for 20 min to inactivate the catalase in the growth medium (Morgulis and Beber, 1928). The total galac-tose concentration in each supernatant was then determined using the Amplex Red Galactose/Galactose Oxidase Kit (Molecular Probes), according to the manufacturer’s instructions. A galactose standard curve was used to convert absorbance readings to moles of galactose.
Galactose metabolism gene expression analysis
Transcription of galactose and lactose-associated genes during mid-exponential growth was assessed by qRT-PCR as previously described (Shainheit et al., 2014). Briefly, cells were grown to mid-exponential phase in CDM with one of the following carbohydrates; 0.5% glucose, galactose or lactose, or 0.5% glucose plus 0.5% galactose, or 0.5% glucose plus 0.5% lactose, the latter two conditions representing carbon catabolite-repressive conditions. Two milliliters samples were stored at −80°C in RNAprotect Cell Reagent (Qiagen) according to the manufacturer’s instructions, for a minimum of 12 h before isolating total RNA by Trizol-chloroform extraction. The resulting RNA was purified further using the RNAeasy Mini Kit (Qiagen), according to kit instructions, and any contaminating DNA was removed by treatment with the TURBO DNA-free kit (Ambion, Life Technologies). No more than 1 μg of the purified RNAwas used to template a single cycle reverse transcriptase reaction with the iScript cDNA Synthesis Kit (Bio-Rad). Reactions lacking reverse transcriptase were included for each sample to assess for residual genomic DNA contamination. The resulting cDNAwas prepared for qRT-PCR reactions in IQ SYBR green Supermix (Bio-Rad) with the primer sets listed in Supporting Information Table S1. The MxP3005P real-time PCR system with MxPro qPCR software (Stratagene) was used to measure cycle threshold (Ct) values, which were subsequently corrected for primer efficiency differences before normalizing to the housekeeping gene, rplI. Expression values are reported relative to WT grown in 0.5% glucose CDM. No less than five to six biological replicates measured in technical duplicate from at least two separate days were tested for each strain in each carbohydrate condition.
For the measurement of galactose-dependent induction of bgaA and lacC over time, mid-exponential THY cultures of WT and HPr S46D were washed twice and resuspended in CDM with no added carbohydrate. A 2 ml sample was collected as the uninduced control. Galactose was added to each such that the final concentration was 0.5%. Additional samples were collected at 30, 60 and 120 min after the addition of galactose. Samples were processed identically as above. Fold induction is reported as the average ratio of each time point over the uninduced control relative to rplI expression within each strain. Five biological replicates of each strain were analyzed from multiple days.
pG+host9-HPr plasmid loss experiment
WT and HPr S46A both carrying the complementing pG+host9-HPr plasmid were grown in THY with erythromycin at a permissible temperature (30°C) for plasmid replication and selection. Upon reaching mid-exponential phase, the cultures were washed and back-diluted into fresh medium (THY and 0.5% glucose CDM were both tested) without selection and allowed to grow at a non-permissive temperature (37°C) in the BioTek Synergy HT plate reader. The cultures were grown for successive generations by back-diluting them into fresh medium after six generations. Samples were plated periodically with and without selection at 30°C to determine the proportion of plasmid-bearing cells remaining. Biological triplicates of each strain were tested on multiple days in both THY and 0.5% glucose CDM.
β-galactosidase assay
β-galactosidase assays in pneumococcus were performed as previously described (Iyer et al., 2005). Briefly, mid-exponential cultures grown in various carbohydrate conditions were collected, washed, permeabilized and assayed for β-galactosidase activity by monitoring the cleavage of o-nitrophenyl-galactopyranoside (ONPG) at absorbance 420 nm. Data reported represent the average of six biological replicates tested on at least two separate days.
LacR-2 putative operator validation in E. coli MG1655
The ʎ red recombineering system was used as previously described to recombineer E. coli MG1655 to serve as a LacZ-based transcriptional validation system for predicted LacR-2 operator sequences (Yu et al., 2000). SOE PCR constructs were created to replace the endogenous regulatory elements of lacZ (from the +1 position of lacI up to the +1 position of lacZ) with a divergently transcribed kanamycin resistance cassette linked to the predicted promoter and regulatory elements of the pneumococcal operons containing predicted LacR-2 operator sequences (Table 2). The lacA-Δoperator strain was created similarly but with an SOE PCR construct containing a 20-base pair deletion encompassing the entirety of the predicted LacR-2 operator at this promoter. The putative repressor, LacR-2, was inducibly expressed in these strains from the pDL993-LacR-2 plasmid conferring tetracy-cline resistance. The lacR-2 coding region from TIGR4 was cloned into pDL993 downstream of the pLac promoter using the SmaI and EcoRI sites. The resulting strains were grown overnight in L broth with tetracycline (2.0 μg ml−1) and varying concentrations of IPTG (0 μM, 1 μM, 10 μM, 100 μM). Cultures were back-diluted 1:100 into identical culture conditions and grown for an additional 2 h before determining LacZ activity by the Miller assay, as described previously (Griffith and Wolf, 2002). Each strain carrying the empty plasmid pDL993 and 0 μM IPTG served as a negative control. Six biological replicates from at least two separate days were tested for each strain.
Supplementary Material
Acknowledgments
The authors thank Dr. Michael Malamy for providing expertise and equipment for the radiolabeled carbohydrate experiments. We would also like to thank Drs. EmilyKate McDonough and Revati Masalami for invaluable discussions of experiments, and EmilyKate McDonough for critical review of the manuscript. A.C. is a Howard Hughes Medical Institute investigator.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Aaberge IS, Eng J, Lermark G, Levik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. 1995:141–152. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- Abranches J, Chen YYM, Burne RA. Characterization of Streptococcus mutans strains deficient in EIIABMan of the sugar phosphotransferase system. Appl Environ Microbiol. 2003;69:4760–4769. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Chen YYM, Burne RA. Galactose metabolism by Streptococcus mutans. Appl Environ Microbiol. 2004;70:6047–6052. doi: 10.1128/AEM.70.10.6047-6052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal M, Shafeeq S, Kuipers OP. LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae. Appl Environ Microbiol. 2014;80:5349–5358. doi: 10.1128/AEM.01370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdić D, Ferretti J. Transcriptional regulation of the Streptococcus mutans gal operon by the GalR repressor. J Bacteriol. 1998;180:5727–5732. doi: 10.1128/jb.180.21.5727-5732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdić D, Pham VTT. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol. 2007;189:5049–5059. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdić D, Sutcliffe I, Russell R, Ferretti J. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene. 1996;180:137–144. doi: 10.1016/s0378-1119(96)00434-9. [DOI] [PubMed] [Google Scholar]
- Alpert C, Siebers U. The lac operon of Lacto-bacillus casei contains lacT, a gene coding for a protein of the Bg1G family of transcriptional antiterminators. J Bacte-riol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V. HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold. FEMS Microbiol Lett. 2003;222:17–23. doi: 10.1016/S0378-1097(03)00242-8. [DOI] [PubMed] [Google Scholar]
- Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, et al. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE. 2012;7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett DL, Anderson RL. Genetic evidence for the physiological significance of the D-tagatose 6-phosphate pathway of lactose and D-galactose degradation in Staphylococcus aureus. J Bacteriol. 1974a;119:698–704. doi: 10.1128/jb.119.3.698-704.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett DL, Anderson RL. Lactose and D-galactose metabolism in group N streptococci: presence of enzymes for both the D-galactose 1-phosphate and D-tagatose 6-phosphate pathways. J Bacteriol. 1974b;117:318–320. doi: 10.1128/jb.117.1.318-320.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker A, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett. 1999;172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- Buckwalter CM, King SJ. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 2012;20:517–522. doi: 10.1016/j.tim.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol. 2008;190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS ONE. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. Genome Res. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- Choy HE, Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc Natl Acad Sci USA. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK. CcpA-independent carbon catabolite repression in Bacillus subtilis. J Mol Microbiol Biotechnol. 2002;4:315–321. [PubMed] [Google Scholar]
- Dalia AB, Standish AJ, Weiser JN. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsono-phagocytic killing by human neutrophils. Infect Immun. 2010;78:2108–2116. doi: 10.1128/IAI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc Natl Acad Sci USA. 2014;111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbon E, Servant P, Poncet S, Deutscher J. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol Microbiol. 2002;43:1039–1052. doi: 10.1046/j.1365-2958.2002.02800.x. [DOI] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Küster E. Protein kinase dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Reizer J, Fischer C, Galinier A, Saier MH, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- Giammarinaro P, Paton JC. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect Immun. 2002;70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Zhou CZ, Receveur-Bréchot V, Collinet B, Declerck N, van Tilbeurgh H. Activation of the LicT transcriptional antiterminator involves a domain swing/ lock mechanism provoking massive structural changes. J Biol Chem. 2005;280:14780–14789. doi: 10.1074/jbc.M414642200. [DOI] [PubMed] [Google Scholar]
- Griffith KL, Wolf RE. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai Z, Ho SN, Pease L. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;80:528–535. [PubMed] [Google Scholar]
- Iyer R, Baliga NS, Camilli A. Catabolite control protein A ( CcpA ) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman GE, Yother J. CcpA-dependent and -independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J Bacteriol. 2007;189:5183–5192. doi: 10.1128/JB.00449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol. 2010;25:15–24. doi: 10.1111/j.2041-1014.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, et al. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol. 2004;54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Bijlsma JJE, Kok J, Kuipers OP. To have neighbour’s fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology. 2006;152:351–359. doi: 10.1099/mic.0.28521-0. [DOI] [PubMed] [Google Scholar]
- Limoli DH, Sladek JA, Fuller LA, Singh AK, King SJ. BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology. 2011;157:2369–2381. doi: 10.1099/mic.0.045609-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner C, Hecker M, Le Coq D, Deutscher J. Bacillus subtilis mutant LicT antiterminators exhibiting Enzyme I- and HPr-independent antitermination affect catabolite repression of the bglPH operon. J Bacteriol. 2002;184:4819–4828. doi: 10.1128/JB.184.17.4819-4828.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca G, Chung Y, Barabote R. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J Bacteriol. 2005;187:7826–7839. doi: 10.1128/JB.187.22.7826-7839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortie LA, Pelletier M, Vadeboncoeur C, Frenette M. The gene encoding IIAB(Man)L in Streptococcus salivarius is part of a tetracistronic operon encoding a phosphoenolpyruvate: mannose/glucose phosphotransferase system. Microbiology. 2000;146:677–685. doi: 10.1099/00221287-146-3-677. [DOI] [PubMed] [Google Scholar]
- Maguin E, Prevost H, Ehrlich D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun. 2006;74:4014–4020. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak BC, Pabijaniak M, de Jong A, Dűhring R, Seidel G, Hillen W, Kuipers OP. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genomics. 2012;13:401. doi: 10.1186/1471-2164-13-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C, Burnaugh AM, Woodiga SA, King SJ. Sialic acid transport contributes to pneumococcal colonization. Infect Immun. 2011a;79:1262–1269. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C, Aten AE, Woodiga SA, King SJ. Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect Immun. 2011b;79:4193–4200. doi: 10.1128/IAI.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C, Stewart JM, Tazi MF, Burnaugh AM, Linke CM, Woodiga SA, King SJ. Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect Immun. 2012;80:1390–1398. doi: 10.1128/IAI.05756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis S, Beber M. Studies on the effect of temperature on the catalase reaction: VI. Heat inactivation of catalase at different hydrogen ion concentrations. J Biol Chem. 1928;77:115–126. [Google Scholar]
- van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskouian B, Stewart GC. Repression and cat-abolite repression of the lactose operon of Staphylococcus aureus. J Bacteriol. 1990;172:3804–3812. doi: 10.1128/jb.172.7.3804-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–147. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Reizer J, Sutrina S, Saier M, Stewart G, Peterkofsky A, Reddy P. Mechanistic and physiological consequences of HPr (ser) phosphorylation on the activities of the phosphoenolpyruvate: sugar phosphotransferase system in gram-positive bacteria: studies with site-specific mutants of HPr. EMBO J. 1989;8:2111–2120. doi: 10.1002/j.1460-2075.1989.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- van Rooijen R, Gasson M, de Vos W. Characterization of the Lactococcus lactis lactose operon promoter: contribution of flanking sequences and LacR repressor to promoter activity. J Bacteriol. 1992;174:2273–2280. doi: 10.1128/jb.174.7.2273-2280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen RJ, Dechering KJ, Niek C, Wilmink J, de Vos WM. Lysines 72, 80 and 213 and aspartic acid 210 of the Lactococcus lactis LacR repressor are involved in the response to the inducer tagatose-6-phosphate leading to induction of lac operon expression. Protein Eng. 1993;6:201–206. doi: 10.1093/protein/6.2.201. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG, Genetik B. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118:731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Servant P, Le Coq D, Aymerich S. CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol Microbiol. 2005;55:1435–1451. doi: 10.1111/j.1365-2958.2005.04473.x. [DOI] [PubMed] [Google Scholar]
- Shainheit MG, Mulé M, Camilli A. The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect Immun. 2014;82:694–705. doi: 10.1128/IAI.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, Yoshida S. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol. 2008;190:3572–3579. doi: 10.1128/JB.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra VS, Homer KA, Rao SG, Andrew PW, Yesilkaya H. Characterization of novel beta-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect Immun. 2010;78:348–357. doi: 10.1128/IAI.00721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeu-wenhoek. 2002;82:59–71. [PubMed] [Google Scholar]
- Tomoyasu T, Imaki H, Masuda S, Okamoto A, Kim H, Waite RD, et al. LacR mutations are frequently observed in Streptococcus intermedius and are responsible for increased intermedilysin production and virulence. Infect Immun. 2013;81:3276–3286. doi: 10.1128/IAI.00638-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadeboncoeur C. Structure and properties of the phosphoenolpyruvate: glucose phosphotransferase system of oral streptococci. Can J Microbiol. 1984;30:495–502. doi: 10.1139/m84-073. [DOI] [PubMed] [Google Scholar]
- Vadeboncoeur C, Brochu D, Reizer J. Quantitative determination of the intracellular concentration of the various forms of HPr, a phosphocarrier protein of the phosphoenolpyruvate: sugar phosphotransferase system in growing cells of oral streptococci. Anal Biochem. 1991;196:24–30. doi: 10.1016/0003-2697(91)90112-7. [DOI] [PubMed] [Google Scholar]
- Vaughan EE, Bogaard PT, van den Catzeddu P, Kuipers OP, de Vos WM. Activation of silent gal genes in the lacgal regulon of Streptococcus thermophilus. J Bacteriol. 2001;183:1184–1194. doi: 10.1128/JB.183.4.1184-1194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R, Monedero V, Dossonnet V, Vadeboncoeur C, Peez-Martinez G, Deutscher J. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol Microbiol. 2000;36:570–584. doi: 10.1046/j.1365-2958.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Society. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, Biswas I, Morrison DA. A self-deleting cre-lox-ermAM cassette, CHESHIRE, for marker-less gene deletion in Streptococcus pneumoniae. J Microbiol Methods. 2009;79:353–357. doi: 10.1016/j.mimet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett. 2008;278:231–235. doi: 10.1111/j.1574-6968.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Burne RA. Multiple sugar: phospho-transferase system permeases participate in catabolite modification of gene expression in Streptococcus mutans. Mol Microbiol. 2008;70:197–208. doi: 10.1111/j.1365-2958.2008.06403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Burne RA. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol Microbiol. 2010;75:1145–1158. doi: 10.1111/j.1365-2958.2009.07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Das S, Burne RA. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol. 2010;192:2434–2444. doi: 10.1128/JB.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Martino NC, Burne RA. Two gene clusters coordinate galactose and lactose metabolism in Streptococcus gordonii. Appl Environ Microbiol. 2012;78:5597–5605. doi: 10.1128/AEM.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.