Abstract
Broad spectrum antiviral drugs targeting host processes could potentially treat a wide range of viruses while reducing the likelihood of emergent resistance. Despite great promise as therapeutics, such drugs remain largely elusive. Here we use parallel genome-wide high-coverage shRNA and CRISPR-Cas9 screens to identify the cellular target and mechanism of action of GSK983, a potent broad spectrum antiviral with unexplained cytotoxicity1–3. We show that GSK983 blocks cell proliferation and dengue virus replication by inhibiting the pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (DHODH). Guided by mechanistic insights from both genomic screens, we found that exogenous deoxycytidine markedly reduces GSK983 cytotoxicity but not antiviral activity, providing an attractive novel approach to improve the therapeutic window of DHODH inhibitors against RNA viruses. Together, our results highlight the distinct advantages and limitations of each screening method for identifying drug targets and demonstrate the utility of parallel knockdown and knockout screens for comprehensively probing drug activity.
Introduction
The development of effective broad spectrum antiviral therapies remains a highly attractive (but equally challenging) goal in drug discovery. Antivirals targeting host cell processes have great potential to demonstrate activity against a range of viruses, reduce the likelihood of mutational resistance, and serve as frontline therapies for rapidly emerging outbreaks of viral disease such as Ebola and influenza4. However, extensive efforts to develop such drugs have been stymied by various factors, including on-target toxicity and limited in vivo activity5. Recently, cell-based phenotypic screens of chemical libraries have generated numerous host-targeting broad spectrum antiviral lead compounds with unidentified targets and mechanisms of action1,6–8. Thus, the development of improved methods for target identification and mechanism elucidation – critical challenges in drug discovery – should facilitate the development of more effective broad spectrum antiviral therapies.
High-throughput yeast deletion and RNAi-based screening approaches have emerged as powerful alternatives to drug target identification methods that utilize affinity-based chemoproteomics or chemical-genetic expression signatures9–13; reviewed in14,15. We recently developed high-coverage shRNA libraries (25 shRNAs/gene) that facilitate pooled genome-wide screening in mammalian cells with dramatically improved hit reliability16–19. While our high-coverage shRNA libraries have demonstrated utility in identifying small molecule drug targets20,21, genome-wide screening is no longer limited to RNAi-mediated gene knockdown. The recent development of the CRISPR-Cas9 system has greatly expanded the scope of genomic screening in mammalian cells by enabling facile interrogation of functional gene deletions22–28. Here, we demonstrate a comprehensive strategy using parallel genome-wide shRNA and CRISPR-Cas9 screens to discover the previously unknown host cell target and mechanism of action of GSK983 (1), a poorly understood broad spectrum antiviral lead compound with unexplained cytotoxicity.
We found that GSK983 blocks virus replication and arrests the growth of rapidly dividing cells by inhibiting the cellular de novo pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (DHODH). Furthermore, we show that exogenous deoxycytidine greatly reduces GSK983 cytotoxicity but not activity against RNA virus replication, providing a novel strategy to improve the therapeutic window of DHODH inhibitors against RNA viruses. Finally, we propose that efficacy of broad spectrum antiviral therapies targeting host pyrimidine metabolism might be improved by pharmacological inhibition of both de novo pyrimidine biosynthesis (via DHODH) and pyrimidine ribonucleoside salvage (via uridine-cytidine kinase, UCK2).
Results
Biological activity of GSK983
We first examined the biological activity of GSK983 (Fig. 1a and Supplementary Results, Supplementary Fig. 1a) in human K562 cells. GSK983 inhibited K562 cell growth with an IC50 of 21 nM (Fig. 1b and Supplementary Fig. 1b), consistent with previous observations1. Cell cycle analysis revealed that 24 h GSK983 treatment caused an accumulation of K562 cells in S phase (Supplementary Fig. 1c,d), while prolonged 72 h treatment induced a dose-dependent increase in K562 cell death by apoptosis (Supplementary Fig. 1e,f).
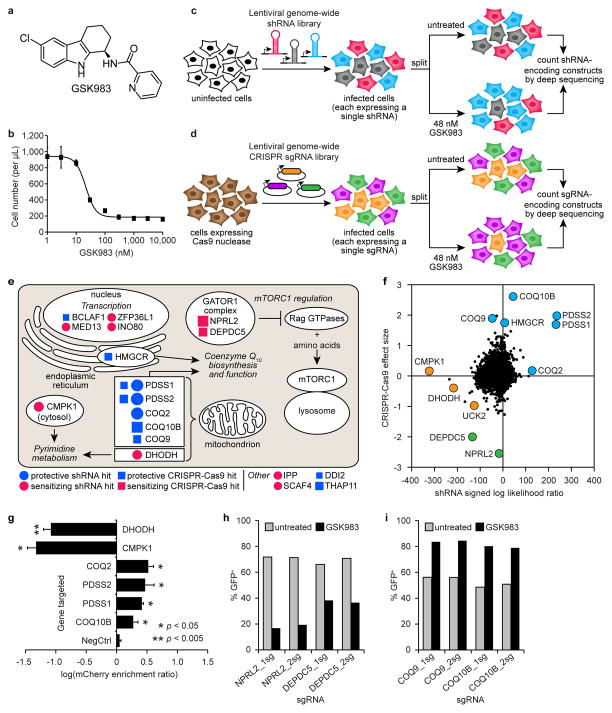
Figure 1.
shRNA and CRISPR-Cas9 screens to identify the cellular target and mechanism of action of GSK983. (a) Structure of GSK983. (b) GSK983 dose response in K562 cells. Viable cells were counted by flow cytometry (FSC/SSC) following 72 h GSK983 treatment at the indicated concentration. Error bars represent ± standard deviation of 8 biological replicates from two independent experiments. Schematic representation of genome-wide shRNA (c) and CRISPR-Cas9 (d) screens. (e) Top ten hits from the shRNA and CRISPR-Cas9 screens in cellular and biological context. Circle size is proportional to MLE score absolute value. Square size is proportional to median fold-enrichment or disenrichment. (f) Comparative analysis of results from shRNA and CRISPR-Cas9 screens. Pyrimidine metabolism (orange), CoQ10 biosynthesis (blue), regulation of mTORC1 activity (green). (g) Validation of selected top hit genes from the shRNA screen using a competitive growth assay. A total of 27 shRNAs were retested (6 targeting DHODH; 4 targeting CMPK1; 3 each targeting COQ2, PDSS2, PDSS1, and COQ10B; and 5 negative controls). Error bars represent ± standard deviation of log(mCherry enrichment ratio) values for all retested shRNAs targeting each gene. P values were calculated by Mann-Whitney U test. Validation of selected sensitizing (h) or protective (i) sgRNAs from the CRISPR-Cas9 screen using a competitive growth assay. Bars represent the average of two biological replicates.
shRNA and CRISPR-Cas9 screens for target identification
We previously established a platform for pooled RNAi screens using ultracomplex shRNA libraries (~25 shRNAs per gene and ~10,000 negative control shRNAs)16–19. More recently, we systematically optimized several features of our shRNA design to create a next-generation shRNA library, which performs comparably to our CRISPRi library29. For the shRNA screen described here, we infected K562 cells with our next-generation shRNA library targeting the entire human protein-coding genome. For the CRISPR-Cas9 screen, we designed a CRISPR single-guide RNA (sgRNA) library targeting the entire human protein-coding genome (~4 sgRNAs per gene and ~2,000 negative control sgRNAs) incorporating previously reported improvements to the sgRNA stem loop30. We stably infected this CRISPR sgRNA library into a K562 cell line constitutively expressing Cas9 endonuclease. For both screens, we split cells expressing the genome-wide shRNA or sgRNA library and cultured them in the presence or absence of 48 nM GSK983 for 10–14 days. We then isolated genomic DNA from untreated and GSK983-treated cells, PCR-amplified shRNA- or sgRNA-encoding DNA constructs, and counted all constructs by deep sequencing.16 (Fig 1c,d).
We used deep sequencing data from the shRNA screen to rank genes according to a maximum likelihood estimator (MLE) metric that we designed to consider the magnitude of sensitization to or protection against GSK983 conferred by the entire set of shRNAs targeting each gene. Similarly, we used deep sequencing data from the CRISPR-Cas9 screen to rank genes according to the median fold-change in sgRNA frequency in the untreated versus GSK983-treated cell populations (Supplementary Datasets 1,2)27.
Analysis of genomic screen results
Inspection of the top 10 hit genes from both screens revealed clear signatures from three major biological pathways (Fig. 1e). Knockdown or knockout of genes whose products are required for coenzyme Q10 (CoQ10) biosynthesis and function (HMGCR, PDSS1, PDSS2, COQ2, COQ9, and COQ10B) protected K562 cells against GSK983 (Fig. 1e and Supplementary Fig. 2a,b). In contrast, shRNA-mediated knockdown of pyrimidine metabolism genes (DHODH and CMPK1) sensitized K562 cells to GSK983 (Fig. 1e and Supplementary Fig. 2b), as did CRISPR-Cas9-mediated knockout of components of the GATOR1 protein complex (NPRL2 and DEPDC5), a recently identified negative regulator of mTORC1 activity (Fig. 1e)31,32. Interestingly, mammalian pyrimidine biosynthesis utilizes CoQ1033 and is subject to regulation by mTORC134,35.
As expected, genes required for essential processes such as nucleotide biosynthesis (e.g., DHODH and CMPK1) that appeared as top hits in the shRNA knockdown screen did not appear as statistically significant hits in the CRISPR-Cas9 deletion screen (Fig. 1f and Supplementary Datasets 1,2). Conversely, certain genes (e.g., mTOR signaling components NPRL2, TSC2, SEH1L, MAPKAP1, RICTOR, RHEB, and TBC1D7) were statistically significant hits in the CRISPR-Cas9 screen but not in the shRNA screen (Fig. 1f and Supplementary Datasets 1,2). A number of genes were statistically significant hits in both screens, including PDSS1, PDSS2, and UCK2. GO enrichment analysis of the top 50 hits from each screen revealed that CoQ10 biosynthesis genes were significantly overrepresented among top hits from both screens, while pyrimidine metabolism genes were significantly overrepresented among top hits from the shRNA screen alone (p < 0.001 in all cases) (Supplementary Tables 1,2)36. Thus, our findings demonstrate the complementary power of parallel shRNA and CRISPR-Cas9 screens to identify connections between biological pathways that may be difficult to detect using either screening approach alone.
We individually retested multiple shRNAs targeting top hit genes to validate genomic screen results (Supplementary Tables 3,4). Using a competitive growth assay, we verified both the sensitized phenotype conferred by DHODH or CMPK1 knockdown and the protected phenotype conferred by knockdown of several CoQ10 biosynthesis genes (Fig. 1g and Supplementary Fig. 3a–c). We also used QPCR to confirm the efficacy of selected shRNAs targeting DHODH and CMPK1 (Supplementary Fig. 3d,e). Similarly, we individually retested top hit sgRNAs from the CRISPR-Cas9 screen to verify both that NPRL2 or DEPDC5 knockout sensitized K562 cells to GSK983 and that knockout of CoQ10 biosynthesis genes protected K562 cells against the drug (Fig. 1h,i and Supplementary Fig. 4a). Additionally, we retested CRISPR sgRNAs targeting selected top hit genes in HeLa cells and obtained similar results, albeit with milder phenotypes, indicating that these genetic modifiers of GSK983 sensitivity are not K562-specific (Supplementary Fig. 4a,b).
GSK983 inhibits cellular dihydroorotate dehydrogenase
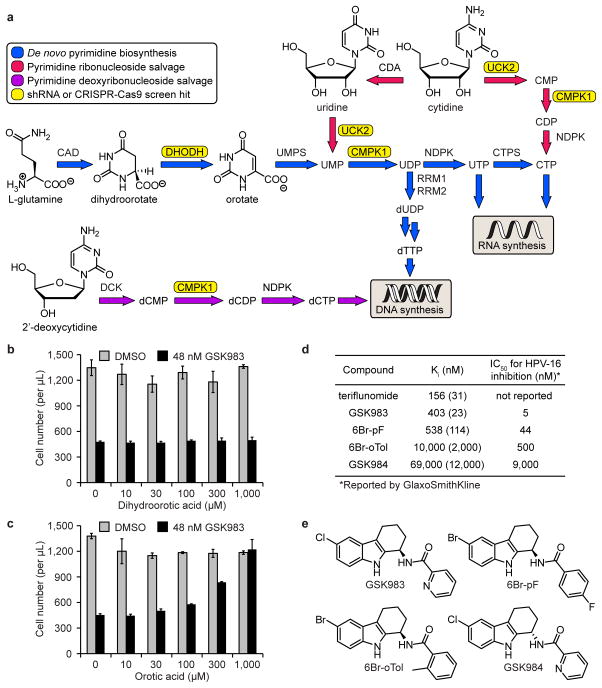
While the CRISPR-Cas9 and shRNA screens each highlighted unique hit genes and together provided a more complete understanding of the biological activity of GSK983, we considered that the highly sensitizing hits in the pyrimidine biosynthesis pathway were among the most likely candidates to be molecular targets of GSK983. We reasoned that cells expressing an shRNA against a protein target of GSK983 should be highly sensitized to GSK983-induced growth inhibition. Consequently, we focused our target identification effort on the pyrimidine metabolism genes DHODH and CMPK1, which were the top sensitizing hits from our genome-wide shRNA screen. Mammalian cells derive pyrimidine (deoxy)ribonucleotide triphosphates either from de novo biosynthesis or pyrimidine salvage, in which intact pyrimidine metabolites are recycled from intracellular nucleic acid degradation or imported into the cell from exogenous sources (Fig. 2a). DHODH (dihydroorotate dehydrogenase) is required for de novo pyrimidine biosynthesis, while CMPK1 plays a critical role in both de novo biosynthesis and pyrimidine salvage (Fig. 2a).
Figure 2.
GSK983 inhibits DHODH to block virus replication and cell proliferation. (a) Schematic representation of mammalian pyrimidine metabolism. Genes that appeared as strong sensitizing hits in the shRNA screen (CMPK1, DHODH, UCK2) and CRISPR-Cas9 screen (UCK2) are highlighted in yellow. (b) Dihydroorotic acid had no effect on GSK983-induced growth inhibition in K562 cells. (c) Orotic acid reversed GSK983-induced growth inhibition in K562 cells. For (b) and (c), viable cells were counted by flow cytometry (FSC/SSC) following 72 h treatment with 48 nM GSK983 or vehicle and the indicated concentration of (dihydro)orotic acid. Error bars represent ± standard deviation of 4 biological replicates. (d) GSK983 and analogues inhibited recombinant human DHODH in vitro. Ki values are averages of two independent Ki determinations at different inhibitor concentrations. The range between independently calculated Ki values for each inhibitor is shown in parentheses. IC50 values for inhibition of episomal HPV-16 replication in cell-based antiviral assays are those reported by GlaxoSmithKline3. (e) Structures of GSK983, 6Br-pF, 6Br-oTol, and GSK984.
To determine whether GSK983 inhibited DHODH, we examined the ability of dihydroorotate and orotate (the substrate and product of DHODH, respectively) to reverse the anti-proliferative effect of GSK983 in K562 cells. Dihydroorotate supplementation had no effect on GSK983-induced growth inhibition (Fig. 2b). In sharp contrast, exogenous orotate reversed the anti-proliferative effect of GSK983 in dose-dependent fashion, with full rescue of K562 cell growth at the highest orotate concentration tested (Fig. 2c). We obtained identical results in HeLa cells (Supplementary Fig. 5a,b). Together, these data strongly suggested that GSK983 is a DHODH inhibitor.
To confirm DHODH as a GSK983 target, we expressed and purified the recombinant human enzyme from E. coli and examined the effect of GSK983 on in vitro enzyme activity, using the known DHODH inhibitor teriflunomide as a positive control (reported Ki = 179 nM)37. GSK983 was a competitive inhibitor of DHODH with respect to decylubiquinone binding (Ki = 403 nM) (Fig. 2d and Supplementary Fig. 5c,d). To determine whether the reported antiviral effect of GSK983 could be attributed to DHODH inhibition, we resynthesized three GSK983 analogues originally prepared and evaluated at GlaxoSmithKline2,3. The extent to which each compound (GSK983, 6Br-pF (2), 6Br-oTol (3), and GSK984 (4)) inhibited DHODH activity in vitro correlated strongly with the reported potency of each compound in cell-based antiviral assays (R2 = 0.993) (Fig. 2d,e and Supplementary Fig. 5d–g)3. Collectively, these findings indicated that the antiviral effect of GSK983 is due to DHODH inhibition.
To determine whether GSK983 inhibited CMPK1, we expressed and purified the recombinant human enzyme from E. coli; however, we observed no effect of GSK983 on in vitro CMPK1 activity. Thus, although CMPK1 was the most sensitizing hit in the shRNA screen (followed by DHODH), the encoded protein is not a direct GSK983 target. Nonetheless, it is logical that shRNA-mediated CMPK1 knockdown is highly toxic in the presence of GSK983, given that CMPK1 knockdown should further impair both de novo pyrimidine biosynthesis and pyrimidine salvage (Fig. 2a).
A new strategy to exploit DHODH as an antiviral target
While there are examples of small molecule DHODH inhibitors that potently block virus replication38–40, prolonged treatment with DHODH inhibitors causes pyrimidine depletion that arrests the growth of rapidly dividing cells. Indeed, the FDA-approved drugs leflunomide (rheumatoid arthritis) and teriflunomide (multiple sclerosis) are DHODH inhibitors that prevent the rapid clonal expansion of activated lymphocytes, a process which requires significantly elevated cellular pyrimidine levels41,42. We were therefore interested in separating the antiviral effect of GSK983 from its anti-proliferative (and cytotoxic) effect on rapidly dividing cells.
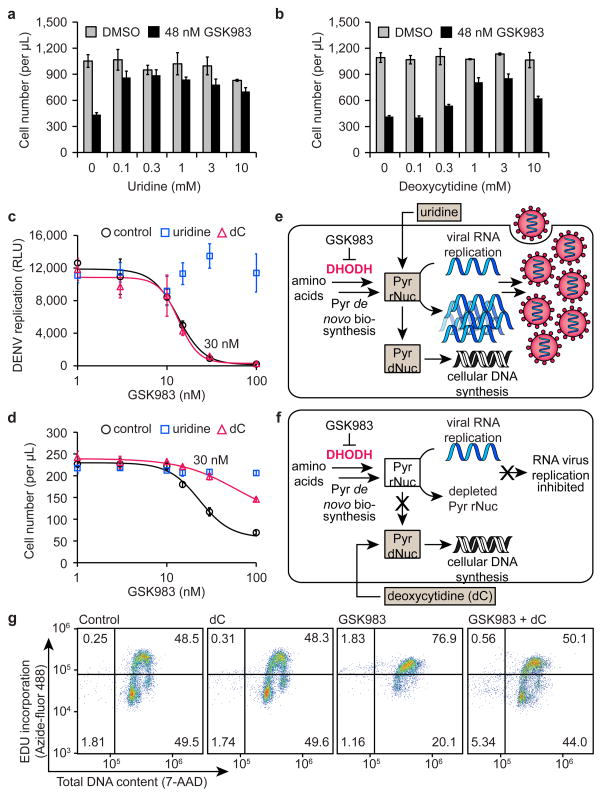
Guided by the appearance of pyrimidine salvage enzymes among the top sensitizing hits from both genomic screens, we examined the ability of pyrimidine salvage metabolites to reverse the anti-proliferative effect of GSK983 on rapidly dividing cells. Uridine, cytidine, or deoxycytidine supplementation reversed GSK983-induced growth inhibition to varying extents in K562 cells (Fig. 3a,b and Supplementary Fig. 6a,b). However, cellular salvage of exogenous ribonucleosides (uridine and cytidine) can sustain RNA virus replication despite DHODH inhibition40,43. We reasoned that deoxycytidine salvage would support DNA but not RNA virus replication given that ribonucleotides cannot be directly biosynthesized from their 2′-deoxy analogues. This raised the intriguing possibility of using a DHODH inhibitor to block RNA virus replication in combination with a deoxycytidine supplement to reverse the anti-proliferative effect on rapidly dividing cells.
Figure 3.
Deoxycytidine (dC) reverses the anti-proliferative effect of GSK983 but not antiviral activity. Uridine (a) and deoxycytidine (b) largely reversed GSK983-induced growth inhibition in K562 cells. For (a) and (b), viable cells were counted by flow cytometry (FSC/SSC) following 72 h treatment with 48 nM GSK983 or vehicle and the indicated concentration of uridine or deoxycytidine. Error bars represent ± standard deviation of 4 biological replicates. (c) GSK983 inhibited replication of luciferase-expressing DENV in A549 cells (black). 1 mM uridine reversed antiviral activity (blue), while 1 mM deoxycytidine did not (red). Error bars represent ± standard deviation of 3 biological replicates. (d) Uridine (blue) and deoxycytidine (red) reversed GSK983-induced growth inhibition in A549 cells. Viable cells were counted by flow cytometry (FSC/SSC) following 72 h treatment with no exogenous pyrimidines (control), 1 mM uridine, or 1 mM deoxycytidine and the indicated concentration of GSK983. Error bars represent ± standard deviation of 3 biological replicates. (e) Ribonucleoside (uridine) salvage sustains both RNA virus replication and cellular DNA synthesis. (f) Deoxyribonucleoside (deoxycytidine) salvage sustains cellular DNA synthesis but not RNA virus replication. For (e) and (f), Pyr = pyrimidine, rNuc = ribonucleotides, dNuc = deoxyribonucleotides. (g) Deoxycytidine reversed GSK983-induced S phase cell cycle arrest in K562 cells. Following 24 h treatment with 48 nM GSK983, cells were treated with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) for 2 h and fixed in 70% EtOH. Cells were stained with Azide-fluor 488 and 7-AAD and analyzed by flow cytometry. Flow cytometry plots depict one of three biological replicates.
To test this therapeutically relevant hypothesis, we studied the antiviral effect of GSK983 on dengue virus (DENV), an RNA virus that has rapidly emerged as the most prevalent mosquito-transmitted virus worldwide and currently causes approximately 100 million infections annually in tropical regions44. We compared the effects of exogenous uridine and deoxycytidine on the ability of GSK983 to inhibit DENV replication in human A549 cells. In the absence of exogenous pyrimidines, GSK983 potently inhibited DENV replication (IC50 = 13.3 nM) (Fig. 3c). As expected, uridine supplementation completely abolished the antiviral activity of GSK983 (Fig. 3c). In sharp contrast, exogenous deoxycytidine did not reverse GSK983-mediated inhibition of DENV replication (IC50 = 13.5 nM) (Fig. 3c). We conducted a cell growth assay conducted in parallel with antiviral experiments to confirm that both uridine and deoxycytidine reduced GSK983 cytotoxicity and rescued A549 cell growth (Fig. 3d), consistent with previous results in K562 cells. Notably, treatment with 30 nM GSK983 and 1 mM deoxycytidine caused a significant (~90%) reduction in DENV replication (Fig. 3c) with a minimal effect on A549 cell growth (Fig. 3d). Thus, deoxycytidine supplementation partially reversed the anti-proliferative effect of GSK983 but did not preclude potent inhibition of DENV replication (Fig. 3e,f).
We additionally tested the effect of GSK983 on the replication of another RNA virus, Venezuelan equine encephalitis virus (VEEV). GSK983 inhibited VEEV replication with an IC50 of 12.8 nM (Supplementary Fig. 6e). As with DENV, we observed that exogenous uridine reversed the antiviral effect of GSK983, while deoxycytidine supplementation had no effect on antiviral activity. Furthermore, we found that exogenous orotic acid, but not dihydroorotic acid, reversed the antiviral effect of GSK983. Collectively, these results indicate that GSK983 potently inhibits replication of RNA viruses by inhibiting cellular DHODH.
Further analysis revealed that exogenous deoxycytidine reversed GSK983-induced S phase cell cycle arrest (Fig. 3g and Supplementary Fig. 7a,b) and cytotoxicity (Supplementary Fig. 7c–e) at 3 days in K562, HeLa, and A549 cells. Deoxycytidine supplementation also markedly reduced teriflunomide toxicity in K562 cells (Supplementary Fig. 7f). These findings suggest that deoxycytidine may reverse the anti-proliferative effect of DHODH inhibitors by sustaining cellular DNA synthesis during S phase. We observed that the capacity of exogenous deoxycytidine to reverse GSK983 toxicity is significant but diminished during extended GSK983 treatment (6 days) (Supplementary Fig. 6c,d), likely due to the inability of deoxycytidine to alleviate the blockade of cellular RNA biosynthesis caused by pharmacological inhibition of DHODH.
Data from our genome-wide shRNA and CRISPR-Cas9 screens also suggested a novel combination chemotherapy to achieve improved activity against RNA viruses by targeting host pyrimidine metabolism. Both shRNA-mediated knockdown and CRISPR-Cas9-mediated deletion of the pyrimidine metabolism enzyme UCK2 (uridine-cytidine kinase) sensitized K562 cells to GSK983, presumably by impairing uridine and cytidine salvage (Fig. 2a). This result suggests that pharmacological inhibition of UCK2 might enhance the antiviral activity of a DHODH inhibitor by preventing flux through the ribonucleoside salvage pathway from sustaining RNA virus replication. Thus, we propose that an effective and specific UCK2 inhibitor might act synergistically with a DHODH inhibitor to achieve improved activity against RNA viruses. Notably, UCK2 inhibition should not impair deoxycytidine salvage, which proceeds via a different pathway (Fig. 2a). Therefore, the possibility of using deoxycytidine to reduce the toxicity of a combination therapy targeting UCK2 and DHODH remains viable.
Discussion
Here, we have demonstrated the utility of parallel genome-wide shRNA and CRISPR-Cas9 screens (Fig. 1c,d) for identifying the target and mechanism of action of a poorly understood therapeutic lead compound, a strategy which should be broadly applicable to other bioactive small molecules. We found that the broad spectrum antiviral compound GSK983 blocks viral replication and arrests the growth of rapidly dividing cells by inhibiting the cellular de novo pyrimidine biosynthesis enzyme DHODH.
Our approach takes advantage of unique properties of each screening technology. shRNA-mediated knockdown of a gene encoding the protein target of a drug is expected to phenocopy pharmacological inhibition of the target. Therefore, we predicted that GSK983 would be more toxic to cells expressing an shRNA against a gene that encoded a protein target of the compound. Furthermore, the broad spectrum of shRNA-mediated knockdown efficiency should facilitate identification of essential genes that modulate GSK983 toxicity. At the same time, CRISPR-Cas9 mediated knockout should enable detection of non-essential genes that require full deletion to produce an observable phenotype.
Indeed, we found that genes required for essential biological processes such as nucleotide biosynthesis and metabolism (e.g., DHODH and CMPK1) appeared as statistically significant hits in the shRNA screen but not in the CRISPR-Cas9 screen. However, other genes (most notably those involved in mTOR signaling and regulation) only appeared as statistically significant hits in the CRISPR-Cas9 screen, presumably because shRNA-mediated knockdown of these genes was insufficient to produce a measurable phenotype. Thus, our findings demonstrate the complementary power of parallel shRNA and CRISPR-Cas9 screens to identify connections between biological pathways that may be difficult to detect using either screening approach alone.
We focused our target identification efforts on the pyrimidine metabolism genes DHODH and CMPK1, which appeared as the most sensitizing hits in our genome-wide shRNA screen and were therefore most likely to be molecular targets of GSK983. DHODH is a 43 kDa mitochondrial flavoprotein that requires a CoQ10 cofactor for activity and catalyzes the fourth and rate-limiting step in the de novo pyrimidine biosynthesis pathway (the oxidation of dihydroorotate to orotate). CMPK1 is a 22 kDa cytosolic nucleoside monophosphate kinase that catalyzes the phosphorylation of uridine 5′-monophosphate (UMP), cytidine 5′-monophosphate (CMP), and 2′-deoxycytidine 5′-monophosphate (dCMP) to the corresponding nucleotide diphosphates (Fig. 2a). Notably, CMPK1 plays a critical role in both de novo pyrimidine biosynthesis and salvage of pyrimidine ribonucleosides and deoxyribonucleosides (Fig. 2a).
Subsequent biochemical assays using recombinant human DHODH and CMPK1 demonstrated that GSK983 inhibited the activity of DHODH but not CMPK1. Thus, we reasoned that we could exploit the pyrimidine salvage pathway to separate the antiviral activity of GSK983 from its cytotoxic effect on rapidly dividing cells. Indeed, exogenously added deoxycytidine alleviated GSK983-induced S phase cell cycle arrest and largely reversed GSK983 cytotoxicity, presumably by facilitating continued cellular DNA synthesis via the deoxyribonucleoside salvage pathway. However, deoxycytidine supplementation did not reverse the potent activity of GSK983 against RNA viruses (DENV and VEEV), because the ribonucleotides required for RNA biosynthesis cannot be directly formed biochemically from their 2′-deoxy analogues. This finding provides an attractive novel approach to improve the therapeutic window of DHODH inhibitors; however, it is important to note that this strategy is likely limited in applicability to RNA viruses. Exogenous deoxycytidine would presumably sustain DNA virus replication via the deoxyribonucleoside salvage pathway notwithstanding pharmacological inhibition of DHODH.
There are numerous examples of small molecule DHODH inhibitors that potently block virus replication in cell-based assays38–40. However, the relevance of DHODH as a target for RNA virus therapies has been questioned because these compounds rarely achieve in vivo efficacy39,40. One plausible explanation for this phenomenon is that circulating plasma uridine levels in mammals often exceed concentrations that have been shown to reverse the antiviral effect of DHODH inhibitors in cell-based assays39,40. Therefore, RNA virus replication can likely be sustained via uridine salvage despite pharmacological inhibition of DHODH.
Mechanistic insights from our genome-wide shRNA and CRISPR-Cas9 screens suggest a novel combination chemotherapy that may improve the in vivo activity of a DHODH inhibitor as an RNA virus therapy. The pyrimidine salvage enzyme UCK2 (uridine-cytidine kinase) strongly sensitized cells to GSK983 in both screens. Thus, an effective and specific UCK2 inhibitor should act synergistically with a DHODH inhibitor to achieve improved activity against RNA viruses by preventing ribonucleoside salvage. Here, deoxycytidine salvage would remain intact, limiting toxicity for host cell DNA replication.
Finally, our dual genomic screening strategy identified other major biological pathways (mTOR regulation and CoQ10 biosynthesis) that affect cellular pyrimidine metabolism. Nonetheless, the mechanisms by which these pathways modulate cellular sensitivity to GSK983 remain poorly understood. Together, our results highlight the importance of genome-wide knockdown and knockout screening as a means of comprehensively interrogating both essential and non-essential genes, and demonstrate the utility of this approach as a functional platform enabling small molecule target identification and rational therapy design.
Online Methods
Chemicals and reagents for biological assays
GSK983 was synthesized as described below. Teriflunomide was obtained from Sigma-Aldrich and was used as received. GSK983 and teriflunomide were dissolved in DMSO to prepare stock solutions which were diluted in the appropriate cell growth medium for biological assays. GSK983 and teriflunomide stock solutions in DMSO were stored at −80 °C. Uridine, cytidine, deoxycytidine, and dihydroorotic acid were obtained from Sigma-Aldrich. Orotic acid was obtained from Fisher Scientific. For pyrimidine supplementation experiments, pyrimidine metabolites (uridine, cytidine, deoxycytidine, orotic acid, and dihydroorotic acid) were dissolved directly in the appropriate growth medium (RPMI or DMEM). 5-Ethynyl-2′-deoxyuridine (EdU) and Azide-fluor 488 were obtained from Sigma-Aldrich and dissolved in DMSO to prepare working stock solutions. Copper (II) sulfate (CuSO4) and ascorbic acid were obtained from Sigma-Aldrich and were used as received. 7-aminoactinomycin D (7-AAD) was obtained from Life Technologies and dissolved in DMSO to prepare a working stock solution.
Cell culture
K562 cells (ATCC) were cultured in RPMI (Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin, and L-glutamine. HeLa cells (HeLa-Kyoto cells, a gift from AA Hyman) and A549 cells (ATCC) were cultured in DMEM (Gibco) supplemented with 10% FBS, penicillin/streptomycin, and L-glutamine. In biological assays, HeLa cells and A549 cells were detached from the growth surface using a trypsin/EDTA solution (Gibco) prior to analysis. Cells were maintained in logarithmic growth during all biological assays. All cell lines were maintained in a humidified incubator (37 °C, 5% CO2), and checked regularly for mycoplasma contamination.
GSK983 dose response and growth time course in K562 cells
To determine the dose response of K562 cells to GSK983, K562 cells were seeded into 24-well plates at a density of 50,000 cells/mL and treated with GSK983 at the indicated concentration for 72 h. Following 72 h treatment, cells were harvested and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer. IC50 values were calculated by fitting a 4-parameter logistic equation to the data. To analyze the time-dependence of GSK983-induced growth inhibition in K562 cells, cells were seeded into 24-well plates at a density of 50,000 cells/mL and treated with the indicated concentration of GSK983. At the indicated time points, cells were harvested and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer.
Cell cycle analysis based on total DNA content (propidium iodide)
K562 cells were seeded into 6-well plates at a density of 100,000 cells/mL. HeLa and A549 cells were seeded into 6-well plates at a density of 200,000 cells/well. Cells were treated with the indicated concentration of GSK983 for 24 h. Where specified, cells were treated for 24 h with deoxycytidine alone (1 mM), GSK983 and deoxycytidine (1 mM), or neither (untreated). Following 24 treatment, cells were harvested and pelleted (300g, 5 min). The supernatant was removed by vacuum and the cells were washed (0.5 mL 1x PBS). Cells were resuspended in 70% EtOH (1 mL, 0 °C) with mild vortexing. Cells were fixed in 70% EtOH overnight at 4 °C. The following day, fixed cells were pelleted (1900g, 5 min), washed (0.5 mL 1x PBS), and resuspended in 200 μL of a total DNA content staining solution (1x PBS with 50 μg/mL RNase A and 10 μg/mL propidium iodide). Samples were incubated for 30 min at 37 °C and analyzed by flow cytometry using a BD Accuri C6 Flow Cytometer. Propidium iodide fluorescence was detected in FL2.
Annexin V-FITC/7-AAD apoptosis assay
K562 cells were seeded into 24-well plates at a density of 50,000 cells/mL and treated with the indicated concentration of GSK983 for 72 h. The final concentration of DMSO in each well was < 0.1%. Following 72 h GSK983 treatment, cells were harvested and pelleted (399g, 5 min). The supernatant was removed by vacuum. Cell death by apoptosis was analyzed using a BioVision Annexin V-FITC Apoptosis Kit (catalog number K101–100). A staining solution was prepared by first diluting the provided Annexin V-FITC solution in binding buffer according to the specifications of the manufacturer. 7-AAD was added from a stock solution in DMSO (0.67 mg/mL) such that the final concentration of 7-AAD in the staining solution was 2.5 μg/mL. Cell samples were resuspended in 0.5 mL of the staining solution. Samples were incubated at room temperature for 10 min in the dark and analyzed by flow cytometry using a BD Accuri C6 Flow Cytometer. FITC fluorescence was detected in FL1 and 7-AAD fluorescence was detected in FL3. Note: The BioVision Annexin V-FITC Apoptosis Kit includes propidium iodide for detection of necrotic cells. However, we found that the use of 7-AAD for necrotic cell staining greatly facilitated flow cytometric analysis in this assay. Compared to the emission spectrum of propidium iodide, the red-shifted emission spectrum of 7-AAD has significantly less overlap with the emission spectrum of FITC.
Genome-wide shRNA screen
We infected our next-generation genome-wide lentiviral shRNA library into K562 cells as described previously16–18. Infected cells were expanded and split into two flasks. In one flask, cells were grown in the presence of 48 nM GSK983 for 14 days, while in the other flask, cells were grown in the absence of GSK983. Untreated cells were diluted to a density of 500,000 cells/mL each day. GSK983-treated cells were diluted to a density of 500,000 cells/mL as needed. After the cell culture period, untreated and GSK983-treated cells were pelleted by centrifugation. Genomic DNA was isolated and shRNA encoding-constructs were counted by deep sequenced as described previously16–18.
Genome-wide CRISPR-Cas9 screen
To conduct the CRISPR-Cas9 screen, we first designed a genome-wide CRISPR sgRNA library with 4 sgRNAs/gene, incorporating previously reported improvements to the sgRNA Cas9 binding region30. Coding sequence models for sgRNA design were based on CCDS. sgRNAs were targeted toward the 5′ end of transcripts, and sgRNAs targeted exons common to all transcripts wherever possible. sgRNAs were 19–25 base pairs long and were adjacent to an NGG at the 3′ end. sgRNAs also contained an endogenous 5′G. Where multiple possible sgRNA lengths existed, only one was picked at random to ensure that no two sgRNAs targeted the same PAM. sgRNAs were scored for off-targets as previously described23 and the most stringent score (e39m1) was used wherever possible. Negative control sgRNAs were designed against scrambled coding sequences and filtered for zero off-targets in the genome.
We generated a K562 cell line stably expressing Cas9 endonuclease by infecting K562 cells with SFFV-Cas9-BFP, which is identical to a vector described previously23 but containing Cas9 with both active catalytic sites. We infected our newly designed lentiviral genome-wide sgRNA library into Cas9-expressing K562 cells according to the procedures described for lentiviral infection of shRNA libraries16–18. Infected cells were expanded and split into two flasks. In one flask, cells were initially treated with 6 and 12 nM GSK983 with little effect; therefore the concentration of GSK983 was increased to 48 nM to ensure strong selection. Cells were grown in the presence of 48 nM GSK983 for 10 days. In the other flask, cells were grown in the absence of GSK983. After the cell culture period, genomic DNA was isolated from the untreated and GSK983-treated cells using a Qiagen DNA Blood Maxi kit according to the manufacturer’s instructions. To prepare the sgRNA sequencing library, the integrated sgRNA-encoding constructs were PCR amplified using Agilent Herculase II Fusion DNA Polymerase with primers oMCB_1562 (5′-AGGCTTGGATTTCTATAACTTCGTATAGCATAC ATTATAC-3′) and oMCB_1563 (5′-ACATGCATGGCGGTAATACGGTTATC-3′). PCR reactions contained 5x Herculase buffer (20 μL), dNTPs (1 μL of 10 mM stock), genomic DNA (10 μg), primer oMCB_1562 (1 μL of 100 μM stock), primer oMCB_1563 (1 μL of 100 μM stock), Herculase II Fusion DNA Polymerase (2 μL), and water (to adjust final reaction volume to 100 μL). The number of PCR reactions was scaled to use all of the isolated genomic DNA from the untreated and GSK983-treated cells. The conditions for the PCR reaction were as follows: 1x 98 °C/2 min, 18x 98 °C/30 s, 59.1 °C/30 s, 72 °C/45 s, 1x 72 °C/3 min. PCR amplicons from genomic DNA isolated from untreated cells were pooled, as were PCR amplicons from genomic DNA isolated from GSK983-treated cells. The pooled PCR amplicons from the untreated and GSK983-treated samples were further amplified in a subsequent PCR reaction using primer oMCB_1349 (5′-CAAGCAGAAGACGGCATACGAGATGCACAA AAGGAAACTCACCCT-3′) and a bar-coded primer (5′-AATGATACGGCG ACCACCGAGATCTACACGATCGGAAGAGCACACGTCTGAACTCAGTCACNNNNNNC GACTCGGTGCCACTTTTTC-3′), where N’s indicate Illumina index barcodes. For the second PCR reaction, the reaction mixtures contained 5x Herculase buffer (20 μL), dNTPs (2 μL of 10 mM stock), an aliquot of the amplicon from the first PCR reaction (5 μL), oMCB_1439 (0.8 μL of 100 μM stock), barcoded primer (0.8 μL of 100 μM stock), Herculase II Fusion DNA Polymerase (2 μL), and water (69.4 μL). The conditions for the second PCR reaction were as follows: 1x 98 °C/2 min, 20x 98 °C/30 s, 59.1 °C/30 s, 72 °C/45 s, 1x 72 °C/3 min. The PCR products from the untreated and GSK983-treated samples were separated by gel electrophoresis (20% TBE-PAGE, 120 V, 50 min), and then gel purified to obtain sgRNA sequencing libraries.
Ranking genes from genome-wide shRNA and CRISPR-Cas9 screens
Given the variance in the efficiency of shRNA-mediated gene knockdown, we developed a novel maximum likelihood estimator (MLE) to approximate the maximum effect size of the collective set of shRNAs targeting each gene. The distribution of shRNAs for a given gene was fit to a mixed model with three distributions: off-target, miss, and on-target. The off-target distribution corresponds to the shRNA knocking down a gene other than its intended target and is estimated from the distribution of all shRNAs other than the negative controls. The missed distribution corresponds to the shRNA having no effect and is estimated from the distribution of all negative control shRNAs. Both estimations were performed by Gaussian kernel smoother with the bandwidth determined by Scott’s rule. Finally, the on-target distribution is the uniform distribution from 0 to I, where I is a fitted parameter and corresponds to the estimated effect size. This allows for the fact that a given shRNA can be anywhere from 0 to 100 percent effective at knocking down the targeted gene product. The relative contribution of the missed and the on-target distribution was also fitted, allowing for different numbers of shRNAs to be considered on-target. The contribution of the off-target distribution was fixed at 10% to allow for outliers. The significance of the MLE was tested using a log likelihood ratio, where the p-values were empirically determined by Monte-Carlo sampling of all shRNAs other than the negative controls.
To call and rank hit genes from the shRNA screen, enrichment values for each shRNA were first calculated as the log ratio of the frequency of the shRNA-encoding construct in genomic DNA isolated from the untreated and GSK983-treated cell populations16. For each gene, the MLE described above was used to estimate both the effect of the set of shRNAs targeting the gene and a p-value representing the significance of that estimate. Genes were then filtered by significance under the Bonferroni correction at p < 0.05 and ranked according to the effect size estimate.
To rank genes from the CRISPR-Cas9 screen, we first filtered genes with fewer than four distinct targeting sgRNAs detected in the deep sequencing data. The effect size was then calculated as the median fold-enrichment value for the set of sgRNAs targeting a given gene. Custom Python scripts for analysis of both screens will be made available upon request.
GO enrichment analysis
GO enrichment analysis was performed using Enrichr (GO Biological Process option)36. Separate GO enrichment analyses were performed on the top 50 hit genes from the shRNA screen and the CRISPR-Cas9 screen. All p values were adjusted using the Benjamini-Hochberg correction for false discovery rate. Supplementary Table 1 shows all enriched GO categories from the shRNA screen with p < 0.01. Supplementary Table 2 shows all enriched GO categories from the CRISPR-Cas9 screen with p < 0.01.
Lentivirus production and lentiviral infections for individual shRNA retests
Pairs of oligonucleotides encoding shRNAs targeting top hit genes were annealed and ligated into the pMCB309 vector backbone, which was previously digested with BstXI and gel-purified. pMCB309 encodes two BstXI cut sites, puromycin resistance, and mCherry. Oligonucleotides were obtained from Integrated DNA Technologies (IDT). The plasmid map will be provided upon request. The sequences of the shRNA-encoding oligonucleotides that were ligated into pMCB309 for individual retesting are shown in Supplementary Table 3.
To produce lentivirus for individual shRNA retests, Mirus transfection reagent (catalog number MIR2305) (2.5 μL) was added to DMEM containing no FBS (97.5 μL) and incubated at room temperature for 5 min. Meanwhile, pMCB309 containing the indicated shRNA-encoding insert (0.75 μg) was combined with 3rd generation lentiviral packaging components (pMDL, pRSV, and pMD2 – available from Addgene) (0.75 μg). The total DNA mixture (pMCB309 with shRNA encoding insert + lentiviral packaging components) was added to the DMEM/Mirus mixture and incubated for 30 min at room temperature. Meanwhile, HEK293T cells were seeded into 6-well plates at a density of 1 million cells/well in 2 mL DMEM. Transfection mixtures were added to HEK293T cells dropwise and cells were incubated for 24 h at 37 °C. The following day, cells were supplemented with 3 mL fresh DMEM. Cells were incubated at 37 °C for an additional 48 h. To harvest the virus, the supernatant was collected and passed through a 0.45 μm syringe filter.
To infect K562 cells with lentivirus, K562 cells were seeded into 24-well plates at a density of 100,000 cells/well in a volume of 100 μL. Polybrene (2 μL of a 4 mg/mL stock solution) was added to the cells, followed by 1 mL of the appropriate lentivirus stock. Cells were spin infected in 24-well plates for 2 h at 1000g at 33 °C. Following spin infection, cells were resuspended and pelleted (300g, 5 min). The supernatant was removed by aspiration and the cells were resuspended in fresh RPMI growth medium and incubated for 72 h at 37 °C. The cells were expanded into 6-well plates and grown in the presence of puromycin (1.0 μg/mL) for 3–5 days to select for infected cells. Following puromycin selection, cells were pelleted (300g, 5 min) and resuspended in fresh RPMI growth medium.
Competitive growth assays for individual shRNA retests
Competitive growth assays for individual shRNA retests were conducted as follows: 250,000 K562 cells expressing a negative control shRNA or an shRNA targeting a gene of interest (mCherry+) and 250,000 uninfected K562 cells (mCherry−) were seeded into 24-well plates. Heterogeneous cell mixtures were cultured in the presence or absence of 48 nM GSK983 for 14 days. As in the genome-wide shRNA screen, untreated cells were diluted to a density of 500,000 cells/mL each day and GSK983-treated cells were diluted to a density of 500,000 cells/mL as needed. Two wells each of untreated cells and GSK983-treated cells were cultured for each shRNA retested. The proportion of mCherry+ cells was monitored by flow cytometry every 72 h using a BD Accuri C6 Flow Cytometer (mCherry fluorescence detected in FL3).
Data was analyzed as follows: for each shRNA retested, the percentage of mCherry+ cells in the two untreated wells was averaged, as was the percentage of mCherry+ cells in the two GSK983-treated wells (Supplementary Figure 3b,c). The ratio of the average percentage of mCherry+ cells in the treated and untreated cell populations was calculated to give an mCherry enrichment ratio for each shRNA retested. Thus, an mCherry enrichment ratio < 1 indicates that mCherry+ cells are depleted in the GSK983-treated cell population versus the untreated cell population. In contrast, an mCherry enrichment ratio > 1 indicates that mCherry+ cells are enriched in the GSK983-treated cell population versus the untreated cell population. For each shRNA, the logarithm of the mCherry enrichment ratio was calculated. A Wilcoxon-Mann-Whitney U Test was performed to compare the log(mCherry enrichment ratio) values for the set of shRNAs targeting each hit gene to the log(mCherry enrichment ratio) values for the set of negative control shRNAs. Using this competitive growth assay, we confirmed that knockdown of DHODH and CMPK1 sensitized K562 cells to GSK983, while knockdown of COQ2, PDSS2, PDSS1, and COQ10B protected K562 cells against GSK983 in good agreement with genomic screen results [Fig. 1g; bar length represents average log(mCherry enrichment ratio) for each gene and error bars represent standard deviation of log(mCherry enrichment ratio) values for each gene].
qPCR to confirm efficacy of individual shRNA reagents
We confirmed the efficacy of shRNAs DHODH_1, DHODH_2, CMPK1_2, and CMPK1_4 using qPCR according to procedures described previously16, and values were normalized first to levels of RPL19, and then to the first of two negative control shRNAs. The following primers were used for qPCR experiments:
hRPL19-5′ ATGTATCACAGCCTGTACCTG
hRPL19-3′ TTCTTGGTCTCTTCCTCCTTG
oMCB1679_CMPK1_qpcr_5′ AAGGTTTCGAGAGCATTCCT
oMCB1680_CMPK1_qpcr_3′ TGAAAGGAAGCAAAGCACCT
oMCB1681_DHODH_qpcr2_5′ AGTCACAGATGCCATTGGAG
oMCB1682_DHODH_qpcr2_3′ GTCCCTCCTCTCATGATCCA
Lentivirus production and lentiviral infections for individual sgRNA retests
Pairs of oligonucleotides encoding sgRNAs targeting top hit genes were annealed and ligated into the pMCB306 vector backbone, which was previously double digested with BstXI and Blp1 and gel-purified. In addition to BstXI and Blp1 cut sites, pMCB306 encodes puromycin resistance and GFP. Oligonucleotides were obtained from Integrated DNA Technologies (IDT). The plasmid map will be provided upon request. The sequences of the sgRNA-encoding oligonucleotides that were ligated into pMCB306 for individual retesting are shown in Supplementary Table 4. Lentivirus production for single sgRNA retests, spin infection into K562 cells, and puromycin selection of infected K562 cells were performed exactly as described in the procedure for individual shRNA retests.
Selected sgRNAs were also retested in HeLa cells. Lentiviral infection and puromycin selection of HeLa cells was performed as follows: HeLa cells were seeded into 24-well plates at a density of 50,000 cells/well. DMEM containing the appropriate lentivirus (1mL) was added to each well and cells were incubated for 72 h at 37 °C. Cells were expanded into 6-well plates and grown in the presence of puromycin (1 μg/mL) for 3–5 days to select for infected cells. DMEM containing puromycin was removed and the cells were re-plated in fresh growth medium.
Competitive growth assays for individual sgRNA retests
For competitive growth assays to retest individual sgRNAs in K562 cells, 250,000 cells expressing an sgRNA targeting a gene of interest (GFP+) and 250,000 cells expressing an sgRNA targeting GFP (GFP−) were seeded into 24-well plates. Cells were cultured in the presence or absence of 48 nM GSK983 for 12 days. Untreated cells were diluted to a density of 500,000 cells/mL each day and GSK983-treated cells were diluted to a density of 500,000 cells/mL as needed. Two wells each of untreated cells and GSK983-treated cells were cultured for each sgRNA retested. The proportion of GFP+ cells was monitored by flow cytometry every 48–72 h using a BD Accuri C6 Flow Cytometer (GFP fluorescence detected in FL1).
For competitive growth assays to retest individual sgRNAs in HeLa cells, 10,000 cells expressing an sgRNA targeting a gene of interest (GFP+) and 10,000 cells expressing an sgRNA targeting GFP (GFP−) were seeded into 24-well plates. Plated cells were incubated for 24 h at 37 °C. Following 24 h incubation, the growth medium in each well was removed and cells were provided fresh growth medium with or without 48 nM GSK983. Cells were cultured in the presence or absence of 48 nM GSK983 for 13 days. Cells were split back 8-fold every 72 h in order to ensure that cell growth remained logarithmic. Two wells each of untreated cells and GSK983-treated cells were cultured for each sgRNA retested. The proportion of GFP+ cells was monitored by flow cytometry every 72 h using a BD Accuri C6 Flow Cytometer (GFP fluorescence detected in FL1).
Cloning, expression, and purification of recombinant human DHODH
Endogenous human DHODH contains a 29 residue N-terminal mitochondrial signal peptide. Here, we expressed and purified a truncated DHODH lacking the N-terminal signal peptide (Δ29DHODH) to facilitate detergent-free purification and increase the solubility of the recombinant enzyme in aqueous buffers. Others have shown that the Δ29DHODH construct retains full catalytic activity despite the N-terminal truncation45. The DNA sequence encoding human DHODH was PCR-amplified from a cDNA template (Origene, SC128197) using Δ29DHODH forward primer: 5′-ACGACAAGCATATGGCCACGGGAGATGAGCG-3′ and Δ29DHODH reverse primer: 5′-GCGACCCGAATTCGGCCGCCGATGATCTGCTCCA ATGGC-3′. The PCR reactions contained 1xGC-Rich Buffer (NEB), DMSO (4%), Phusion High-fidelity DNA Polymerase (NEB) (0.05 unit/μL), dNTPs (2 mM), MgCl2 (1.5 mM), cDNA template (20 ng) and primers (1 μM each). An annealing temperature of 53.9 °C was used for PCR amplification. The amplified insert was cloned into pET21a using NdeI and EcoRI restriction sites to form a construct encoding Δ29DHODH with a C-terminal His6 tag.
Expression of recombinant human DHODH was performed as described previously33. Briefly, E. coli BL21 (DE3) cells were transformed with a pET21a plasmid containing an insert encoding Δ29DHODH. Single colony transformants were grown in a 37 °C shaker for 12–15 h in 50 mL of 2xYT medium (Sigma Aldrich) supplemented with 100 μg/mL ampicillin. A 20 mL aliquot of bacterial culture was centrifuged at 5000g for 10 min. The supernatant was discarded, and the cell pellet was resuspended in fresh 2xYT medium (20 mL). The resuspended cells were added to 2 L of 2xYT medium containing 100 μg/mL ampicillin, and the culture was grown aerobically at 37 °C until A600 was ~ 0.6 to 0.8. IPTG (0.1 mM) and flavin mononucleotide (100 μM) were added and the culture was grown aerobically at 25 °C for an additional 20 h. Cells were harvested by centrifugation at 5000g for 20 min at 4 °C. The resulting cell paste was stored at −80 °C prior to protein purification.
Recombinant human Δ29DHODH was purified as follows: cell paste (30 g) was thawed and resuspended in ~100 mL start buffer containing HEPES (50 mM, pH 7.7), NaCl (300 mM), glycerol (10% v/v), and Triton X-100 (0.5% v/v). The cells were lysed by sonication (35% amplitude, 4.0 s pulse, 9.0 s pause, 15 min). The sample was centrifuged at 23,700g for 45 min (Beckman JA-20 rotor) and the supernatant was passed through a 0.45 μm syringe filter. The sample was loaded onto a 5 mL HisTrap HP column (GE Healthcare Life Sciences). The column was washed with 20 mL each of start buffer containing 5 and 10 mM imidazole. The desired Δ29DHODH protein was eluted by gradually increasing the imidazole concentration to 500 mM. The eluate was collected in several fractions, which were analyzed by Tris-SDS-PAGE (4–20% polyacrylamide gradient). The fractions containing the desired protein were combined and concentrated to a volume of less than 2 mL using an Amicon Ultra-15 centrifugal filter unit with a 10 kDa molecular mass cut off (Millipore). An Amicon filter was used to exchange the concentrated protein into a storage buffer containing HEPES (50 mM, pH 7.7), KCl (300 mM), and glycerol (10% v/v). Following buffer exchange, Δ29DHODH was aliquoted, flash frozen with liquid nitrogen, and stored at −80 °C.
In vitro enzyme activity assays with recombinant human Δ29DHODH
Δ29DHODH activity was measured in the presence of teriflunomide, GSK983, and GSK983 analogues using a coupled assay in which the oxidation of dihydroorotic acid (DHO) and subsequent reduction of ubiquinone is stoichiometrically equivalent to the reduction of 2,6-dichlorophenolindophenol (DCPIP)46. The reduction of DCPIP can be monitored by the loss of absorbance at 610 nm (ε = 21,500 M−1cm−1). Kinetic constants were calculated from measurements in which Δ29DHODH (10 nM) was either titrated with different concentrations of decylubiquinone (QD, 10–60 μM) at a fixed concentration of DHO (200 μM) or titrated with different concentrations of DHO (10–60 μM) at a fixed concentration of QD (100 μM). Enzymatic assays were conducted at 25 °C in a reaction buffer containing Tris (100 mM, pH 8.0), NaCl (150 mM), DCPIP (60 μM), glycerol (10% v/v), and Triton X-100 (0.1% v/v). The Km values for Δ29DHODH (15 μM for DHO and 14 μM for QD) were in good agreement with previously reported values for both the full-size and N-terminally truncated enzyme45. To determine Ki values for inhibitor compounds (teriflunomide, GSK983, 6Br-pF, 6Br-oTol, and GSK984), Δ29DHODH (10 nM) and the appropriate inhibitor compound were added to the previously described reaction buffer. For each inhibitor compound, three reactions were prepared: a control reaction containing no inhibitor and two reactions containing different inhibitor concentrations. Δ29DHODH was incubated with inhibitor compounds at room temperature for 5 min. The indicated concentration of QD and DHO (200 μM) were added to the reaction mixtures and absorbance at 610 nm was monitored using a PerkinElmer Lamda 25 UV-Vis spectrophotometer.
For enzyme activity assays, inhibitor compounds were prepared as 1 mM stock solutions in DMSO. A QD stock solution (10 mM) was prepared in a buffer containing Tris (100 mM, pH 8.0) and Triton X-100 (1.0 % v/v). A DHO stock solution (10 mM) was prepared in a buffer containing Tris (100 mM, pH 8.0). Concentrated stocks of inhibitor compounds, QD, and DHO were further diluted as needed with a buffer containing Tris (100 mM, pH 8.0) and Triton X-100 (0.1 % v/v) to prepare working stocks. The total volume of the reaction mixture for each assay was 100 μL, and the final concentration of DMSO was 0.01% (v/v).
Cloning, expression, and purification of recombinant human CMPK1
The DNA sequence encoding human CMPK1 (196 amino acid residues) was PCR-amplified from a cDNA template (GE Healthcare Dharmacon, catalog number MHS6278-202832682) using the forward primer 5′-AAAAAACATATGAAGCCGCTG GTCGTGTTC-3′ and the reverse primer 5′-CACGTCTAAAAACTGTTCCTTCC GATTCCTAGGTTTTTT-3′ and gel-purified. The PCR-amplified CMPK1-encoding construct was double digested with NdeI and BamHI-HF (New England BioLabs) in CutSmart buffer for 4h at 37 °C and PCR-purified (Thermo GeneJet PCR Purification Kit). Similarly, vector pET28 was double digested with NdeI and BamHI-HF for 3 h at 37 °C and gel-purified. The CMPK1-encoding insert was ligated into digested pET28 using T4 DNA Ligase (Invitrogen).
E. coli BL21 (DE3) cells were transformed with a pET28 plasmid containing a CMPK1-encoding insert. Single colony transformants were grown in 5 mL of LB growth medium containing 50 μg/mL kanamycin for 16 h at 37 °C. Overnight starter culture (2.5 mL) was added to 1 L of LB growth medium containing 50 μg/mL kanamycin and grown at 37 °C for ~3 h. IPTG (150 μM) was added when A600 was ~0.65. Cells were grown at 18 °C for an additional 16 h. Cells were harvested by centrifugation at 4000g for 20 min at 4 °C. The resulting cell paste was stored at −80 °C prior to protein purification.
Recombinant human CMPK1 was purified as follows: cell paste was thawed and resuspended in 50 mL of lysis buffer containing Tris (40 mM, pH 7.5), NaCl (10 mM), NaF (5 mM), and DTT (1 mM). Cells were lysed by sonication and centrifuged at 25,000g for 1 h. The supernatant was incubated with a slurry of Ni-NTA resin for 1 h at 4 °C and loaded onto a column. The column was washed with 40 mL lysis buffer, then 20 mL each of buffers containing Tris (40 mM, pH 7.5), NaCl (10 mM), DTT (1 mM), and imidazole (10, 40, or 200 mM). The eluate was examined by SDS-PAGE and fractions containing CMPK1 were combined. Isolated CMPK1 was further purified by anion exchange chromatography using an Äkta Pure 25 FPLC instrument equipped with a 5 mL Hi Trap Q anion exchange column. Prior to loading onto the anion exchange column, the combined Ni-NTA column eluate containing CMPK1 was diluted in Buffer A containing Tris (50 mM, pH 8.0) and DTT (1 mM) such that the imidazole concentration in the diluted sample was less than 50 mM. The desired protein was eluted from the anion exchange column using a linear gradient from 0 to 95% Buffer B containing Tris (50 mM, pH 8.0), DTT (1 mM), and NaCl (500 mM) over 15 column volumes. Fractions containing the desired protein were spin-concentrated to a volume of less than 1 mL using an Amicon Ultra-15 centrifugal filter unit with a 10 kDa molecular mass cut off (Millipore). An Amicon filter was used to exchange the concentrated CMPK1 into a storage buffer containing Tris (50 mM, pH 7.5) and glycerol (10% v/v). Following buffer exchange, purified CMPK1 was aliquoted, flash frozen with liquid nitrogen, and stored at −80 °C.
In vitro enzyme activity assays with recombinant human CMPK1
The activity of recombinant human CMPK1 was measured in the presence of GSK983 using an ATP consumption assay. CMPK1 (10 nM) was added to a reaction buffer containing Tris (50 mM, pH 7.5), MgCl2 (2 mM), DTT (2 mM), ATP (100 μM), CMP (100 μM), and GSK983 (0–10 μM, from a DMSO stock solution). The final concentration of DMSO in all reaction mixtures was 2% (v/v). Enzyme activity assays were conducted at 37 °C. The consumption of phosphoryl donor ATP was monitored using a Promega Kinase-Glo Luminescent Kinase Assay kit according to the manufacturer’s instructions. We observed no effect of GSK983 on CMPK1 activity in vitro at GSK983 concentrations up to 10 μM.
Pyrimidine metabolite supplementation experiments
For pyrimidine metabolite supplementation experiments in K562 cells, cells were seeded into 24-well plates at a density of 50,000 cells/mL in RPMI growth medium containing the indicated concentration of pyrimidine metabolite and the indicated concentration of DHODH inhibitor (GSK983 or teriflunomide) where applicable. Pyrimidine metabolites were dissolved directly in the growth medium. GSK983 and teriflunomide were delivered from stock solutions in DMSO as described above. Cells were incubated in the presence of exogenous pyrimidines and the indicated DHODH inhibitor at 37 °C for 72 h. Following 72 h treatment, cells were harvested and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer. For 6 day experiments, cells were plated as described above and diluted to a density of 50,000 cells/mL after 3 days using fresh growth medium containing the appropriate concentrations of GSK983 and pyrimidine metabolites, and cell viability was assessed after an additional 3 days (6 days total) as described above.
For pyrimidine metabolite supplementation experiments in HeLa and A549 cells, cells were seeded into 24-well plates at a density of 20,000 cells/well and incubated for 24 h at 37 °C. Following 24 h incubation, the growth medium in each well was removed and replaced with fresh DMEM containing the indicated concentration of pyrimidine metabolite and GSK983. Pyrimidine metabolites were dissolved directly in the growth medium. GSK983 was delivered from a stock solution in DMSO as described above. Cells were incubated in the presence of exogenous pyrimidines and GSK983 at 37 °C for an additional 72 h. Following 72 h treatment, cells were harvested and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer.
Cell cycle analysis based on 5-ethynyl-2′-deoxyuridine (EdU) incorporation
K562 cells were treated with 48 nM GSK983, 1 mM deoxycytidine, both 48 nM GSK983 and 1 mM deoxycytidine, or neither (untreated) for 24 h. Cells were seeded into 6-well plates at a density of 200,000 cells/mL. The final concentration of DMSO in each well was < 0.1%. After 24 h incubation with GSK983/deoxycytidine, cells were treated with 10 μM EdU for 2 h. EdU-treated cells were harvested, pelleted (300g, 5 min), washed (0.5 mL 1x PBS), and resuspended in 70% EtOH (1 mL, 0 °C) with mild vortexing. Cells were fixed in 70% EtOH overnight at 4 °C. The following day, fixed cells were pelleted (1900g, 5 min), washed (0.5 mL 1x PBS) and resuspended in 200 μL of a freshly prepared EdU labeling solution containing Tris-HCl (100 mM, pH 8.5), CuSO4 (1mM), ascorbic acid (100 mM), and the azide-functionalized fluorophore Azide-fluor 488 (10 μM). Cells were incubated in the EdU labeling solution in the dark at room temperature for 30 min. The cells were pelleted (475g, 5 min), and the supernatant was removed by vacuum. Cells were washed twice (0.5 mL 1x PBS for each wash). Following the second PBS wash, cells were resuspended in 200 μL of a freshly prepared total DNA content staining solution containing 7-AAD (25 μg/mL in 1x PBS; 7-AAD diluted in PBS from a stock solution in DMSO). Cells were incubated in the total DNA content staining solution in the dark at room temperature for 20 min. Cells were analyzed by flow cytometry using a BD Accuri C6 Flow Cytometer. Azide-fluor 488 fluorescence was detected in FL1. 7-AAD fluorescence was detected in FL3. In a first control experiment, cells that had not been treated with EdU were subjected to the dual staining protocol described above. In the absence of EdU treatment, we observed no specific labeling of S phase cells (data not shown). In a second control experiment, cells that had been incubated with 10 μM EdU for 2 h were stained with 7-AAD alone. In the absence of Azide-fluor 488, we observed no specific labeling of S phase cells (data not shown).
Construction of pDENV-Luc infectious clone
The design of the DENV reporter used here was based on the previously described mDV-R construct47. Briefly, the viral 5′UTR was followed by a duplication of the first 104 nucleotides of the C coding region, which contains cis-acting elements required for replication (CAE). The CAE was fused to the Renilla luciferase coding region followed by the complete DENV open reading frame (ORF). Between the Renilla luciferase and DENV structural protein coding sequences, a foot and mouth disease virus (FMDV) 2A sequence was introduced to provide co-translational cleavage and luciferase release. The construct was based on pD2/IC-30P, which contains a full-length infectious clone encoding dengue virus serotype 2 strain 1668148. We introduced an Envelope Q399H mutation to enhance viral infection in mammalian cells using a QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) (primers: 5′-GGAAGTTC TATCGGCCACATGTTTGAGACAAC-3′ and 5′-GTTGTCTCAAACATGTGGCCGATAGA ACTTCC-3′). We gene-synthesized a fragment containing the T7 polymerase promoter sequence followed by the first 102 nucleotides of the C coding region in frame with the Renilla luciferase and FMDV 2A sequences. This fragment was PCR-amplified, introducing a SacI site at the 5′ end and a NheI site (present in the FMDV 2A sequence) at the 3′ end using primers: 5′-CGAAATTCGAGCTCACGCG-3′ and 5′-TCCTGCTAGCTTGAGCAAATCAAAGTTC-3′. To create an in-frame fusion of FMDV 2A with the DENV-ORF, a second DNA fragment was PCR-amplified from a pD2/IC-30P template (primers: 5′-TCAAGCTAGCA GGAGACGTTGAGTCCAACCCCGGGCCCATGAATAACCAACGGAAAAAGGCG-3′ and 5′-GGAAGAGCATGCAG TCGGAAATG-3′), thus introducing 5′ NheI and 3′ SphI restriction sites. The two fragments were cut with the respective restriction enzymes and ligated into pD2/IC-30P (previously digested with SacI and SphI) to create pDENV-Luc. DENV-Luc virus was produced via in-vitro transcription of pDENV-Luc and transfection into BHK cells as described previously48.
DENV antiviral assays
For DENV inhibition assays (Fig. 3c), human A549 cells were seeded into 24-well plates at a density of 20,000 cells/well and incubated for 24 h. The cells were treated with DMEM containing GSK983 at the indicated concentrations for 4 h at 37 °C. Growth medium containing GSK983 was removed and cells were incubated for 1 h with DENV-Luc (no GSK983) at 37 °C. Following DENV-Luc incubation, cells were washed with 1x PBS and treated with fresh DMEM containing GSK983 at the indicated concentrations. Cells were incubated at 37 °C for an additional 72 h. Where specified, the growth medium was supplemented with 1 mM uridine or 1mM deoxycytidine. DENV-Luc replication was monitored by the production of Renilla luciferase, which was measured using the Renilla-Glo Luciferase Assay System (Promega) according to the specifications of the manufacturer.
For the accompanying cell viability assay (Fig. 3d), A549 cells were seeded into 24-well plates at a density of 20,000 cells/well incubated for 24 h at 37 °C. Cells were then treated with GSK983 at the indicated concentration for 72 h. Where specified, the growth medium was supplemented with 1 mM uridine or 1 mM deoxycytidine. Following 72 h treatment, cells were harvested and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer.
VEEV antiviral assays
A plasmid encoding the recombinant VEEV-GFP genome (vaccine strain TC-83) was kindly provided by Professor Frolov (University of Alabama at Birmingham)49. This plasmid was used to generate replication competent VEEV (capable of expressing GFP upon infection) in Huh7 cells. For the VEEV inhibition assay (Supplementary Fig. 6e), human A549 cells were seeded into 96-well plates at a density of 10,000 cells per well and incubated for 24 h at 37 °C in DMEM containing GSK983 only (at the indicated concentrations), or supplemented with 1 mM uridine, 1mM deoxycytidine, 1 mM dihydroorotic acid, or 1 mM orotic acid. Following this incubation, VEEV-GFP (at an MOI of 20 plaque forming units/cell) was added to the media and cells were further incubated for 16 hours at 37 °C. GFP expression was then used to measure VEEV-GFP replication in the infected samples using flow cytometry. All flow cytometry was performed using a BD LSRFortessaTM cell analyzer (BD Biosciences) and data was analyzed and assembled using FlowJo software (TreeStar, Inc.).
Supplementary Material
Acknowledgments
The authors wish to thank Jonathan Weissman, Owen Chen, Kyuho Han, Brian Lowry, James Kuo, Nicholas Plugis, and Khanh Nguyen for helpful discussions. We would also like to thank Anne Brunet, Julien Sage, and Douglas Vollrath for critical reading of this manuscript. R.M.D. was supported by a National Science Foundation Graduate Research Fellowship under Grant No. DGE-114747 and a Burt and DeeDee McMurtry Stanford Graduate Fellowship. This work was funded by NIH grants U19-AI109662 and Director’s New Innovator Award Program 1DP2HD084069-01, and a seed grant from Stanford ChEM-H. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Author Contributions
R.M.D. synthesized and characterized GSK983 and analogues, performed the genome-wide shRNA and CRISPR-Cas9 screens, assisted with statistical analysis of genomic screen results, validated genomic screen hits, designed and conducted GSK983 biological activity assays, pyrimidine supplementation experiments, and cell cycle analyses, and assisted with DENV and VEEV antiviral assays. D.W.M. designed the maximum likelihood estimator metric and conducted statistical analysis of genomic screen results. A.Ö. expressed and purified DHODH and performed in vitro enzymatic assays. R.M.D. and A.Ö. expressed and purified CMPK1 and performed in vitro enzymatic assays. S. P. assisted with DENV and VEEV antiviral assays. J.E.C. and R.M. constructed the DENV luciferase reporter. M.K. designed the shRNA library with assistance from M.A.H. and M.C.B. M.A.H. designed the CRISPR-Cas9 sgRNA library with assistance from L.A.G. and M.C.B. A.L. provided technical support. J.E.C. provided guidance for DENV antiviral assays. M.S., J.S.G., C.K., and M.C.B. conceived of the study. R.M.D., C.K., and M.C.B. wrote the manuscript and prepared the figures.
References
- 1.Harvey R, et al. GSK983: a novel compound with broad-spectrum antiviral activity. Antiviral Res. 2009;82:1–11. doi: 10.1016/j.antiviral.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boggs SD, et al. Efficient asymmetric synthesis of N-[(1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazol-1-yl]-2-pyridinecarboxamide for treatment of human papillomavirus infections. Org Process Res Dev. 2007;11:539–545. [Google Scholar]
- 3.Gudmundsson KS, et al. Tetrahydrocarbazole amides with potent activity against human papillomaviruses. Bioorg Med Chem Lett. 2009;19:4110–4114. doi: 10.1016/j.bmcl.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Lou Z, Sun Y, Rao Z. Current progress in antiviral strategies. Trends Pharmacol Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debing Y, Jochmans D, Neyts J. Intervention strategies for emerging viruses: use of antivirals. Curr Opin Virol. 2013;3:217–224. doi: 10.1016/j.coviro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aman MJ, et al. Development of a broad-spectrum antiviral with activity against Ebola virus. Antiviral Res. 2009;83:245–251. doi: 10.1016/j.antiviral.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Panchal RG, et al. Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antiviral Res. 2012;93:23–29. doi: 10.1016/j.antiviral.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Kinch MS, et al. FGI-104: a broad-spectrum small molecule inhibitor of viral infection. Am J Transl Res. 2009;1:87–98. [PMC free article] [PubMed] [Google Scholar]
- 9.Giaever G, et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregori-Puigjane E, et al. Identifying mechanism-of-action targets for drugs and probes. Proc Natl Acad Sci USA. 2012;109:11178–11183. doi: 10.1073/pnas.1204524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 12.Parsons AB, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 13.Petrone PM, et al. Rethinking molecular similarity: comparing compounds on the basis of biological activity. ACS Chem Biol. 2012;7:1399–1409. doi: 10.1021/cb3001028. [DOI] [PubMed] [Google Scholar]
- 14.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 15.Schenone M, Dancik V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassik MC, et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc Natl Acad Sci USA. 2013;110:E2317–2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc. 2014;9:1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matheny CJ, et al. Next-generation NAMPT inhibitors identified by sequential high-throughput phenotypic chemical and functional genomic screens. Chem Biol. 2013;20:1352–1363. doi: 10.1016/j.chembiol.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc Natl Acad Sci USA. 2015;112:E3384–3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, et al. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure. 2000;8:25–33. doi: 10.1016/s0969-2126(00)00077-0. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 36.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JP, et al. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry. 1996;35:1270–1273. doi: 10.1021/bi952168g. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann HH, et al. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci USA. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang QY, et al. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol. 2011;85:6548–6556. doi: 10.1128/JVI.02510-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonavia A, et al. Identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV) Proc Natl Acad Sci USA. 2011;108:6739–6744. doi: 10.1073/pnas.1017142108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox RI, et al. Mechanism of action for leflunomide in rheumatoid arthritis. Clin Immunol. 1999;93:198–208. doi: 10.1006/clim.1999.4777. [DOI] [PubMed] [Google Scholar]
- 42.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 43.Qing M, et al. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob Agents Chemother. 2010;54:3686–3695. doi: 10.1128/AAC.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullrich A, Knecht W, Fries M, Loffler M. Recombinant expression of N-terminal truncated mutants of the membrane bound mouse, rat and human flavoenzyme dihydroorotate dehydrogenase. A versatile tool to rate inhibitor effects? Eur J Biochem. 2001;268:1861–1868. [PubMed] [Google Scholar]
- 46.Bader B, Knecht W, Fries M, Loffler M. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr Purif. 1998;13:414–422. doi: 10.1006/prep.1998.0925. [DOI] [PubMed] [Google Scholar]
- 47.Samsa MM, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinney RM, et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 49.Atasheva S, et al. Interplay of acute and persistent infections caused by Venezuelan equine encephalitis virus encoding mutated capsid protein. J Virol. 2010;84:10004–10015. doi: 10.1128/JVI.01151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.