Abstract
Most patient care questions raised by clinicians can be answered by online clinical knowledge resources. However, important barriers still challenge the use of these resources at the point of care.
Objective
To design and assess a method for extracting clinically useful sentences from synthesized online clinical resources that represent the most clinically useful information for directly answering clinicians’ information needs.
Materials and methods
We developed a Kernel-based Bayesian Network classification model based on different domain-specific feature types extracted from sentences in a gold standard composed of 18 UpToDate documents. These features included UMLS concepts and their semantic groups, semantic predications extracted by SemRep, patient population identified by a pattern-based natural language processing (NLP) algorithm, and cue words extracted by a feature selection technique. Algorithm performance was measured in terms of precision, recall, and F-measure.
Results
The feature-rich approach yielded an F-measure of 74% versus 37% for a feature co-occurrence method (p<0.001). Excluding predication, population, semantic concept or text-based features reduced the F-measure to 62%, 66%, 58% and 69% respectively (p<0.01). The classifier applied to Medline sentences reached an F-measure of 73%, which is equivalent to the performance of the classifier on UpToDate sentences (p=0.62).
Conclusions
The feature-rich approach significantly outperformed general baseline methods. This approach significantly outperformed classifiers based on a single type of feature. Different types of semantic features provided a unique contribution to overall classification performance. The classifier’s model and features used for UpToDate generalized well to Medline abstracts.
Keywords: text summarization, clinical decision support, natural language processing, machine learning
Graphical abstract
1. INTRODUCTION
Online clinical knowledge resources contain answers to most clinical questions raised by clinicians in patient care.[1] Yet, over 60% of these questions go unanswered.[2] Despite significant adoption of online resources and advances in information retrieval technology, important barriers, such as lack of time and efficient access to answers, still challenge clinicians’ use of clinical knowledge resources. Previous efforts to address this problem include methods such as question answering [3] and text summarization.[4]
Previous efforts have focused on processing abstracts or full-text articles from the primary biomedical literature, with promising early results in laboratory settings.[3, 4] Yet, consuming the primary literature is labor intensive and not compatible with busy clinical workflows. Rather, clinicians prefer online resources, such as UpToDate and Dynamed, that are written by experts who synthesize the latest clinical evidence on a specific topic.[5–9] These resources are very different than the primary literature, both in terms of discourse and structure. While the primary literature focuses on presenting the results of clinical studies, synthesized resources provide clinically actionable recommendations that can be applied to specific patients. Previous studies in clinical decision support (CDS) systems have shown that clinically actionable recommendations are more effective in improving providers’ performance and patient outcomes.[10] For example, a meta-analysis investigating the use of abatacept for rheumatoid arthritis concluded that “there is moderate-level evidence that abatacept is efficacious and safe in the treatment of rheumatoid arthritis.” On the other hand, UpToDate provides a patient-specific recommendation that synthesizes the evidence in the primary literature: “abatacept may be used as an alternative to a TNF inhibitor in patients in whom MTX plus a TNF inhibitor would otherwise be appropriate, particularly in patients unable to use a TNF inhibitor and in patients with a high level of disease activity.”[11]
An important step in question answering and text summarization is the extraction of key sentences, typically based on a sentence ranking algorithm.[4] We propose that, to provide effective CDS, question answering and text summarization tools should focus on extracting clinically actionable recommendations from online information resources, particularly from synthesized resources such as UpToDate. In a previous exploratory study, we tested the feasibility of addressing this problem with a simple semantic-based approach.[12] In the present study, we developed and assessed a feature-rich (i.e., semantic and text-based) classification model that extracts, from synthesized resources, the most useful sentences to support patient-specific treatment decisions. We also conducted an exploratory assessment of the model’s generalizability to sentences from the primary literature.
2. BACKGROUND AND SIGNIFICANCE
In question answering and text summarization systems, researchers have used a variety of salient sentence extraction techniques. The first technique was proposed by Edmundson in 1969, in which a score is calculated for each sentence using a statistical function based on cue phrases, keywords, and sentence location.[13] Since then, several variations of the “Edmundsonian paradigm” have been investigated. Lin proposed modeling relevant keywords in specific domains as a mechanism to assess sentence relevance.[14] Sentence position has also been used as a predictor of salient sentences, such as the first paragraph of news articles and the conclusion section for scientific articles.[15] Another common technique is to look for specific cue words, such as “in conclusion” or “in summary.” Machine learning approaches have been proposed using Edmundsonian features as input for the binary classification of sentence relevance.[16, 17] Finally, popular graph-based approaches, such as TextRank, use graph algorithms to compute the similarity between a topic and sentences as well as among sentences themselves.[18–22]
According to a systematic review of recent biomedical text summarization techniques, researchers have employed variations of the sentence extraction methods described above.[4] The most notable differences are the ubiquitous preference for knowledge rich methods that leverage domain-specific tools, such as the Unified Medical Language System (UMLS), MetaMap,(50) and SemRep;(51) as well as the increased adoption of hybrid approaches.[3, 23–25]
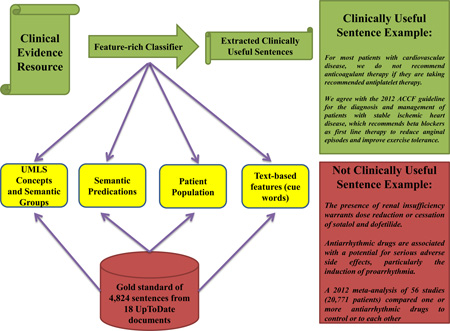
Despite substantial work on sentence extraction for biomedical text summarization and question answering, previous studies have focused largely on the primary literature.[4] A common goal has been to generate summaries that resemble article abstracts written by study authors. However, previous research has shown that, for clinical decision-making, clinicians prefer synthesized resources to the primary literature. In addition, clinicians prefer patient-specific, actionable recommendations, as opposed to a general overview.[5–9] Specifically, useful sentences (see Table 1 for definitions and examples) should contain an explicit assertion about a specific treatment and for a specific type of patient or patient population. Algorithms need to consider these attributes in order to extract clinically useful sentences. On the other hand, general sentence extraction approaches aim to identify topic-relevant sentences or representative sentences to produce an overview. Therefore, these approaches are not optimal for clinical decision support.
Table 1.
Summary of the criteria used to rate sentences according to clinical usefulness.
| Rating | Definition | Examples |
|---|---|---|
| 1 | Not relevant for clinical decision making, such as sentences that introduce the scope of a document or provide navigation to related documents. |
“The management of children with heart failure will be presented here.” “Psychosis and treatment of psychotic depression are discussed separately.” |
| 2 | Provide background information, such as the epidemiology and physiopathology of a condition, mechanism of action of a drug, and description of the design of a research study. |
“A 2012 meta-analysis of 56 studies (20,771 patients) compared one or more antiarrhythmic drugs to control or to each other.” “Heart failure is estimated to affect 12,000 to 35,000 children below the age of 19 years in the United States each year.” |
| 3 | Describe potentially useful details about a specific treatment, but at the level of detail that would not be useful for text summaries. Examples include: 1) treatment adverse effects, contraindications, precautions, and monitoring; 2) results or conclusions of clinical trials or meta-analyses; and 3) overall treatment efficacy, but not the specific patients for which the treatment should be used. |
“The presence of renal insufficiency warrants dose reduction or cessation of sotalol and dofetilide.” “Antiarrhythmic drugs are associated with a potential for serious adverse side effects, particularly the induction of proarrhythmia.” “In the EMERALD trial, Dofetilide had a somewhat better efficacy than sotalol.” |
| 4 | Include a recommendation for or against a specific intervention for a specific patient population. |
In patients with inadequate glycemic control on sulfonylureas, with A1C >8.5 percent, we suggest switching to insulin.” “For patients with adrenergically-mediated AF, we suggest beta blockers as first-line therapy, followed by sotalol and amiodarone.” |
| 5 | Same as 4, but include an explicit attribution to a source of evidence (e.g., meta-analysis, medical society guideline). | We agree with the 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and management of patients with stable ischemic heart disease, which recommends beta blockers as first line therapy to reduce anginal episodes and improve exercise tolerance.” |
In the present study, we investigate a hybrid method to extract clinically useful sentences from synthesized evidence resources such as UpToDate, a popular knowledge resource written by clinical experts in various medical specialties. We employ a feature-rich approach based on supervised machine learning techniques and a set of Edmundsonian and semantic NLP features. Similar hybrid approaches have been employed in previous sentence extraction research.[4] Our main contribution consists of identifying a rich set of features that serve as predictors of clinically useful sentences and can be effectively used for the extraction of those sentences for clinical decision support. The following sections describe the NLP tools used to extract these features.
2.1 MedTagger
MedTagger, an extension of the cTAKES NLP pipeline [26], is a modular system of pipelined components that combine rule-based and machine learning techniques to extract semantically viable information from clinical documents.[27] For each sentence in a document, MedTagger extracts a set of concepts from the Unified Medical Language System (UMLS). The process includes OpenNLP tokenization [28], lexical normalization [29], and dictionary-based concept extraction according to the Aho-Corasick algorithm [21] using both the UMLS Metathesaurus and MeSH. Overall, the precision and recall for MedTagger on the CLEF 2013 shared task were 80% and 57% respectively for strict evaluation and 94% and 77% respectively for relaxed evaluation.[27] The accuracy of MedTagger on a corpus depends on the coverage of the UMLS Metathesaurus in the specific domain of interest and on the accuracy of the lexical normalization resource, both of which have been extensively used in several text mining systems. While the system has not been intrinsically evaluated for biomedical abstracts, MedTagger has been used in previous studies as a component of literature-mining pipelines.[19, 30]
2.2 SemRep
SemRep is a semantic NLP parser that uses underspecified syntactic analysis and structured domain knowledge from the UMLS to extract semantic predications.[31] Semantic predications are relations that consist of a subject, a predicate, and an object. The subject and object of predications are represented with UMLS concepts. Predicates consist of semantic relations such as IS_A, TREATS, and AFFECTS. For example, from the sentence below:
“Quinidine, procainamide, and disopyramide are recommended for patients with atrial fibrillation”
SemRep extracts the following predications:
Atrial Fibrillation PROCESS_OF Patients
Disopyramide TREATS Atrial Fibrillation
Procainamide TREATS Atrial Fibrillation
Quinidine TREATS Atrial Fibrillation
In an exploratory study, we concluded that the number of semantic predications in a sentence is correlated with clinically useful sentences.[12] A subset of SemRep predicates is more relevant to sentences that describe disease treatment, such as TREATS and the comparative predicates COMPARED_WITH, HIGHER_THAN, LOWER_THAN, and SAME_AS. Comparative predicates are extracted in sentences that contrast two treatment alternatives.[32] For example, from the sentence below:
“Etanercept and adalimumab might be safer than infliximab.”
SemRep extracts the following comparative predications:
Adalimumab HIGHER_THAN infliximab
Etanercept HIGHER_THAN infliximab
In a previous study, SemRep's precision and recall for treatment-related predications (i.e., TREATS and comparative predications) were 78% and 50% respectively.[33]
3. METHODS
The study method consisted of: 1) extension of a gold standard of labeled sentences from UpToDate; 2) extraction of semantic and text-based features for sentence classification; 3) development of classification models for identifying clinically useful sentences; and 4) testing of a set of hypotheses regarding the performance of these classification models.
3.1 Gold Standard
We extended a gold standard developed in a previous feasibility study.[12] The resulting gold standard consisted of all 4,824 sentences from 18 UpToDate documents on the treatment of six chronic conditions: coronary artery disease, hypertension, depression, heart failure, diabetes mellitus, and prostate cancer. These documents were selected through a manual search using UpToDate’s search engine. From the search results, we selected the documents most frequently accessed by clinicians according to UpToDate’s usage log.
Sentences were rated independently by three raters with clinical background (two physicians and one dentist) according to a 5-point clinical usefulness scale (Table 1). The rationale supporting this rating approach is based on evidence that clinical decision support tools that provide patient-specific, actionable recommendations are more likely to produce positive outcomes.[10, 34] The rating scale with instructions was developed iteratively in three stages. In the first stage, two raters independently rated sentences from one document reaching an inter-rater agreement of 0.52 (linear weighted kappa). After reconciling disagreements, the instructions were refined and another set of sentences were rated reaching an inter-rater agreement of 0.74. A third rater was included for the remainder of the documents after further refinement of the rating instructions. The final inter-rater agreement obtained a linear weighed kappa of 0.82. To test algorithm generalizability, a similar rating process was followed to produce a gold standard of all 1,072 sentences from a random set of 140 recent PubMed abstracts that reported results of randomized clinical trials.
3.2 Overview of Actionable Sentence Classification Method
To extract clinically useful treatment sentences we built a classification model based on a set of features extracted from the document’s sentences. These features were selected based on domain knowledge derived from experimental research.[10, 34, 35] More specifically, we followed three underlying principles to guide the selection of potential features: (i) clinically useful sentences should have one or more actionable treatment recommendations; (ii) candidate sentences should define the types of patients (i.e., population) that qualify for a particular recommendation; and (iii) recommendation statements should be assertive, with constructs such as deontic terms (e.g., “we recommend”, “we suggest”), and should include attribution to an evidence source (e.g., “according to the ACC guideline…”).
To capture these attributes, we extracted a combination of semantic and text-based features (cue words). Semantic features included UMLS concepts, UMLS semantic groups [36], semantic predications, and patient population. Text-based features were extracted directly from the UpToDate dataset using text-based feature selection techniques.
3.3 UMLS Concepts and Semantic Groups
Since the focus of the present study was to extract treatment recommendations, we narrowed MedTagger’s output to treatment-related concepts. Each extracted UMLS concept was mapped to one of four UMLS semantic groups: Chemicals & Drugs (CHEM), procedures (PROC), physiology (PHYS), and disorders (DISO).[36] Moreover, in order to avoid recommendations that are too general and therefore not clinically useful, concepts with the following general semantic types were removed from our dataset: “Body Part, Organ, or Organ Component”, “Neuroreactive Substance or Biogenic Amine”, “Nucleic Acid, Nucleoside, or Nucleotide”, “Amino Acid, Peptide, or Protein”, and “Functional Concept“. These semantic types were selected from a subset of the root semantic types based on domain knowledge. We derived five features from the concepts extracted from each sentence: total number of concepts in the sentence (one feature) and number of concept instances per UMLS semantic group (four features). For instance, the following sentence contains two concepts in the semantic group procedures and one concept in the semantic group disorders:
“For most patients with cardiovascular disease (DISO), we do not recommend anticoagulant therapy (PROC) if they are taking recommended antiplatelet therapy (PROC).”
3.4 Semantic Predications
To extract semantic predications, we processed UpToDate documents available in the gold standard with SemRep. We derived seven features from semantic predications: total number of predications with a treatment-related predicate (one feature) and number of predication instances per treatment-related predicate, including negated predicates when applicable, (six features): TREATS/NEG_TREATS, ADMINISTERED_TO /NEG_ADMINISTERED_TO, AFFECTS/NEG_AFFECTS, PROCESS_OF / NEG_PROCESS_OF, PREVENTS / NEG_PREVENTS, and COMPARED_WITH / HIGHER_THAN / LOWER_THAN / SAME_AS. For instance, from the sentence below:
“We suggest that methotrexate (MTX) be used as the initial DMARD for patients with moderately to severely active RA, rather than another single nonbiologic or biologic DMARD or combination therapy.”
SemRep produces the following output:
Rheumatoid Arthritis PROCESS_OF patients
Antirheumatic Drugs, Disease-Modifying (DMARD) TREATS Rheumatoid Arthritis
Combined Modality Therapy TREATS Rheumatoid Arthritis
which yields the following features:
Total number of predications: 3
PROCESS_OF instances: 1
TREATS instances: 2
3.5 Patient Population
Patient population determines whether a sentence includes a description of the types of patients who are eligible to receive a certain treatment. For instance:
“In patients with inadequate glycemic control on sulfonylureas, with A1C >8.5 percent, we suggest switching to insulin.”
“For patients with adrenergically-mediated AF, we suggest beta blockers as first-line therapy, followed by sotalol and amiodarone.”
To identify the population of interest, we developed a pattern-based method that returns a population phrase. The method uses two NLP parsers, the Stanford lexical parser [37] and Tregex [38]; 130 population-related concepts obtained from the patient or disabled group UMLS semantic type; and 22 terms that were manually identified in UpToDate sentences not included in the gold standard. First, sentences with population-related concepts or terms are filtered. Second, each filtered sentence is processed with the Stanford lexical parser to generate a constituent tree of verb and noun phrases. Third, the node labels are queried using Tregex, a tree query language for querying expressions of a parse tree. Finally, the algorithm extracts the population phrase identified. The Tregex patterns are similar to regular expressions, but more advanced and easier to use (Table 2). A binary feature was produced to indicate whether a sentence includes a population or not.
Table 2.
Tregex Patterns for population phrase extraction.
| Query for parse tree | Tregex Pattern | Example (phrase matching Tregex Pattern is underlined) |
|---|---|---|
| Noun phrase | @NP | “For patients with atrial fibrillation (AF), there are two main strategies to manage the irregular rhythm and its impact on symptoms: rhythm control (restoration followed by maintenance of sinus rhythm with either antiarrhythmic drugs or radiofrequency catheter ablation).” |
| Noun phrase with two consecutive prepositional phrases | @NP, PP $ PP | “The need for and appropriate frequency of routine dental scaling and polishing in patients at low risk for periodontal disease is uncertain.” |
| Verb phrase | @VP | “Inotropic agents are used during acute exacerbations of heart failure to improve cardiac output and to stabilize patients awaiting heart transplantation.” |
| Verb phrase with two consecutive prepositional phrases | @VP, PP $ PP | “An exception to this need for maintenance anticoagulation occurs in patients without structural heart disease who are at low risk for embolization.” |
| Noun phrase preceding subordinating conjunction | NP < SBAR | “Nonpharmacologic methods to maintain sinus rhythm (including surgery and radiofrequency ablation) in selected patients who are refractory to conventional therapy are discussed elsewhere.” |
| Noun phrase preceding adjective phrase | NP < ADJP | “ARBs are usually reserved for patients unable to tolerate ACE inhibitors due to cough or angioedema.” |
| Noun phrase succeeding prepositional phrase | NP > PP | “Clinical outcomes after several years of treatment are similar in trials that compared patients initially receiving MTX, who then stepped up to combination therapy after an inadequate response, with patients initially treated with combination therapy.” |
| Noun phrase starting with noun, i.e. population group | NP $ NNS | “Subgroup analyses of diabetics randomized in the Heart Protection Study contributed to the formulation of the hypothesis of lipid lowering with 40 mg of simvastatin in diabetic patients without prior CHD to reduce risks of subsequent CVD events.” |
| Prepositional phrase succeeding noun phrase | PP > NP | “The benefits of glycemic control on macrovascular and microvascular disease in patients with type 1 and type 2 diabetes are discussed elsewhere.” |
| Two consecutive prepositional phrases | PP $ PP | The ATP III guidelines published in 2001 recommended that the goal LDL-cholesterol should be less than 100 mg/dL (2.6 mmol/L) in all high-risk patients including secondary prevention and in those with a coronary equivalent including diabetes.” |
A preliminary analysis of the algorithm with a gold standard of 1825 sentences from UpToDate yielded a precision and recall of 91% and 97% in identifying sentences that mentioned a patient population (unpublished data). A formal experiment to assess the performance of this approach is underway. Given the optimal performance and the simplicity of the algorithm (e.g., it does not depend on availability of training data), we opted not to experiment with other alternatives for population identification, such as supervised learning techniques.
3.6 Text-based Features (cue words)
Text-based features consisted of a set of potentially useful cue terms, such as deontic terms (e.g., suggest, recommend). To identify these terms we selected from the gold standard a training set of three random documents, which were excluded from later experiments. All bigram terms from these documents were extracted and the top 15 terms with the highest Pearson correlation values were selected. This approach is aligned with the method proposed by Hall et al.[39] Other feature weighting methods such as information gain [40] and gini index [41] returned the same 15 terms. These terms were manually inspected and grouped into four term categories based on domain knowledge (four features). From these categories, we derived four features that consisted of the number of cue terms per term category in a sentence (Table 3): (i) references to other documents, such as “is discussed elsewhere”; (ii) terms related to study design, such as random and placebo; (iii) terms used in deontic modality, which overlap with terms identified by Lomotan et al. [42] (e.g., recommend, suggest), and terms that indicate evidence sources (e.g., guideline); and (iv) terms that denote “treatment” (e.g., therapy). The first and second categories correlated with sentences that are not clinically useful, while terms in the third and fourth groups correlated with clinically useful sentences.
Table 3.
List of term categories and cue terms.
| Term categories | Cue terms | Examples |
|---|---|---|
| References to external content | Discuss, detail, separate, following | “The use of these agents in patients with SIHD is discussed in detail separately.” |
| Study design | Random, trial, placebo, effect | The effectiveness of instructions for CHD in older adults is often unrecognized or underestimated, in part because older adults are usually underrepresented in randomized controlled trials.” |
| Deontic terms and evidence source attributions | Recommend, suggest, should, advise, guideline | “We agree with the 2012 ACCF / AHA / ACP / AATS / PCNA / SCAI / STS guideline for the diagnosis and management of patients with stable ischemic heart disease, which recommends that all patients with SIHD should receive education and counseling about issues such as medication compliance, control of risk factors, and regular exercise.” |
| Treatment | Treatment, therapy | “Nitrates, usually in the form of a sublingual preparation, are the first-line therapy for the treatment of acute anginal symptoms.” |
3.7 Classification Model
From the approach above, 17 features were selected for sentence classification (Table 4). The distribution of sentences in the gold standard according to each feature category is available on Table s1 of the online supplement.
Table 4.
Features used to develop the classification model.
| Feature Category |
Feature Type | Number of features |
Description |
|---|---|---|---|
| Semantic | Predication | 7 | Total number of predications with a treatment-related predicate (1 feature) and number of predication instances per treatment-related predicate (i.e., TREATS / NEG_TREATS, ADMINISTERED_TO / NEG_ADMINISTERED_TO, AFFECTS / NEG_AFFECTS, PROCESS_OF / NEG_PROCESS_OF, PREVENTS / NEG_PREVENTS, and COMPARED_WITH / HIGHER_THAN / LOWER_THAN / SAME_AS) - 6 features. |
| Semantic | Population | 1 | Whether or not a sentence includes a description of the types of patients who are eligible to receive a certain treatment. |
| Semantic | Concept | 5 | Total number of concepts in the sentence (1 feature) and number of concept instances per UMLS semantic group (i.e., Chemicals & Drugs (CHEM), procedures (PROC), physiology (PHYS), and disorders (DISO) - 4 features. |
| Syntactic | Text-based | 4 | Four categories of potentially useful cue terms mentioned in Table 3 including “References to external content”, “Study design”, “Deontic terms and evidence source attributions” and “Treatment”. |
To select an optimal classifier, we evaluated six different classification algorithms: Kernel-based Bayesian Network, Naïve Bayes, Neural Network, Support Vector Machine (LibSVM), K-Nearest Neighbor, and Logistic Regression. Algorithms were evaluated with the following parameter settings: kernel type, estimation mode, and number of kernels were varied for the Kernel-based Bayesian Network; number of hidden layers, number of nodes in each layer, learning rate, and momentum were varied for the Neural Network; and Kernel type along with the corresponding parameters of each kernel type were varied for the Support Vector Machine and Logistic Regression. The value of k and the weighted voting approach were changed for the K-Nearest Neighbor algorithm. Since our gold standard is unbalanced (87% negative vs. 13% positive cases), probability threshold adjusting was applied to all algorithms. The same three documents used for selecting text-based features were used for finding the best parameter setting for each classifier.
A Kernel-based Bayesian Network classifier with 50 Gaussian kernel density greedy estimators performed best and was used in subsequent experiments. This classifier is a Bayesian Network that estimates the true density of the continuous variables using kernels. A kernel is a weighting function, which is generally used in non-parametric estimation techniques. It is employed in kernel density estimation to estimate the density function of random variables. More details about this algorithm can be found elsewhere. [43] As shown in previous research, the Kernel-based Bayesian Network is robust to highly imbalanced datasets [44, 45] (i.e., the number of positive cases is much smaller than the negative cases), such as the one in the present study.
3.8 Assessment of Classification Performance
We conducted four experiments to test the following hypotheses:
Hypothesis 1: A feature-rich classifier that benefits both from semantic and syntactic features performs better than a feature co-occurrence classifier. We compared our specialized hybrid sentence classifier, which uses both semantic and text-based features, with a feature co-occurrence classifier, which takes the binary occurrence of all UMLS concepts, predications, population, and cue word features in a sentence as input and predicts the sentence usefulness using a Kernel-based Bayesian Network classifier. This classifier is similar to a unigram text-based classifier but with concepts, predications, population, and cue words as input rather than words.
Hypothesis 2: A feature-rich classifier that uses both semantic and syntactic features performs better than classifiers that use only one of them. This experiment assessed the contribution of each category of features to classification performance. The semantic features include Predication, Population and Concept feature types (Table 4). Text-Based feature types (i.e., cue words) are listed on Table 3.
Hypothesis 3: Each type of semantic feature provides a significant contribution to the overall performance of the feature-rich classifier. We assessed the contribution of three types of semantic features to overall classification performance: predication-based, concept-based and population-based (Table 4). The feature-rich sentence classifier was compared to three other classifiers, each of which containing all but one semantic feature type.
Hypothesis 4: The feature-rich classifier is generalizable to other types of online clinical resources. To test this exploratory hypothesis, we conducted a pilot evaluation of the feature-rich classifier on a random set of Medline sentences described in the “Gold Standard” section.
3.8.1 Experiment Procedures
In each of the experiments above, we used the same gold standard excluding the 3 documents that were used to extract text-based features. Ordinal ratings were converted into binominal values: sentences rated as 4 and 5 were considered as the positive class (i.e., clinically useful sentences) and the remaining sentences were considered as the negative class. As a result, 13% of the sentences in the gold standard were labeled as positive versus 87% as negative.
All experiments employed a leave-one-out strategy with 15 iterations and were implemented using RapidMiner (www.rapidminer.com). In each iteration, 14 documents were used for classifier training and one was left out for testing classification performance. To test Hypothesis #4 on Medline sentences, we employed a 20-fold cross-validation strategy with each fold containing 7 abstracts.
3.8.2 Data Analysis
Classification performance was measured according to the average precision, recall, and F-measure across 15 iterations. F-measure was defined a priori as the primary outcome for hypotheses testing. For statistical significance, first we applied the Friedman’s test to verify differences among multiple classifiers. If significant at an alpha of 0.05, pairwise comparisons were made with the Wilcoxon Signed-Rank test. This statistical approach is aligned with the method recommended by Demsar [46], which accounts for intra-class correlation in cross-validation experiments.
4. RESULTS
Descriptive statistics of the sentences and features in the gold standard show that all feature types that were assumed to be predictive of useful sentences were more frequent in useful sentences than in non-useful sentences (Table s1 of the online supplement). Detailed results for all experiments are reported in Tables s2 to s5 of the online supplement. The Bayesian Network algorithm outperformed the other alternatives (Table s6 in the online supplement). Different parameter settings did not significantly change the performance of any of the algorithms.
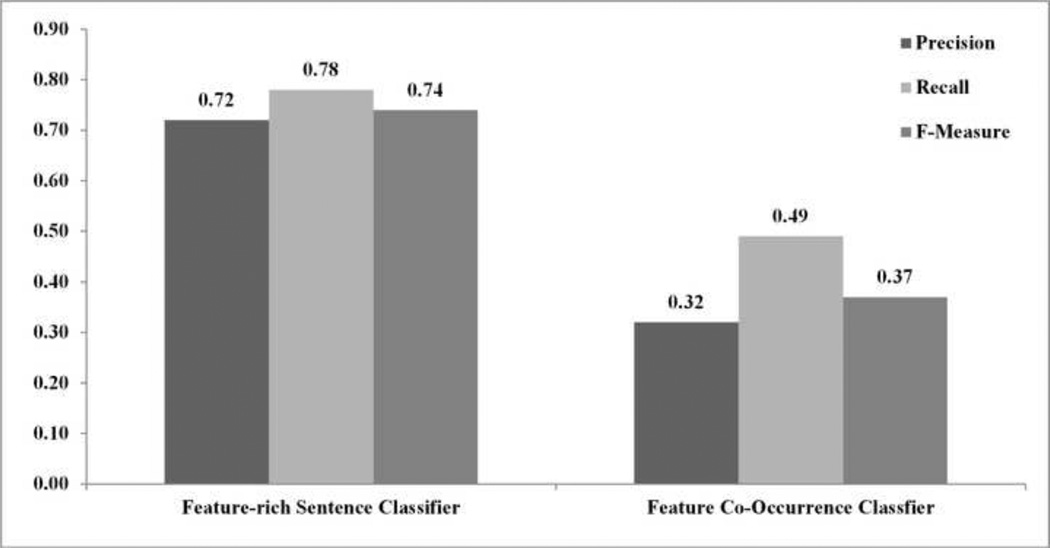
Experiment #1: A feature-rich classifier that benefits both from semantic and syntactic features performs better than a feature co-occurrence classifier. Figure 1 summarizes the results. The feature-rich sentence classifier performed significantly better than the feature co-occurrence classifier (F-measure = 74% versus 37%; p<0.001).
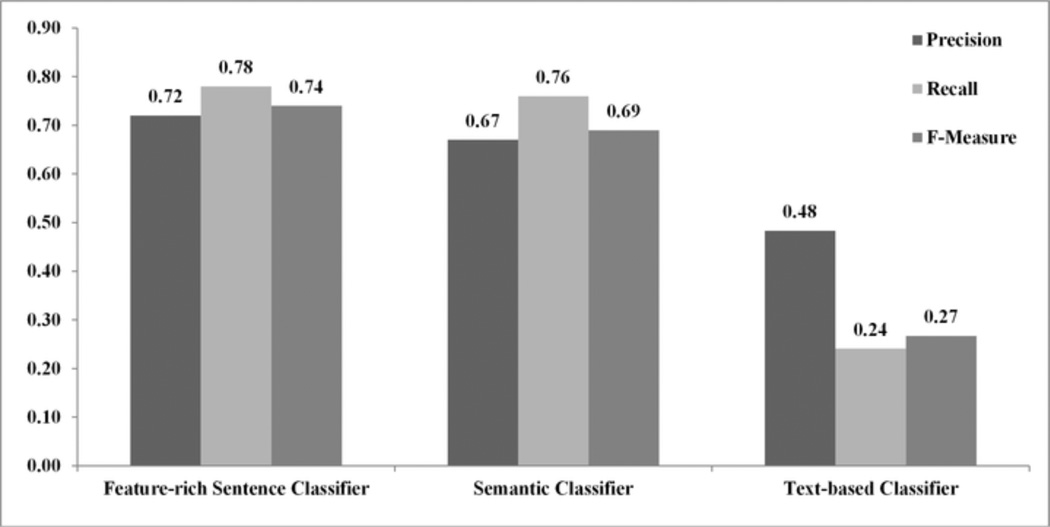
Experiment #2: A feature-rich classifier that uses both semantic and syntactic features performs better than classifiers that use only one of them. Figure 2 summarizes the results. The feature-rich sentence classifier performed significantly better than the semantic classifier and the text-based classifier (F-measure = 74% versus 69% and 27%; p = 0.002 and p = 0.001 respectively). The semantic classifier performed significantly better than the text-based classifier (p = 0.003).
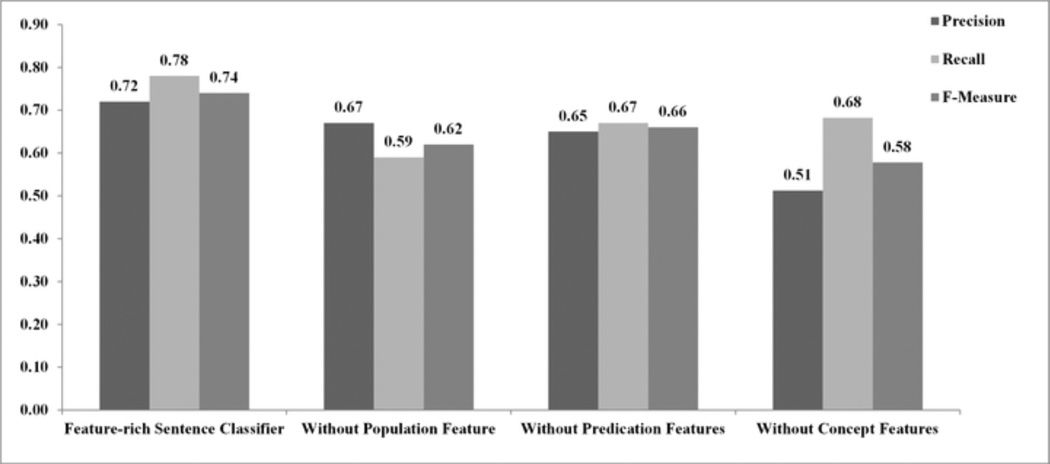
Experiment #3: Each type of semantic feature provides a significant contribution to the overall performance of the feature-rich classifier. The results are summarized in Figure 3. The feature-rich classifier performed significantly better than the classifiers without population, predication, and concept features (F-measure = 74% versus 62%, 66% and 58% respectively; p < 0.01 for all comparisons). The difference among the three classifiers without one of the semantic feature types was not significant (p > 0.5 for all comparisons).
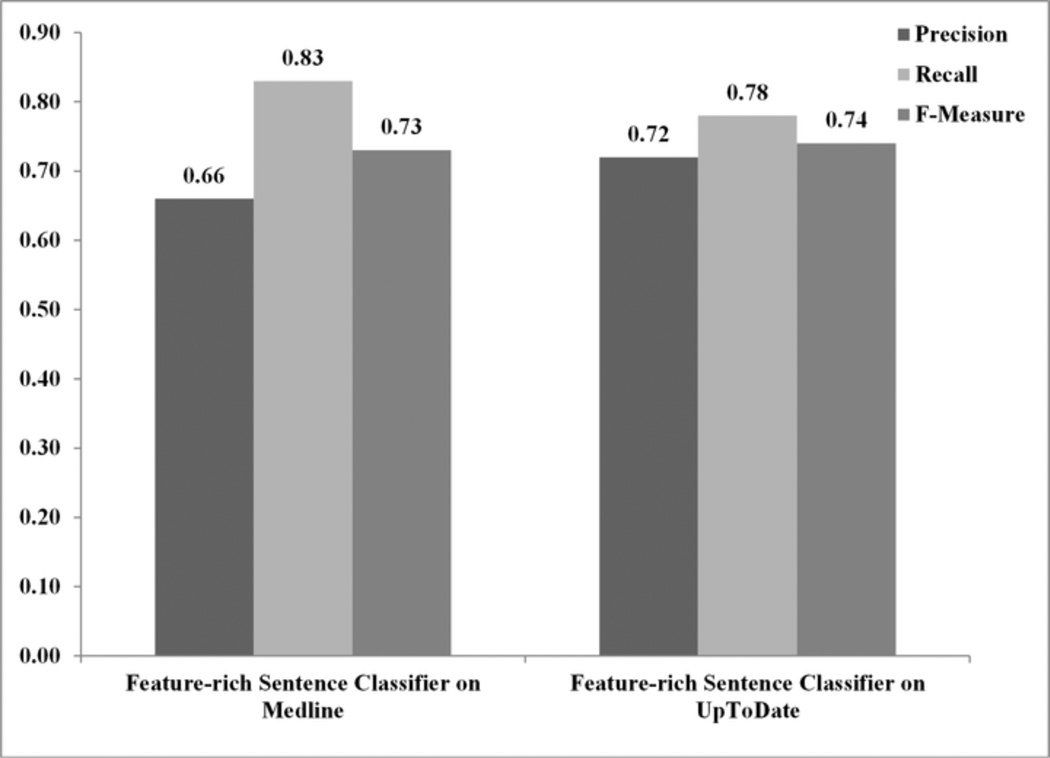
Experiment #4: The feature-rich-classifier is generalizable to other types of online clinical resources. Figure 4 summarizes the results. The performance of the feature-rich classifier on Medline sentences was equivalent to the performance of the same classifier on UpToDate sentences (F-measure = 73% versus 74%; p=0.62).
Figure 1.
Average precision, recall and F-measure of the feature-rich sentence classifier, term frequency, and bigram classifier (Experiment #1).
Figure 2.
Average precision, recall and F-measure of the feature-rich sentence classifier, semantic classifier, and text-based classifier (Experiment #2).
Figure 3.
Average precision, recall and F-measure of the feature-rich sentence classifier and feature-rich classifiers with one of the feature types excluded (Experiment #3).
Figure 4.
Average precision, recall and F-measure of the feature-rich sentence classifier on UpToDate and Medline sentences (Experiment #4).
5. DISCUSSION
In this study we investigated an automated method for extracting clinically useful sentences from synthesized online clinical resources such as UpToDate. Such a method is an important component for clinical evidence summarization and question answering systems aimed at assisting clinicians with patient-specific clinical questions and decision-making. Based on a recent systematic review of biomedical text summarization, this is the first study to investigate methods to extract clinically useful sentences from synthesized evidence resources.[4] Also, we conducted an exploratory investigation of the generalizability of the method to the primary literature.
Overall, the feature-rich classifier had an F-measure of 74%, with a recall of 78% and precision of 72%. This precision is much higher than the overall rate of clinically useful sentences in the UpToDate documents in the dataset, which is 13%. Therefore, the feature-rich classifier could be used to enable a more efficient alternative or complementary mechanism for clinicians to peruse clinical evidence from clinical knowledge resources. One advantage is that in addition to classifying sentences, our method generates rich sentence-level metadata, which could be leveraged by interactive text summarization tools and to enable semantic integration with electronic health record (EHR) systems.[47, 48] We are currently designing a context-aware clinical knowledge summarization tool that employs the feature-rich sentence classifier algorithm along with a sentence ranking algorithm based on a clinician’s information needs. The tool is designed to integrate with EHR systems via OpenInfobutton,[49] an open source Web service compliant with the Health Level Seven (HL7) Infobutton Standard.[47] A description of the tool along with results of a formative evaluation are available elsewhere.[50, 51]
We conducted four experiments to test four hypotheses. The first experiment showed that the domain-specific method, which is based on semantic and syntactic features of sentences, significantly outperformed the feature co-occurrence method. This is an expected outcome, since the domain-specific method looks for specific characteristics that contribute to the clinical usefulness of sentences. Specifically, the features used in the feature-rich classifier were fine-tuned based on domain knowledge. This finding also highlights the importance of state-of-the-art, biomedical semantic understanding methods and tools, such as MetaMap [52] and SemRep,[31] in the context of biomedical text summarization.
The second and third experiments showed that each feature type provides additional contribution to the overall classification performance. This may be explained by the different strengths and weaknesses of each feature type. For example, semantic predications and concepts identify relations between treatment interventions and conditions; the population algorithm identifies the definition of a specific patient population; and cue words identify patterns associated with useful sentences (e.g., deontic terms, evidence attributions) and non-useful sentences (e.g., study design terms). Moreover, the results showed that predication and concept-based features improved recall, while the population-based feature improved precision. Although individual text-based features performed significantly worse than other feature types, text-based features improved overall precision because cue words, such as deontic terms, could not be detected by the other feature types used in our study. The fourth experiment suggests that the classifier’s model and features used for UpToDate are generalizable to Medline, possibly because the structure and semantics of clinically useful sentences are similar among different online clinical resources. We recently completed a more thorough analysis that confirms these exploratory findings.[53]
Error analysis identified two main categories of misclassification errors. False-positive cases were caused by a wide range of problems. We describe three categories that occurred most frequently. The first one (13% of the cases) consisted of recommendations that were too general. For example, in “All patients with CVD should have measurement of waist circumference and calculation of body mass index,” both the population (“all patients”) and the intervention (“waist circumference and body mass index”) are too general. Our algorithm includes a mechanism to exclude general concepts, but a more sophisticated approach is needed help to further improve performance. The second category (16% of the cases) consisted of sentences that present details about the implementation of a certain treatment, such as “In comparison, amiodarone is metabolized in the liver and dose adjustment is probably necessary in patients with hepatic dysfunction.” While these sentences may be useful once a clinician identifies a recommendation that applies to her patient, they present details that would not be important in the first tier of a text summary. The third category (6% of the cases) consisted of sentences that contained all the desired features, except that they lacked a specific patient population, such as “Although some have suggested that combination antiarrhythmic drug therapy may be an alternative, there are limited data to support such an approach and the patient may be exposed to a greater risk of proarrhythmia and other side effects.”
False-negative cases were due to two main categories. The first one (56% of the cases) consisted of useful sentences from which SemRep and MedTagger failed to extract useful treatment predications and concepts, such as “non-pharmacologic therapies that may benefit patients with angina refractory to the above medical therapies include enhanced external counterpulsation, spinal cord stimulation, and transmyocardial revascularization.” Improvements in the coverage of the underlying controlled terminologies used by SemRep and MedTagger would address most of these problems. The second category (9% of the cases), consisted of deontic and population expressions that our algorithm did not account for (e.g., “we consider,” “we use”), such as in “we use calcium channel blockers and nitrates routinely to relieve symptoms when initial treatment with beta blockers is not successful or if beta blockers are contraindicated or cause side effects.”
Our study had a few limitations. First, our approach was tuned to extract treatment recommendations and most likely needs to be adapted to extract diagnostic recommendations. For example, a different set of predicates and semantic groups would be necessary. Second, the experiments were conducted in a relatively small subset of UpToDate documents. Yet, the sample size had enough statistical power to detect differences among the various approaches tested. In addition, documents were chosen based on a representative sample of common and complex chronic conditions that affect a large patient population. Therefore, it is expected that our experimental results will generalize to other documents on the treatment of similar conditions.
6. CONCLUSION
We investigated domain-specific supervised machine learning methods using a rich set of semantic and syntactic features to classify clinically useful sentences in UpToDate articles. The feature-rich approach significantly outperformed classifiers based on a single type of feature. Different types of semantic features provided a unique contribution to overall classification performance. The Kernel-based Bayesian Network method outperformed other machine learning algorithms. In future studies, the resulting sentence classifier can be used as a component in text summarization and question answering systems to help clinicians’ decision-making.
Supplementary Material
Highlights.
We investigated automated extraction of clinically useful sentences from UpToDate.
A Kernel-based Bayes Network classifier was applied on domain-specific features.
Features: UMLS concepts, semantic predications, patient population and cue words.
The proposed approach outperformed baselines and alternate classifiers.
The classifier’s model and features generalized to Medline abstracts.
Acknowledgments
This project was supported by grants 1R01LM011416-01 and 4R00LM011389-02 from the National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hoogendam A, Stalenhoef AF, Robbe PF, Overbeke AJ. Analysis of queries sent to PubMed at the point of care: observation of search behaviour in a medical teaching hospital. BMC Med Inform Decis Mak. 2008;8:42. doi: 10.1186/1472-6947-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Fiol G, Workman TE, Gorman PN. Clinical questions raised by clinicians at the point of care: a systematic review. JAMA Intern Med. 2014;174:710–718. doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 3.Cao Y, Liu F, Simpson P, Antieau L, Bennett A, Cimino JJ, et al. AskHERMES: An online question answering system for complex clinical questions. J Biomed Inform. 2011;44:277–288. doi: 10.1016/j.jbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra R, Bian J, Fiszman M, Weir CR, Jonnalagadda S, Mostafa J, et al. Text summarization in the biomedical domain: a systematic review of recent research. J Biomed Inform. 2014;52:457–467. doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoogendam A, Stalenhoef AF, Robbe PF, Overbeke AJ. Answers to questions posed during daily patient care are more likely to be answered by UpToDate than PubMed. Journal of medical Internet research. 2008;10:e29. doi: 10.2196/jmir.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayyah Ensan L, Faghankhani M, Javanbakht A, Ahmadi SF, Baradaran HR. To compare PubMed Clinical Queries and UpToDate in teaching information mastery to clinical residents: a crossover randomized controlled trial. PLoS One. 2011;6:e23487. doi: 10.1371/journal.pone.0023487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shariff SZ, Bejaimal SA, Sontrop JM, Iansavichus AV, Weir MA, Haynes RB, et al. Searching for medical information online: a survey of Canadian nephrologists. Journal of nephrology. 2011;24:723–732. doi: 10.5301/JN.2011.6373. [DOI] [PubMed] [Google Scholar]
- 8.Sheets L, Callaghan F, Gavino A, Liu F, Fontelo P. Usability of selected databases for low-resource clinical decision support. Applied clinical informatics. 2012;3:326–333. doi: 10.4338/ACI-2012-06-RA-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiele RH, Poiro NC, Scalzo DC, Nemergut EC. Speed, accuracy, and confidence in Google, Ovid, PubMed, and UpToDate: results of a randomised trial. Postgraduate medical journal. 2010;86:459–465. doi: 10.1136/pgmj.2010.098053. [DOI] [PubMed] [Google Scholar]
- 10.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. Bmj. 2005;330:765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell L, Singh JA. Abatacept for rheumatoid arthritis. The Cochrane database of systematic reviews. 2009:CD007277. doi: 10.1002/14651858.CD007277.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra R, Del Fiol G, Kilicoglu H, Jonnalagadda S, Fiszman M. Automatically extracting clinically useful sentences from UpToDate to support clinicians' information needs. AMIA Annu Symp Proc. 2013;2013:987–992. [PMC free article] [PubMed] [Google Scholar]
- 13.Edmundson HP. New methods in automatic extracting. Journal of the ACM (JACM) 1969;16:264–285. [Google Scholar]
- 14.Lin C-Y, Hovy E. The automated acquisition of topic signatures for text summarization; Proceedings of the 18th conference on Computational linguistics-Volume 1: Association for Computational Linguistics; 2000. pp. 495–501. [Google Scholar]
- 15.Plaza L, Carrillo-de-Albornoz J. Evaluating the use of different positional strategies for sentence selection in biomedical literature summarization. BMC bioinformatics. 2013;14:71. doi: 10.1186/1471-2105-14-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu Y, Zhu X, Hirst G. Using outcome polarity in sentence extraction for medical question-answering. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2006:599–603. [PMC free article] [PubMed] [Google Scholar]
- 17.Kupiec J, Pedersen J, Chen F. A trainable document summarizer; Proceedings of the 18th annual international ACM SIGIR conference on Research and development in information retrieval: ACM; 1995. pp. 68–73. [Google Scholar]
- 18.Mihalcea R, Tarau P. TextRank: Bringing order into texts. Association for Computational Linguistics. 2004 [Google Scholar]
- 19.Jonnalagadda SR, Del Fiol G, Medlin R, Weir C, Fiszman M, Mostafa J, et al. Automatically extracting sentences from Medline citations to support clinicians' information needs. J Am Med Inform Assoc. 2013;20:995–1000. doi: 10.1136/amiajnl-2012-001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales LP, Esteban AD, Gervás P. Concept-graph based biomedical automatic summarization using ontologies. 3rd Textgraphs Workshop on Graph-Based Algorithms for Natural Language Processing: Association for Computational Linguistics. 2008:53–56. [Google Scholar]
- 21.Plaza L, Díaz A, Gervás P. A semantic graph-based approach to biomedical summarisation. Artificial intelligence in medicine. 2011;53:1–14. doi: 10.1016/j.artmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Fiszman M, Shin D, Wilkowski B, Rindflesch TC. Clustering cliques for graph-based summarization of the biomedical research literature. BMC bioinformatics. 2013;14:182. doi: 10.1186/1471-2105-14-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeve L, Han H, Brooks AD. BioChain: lexical chaining methods for biomedical text summarization. Proceedings of the 2006 ACM symposium on Applied computing: ACM. 2006:180–184. [Google Scholar]
- 24.Shi Z, Melli G, Wang Y, Liu Y, Gu B, Kashani MM, et al. Question answering summarization of multiple biomedical documents. Advances in Artificial Intelligence: Springer. 2007:284–295. [Google Scholar]
- 25.Yoo I, Hu X, Song IY. A coherent graph-based semantic clustering and summarization approach for biomedical literature and a new summarization evaluation method. BMC bioinformatics. 2007;8(Suppl 9):S4. doi: 10.1186/1471-2105-8-S9-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Wagholikar K, Jonnalagadda S, Sohn S. Integrated cTAKES for Concept Mention Detection and Normalization. 2011 [Google Scholar]
- 28.Baldridge J, Morton T. OpenNLP. 2004 [Google Scholar]
- 29.Divita G, Browne AC, Rindflesch TC. Evaluating lexical variant generation to improve information retrieval. Proceedings of the AMIA Symposium. 1998:775. [PMC free article] [PubMed] [Google Scholar]
- 30.Moosavinasab S, Rastegar-Mojarad M, Liu H, Jonnalagadda SR. Towards Transforming Expert-based Content to Evidence-based Content. AMIA Summits on Translational Science Proceedings. 2014;2014:83. [PMC free article] [PubMed] [Google Scholar]
- 31.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Fiszman M, Demner-Fushman D, Lang FM, Goetz P, Rindflesch TC. Interpreting comparative constructions in biomedical text. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing. 2007:137–144. [Google Scholar]
- 33.Rindflesch TC, Fiszman M, Kilicoglu HBL. Semantic Knowledge Representation Project; A report to the Board of Scientific Counselors. U.S. National Library of Medicine, LHNCBC. 2003 [Google Scholar]
- 34.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians' clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005;12:217–224. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sackett S, Richardson R. Evidence-based practice. Foundations of Evidence-Based Social Work Practice. 2006:35. [Google Scholar]
- 36.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Studies in health technology and informatics. 2001;84:216–220. [PMC free article] [PubMed] [Google Scholar]
- 37.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics. 2003;1:423–430. [Google Scholar]
- 38.Levy R, Andrew G. Tregex and Tsurgeon: tools for querying and manipulating tree data structures; Proceedings of the fifth international conference on Language Resources and Evaluation; 2006. pp. 2231–2234. [Google Scholar]
- 39.Hall MA. Correlation-based feature selection for machine learning: The University of Waikato. 1999 [Google Scholar]
- 40.Azhagusundari B, Thanamani AS. Feature selection based on information gain. International Journal of Innovative Technology and Exploring Engineering (IJITEE) ISSN. 2013:2278–3075. [Google Scholar]
- 41.Singh SR, Murthy HA, Gonsalves TA. Feature Selection for Text Classification Based on Gini Coefficient of Inequality. FSDM: Citeseer. 2010:76–85. [Google Scholar]
- 42.Lomotan EA, Michel G, Lin Z, Shiffman RN. How “should” we write guideline recommendations? Interpretation of deontic terminology in clinical practice guidelines: survey of the health services community. Quality and Safety in Health Care. 2010;19:509–513. doi: 10.1136/qshc.2009.032565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez A, Larrañaga P, Inza I. Bayesian classifiers based on kernel density estimation: Flexible classifiers. International Journal of Approximate Reasoning. 2009;50:341–362. [Google Scholar]
- 44.He Y-L, Wang R, Kwong S, Wang X-Z. Bayesian classifiers based on probability density estimation and their applications to simultaneous fault diagnosis. Information Sciences. 2014;259:252–268. [Google Scholar]
- 45.Murakami Y, Mizuguchi K. Applying the Naïve Bayes classifier with kernel density estimation to the prediction of protein–protein interaction sites. Bioinformatics. 2010;26:1841–1848. doi: 10.1093/bioinformatics/btq302. [DOI] [PubMed] [Google Scholar]
- 46.Demšar J. Statistical comparisons of classifiers over multiple data sets. The Journal of Machine Learning Research. 2006;7:1–30. [Google Scholar]
- 47.Del Fiol G, Huser V, Strasberg HR, Maviglia SM, Curtis C, Cimino JJ. Implementations of the HL7 Context-Aware Knowledge Retrieval ("Infobutton") Standard: challenges, strengths, limitations, and uptake. J Biomed Inform. 2012;45:726–735. doi: 10.1016/j.jbi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Fiol G, Haug PJ, Cimino JJ, Narus SP, Norlin C, Mitchell JA. Effectiveness of topic-specific infobuttons: a randomized controlled trial. J Am Med Inform Assoc. 2008;15:752–759. doi: 10.1197/jamia.M2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Fiol G, Curtis C, Cimino JJ, Iskander A, Kalluri AS, Jing X, et al. Disseminating context-specific access to online knowledge resources within electronic health record systems. Studies in health technology and informatics. 2013;192:672–676. [PMC free article] [PubMed] [Google Scholar]
- 50.Del Fiol G, Mostafa J, Pu D, Medlin R, Slager S, Jonnalagadda S, Weir CR. Int J Med Inform. 2016;86:126–134. doi: 10.1016/j.ijmedinf.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Del Fiol G, Pu D, Weir CR, Medlin R, Jonnalagadda S, Mishra R, et al. Workshop of Interactive Systems in Healthcare. Washington, DC: 2014. Iterative design of an Interactive Clinical Evidence Summarization Tool. [Google Scholar]
- 52.Aronson AR. Metamap: Mapping text to the umls metathesaurus. Bethesda, MD: NLM, NIH, DHHS; 2006. [Google Scholar]
- 53.Morid MA, Jonnalagadda S, Fiszman M, Raja K, Del Fiol G. Classification of Clinically Useful Sentences in MEDLINE. American Medical Informatics Association (AMIA) Annual Symposium 2015. 2015 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.