Abstract
Gold nanoparticles have been labeled with various radionuclides and extensively explored for single photon emission computed tomography (SPECT) in the context of cancer diagnosis. The stability of most radiolabels, however, still needs to be improved for accurate detection of cancer biomarkers and thereby monitoring of tumor progression and metastasis. Here we report the first synthesis of Au nanoparticles doped with 199Au atoms for targeted SPECT tumor imaging in a mouse triple negative breast cancer (TNBC) model. By directly incorporating 199Au atoms into the crystal lattice of each Au nanoparticle, the stability of the radiolabel could be ensured. The synthetic procedure also allowed for a precise control over both the radiochemistry and particle size. When conjugated with D-Ala1-peptide T-amide, the Au nanoparticles doped with 199Au atoms could serve as a CCR5-targeted nanoprobe for the sensitive and specific detection of both TNBC and its metastasis in a mouse tumor model.
Keywords: Gold nanoparticles, single photon emission computed tomography, breast cancer, CCR5 targeting, radiolabeling
Graphical Abstract
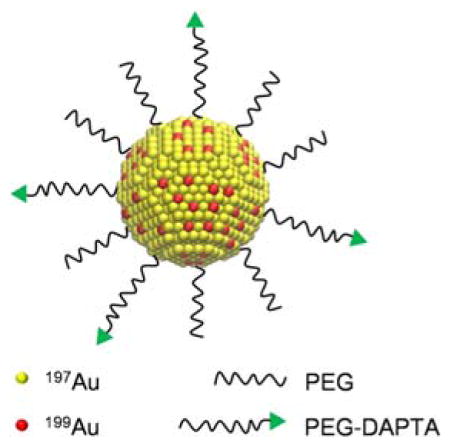
Radioactive Au nanoparticles doped with 199Au atoms were conjugated with D-Ala1-peptide T-amide (DAPTA) to serve as a CCR5-targeted nanoprobe for the sensitive and specific detection of both triple negative breast cancer and its metastasis in a mouse model by SPECT.
1. Introduction
Gold nanoparticles (AuNPs) have been extensively explored for biomedical applications owing to their unique properties, including bio-inertness, controllable surface chemistry, straightforward synthesis, and tunable optical scattering/absorption.[1] Among various demonstrations, their use as a platform material for cancer diagnosis has been of particular interest. To this end, AuNPs have been labeled with a number of radionuclides for use with single photon emission computed tomography (SPECT) or positron emission tomography (PET), where only a trace amount of the probe needs to be administrated in vivo.[2] Typically, the AuNPs are labeled with a SPECT or PET radionuclide through the use of a macrocyclic chelating ligand. Chelator-free strategies such as nuclear bombardment, radiochemical synthesis, and post-synthetic radiolabeling, have also been reported.[3] Despite its popularity and versatility, the stability of a chelator-based radiolabel still needs to be greatly improved before it can be applied to image-guided cancer treatment, which relies on the accurate determination of tumor-related biomarkers for staging the disease, designing the treatment plan, monitoring the treatment response, and ultimately improving the patient care. For this particular application, it is of critical importance to ensure full integration of radionuclide and nanoparticle in the dynamic in vivo environment.
Of the radioisotopes of Au, both 198Au and 199Au are medically useful because of their desirable nuclear properties.[4] Although 198Au has been used for both SPECT and Cerenkov luminescence imaging,[3m, 5] its abundant moderate-energy β-emission and high-energy γ emission make it more suitable for therapeutic applications rather than nuclear imaging.[3i, 6] In comparison, 199Au (t1/2 = 3.2 day) can be produced carrier-free and with high specific activity by irradiating enriched 198Pt with neutrons. A recent study involving graphene oxide radiolabeled with a mixture of 199Au and 198Au ions through a chelating ligand showed great potential for SPECT tumor imaging.[7] However, both the stability of the chelated radiolabels and the image quality need to be further improved.
In comparison with other imaging modalities, radionuclide-based molecular imaging probes are more favored for evaluating breast cancer (BC) progression and metastasis due to the high sensitivity and minimal pharmacodynamics effect in vivo.[8] However, it is still an unmet clinical need to develop a molecular probe, especially a multivalent, nanoparticle-based agent, for detecting the prognostic biomarkers and thus monitoring the progression and metastasis of BC with improved specificity and sensitivity.[8a, 9] Among various biomarkers reported, the chemokine receptor CCR5 has emerged as a promising prognostic biomarker for tracking BC progression in both preclinical and clinical studies, especially for triple negative breast cancer (TNBC).[10] Significantly, the expression of CCR5 is closely associated with TNBC metastasis and has started to show promise as a therapeutic target for dealing with metastasis.[11]
In this work, we have demonstrated the synthesis of AuNPs doped with 199Au atoms using a one-step procedure for biodistribution and SPECT imaging in an orthotopic 4T1 TNBC mouse model. The direct incorporation of 199Au atoms into the crystal lattice of each AuNP ensured the highest possible stability for the radiolabel. The straightforward preparation also allowed for a tight control over the size of the AuNPs, as well as the concentration of 199Au and thus the specific activity of resultant AuNPs, which is critical to trace amount administration with high sensitivity. When conjugated with D-Ala1-peptide T-amide (DAPTA),[12] the targeted probe showed both favorable pharmacokinetics and specific detection of CCR5 in the 4T1 tumor model.
2. Results and Discussion
To precisely control the size and shape of the AuNPs, we employed a seed-mediated procedure for the synthesis of single-crystal Au nanospheres as we reported previously.[13] We introduced the radioactive 199Au3+ into a synthesis during the growth step to minimize the radiation exposure and radiolysis effects. In a typical process, a seed solution containing Au clusters was prepared first by adding a freshly prepared NaBH4 solution into a solution containing cetyltrimethylammonium bromide (CTAB) and HAuCl4 under vigorous magnetic stirring. The reaction mixture was then left undisturbed at room temperature for at least 3 h to ensure the complete decomposition of remaining NaBH4. For the preparation of 5-nm AuNPs, a solution of cetyltrimethylammonium chloride (CTAC) was gently mixed with a solution of HAuCl4, followed by the introduction of ascorbic acid and the cluster solution. The resultant AuNPs were collected by centrifugation. The 5-nm Au NPs could also be grown into larger sizes (e.g., 10 nm) by increasing the amount of HAuCl4 added into the reaction mixture. For 18-nm AuNPs, they were grown from 10-nm AuNPs by following a similar seeded growth procedure. Their UV-vis spectra show a strong absorption peak in the range of 520–530 nm (Figure S1), in agreement with our previous report.[3k]
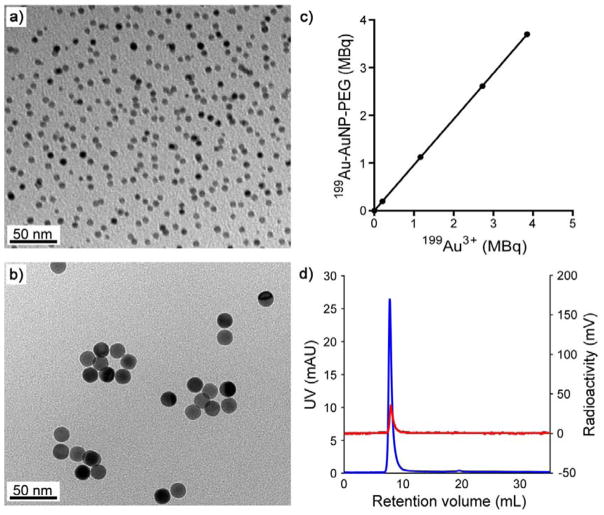
The AuNPs doped with 199Au atoms (199Au-AuNPs) were prepared using the same protocol as described above, except for the introduction of 199Au3+ ions along with the non-radioactive Au3+ ions at a molar ratio (199Au:197Au) of 1:1.65 ×105. The transmission electron microscopy (TEM) images shown in Figure 1, a and b, indicate that the 199Au-AuNPs in both samples had a more or less spherical shape, together with a uniform size distribution: 4.7 ± 1.3 nm and 17.6 ± 0.9 nm. As shown in Figure 1c, the specific activity of the 199Au-AuNPs could be readily controlled by varying the amount of 199Au3+ ions added into the reaction mixture, ensuring trace amount administration for in vivo SPECT imaging. Importantly, due to the direct introduction of 199Au3+ ions into the reaction solution, the incorporation yield could reach a level as high as 96.2 ± 0.2% (n = 8). After purification, the fast protein liquid chromatography (FPLC) only showed one single peak in the UV and radioactivity traces (Figure 1d), respectively, indicating 100% chemical and radiochemical purity. Since 199Au is chemically identical to other Au isotopes, the direct incorporation strategy also warrants the stability needed for accurate measurement in vivo, as we have demonstrated for 198Au-doped Au nanostructures.[5a]
Figure 1.
(a, b) TEM image of the 199Au-doped AuNPs of (a) 5 nm and (b) 18 nm in diameter, respectively, after 90 days of decay. (c) Correlation between the radioactivity of the 199Au3+ precursor used in a synthesis and the resultant 5-nm 199Au-doped AuNPs, whose surfaces were covered by PEG. (d) FPLC analysis of the 5-nm 199Au-AuNP-PEG in both UV (blue) and radioactivity (red) traces.
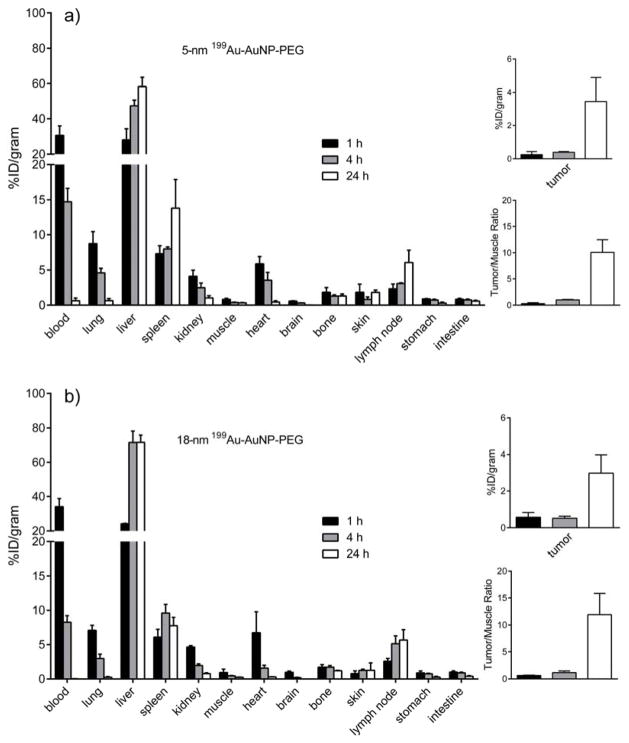
After separation by ultrafiltration, the 199Au-doped AuNPs were PEGylated and then subjected to pharmacokinetic evaluation in a mouse 4T1 tumor model. The PEGylation was carried out with poly(ethylene glycol) methyl ether thiol (HS-PEG5000-OMe, MW 5000) according to our previously reported procedures.[3j, 3k] As shown in Figure 2, the two samples of 199Au-AuNPs showed very similar biodistribution profiles at the three time points. At 1 h post injection, the 5-nm and 18-nm AuNPs showed comparable high blood pool retention (45.2 ± 5.8 %ID/g and 48.0 ± 5.5 %ID/g, respectively, for blood, lung, and heart in total). Because of a smaller size, the clearance of 5-nm AuNPs from blood circulation was ~35% slower than that of the 18-nm counterpart at 4 h post injection. Although the liver accumulation was slightly lower, the spleen uptake was 78% higher. This trend was consistent with what was reported in previous publications.[2a, 3k] As the retention in blood circulation dropped, the tumor accumulation of the AuNPs gradually increased in an opposite trend. At 24 h post injection, the tumor/muscle contrast ratio for the 5-nm AuNPs was 10.1 ± 2.3 (n = 4), comparable to what (11.9 ± 3.9, n = 4) was observed for the 18-nm AuNPs. Again, this trend was in agreement with what was reported in a previous publication.[3k]
Figure 2.
(left panel) Biodistributions of the 199Au-doped AuNPs in 4T1 tumor-bearing mice at 1, 4, and 24 h post injection (n = 4 per group). The AuNPs were 5 nm and 18 nm in diameter, respectively, and their surfaces were covered by PEG chains: a) 5-nm 199Au-AuNP-PEG and b) 18-nm 199Au-AuNP-PEG. (right panel) Plots showing the levels of accumulation of the AuNPs in the tumor relative to muscle at 1, 4, and 24 post injection.
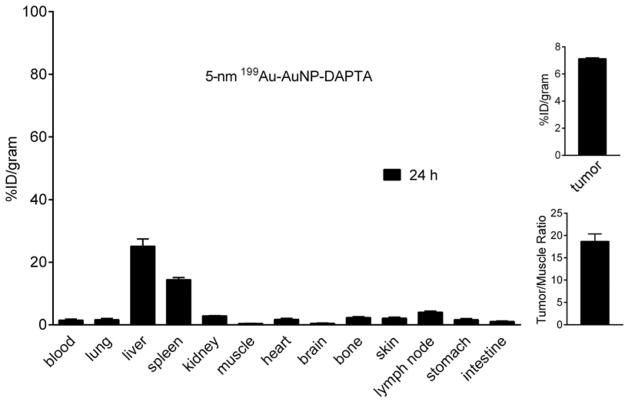
In a prior study, we confirmed the binding specificity of DAPTA (D-A1STTTNYT-NH2) toward the CCR5 receptor in vivo.[12] In the present work, we conjugated orthopyridyl disulfide-polyethylene glycol-succinimidyl valerate (OPSS-PEG5000-SVA, MW ≈ 5000) with DAPTA, and then mixed the product with HS-PEG5000-OMe at a molar ratio of 1:3 to functionalize the surface of the 5-nm 199Au-AuNPs (see Scheme 1). Interestingly, the biodistribution profile of the DAPTA-conjugated AuNPs was substantially improved relative to the non-targeted AuNPs of the same size (Figure 3). The blood pool retention (4.77 ± 0.65 %ID/g) at 24 h post injection was almost three times as high as what was obtained for the non-targeted AuNPs. Although the spleen accumulation was similar, the liver uptake was approximately 60% less than the non-targeted sample. It is clear that the presence of DAPTA on the surface of AuNPs could greatly reduce their uptake by liver and thus affect their biodistribution profile. This observation is consistent with our recent study involving polymeric nanoparticles, in which the presence of DAPTA on the surface was also found to greatly affect the surface charges of the particles, reducing their uptake by liver and altering the biodistribution profile.[12] Importantly, the tumor accumulation (7.13 ± 0.08 %ID/g, n = 4 per group, p < 0.05) was more than doubled, which also led to significantly enhanced tumor/muscle ratio (18.7 ± 1.69, n = 4 per group, p < 0.01), clearly demonstrating the advantage of CCR5-targeted imaging in the 4T1 TNBC mouse model. Given the fact that the tumor uptake of non-targeted 5-nm AuNPs (3.45±1.44 %ID/g) at 24 h was mainly through the non-specific enhanced permeability and retention (EPR) effect, the more than doubled accumulation in tumor for the targeted 199Au-AuNP-DAPTA should be attributed to the binding of DAPTA to CCR5 receptor and thus a better retention effect. There is a great potential to further increase the targeting efficiency by adding more DAPTA peptide to the surface of the nanoparticles.
Scheme 1.
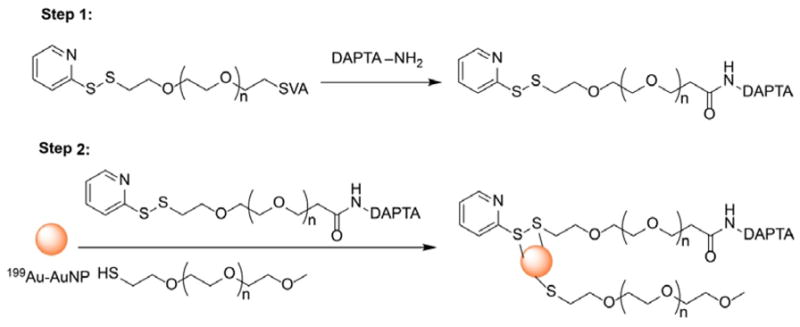
Schematic illustration showing the synthesis of 199Au-AuNP-DAPTA, whose surface was covered by a mixture of PEG-DAPTA and PEG.
Figure 3.
(left panel) Biodistribution of the 5-nm 199Au-AuNP-DAPTA probe in 4T1 tumor-bearing mice at 24 h post injection (n = 4 per group). (right panel) Plots showing the levels of accumulation of the AuNPs in the tumor at 24 post injection.
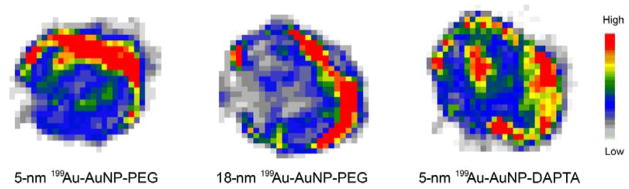
We further assessed the potential of the 5-nm 199Au-AuNP-DAPTA conjugate as a probe for NanoSPECT/CT imaging in a mouse 4T1 TNBC model. As revealed in Figure 4, 199Au-AuNP-DAPTA clearly shows a heterogeneous pattern of penetration and retention inside the tumor. Additionally, the SPECT/CT image shows high liver and spleen accumulation, in consistent with the biodistribution data. Compared to the unsatisfactory resolution of the SPECT image obtained from graphene oxide labeled with a mixture of 199Au and 198Au ions and the potentially high radiation exposure due to the high energy of 198Au,[7] the image acquired using 199Au-AuNP-DAPTA as a probe was significantly improved in quality. Our result clearly demonstrates the suitability of 199Au for SPECT imaging and the potential of 199Au-AuNP-DAPTA for accurately detecting CCR5 in TNBC due to the exceptional stability of the radiolabel and the in vivo targeting specificity.[5a]
Figure 4.
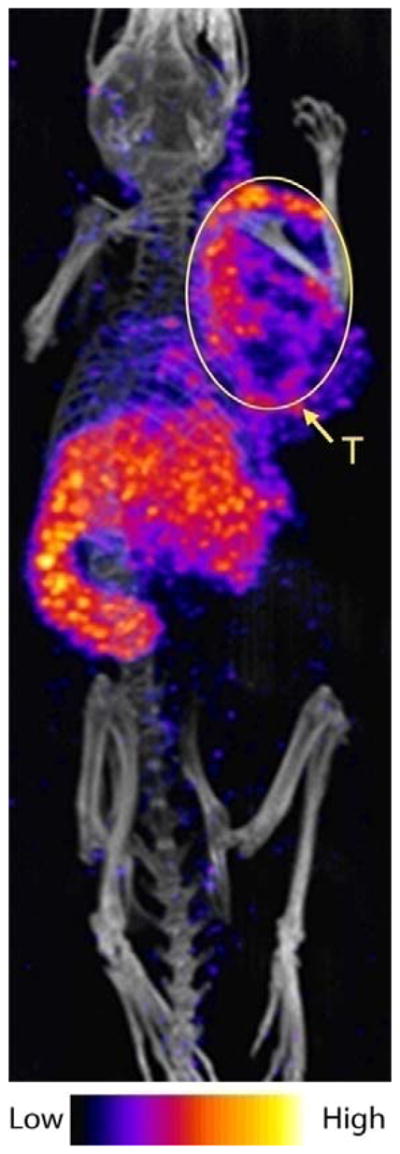
NanoSPECT/CT image of a 4T1 tumor-bearing mouse 24 h post injection of the 5-nm 199Au-AuNP-DAPTA probe (the tumor is labeled by an ellipse in yellow color. T: tumor).
We also performed autoradiography of the mice injected with the three different types of probes (5-nm 199Au-AuNP-PEG, 5-nm 199Au-AuNP-DAPTA, and 18-nm 199Au-AuNP-PEG) to compare their intratumoral distributions in the extracted and sectioned tissues. As indicated by the images in Figure 5, all the probes show heterogeneous distributions, with most of the particles being accumulated around the surface of the tumor. The distribution of the 5-nm 199Au-AuNP-DAPTA was slightly more homogeneous than the other two samples, in agreement with the SPECT images.
Figure 5.
Representative autoradiography showing the distributions of three different types of 199Au-doped probes in the 4T1 tumors at 24 h post injection: 5-nm AuNP-PEG, 18-nm AuNP-PEG, and 5-nm AuNP-DAPTA. All these probes showed a heterogeneous pattern for the intratumoral distribution.
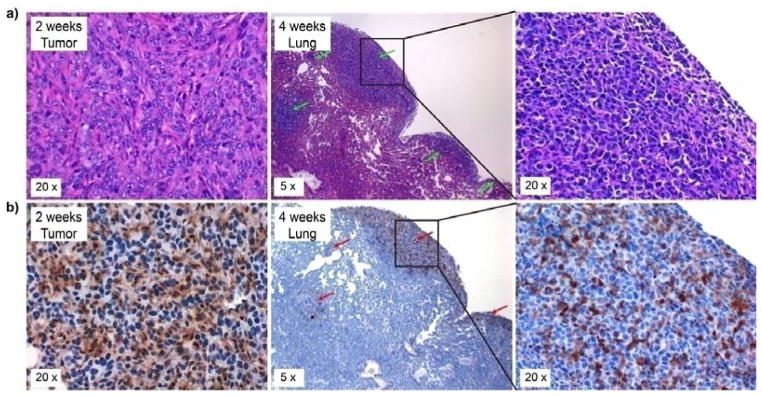
It has been reported that chemokine CCL5 promotes BC aggressiveness and progression mainly through CCR5, which shows significant association between increased expression in tumor cells and advancement of the tumor.[11a, 11b] As shown in Figure 6, CCR5 was up-regulated in the primary tumor cells at two weeks post tumor implant, further supporting the use of 199Au-AuNP-DAPTA for CCR5-targeted molecular imaging. More importantly, at four weeks post-implant, significant metastasis in lung was observed, and the expression of CCR5 was found to be specifically associated with the metastatic tumor cells but not the normal lung tissue. This observation is in agreement with previous reports,[11c, 14] in which CCR5-positive tumor cells were found to display increased invasiveness for metastasis. Taken together, these studies validate the great potential of CCR5 as a specific target for TNBC metastasis imaging and therapy although the current work is mainly focused on the preparation of 199Au-AuNPs for SPECT imaging of primary tumor. Future studies are underway to fully interrogate the potential of the CCR5-targeted probe for sensitive and specific detection of lung metastasis in the 4T1 mouse model.
Figure 6.
(a) H&E and (b) immunohistochemistry of the 4T1 mouse tumor and its lung metastasis at 2 and 4 weeks post tumor implant, showing the up-regulation of CCR5 and its specific expression in lung metastasis (Green arrow: metastasis; Red arrow: CCR5 stained in the brown color).
3. Conclusion
In summary, we have successfully demonstrated the synthesis of 199Au-doped AuNPs with two different sizes and assessed their potential as probes for SPECT imaging of TNBC. The specific activity of the 199Au-AuNPs could be controlled by varying the amount of 199Au3+ precursor added into the reaction system. The direct incorporation of 199Au atoms into the crystal lattice of each AuNP ensured stable radiolabeling for accurate imaging of cancer biomarkers in vivo. Indeed, the radioactive 199Au enabled us to use the AuNPs for high-quality SPECT imaging and longitudinal investigation. When conjugated with DAPTA for CCR5 targeting, the 199Au-AuNPs showed improvement in biodistribution profile, as well as great enhancement in tumor SPECT/CT imaging sensitivity and specificity. Taken together, this study has demonstrated the merit of 199Au-doped Au nanostructures as a platform for oncological imaging and may serve as a point of entry for a broader range of biomedical applications in the pre-clinical or translational setting. Our current effort is focused on the development of 199Au-doped AuNPs with optimized sizes and shapes to reduce their accumulation in both liver and spleen, as well as the improvement in targeting specificity for early and sensitive detection of tumor metastasis.
4. Experimental Section
Chemicals
Gold(III) chloride trihydrate (HAuCl4·3H2O, ≥99.9%), ascorbic acid (AA, ≥99.0%), sodium borohydride (NaBH4, 98%), cetyltrimethylammonium bromide (CTAB, ≥99%), and cetyltrimethylammonium chloride (CTAC, ≥98.0%) were all obtained from Sigma-Aldrich (St. Louis, MO) and used as received. Water with a resistivity of 18.2 MΩ/cm was prepared using an E-Pure filtration system from Barnstead International (Dubuque, IA). Poly(ethylene glycol) methyl ether thiol (HS-PEG5000-OMe, Mw≈5,000) was purchased from Nanocs (Boston, MA) and orthopyridyl disulfide-polyethylene glycol-succinimidyl valerate (OPSS-PEG5000-SVA) was purchased from Laysan Bio (Arab, AL). D-Ala1-peptide T-amide (DAPTA) was customer-synthesized by CPC scientific (Sunnyvale, CA). The H199AuCl4 solution was supplied by the Missouri University Research Reactor (Columbia, MO).
Preparation of Au nanoparticles
Gold nanoparticles (AuNPs) were synthesized using a method based on seed-mediated growth. Specifically, a seed solution was prepared first by adding 300 μL of freshly prepared ice-cold NaBH4 aqueous solution (10 mM) into a mixture of 2.5 mL of aqueous CTAB (200 mM) and 2.5 mL of aqueous HAuCl4 (0.5 mM) under magnetic stirring. The mixturewas then left undisturbed for 3 h to ensure complete decomposition of remaining NaBH4. For the synthesis of AuNPs 5 nm in diameter, 2 mL of aqueous CTAC (200 mM) was mixed with 2 mL of aqueous HAuCl4 (0.5 mM), followed by the addition of 1.5 mL of aqueous AA (100 mM). Finally, 1000 μL of the seed solution (Au clusters) was added and the mixture was magnetically stirred for 1 h. The AuNPs were collected and purified by ultrafiltration (MWCO≈100 K, Amicon, 10,000 g, 5 min). The particles were washed three times with 4 mL of ultrapure H2O.
To synthesize AuNPs of 18 nm in diameter, 1 mL of aqueous CTAC (200 mM) was mixed with 1 mL of H2O. After gentle mixing, 130 μL of aqueous AA (10 mM) and 160 μL of the seed solution (10 nm AuNPs) were introduced. Finally, 2 mL of aqueous HAuCl4 (0.5 mM) was added under magnetic stirring using a syringe pump at a rate of 2 mL/h. After 1 h, the resultant AuNPs were collected by centrifugation (13,200 rpm, 10 min). The particles were washed three times with 1 mL of ultrapure H2O.
Preparation of Au nanoparticles doped with 199Au atoms (199Au-AuNPs)
The 199Au-doped AuNPs were prepared using a protocol similar to what was used for cold AuNPs except for the introduction of 199Au3+ into the system. All the experiments involving radioactive species were conducted behind a lead shield or under appropriate protection according to the guidance from the Radiation Safety Office of Washington University in St. Louis. For 199Au-AuNPs of 5 nm in diameter, 2 mL of aqueous CTAC (200 mM) was mixed with 2 mL of aqueous HAuCl4 (0.5 mM) containing 69.9 MBq of H199AuCl4. After gentle mixing, 1.5 mL of aqueous AA (100 mM) was added. Finally, 1000 μL of seed solution (Au clusters) was added and the solution was magnetically stirred for 1h. The as-obtained AuNPs were purified by ultrafiltrtion (MWCO≈100 K, Amicon, 10,000 g, 5min). The particles were washed three times with 4 mL of ultrapure H2O. To synthesize 199Au-AuNPs of 18nm in diameter, 2 mL of aqueous CTAC (200 mM) was mixed with 2 mL of H2O. After gentle mixing, 260 μL of aqueous AA (10 mM) and 320 μL of seed solution (10 nm AuNPs) were added. Finally, 4 mL of aqueous HAuCl4 (0.5 mM) containing 81.6 MBq of H199AuCl4 was added under stirring using a syringe pump at a rate of 2 mL/h. After stirring for 1 h, the as-prepared 199Au-AuNPs were collected by centrifugation (13,200 rpm, 10 min) and washed three times with 1 mL of ultrapure H2O.
PEGylation of Au nanoparticles
The OPSS-PEG5000-DAPTA molecules were prepared by coupling the primary amine group on DAPTA with the PEG NHS ester derivative of OPSS-PEG5000-SVA. The amine-reactive OPSS-PEG5000-SVA was mixed with DAPTA in PBS (pH 7.0) at a molar ratio of 1:5 and allowed to react at 4 °C overnight. The AuNPs were then incubated with a solution of HS-PEG5000-OMe (AuNP-PEG) or a mixture of OPSS-PEG5000-DAPTA and HS-PEG5000-OMe (1:3) to obtain AuNP-DAPTA. The conjugation was conducted by reacting the AuNPs with PEG molecules at a molar ratio of 1:100,000 (AuNP:PEG). The mixture was shaken at 950 rpm at room temperature overnight. The as-prepared AuNPs were purified by ultrafiltration (MWCO≈100 K, Amicon, 10,000g, 5 min). The AuNPs were washed three times with 4 mL of ultrapure H2O and then dispersed in saline (APP Pharmaceuticals, Schaumburg, IL) for animal study.
For the AuNPs of 18 nm in diameter, PEGylation was conducted by following the same procedure mentioned above. The as-prepared AuNPs were collected by centrifugation (13,200 rpm, 10 min). After conjugation, the AuNPs were washed three times with 1 mL of ultrapure H2O and reconstituted in saline for animal study.
Characterization of Au nanoparticles
The UV-vis spectra were recorded using a Lambda 750 spectrometer (Perkin-Elmer, Wellesley, MA). The size and morphology were examined using a Hitachi HT7700 microscope operated at 120 kV.
Animal biodistribution studies
All animal studies were performed in compliance with guidelines set forth by the NIH Office of Laboratory Animal Welfare and approved by the Washington University Animal Studies Committee. The 4T1 tumor model was generated by subcutaneous injection of 5 × 106 cancerous cells in 100 μL saline under the right front legs of 7-week old female BALB/c mice weighing 15–20 g. Tumors were allowed to grow for 8–10 days to reach a size between 200–300 mm3 before the animals were subjected to in vivo biodistribution and PET imaging studies. About 111–185 KBq of 199Au-AuNPs in 100 μL saline was injected via the tail vein. The mice were anesthetized with inhaled isoflurane and re-anesthetized before euthanizing them by cervical dislocation at each time point (1 h, 4 h, and 24 h post injection, n = 4 per group). Organs of interest were collected, weighed, and counted in a Beckman 8000 gamma counter (Beckman, Fullerton, CA). Standards were prepared and measured along with the samples to calculate the percentage of the injected dose per gram of tissue (%ID/g) and the percentage of injected dose per organ (%ID/organ).
SPECT/CT imaging
The 4T1 tumors were allowed to grow for up to 2 weeks prior to the SPECT/CT imaging study. Mice were anesthetized with isoflurane and injected with 29.6 MBq of 199Au-AuNP-DAPTA in 100 μL of saline via the tail vein. SPECT/CT scans were performed on NanoSPECT/CT (Bioscan, Washington, D.C.) at 24 h post injection. CT scan was performed first (using a 45 KVP energy tube at 177 mA and 180 projections with 400 ms exposure), followed by helical SPECT of 60 projections with 60 s each. CT and SPECT projections were reconstructed using InvivoScope software (Bioscan, Inc., Washington, DC). The high-resolution NanoSPECT/CT anatomical and radioactive images have isotropic voxel size of 0.4 and 0.6 mm respectively.
Autoradiography studies
After SPECT/CT scan, tumors were excised and frozen in an Optimal Cutting Temperature (O.C.T.) compound and cut into 8–20 slices (50 μm in thickness) using a Vibratome 8850 whole body cryo-microtome (SIMS Co., Ltd, Tokyo, Japan). The tumor slices were placed on glass slides for 2D autoradiography using an InstantImager Electronic Autoradiography instrument (Packard, Meriden, CT). Images were acquired and analyzed using Imager software (Packard, Meriden, CT).
Immunohistochemistry and histology quantification
Serial sections of 5 μm in thickness were cut from paraformaldehyde-fixed (24 h) and paraffin-embedded specimens. The sections were deparaffinized and rehydrated through a series of xylenes and graded alcohols before undergoing antigen retrieval pre-treatment (10 mM Tris, 1 mM ethylenediaminetetraacetic acid, 0.05% Tween, and at pH = 9.0 for 10 min). They were then treated with 0.3% H2O2 for 30 min, followed by blocking serum for 1 h to prevent non-specific binding (Vectastain, Vector Laboratories, Burlingame, CA). The sections were finally incubated overnight at 4 °C with a primary antibody (anti-CCR5, 1:100 in blocking serum, Santa Cruz Biotechnology, Dallas, TX). A secondary antibody was applied (Vector Laboratories) and brown color development was achieved through the use of a horse radish peroxidase-based immunostaining kit (Vector Laboratories). Nuclei were counterstained blue with hematoxylin. Digital images of the stained sections were obtained using a light microscope (Leica Microsystems). In addition, hematoxylin and eosin (H&E) stain was performed on consecutively cut specimens to analyze the morphology of the tissue.
Statistical analysis
Group variation is described as mean ± SD. Groups were compared using 1-way ANOVA with a Bonferroni post-test. Individual group differences were determined with the use of a 2-tailed student t-test. The significance level in all tests was p 0.05. Prism, version 6.04 (GraphPad), was used for all statistical analyses.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from NIH (R01 CA138527) and startup funds from the Georgia Institute of Technology. As a visiting Ph.D. student from Peking University, B.P. was also partially supported by China Scholarship Council.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Yongfeng Zhao, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Bo Pang, The Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, Georgia 30332, United States. Department of Biomedical Engineering, Peking University, Beijing, 100871, P. R. China.
Hannah Luehmann, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Lisa Detering, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Dr. Xuan Yang, The Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, Georgia 30332, United States
Deborah Sultan, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Scott Harpstrite, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Prof. Vijay Sharma, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States
Prof. Cathy S. Cutler, Research Reactor Center, University of Missouri, Columbia, Missouri 65211, United States
Prof. Younan Xia, The Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, Georgia 30332, United States.
Prof. Yongjian Liu, Department of Radiology, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
References
- 1.a) Cobley CM, Chen J, Cho EC, Wang LV, Xia Y. Chem Soc Rev. 2011;40:44. doi: 10.1039/b821763g. [DOI] [PubMed] [Google Scholar]; b) Kumar D, Saini N, Jain N, Sareen R, Pandit V. Expert Opin Drug Deliv. 2013;10:397. doi: 10.1517/17425247.2013.749854. [DOI] [PubMed] [Google Scholar]; c) Meir R, Motiei M, Popovtzer R. Nanomedicine (Lond) 2014;9:2059. doi: 10.2217/nnm.14.129. [DOI] [PubMed] [Google Scholar]; d) Webb JA, Bardhan R. Nanoscale. 2014;6:2502. doi: 10.1039/c3nr05112a. [DOI] [PubMed] [Google Scholar]; e) Trouiller AJ, Hebie S, El Bahhaj F, Napporn TW, Bertrand P. Eur J Med Chem. 2015;99:92. doi: 10.1016/j.ejmech.2015.05.024. [DOI] [PubMed] [Google Scholar]; f) Wang Y, Black KC, Luehmann H, Li W, Zhang Y, Cai X, Wan D, Liu SY, Li M, Kim P, Li ZY, Wang LV, Liu Y, Xia Y. ACS Nano. 2013;7:2068. doi: 10.1021/nn304332s. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Yang X, Yang M, Pang B, Vara M, Xia Y. Chem Rev. 2015;115:10410. doi: 10.1021/acs.chemrev.5b00193. [DOI] [PubMed] [Google Scholar]
- 2.a) Liu Y, Welch MJ. Bioconjug Chem. 2012;23:671. doi: 10.1021/bc200264c. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guo Y, Aweda T, Black KC, Liu Y. Curr Top Med Chem. 2013;13:470. doi: 10.2174/1568026611313040007. [DOI] [PubMed] [Google Scholar]
- 3.a) Gibson N, Holzwarth U, Abbas K, Simonelli F, Kozempel J, Cydzik I, Cotogno G, Bulgheroni A, Gilliland D, Ponti J, Franchini F, Marmorato P, Stamm H, Kreyling W, Wenk A, Semmler-Behnke M, Buono S, Maciocco L, Burgio N. Arch Toxicol. 2011;85:751. doi: 10.1007/s00204-011-0701-6. [DOI] [PubMed] [Google Scholar]; b) Sun X, Cai W, Chen X. Acc Chem Res. 2015;48:286. doi: 10.1021/ar500362y. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lipka J, Semmler-Behnke M, Sperling RA, Wenk A, Takenaka S, Schleh C, Kissel T, Parak WJ, Kreyling WG. Biomaterials. 2010;31:6574. doi: 10.1016/j.biomaterials.2010.05.009. [DOI] [PubMed] [Google Scholar]; d) Yang S, Sun S, Zhou C, Hao G, Liu J, Ramezani S, Yu M, Sun X, Zheng J. Bioconjug Chem. 2015;26:511. doi: 10.1021/acs.bioconjchem.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gao F, Cai P, Yang W, Xue J, Gao L, Liu R, Wang Y, Zhao Y, He X, Zhao L, Huang G, Wu F, Zhao Y, Chai Z, Gao X. ACS Nano. 2015;9:4976. doi: 10.1021/nn507130k. [DOI] [PubMed] [Google Scholar]; f) Goel S, Chen F, Ehlerding EB, Cai W. Small. 2014;10:3825. doi: 10.1002/smll.201401048. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Sun X, Huang X, Yan X, Wang Y, Guo J, Jacobson O, Liu D, Szajek LP, Zhu W, Niu G, Kiesewetter DO, Sun S, Chen X. ACS Nano. 2014;8:8438. doi: 10.1021/nn502950t. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Shaffer TM, Wall MA, Harmsen S, Longo VA, Drain CM, Kircher MF, Grimm J. Nano Lett. 2015;15:864. doi: 10.1021/nl503522y. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Chanda N, Kan P, Watkinson LD, Shukla R, Zambre A, Carmack TL, Engelbrecht H, Lever JR, Katti K, Fent GM, Casteel SW, Smith CJ, Miller WH, Jurisson S, Boote E, Robertson JD, Cutler C, Dobrovolskaia M, Kannan R, Katti KV. Nanomedicine. 2010;6:201. doi: 10.1016/j.nano.2009.11.001. [DOI] [PubMed] [Google Scholar]; j) Wang Y, Liu Y, Luehmann H, Xia X, Brown P, Jarreau C, Welch M, Xia Y. ACS Nano. 2012;6:5880. doi: 10.1021/nn300464r. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Zhao Y, Sultan D, Detering L, Cho S, Sun G, Pierce R, Wooley KL, Liu Y. Angew Chem. 2014;126:160. doi: 10.1002/anie.201308494. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2014;53:156. [Google Scholar]; l) Zhao Y, Sultan D, Detering L, Luehmann H, Liu Y. Nanoscale. 2014;6:13501. doi: 10.1039/c4nr04569f. [DOI] [PubMed] [Google Scholar]; m) Zhou C, Hao G, Thomas P, Liu J, Yu M, Sun S, Öz OK, Sun X, Zheng J. Angew Chem. 2012;124:10265. doi: 10.1002/anie.201203031. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2012;51:10118. [Google Scholar]
- 4.Cutler CS, Hennkens HM, Sisay N, Huclier-Markai S, Jurisson SS. Chem Rev. 2013;113:858. doi: 10.1021/cr3003104. [DOI] [PubMed] [Google Scholar]
- 5.a) Wang Y, Liu Y, Luehmann H, Xia X, Wan D, Cutler C, Xia Y. Nano Lett. 2013;13:581. doi: 10.1021/nl304111v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Black KC, Wang Y, Luehmann HP, Cai X, Xing W, Pang B, Zhao Y, Cutler CS, Wang LV, Liu Y, Xia Y. ACS Nano. 2014;8:4385. doi: 10.1021/nn406258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhry IA, Liu M, Shamsi FA, Arat YO, Shetlar DJ, Boniuk M. Retina. 2009;29:73. doi: 10.1097/IAE.0b013e3181863f7c. [DOI] [PubMed] [Google Scholar]
- 7.Fazaeli Y, Akhavan O, Rahighi R, Aboudzadeh MR, Karimi E, Afarideh H. Mater Sci Eng C Mater Biol Appl. 2014;45:196. doi: 10.1016/j.msec.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 8.a) Nienhuis HH, Gaykema SB, Timmer-Bosscha H, Jalving M, Brouwers AH, Lub-de Hooge MN, van der Vegt B, Overmoyer B, de Vries EG, Schroder CP. Pharmacol Ther. 2015;147:63. doi: 10.1016/j.pharmthera.2014.11.004. [DOI] [PubMed] [Google Scholar]; b) Alcantara D, Leal MP, Garcia-Bocanegra I, Garcia-Martin ML. Front Chem. 2014;2:112. doi: 10.3389/fchem.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Bardhan R, Lal S, Joshi A, Halas NJ. Acc Chem Res. 2011;44:936. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) van Uden DJ, van Laarhoven HW, Westenberg AH, de Wilt JH, Blanken-Peeters CF. Crit Rev Oncol Hematol. 2015;93:116. doi: 10.1016/j.critrevonc.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 10.a) Yaal-Hahoshen N, Shina S, Leider-Trejo L, Barnea I, Shabtai EL, Azenshtein E, Greenberg I, Keydar I, Ben-Baruch A. Clin Cancer Res. 2006;12:4474. doi: 10.1158/1078-0432.CCR-06-0074. [DOI] [PubMed] [Google Scholar]; b) Niwa Y, Akamatsu H, Niwa H, Sumi H, Ozaki Y, Abe A. Clin Cancer Res. 2001;7:285. [PubMed] [Google Scholar]; c) Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hebert CA. Biochemistry. 1997;36:9642. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]; d) Soria G, Ben-Baruch A. Cancer Lett. 2008;267:271. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]; e) Schmadeka R, Harmon BE, Singh M. Am J Clin Pathol. 2014;141:462. doi: 10.1309/AJCPQN8GZ8SILKGN. [DOI] [PubMed] [Google Scholar]; f) Trop I, LeBlanc SM, David J, Lalonde L, Tran-Thanh D, Labelle M, El Khoury MM. Radiographics. 2014;34:1178. doi: 10.1148/rg.345130049. [DOI] [PubMed] [Google Scholar]; g) Azenshtein E, Luboshits G, Shina S, Neumark E, Shahbazian D, Weil M, Wigler N, Keydar I, Ben-Baruch A. Cancer Res. 2002;62:1093. [PubMed] [Google Scholar]; h) Banin Hirata BK, Oda JM, Losi Guembarovski R, Ariza CB, de Oliveira CE, Watanabe MA. Dis Markers. 2014;2014:513158. doi: 10.1155/2014/513158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Fertig EJ, Lee E, Pandey NB, Popel AS. Sci Rep. 2015;5:12133. doi: 10.1038/srep12133. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Norton KA, Popel AS, Pandey NB. Am J Cancer Res. 2015;5:1295. [PMC free article] [PubMed] [Google Scholar]; c) Velasco-Velazquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, Pestell RG. Cancer Res. 2012;72:3839. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 12.Luehmann HP, Pressly ED, Detering L, Wang C, Pierce R, Woodard PK, Gropler RJ, Hawker CJ, Liu Y. J Nucl Med. 2014;55:629. doi: 10.2967/jnumed.113.132001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Zheng Y, Zhong X, Li Z, Xia Y. Part Part Syst Charact. 2014;31:266. [Google Scholar]; b) Zheng Y, Ma Y, Zeng J, Zhong X, Jin M, Li ZY, Xia Y. Chem Asian J. 2013;8:792. doi: 10.1002/asia.201201105. [DOI] [PubMed] [Google Scholar]
- 14.Velasco-Velazquez M, Pestell RG. Oncoimmunology. 2013;2:e23660. doi: 10.4161/onci.23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.