Introduction
The heart has a geometrically complex structure containing anisotropic laminar myofibrils within a dense vascular bed, that support directional synchronous contractions necessary for the circulation of blood.[1] Changes in the heart structure are associated with functional decline,[2–4] and the control of tissue architecture would help elucidate the dynamics of disease progression.[5] Here, we propose a bottom up approach to micromolding cardiovascular tissue composites consisting of independently fabricated cardiac and endothelial modules. This approach enabled structure-function studies both at the cellular level, where cardiac anisotropy leads to cardiomyocyte alignment and directional contraction, and at the tissue level, where cardiac density within a vascular bed controls synchronicity of beating. This paradigm for microfabricating functional cellular subunits and assembling them into tissue composites enables correlation of microarchitecture to cell and tissue function, to define principles for engineering of cardiac tissue structures.
Multiple cellular components, including cardiac myofibrils and microvascular networks, are assembled within a fibrous extracellular matrix (ECM) of the heart into precise geometric patterns that determine the pumping function.[1] Alterations in the assembly of structural components can lead to the heart failure.[2–4] Cardiac myofibrils align to the ECM providing contraction and conduction in a directional manner, important for the synchronous pumping of the heart.[1] A misalignment of cardiomyocytes, common in cardiac hypertrophy, can cause non-directional conduction within cardiac tissue, leading to arrhythmias and impaired contractile function.[6, 7] Under all conditions, microvascular networks in the heart provide energy and nutrients to this metabolically demanding organ.[8]
The dense extracellular matrix of the heart undergoes constant turnover by cardiac fibroblasts and remodels in response to disease.[9] Increases in the amounts of non-contractile or non-conductive tissue, such as the fibrotic tissue formed after ischemic damage to the heart, can also lead to arrhythmia, block of conduction and decreased muscle strength.[4] The modular structure of the heart is therefore essential for how the specialized function of each subunit contributes to the organ function. In order to study the structure-function relationships in the heart, each component - cardiac myofibrils, microvascular networks, and extracellular matrix – can be independently engineered and then assembled into a composite tissue.
Attempts to create composite cardiovascular tissues have primarily focused on three major strategies: (i) simple mixing of cardiomyocytes with endothelial cells,[10, 11] (ii) incorporation of endothelial cells into channeled cardiac scaffolds,[12, 13, 14] and (iii) patterning of cardiomyocytes atop the vascular networks.[15] However, the cardiomyocytes and endothelial cells interact differently with their surrounding.. For example, cardiomyocytes can align inside anistropic scaffolds, including poly(glycerol sebacate) (PGS),[14, 16] fibrin/Matrigel composites,[17] and methacrylated tropoelastin,[18] leading to directional contraction. In turn, endothelial cells form microvascular networks within gelatin methacrylate[19] and fibrin-PLLA/PLGA composites,[20] and may be positively influenced either by Matrigel[21] or incorporation of supporting cells.[20]
To this end, we hypothesized that cardiovascular tissue composites could be fabricated from independent cardiac and endothelial modules, and that cellular functions could be controlled by both individual module and overall tissue architectures. Each module was customized to match specific geometric requirements, and the cardiac and vascular modules were then combined into precise architectures of cardiovascular tissue composites that were studied for their emergent functions. The endothelial modules consisted of endothelial cells within a biodegradable fibrin hydrogel capable of forming vascular networks. The cardiac module consisted of cardiomyocytes within hydrogels of varying isotropies, where elongated substrates promoted troponin fiber alignment, anisotropic contraction, and fast calcium cycling.
The ability to precisely control the tissue geometry using this technique allowed independent variation of the cardiac module anisotropy and of the density of contractile cardiac units within the endothelial module, resulting in 8 different cardiovascular tissue states that were studied. We explored the contraction of the cardiovascular tissue composite and found that the cardiac modules were capable of beating synchronously only within certain geometric configurations. We generated a range of cardiovascular tissue organizations by assembling cardiac and endothelial modules, and studied the effects of patterning geometries cellular function in the individual module and the assembled composite structures.
Results
Cardiovascular tissue composite fabrication method and design
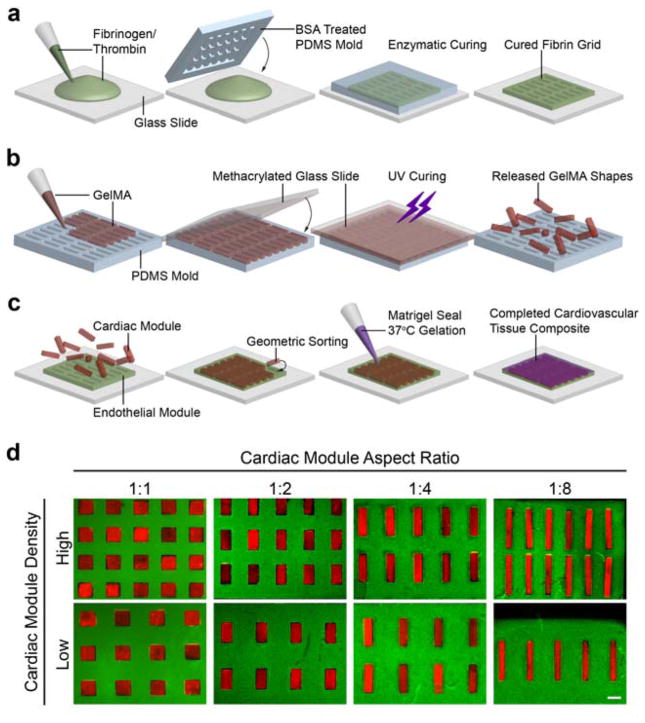
Cardiovascular tissue composites were created using the micromolding and geometric sorting techniques. These methods were developed by building upon our previous work.[22] The cell-containing hydrogel shapes were micromolded, sorted into a geometrically precise hydrogel template containing the negative relief of the shapes, and locked into the template by gelation of a sealant. Specifically, the cardiac modules (rectangular shapes containing cardiomyocytes) were sorted into the endothelial module (a hydrogel grid containing endothelial cells), resulting in a cardiovascular tissue composite designed to mimic the lamellar structure of the heart (Figure 1).
Figure 1. Method for fabricating composite cardiovascular tissue.
(a) Endothelial modules were fabricated by pipetting a fibrinogen thrombin mixture between a glass slide and a PDMS template mold that was allowed to cure for 5 min prior to endothelial cell seeding. (b) Cardiac modules were also fabricated by micromolding, using GelMA, a UV-light curable hydrogel, and a PDMS mold containing an array of any single type of rectangle. (c) To form tissues, cardiac modules were sorted within the endothelial module and sealed with a thin layer of Matrigel. (d) An array of cardiovascular tissue composite designs, where the cardiac modules are represented by red fluorescent beads encapsulated within hydrogels and the endothelial moduless are represented by green fluorescent beads encapsulated within hydrogels. Aspect ratio and loading density of the cardiac module were varied independently (scale: 500 μm).
Fibrin, gelatin methacrylate (GelMA), and Matrigel hydrogels were chosen as extracellular matrix materials based on their biodegradability, promotion of cardiovascular development, and ability to form high fidelity patterns.[17, 19, 20, 23] To generate the endothelial module, we first created a fibrin grid, molded within a polydimethylsiloxane (PDMS) template that was coated with bovine serum albumin (BSA) to prevent hydrogel adhesion (Figure 1a). The hydrogel was seeded with endothelial cells and grown for 24 hours before subsequent geometric sorting and sealing. The cardiac module fabricated using GelMA was molded into a PDMS template containing a grid of rectangular shapes, covered with a methacrylated glass slide, and cured using ultraviolet light. The shapes were then released from the mold (Figure 1b), seeded with cardiomyocytes, and grown for 4 days. After sorting the cardiac and endothelial modules, a thin layer of Matrigel was added to seal the resulting cardiovascular composite (Figure 1c).
Cardiovascular tissue composite designs were optimized using a MATLAB algorithm. Cardiac tissue geometry was deconstructed into parameters salient to tissue function. The aspect ratio of the cardiac module was varied from 1:1 to 1:8, while keeping the overall area constant, under the hypothesis that high aspect ratio could promote cardiac alignment.[24] A minimum dimension of 150 μm ensured that each hydrogel was large enough for assessing multicellular function. We further constrained the assembled composite designs to shapes organized in a tessellating grid patterns. Our final parameter was the relative fraction of cardiac tissue within the entire tissue, measured by the shape to scaffold area ratio, and varied to mimic structural changes that occur over the course of cardiac disease.[4]
We implemented a grid search algorithm in MATLAB with multiple design constraints and practical fabrication constraints to obtain distinct tissue designs (Figure 1d). Ultimately, two parameters - the aspect ratio of the cardiac modules and loading density of these units within the endothelial module, were varied independently. This resulted in four aspect ratios ranging from 1:1 (481 μm × 481 μm) to 1:8 (170 μm × 1,360 μm) loaded in tissue composites at high density (~1:3 by area) or low density (~1:6 by area) to obtain eight distinct types of tissue composites, each measuring 5.1 mm × 5.1 mm (Figure 1d).
Endothelial module vascular network formation
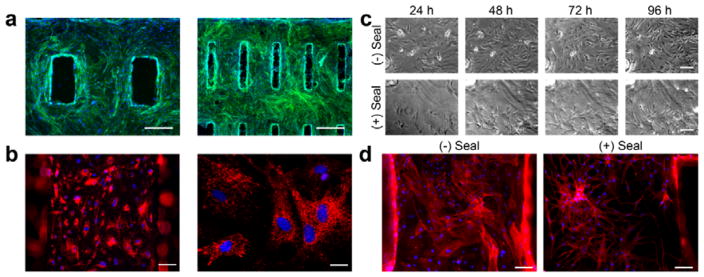
Endothelial modules were capable of generating vascular networks (Figure 2). Rat aortic endothelial cells were grown on fabricated fibrin hydrogels for four days, resulting in the complete coverage of the scaffold. Importantly, degradation of the fibrin was minimal during this time period such that the designed geometric patterns were retained even after incorporation of cells (Figure 2a). Endothelial cells were functional in this environment, as evidenced by the uptake of acetylated LDL (Figure 2b) that is characteristic for healthy endothelium.[25] We tested the ability of Matrigel seal to induce vascular network formation. Without the seal, endothelial cells maintained flat morphology, growing to cover the entire surface. In contrast, endothelial cells within Matrigel-sealed hydrogels developed spindle-like cell morphology, with branches extending from the endothelial cell nodes and connecting to surrounding cells (Figure 2c). Staining for the cytoskeletal protein actin using Phalloidin confirmed the formation of extensive vascular network (Figure 2d).
Figure 2. Endothelial module supports vascular network formation.
(a) Fluorescence micrographs of endothelial cells on fibrin hydrogels where green represents actin (Phalloidin) and blue represents nuclei (DAPI) (Scale = 500 μm). (b) Acetylated LDL uptake (red) in endothelial cells on fibrin hydrogels, nuclei counterstained with DAPI (blue). Low power (left, scale = 100 μm) and high power (right, scale = 25 μm). (c) Time course micrographs of endothelial cells grown on fibrin without or with the Matrigel seal over the course of 4 days, where the Matrigel promotes the formation of a network-like pattern (scale = 100 μm). (d) Actin staining with Phalloidin (red) at the end of the 4-day culture period further confirms the presence of dense endothelial networks in the sealed scaffolds (scale = 100 μm).
Cardiac modules promoted cellular alignment
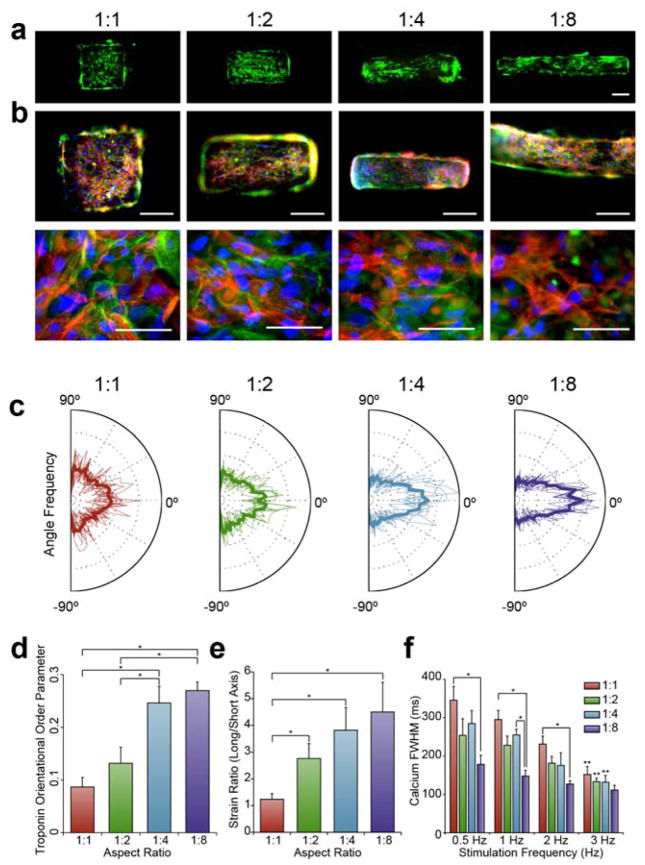
The cardiac module consisted of cardiomyocytes grown for four days on gelatin methacrylate (GelMA) hydrogels, where the high aspect ratio of the substrate resulted in cellular alignment and anisotropic contraction (Figure 3). After four days of culture, the GelMA hydrogel contained viable cells as demonstrated by live/dead staining (Figure 3a). Immunostaining for troponin (red) and phalloidin (green) revealed that nearly all cells were cardiomyocytes. While Troponin fibers were randomly oriented in the 1:1 aspect ratio group, higher aspect ratios showed a geometry-induced response, with troponin fibers aligning in parallel to the long axis of the substrate (Figure 3b).
Figure 3. Cardiac module aspect ratio drives cardiomyocyte anisotropy and function.
(a) Live (green) and dead (red) staining of cardiomyocyte-laden GelMA shapes demonstrates high fidelity of patterning and high density of live cardiomyocytes (scale = 200 μm). (b) Troponin (red) staining reveals that most cells on the GelMA are cardiomyocytes, counterstained with Phalloidin (green) and DAPI (blue). Troponin fibers are visibly disorganized in the 1:1 aspect ratio group, and become more aligned to the long axis as aspect ratio increases. Low power (top, scale: 200 μm) and high power (bottom, scale: 50 μm) views. (c) Distribution of calculated troponin fiber angles in individual analyzed images with the average in bold. A greater preponderance of angles near 0° occurred with cardiomyocytes on higher aspect ratios substrates. (d) The orientational order parameter (OOP, average Average ± SEM), measured from the distribution of angles, shows greater troponin alignment in the high aspect ratio groups (*p<0.05, n=8). (e) Long axis strain/short axis strain (strain ratio) (average ± SEM) generated in cardiac modules (*p<0.05, n≥14). (f) Full-width half maximum (FWHM) (average ± SEM) calcium transient all four aspect ratio groups stimulated at 0.5, 1, 2, and 3 Hz frequencies (*p<0.05, **p<0.05 between 3 Hz group and corresponding 0.5 Hz group with the same aspect ratio, n=3).
Troponin alignment was quantitatively assessed using an image processing algorithm. In ImageJ, images were background reduced, thresholded, and skeletonized, resulting in a single one-dimensional line for each troponin fiber segment (Figure S1). The angle of each troponin fiber ranged from −90° to 90°, with each extreme representing a fiber perpendicular to the long axis of the GelMA substrate, and 0° representing a fiber aligned perfectly with the long axis. Distributions of the individual images were overlaid in Figure 3c with the average distribution displayed in bold.
The 1:1 aspect ratio group yielded a roughly even distribution of troponin in all directions, while the higher aspect ratios yielded approximately twice the frequency of angles parallel than perpendicular to the long axis. Each distribution was further reduced to an orientational order parameter (OOP), where a high OOP represented more aligned fibers.[26] Cardiomyocytes were sensitive to the aspect ratio of the underlying substrate, with the 1:4 and 1:8 aspect ratio shapes demonstrating significantly greater alignment than the 1:1 and 1:2 aspect ratio shapes (Figure 3d). Overall, patterning cardiomyocytes on high-aspect ratio cardiac modules promoted cell alignment, an important feature of cardiac tissue structure.
Cardiac module mechanical and electromechanical coupling function
Strain analysis revealed that the geometrically induced alignment of cardiomyocytes contributed to anisotropic contraction (Figure 3e). We analyzed high-speed videos of the contraction by applying a motion tracking algorithm that determined pixel velocities and was further evaluated for strains developed in the long or short axis directions. 1:1 aspect ratio modules developed strain uniformly in both directions, while high aspect ratio modules developed strains favoring the long axis (Figure S2). The long to short axis strain ratio was approximately 1 for the 1:1 aspect ratio group, and higher at higher aspect ratios (Figure 3e). Thus, the aspect ratio defined the direction and strength of contractions in the cardiac module.
Higher aspect ratio cardiac modules exhibited more efficient electromechanical coupling. We used the calcium sensitive dye Fluo-4 to measure the transients in cardiac modules when they were electrically induced to beat at 0.5, 1, 2, and 3 Hz. We measured three transients: the time to peak, the rate constant of calcium decay (Tau), and the full-width-half-max time (FWHM), to determine how efficiently the cardiac modules cycled calcium. In general, cardiomyocytes displayed physiologic, frequency-dependent shortening of calcium transients (Figure 3f and Figure S3). Interestingly, calcium transients were also affected by the aspect ratio of the substrate. Cardiomyocytes in 1:8 aspect ratio modules had a significantly lower FWHM and time to peak at all stimulation frequencies except for 3 Hz, indicating more rapid electromechanical coupling.
Cardiac and endothelial module assembly to cardiovascular tissue composites
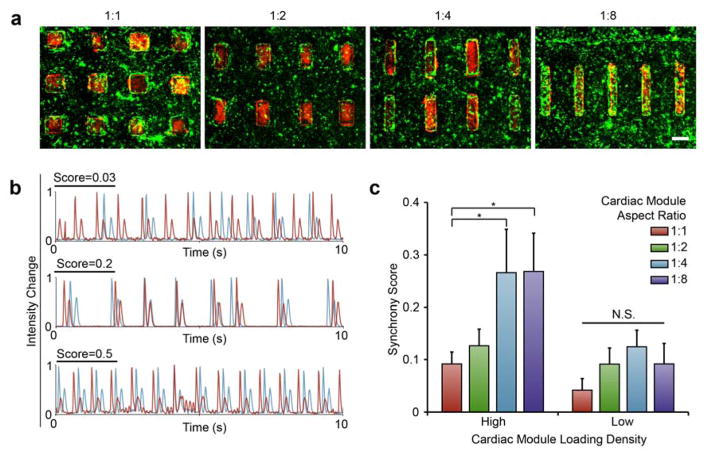
We co-cultured endothelial and cardiac modules as assembled cardiovascular tissue composites (Figure 4). The sorting resulted in composite tissues that maintained the high geometric specificity pre-determined by micropatterning design (Figure 4a). The cardiomyocytes and endothelial cells were stained with a red and green fluorescent live cell dye, respectively. After sorting, cells were cultured for 48 hours prior to imaging and assessment. With all four aspect ratios, composites maintained high level of cellularity, and the boundaries of most modules were clearly appreciated.
Figure 4. Composite tissue architecture defines cardiac beat synchrony.
(a) Cardiac modules (red) within sorted within endothelial modules (green). (scale: 500 μm). (b) Traces of adjacent shapes from a range of synchrony scores where 0.03 shows no visible synchrony, and scores of 0.2 and 0.5 show high levels of synchrony between the shapes. (c) Synchrony score (average ± SEM) from all eight composite tissue designs, four shape aspect ratios in two loading densities. (*p<0.05, NS = not significant, n≥3).
Cardiovascular tissue composite synchrony analysis
Cardiac modules within tissue composites showed geometry-dependent beat synchronization. We explored the ability of the adjacent cardiac modules within fully formed composite tissues to synchronize their beating, an important property of engineered tissues, the lack of which is a predictor of cardiac arrhythmia.[27] A synchrony metric was developed, where two synchronous cardiac modules were defined as those with the same frequency and without any phase shift. Phase contrast videos of the tissues were analyzed resulting in traces that displayed two peaks per beat, where the first, higher peak, represented contraction, and the second, more shallow and broad peak, represented relaxation (Figure S4). A Fourier transform of the contraction peaks enabled analysis of the frequency of beating.
We searched frequencies near the beating frequency of the cardiac modules, and determined the maximum coherence, a measure of similarity between the frequencies of contraction, where 1 represented identical similarity and 0 represented no similarity. This frequency of maximum coherence was matched to the corresponding phase shift between the two traces (Figure S4), where 0 represented no phase difference and ranged to π, which represented maximally different phases. We thus defined synchrony as two adjacent cardiac shapes with a high coherence and low phase shift:
| (1) |
where σ represented the synchrony score, κmax represented the maximum coherence near the frequency of beating, and φ represented the correlated phase shift.
All cardiovascular tissue types were capable of synchronization (Figure S5), with the electrical stimulation at 2 Hz and 3 Hz forcing simultaneous contraction of all cardiac modules. Traces generated from adjacent modules overlapped (Figure S5a) and all tissue composites, regardless of the aspect ratio or loading density, had synchronization scores approaching 1 (Figure S5b).
Spontaneous beating synchrony emerged from the cooperation of cardiac module anisotropy and high loading density. We captured spontaneous contractions from different cardiovascular tissue composites. In general, the synchrony scores ranged from 0 to 0.5, where a score of 0 displayed no visual relationship between beat traces, and scores of 0.2 or 0.5 displayed strong similarities in the peak intensity changes, suggesting synchronization of adjacent cardiac modules (Figure 4b). Remarkably both the aspect ratio and patterning density affected synchronicity (Figure 4c). While the 1:1 and 1:2 aspect ratio cardiac modules were unable to synchronize at either patterned density, the 1:4 and 1:8 aspect ratio cardiomyocyte modules synchronized with surrounding modules, but only if organized densely in the endothelial module. Therefore, synchrony - a function of healthy cardiac tissue, depends on both cardiomyocyte organization and overall tissue structure.
Discussion
Overall, we present an approach to generating and assessing composite cardiovascular tissues built from individual cardiac and endothelial modules. A modular approach allowed precise microscale definition of cardiomyocyte anisotropy and organization into various endothelialized tissue architectures. In particular, the generation of a coherent set of geometrically varied composites permitted the study of a spectrum of architectural states and how these may lead to healthy or diseased-like contractile function. Nearly all specialized tissues contain multiple cell types placed into precise geometric locations. The tissue-engineering framework described here, with parameterization of tissue modules and their on-demand assembly, could be extended to other tissues with high level of spatial organization. The proposed approach enables study of how tissue geometry can affect tissue function.
At the level of the individual modules, we successfully formed vascular networks within the endothelial module, a component important for engineering nearly all types of tissues.[28] Cardiac module anisotropy enhanced cardiomyocyte alignment and electromechanical function, including anisotropic contractions and shorter calcium transients. The higher strains in high-aspect ratio shapes were due to the cardiomyocyte alignment. When coupled with the aspect ratio, these higher strains result in markedly increased movement in the direction of the long axis, demonstrating that aspect ratio can contribute to anisotropic contraction in two ways: (i) by geometry alone, and (ii) by alignment of cardiomyocytes. Faster calcium transients in the high aspect ratio groups suggest that cardiomyocytes have more developed calcium handling machinery and more efficient coupling to neighboring cardiomyocytes. Our ability to tune cardiomyocyte alignment could support mechanistic studies on how alignment and cardiomyocyte connection can affect calcium conduction as well as overall mechanical contractile behavior on the organoid level.
On the tissue composite level, we studied cardiac beating synchrony, an emergent property of cardiomyocyte contraction. This is, to our knowledge, the first demonstration in an engineered system that cardiomyocyte alignment and overall tissue architecture act synergistically to generate synchronous tissue. Such studies are of interest based on clinical observations that cardiomyocyte disarray, as in hypertrophic cardiomyopathy, or local reduction in cardiomyocyte mass, as in cardiac fibrosis following myocardial infarction, can lead to arrhythmias.[3, 4, 7] We envision that this approach will have utility in studying specific geometric parameters for their functional and mechanistic effects, as in studies of heart failure, where various tissue patterns can mimic disease progression and functional decline.[3]
In summary, we show a modular approach to generating and studying cardiovascular tissue composite architectures in which cellular function can be assessed as both individual modules and assembled tissue composites. We demonstrate the importance of two geometric parameters: the aspect ratio of the cardiac module and the density of contractile units within the endothelial module. Aspect ratio of the cardiac modules guided cardiomyocyte alignment, anisotropic contractile function, and electromechanical coupling. The ability of cardiac modules to synchronize their contractile behavior was determined by the cooperation of the cardiac module aspect ratio and the overall architecture of the cardiovascular tissue composite. Therefore, the proposed modular approach to fabricate and assess composite tissue structures will help define how cardiac tissue structure may lead to changes in function on multiple scales, and how these changes may contribute to heart health and disease.
Experimental Section
Experimental details can be found in the Supporting Information.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding of this research by NIH (grants HL076485, EB17103 and EB002520 to GVN; F30 HL112505 to GE) and New York State (grants CO26449 and C028119 to GVN).
Footnotes
Authors contributions
B. W. Lee, G. Eng, and G. Vunjak-Novakovic conceived the study; G. Vunjak-Novakovic supervised the project; B. W. Lee, B. Liu, A. Pluchinsky, and N. Kim, and G. Eng conducted experiments and analyzed data; B. W. Lee, B. Liu, G. Eng, and G. Vunjak-Novakovic wrote the manuscript.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Benjamin W. Lee, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA. College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA
Bohao Liu, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA. College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA.
Adam Pluchinsky, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Nathan Kim, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Dr. George Eng, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA. College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA
Prof. Gordana Vunjak-Novakovic, Laboratory for Stem Cells and Tissue Engineering, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA. Department of Medicine (in Medical Sciences), Columbia University, New York, NY 10032, USA
References
- 1.a) Gilbert SH, Benoist D, Benson AP, White E, Tanner SF, Holden AV, Dobrzynski H, Bernus O, Radjenovic A. Am J Physiol Heart Circ Physiol. 2012;302:H287. doi: 10.1152/ajpheart.00824.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pope AJ, Sands GB, Smaill BH, LeGrice IJ. Am J Physiol Heart Circ Physiol. 2008;295:H1243. doi: 10.1152/ajpheart.00484.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton MG, Sharpe N. Circulation. 2000;101:2981. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 3.Mann DL, Bristow MR. Circulation. 2005;111:2837. doi: 10.1161/CIRCULATIONAHA.104.500546. [DOI] [PubMed] [Google Scholar]
- 4.Jessup M, Brozena S. N Engl J Med. 2003;348:2007. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 5.a) Kolewe ME, Park H, Gray C, Ye X, Langer R, Freed LE. Adv Mater. 2013;25:4459. doi: 10.1002/adma.201301016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Proc Natl Acad Sci USA. 2013;110:9770. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey N, Luedde M, Katus HA. Nat Rev Cardiol. 2012;9:91. doi: 10.1038/nrcardio.2011.159. [DOI] [PubMed] [Google Scholar]
- 7.Varnava AM, Elliott PM, Mahon N, Davies MJ, McKenna WJ. Am J Cardiol. 2001;88:275. doi: 10.1016/s0002-9149(01)01640-x. [DOI] [PubMed] [Google Scholar]
- 8.Rakusan K. Adv Organ Biol. 1999;7:129. [Google Scholar]
- 9.a) Porter KE, Turner NA. Pharmacol Ther. 2009;123:255. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]; b) MacKenna D, Summerour SR, Villarreal FJ. Cardiovasc Res. 2000;46:257. doi: 10.1016/s0008-6363(00)00030-4. [DOI] [PubMed] [Google Scholar]
- 10.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Circ Res. 2011;109:47. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Narmoneva DA, Vukmirovic R, Davis ME, Kamm RD, Lee RT. Circulation. 2004;110:962. doi: 10.1161/01.CIR.0000140667.37070.07. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stevens KR, Kreutziger KL, Dupras SK, Korte FS, Regnier M, Muskheli V, Nourse MB, Bendixen K, Reinecke H, Murry CE. Proc Natl Acad Sci USA. 2009;106:16568. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vollert I, Seiffert M, Bachmair J, Sander M, Eder A, Conradi L, Vogelsang A, Schulze T, Uebeler J, Holnthoner W, Redl H, Reichenspurner H, Hansen A, Eschenhagen T. Tissue Eng Part A. 2014;20:854. doi: 10.1089/ten.TEA.2013.0214. [DOI] [PubMed] [Google Scholar]
- 13.a) Ye X, Lu L, Kolewe ME, Park H, Larson BL, Kim ES, Freed LE. Biomaterials. 2013;34:10007. doi: 10.1016/j.biomaterials.2013.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zieber L, Or S, Ruvinov E, Cohen S. Biofabrication. 2014;6:024102. doi: 10.1088/1758-5082/6/2/024102. [DOI] [PubMed] [Google Scholar]; c) Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, Hauch KD, Laflamme MA, Murry CE, Ratner BD. Proc Natl Acad Sci USA. 2010;107:15211. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye X, Lu L, Kolewe ME, Hearon K, Fischer KM, Coppeta J, Freed LE. Adv Mater. 2014;26:7202. doi: 10.1002/adma.201403074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Sekine H, Shimizu T, Hobo K, Sekiya S, Yang J, Yamato M, Kurosawa H, Kobayashi E, Okano T. Circulation. 2008;118:S145. doi: 10.1161/CIRCULATIONAHA.107.757286. [DOI] [PubMed] [Google Scholar]; b) Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, Kobayashi E, Umezu M, Okano T. Nat Commun. 2013;4:1399. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Iyer RK, Chiu LL, Vunjak-Novakovic G, Radisic M. Biofabrication. 2012;4:035002. doi: 10.1088/1758-5082/4/3/035002. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Dvir T, Kedem A, Ruvinov E, Levy O, Freeman I, Landa N, Holbova R, Feinberg MS, Dror S, Etzion Y, Leor J, Cohen S. Proc Natl Acad USA. 2009;106:14990. doi: 10.1073/pnas.0812242106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelmayr GC, Jr, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhas M, Engelmayr GC, Jr, Fontanella AN, Palmer GM, Bursac N. Proc Natl Acad Sci USA. 2014;111:5508. doi: 10.1073/pnas.1402723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Adv Funct Mater. 2013;23 doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Adv Funct Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesman A, Koffler J, Atlas R, Blinder YJ, Kam Z, Levenberg S. Biomaterials. 2011;32:7856. doi: 10.1016/j.biomaterials.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Melero-Martin JM, De Obaldia ME, Kang SY, Khan ZA, Yuan L, Oettgen P, Bischoff J. Circ Res. 2008;103:194. doi: 10.1161/CIRCRESAHA.108.178590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng G, Lee BW, Parsa H, Chin CD, Schneider J, Linkov G, Sia SK, Vunjak-Novakovic G. Proc Natl Acad Sci USA. 2013;110:4551. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang KM, Annabi N, Ercole F, Zhou K, Karst D, Li F, Haynes JM, Evans RA, Thissen H, Khademhosseini A, Forsythe JS. Adv Funct Mater. 2015;25:977. doi: 10.1002/adfm.201403124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro AJ, Ang YS, Fu JD, Rivas RN, Mohamed TM, Higgs GC, Srivastava D, Pruitt BL. Proc Natl Acad Sci USA. 2015;112:12705. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voyta JC, Via DP, Butterfield CE, Zetter BR. J Cell Biol. 1984;99:2034. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feinberg AW, Alford PW, Jin H, Ripplinger CM, Werdich AA, Sheehy SP, Grosberg A, Parker KK. Biomaterials. 2012;33:5732. doi: 10.1016/j.biomaterials.2012.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a) Cho GY, Song JK, Park WJ, Han SW, Choi SH, Doo YC, Oh DJ, Lee Y. J Am Coll Cardiol. 2005;46:2237. doi: 10.1016/j.jacc.2004.11.074. [DOI] [PubMed] [Google Scholar]; b) McDevitt TC, Angello JC, Whitney ML, Reinecke H, Hauschka SD, Murry CE, Stayton PS. J Biomed Mater Res. 2002;60:472. doi: 10.1002/jbm.1292. [DOI] [PubMed] [Google Scholar]; c) Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE. Nature. 2014;510:273. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Hirt MN, Hansen A, Eschenhagen T. Circ Res. 2014;114:354. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]; b) Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens TP, Radisic M. Tissue Eng Part B Rev. 2010;16:169. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.