Abstract
Hebbian plasticity, including LTP and LTD, has long been regarded as important for local circuit refinement in the context of memory formation and stabilization. However, circuit development and stabilization additionally relies on non-Hebbian, homoeostatic, forms of plasticity such as synaptic scaling. Synaptic scaling is induced by chronic increases or decreases in neuronal activity. Synaptic scaling is associated with cell-wide adjustments in postsynaptic receptor density, and can occur in a multiplicative manner resulting in preservation of relative synaptic strengths across the entire neuron's population of synapses. Both active DNA methylation and de-methylation have been validated as crucial regulators of gene transcription during learning, and synaptic scaling is known to be transcriptionally dependent. However, it has been unclear whether homeostatic forms of plasticity such as synaptic scaling are regulated via epigenetic mechanisms. This review describes exciting recent work that has demonstrated a role for active changes in neuronal DNA methylation and demethylation as a controller of synaptic scaling and glutamate receptor trafficking. These findings bring together three major categories of memory-associated mechanisms that were previously largely considered separately: DNA methylation, homeostatic plasticity, and glutamate receptor trafficking.
Keywords: TET, methylation, active demethylation, cancer, development, learning, memory, transcription factor, gene expression, chromatin, DNA, histone, epigenetic, metaplasticity, homeostatic plasticity, neuroepigenetics, AMPA receptor, neuron, synaptic scaling, place cell, place field, consolidation, amnesia, Rett syndrome
Introduction
The molecular and genetic basis of memory formation and stabilization has been a vigorous area of research in biomedicine for the past three decades. Despite that, unifying models for the means through which transcriptional regulation controls neural function have yet to be achieved. In this review and commentary, I will highlight a recent convergence of three previously separate areas of memory research that appear likely to allow a new, more unified, model for the role of transcriptional regulation in memory formation and stabilization. The emerging field of neuroepigenetics deals in part with the role of chromatin structure and DNA methylation in learning and memory. A more established set of mechanisms pertaining to glutamate receptor trafficking is accepted as being important in synaptic plasticity and memory formation. Finally, non-Hebbian forms of synaptic and neural plasticity such as homeostatic plasticity, plasticity of intrinsic membrane properties, and synaptic scaling have a solid foundation of in vitro and theoretical/computational studies, suggesting an important role in memory stabilization. However, the underlying molecular genetic mechanisms have been undetermined. Recent studies by a number of groups now delineate a linkage among the previously disparate areas of neuroepigenetics, receptor trafficking, and synaptic scaling that afford the possibility of a unified perspective on the interacting roles of these processes in memory formation and long-term memory storage in the mammalian CNS.
Dynamic regulation of DNA methylation in memory
An active area of research in the memory field from a very early stage of molecular investigation has been the role of signal transduction mechanisms in hippocampal neuronal plasticity and memory formation. A more recent emerging idea has been that epigenetic mechanisms, especially histone modifications and DNA cytosine methylation, contribute to long-lasting behavioral change (Tsankova et al. 2006, Kumar et al. 2005, Meaney & Szyf 2005b, Meaney & Szyf 2005a, Weaver et al. 2005, Alarcon et al. 2004, Wood et al. 2006, Korzus et al. 2004, Levenson et al. 2004, Chwang et al. 2006, Feng et al. 2010, Miller et al. 2010, Miller & Sweatt 2007, Miller et al. 2008, Lubin & Sweatt 2007, Lubin et al. 2008, Roth et al. 2009, Zovkic et al. 2014, Levenson et al. 2006, Ooi & Bestor 2008, Kangaspeska et al. 2008, Metivier et al. 2008, Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Guo et al. 2011a, Rudenko et al. 2013, Zhang et al. 2013, Roberson & Sweatt 1999, Levenson & Sweatt 2005, Crick 1984, Holliday 1999, Jiang et al. 2008, Sweatt 2009, Graff & Mansuy 2008, Barrett & Wood 2008, Feng & Fan 2009, Day et al. 2013, Day & Sweatt 2010, Kriaucionis & Heintz 2009, Tahiliani et al. 2009, Niehrs 2009, Engel et al. 2009, Schmitz et al. 2009, Gehring et al. 2009, Wu & Sun 2009, Sweatt 2013). Epigenetic mechanisms, such as histone acetylation and DNA cytosine methylation, are powerful regulators of gene transcription even in non-dividing cells such as neurons. In general, histone acetylation is associated with transcriptional activation, and cytosine methylation in gene promoter regions leads to transcriptional suppression. It has been known for about a decade that epigenetic mechanisms, including histone acetylation and DNA Methyltransferase (DNMT) activity, are necessary for NMDA receptor-dependent LTP for associative fear conditioning, spatial learning, and for more complex operant memory types such as reward conditioning and drug addiction (Tsankova et al. 2006, Kumar et al. 2005, Meaney & Szyf 2005b, Meaney & Szyf 2005a, Weaver et al. 2005, Alarcon et al. 2004, Wood et al. 2006, Korzus et al. 2004, Levenson et al. 2004, Chwang et al. 2006, Feng et al. 2010, Miller et al. 2010, Miller & Sweatt 2007, Miller et al. 2008, Lubin & Sweatt 2007, Lubin et al. 2008, Roth et al. 2009, Zovkic et al. 2014, Levenson et al. 2006, Ooi & Bestor 2008, Kangaspeska et al. 2008, Metivier et al. 2008). Studies from a wide variety of laboratories have now shown that epigenetic signaling is involved in many forms of synaptic plasticity and learning in essentially every species that has so far been examined, including humans. Many investigators find this idea fascinating because the same mechanisms are used for triggering and storing long-term “memory” at the cellular and developmental level, for example when cells differentiate. In this vein, over the last decade a variety of groups contributed to making an important discovery - that regulation of DNA methylation is a dynamic process in the adult CNS, involved in controlling long-lasting changes in synaptic function and behavior (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Guo et al. 2011a, Rudenko et al. 2013, Zhang et al. 2013, Roberson & Sweatt 1999, Levenson & Sweatt 2005, Crick 1984, Holliday 1999, Jiang et al. 2008, Sweatt 2009, Graff & Mansuy 2008, Barrett & Wood 2008, Feng & Fan 2009, Day et al. 2013, Day & Sweatt 2010, Kriaucionis & Heintz 2009, Tahiliani et al. 2009, Niehrs 2009, Engel et al. 2009, Schmitz et al. 2009, Gehring et al. 2009, Wu & Sun 2009, Sweatt 2013, Swank & Sweatt 2001).
Regarding the role of active DNA demethylation in control of learning and memory, the few early studies that began to test this hypothesis relied on either acute DNMT inhibition with small molecules, or conditional knockout approaches that deleted specific DNMT isoforms (Feng et al. 2010, Miller et al. 2010, Miller & Sweatt 2007, Miller et al. 2008, Lubin et al. 2008, Feng & Fan 2009). In one of the most recent phases of studies of the physiologic and behavioral role of active DNA demethylation in the CNS, several groups have been exploring the role of the TET dioxygenase family of signal transduction cascades in hippocampal neuronal plasticity and learning (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013). These studies focused on the mechanisms through which the TET dioxygenase pathway controls memory-associated active cytosine demethylation and neuronal gene transcription. As active, stimulus-dependent regulation of active DNA methylation by DNMTs in the adult CNS has been extensively reviewed in prior publications, I will not reiterate those mechanisms here (Day & Sweatt 2010). Rather, I will focus on recent discoveries that TET dioxygenases and an attendant complex set of complementary biochemical pathways control active DNA cytosine demethylation in non-dividing neurons in the adult brain, and describe the role of those pathways in learning and memory in vivo.
Ten-eleven translocation (TET) proteins are Fe(II)- and 2-oxoglutarate (2OG)-dependent dioxygenases that successively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Figure 1A). TET family proteins were first identified as cancer-related genes associated with an aberrant chromosomal “ten-eleven translocation”, hence the name, and the TET proteins are significant players in cellular carcinogenesis (Tahiliani et al. 2009, Niehrs 2009, Gehring et al. 2009).
Figure 1. Biochemical pathway for modification of cytosine within DNA by TET.
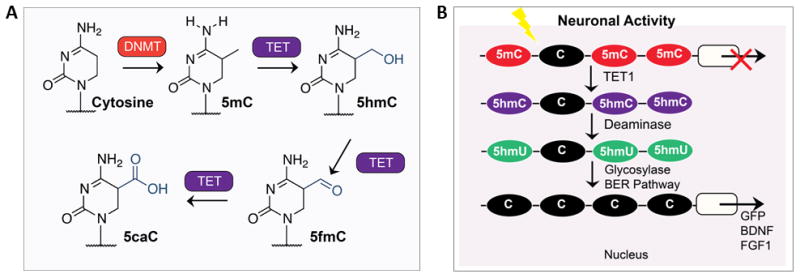
(A) 5-methylCytosine (5mC) bases, introduced by DNA methyltransferase (DNMT) enzymes, can subsequently be oxidized iteratively by the TET family of dioxygenases to 5-hydroxymethylCytosine (5hmC), 5-formylCytosine (5fC) and 5-carboxylated Cytosine (5caC). Figure and legend adapted from R.M. Kohli and Y Zhang, ‘TET enzymes, TDG and the dynamics of DNA demethylation’ Nature 502, 472–479 (24 October 2013). (B) TET1 dioxygenase coverts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in mammalian cells. Subsequent demethylation occurs through a process that requires deamination and the base-excision pathway (BER) mechanism. Demethylation of both exogenously introduced and endogenous 5hmCs is promoted by the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases. Furthermore, Tet1 and Apobec1 are involved in neuronal activity-induced, region-specific, active DNA demethylation and subsequent gene expression in the adult CNS. Figure and legend adapted from (Kohli & Zhang 2013, Guo et al. 2011c).
A new and emerging area of research emphasis in terms of investigating TET protein function is focused on understanding the molecular mechanisms whereby TETs contribute to long-term memory consolidation and storage. One of the most exciting findings to come out of the last phase of this new area of investigation was the identification of the “leukemia associated gene” TET1 dioxygenase as a driver of active DNA demethylation in hippocampal neurons (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013), and the discovery that this signaling pathway also controls long-term and remote memory consolidation in the contextual fear conditioning paradigm (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001). These results identified the heretofore mysterious mechanism for activity- and experience-dependent active cytosine demethylation in the CNS. Given the now-clear importance of understanding the roles and regulation of TET dioxygenase-dependent epigenetic mechanisms in neural plasticity and learning, a number of labs are currently pursuing investigations into the genomic, epigenomic, and functional targets of TET dioxygenase regulation in the hippocampus, cerebral cortex, and in other memory-associated parts of the CNS.
TET enzymes are thereby a relatively new addition to a large number of cancer-related genes and regulatory pathways that have been implicated in synaptic plasticity, learning, memory, and intellectual disabilities syndromes (see Table 1). This is not merely a coincidence but rather a manifestation of conservation of function of these pathways in the broad sense of cell biology (Sweatt 2001). Neural plasticity is most frequently thought of as a sine qua non of nervous system development and of ongoing experience-driven change in the adult nervous system, but cellular plasticity is a critical capacity for most types of cells in an organism. Tissues with regenerative capacity such as blood, liver, and skin require plasticity of the regulation of cell division. Immune cells require the capacity for rapid and highly precise adaptive responses to infection and injury. The cardiovascular and endocrine systems are highly responsive in order to achieve dynamic regulation of their component cells and tissues. Thus oncogenesis, development, homeostasis, regeneration, and neural plasticity are different facets of the same underlying evolutionary core requirement for cellular adaptation and the subsequent perpetuation of acquired change. In retrospect, it is not surprising that shared mechanisms such as mitogen activated protein kinase (MAPK) signaling, chromatin regulation, and DNA methylation/demethylation underlie all of these organismal processes (Levenson et al. 2004, Sweatt 2001, Roberson & Sweatt 1999, Swank & Sweatt 2001). These disparate forms of cellular plasticity are not just loose analogs of each other, it is now clear that they are molecular homologs of each other.
Table 1. Oncogenic Pathways and Regulators Implicated in Learning, Memory and Intellectual Disabilities.
| Mitogen-Activated Protein Kinase Cascades and their Targets | |
| ERK, CREB, Calcineurin, MAPKs, Raf, Ras, RSKs | AD, Coffin-Lowry Syndrome, Costello Syndrome, NFMR |
| Immediate Early Genes and Transcriptional Regulators | |
| Fmr1, Fmr2, Fos, Fyn, Jun, MAF, PS-1, Src,Tcf4, Wnt | AD, Drug addiction, Fragile X Syndrome, Pitt-Hopkins Syndrome |
| Protein Synthesis and Stability | |
| Pl3K/AKT, p53, mTOR, Plks, NF-kB, Usp9x | AD, Angelman Syndrome, Down Syndrome |
| Epigenetic Pathways | |
| BER, DNMTs, GAD45,TETs, Histone subunit exchange, Histone PTMs (e.g. acetylation, methylation, phosphorylation) | Cognitive Aging, Rett Syndrome, Rubinstein-Taybi Syndrome |
Note: These examples are illustrative, and this is not a comprehensive list.
However, for the majority of neuroscientists and neuroepigeneticists, the intrigue of TET family proteins and their principal enzymatic product 5-hydroxymethylcytosine resides not in their oncogenic capacity but rather in their capacity to regulate neuronal activity-, plasticity-, and memory-associated processes. As mentioned above, an exciting recent development related to these functions is the discovery that TET dioxygenases regulate active DNA demethylation in fully differentiated non-dividing neurons in the adult nervous system. This review presents an overview of TET regulation of DNA (de)methylation in neurons, the role of this process in memory formation, and recent discoveries concerning a likely target of TET-driven active DNA demethylation in neurons – glutamate receptor trafficking and neuronal homeostatic synaptic scaling.
An early clue concerning the physiologic, non-oncogenic function of TET proteins arose from analytical chemical analysis of the tissue and cellular distribution of the biochemical species it produces. As already described, the predominant product of TET enzymatic activity is 5 –hydroxymethylcytosine (5hmC). Intriguingly, 5hmC is most abundantly produced in terminally differentiated non-dividing neurons in the adult CNS and in the totipotent newly fertilized zygote (and its daughter embryonic stem cells). What do these two highly disparate cell types have in common? Their raison d'etre is to be plastic. Indeed, these two types of cells (neurons and embryonic stem cells) could be characterized as hyperplastic relative to the vast majority of cell types in mammals. This relative overabundance of 5hmC in hyperplastic cells strongly suggested that one role of TET proteins might be conferring transcriptional plasticity on cells, attendant with triggering lasting functional change. This concept was also consistent with the already-described oncogenic potential of TET proteins. Subsequent work has strongly supported this supposition of a role for TETs in neural plasticity (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013).
The remainder of this review will focus on the mechanisms through which TET proteins, specifically, and the regulation of DNA cytosine methylation, more generally, contribute to neural plasticity and memory formation. First I will describe recent discoveries demonstrating that TET dioxygenases control active cytosine demethylation in the nervous system, and briefly review the fairly complex set of biochemical steps involved in that process. I will then move on to describing a recently formulated hypothesis for how DNA (de)methylation might control neuronal function, namely the concept that demethylation pathways might control cell-wide homeostatic plasticity of glutamate receptor function. For the final sections I will review findings implicating TET proteins and active DNA demethylation in controlling cell-wide glutamatergic receptor plasticity and memory formation in the hippocampus, and multiplicative glutamatergic synaptic scaling and memory storage in the neocortex.
TET dioxygenases control active cytosine demethylation in the nervous system
The idea of the occurrence of active DNA demethylation in cells, particularly in non-dividing cells such as neurons, has been contentious. Traditional epigenetic mechanisms and studies have posited only passive DNA demethylation as a result of cell division and failure to replicate DNA methylation marks. Active demethylation through direct chemical removal of methyl groups on cytosines had been proposed earlier, but this concept has been controversial and some early findings have not been reproducible (Ooi & Bestor 2008, Day & Sweatt 2010, Niehrs 2009, Engel et al. 2009, Schmitz et al. 2009, Gehring et al. 2009, Wu & Sun 2009). However, a number of key discoveries independently replicated by several groups demonstrated the existence of active DNA demethylation in non-dividing cells in the mature CNS (Miller & Sweatt 2007, Miller et al. 2008, Lubin & Sweatt 2007, Lubin et al. 2008, Roth et al. 2009, Zovkic et al. 2014, Levenson et al. 2006, Ooi & Bestor 2008, Kangaspeska et al. 2008, Metivier et al. 2008, Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013). For example, several early studies provided indirect evidence for active DNA demethylation in the adult CNS in response to behavioral training (fear conditioning) using non-quantitative methods, such as PCR-based methods and methylation-dependent immunoprecipitation (Miller & Sweatt 2007, Miller et al. 2008, Lubin et al. 2008, Lubin & Sweatt 2007). This led to hypothesizing the existence of rapid, active DNA demethylation in the adult CNS. Consistent with this idea, two publications (Kangaspeska et al. 2008, Metivier et al. 2008) demonstrated rapid DNA demethylation and re-methylation, referred to as “cycling” of methyl-cytosine (hereafter abbreviated MeC) in cultured cells. This “cycling” demethylation occurred too rapidly to be explained by passive demethylation through cell division and was therefore proposed to be due to an active demethylation process.
Early investigators also proposed a specific demethylation mechanism: C-to-T conversion of MeC, followed by base-excision repair of the resulting nucleotide mismatch [(Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010), see Figure 1B]. Moreover, additional recent studies (Kriaucionis & Heintz 2009, Tahiliani et al. 2009) have shown that a novel DNA base, hydroxymethyl-cytosine, occurs in the CNS and could potentially serve as a specific precursor nucleoside for active demethylation. Most recently, exciting work from Song and colleagues supported this idea (Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010) – these investigators demonstrated that DNA repair mechanisms are utilized to demethylate DNA in non-dividing neurons, specifically through base-excision repair mechanisms controlled by the growth and DNA damage 45 (GADD45-beta) and TET regulatory systems. These findings conclusively demonstrated that demethylation can occur independently of DNA replication, and in terminally differentiated neurons.
Finally, recent studies of the CNS from the Song, Sweatt and Tsai groups have directly demonstrated active DNA demethylation in the adult CNS, driven by TET1 oxygenase, and that this mechanism controls memory capacity (Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013). Thus, there is now a substantial body of evidence supporting the idea that active DNA demethylation can occur in non-dividing neurons, findings which make viable the idea that active control of DNA demethylation plays a role in memory-associated transcriptional regulation in the CNS.
A hypothesis for how DNA (de)methylation might regulate neuronal function: control of cell-wide homeostatic plasticity of glutamate receptor function
Given the exciting discovery that TETs regulate active DNA demethylation in neurons, a compelling question then becomes: what is the role of active demethylation in controlling neuronal function? In other words, what is it that TET-driven demethylation does in order to control memory formation and neuronal plasticity? With these questions in mind, neuroscientists' interest in TETs should be piqued by recent discoveries that TET enzymes and active DNA demethylation control one of the major areas of research in the neural plasticity field over the last two decades, that is synaptic glutamate receptor trafficking (Guzman-Karlsson et al. 2014, Abbott & Nelson 2000, Cooper & Bear 2012, Turrigiano & Nelson 2000, Vitureira & Goda 2013, Nelson & Turrigiano 2008, Kim & Tsien 2008, Turrigiano et al. 1998, Ibata et al. 2008, Soares et al. 2013, Wierenga et al. 2005, Qiu et al. 2012, Wang et al. 2012, Gong et al. 2007, Correa et al. 2012, Zhong et al. 2012, Blackman et al. 2012, Baker-Andresen et al. 2013, Nelson et al. 2008, Turrigiano & Nelson 2004, Keck et al. 2013, Bredt & Nicoll 2003, Malinow & Malenka 2002, McCormack et al. 2006, Kessels & Malinow 2009, Clark & Nelson 2015). In this section I will provide a summary and overview of these recent results.
One of the defining aspects of epigenetic mechanisms is that they can drive cell-wide changes in gene expression (Day & Sweatt 2010). Therefore, an intriguing possibility regarding the role of active demethylation in neurons is that this mechanism plays to its strengths and is utilized for driving cell-wide functional changes (Sweatt 2013, Guzman-Karlsson et al. 2014). Cell-wide changes differ from the more widely studied “Hebbian” forms of plasticity, such as LTP and LTD, because they are not synapse-specific. Rather, homeostatic forms of neural plasticity regulate all the synapses on a neuron (typically 10,000 or so in a mammalian CNS pyramidal neuron) in unison in an orchestrated fashion. Thus, these forms of plasticity serve to coordinately regulate the entire neuronal complement of glutamate receptors (GluRs) such that the entire population of synapses is scaled up or down to keep total glutamate-gated conductance within a healthy and stable range that does not exceed the homeostatic capacity of the neuron. Moreover, this type of “multiplicative” scaling maintains the relative contribution of each individual synapse constant relative to the other 9,999 synapses in the neuron. Multiplicative scaling thereby keeps the relative ionic conductance or strength of a specific synapse at a constant proportional representation of the total synaptic weight of the cell. This maintains synapse A's relative contribution to bringing the neuron to threshold for firing an action potential the same, even as the entire 10,000 or so synapses on the neuron are coordinately moved up and down.
One can see how such a mechanism would confer stability of operation of a neuron within a functioning circuit (say, for stably storing a specific memory), by keeping constant the relative fractional contribution of each specific synapse to driving the cell and circuit. Moreover, homeostatic compensation of this sort simultaneously allows the neuron to down-adjust (or up-adjust) to the total metabolic burden of neuron-wide synaptic activity on the fly as individual synapses become stronger or weaker due to synapse-specific Hebbian mechanisms.
Contemplating multiplicative synaptic scaling in the abstract, such as we have just done, helps make clear the immense molecular complexity of implementing such a mechanism. Both membrane intrinsic properties and synaptic glutamatergic receptor function and trafficking would need to be coordinately regulated, with each of those components being subserved by dozens or even hundreds of individual gene products. What type of transcription-regulating mechanism would be capable of such coordinated regulation of cell-wide functional state? Epigenetic mechanisms are one category of mechanism with the necessary “computational” capacity and breadth of targeting across the genome to achieve such a feat. After all, such a type of complex genome-wide regulation would simply recapitulate epigenetic mechanisms' well-established capacity to perform similar coordinated operations during neural development and cell-fate determination.
Thus, as I and others have previously described, a speculative notion is that the neuronal epigenome may be preferentially involved in regulating and mediating non-Hebbian plasticity (Day & Sweatt 2010, Sweatt 2013, Guzman-Karlsson et al. 2014, Baker-Andresen et al. 2013, Clark & Nelson 2015). For example, epigenetic molecular mechanisms may be particularly relevant to various forms of metaplasticity, operating to establish a set-point for biasing the entire cell toward or against being susceptible to synapse-specific plasticity mechanisms such as LTP. Similarly, the neuronal or glial epigenome might be allocated to controlling intrinsic properties that are themselves cell-wide, such as excitability and activity-dependent synaptic scaling. Conceptually, the epigenome, having the capacity to control the entire genomic output and sense pan-cellular signaling mechanisms, such as MAPK signaling (Levenson et al. 2004, Chwang et al. 2006, Swank & Sweatt 2001), might be the ideal control point for achieving coordinated orchestration of the readout of a plethora of ion channels, receptors, and trafficking mechanism in order to achieve homeostatic plasticity.
In terms of background studies, this hypothesis is particularly appealing for two reasons. First, exciting recent results specifically implicated neuronal epigenetic regulatory pathways in regulating homeostatic plasticity – especially relevant are recent data directly implicating the methyl-cytosine binding protein MeCP2 in regulating synaptic scaling (Qiu et al. 2012, Wang et al. 2012, Gong et al. 2007, Correa et al. 2012, Zhong et al. 2012, Blackman et al. 2012, Baker-Andresen et al. 2013) and the discovery that DNMT inhibition can affect excitatory synaptic transmission (Nelson et al. 2008, Levenson et al. 2006). Second, a substantial body of prior literature indicated a role for homeostatic plasticity such as synaptic scaling in memory formation and stabilization. This role for synaptic scaling is over-and-above synapse-specific forms of plasticity such as LTP and LTD. Moreover, intrinsic plasticity, that is changes in the neuronal membrane's biophysical properties and changes in cellular excitability, has been implicated in memory network homeostasis (Abbott & Nelson 2000, Cooper & Bear 2012, Turrigiano et al. 1998, Turrigiano & Nelson 2000, Turrigiano & Nelson 2004, Vitureira & Goda 2013, Nelson & Turrigiano 2008, Kim & Tsien 2008, Ibata et al. 2008, Soares et al. 2013, Wierenga et al. 2005, Qiu et al. 2012, Wang et al. 2012, Gong et al. 2007, Keck et al. 2013, Zhang & Linden 2003, Desai et al. 1999).
Thus, there was a compelling body of prior work supporting the proposed model that DNA methylation controls synaptic scaling, not only from direct experimental observations but also based on more inferential associations between the effects of DNMT inhibition on cognition compared with known roles of intrinsic plasticity, synaptic scaling, and excitatory/inhibitory balance in controlling neural plasticity and cognition. With these background observations in mind, two recent papers specifically tested the idea that active DNA methylation and the TET dioxygenase DNA demethylation system are drivers of homeostatic neuronal regulation. I will describe selected results from these two papers in the next section.
Studies of synaptic scaling in vitro
Synaptic scaling is an activity-dependent form of plasticity regulating neuronal synaptic connectivity cell-wide. In experimental preparations to study homeostatic plasticity, synaptic scaling is typically induced using 24-hour treatments of tetrodotoxin (TTX) or KCl to model chronically decreased or increased activity, respectively. Whole-cell, voltage-clamp recordings are used to record miniature excitatory postsynaptic currents (mEPSCs) in visually identified cells, typically glutamatergic pyramidal neurons maintained in vitro (Figure 2Ai). EPSCs mediated by gluatamatergic transmission are pharmacologically isolated by blocking GABA-mediated IPSCs with bicuculline. Under conditions such as these, TTX induces synaptic up-scaling, seen as an increase in the efficacy of glutamatergic synapses present on the neuronal surface membrane (Figure 2Aii and 2Aiii).
Figure 2. DNA methylation regulates synaptic scaling.
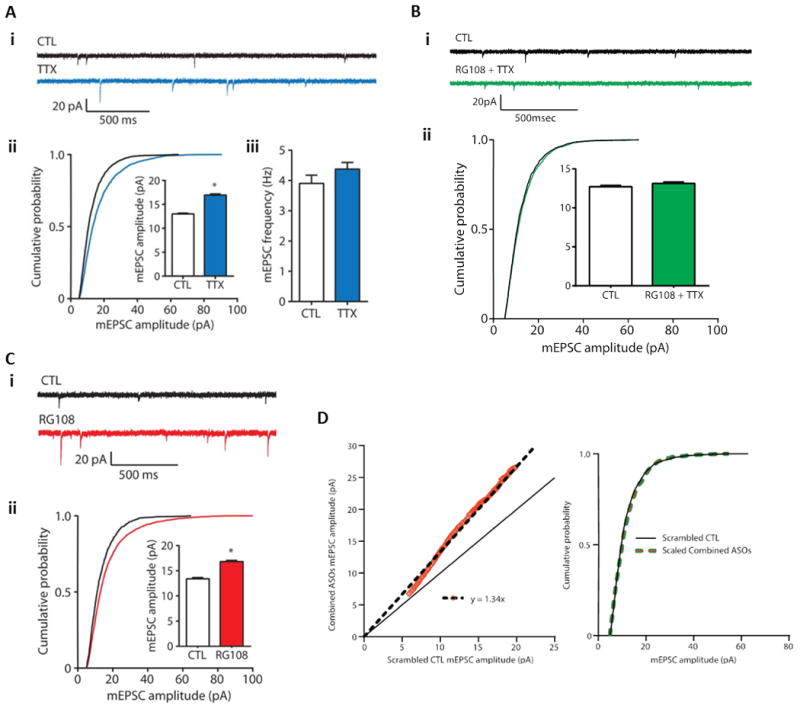
(A) Homeostatic upscaling of excitatory synaptic strength due to chronic inhibition of sodium channels with tetrodotoxin (TTX). (i) Sample spontaneous miniature excitatory post-synaptic current traces (mEPSCs) – a sample recording epoch illustrating mEPSC records from cortical pyramidal neurons after 24 hours of exposure to vehicle control (CTL) or TTX (blue) is shown. Downward spikes are postsynaptic mEPSCs triggered by glutamate release presynaptically. (ii) Cumulative probability distributions of mEPSC amplitude and overall mean mEPSC amplitudes in control and TTX-treated pyramidal neuron cultures are shown – the rightward shift of the blue line and increase in total average mEPSC amplitude are indicative of synaptic upscaling. P < 0.001, Kolmogorov-Smirnov (K-S) test. Inset: *P < 0.001, Mann-Whitney (M-W) test. (iii) Mean mEPSC frequencies do not increase in cortical pyramidal neurons treated with TTX, suggesting (as has been previously demonstrated) that the increase in (ii) is not due to an increase in total number of synaptic connections on the postsynaptic neuron. Data from cells pooled from at least six experiments for each condition (CTL, n = 21 cells; TTX, n = 15 cells). (B) Induction of excitatory synaptic scaling by TTX is blocked by DNMT inhibition. (i) Sample mEPSC traces from cortical pyramidal neurons after 24 hours of exposure to control (CTL) or TTX + the competitive DNMT inhibitor RG108 (green). (ii) Cumulative probability distributions and mean mEPSC amplitudes from cortical pyramidal neurons treated with TTX in the presence of RG108. Data are cumulative of cells pooled from at least three experiments for each condition (CTL, n = 17 cells; RG108 + TTX, n = 12 cells). These results compared to the data presented in (A) indicate that DNMT activity is necessary for TTX-induced synaptic scaling. (C) DNMT inhibition with RG108 or via DNMT mRNA knockdown upscales excitatory synaptic strength. (i) Sample mEPSC records from cortical pyramidal neurons after 24-hour treatment with control (CTL) or the DNMT inhibitor RG108 (red). (ii) Cumulative probability distributions and mean mEPSC amplitudes from cortical pyramidal neurons treated with RG108. These data indicate that DNA cytosine demethylation (known to occur secondary to DNMT inhibition) is capable of triggering synaptic upscaling. Please note that the data presented in (B) also indicate that the RG108-triggered synaptic upscaling is dependent on neuronal action potential firing, that is, that the RG108 effect is dependent on neuronal activity. Data from cells pooled from at least four experiments for each condition (CTL, n = 12 cells; RG108, n = 12 cells). P < 0.001, K-S test. Inset: *P < 0.001, M-W test. (D) Combined Dnmt1 and Dnmt3a knockdown multiplicatively upscales excitatory strength. Left Panel - Rank Order Plot of one thousand randomly selected mEPSC amplitudes from scrambled CTL and combined ASO treatment (orange values). Linear regression yielded a scaling factor of 1.34. Right Panel – Scaled-down amplitudes from combined ASO treatment were not different than scrambled CTL (K – S test, P = 0.1323). This graphical analysis illustrates that the synaptic upscaling induced by DNMT knockdown is multiplicative in nature. This finding is consistent with the hypothesis that the upscaling maintained relative individual synaptic strengths across the entire population of neuronal synapses, that is that the fractional contribution of each individual synapse was preserved and the entire neuronal synaptic population was regulated in a proportional, coordinated fashion. Bar graphs are means ± SEM. Figure and legend adapted from (Meadows et al. 2015).
Several laboratories have begun to directly test the hypothesis that active control of DNA methylation and demethylation regulate homeostatic synaptic scaling. Initial molecular studies demonstrated that synaptic scaling due to TTX application is associated with altered TET dioxygenase gene expression (Meadows et al. 2015, Yu et al. 2015). This molecular finding put TET dioxygenase-driven demethylation “in play” as a potential regulator of homeostatic synaptic scaling. Moreover, in these same recent experiments, the Sweatt and Hablitz labs observed that RG108, a DNMT inhibitor that was previously shown to block activity-induced DNA methylation in neurons (Miller et al. 2010, Miller & Sweatt 2007, Lubin et al. 2008), blocks TTX-induced shifts in synaptic scaling in a cultured cortical neuron system (Meadows et al. 2015)(Figure 2B). DNMT inhibition by RG108 or DNMT3a/b knockdown also triggered synaptic scaling in an activity-dependent and transcription-dependent fashion, further consistent with a role for active methylation/demethylation in regulating synaptic scaling (see Figure 2C and (Meadows et al. 2015)). Finally, DNMT inhibition/knockdown caused upscaling that was “multiplicative” in nature, indicating that the entire cellular complement of synapses was regulated in a coordinated fashion that preserved the relative synaptic weights across the entire population of synapses (Meadows et al. 2015)(Figure 2D). Overall, these data are strongly supportive of the hypothesis that DNA demethylation has the capacity to regulate homeostatic plasticity in CNS neurons, specifically that TTX-induced synaptic scaling depends on active changes in DNA methylation.
In contemporaneous studies, this hypothesis was also developed, refined, and more thoroughly tested in independent experiments by the Song lab, who directly evaluated the capacity of TET dioxygenase to regulate synaptic scaling (Yu et al. 2015)(Figure 3). These investigators found that increased DNA methylation, as produced by TET3 dioxygenase inhibition in the presence of bicucculine, triggered synaptic upscaling (Figure 3A, right-hand panel). This effect was not mimicked by either TET1 or TET2 knockdown (Figure 3A left and middle panels). The effects of TET3 knockdown were blocked by TTX treatment, and indeed TTX caused TET3 knockdown to elicit synaptic downscaling (Figure 3A right-hand panel). Moreover, causing active DNA demethylation in neurons by over-expressing TET1 dioxygenase catalytic domain led to synaptic downscaling (Figure 3B upper panel), TET1 overexpression blocked TTX-induced synaptic upscaling (Figure 3B upper panel), and TET3 overexpression directly caused synaptic downscaling (Figure 3B lower panels). In additional experiments, the Song lab demonstrated that other crucial components of the neuronal active demethylation pathway, such as the base excision repair (BER) pathway, also control synaptic scaling (Yu et al. 2015).
Figure 3. Tet3 expression regulates glutamatergic synaptic transmission and synaptic scaling.
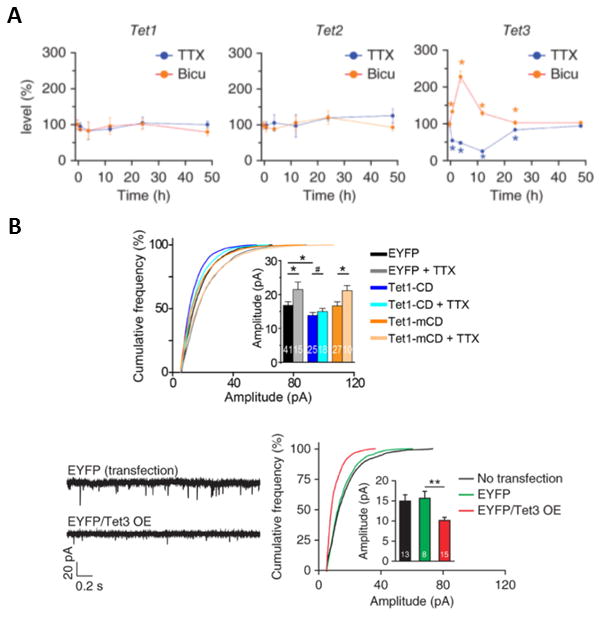
(A) Synaptic activity-dependent expression of Tet3 regulates glutamatergic synaptic transmission. In these experiments the effects of TET1 knockdown (left panel), TET2 knockdown (middle panel) and TET3 knockdown (right panel) were evaluated, and overall glutamatergic synaptic activity was quantitated as a percent of control. In all cases the effects of TET isoform knockdown were evaluated ether in the presence of sodium channel blockade (TTX, blue graphs) or GABA receptor blockade (Bicuculline, orange graphs). Tet3-knockdown neurons exhibited elevated glutamatergic synaptic transmission when GABA receptors were blocked (right panel). (B) Neurons overexpressing TET3 catalytic domain exhibit decreased glutamatergic synaptic transmission, and TET3 catalytic domain overexpression blocks TTX-induced synaptic upscaling. The left and right bottom panels illustrate that TET3 catalytic domain overexpression (EYFP/Tet3 OE) down-scales glutamatergic synaptic transmission relative to enhanced yellow fluorescent protein (EYFP) controls (red versus green bars). The upper panel presents cumulative probability curves demonstrating that TTX by itself triggers upscaling (gray bars), while TET1 catalytic domain (Tet1-CD) triggers downscaling (blue bars) and blocks the TTX-induced upscaling (aqua bars). The orange bars illustrate that the effects of TET1 catalytic domain are dependent on the activity of the enzyme, because they are not mimicked by transfection of a mutant catalytically inactive catalytic domain (Tet1-mCD). Values represent mean ± s.e.m. (*P < 0.05, **P < 0.01). Figure and legend adapted from (Yu et al. 2015). Please see (Yu et al. 2015) for additional details.
Taken together, these studies strongly suggest that DNA methylation is a mechanism that drives neuronal homeostatic stability. Specifically, the observation that TET1 inhibition causes upscaling is indicative that increasing DNA methylation is sufficient to trigger homeostatic upscaling. This interpretation is consistent with the additional observation that DNMT inhibition blocks the capacity of TTX to cause synaptic upscaling. More broadly, these findings are also consistent with the hypothesis that TET-driven active demethylation mediates synaptic down-scaling cell-wide in a catalysis-dependent fashion. Thus, DNMTs and TETs do not appear to control identical processes because DNMT inhibition is linked only with upscaling (at least thus far) while TETs appear to regulate both upscaling and downscaling. Nevertheless, this linkage of DNA (de)methylation to homeostatic forms of synaptic plasticity provides an important new insight into a major mechanism whereby epigenetic mechanisms contribute to memory formation and stabilization.
Future, more detailed mechanistic studies will hopefully illuminate the specific roles of cytosine methylation versus demethylation vis-à-vis homeostatic plasticity. One speculative possibility is that increased DNA methylation via the DNMTs serves as an activity-induced trigger for upscaling, while cytosine demethylation via the TETs serves to actively and dynamically regulate overall synaptic strength, both up and down, cell-wide in an ongoing fashion. However, this is but one of a large number of possible scenarios that might deserve future attention. In addition, the role of non-CpG methylation and the role(s) of specific isoforms of DNMTs await further study. Neurons exhibit extensive cytosine methylation at non-CpG sites in the genome, which appears to be principally regulated by the de novo methyltransferase DNMT3a (Guo et al. 2014). Therefore, it will be very interesting to investigate the role of “non-canonical” DNA methylation sites in novel forms of neural plasticity as part of this expanding understanding of the contribution of epigenetic mechanisms in cognitive function.
AMPA receptor trafficking and synaptic scaling
Given that DNA methylation controls synaptic scaling, what is the functional target of altered gene methylation? A considerable body of prior work had directly implicated trafficking of glutamate receptors to the neuronal cell surface membrane and into synapses as being a critical mediator of synaptic scaling (Abbott & Nelson 2000, Cooper & Bear 2012, Turrigiano & Nelson 2000, Vitureira & Goda 2013, Nelson & Turrigiano 2008, Kim & Tsien 2008, Turrigiano et al. 1998, Ibata et al. 2008, Soares et al. 2013, Wierenga et al. 2005, Qiu et al. 2012, Wang et al. 2012, Gong et al. 2007, Correa et al. 2012, Zhong et al. 2012, Blackman et al. 2012, Baker-Andresen et al. 2013, Nelson et al. 2008). Specifically, previous studies had shown that trafficking of the AMPA subtype of glutamate receptor plays a central role in synaptic scaling (Soares et al. 2013). With this in mind, in their recent studies Song and colleagues specifically investigated the hypothesis that the TET dioxygenase active demethylation pathway controls AMPA receptor trafficking to the neuronal surface. This hypothesis was borne out in an impressive fashion in their studies (Yu et al. 2015). Thus, Song and colleagues found that TET3 regulates neuronal AMPA receptor surface expression, and also that TET3 knockdown blocked TTX-induced increases in cell surface glutamate receptor expression. These results directly implicate DNA methylation/demethylation in controlling neuronal AMPA receptor trafficking. These data also strongly support the hypothesis that one component of the mechanisms whereby DNA methylation controls synaptic scaling is via regulation of glutamate receptor cell surface expression.
Finally, an additional alternative hypothesis that has not yet been explored is that DNA methylation/demethylation pathways, including TETs, might control other forms of neuron-wide plasticity, such as altering intrinsic properties, excitability, and action potential firing (Guzman-Karlsson et al. 2014, Zhang & Linden 2003, Desai et al. 1999). In ways reminiscent of DNA methylation controlling glutamate receptor expression and trafficking, specific modulation of Kv channels, SK channels, or HCN channels might occur in response to TET1 or DNMT activity. These additional alternative hypotheses can be tested using the same neurophysiology methods utilized for investigating synaptic scaling.
Conceptual innovation: memory storage at the cellular level involves cell-wide changes
The overarching hypothesis arising from the studies described above is that, during memory formation and storage, DNA methylation controls the homeostatic stabilization of activity- and experience-induced plastic changes at the circuit and behavioral levels. If this idea is true, it will require a redefinition of several of the fundamental tenets of current thinking among memory neuroscientists. Currently a central organizing concept of physiological psychology, at least regarding behavioral change, is the principle of Hebbian synaptic plasticity as the central mechanism underlying behavioral modification. The discoveries described above indicate a necessity to re-think certain aspects of this foundational hypothesis. A single adult neuron can have 10,000 synapses, but has only one nucleus. Within the nucleus there are only one or two copies of a given specific gene. Thereby, chemical modification of DNA implies a state change for the entire neuron. Chemically modifying and silencing or activating a given gene can alter the genomic complement of the entire cell. If chemical modification of genes underlies behavioral plasticity, this implies a re-thinking of the fundamental assumption that synaptic plasticity is the only locus driving behavioral modification. At a minimum, there appear to be two levels of mechanism in play – one residing at the individual synapse and one residing at the epigenome and operating cell-wide. This new conceptualization of the molecular neurobiology of mammalian memory formation allows a rationale for how the well-established role for synapse-specific plasticity in these phenomena (e.g. LTP) can interdigitate with the emerging discovery of epigenetically driven genome-wide state changes for the neuron being involved in memory – cell-wide homeostatic plasticity being the bridge between these two seemingly disparate cellular mechanisms.
Are there any data supporting this supposition at the in vivo level? Given the recently discovered capacity of DNMT inhibition to regulate homeostatic plasticity in vitro, one can now go back and re-consider existing literature showing effects of these same agents on memory stability and hippocampal circuit function in vivo. In other words, we now realize that DNMT inhibition has effects on homeostatic plasticity, so what prior data are available assessing the effects of this means of disrupting homoeostatic plasticity on memory formation and storage in vivo? And can the documented effects of DNMT inhibition on memory consolidation and stabilization be rationalized as potentially being due to disruption of homeostatic plasticity? In this final section I will review findings indicating that DNMT inhibitors disrupt stabilization of hippocampal circuit function in spatial memory formation and that DNMT inhibition destabilizes memory storage in the cortex, and place these observations into a context of potentially being due to disruption of homeostatic plasticity.
DNA methylation controls hippocampal circuit stabilization and cortical memory storage in vivo
An understanding of the in vivo role of homeostatic plasticity mechanisms, such as synaptic scaling, has been hampered by experimenters not having tools available to them to manipulate synaptic homeostasis. However, the results described above clearly illustrate that manipulating the active DNA methylation/demethylation pathways perturbs synaptic scaling, synaptic homeostasis, and glutamate receptor trafficking. Thus, it is now apparent (in retrospect) that acute application of DNMT inhibitors causes alterations in synaptic scaling. This allows a re-interpretation of previously published results pharmacologically or genetically perturbing the DNA methylome in vivo in the light of their effects on neuronal synaptic homeostasis. In other words, it is now clear that previous experiments wherein DNMT inhibitors were acutely administered in vivo were experiments wherein neuronal homeostatic plasticity mechanisms were being disrupted. With this in mind, what then are the effects of DNMT inhibition on memory storage and hippocampal circuit stabilization in the behaving animal?
DNA methylation controls the storage of remote memory in the anterior cingulate cortex
Work from several laboratories, including those of Frankland and colleagues and Silva and colleagues, have demonstrated that remote, i.e. very long-lasting, contextual memories in rodents are consolidated and stored in the anterior cingulate cortex (Frankland et al. 2004, Restivo et al. 2009, Wang et al. 2009). It is intriguing to consider that lasting changes in DNA methylation might contribute to stabilization of remote memory in the cortex, as has been previously speculated upon by several authors (Miller et al. 2010, Levenson & Sweatt 2005, Crick 1984, Holliday 1999, Jiang et al. 2008, Sweatt 2009, Graff & Mansuy 2008). Relevant prior studies indicated that DNA methylation contributes to memory maintenance in the cortex (Miller et al. 2010), specifically testing the hypothesis that DNA methylation contributes to regulating long-term associative fear conditioning memory stability in the cortex. In addition, while this manuscript was in final revision, an exciting and exceptionally thorough new publication from the Fischer and Bonn groups (Halder et al. 2016) directly demonstrated lasting memory-associated changes in DNA methylation in the cortex. This comprehensive genome-wide assessment strongly supports the hypothesis that DNA cytosine methylation contributes to long-term memory stabilization and storage in vivo.
These prior experiments by Miller et al. and the Bonn/Fischer groups investigated the dorsomedial prefrontal cortex, a brain region known to support long-lasting memories (Frankland et al. 2004, Restivo et al. 2009, Wang et al. 2009). Contextual fear conditioning in rodents results in a memory that persists for many months, during which time the associative memory transitions from “recent” to “remote”. This change is thought to represent system consolidation, in which the memory shifts from the hippocampus (HPC) to a long-term dependence on the dorsomedial prefrontal cortex (dmPFC).
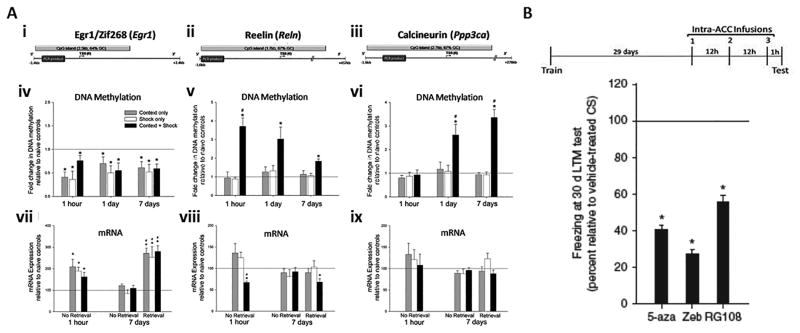
In the following few paragraphs, I will discuss the results of the Miller et al. experiments in some detail, because these investigators tested the effects of DNMT inhibitors on memory stability, apropos to the main thrust of this review. To begin characterizing the role of DNA methylation in the dmPFC (anterior cingulate [ACC] and prelimbic cortices) following hippocampus-dependent learning, Miller et al. examined the pattern of methylation in the promoter of three memory-associated genes with large, CG-rich CpG islands (Egr1/zif268, reelin and calcineurin;) at 1 hour, 1 day and 7 days after training animals for contextual fear conditioning (Miller et al. 2010)(Figure 4A). Importantly, in one specific group of animals, no retrieval test was given before sacrificing them, so that any molecular changes could be assumed to be due to the earlier training.
Figure 4. Persisting changes in DNA methylation support long-term memory stabilization in the dorsomedial prefrontal cortex.
(A) Both DNA methylation and gene transcription change in the dmPFC, in a time-dependent fashion, following training for contextual fear conditioning. (i-iii), location of promoter CpG islands analyzed in mDIP assay for Egr1/Zif268 (i), reelin (ii) and calcineurin (iii). (iv) Relative to naïve controls, all treatment groups showed demethylation of Egr1/zif268 at all time points (1h: F(3, 18) = 22.99, P ≤ 0.001; 1 day: F(3, 19) = 14.99, P ≤ 0.001; and 7 days post-training: F(3, 25) = 15.50, P ≤ 0.001). * post hoc comparisons, P ≤ 0.001. (v) Relative to all treatment groups, reelin's promoter is hypermethylated in trained animals at all time points (1h: F(3, 18) = 29.05, P ≤ 0.001; 1 day: F(3, 19) = 6.63, P ≤ 0.005; and 7 days post-training: F(3, 25) = 10.58, P ≤ 0.001). * post hoc comparisons, P ≤ 0.05. 7 days post-training, reelin's hypermethylation is significantly less than at 1 hour (F(2, 20) = 6.36, P ≤ 0.01; #). (vi) Relative to all treatment groups, calcineurin's promoter is hypermethylated in trained animals 24 hours and 7 days post-training (1h: F(3, 18) = 0.27, P ≥ 0.05; 1 day: F(3, 19) = 5.73, P ≤ 0.01; and 7 days post-training: F(3, 25) = 33.52, P ≤ 0.001). * post hoc comparisons, P ≤ 0.05. 1 and 7 days post-training, calcineurin's hypermethylation is significantly greater than at 1 hour (F(2, 20) = 13.96, P ≤ 0.01; #). (vii) Egr1/zif268 transcript is elevated in all treatment groups 1 hour after training (relative to naïve controls—context: t4 = 3.11, shock: t4 = 5.10, context + shock: t4 = 3.02; * P ≤ 0.05) and following a retrieval test 7 days after training (relative to naïve controls—context: t4 = 6.95, shock: t4 = 3.12, context + shock: t6 = 6.59; * P ≤ 0.05) (relative to 7 day no retrieval groups—F(5, 27) = 11.97, # post hoc comparisons P ≤ 0.005). (viii) Relative to all treatment groups, reelin transcript is significantly lower in trained animals 1 hour after training (relative to naïve controls—t4 = -9.53, * P ≤ 0.005; relative to context and shock controls—F(2, 12) = 6.14, # P ≤ 0.05). Following a retrieval test 7 days after training, trained animals' reelin transcript is significantly lower than naïve controls (t6 = -4.00, * P ≤ 0.01). (ix) Calcineurin transcript is equivalent across all groups. N's = 4-8 animals per group. Figure and legend adapted from (Miller et al. 2010). Please see (Miller et al. 2010) for additional details.
In these studies, Miller et al. observed that the immediate early gene Egr1/zif268's promoter was demethylated in all groups (context only, shock only and context plus shock trained animals), at all time points (Miller et al. 2010)(Figure 4Aiv). Thus, a variety of environmental stimuli (exploration of a novel environment, mild foot shock and associative training) are capable of producing a lasting change in the methylation state of Egr1/zif268 in the dmPFC. In addition, Egr1/zif268 transcript was elevated above naïve control levels in all animals (Figure 4Avii), except those sacrificed without a retrieval test 7 days after training. This result is consistent with the gene's pattern of demethylation at both the one-hour and seven-day time points, the dmPFC's response to novelty and learning, and the rapid reflection of cellular activation provided by immediate early genes (IEGs). The promoter region of reelin, a positive regulator of memory (Weeber et al. 2002, Beffert et al. 2005), was hypermethylated in trained animals within an hour of training and remained methylated for at least 7 days, though levels shifted toward controls at the later time points (Figure 4Av). In concordance with the increase in transcription-repressing cytosine methylation, levels of reelin transcript were reduced in trained animals one hour after training and one hour after a 7 day post-training retrieval test (Figure 4Aviii). Methylation of the phosphatase and memory suppressor (Malleret et al. 2001), calcineurin, was unchanged in the prefrontal cortex an hour after training. However, the gene's promoter became hypermethylated within 1 day of training and continued to increase over the next 7 days (Figure 4Avi), though transcript levels were equivalent to controls (Figure 4Aix).
To determine if the lasting cortical methylation is truly a reflection of associative learning, Miller et al. gave animals pre-training injections of the NMDA receptor antagonist MK-801. A subgroup of animals, tested 7 days post-training for contextual fear memory, confirmed the ability of NMDA receptor antagonism to interfere with acquisition of a fear memory (Benvenga & Spaulding 1988). MK-801 also prevented the 7 day reelin and calcineurin hypermethylation, without affecting Egr1/zif268, providing further support that the reelin and calcineurin hypermethylation is in specific response to associative environmental signals. These data are in agreement with the hypothesis that gene-specific DNA methylation occurs in response to associative learning, and further demonstrates that this process is engaged in multiple brain regions.
In a 2004 study, Frankland et al. investigated what effect pharmacologically inactivating the ACC, a subregion of the dmPFC, would have on fear memory retrieval at various post-training time points (Frankland et al. 2004). Interestingly, intra-ACC infusions of lidocaine at both 18 and 36 days (remote memory), but not 1 or 3 days post-training (recent memory), interfered with retrieval. This suggests that system consolidation of contextual fear memory occurs between 3 and 18 days and further suggests that the cortical DNA methylation events Miller et al. observed during the first week post-training are appropriately timed to participate in the initial laying down of a memory trace in the cortex. Thus, to further characterize the source of learning-induced cortical methylation, Miller et al. infused the NMDA receptor antagonist APV directly into the HPC (CA1) immediately before training (Sanders & Fanselow 2003). APV not only interfered with learning, but also blocked the reelin and calcineurin methylation present in the dmPFC 7 days after training, indicating that a single hippocampus-dependent learning experience is sufficient to drive lasting, gene-specific DNA hypermethylation in the cortex. These findings are in agreement with the system consolidation view of contextual fear memory, and present a tangible marker (DNA methylation) that can be used for a learning-induced dialogue between the HPC and dmPFC.
In order to support memory persistence, DNA methylation in the cortex would need to be long-lasting. Therefore, in an additional experiment, Miller et al. examined the persistence of a fear memory and the methylation status of these genes 30 days after training. Increased methylation of calcineurin's promoter in trained animals was observed at this later time point, as was a preservation of decreased zif268 methylation (Miller et al. 2010). Reelin, however, exhibited a declining pattern of methylation, such that it was equal to controls by 30 days. These results indicate that a single hippocampus-dependent learning event epigenetically marks genes in the prefrontal cortex for a minimum of 30 days, well into the time period that memories are thought to become dependent upon this region. Miller et al. also examined transcript levels of these three genes following a one month post-training retrieval test. Memory retrieval-associated Calcineurin mRNA was specifically reduced in trained animals, while Egr1/zif268 transcription increased in all groups. These observations are in good agreement with the alterations in DNA methylation that were observed at these gene sites.
Is DNA methylation in the dmPFC necessary for the maintenance of a remote memory in vivo? To answer this question, Miller et al. examined the effect on memory of inhibiting DNMTs 30 days after training. In these experiments, beginning on post-training day 29, animals received an intra-ACC infusion of the DNMT inhibitor (DNMTi) 5-azadeoxycytidine (5-aza) every 12 hours for a total of three infusions (Fig. 10a). DNMTi animals failed to display normal fear memory in the retrieval test given one hour after the final infusion (Figure 4B), supporting the idea that methylation in the ACC is critical to the support of remote memory.
As discussed in the introduction to this commentary, the discovery that DNMT inhibition both disrupts normal synaptic scaling and is by itself capable of driving multiplicative changes in synaptic homeostasis in an activity-dependent fashion allows a renewed assessment of these prior results using DNMT inhibition to probe cortical memory formation and stabilization. These results from in vivo behavioral studies in the cortex demonstrate post-consolidation effects of DNMTi infusion on memory stability. This observation is not readily explained by effects on Hebbian plasticity, by virtue of the fact that the memory has already been consolidated at the 30-day time point. However, disrupting homeostatic mechanisms can readily explain this effect, implying that the previously stable Hebbian plasticity degrades due to a post-hoc loss of multiplicative synaptic scaling after DNMT inhibitor infusion. This is especially appealing considering that prior modeling and neurophysiologic studies have demonstrated a necessity for homeostatic synaptic plasticity to confer memory stabilization for neurons within functional memory circuits (Abbott & Nelson 2000, Cooper & Bear 2012, Turrigiano & Nelson 2000, Vitureira & Goda 2013, Nelson & Turrigiano 2008, Kim & Tsien 2008, Turrigiano et al. 1998, Ibata et al. 2008, Soares et al. 2013, Wierenga et al. 2005, Qiu et al. 2012, Wang et al. 2012, Gong et al. 2007, Correa et al. 2012, Zhong et al. 2012, Blackman et al. 2012, Baker-Andresen et al. 2013, Nelson et al. 2008, Turrigiano & Nelson 2004, Keck et al. 2013). Moreover, this concept addresses the conundrum of how cell-wide changes (driven by epigenetic marks) can be participating in memory storage in the face of the necessity of a role for synapse specificity in memory circuits. Perhaps most importantly, these studies suggest that one crucial component of the engram is DNA methylation-dependent stabilization of neuronal synaptic weights by multiplicative scaling mechanisms in vivo.
DNA methylation controls hippocampal place field stability in vivo
While it is not fully known how deficits in hippocampal plasticity lead to memory deficits in the behaving animal, prior results have demonstrated that application of a variety of DNMT inhibitors, and conditional deletion of the DNMT 1 and 3A genes, leads to deficits in hippocampal LTP and deficits in hippocampus-dependent long-term contextual learning (Feng et al. 2010, Miller et al. 2010, Miller & Sweatt 2007, Miller et al. 2008, Lubin et al. 2008, Lubin & Sweatt 2007, Roth et al. 2009, Zovkic et al. 2014, Levenson et al. 2006). In a recent series of studies, Roth et al. investigated the capacity of DNA methylation to regulate hippocampal circuit function in behaving animals, and found that hippocampal place cells are able to form place fields in a normal fashion in the presence of DNMT inhibition (Roth et al. 2015). So, it appears that the initial (presumably Hebbian) mechanisms that drive place field establishment are operating normally in the hippocampus in the presence of DNMT inhibitors. However, Roth et al. observed in their experiments that the stability of the place field degrades with DNMT inhibition. This could be due to DNMT inhibitors disrupting late-phase LTP, which is known to be transcription dependent and is disrupted by DNMTi. However, the observation may be equally well explained as being due to a mechanism such as disrupted homeostatic scaling with DNMT inhibition and the resultant degradation of place field fidelity in the face of continued hippocampal circuit operation post-learning.
These recent studies took a systems neuroscience approach by investigating the capacity of DNA methylation to control the formation and stabilization of hippocampal place cell firing patterns (Barnes et al. 1997, Mehta et al. 1997, Shen et al. 1997, Skaggs 1993) and thus are interesting to consider in light of the recent discovery that DNMT inhibitors disrupt homeostatic synaptic scaling mechanisms. Place cells are studied, in part, as an in vivo readout of hippocampal circuit function and ongoing hippocampus-dependent cognition in the behaving animal. Place cells in rats maintain environmentally specific short and long-term spatial representations of a familiar environment, and generate new, uniquely located place fields in novel environments (Figure 5, top panels). If DNA methylation-dependent homeostatic scaling mechanisms are involved in maintaining spatial representations, alterations in DNA methylation levels should disrupt place cell firing patterns, evoking instability among place fields across repeated exposures to familiar or similar environments. In addition, DNMT inhibition could disrupt plasticity or pattern separation mechanisms when rats are transitioning to a novel or dissimilar environment. In recent studies, Roth et al. used intracerebroventricular (ICV) infusions to pharmacologically manipulate DNA methylation levels in the CNS (Miller & Sweatt 2007, Lubin et al. 2008, Roth et al. 2015), and then applied in vivo single-unit neurophysiology techniques to record place cell firing patterns in CA1 and CA3 as male rats explored a circular track “maze” in different environments. To assess the stability of place fields, Roth et al. compared spatial correlations of place cell firing rate maps generated during sequential exposures to similar and dissimilar environments over a 3 day period for each control and drug treatment condition.
Figure 5. DNMT inhibition disrupts hippocampal place field stability in vivo. Top panels.
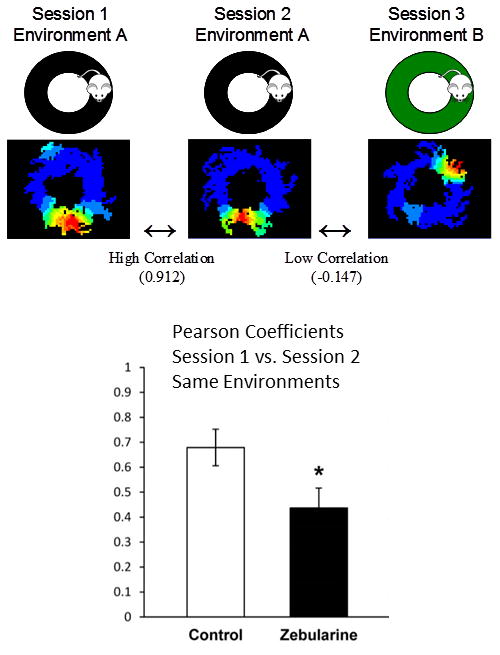
illustrate data obtained using in vivo multi-electrode recording techniques to quantitate place cell firing patterns, and to evaluate consistency of firing of a single hippocampal (dorsal CA1/CA3) place cell. For illustrative purposes data from a single control animal (and a single place cell) are shown. Pseudo-color images illustrate firing rates (blue = low, red = high). In this experiment the animal was placed sequentially onto a round circular maze, with each session separated by 10 minutes. As shown, there is a strong correlation of place cell firing between session 1 and session 2, when the animal is simply re-placed into the same familiar environment. However, when the same animal is placed into a new different environment (session 3), the place cell shows a different place field firing pattern. The dissimilarities in place cell firing can be quantitated using a Pearson correlation coefficient. The bar graph in the Bottom Panel presents Pearson correlation coefficient data testing the effects of the DNMT inhibitor zebularine on place cell firing pattern consistency in this type of place cell experiment (n=5 Long-Evans rats, p<0.05). Zebularine infusion ICV leads to a significant decrease in Pearson correlation coefficients for animals when they are re-placed into the familiar environment. These data are consistent with the idea that DNMT inhibition leads to a destabilization of place cell firing patterns. For a more complete description of these results please see (Roth et al. 2015).
Their results, summarized in Figure 5, indicate that DNA methyltransferase inhibition disrupts hippocampal place field stability. These studies quantitated place field firing pattern correlations for single animals, assessing firing patterns when an animal was exposed to an environment (a circular maze), and then re-placed into the circular maze after a 10-minute time gap (Figure 5). Zebularine infusions led to significant decreases (p<0.05) in place cell firing pattern consistency between one maze exposure to the next (Figure 5, lower panel). These results provide support for the hypothesis that DNA methylation is involved in maintaining temporal constancy in the output of hippocampal circuit function, specifically, spatial representation as manifested in place field firing patterns. Equally importantly in the context of the present discussion, these data strongly support the hypothesis of a role for methylation-dependent control of homeostatic plasticity mechanisms as a process contributing to stabilization of place cell firing patterns.
Multiple Roles for DNA Methylation in Neural Plasticity
This review and commentary has focused on exciting recent results implicating DNA methylation in regulating homeostatic synaptic scaling, and the ways in which these new findings might be integrated into earlier studies of the effects of DNMT inhibition on memory stability. However, as a final point I also should emphasize that a fairly large number of prior studies have demonstrated the capacity of DNMT inhibition to block learning and memory when the inhibitors are applied before or immediately after training (Miller et al. 2010, Miller et al. 2008, Miller & Sweatt 2007, Lubin et al. 2008, Lubin & Sweatt 2007). For the purposes of this review, I have focused thus far on studies wherein stabilization or storage of experience-induced functional or behavioral change was assessed. That is because these time-dependent phenomena are particularly intriguing from the point of view of considering DNA methylation-dependent control of homeostatic synaptic scaling. However, it is certainly important to note that cytosine methylation-dependent regulation of a variety of neuronal plasticity mechanisms (with or without attendant disruption of Hebbian mechanisms such as LTP) could also contribute to disruption of initial stages of memory formation and the maintenance of those changes once established.
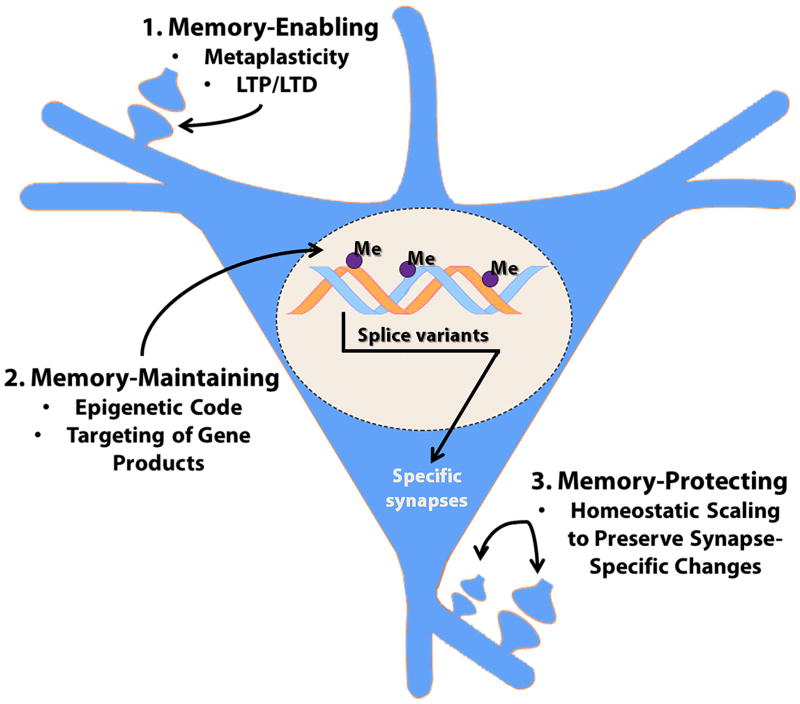
Therefore, in this final section I would like to provide a broader summary of the likelihood that DNA methylation may control multiple sites of neural plasticity, in the context of memory formation (i.e. learning), storage, and stabilization (see Figure 6). In broad overview, DNA methylation might contribute to memory by any one of three mechanisms: by being 1) memory-enabling, 2) memory-maintaining, or 3) memory-protecting. Memory-enabling mechanisms would include epigenetic regulation of phenomena such as metaplasticity, wherein methylation might control the set-point for plasticity in the cell, and thereby regulate the overall likelihood of synapse-specific forms of plasticity being induced in that cell. Alternatively, there is good evidence that DNMT inhibition actually blocks LTP induction in hippocampal slices (Levenson et al. 2006), implying that DNA methylation might directly control synapse-specific enhancement of synaptic strength that contributes to the initial establishment of the engram. Memory-maintaining mechanisms would invoke a role for DNA methylation as a specific and persisting molecular component of the engram per se, i.e. that methylation was involved in encoding information in the neuron at the molecular level. For example, persisting changes in cytosine methylation at specific gene loci might participate as part of a methylation code for preserving functional change. To illustrate this idea, site-specific DNA methylation might mark a unique gene subregion and direct the production of a specific splice variant that targets the gene's RNA or protein product to specific synapses or neuronal subregions. For example, one could conceive of this as being part of, or interacting with, a “synaptic tag.” This potential memory-maintaining role for DNA methylation is an open question and is quite speculative at this point, but is important to consider as part of a comprehensive model. The third category, memory-protecting roles of DNA methylation, has been the central theme of this review, focused on the newly discovered role of DNA methylation in controlling homeostatic synaptic scaling. This category of mechanism involves methylation regulating the cell-wide plasticity that is necessary for stabilizing synapse-specific mechanisms over time, contributing to memory storage. As already discussed, this hypothesis of a memory-protecting role for DNA methylation is now garnering direct experimental support.
Figure 6. Potential mechanisms through which DNA methylation might control memory formation and stabilization.
This diagram summarizes the three main categories of mechanisms whereby cytosine methylation might contribute to memories being established and maintained, termed: 1. Memory-Enabling, 2. Memory-Maintaining, and 3. Memory-Protecting. Please see the main text for further discussion.
Summary and Closing Comments
The advent of sophisticated molecular, genetic and cellular techniques has lent itself to a relatively deep understanding of how memories are initially formed. In stark contrast, however, is our limited understanding of how these same memories are maintained by the brain (Dudai 2004, Medina et al. 2008). A variety of recent studies have used contextual fear conditioning, a form of hippocampal associative learning, to explore the hypothesis that changes in the cortex support long-lasting memories (Frankland et al. 2004, Restivo et al. 2009, Wang et al. 2009). These recent studies have indicated that contextual fear conditioning in rodents results in a memory that persists for many months, during which time the associative memory transitions from “recent” to “remote”. This is an example of systems consolidation, in which the memory shifts from the hippocampus to a long-term dependence on the dorsomedial prefrontal cortex. However, the cellular mechanisms underlying remote memory stabilization for enduring high-fidelity storage have only been sparsely studied. This review considers the hypothesis that DNA methylation, specifically through regulating homeostatic synaptic scaling, participates in stabilization of remote memory after it has been formed and embedded in the cortex.
In terms of neurophysiologic mechanisms, the studies described in this commentary strongly implicate control of homeostatic synaptic scaling as one critical role for DNA methylation regarding its role in memory stabilization. Moreover, these findings indicate that chemical modification of DNA by TET1 dioxygenase is an integral component of the molecular processes underlying long-term behavioral change. These various results support the emerging idea that DNA methylation is dynamically regulated in the adult CNS in response to experience, and that this cellular mechanism is a crucial step in memory formation and stabilization. In other words, that these mechanisms constitute one component of the engram.
In terms of molecular epigenetics, broadly speaking these results, along with many others (Meaney & Szyf 2005b, Meaney & Szyf 2005a, Weaver et al. 2005, Zovkic et al. 2014, Levenson et al. 2006, Ooi & Bestor 2008, Kangaspeska et al. 2008, Metivier et al. 2008, Kaas et al. 2013, Sultan et al. 2012, Sweatt 2001, Guo et al. 2011a, Guo et al. 2011b, Guo et al. 2011c, Ma et al. 2009a, Ma et al. 2009b, Ito et al. 2010, Rudenko et al. 2013, Zhang et al. 2013, Bird 1986, Clark & Nelson 2015), clearly indicate that we can no longer think of DNA methylation as being static: set up during development as a permanent change. Instead, DNA methylation is subject to active and ongoing regulation in the adult nervous system, controlled by behavioral experience. The current gaps in our knowledge include addressing which genes are regulated by active demethylation in the adult CNS, determining whether active demethylation is sufficient to cause alterations in behavior, and identifying the neural mechanisms through which active demethylation acts in order to trigger behavioral change. It is important to note that these findings suggest that memory formation involves both increased methylation at memory suppressor genes and decreased methylation at memory promoting genes. Thus, memory function might be driven by either hypermethylation or hypomethylation.
Overall, these observations suggest that active DNA demethylation is necessary for memory and that active alterations in DNA methylation mediate memory formation and storage in the adult rat hippocampus and cortex. In my opinion, these findings significantly advance our understanding of the core mechanisms underlying the transcriptional regulation necessary for long-term memory formation and stabilization. It is reasonable to hypothesize, based on recent discoveries of DNA methylation controlling synaptic scaling, that regulation of DNA methylation and its attendant control of synaptic scaling and glutamate receptor trafficking is an integral component of long-term memory consolidation.
The findings described in this commentary also have important implications regarding the learning and memory dysfunction associated with Rett syndrome (Qiu et al. 2012, Zhong et al. 2012, Blackman et al. 2012, Baker-Andresen et al. 2013, Nelson et al. 2008, Chahrour et al. 2008, Moretti et al. 2006, Collins et al. 2004, Chapleau et al. 2009, Clark & Nelson 2015). The gene for the methyl-cytosine binding protein MeCP2 is disrupted in Rett syndrome, implicating a priori that mis-reading of DNA methylation marks contributes to Rett pathology. In this context, as has been previously hypothesized (Zhong et al. 2012, Blackman et al. 2012, Nelson et al. 2008), the recent findings of DNA methylation controlling multiplicative synaptic scaling (Meadows et al. 2015, Yu et al. 2015) strongly support the idea of a critical role for disruption of homeostatic synaptic plasticity in Rett syndrome. It is specifically intriguing to consider that disruption of homeostatic plasticity contributes to the age-dependent regression of neural function that is observed in Rett patients, as a loss of homeostatic synaptic stabilization would be likely to drive deleterious long-term time-dependent processes in the CNS.
The principal limitation of studies thus far in the neuroepigenetics field is that they have not determined in a comprehensive fashion how DNA demethylation at the cellular level gets translated into altered neural and behavioral function. Previous results, including the recent work of the Song lab, have implicated methylation-dependent regulation of arc transcription as one specific contributing mechanism (Yu et al. 2015, Chowdhury et al. 2006, Rial Verde et al. 2006, Shepherd et al. 2006, Guzowski et al. 1999). However, it is difficult to generate a comprehensive molecular model to tie specific changes at a single gene (such as arc) to complex multicellular, multicomponent processes like synaptic scaling, memory stabilization, and behavioral memory at this point, because of the current limitations in our understanding of the means by which even alteration of a single gene transcript is transduced into functional synaptic changes. A full understanding of how transcriptional changes driven by DNA demethylation gets transformed into functional changes in the cell and synapse will require further investigation in the future.
Nevertheless, the studies described in this review serve as a foundation for formulating future specific hypotheses concerning how DNA demethylation might control persisting changes in synaptic and neural circuit function at the molecular level, and ultimately understanding how transient or persisting changes in DNA methylation manifest themselves in altered neuronal function. They also serve as a set of first steps toward demonstrating how active DNA demethylation is a signal utilized by the brain to form and preserve transcription-dependent long-term and remote memories, specifically that cytosine methylation/demethylation controls homeostatic neural plasticity, a finding which has important implications for the question of how memories are maintained in vivo.
Acknowledgments
The author thanks Hongjun Song, John Hablitz, Garrett Kaas, Courtney Miller, Eric Roth, Tania Roth and Jarrod Meadows for many helpful discussions and for sharing data figures of their work. I also thank Kimberly Hawkins, Sarah Strange, and Cristin Gavin for help in preparing the figures and manuscript. The comments of the two anonymous reviewers were particularly helpful in revising and clarifying the manuscript for final publication. All data figures and corresponding legends were adapted directly from previously published work. I apologize to the many authors whose primary work was not directly cited, owing to limitations of space. Research in the author's laboratory is supported by funds from the NINDS, NIMH, NINR, DARPA, the Pitt-Hopkins Syndrome Foundation, the Civitan International, and the Evelyn F. McKnight Brain Research Foundation.
List of abbreviations
- CNS
central nervous system
- LTP
long-term potentiation
- DNMT
DNA methyltransferase
- TET
ten-eleven translocation
- MAPK
mitogen activated protein kinase
- 5hmC
5-hydroxymethylcytosine
- MeC
methyl-cytosine
- GADD45
growth and DNA damage 45
- LTD
long-term depression
- GluR
glutamate receptor
- IP
intrinsic plasticity
- TTX
tetrodotoxin
- mEPSC
miniature excitatory postsynaptic currents
- BER
base excision repair
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- SK channel
Small conductance calcium-activated potassium channel
- HCN channel
Hyperpolarization-activated cyclic nucleotide-gated channel
- HPC
hippocampus
- dmPFC
dorsomedial prefrontal cortex
- ACC
anterior cingulate cortex
- IEG
immediate early gene
- NMDA
N-methyl-D-aspartate
- ICV
intracerebroventricular
References
- Abbott LF, Nelson SB. Synaptic plasticity: taming the beast. Nat Neurosci. 2000;3(Suppl):1178–1183. doi: 10.1038/81453. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Barnes CA, Suster MS, Shen J, McNaughton BL. Multistability of cognitive maps in the hippocampus of old rats. Nature. 1997;388:272–275. doi: 10.1038/40859. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Benvenga MJ, Spaulding TC. Amnesic effect of the novel anticonvulsant MK-801. Pharmacol Biochem Behav. 1988;30:205–207. doi: 10.1016/0091-3057(88)90445-5. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Blackman MP, Djukic B, Nelson SB, Turrigiano GG. A critical and cell-autonomous role for MeCP2 in synaptic scaling up. J Neurosci. 2012;32:13529–13536. doi: 10.1523/JNEUROSCI.3077-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapleau CA, Calfa GD, Lane MC, Albertson AJ, Larimore JL, Kudo S, Armstrong DL, Percy AK, Pozzo-Miller L. Dendritic spine pathologies in hippocampal pyramidal neurons from Rett syndrome brain and after expression of Rett-associated MECP2 mutations. Neurobiol Dis. 2009;35:219–233. doi: 10.1016/j.nbd.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Nelson SB. Synapse and genome: An elusive tete-a-tete. Sci Signal. 2015;8:pe2. doi: 10.1126/scisignal.aad2441. [DOI] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Cooper LN, Bear MF. The BCM theory of synapse modification at 30: interaction of theory with experiment. Nat Rev Neurosci. 2012;13:798–810. doi: 10.1038/nrn3353. [DOI] [PubMed] [Google Scholar]
- Correa SA, Hunter CJ, Palygin O, et al. MSK1 regulates homeostatic and experience-dependent synaptic plasticity. J Neurosci. 2012;32:13039–13051. doi: 10.1523/JNEUROSCI.0930-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD. DNA methylation regulates associative reward learning. Nat Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai NS, Rutherford LC, Turrigiano GG. Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nat Neurosci. 1999;2:515–520. doi: 10.1038/9165. [DOI] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, Hoffman B, Liebermann DA. Conserved DNA methylation in Gadd45a(-/-) mice. Epigenetics. 2009;4:98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science. 2004;304:881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Gong B, Wang H, Gu S, Heximer SP, Zhuo M. Genetic evidence for the requirement of adenylyl cyclase 1 in synaptic scaling of forebrain cortical neurons. Eur J Neurosci. 2007;26:275–288. doi: 10.1111/j.1460-9568.2007.05669.x. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011a;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011b;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011c;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Karlsson MC, Meadows JP, Gavin CF, Hablitz JJ, Sweatt JD. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19:102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- Holliday R. Is there an epigenetic component in long-term memory? J Theor Biol. 1999;200:339–341. doi: 10.1006/jtbi.1999.0995. [DOI] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Langley B, Lubin FD, et al. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Keck T, Keller GB, Jacobsen RI, Eysel UT, Bonhoeffer T, Hubener M. Synaptic scaling and homeostatic plasticity in the mouse visual cortex in vivo. Neuron. 2013;80:327–334. doi: 10.1016/j.neuron.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tsien RW. Synapse-specific adaptations to inactivity in hippocampal circuits achieve homeostatic gain control while dampening network reverberation. Neuron. 2008;58:925–937. doi: 10.1016/j.neuron.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009a;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009b;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, et al. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- McCormack SG, Stornetta RL, Zhu JJ. Synaptic AMPA receptor exchange maintains bidirectional plasticity. Neuron. 2006;50:75–88. doi: 10.1016/j.neuron.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, Hablitz JJ, Sweatt JD. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci Signal. 2015;8:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005a;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005b;28:456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Medina JH, Bekinschtein P, Cammarota M, Izquierdo I. Do memories consolidate to persist or do they persist to consolidate? Behav Brain Res. 2008;192:61–69. doi: 10.1016/j.bbr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Mehta MR, Barnes CA, McNaughton BL. Experience-dependent, asymmetric expansion of hippocampal place fields. Proc Natl Acad Sci U S A. 1997;94:8918–8921. doi: 10.1073/pnas.94.16.8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Sylwestrak EL, Lieberman DN, Zhang Y, Liu XY, Ghosh A. The Rett syndrome protein MeCP2 regulates synaptic scaling. J Neurosci. 2012;32:989–994. doi: 10.1523/JNEUROSCI.0175-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo L, Vetere G, Bontempi B, Ammassari-Teule M. The formation of recent and remote memory is associated with time-dependent formation of dendritic spines in the hippocampus and anterior cingulate cortex. J Neurosci. 2009;29:8206–8214. doi: 10.1523/JNEUROSCI.0966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Sweatt JD. A biochemical blueprint for long-term memory. Learn Mem. 1999;6:381–388. [PMC free article] [PubMed] [Google Scholar]
- Roth ED, Roth TL, Money KM, SenGupta S, Eason DE, Sweatt JD. DNA methylation regulates neurophysiological spatial representation in memory formation. Neuroepigenetics. 2015;2:1–8. doi: 10.1016/j.nepig.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MJ, Fanselow MS. Pre-training prevents context fear conditioning deficits produced by hippocampal NMDA receptor blockade. Neurobiol Learn Mem. 2003;80:123–129. doi: 10.1016/s1074-7427(03)00040-6. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Shen J, Barnes CA, McNaughton BL, Skaggs WE, Weaver KL. The effect of aging on experience-dependent plasticity of hippocampal place cells. J Neurosci. 1997;17:6769–6782. doi: 10.1523/JNEUROSCI.17-17-06769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Gothard KM, Markus EJ. An information-theoretic approach to deciphering the hippocampal code. In: Hanson JDCSJ, Giles CL, editors. Advances in Neural Information Processing Systems 5. Morgan Kaufman; San Mateo, CA: 1993. pp. 1030–1037. [Google Scholar]
- Soares C, Lee KF, Nassrallah W, Beique JC. Differential subcellular targeting of glutamate receptor subtypes during homeostatic synaptic plasticity. J Neurosci. 2013;33:13547–13559. doi: 10.1523/JNEUROSCI.1873-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Protooncogenes subserve memory formation in the adult CNS. Neuron. 2001;31:671–674. doi: 10.1016/s0896-6273(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Hebb and homeostasis in neuronal plasticity. Curr Opin Neurobiol. 2000;10:358–364. doi: 10.1016/s0959-4388(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Vitureira N, Goda Y. Cell biology in neuroscience: the interplay between Hebbian and homeostatic synaptic plasticity. J Cell Biol. 2013;203:175–186. doi: 10.1083/jcb.201306030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Gilbert J, Man HY. AMPA receptor trafficking in homeostatic synaptic plasticity: functional molecules and signaling cascades. Neural Plast. 2012;2012:825364. doi: 10.1155/2012/825364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Teixeira CM, Wheeler AL, Frankland PW. The precision of remote context memories does not require the hippocampus. Nat Neurosci. 2009;12:253–255. doi: 10.1038/nn.2263. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- Wierenga CJ, Ibata K, Turrigiano GG. Postsynaptic expression of homeostatic plasticity at neocortical synapses. J Neurosci. 2005;25:2895–2905. doi: 10.1523/JNEUROSCI.5217-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun YE. Reversing DNA methylation: new insights from neuronal activity-induced Gadd45b in adult neurogenesis. Sci Signal. 2009;2:pe17. doi: 10.1126/scisignal.264pe17. [DOI] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Linden DJ. The other side of the engram: experience-driven changes in neuronal intrinsic excitability. Nat Rev Neurosci. 2003;4:885–900. doi: 10.1038/nrn1248. [DOI] [PubMed] [Google Scholar]
- Zhong X, Li H, Chang Q. MeCP2 phosphorylation is required for modulating synaptic scaling through mGluR5. J Neurosci. 2012;32:12841–12847. doi: 10.1523/JNEUROSCI.2784-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature. 2014;515:582–586. doi: 10.1038/nature13707. [DOI] [PMC free article] [PubMed] [Google Scholar]