Abstract
Epigenetic gene regulation is a dynamic process orchestrated by chromatin-modifying enzymes. Many of these master regulators exert their function through covalent modification of DNA and histone proteins. Aberrant epigenetic processes have been implicated in the pathophysiology of multiple human diseases. Small-molecule inhibitors have been essential to advancing our understanding of the underlying molecular mechanisms of epigenetic processes. However, the resolution offered by small molecules is often insufficient to manipulate epigenetic processes with high spatio-temporal control. Here, we present a novel and generalizable approach, referred to as ‘Chemo-Optical Modulation of Epigenetically-regulated Transcription’ (COMET), enabling high-resolution, optical control of epigenetic mechanisms based on photochromic inhibitors of human histone deacetylases using visible light. COMET probes may translate into novel therapeutic strategies for diseases where conditional and selective epigenome modulation is required.
Introduction
Epigenetic mechanisms are critical to the dynamic regulation of activity-dependent gene expression and play key roles in many physiological processes including cell proliferation, neuroplasticity, and regulation of the circadian clock 1–3. At the biochemical level, these mechanisms involve the interplay of histone-modifying complexes, which alter the N-terminal tails of histone proteins through post-translational modifications, ATP-dependent nucleosome remodeling complexes, and DNA methylation4.
The utilization of small-molecule probes as tools to interrogate the biology of epigenetic gene regulation has tremendously increased our knowledge and provided detailed insights into mechanisms essential to the dynamic regulation of the human genome 5. Small molecules provide an orthogonal and complementary approach to genetic methods 6,7. However, the precision offered by small molecules is generally confined by pharmacokinetic principles and is often insufficient when high spatio-temporal resolution is required.
Over the last decade, extensive efforts have been dedicated to develop technologies that enable the use of light to modulate biological functions. Light is a reagent that can be applied and controlled with unparalleled precision both temporally and spatially. Optogenetic methods, which have revolutionized neuroscience, are based on transgenic neurons with photo-responsive ion channels that allow for real-time manipulation of individual neurons ex vivo and in live animals 8. More recently, the development of genetically encoded light-inducible transcriptional effectors has enabled targeted gene activation 9.
Similarly, small molecules with unique chemical motifs, which can be switched reversibly between two distinct geometries upon exposure to light of a specific wavelength, have attracted attention in protein engineering and small-molecule probe design 10,11. This methodology holds tremendous potential for improving our ability to precisely control key biological systems that can overcome many of the limitations of currently available pharmacological inhibitors, without the need for genetically engineered cell lines or organisms as required by optogenetic approaches 12. However, to date, successful implementations have been limited to applications that are characterized by threshold-triggered responses, such as the inhibition of ion channels 13,14, while efficient inhibition of enzyme functions with high spatio-temporal control has not been accomplished.
Here, we report the development of a novel concept, referred to as Chemo-Optical Modulation of Epigenetically-regulated Transcription (COMET), which combines fast-relaxing photochromic ligands and small-molecule inhibitors with long target-residence time to enable the development of tool compounds that allow for high spatio-temporal control of the epigenome. Based on this strategy, we have designed class and isoform-selective inhibitors of human histone deacetylases (HDACs). We demonstrated that the COMET probes exhibit up to three orders of magnitude increased potency when exposed to blue light and allow for optically controlled HDAC-dependent modulation of gene expression in living human cells.
Results
Conceptual Approach
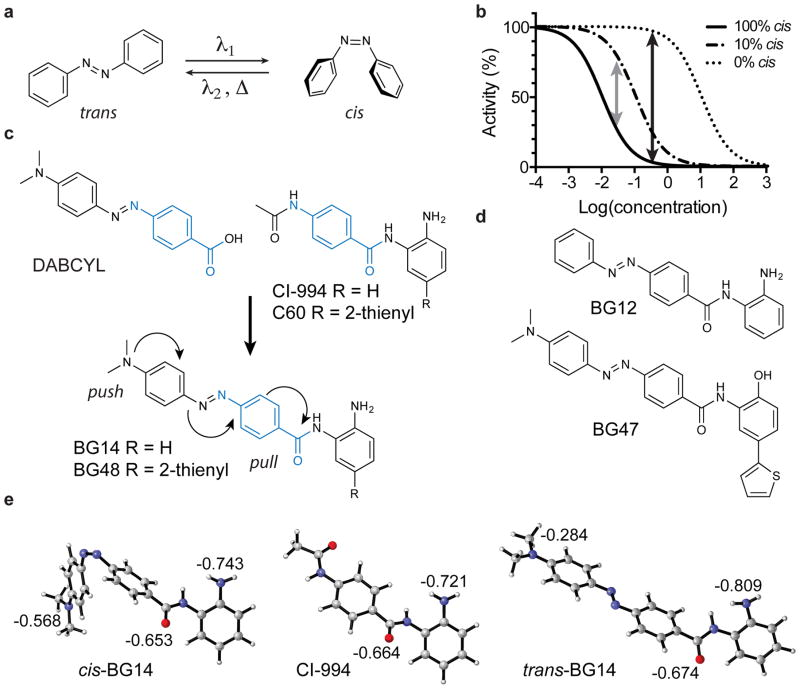
The limited success in developing light-controlled enzyme modulators is in part inherent to the molecular properties of currently employed photoswitches. Generally, a photochromic ligand can adopt two distinct geometries, which represent the thermodynamic ground state and a metastable higher-energy state 10. With azobenzenes, the most widely used photochromic ligand for biological studies to date, the respective states correspond to the trans and cis isomers (Fig. 1a). Trans/cis isomerization can be induced with ultraviolet (UV)/visible (Vis) light, and the ratio of both isomers obtained after irradiation under equilibrium conditions inversely correlates to the absorbance coefficient at the wavelength used for isomerization. In virtually all reported examples, both isomers have absorbance overlap at any given wavelength, and the ratio of the respective absorption coefficients is generally less than 10. Therefore, light-induced isomerization can never be quantitative in terms of complete transformation as it does not allow for >10:1 enrichment, and consequently light-induced “deactivation” will generally retain >5 % of the active isomer 15. While this can be sufficient for modulating a threshold-based biological function, such as an ion channel opening, it is generally not suitable to study differential enzymatic activity in a cellular context (Fig. 1b). In contrast, thermal relaxation from the metastable state (cis) to the thermodynamic ground state (trans) is quantitative. However, the rate constants depend highly on the electronic nature of the substituents, and half-lives can vary from microseconds to weeks15.
Figure 1. Overview of photochromic HDAC inhibitor design for use in COMET.
(a) Schematic of azobenzene trans/cis isomerization. (b) Theoretical model of the dose-dependent activity of a photochromatic inhibitor as a function of the trans/cis isomer ratio. (c) Azobenzene-based COMET HDAC inhibitor design combining elements of DABCYL and the class I HDAC-selective HDAC inhibitors CI-994 and compound C60 to produce the hybrid structure of BG14 and BG48, respectively. (d) Structures of control compound BG12 and HDAC1/2-biased COMET probe BG47. (e) Density functional theory (DFT) calculations (Gaussian 09) of the electrostatic potentials of CI-994, cis-BG14 and trans-BG14 showed that the electrostatic potential of the zinc-chelating moieties (carbonyl oxygen and aniline nitrogen) in cis-BG14 are very similar to CI-994 and less negative than trans-BG14.
Most efforts using azobenzenes in biological settings have focused on compounds with medium to long thermal relaxation half-lives (minutes to weeks), while compounds with short and very short (sub-milliseconds to seconds) thermal relaxation half-lives are generally considered less attractive 10,11,15. The rationale is based on the notion that maintaining a significant population of the cis isomer of a photoswitchable compound is required for sustained inhibition of the target. Compounds with fast thermal relaxation would consequently require very high light intensity and continuous exposure, which in turn could result in thermal stress or damage to the biological specimen. We hypothesized that this concern, while valid for target-inhibitor pairs that follow the standard Michaelis-Menten model with fast equilibrium kinetics, would not apply for small-molecule ligands with long target-residence times 16. Conversely, the seemingly unattractive feature of rapid thermal relaxation would become an advantage when applied to probes with long residence times. That is, provided that the metastable isomer corresponds to the active isomer, such ligands should be stabilized and remain engaged with the target following light exposure, thus eliminating the requirement for constant exposure 17,18. Unbound ligands will rapidly revert to the ground state in the absence of light, which would allow for precise confinement of small molecule activity. Here, we demonstrated that this can indeed be the case using azobenzene-based photochromic compounds designed to target HDAC family members that enable optical control of epigenetic mechanisms for the first time without the need for genetically engineered responsive cells.
Inhibitor design
Thermally fast relaxing azobenzenes commonly feature an electron-donating substituent on one benzene ring and an electron-withdrawing substituent on the other. These “push-pull” azobenzenes strongly absorb light in the visible electromagnetic spectrum, which allows for photoisomerization using well-tolerated blue light, rather than less biocompatible UV light. Furthermore, changes of overall electronic properties as result of cis isomerization are more pronounced compared to azobenzenes with symmetric substitution patterns. We have exploited this feature in our design of photochromic HDAC inhibitors that are formal hybrids of para-methyl red (DABCYL), which is commonly used as a biocompatible fluorescence quencher, and an ortho-amino anilide (OAA) HDAC inhibitor such as CI-994 (Tacedinaline), Merck Compound-60 (C60) (Fig. 1c) and MS-275 (Entinostat) 19. OAA and the corresponding hydroxyl analogs are characterized by long target-residence time and show remarkable preference for inhibiting the deacetylase activity of class I HDACs1-3 over the other eight zinc-dependent HDAC isoforms 20,21. OAA (or hydroxyl analogs) chelate the catalytic zinc in the active site of HDAC enzymes 19. The zinc binding affinity of the OAA is sensitive to electronic effects, and electron rich substituents are generally not well tolerated 22. Following this design strategy, we synthesized BG14 (1), BG47 (2), and BG48 (3) as prototype COMET HDAC inhibitors, and BG12 (4) as control compound that lacks the dimethylamino substituent (Fig. 1c, d). BG47 and BG48 feature a 4-(2′-thienyl)-substituted benzamide analogous to C60, which was predicted to provide selectivity for HDAC1/2 over HDAC3 19.
Based on the published crystal structures of HDAC2 with benzamide inhibitors (3MAX and 4LY1) 19,23 suggesting that the 4-dimethylamino phenyl moiety of these prototypical COMET HDAC inhibitors would be solvent exposed, as well as the published structure-activity-relationship of OAA series, we considered that the steric change as result of trans-to-cis isomerization would not significantly alter the affinity for the targeted HDAC isoforms. As an alternative, we hypothesized that disruption of the electron-donating effect of the DABCYL moiety as a result of the trans-to-cis isomerization would significantly change the overall electronic properties of the HDAC-targeting anilide and as a consequence increase the zinc-chelating ability of the cis isomers. To test this hypothesis, we performed density functional theory (DFT) calculations (Gaussian 09) using B3LYP with the 6-31+G(d) basis set to compute the electrostatic potential of the prototypic inhibitor BG14 in the trans and cis geometries in comparison to the reference inhibitor CI-994. As shown in Figure 1e, the electronic potentials of the benzamide carbonyl-oxygen and the aniline-nitrogen, which together chelate the active site zinc in the ligand-HDAC complex 19,23, were less negatively charged and more similar to CI-994 in the cis isomer than the thermodynamically favored stable trans isomer. These computational data supported the hypothesis that optical modulation of the electronic properties of zinc-chelating HDAC inhibitors provides a novel means to control HDAC activity using COMET probes.
Inhibitor characterization and biochemical profiling
Physical characterization of the prototype COMET HDAC inhibitors in PBS confirmed that all compounds strongly absorb light in the blue spectrum with maximum absorbance around 470 nm (Supplementary Results, Supplementary Fig. 1), which renders them suitable for biological exploration.
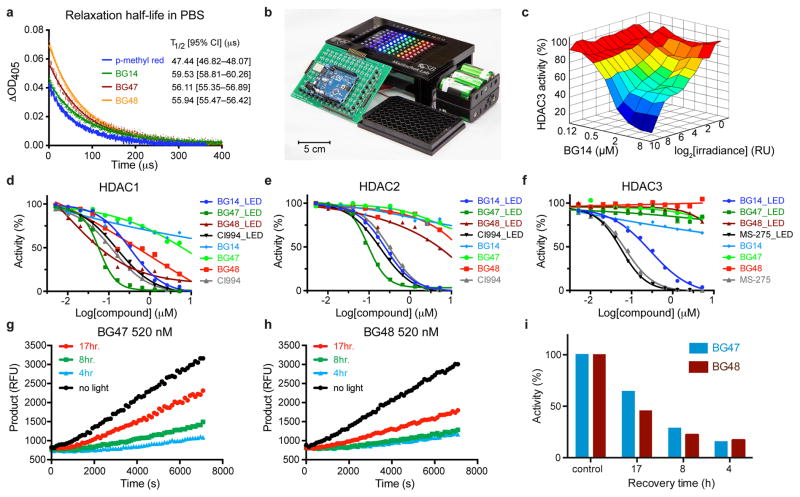
Thermal relaxation half-lives of push-pull azobenzene analogs similar to the presented compounds in aqueous solution at physiological pH have, to the best of our knowledge, not been reported. Previous approaches to study fast cis-to-trans isomerization are based on laser flash photolysis to measure the transient absorbance change following excitation with a short laser pulse24. Such experimental setups offer picosecond resolution; however, they are expensive and not commonly accessible. As an alternative, to measure relaxation kinetics with microsecond resolution, we designed a readily adaptable instrumentation setup (Supplementary Fig. 2 and Supplementary Fig. 3). Using this approach, we determined the thermal relaxation half-lives of HDAC ligands to range from 55–60 μs, relative to 47 μs for DABCYL, which is approximately 120× faster than previous measurements for DABCYL in chloroform (Fig. 2a) 17. The estimated distance of diffusion of small molecules (diffusion coefficient D = 10−6–10−7cm2/s) at 10×T1/2 (500 μs) in water is 0.1–0.3 μm, and is therefore expected to limit diffusion of “activated” inhibitors well within subcellular dimensions.
Figure 2. Optical properties and validation of light-dependent inhibitory activity toward recombinant HDACs of COMET probes.
(a) Measurement of thermal relaxation half-life. Each data point represents the mean values of 128 independent measurements. A least-squared fit modeling a one-phase decay (Graphpad Prism) was used to determine the relaxation half-lives. T1/2 values are reported with 95% Confidence Intervals. (b) 8x12 LED array for high-throughput COMET probe profiling. (c) Light intensity and concentration-dependent inhibition of recombinant HDAC3 deacetylase activity by BG14 (470 nm, Imax = 17 mW/cm2). Mean values of duplicate experiments. Dose dependent and light dependent (470 nm, 17 mW/cm2) activity of COMET probes and reference HDAC inhibitors against HDAC1(d), HDAC2(e) and HDAC3(f). Data represent mean values of duplicate experiments. Dose response estimation of HDAC target residence time of COMET probes. HDAC1 was incubated with BG47 (520 nM) (g) or BG48 (520 nM) (h) and exposed to blue light (470 nm, 17 mW/cm2) for 1 h and then kept for varying time in the dark at room temperature to allow HDAC bound inhibitor to dissociate. After the indicated time (4, 8, 17 h), HDAC substrate was added, and enzymatic activity was measured in kinetic mode over 2 h.(i) Enzymatic activity relative to control following light exposure and dark phase recovery. RU – relative units, RFU – relative fluorescence units. (g–i) Data represent individual measurements of representative biological replicate.
To allow for high-throughput profiling of biochemical and cellular activity with COMET, we designed and optimized microprocessor-controlled (using the open-source Arduino platform) 12×8 light-emitting diode (LED)-arrays that are compatible with 96-well microtiter plates and may be used in standard tissue culture incubators due to their self-contained nature and small footprint (Fig. 2b, Supplementary Fig. 4, Supplementary Fig. 5, Supplementary Video 1).
Using an LED array equipped with blue (470 nm) LEDs, which we identified to be best suited for the tested compounds (Supplementary Fig. 6), we were able to accurately profile the light-dependent HDAC inhibitory activity of the COMET probes using a biochemical assay measuring the deacetylation of a synthetic acetyl-lysine tripeptide substrate based on histone H4 Lysine 12 (Fig. 2c–f) 20. Following exposure to blue light (470 nm), but not in the absence of light, BG14, BG47 and BG48 strongly inhibited the enzymatic activity of HDAC1 (Fig. 2d) and HDAC2 (Fig. 2e) in dose-dependent fashion while no differential activity upon light exposure was observed for the reference HDAC inhibitors CI-994 and MS-275, respectively 20. As expected, profiling of the COMET probe set against HDAC3 (Fig. 2f) revealed that only BG14 potently inhibited enzymatic activity in a light-dependent fashion, while BG47 and BG48 did not inhibit HDAC3 in either the presence or absence of light, which also indicated that the inhibitory activity was not the result of non-specific inhibition. Furthermore, as shown for BG14 and HDAC3, the inhibitory activity was dependent on light intensity (Fig. 2c), demonstrating that the degree of target inhibition can be optically controlled.
In addition, BG12, a close analogue of BG14 that does not feature the dimethylamino substituent, which we deemed to be responsible for the low affinity of the COMET probes in trans-geometry because of its electron-donating effect, potently inhibited both HDAC isoforms in trans-geometry and did not exhibit differential potency upon photoisomerization (Supplementary Fig. 7). These results supported our hypothesis that electronic, rather than steric effects, are the major drivers for the differential activity of the COMET probes upon light activation. Consistent with our mechanistic predictions, the COMET probes exhibited long target residence times, which is characteristic for benzamide-based HDAC inhibitors. As illustrated in Figure 2 g–i, the enzymatic activity recovered in a time-dependent fashion following light exposure. Specifically, HDAC1, which we found to be most stable under longer biochemical assay condition, was incubated with varying concentrations of BG47 or BG48 and exposed for 1 h to 470 nm light of constant intensity (470 nm, 17 mW/cm2) followed by a variable period without light. In the absence of light, unbound COMET probes were inactive, and enzyme-bound inhibitors dissociated at a rate governed by koff and thereby liberated catalytically active HDAC. Because unbound COMET probes quickly relaxed to the inactive trans-isomer, dissociation was essentially irreversible.
Inhibitor In Vitro Profiling
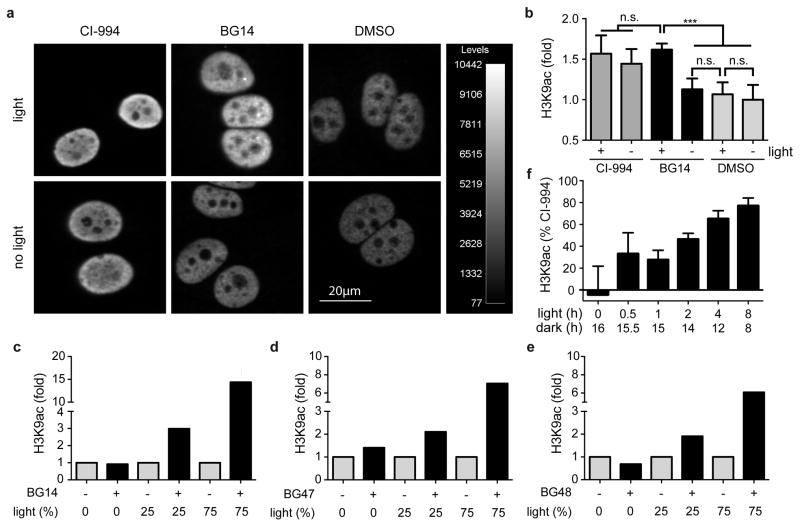
Following the biochemical characterization of the COMET probes, we profiled BG14, BG47, and BG48 in cellular assays using acetylation of histone H3 Lysine 9 (H3K9ac) as a pharmacodynamic marker for HDAC inhibition. As shown in Figure 3, treatment of MCF-7 cells (a human breast cancer cell line) with BG14 in the presence of light for 16 hours strongly induced acetylation of H3K9 relative to the vehicle control comparable to CI-994 treatment in an immunofluorescence assay (Fig. 3 a, b). H3K9 acetylation levels directly correlated with total light exposure (Fig. 3 c–e, Supplementary Fig. 8) as quantified by western blot analysis. Independently of the assay method, we did not observe significant increase in histone acetylation in the absence of light. As expected, BG14, which also inhibited HDAC3, induced the strongest increase of H3K9ac (Fig. 3c) compared to the HDAC1/2-selective inhibitors BG47 (Fig. 3d) and BG48 (Fig. 3e). These results with COMET probes demonstrated, for the first time, light-dependent control of the human epigenome.
Figure 3. Light-dependent control of the human epigenome with COMET probes.
(a) Immunofluorescence analysis of H3K9ac levels in MCF-7 cells treated with CI-994 (25 μM) and BG14 (25 μM) in presence and absence of light (470 nm, 8.5 mW/cm2, 1Hz, 75% duty cycle). Scale bar = 20μm. (b) Quantification of H3K9ac intensity changes by automated confocal microscopy. Mean values +s.d. are shown (n=3). Significance values (n.s. – not significant, *** p < 0.001) were calculated by one-way ANOVA with Tukey’s post-hoc test (Graphpad PRISM). (c–e) Western blot analysis of H3K9ac levels in MCF-7 cells after treatment with COMET probes at 25 μM and varying amounts of light exposure (Supplementary Fig. 8). Specifically, cells were exposed to no light or pulsing light (470 nm, 8.5 mW/cm2) modulated at a frequency of 1 Hz (the duty cycle is expressed in % ON time). Values were normalized to vehicle treated cells and represent duplicate means of biological duplicates. (f) Persistence of HDAC inhibition. Quantification of relative immunofluorescence intensity of H3K9ac upon treatment of MCF-7 cells with DMSO, CI-994 (25 μM) and BG14 (25 μM) for a total of 16 h experimental time under varying duration of light exposure (470 nm, 8.5 mW/cm2, 1Hz, 50% duty cycle) at the beginning of the treatment period, followed by varying length of dark phases. Cellular H3K9 acetylation levels were normalized within each group to the positive control CI-994 = 100% and DMSO = 0. Mean values +s.d. are shown (n≥3). Detailed statistical values are provided in Supplementary Figure 8.
Next, we investigated the time dependence and persistence of HDAC inhibition following light exposure and measured H3K9ac, which is a highly dynamic histone modification, 25 after varying periods of light exposure and dark recovery. As shown in Figure 3f (see Supplementary Fig. 9 for detailed analysis), H3K9ac levels increased with prolonged exposure to 470 nm light and plateaued around 4–8 h at an intensity level that is comparable to the effect observed with CI-994. Importantly, H3K9 acetylation levels were persistent for over 8 h without light exposure, consistent with prolonged inhibition of HDAC activity as a consequence of the long residence time of our inhibitors. These results were in good agreement with the biochemical profiling data demonstrating optical control of HDAC inhibition.
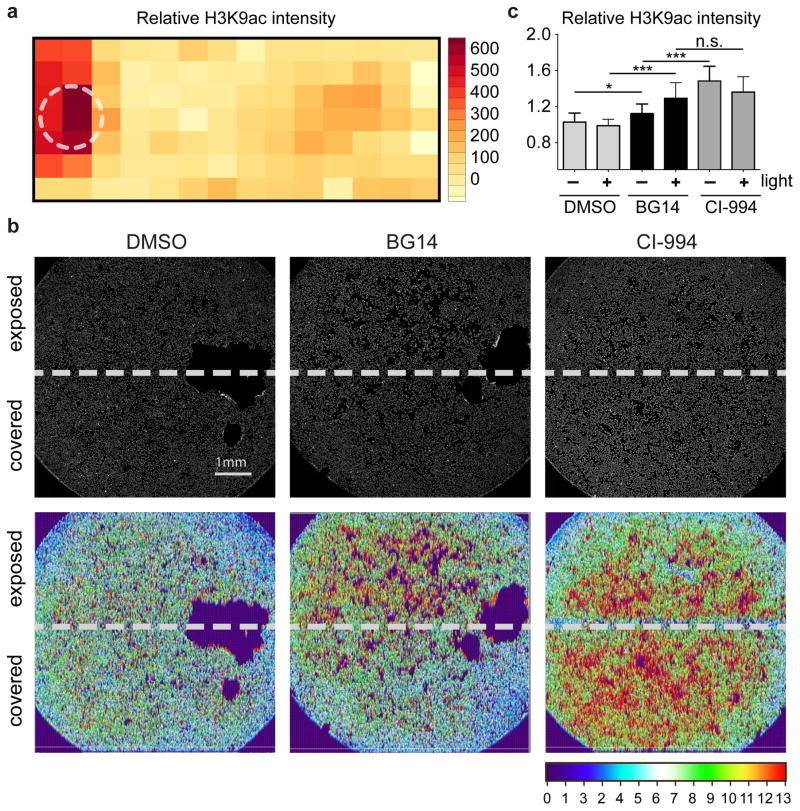
To demonstrate spatial control within a cell population, we exposed MCF-7 cells grown in a 1-well chamber slide in the presence of BG14. Using our 96-well LED array, we exposed one side to blue light from a single LED for 6 h followed by 10 h in the dark and quantified histone H3K9ac by immunofluorescence staining. As shown in Figure 4a, significant increase of H3K9 acetylation was limited to the light exposed areas within the same chamber.
Figure 4. Light-dependent spatial control of epigenetic regulation with the COMET probe BG14.
Quantification of relative immunofluorescence intensity of H3K9ac upon treatment of MCF-7 cells with BG14 and light. (a) MCF-7 cells in a 1-well chamber slide were placed over a 5 mm LED (470 nm, 8.5 mW/cm2, 0.9 Hz, 33% duty cycle for 6 h, followed by 10 h without light exposure). The heatmap represents relative levels of histone H3K9ac (red indicates elevated H3K9ac levels). The white circle indicates position of LED. (b) Wells of a 96-well culture plate were partially covered at the bottom of the plate prior to treatment of MCF-7 cells with DMSO control, BG14 and CI-994 in presence of light for 16 h followed by immunofluorescence staining for H3K9ac. Heatmaps of H3K9ac intensity throughout wells. Scale bar = 1mm. (c) Quantification of H3K9ac spatial intensity differences by automated, confocal microscopy. Mean values +s.d. (n=5). Significance values (n.s. – not significant, * p < 0.05, *** p < 0.001) were calculated by one-way ANOVA with Tukey’s post-hoc test (Graphpad PRISM).
To further explore the degree of spatial control that is achievable with our current LED matrix, we repeated the experiment in 96-well plates—a format that will enable high-throughput discovery and facilitate exploration of the biological effects of COMET probes under a range of experimental conditions. Light exposure was restricted to one-half of each well that was covered on the bottom with black tape, and MCF-7 cells were treated as described above. As shown in Figure 4b, neither DMSO nor CI-994 treated wells showed differential histone H3K9ac in light-exposed and non-exposed areas. In contrast, BG14 strongly induced histone H3K9ac in the light-exposed portion of the well to levels comparable to CI-994. Only a slight increase of histone H3K9ac was detectable in the covered portion of the well (Fig. 4c), which we speculated was not caused by diffusion of cis-BG14, which would have resulted in an activity gradient, but by partial internal light reflection and light scattering on the air/liquid and liquid/plastic interface that can be overcome in the future by improved optical delivery methods. Nonetheless, our results demonstrated that the HDAC inhibitory activity of BG14 can at least be confined within < 1 mm.
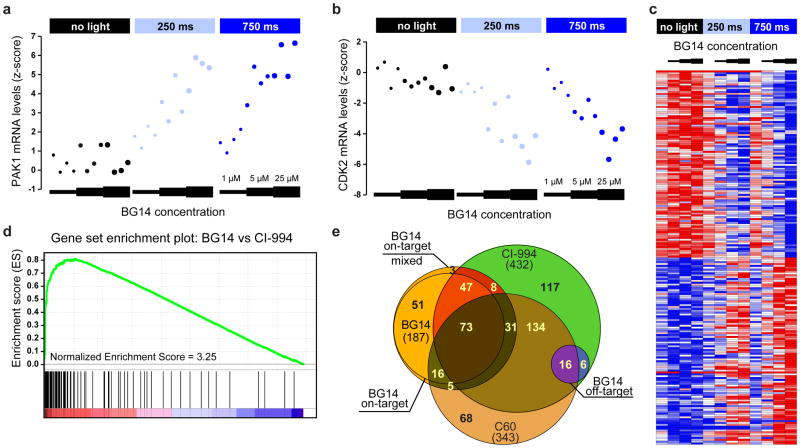
Having demonstrated light-dependent control of the acetylation state of the epigenome using COMET probes, we next sought to determine the biological consequence of HDAC inhibition at the level of the transcriptome in comparison to conventional light-independent HDAC inhibitors. To gain quantitative insights into the specific genes regulated by the COMET probes under a wide range of experimental conditions, including variation of compound dose, treatment time, and light exposure, we performed high-throughput gene expression profiling using the L1000 method from Genometry (Genometry, Inc.). The L1000 assay is a novel, high-throughput, multiplexed mRNA expression profiling technique developed as part of the NIH Common Fund supported LINCS project. It is based on the direct measurement of a reduced representation of the transcriptome (978 ‘landmark’ transcripts) and inference of the portion of the transcriptome not explicitly measured using an algorithm trained on tens of thousands of historical microarray-derived gene expression profiles. Eighty invariant transcripts are also explicitly measured to enable scaling and normalization. Specifically, we treated human MCF-7 cells and measured transcriptional changes as a function of compound concentration, light exposure, and treatment duration relative to the reference HDAC inhibitor CI-994. Light treatment alone or treatment with COMET probes in the absence of light had little to no effect on gene expression, while numerous light-dose and compound-concentration dependent transcriptional changes were observed in the presence of light. As expected for HDAC inhibition and consistent with previous reports, BG14 induced both up- and down-regulation of specific transcripts as illustrated by the examples of the directly measured landmark genes PAK1 and CDK2 (Fig. 5a, b).
Figure 5. Characterization of COMET-mediated gene expression signatures in human cells.
BG14 induces a light dose and compound concentration-dependent change in gene transcription in MCF-7 cells that is highly correlated with the gene expression signature of class I HDAC inhibitors. Specific examples of light-dependent up- and down-regulation of the HDAC inhibitor responsive landmark genes PAK1 (a) and CDK2 (b) in the presence of varying concentrations of BG14 and varying light exposure (no light, 250ms/750ms on/off, 750ms/250ms on/off (470 nm, 8.5 mW/cm2, 1 Hz) for 1 6 h. Data points represent values of 4 independent biological replicates. (c) Heat map of top 100 up- and down- regulated transcripts (landmark and inferred genes) that were specifically modulated by BG14 in light-dependent fashion. No meaningful transcriptional changes were observed by BG14 treatment alone or by light in the absence of inhibitor (control, 1 μM, 5 μM, 25μM; data represent average of n=4 measurements). (d) Gene Set Enrichment Analysis (GSEA) confirms the strongly correlated transcriptional response of CI-994 and BG14 (25 μM, 470 nm, 8.5 mW/cm2, 1 Hz, 75% duty cycle, 1 6 h). p < 0.001 and FDR q < 0.001 (e) Venn diagram showing the overlap between CI-994 and C60 response genes and BG14 regulated light-dependent “on-target” and light-independent “off-target” genes, respectively.
To obtain a more comprehensive assessment of the light-dependent activity of BG14, we performed a global gene expression analysis (using directly measured and inferred genes) and ranked the top 100 up- and down-regulated genes, respectively (Fig. 5c, Supplementary Fig. 10). The relative expression changes directly correlated with average light exposure and inhibitor concentration. Importantly, light or inhibitor treatment alone did not induce meaningful changes in the respective transcript levels for the majority of transcripts. Use of Gene Set Enrichment Analysis (GSEA) 26 to compare the expression patterns of BG14 with CI-994 demonstrated a high correlation between the respective expression profiles under light treatment conditions, establishing strong support for similar biological activity (Fig. 5d, Supplementary Fig. 11).
Interestingly, albeit fewer genes, a more significant correlation was observed between COMET HDAC inhibitors and reference HDAC inhibitors in the absence of light. Given the low HDAC inhibitory activity of BG14 and the absence of measurable induction of histone acetylation under these conditions in the biochemical assays described above, these results suggested that the observed transcriptional changes may be caused by off-target (i.e. non HDAC related) effects that are common to BG14 and CI-994, which share high chemical similarity. To explicitly differentiate off- and on-target effects of BG14, we analyzed the L1000 gene expression profiles for light-induced, dose-dependent changes. Following a 16 h treatment with and without light, BG14 significantly (normalized expression score > 1.5) changed the expression of 234 and 69 landmark genes, respectively (Fig. 5e, Supplementary Fig. 12, Supplementary Data Set 1). Of the 69 genes that were differentially expressed by BG14 treatment alone (without light), 22 genes (“off-target genes”) were not further modulated in the presence of light, while 47 genes (“mixed on-target”) showed significant additional expression changes in the presence of light. Strikingly, the two sets showed 100% and 83% overlap with the CI-994 signatures. Notably, 187 (“on-target”) of 234 genes (80%) were only differentially expressed following BG14 exposure in the presence of light, and 120 (65%) of these on-target genes (“Core Gene Set”) overlapped with the 432 CI-994 responsive genes. A network analysis of the 120 Core Gene Set identified pathways associated with cell cycle regulation and mitochondrial function (Supplementary Fig. 13a), consistent with the well-established biological consequence of HDAC inhibition 27,28. Furthermore, the genes in the cell cycle-related networks comprised a large proportion of the top 25 down-regulated genes within the Core Gene Set (Supplementary Fig. 13b). Similar results were obtained for the analysis of all BG14 on-target genes. In contrast, the network analysis of off-target genes did not identify any specific biological pathways with any statistical significance (FDR < 0.05). Additionally, we identified 343 responsive genes for the HDAC 1/2 inhibitor C60, of which 59% overlapped with CI-994. Comparably, 53% of BG14 on-target and 73% off-target genes corresponded to the C60 gene signature, further strengthening the observed HDAC inhibitory function of our COMET probe.
Discussion
The ability to modulate the function of chromatin-modifying enzymes with high spatio-temporal control in cells and tissues will be critical to enable a deeper understanding of the molecular basis and physiological consequences of epigenetic gene regulation. In the studies presented here, we have established COMET as a novel approach to modulate the activity of specific HDAC isoforms using small molecules with high precision, and have demonstrated that this inhibition of HDACs can be correlated to changes in gene expression, providing a novel means to control the transcriptome without the need to generate genetically-engineered cell lines or transgenic organisms. Given the optical and pharmacological properties of the COMET probes, our data suggest that these tools will enable inhibition of HDAC function with subcellular resolution in living cells and potentially whole organisms, with previously unmatched precision. Future applications in vivo of COMET HDAC probes may yield novel insight into complex biological processes where the afforded spatio-temporal control will be invaluable, such as memory formation 1, as well as novel therapeutic strategies for disorders with epigenetic dysregulation, including neurodegeneration 29,30, stress responses 31,32, and oncogenesis 33. Of particular interest in this context is to selectively target defined regions of tissues, for example to establish a dose-response study within the same tumor sample or regions of a tissue in the nervous system that are known to exhibit different effects on a neurocircuit, in order to better understand the spatial-temporal regulation of biological function.
Polypharmacology of small molecules is well recognized, and specific or non-specific binding to secondary targets can contribute to the global biological response 34. Off-target effects mediated by unknown or unintended targets can complicate the use of pharmacological probes to dissect protein function. Commonly used “best practices” most often rely on comparative analysis of active versus inactive molecules differing in structural elements. Since structurally distinct molecules can exhibit significantly altered physicochemical and other properties (e.g. cell permeability, metabolism, stability), such differences may confound the interpretation of the resulting biological effects. Alternatively, as we demonstrated here with COMET HDAC inhibitors, exploiting the photochromic properties of molecules offers the compelling advantage of allowing comparison of the same molecular entity in inactive versus active states. In particular for photochromic ligands with fast thermal relaxation kinetics, the fraction of free ligand in the thermodynamically unfavored state will be very small and only exist during periods of light exposure. In addition, off-target interactions caused by such photochromic ligands that are specific to the thermodynamically unfavored state are expected to be transient except for off-target interactions that are also characterized by very slow dissociation kinetics. However, differential membrane diffusion kinetics altering cell permeability may be observed as a consequence of the distinct molecular geometries and consequently alter the kinetics of cellular uptake kinetics. This concern will especially apply for measurements performed within minutes of ligand addition in a pre-equilibrium state and should be characterized by the absence of off-target effects in the non-light control.
Having developed a readily accessible and extensible platform for optoepigenetic profiling using LED arrays, we expect that this approach will furthermore be compatible with instrumentation setups that are commonly already used for optogenetic studies and will thus be readily implemented. We note that while the very long target residence time can represent an advantage for applications that require prolonged target inhibition, it will nonetheless be desirable in the future to develop analogs with shorter residence time for experimental designs that require tight temporal control over target-ligand dissociation. Here our Arduino-based, microprocessor-controlled LED-array platform will greatly improve the throughput of discovering novel photochromic probes and optimizing their use in biological systems. Thus, upon the identification of ligands with optimized residence times for other targets, as well as the future development of photochromic scaffolds specifically tuned for in vivo use, a novel optically-based pharmacology for biological investigation and therapeutic applications can be envisioned.
Online Methods
Measurement of cis-to-trans Relaxation
The lifetime of the active molecule was measured by recording the time evolution of the optical absorbance of the solution. Conventional instruments are poorly suited for making this measurement because the change happens relatively quickly (on the order of 100 μs) and the percentage change in absorbance is small. We developed our instrumentation setup specifically to measure the rate at which the absorbance returns to its resting rate following photo activation.
Optical Excitation System
A single 405 nm laser diode (US Lasers D405-120) was used both to activate the sample and to probe the optical density. During activation, 8 V was applied across the laser diode for 100 μs with 10 μs rise and fall times by an arbitrary waveform generator (Tektronix AFG3252). The activation pulse was repeated at a frequency of 1 kHz. Between activation pulses, 3.6 V was applied across the laser diode to produce a beam for probing the optical density of the sample. The laser power was measured independently in activation and probing mode by alternating between the mode of the interest and turning the laser completely off. The average power was then measured using a laser power meter (Edmund Optics Lasercheck). The transient power during each mode was then calculated as 13.8 mW during activation and 17.4 μW during probing by virtue of the known duty cycle.
Optical Measurement System
The optical signal was converted to charge using an avalanche photodiode (Hamamatsu Si APD S2382) operating well below its avalanche threshold to achieve high linearity and high speed. The cathode was biased at 30V from a bench top power supply (Keithley 2402 Sourcemeter). The anode was connected to a transimpedance amplifier built out of an operational amplifier (Analog Devices AD8532) with resistive feedback as shown in Supplementary Fig. 2. This operational amplifier was chosen primarily based on its low input bias current of 5pA along with sufficient speed (3 MHz bandwidth) and rail-to-rail operation. A 500kΩ resistor was used to convert the current from the photodiode to a voltage at 0.5V per μA.
The output signal from the transimpedance amplifier contained an offset of approximately 4 Volts. This offset was reduced in order to digitize the signal at the maximum gain on the oscilloscope, and a second operational amplifier was used to adjust this offset. The same type of operational amplifier was used in the second stage, here configured as an inverting stage. A potentiometer was used to control the constant voltage that was subtracted from the output of the first stage (V1). In order to accurately place the final output (V2) around earth ground, the circuit used a negative supply at −0.8 V.
The output of the measurement circuit was digitized using a 1GHz real time oscilloscope (Tektronix MSO 4104). The channel was configured for 1MΩ input with a 200μs time base and a 20mV vertical scale. Averaging was performed on the oscilloscope over 128 traces. The data was then transferred from the oscilloscope over a universal serial bus (USB) connection to a computer running Matlab (Mathworks). Ten such traces were then acquired and averaged. An illustration of the entire measurement system is depicted in Supplementary Fig. 3.
Optical Measurement Procedure
Stock solutions (0.5 mM) of test compounds were prepared in dimethylformamide (DMF) (Fisher) and diluted 100:1 in phosphate buffered saline (PBS) (Fisher). 100 μL of the solution was then transferred into a semi-micro 10 mm cuvette (Fisher 14-955-127). A blank cuvette was prepared with 100 μL of PBS.
For each sample, ten measurements were made of the blank as well as the sample itself. The light absorbed by the sample was calculated by taking the difference between the data measured for the blank and the data measured for the sample. The difference signals for all ten runs were then averaged together. The averaged signal was filtered through a 100 kHz lowpass filter (5-tap infinite-impulse response Butterworth) to remove high frequency noise. A least-squared fit modeling a one-phase decay (Graphpad Prism) was used to determine the relaxation half-lives. Values are reported as 95% Confidence Intervals.
Microplate-compatible 8x12 LED Array
To allow for high-throughput profiling, we have developed microprocessor controlled 12 x 8 LED arrays that match the standardized 96-well microplate layout (figure 4). The experimental setups were assembled from four modular units (PCB board with LEDs, microplate stage, microcontroller, power source) (Supplementary Fig. 4 and Supplementary Fig. 5).
PCB board with LEDs
PCB boards to hold LEDs, transistors, resistors, and microcontroller interface were designed using ExpressPCB Software and manufactured by ExpressPCB (ExpressPCB, www.expresspcb.com). All electronic components were purchased from Mouser Electronics (www.mouser.com) and assembled in house. LED arrays either displayed columns with different color LEDs (with identical LEDs within each column) to allow for evaluation of wavelength dependent activity or 96-LED arrays with identical LEDs (385 nm, Mouser#_593-VAOL5GUV8T4_385 nm; 390 nm, Mouser#_749-UV5TZ-390-30_390 nm; 405 nm, Mouser#_749-UV5TZ-405-30_405 nm; 430 nm, Mouser#_638-MV8B01_430 nm; 470 nm, Mouser#_941-C503BBCNCV0Z0462_470 nm; 502 nm, Mouser#_638-MV8G03_502 nm; 527 nm, Mouser#_941-C503BGCNCY0C0792_527 nm; 562 nm, Mouser#_638-MV64521_562 nm; 591 nm, Mouser#_941-C503BACNCY252030_591 nm; 624 nm, Mouser#_941-C503BRCNCW0Z0AA2_624 nm; 660 nm, Mouser#_696-LX5093SRC/F_660 nm). Per column current was set at 20 mA by Temperature Compensated Constant Current LED Driver ICs (Supertex Inc., Mouser# 689-CL25N3-G) and switched using NPN transistors (Mouser# 512-PN2222ABU).
Microplate Stage
Microplate stages to hold LED arrays were designed using Adobe Illustrator and manufactured by laser cutting from black acrylic resin (Ponoko). The stages were designed with exchangeable faceplates to match top and bottom profiles of 96-well microplates, enabling illumination from top or bottom, respectively. For cell culture experiments, we included a matching 96-well mask that was manufactured by water jet cutting (ACP Waterjet) from 0.25” copper as heat sink to limit temperature changes within < 0.5 °C. The LED arrays were connected to an Arduino microcontroller (Arduino Uno or Arduino Mega, www.arduino.cc) that was programmed to control individual LEDs, LED rows or LED columns within the array. The controller allowed modulation of LED intensity by modulating the on/off timing (PWM) or by directly modulating the voltage supplied. Exposure of each well can be individually programmed, enabling precise modulation of the light exposure with <10 ms resolution (limited by LED response). We have designed the integrated circuit of LEDs and microcontroller to be powered optionally by a power supply unit connected to a standard 110 V outlet, computer USB-port or by a battery-pack of 6 conventional rechargeable C-cell batteries, which allows for use in a cell culture incubator for up to 16 h.
Biological Profiling
Biochemical HDAC assay
The light-dependent inhibitory activity of the test and reference compounds was determined as reported previously with minor modifications.20 Full length recombinant HDAC proteins (BPS Biosciences) were diluted into HDAC assay buffer (50 mM HEPES, 100 mM KCl, 0.001% (v/v) Tween-20, 0.05% (w/v) bovine serum albumin (BSA), pH 7.4) and 100 μL/well dispensed (Multidrop, Thermofisher) into white 96-well plates (Corning) followed by dispensing of test compounds from 10 mM stock solutions using an HP D300 Digital Dispenser (Hewlett-Packard). The LED-array was then placed on top of the 96-well plate and enzyme inhibitor solutions were exposed to light for 1 h (470 nm, 17 mW/cm2). Light exposure was either continues at fixed intensity or modulated by on/off cycles at a frequency of 1 Hz. Following light exposure, HDAC substrate (final concentration HDAC1 (6 μM); HDAC2 (4.5 μM); HDAC3 (9.5 μM) and 10 x trypsin were added in 10 μL, and substrate turnover was measured in continuous kinetic mode (1 read/min) for 30 min on a TECAN Safire 2 multi-well plate reader (λex=345 nm, 20 nm bandwidth, λem=440 nm, 20 nm bandwidth). Data were analyzed on a plate-by-plate basis for the linear range of fluorescence over time. The first derivative of data obtained from the plate capture corresponding to the mid-linear range was imported into analytical software (GraphPad Prism). Azobenzene-functionalized inhibitors show strong absorbance in the spectral range of the fluorescence signal, which results in an inhibitor concentration-dependent decrease of fluorescence signal. The relative signal decrease is linearly correlated with the azobenzene concentration, but independent from the concentration of the substrate and the amount of the generated fluorescent product 7-AMC. We have therefore acquired standard curves to corrected raw intensity values (Supplemental Fig. 14). Replicate experimental data from incubations with inhibitor were normalized to controls. All reported biochemical experiments are representative examples of at least two independent biological replicates.
Cell Culture
Human MCF-7 cells (ATCC) were propagated in RPMI media (+10%FBS +1% L-glutamine +1% Pen/Strep) at 37 °C and 5% CO2. Cell culture was routinely checked for mycoplasma contamination using the MycoAlert™ Mycoplasma Detection Kit (Lonza) according to the manufacturer’s protocol. Prior to assays, cells were transferred to phenol red free RPMI medium and seeded into black/clear bottom 96-well plates (BD Biosciences #BD356692) at 7,000 cells/well density for immunofluorescent assays and 10,000 cells/well for western blot analysis and L1000 gene expression experiments. One day post plating, COMET probes were added to cells using an HP D300 Digital Dispenser (Hewlett-Packard), and cells were exposed to light as indicated.
Western blot assay
Following the addition of vehicle control or COMET probes (BG14, BG47, BG48) at a final concentration of 25 μM, MCF-7 cells were exposed to varying amounts of 470 nm light (8.5 mW/cm2) for 16 h in a tissue culture incubator (37 °C, 5% CO2). The light exposure was modulated at 1 Hz (no light, 250 ms on/750 ms off, 750 ms on/250 ms off). Subsequently, cells were lysed and the relative amount of acetylated H3K9 was analyzed by standard western blot analysis (rabbit anti-H3K9ac antibody, Millipore #07-352 1:10,000, anti-rabbit-horseradish peroxidase (HRP) 1:2,000, Cell Signaling #7074) using anti β-Actin as loading control (Sigma A1978, anti-mouse-HRP Cell Signaling #7076 1:10,000). Fold increase over DMSO treated samples were quantified with ImageJ software (http://imagej.nih.gov/ij). Western blot assays were performed in triplicate with each treatment done in duplicate. Differences in mean acetylation levels comparing presence and absence of light were assessed using a two-sided t-test with p<0.05 considered significant.
Immunofluorescent Assay
Following the addition of vehicle control, CI-994, and BG14 at a final concentration of 25 μM, MCF-7 cells were exposed to no light or cycles of 470 nm light (8.5 mW/cm2, 300 ms on/900 ms off) for 16 h in a tissue culture incubator (37 °C, 5 % CO2). Afterwards, cells were fixed with 4 % formaldehyde for 1 h, followed by a 20 min treatment with 100 % MeOH. Permeabilization and blocking was performed in PBS containing 0.1 % Triton X-100 and 2 % BSA. For immunofluorescent quantification of histone modification, cells were incubated overnight with anti-acetylated-H3K9 antibody (Millipore #07-352, 1:2,000), followed by a 1 h incubation with Alexa-594 conjugated secondary antibody (Molecular Probes A11012, 1:500). Images were captured by high-content, automated, confocal microscopy using the IN Cell Analyzer 6000 (GE Healthcare), both with a 40x objective for high-resolution images and a 10x objective for quantification of acetylation signal intensity. Nuclear intensity of acetylated H3K9 was quantified in 4 fields/well (> 3500 cells/well) and normalized to DMSO control wells. Immunofluorescent assays were performed in 3 independent biological replicates with treatments done in duplicate. A total of four fields per well were captured and >3500 cells per field were quantified. Error bars = s.d. . Comparisons of treatments were assessed using one-way analysis of variance (ANOVA) followed-up with a Tukey's Honest Significant Difference (HSD) post-hoc test with p<0.05 considered significant.
Time dependence and persistence of HDAC inhibition
Upon addition of BG14, CI-994 (both at 25 μM final concentration) or vehicle, MCF-7 cells were exposed to varying amounts of light during a total assay time of 16 hours: 16h no light, 0.5h light & 15.5 h no light, 1 h light & 15 h no light, 2 h light & 14 h no light, 4 h light & 12 h no light, 8 h light & 8 h no light. Light cycles consisted of 470 nm light (8.5 mW/cm2) modulated at 1Hz (500 ms on/off). After 16 hours, cells were fixed, stained, imaged, and quantified as described earlier (Immunofluorescent Assay). Nuclear intensity of acetylated H3K9 was quantified for 9 fields/well with n≥3 per treatment. Acetylation values were normalized within each group to DMSO = 0 and CI-994 = 100%. Mean and s.d. values are shown. Two-sided t-test performed for each experimental condition.
Spatial control of HDAC inhibition
A) MCF-7 cells were plated in a 1-well chamber slide (Lab Tek Chamber slide #177372) at 750k cells in 5 mL media/chamber. One day post plating, cells were treated either with DMSO or BG14 at 25 μM and the chambers were placed over a single 5 mm LED (470 nm, 8.5 mW/cm2, pulsed at 300 ms on/600 ms off). Light exposure was conducted for 6 hours, followed by an additional 10 h treatment without light exposure before cells were fixed. Slides were stained as described earlier (Immunofluorescent Assay). The entire 1-well chamber slide was imaged with the IN Cell Analyzer 6000 (7 x 14 captures). The mean nuclear intensity for acetylated H3K9 was determined for each captured field and normalized to its site-specific DMSO field. The normalized mean H3K9ac values/field was used to create a heatmap. B) Wells of a 96-well culture plate were partially covered (50%) at the bottom with black tape prior to plating MCF-7 cells at 7.5k cells/well. Cells were treated for a total of 16 hours one day post plating with DMSO, CI-994, and BG14 (25 μM for both compounds) and exposed to light (470 nm, 8.5 mW/cm2, 300 ms on/900 ms off cycles). Cells were fixed, stained, and imaged as previously described (Immunofluorescent Assay). To capture entire wells, 4 x 4 images were taken. For quantification, covered and light exposed halves in each well were treated independently. Nuclear intensity of acetylated H3K9 was determined and the fold change in comparison to appropriate (covered/light exposed well half) DMSO control was calculated. For visualization, shading correction was performed on all images. 4x4 images per well were used to create whole well montages in ImageJ. Subsequently, the Surface plot analysis plugin in ImageJ was used to create heatmaps for H3K9ac in each well. For each treatment, we used n=5. Mean and s.d. values shown. One way analysis of variance (ANOVA) followed-up with Tukey’s post-hoc test performed with p<0.05 considered significant.
L1000 Sample Production
MCF-7 cultures were treated with BG14, BG47, BG48 (1 μM, 5 μM, and 25 μM), and CI-994 (1 μM and 25 μM), by transfer of compounds using an HP D300 Digital Dispenser (Hewlett-Packard). Subsequently, cells were exposed for 0%, 25%, or 75% of time per 1 sec light cycle (470 nm, 8.5 mW/cm2) for a total of 16 h, and cell lysates were harvested according to the protocol provided by Genometry Inc. and stored at −80 °C. L1000 analysis was performed by Genometry Inc. All experiments were performed in quadruplicate.
Transcriptional Analysis
L1000 data consisting of landmark (LM) and inferred (INF) data that has been log2 transformed was obtained from Genometry, Inc. (full gene expression profiles are available upon request from data@genometry.com). All analysis of L1000 data used treatments that were represented relative to corresponding controls with the robust z-scores. The robust z-scores for each feature calculated using the median and median absolute deviation (MAD) of the vehicle controls specific to the batch (4 to 6 vehicle controls in each of 6 batches total) of each sample. Because of the limited number of vehicle controls in each batch, MAD was thresholded to a minimum of 0.1 and the robust z-score was set to 0 if MAD was 0.
Gene Set Enrichment Analysis (GSEA) (http://www.broadinstitute.org/gsea/index.jsp) was used to assess whether signatures were enriched in treatments by the experimental compounds. GSEA was used in a pre-ranked list mode where enrichment is determined by the positions of features from the gene sets in the ranked lists of all the features. P-values for the significance of enrichment scores were found by permuting the gene sets (1000 permutations). The ranked lists used by GSEA were created by calculating scores for each feature using either signal-to-noise ratio (SNR) for two class distinctions or cosine distance to a template for multi-class distinctions. SNR were calculated by finding the difference of the means of the two classes of samples divided by the sum of the standard deviations of the two classes of samples. SNR was used to create ranked lists of features that compared DMSO to a treatment for a specific compound, treatment time, light level, and dose (where the DMSO control had the same treatment time and light level). Cosine distance was used to rank features according to their similarity to a normalized ideal template where the template tries to select features that show increased effects with increasing dose and increasing light levels. Supplementary Fig. 15a shows the template prior to normalization (to zero mean and standard deviation of one) used for ranking features according to light and chemical concentration dose with each of the compounds. Each compound also had a no light dose response template as shown in Supplementary Fig. 15b to discover off-target effects for the compounds. Reference compounds used a three-value template (0,1,2 prior to normalization for DMSO, compound at low-dose, and compound at high dose) and were evaluated at zero light and maximum light. The gene sets that were used in GSEA came from the literature or were created by choosing the 100 highest and lowest scoring features associated with each ranked list (each treatment).
Identification of Light-Dependent Gene Signatures
SNR-based lists for BG14, CI-994 and C60 were reduced to landmark genes. A gene was scored as ‘active’ when its SNRs fulfilled either one of the following criteria:
abs(SNR) > 1.5;
abs(SNR[25 μM, 750 ms] - SNR[1 μM, 0 ms] ) > 1.5.
Genes were ranked by their score differences: SNR[25 μM, 750 ms] - SNR[1 μM, 0 ms] in descending order. CI-994 response gene list (in absence of light) is comprised of 432 genes. C60 response gene list (in absence of light) is comprised of 343 genes. BG14 response gene list contains a total of 256 genes.
The BG14 response gene list was subdivided into 3 categories:
-
a BG14 response gene was scored as off-target when it fulfilled the following criteria:
abs(SNRNoLight)>1.5
abs(SNRLight –SNRNoLight) ≤1.5
22 BG14 off-target genes were identified.
-
a BG14 response gene was scored as mixed on-target when it fulfilled the following criteria:
abs(SNRNoLight) > 1.5
abs(SNRLight –SNRNoLight) > 1.5
47 BG14 mixed on-target genes were identified.
-
a BG14 response gene was scored as clean on-target when it fulfilled the following criteria:
abs(SNRNoLight) ≤1.5
abs(SNRLight –SNRNoLight) > 1.5
187 BG14 clean on-target genes were identified.
Lastly, we used Venn diagram analysis (www.cmbi.ru.nl/cdd/biovenn) to identify the Core Gene list (120) consisting of the overlap between the CI-994 response gene list and the BG14 clean on-target genes.
Density functional theory calculations
To find the lowest energy conformers, a solution phase conformational search using MacroModel (version 10.6, Schrödinger, LLC) was performed with OPLS_2005 35 as force field and Polak-Ribiere Conjugate Gradient (PRCG) as minimization method. The twenty (eight for CI-994 derivatives) lowest energy conformers were geometry optimized using density functional theory (DFT) calculations with B3LYP37 as functional and the 6-31+G(d) basis set. The vibrational frequencies were computed at the same level to verify that optimized structures are energy minima and to evaluate zero-point vibrational energies (ZPVE) and thermal corrections at 298 K. All calculations were performed using the Solvation Model Based on Density (SMD) solvent continuum as solvent method. 38 The electrostatic potentials (ESP) were calculated by performing a population analysis in solvent concerning CHelp scheme 36. All calculations were performed using Gaussian09 software. The ESP for the Zn-binding nitrogen and oxygen atoms for the lowest energy conformers, as well as the Boltzmann weights ESP values based on the twenty (eight for CI-994 derivatives) lowest energy conformers, are provided in Supplementary Fig. 16.
Supplementary Material
Acknowledgments
We would like to thank members of the Haggarty and Mazitschek laboratories for their constructive feedback throughout the project. We gratefully acknowledge financial support from the National Institute of Health (R01NS088209 R.M and S.J.H., P50CA086355 R.M, R01DA028301 S.J.H., T32-CA079443 J.A.H). S.J.H. is supported through funding from the Tau Consortium, Bluefield Consortium for FTD, and Pitt-Hopkins Research Foundation. The computations in this paper were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We would like to thank Stephen Johnston (The Broad Institute of Harvard and MIT, Analytical Chemistry Group) for acquiring the high-resolution mass spectra. Special thanks to the Arduino development team and the open-source Maker Movement.
Footnotes
Author Contributions
R.M and S.J.H. conceived of the idea for the study; designed, directed and interpreted experiments, and wrote the manuscript; R.M. designed, synthesized and characterized inhibitors, and designed and built LED array; S.A.R. planned and performed cell based assays, high-content image analysis, analyzed data and helped prepare the manuscript; B.G. synthesized inhibitors, planned and performed cell based assays, analyzed data and helped prepare the manuscript; J.A.H and D.M.S-K. performed biochemical assays and analyzed data; L.T. performed DFT calculation studies and analyzed data; K.N.R. analyzed gene expression profiling data; J.L, W.R-B. and B.Z. performed gene expression analysis and analyzed data; H.W. and C.S. designed experimental set-up for relaxation experiments, performed relaxation experiments and analyzed data.
Competing financial interests
R.M. has financial interests in SHAPE Pharmaceuticals and Acetylon Pharmaceuticals. He is also the inventor on IP licensed to these two entities. S.J.H. has financial interests in Rodin Therapeutics and is an inventor on IP licensed to this entity. None of these entities were involved in the present study and the licensed IP does not include any of the work presented here. A patent application has been filed on the reported invention WO 2014160221.
References
- 1.Haggarty SJ, Tsai LH. Probing the role of HDACs and mechanisms of chromatin-mediated neuroplasticity. Neurobiol Learn Mem. 2011;96:41–52. doi: 10.1016/j.nlm.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orozco-Solis R, Sassone-Corsi P. Epigenetic control and the circadian clock: Linking metabolism to neuronal responses. Neuroscience. 2014;264C:76–87. doi: 10.1016/j.neuroscience.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 6.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beharry AA, Woolley GA. Azobenzene photoswitches for biomolecules. Chem Soc Rev. 2011;40:4422–4437. doi: 10.1039/c1cs15023e. [DOI] [PubMed] [Google Scholar]
- 11.Fehrentz T, Schonberger M, Trauner D. Optochemical genetics. Angew Chem Int Ed Engl. 2011;50:12156–12182. doi: 10.1002/anie.201103236. [DOI] [PubMed] [Google Scholar]
- 12.Kramer RH, Mourot A, Adesnik H. Optogenetic pharmacology for control of native neuronal signaling proteins. Nat Neurosci. 2013;16:816–823. doi: 10.1038/nn.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schonberger M, Althaus M, Fronius M, Clauss W, Trauner D. Controlling epithelial sodium channels with light using photoswitchable amilorides. Nat Chem. 2014;6:712–719. doi: 10.1038/nchem.2004. [DOI] [PubMed] [Google Scholar]
- 14.Frank JA, et al. Photoswitchable fatty acids enable optical control of TRPV1. Nat Commun. 2015;6:7118. doi: 10.1038/ncomms8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Light-controlled tools. Angew Chem Int Ed Engl. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- 16.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 17.Niino H, et al. Stabilization of a labile cis-azobenzene derivative with amphiphilic cyclodextrins. Chem Lett. 1988:1227–1230. [Google Scholar]
- 18.Shinkai S, Minami T, Kusano Y, Manabe O. Photoresponsive crown ethers. 8 Azobenzenophane-type switched-on crown ethers which exhibit an all-or-nothing change in ion-binding ability. J Am Chem Soc. 1983;105:1851–1856. [Google Scholar]
- 19.Bressi JC, et al. Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg Med Chem Lett. 2010;20:3142–3145. doi: 10.1016/j.bmcl.2010.03.091. [DOI] [PubMed] [Google Scholar]
- 20.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 22.Andrews DM, et al. Design and campaign synthesis of piperidine- and thiazole-based histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:2580–2584. doi: 10.1016/j.bmcl.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Lauffer BE, et al. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem. 2013;288:26926–26943. doi: 10.1074/jbc.M113.490706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Amoros J, Martinez M, Finkelmann H, Velasco D. Kinetico-mechanistic study of the thermal cis-to-trans isomerization of 4,4’-dialkoxyazoderivatives in nematic liquid crystals. J Phys Chem B. 2010;114:1287–1293. doi: 10.1021/jp909557h. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montojo J, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckers T, et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int J Cancer. 2007;121:1138–1148. doi: 10.1002/ijc.22751. [DOI] [PubMed] [Google Scholar]
- 29.Graff J, Tsai LH. The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- 30.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder FA, et al. selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS ONE. 2013;8:e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graff J, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 34.Peters JU. Polypharmacology - foe or friend? J Med Chem. 2013;56:8955–8971. doi: 10.1021/jm400856t. [DOI] [PubMed] [Google Scholar]
- 35.Banks JL, et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT) J Comput Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chirlian LE, Francl MM. Atomic charges derived from electrostatic potentials: a detailed study. Journal of Computational Chemistry. 1987;8:894–905. [Google Scholar]
- 37.Lee Yang, Parr Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 38.Marenich AV, Cramer CJ, Truhlar DG. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J Phys Chem B. 2009;113:6378–6396. doi: 10.1021/jp810292n. [DOI] [PubMed] [Google Scholar]
- 39.Moradei OM, et al. Novel aminophenyl benzamide-type histone deacetylase inhibitors with enhanced potency and selectivity. J Med Chem. 2007;50:5543–5546. doi: 10.1021/jm701079h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.