Abstract
The impact of alterations in intestinal microbiota on microbial metabolites and on disease processes, such as graft-versus-host disease (GVHD), is not known. Here we performed unbiased analysis to identify novel alterations in gastrointestinal microbiota-derived short chain fatty acids (SCFA) after allogeneic bone marrow transplant (allo-BMT). Alterations in the amounts of only one SCFA, butyrate, were observed only within the intestinal tissue. The reduced butyrate in CD326+ intestinal epithelial cells (IECs) after allo-BMT resulted in decreased histone acetylation, which was restored upon local administration of exogenous butyrate. Butyrate restoration improved IEC junctional integrity, decreased apoptosis, and mitigated GVHD. Furthermore, alteration of the indigenous microbiota with 17 rationally selected strains of high butyrate producing Clostridia also decreased GVHD. These data demonstrate a heretofore unrecognized role of microbial metabolites and suggest that local and specific alteration of microbial metabolites has direct salutary effects on GVHD target tissues and can mitigate its severity.
INTRODUCTION
Alterations in the intestinal microbiome are associated with several disease processes1-5. However, the effect that changes in the community structure of the microbiome have on the production of microbial-derived metabolites is poorly explored. Microbial metabolites influence disease severity, but whether these alterations in microbial metabolites can impact outcomes after allogeneic bone marrow transplant (allo-BMT) are not known. Allo-BMT is a critical interventional therapy for patients with aggressive hematological malignancies6,7. Although allo-BMT is a curative and widely used treatment, approximately 40-50% of patients experience severe gastrointestinal damage from graft-versus-host disease (GVHD), which leads to high transplant-related mortality7,8.
Studies have revealed that the intestinal microbiota is significantly altered in patients with GVHD and the alterations correlate with GVHD severity and pathogenesis4,9. Nevertheless, the direct causality of the changes in the host microbiota on GVHD severity is unclear. More relevantly, whether changes in the microbiota result in alterations in levels of microbial metabolites and by-products that have biological impact on allogeneic-BMT remain unknown. Microbial metabolites such as short chain fatty acids (SCFA) are exclusively derived from the GI microbiota and are not made by the host. Some of these fatty acids (FAs), specifically the histone deacetylase inhibitor (HDACi) butyrate, is a preferred energy source for intestinal epithelial cells (IECs)10-13 and administration of exogenous HDACi regulates GVHD14-16. But the impact that host indigenous microbial metabolites that function as HDACi have on GVHD remains unknown11-13.
Here, we performed unbiased profiling of the microbial metabolome with a specific focus on targeted FAs after experimental allo-BMT. We found that only one SCFA, namely butyrate was significantly reduced only in the intestinal tissue of allo-BMT recipients resulting in decreased acetylation of histone H4 within IECs. Increasing intestinal butyrate restored acetylation of histone H4, protected IECs, and decreased the severity of GVHD. Furthermore, rationally altering host GI microbiota to high butyrate producers17 mitigated GVHD.
RESULTS
Targeted microbial metabolite profiling
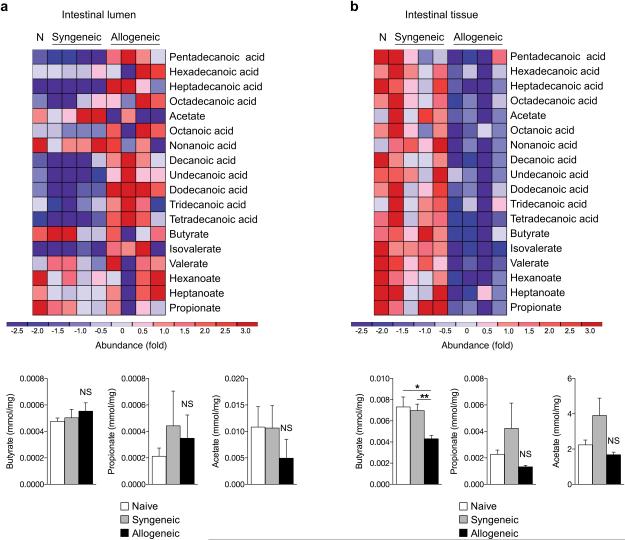
We hypothesized that alterations in the composition of the microbiota in the GI lumen would result in an altered microbial metabolome after GVHD4,18. We determined the concentration of microbial FA metabolites, both short-chain FAs and long-chain FAs up to 18 carbons in length, from several sites seven days (day +7) after BMT. We analyzed the serum, spleen, liver, intestines, and luminal contents (stool) of the intestines with gas chromatography mass spectrometry (GC/MS) (Supplementary Fig. 1a). We utilized a well-established, clinically relevant model of MHC-mismatched BMT with C57BL/6J (H-2b) cells transferred to lethally irradiated Balb/c (H-2d) mice and compared it to syngeneic transplant and naive animals. The animal cages were exchanged on day 3 to take any alterations in the microbial environment into account, and analysis following GC/MS was performed in a blinded manner. The concentrations of the FAs were not significantly different in the luminal contents of the intestines between any of the groups (Fig. 1a and Supplementary Fig. 1b). They were also not significantly different in the serum or the tissues such as the spleen and liver of allogeneic animals (Supplementary Fig. 1c and Supplementary Table 1) compared with syngeneic animals and naive controls. However, the greatest and the only statistically significant difference was observed in just one SCFA, butyrate, which was significantly decreased only in the intestinal tissue at day 7 (Fig. 1b and Supplementary Fig. 1d). We once again observed similar results on day 21 as on day 7 (Supplementary Fig. 2). Collectively these data demonstrate that butyrate levels are consistently reduced only in the intestinal tissue after allo-BMT.
Figure 1. Allogeneic BMT reduces IEC intracellular butyrate.
(a) Fatty acid levels (short and long chain) on day 7 in the intestinal luminal contents (stool) and (b) in the intestinal tissue of recipients of syngeneic BMT (BALB/c → BALB/c), allogeneic BMT (C57BL/6J → BALB/c), or no BMT (N - naive). Graphical results of all animals combined are shown below heatmaps for SCFAs butyrate, propionate, and acetate. Representative heatmaps are shown of n = 10 animals in naive and syngeneic groups and n = 9 in allogeneic group. *P < .05; **P < .01 of ANOVA test. Bars and error bars represent the means and standard errors of the mean, respectively.
Functional impact of altered levels of SCFA in the IECs
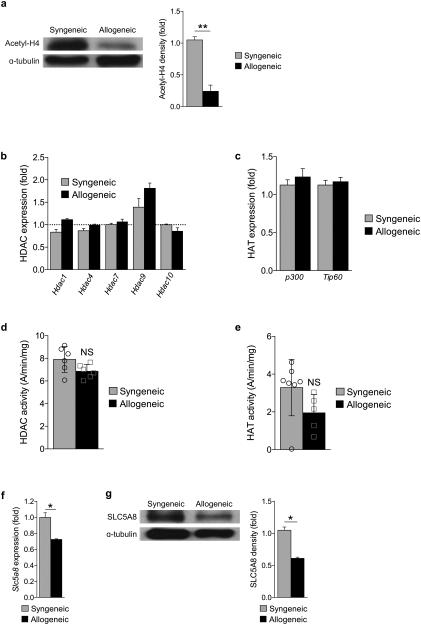
In light of the reduction of butyrate in allogeneic animals only in the intestinal tissue, we next analyzed the potential functional impact of reduced butyrate in IECs. Because butyrate is an HDACi10,12,19, we examined the degree of histone acetylation by immunoblotting purified CD326+ IECs after BMT. The degree of acetylation of histone H4 was significantly decreased on day 7 (Supplementary Fig. 3a) and day 21 (Fig. 2a) following allo-BMT demonstrating that reduced butyrate resulted in decreased histone acetylation. Therefore to confirm if the decreased acetylation is secondary to decreased HDAC inhibition from reduction in butyrate and not due to potential alterations in HDAC and HAT enzyme levels20 following transplant, we analyzed the expression of HDACs and HATs in IECs after BMT. We observed similar levels of several HDACs (Hdac 1,4,7,9, and 10) (Fig. 2b) and HATs (p300 and TIP60) (Fig. 2c) by qPCR in the IECs (CD326+) of both syngeneic and allogeneic BMT recipients. Furthermore both HDAC (Fig. 2d) and HAT (Fig. 2e) enzyme activity were not different in these animals. These data suggest that reduction in histone acetylation in the IECs after allo-BMT is from reduced levels of butyrate.
Figure 2. Loss of butyrate decreases histone acetylation in IECs.
(a) Protein expression of acetyl-histone H4 (top blot) and densitometric analysis normalized to presence of α-tubulin (shown right of blot) 21 days following syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) BMT. Densitometric analysis of 3 experiments combined; each experiment had n=5-6 mice per group. (b) Gene expression of representatives of class I, II, and IV HDAC enzymes, (c) histone acetyltransferase (HAT) enzyme levels, (d) HDAC activity, and (e) HAT activity in syngeneic and allogeneic CD326+ IECs. A = absorbance. (f) Gene expression and (g) protein levels of SLC5A8 (monocarboxylate transporter of butyrate) in IECs (CD326+) of syngeneic and allogeneic transplant recipients 21 days following BMT. Representative immunoblots; densitometric analysis of three similar experiments combined is shown right of blots. *P < .05; **P < .01 of students t-test. Bars and error bars represent the means and standard errors of the mean, respectively.
Reduced uptake of butyrate by the IECs
We next explored whether the diminished concentration of butyrate observed in the intestinal tissue was from impaired uptake of butyrate following allo-BMT. To this end, we examined the expression of the known butyrate monocarboxylate transporter (SLC5A8) and the receptor of butyrate (GPR43) in IECs following allo-BMT10,21. Decreased gene expression (Fig. 2f) and protein (Fig. 2g) of both SLC5A8 and GPR43 (Supplementary Fig. 3b-c) were observed in IECs from allogeneic animals following transplant on day +21 and also on day +7 (Supplementary Fig. 3d) suggesting that reduction in butyrate concentration in the IECs is due to reduced uptake of the microbiota-derived luminal butyrate. We next cultured primary IECs with proinflammatory mediators (IFN-γ and/or TNF) and analyzed the expression of the butyrate transporter SLC5A8. Exposure of IECs to inflammatory cytokines significantly decreased the expression of Slc5a8 (Supplementary Fig. 3e). These data indicate that the intense inflammatory milieu following allo-BMT causes reduced expression of butyrate transporters and receptors leading to reduction in butyrate and histone acetylation in IECs.
Rescuing the cellular effects of reduced butyrate
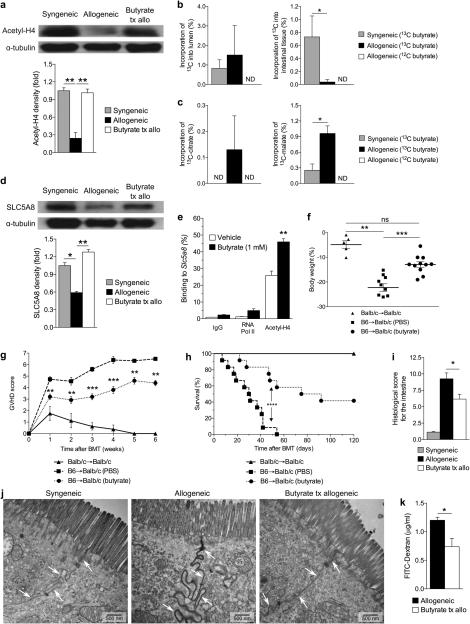
We next determined if the reduced amount of butyrate in IECs could be restored in vivo and further, whether this would have a functional impact on histone acetylation. In addition to utilizing transporters, butyrate can also diffuse across the mucosal barrier into IECs when present in high concentrations10,22. Therefore, we hypothesized that administration of high amounts of butyrate locally would restore histone acetylation of IECs, in vivo. To test this, C57BL/6J cells were transferred to Balb/c mice and administered vehicle or butyrate via daily intragastric gavage. Daily butyrate administration for 21 days significantly restored acetylation of histone H4 compared with untreated allo-BMT recipients (Fig. 3a).
Figure 3. Intragastric butyrate gavage enhances its uptake and histone acetylation in IECs.
Immunoblot of CD326+ IECs from syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) BMT recipients that received vehicle or butyrate (10mg/kg). (a) Acetylated histone-H4 21 days post-BMT with densitometric analysis of three experiments combined, shown below blots; representative immunoblots shown. (b) 13C label incorporation of butyrate in luminal contents and intestinal tissue of the large intestine in mice gavaged with 13C-Butyrate or non-labeled 12C-Butyrate, n=6 mice per group. (c) 13C label incorporation in tricarboxylic acid metabolite pools of the large intestine of mice fed a bolus (2 g/kg) of 13C-Butyrate or 12C-Butyrate, n=6 mice per group. ND = not detected. (d) Levels of SLC5A8 in IECs of syngeneic and allogeneic BMT recipients 21 days following transplant, from a. (e) ChIP of butyrate treated IECs (CD326+) and binding of acetylated histone H4 in the promoter region of Slc5a8. (f) Weight loss on day 21 with (g) GVHD clinical score, (h) survival, and (i) intestinal histopathology 21 days post-BMT of recipients treated with intragastric vehicle or butyrate; syngeneic n=6, n=12 mice per allogeneic group. (j) Transmission electron microscopy (TEM) of intestines, isolated from BMT recipients with or without intragastric gavage of butyrate for duration of experiment; all samples were isolated 7 days following BMT and stained with ruthenium red (0.1%); arrows indicate cell-cell interface. (k) Level of FITC-dextran translocation across the GI-barrier into blood serum in butyrate treated allogeneic BMT recipients, compared to vehicle control 21 days post BMT. *P < .05; **P < .01; ***P < .0001 of ANOVA a - d, i, k; students t-test e; Mantel-Cox log-rank test g, h. Bars and error bars represent the means and standard errors of the mean, respectively.
We next determined whether there was a difference in the uptake and metabolism of the exogenously administered butyrate between the syngeneic and allogeneic recipients. To this end we performed metabolic flux analysis (MFA) assessing label incorporation of heavy 13C labeled butyrate into luminal and intestinal tissue butyrate pools by utilizing GC/MS as above. We treated recipients of syngeneic and allogeneic transplant 7 days after BMT with a bolus of either 13C-butyrate or regular 12C-butyrate, which served as the control. The IECs and luminal contents were harvested 6 hours later and analyzed for incorporation of 13C. Although we saw similar amounts of heavy 13C butyrate in the lumen of the large intestine, we saw significantly decreased 13C heavy butyrate in the intestinal tissue of recipients of allo-BMT, suggesting reduced uptake (Fig. 3b). To determine whether there were any differences in the metabolism of butyrate, we further examined the presence of 13C-butyrate in different stages of the tricarboxylic acid (TCA) cycle. Specifically, we determined the incorporation of 13C-butyrate into citrate, succinate and further downstream, into malate (Supplementary Fig. 3f). There was a significant difference in the amount of heavy carbon in citrate and malate (Fig. 3c) and a trend towards greater incorporation into succinate (Supplementary Fig. 3g) within the IECs from the large intestine. Similarly, examination of IECs from both small and large intestines combined also revealed an overall increased level of 13C incorporation in the downstream metabolite of the TCA cycle, malate, in allo-BMT recipients compared with syngeneic animals (Supplementary Fig. 3h), suggesting an increased rate of metabolism in the IECs of these animals.
Furthermore, daily intragastric gavage of butyrate resulted in an increase in butyrate transporter SLC5A8 (Fig. 3d), suggesting that butyrate has a positive feedback mechanism resulting in an increase of its own transporter. To determine whether butyrate was directly responsible for induction of its own transporter, we analyzed the degree of histone acetylation at the promoter of SLC5A8 with chromatin immunoprecipitation (ChIP). We found an increased association of acetylated histone H4 in the promoter region of Slc5a8 in IECs (CD326+) treated with butyrate (Fig. 3e). These data demonstrate that reduced butyrate following allo-BMT has functional effects on IECs.
Increase in intestinal butyrate mitigated GVHD
Systemic administration of HDACi decreases acute GVHD14,15,23. We therefore determined if increasing local levels of endogenous HDACi, butyrate, would impact GVHD severity. Again using the C57BL/6J into Balb/c model, we administered vehicle control or butyrate via daily intragastric gavage for one week, followed by administration every other day for the remainder of the experiment.
Administration of butyrate resulted in decreased weight loss (Fig. 3f), GVHD clinical scores (Fig. 3g), and increased survival (Fig. 3h). We found similar improved survival using a second clinical model of BMT, a MHC-matched, minor antigen mismatched model. C3H.SW (H-2b) cells were transferred to C57BL/6J (H-2b) mice, thus demonstrating strain-independent results (Supplementary Fig. 4a). Furthermore, histopathological analysis 21 days following BMT exhibited decreased histological scores in the intestines of the major MHC mismatch model (Fig. 3i) and decreased histological scores in both the intestines and liver in the model of minor antigen mismatch (Supplementary Fig. 4b). To determine whether irradiation related inflammation was critical for butyrate induced protection, we next examined the butyrate induced protective effects in a non-irradiated model of parent (C57BL/6J, H-2b) into F1 (B6D2F1, H-2b/d) BMT. We once again observed significantly less weight loss (Supplementary Fig. 4c) and improved survival (Supplementary Fig. 4d) in allogeneic recipients that were treated with intragastric butyrate. Next, to further evaluate the magnitude of the butyrate-induced protective effect, we determined GVHD mortality once again utilizing the MHC disparate B6 into BALB/c model. Intragastric gavage of butyrate induced significant GVHD survival benefit in allogeneic animals that received higher doses of T cells (Supplementary Fig. 4e). These data collectively show that butyrate induced GVHD protective effects regardless of strain combinations, conditioning, or higher alloreactive T cell doses.
Increase in intracellular butyrate protects GI epithelium
We next determined if the decreased GI GVHD resulted in reduced translocation of luminal contents and improved epithelial integrity24-26, we utilized transmission electron microscopy (TEM) to examine the ability of butyrate to preserve cellular junctions following allo-BMT. Significantly, intense leakage of the electron dense stain ruthenium red25 was found in allo-BMT recipients treated with vehicle alone (Fig. 3j, middle panel). However, TEM studies demonstrate that the integrity of the IEC junction was preserved at both day 7 (Fig. 3j, right panel) and day 21 (Supplementary Fig. 4f) in allo-BMT recipients that received local intragastric administration of butyrate.
We further assessed the intestinal permeability after allo-BMT by intragastric administration of FITC-dextran, a non-metabolized carbohydrate27. Butyrate-treated allo-BMT recipients exhibited significantly less detectable FITC-dextran in the serum at 21 days following transplant (Fig. 3k).
Reduction in GVHD is independent of donor Treg cells
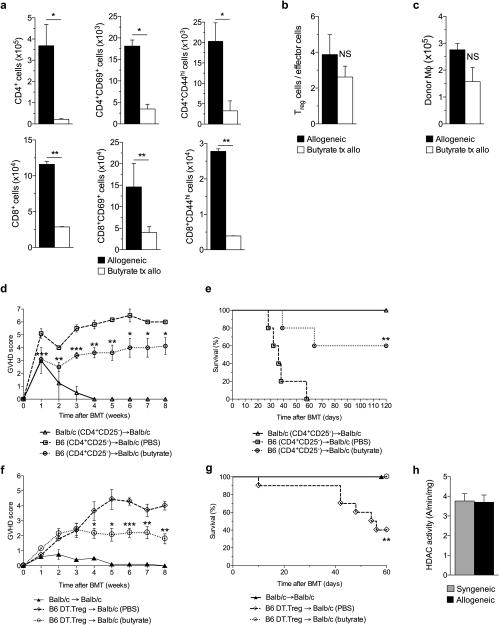
Treg cells mitigate GVHD and butyrate has been shown to increase intestinal Treg cells17,28. We therefore analyzed the cellular contents of the intestine 21 days after allo-BMT to determine whether butyrate had an impact on local Treg cells. The total numbers of CD45.1+ cells recovered from the intestinal lamina propria were not different between vehicle- and butyrate-treated allo-BMT recipients (Supplementary Fig. 4g). By contrast, intestinal infiltration of donor CD4+ and CD8+ T cells and activated T cells (CD69+ or CD44hi) was decreased in animals that received local intragastric butyrate administration (Fig. 4a). However, the ratio of donor Treg cells to effector T cells was not different in the intestines of these animals (Fig. 4b). Microbiota-derived butyrate has been shown to increase immune-regulatory macrophages in the GI tract which increased Treg cells29. However we observed no difference in the total number of donor macrophages in the intestine of allogenenic recipients that were treated with either vehicle or butyrate (Fig. 4c).
Figure 4. Donor Treg cells are dispensable for GVHD protection.
(a)-(c) Intestinal immunophenotypical analysis of recipients 21 days following allogeneic (C57BL/6J → BALB/c) BMT treated with either butyrate or vehicle via intragastric gavage. (a) Total cell numbers of intestinal CD4+ & CD8+ T cells (left column) and activated T cells: CD69+ T cells (middle column) & CD44hi T cells (right column). (b) Ratio of intestinal Treg cells (CD4+CD25+FoxP3+) to effector cells (CD4+FoxP3−) and (c) total cell number of intestinal donor macrophages (CD11b+F4.80+) in recipients of allo-BMT treated with vehicle and butyrate. (d)-(e) Recipients received CD4+CD25− (Treg depleted) 1.5 × 106 donor T cells and T cell depleted (TCD) bone marrow. (d) GVHD score and (e) survival in animals treated with vehicle or butyrate (10mg/kg); syngeneic n=6, n=12 mice per allogeneic group. (f)-(g) Recipients received Treg depleted (C57BL/6J DT.Treg) 0.5 × 106 donor cells and TCD bone marrow with resulting (f) GVHD score and (g) survival in butyrate treated BMT recipients. (h) HDAC activity in T cells of the spleen 21 days following syngeneic or allogeneic BMT. Syngeneic n=5, n = 10 mice per allogeneic group. *P < .05; **P < .01; ***P < .0001 of students t-test a - c; Mantel-Cox log-rank test d - g. Bars and error bars represent the means and standard errors of the mean, respectively.
To further determine whether the salutary effects of local treatment with butyrate on GI GVHD were dependent on donor Treg cells, we next performed a BMT, utilizing the same MHC-mismatched BMT model in which C57BL/6J (H-2b) cells are transferred to Balb/c (H-2d) mice. We transferred T cell-depleted (TCD) bone marrow and purified CD4+ CD25− T cells from donors and administered vehicle or butyrate to the recipients daily for 1 week, then every other day thereafter as above. We still observed decreased GVHD (Fig. 4d-e) indicating that donor Treg cells may be dispensable for the reduction of GVHD .
To further confirm the donor Treg cell-independent protective effects of butyrate on GVHD, we next utilized donor C57BL/6J mice with a knock-in of human diphtheria toxin receptor (DTR) expressed only on Treg cells (DREG mice)30,31. We injected DREG donor mice with diphtheria toxin (DT) (10 µg/kg) on day −2 and day −1 and confirmed loss of Foxp3-Treg expression (Supplementary Fig. 4h). The Treg depleted T cells were then used as donor cells in the major MHC mismatch model used above. We again saw significantly improved clinical GVHD (Fig. 4f), less weight loss (Supplementary Fig. 4i), and improved survival (Fig. 4g).
To further examine whether the local administration of butyrate impacted donor T cells directly, we determined the HDAC and HAT enzymatic activity in donor T cells harvested from the recipient animals. Both the HDAC activity (Fig. 4h) or HAT activity (Supplementary Fig. 4j) in the donor T cells harvested from the recipient spleen were similar between butyrate and vehicle treated allogeneic animals. These collectively demonstrate that the reduction in GI GVHD upon intragastric administration of butyrate is independent of its potential effects from donor Treg cells.
Butyrate protects IECs from allo-T cell mediated damage
We next sought to explore the potential mechanisms that contribute to butyrate-induced protection from severe GVHD. Because (a) butyrate is decreased in IECs, (b) administration of butyrate mitigated GI GVHD independent of Treg cells, but (c) improved junction integrity, we therefore explored whether butyrate had direct effects on protecting IECs from allo-T cell mediated damage and conditioning. We treated IECs ex vivo with vehicle or butyrate for 24 hours and withheld or subjected the cells to irradiation (6 Gy), followed by 24 hours of additional incubation with butyrate. We observed that butyrate was not toxic to IECs (Fig. 5a, left) and more importantly conferred protection from irradiation-induced apoptosis (Fig. 5a, right).
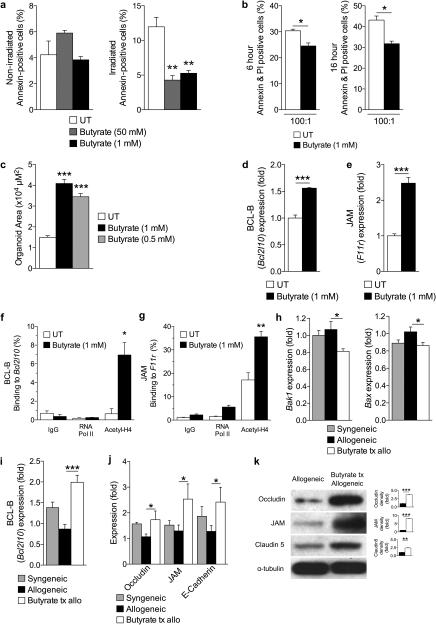
Figure 5. Butyrate treatment enhances IEC barrier.
(a) CD326+ purified intestinal epithelial cells (IECs) cultured in the presence or absence of butyrate and withheld (left panel) or subjected to (right panel) irradiation (6 Gy). (b) Allogeneic CD8+ T cell killing assay. CD326+ IECs incubated with or without butyrate overnight followed by co-culture with allo-primed CD8+ T cells. (c) Size (µm2) of cultured primary intestinal organoids following butyrate treatment (0.5 mM and 1 mM), compared to control. Gene expression of (d) anti-apoptotic protein BCL-B (Bcl2l10) and (e) junctional protein JAM (F11r) in primary CD326+ IECs in the presence of butyrate 1mM. (f)-(g) Chromatin immunoprecipitation assay (ChIP) of butyrate treated IECs (CD326+) binding acetylated histone H4 in the promoter region of (f) Bcl2l10 and (g) F11r. (h)-(i) Analysis of IECs (CD326+) isolated from recipients of syngeneic (BALB/c → BALB/c) and allogeneic (C57BL/6J → BALB/c) BMT treated with butyrate or vehicle daily via intragastric gavage for 21 days. (h) Gene expression of pro-apoptotic proteins Bak1 (left) and Bax (right), (i) anti-apoptotic protein BCL-B (Bcl2l10), and (j) junctional proteins in CD326+ purified IECs isolated 21 days following BMT. (k) Immunoblot of CD326+ purified intestinal epithelial cells from allogeneic (C57BL/6J → BALB/c) BMT recipients treated daily with intragastric vehicle or butyrate (10mg/kg) for occludin, JAM, and claudin 5; isolated 21 days following BMT. Densitometric analysis of two experiments combined shown to the right of each blot, compared to α-tubulin loading control; representative immunoblots shown. Syngeneic n=5, n = 10 mice per allogeneic group. *P < .05; **P < .01; ***P < .0001 of students t-test a – g, k; ANOVA h – j. Bars and error bars represent the means and standard errors of the mean, respectively.
We next determined the ability of butyrate-treated IECs to withstand damage mediated by alloreactive T cells. We isolated and cultured primary IECs with butyrate or vehicle control, overnight. The pre-treated IECs were next co-cultured with primed allogeneic CD8+ T cells, in the absence of butyrate. Fewer butyrate pre-treated IECs succumbed to CD8+ T cell killing within 6 hours (Fig. 5b, left) and 16 hours (Fig. 5b, right) following co-culture, compared with control.
Because butyrate is a primary energy source for IECs11-13, we next determined whether butyrate enhances survival and growth of IECs in vitro. To this end, we cultured intestinal organoids in the presence or absence of butyrate. We observed that culture in the presence of butyrate significantly increased organoid size (Fig. 5c). Next we confirmed the impact of butyrate on IEC junctional function in the organoid cultures by determining the mRNA expression of claudins. We observed that culture of organoids with butyrate significantly increased the mRNA expression of claudins (Cldn1, Cldn5, Cldn6, Cldn10, Cldn11, Cldn13, Cldn14, Cldn17, and Cldn18) (Supplementary Fig. 5a).
Molecular mechanisms of IEC protection
We hypothesized that butyrate would increase anti-apoptotic genes by modulation of histone acetylation. We analyzed pro- and anti-apoptotic mRNA expression levels32 and found that Bak1 and Bax were significantly decreased (Supplementary Fig. 5b), whereas transcripts of the anti-apoptotic protein BCL-B (Bcl2l10) were significantly increased (Fig. 5d) in butyrate treated IECs. We next examined mRNA expression of junctional proteins such as occludin (Ocln) (Supplementary Fig. 5c) and JAM (F11r) (Fig. 5e) in IECs following butyrate treatment, which significantly increased their expression. We also determined if the restored acetylation of histone H4 observed in butyrate-treated allo-BMT recipients was responsible for increased BCL-B (Bcl2l10) and JAM (F11r) expression via ChIP. We found that acetylation of histone H4 was indeed associated with the promoter region of Bcl2l10 (Fig. 5f) and F11r (Fig. 5g) in butyrate treated IECs (CD326+).
These data thus collectively suggest butyrate has several salutary effects on IECs that may or may not be mutually exclusive, such as regulating the expression of genes involved in decreased IEC apoptosis and increased junctional proteins in IECs. To determine if these are involved in in vivo protection from GVHD, we determined the expression of pro- and anti-apoptotic proteins as well as junctional proteins in IECs isolated 21 days following allo-BMT. Pro-apoptotic transcripts of Bak1 and Bax were significantly decreased in allo-BMT recipients that received intragastric butyrate treatment (Fig. 5h) while the anti-apoptotic BCL-B (Bcl2l10) expression was increased (Fig. 5i). Further, gene expression of junctional proteins were, again, similarly increased in butyrate treated animals (Fig. 5j). To determine if these results have biological consequences on protein expression, we also examined the protein amounts of Occludin, JAM, and Claudin 5. Indeed, immunoblot analysis revealed increased junctional proteins in recipients of allo-BMT treated with intragastric butyrate (Fig. 5k). Overall, our results identify several ways in which butyrate can directly enhance epithelial cell function ranging from protection from irradiation and allo-T cell mediated apoptosis to proliferation and junctional protein expression, both in vitro and in vivo.
High butyrate producing microbiota mitigate GVHD
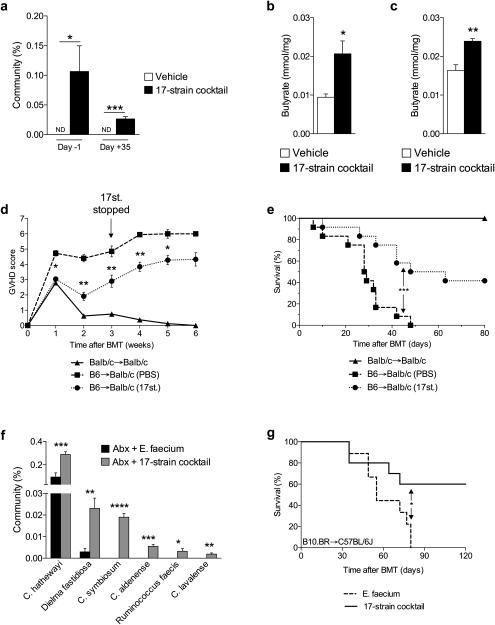
The endogenous HDACi butyrate is a by-product of microbial fermentation33. Therefore, we next tested the hypothesis that altering the composition of indigenous GI microbiota in hosts to those that can produce high levels of butyrate will mitigate GVHD. We utilized 17 rationally selected strains of Clostridia that have been shown to increase butyrate both in vitro and in vivo17,34. We administered these strains via intragastric gavage every other day to naive mice starting 14 days prior to allo-BMT and continued administration of the 17-strain cocktail for 21 days post-BMT. We characterized the microbiota in feces collected from animals that received vehicle and 17-strain administration by 16S rRNA-encoding gene sequencing. In animals that received the 17 Clostridial strains, 16S analysis35-37 revealed an important biologically significant shift in the microbiota indicating that these organisms could be detected (Fig. 6a and Supplementary Fig. 6a). Furthermore, GC/MS analysis 21 days following allo-BMT revealed a significant increase of butyrate in the luminal contents (Fig. 6b) and significantly increased butyrate in the intestinal tissues (Fig. 6c) of animals that received intragastric gavage of the 17 Clostridial strains. The recipients of intragastric gavage of the 17 strains and allo-BMT exhibited significantly decreased GVHD (Fig. 6d-e). Detectable levels of the 17 Clostridial strains were diminished within 2 weeks (day +35) of ceasing intragastric administration (Fig. 6a).
Figure 6. Rationally altering the commensal microbiota reduces GVHD.
(a) 16S rRNA-encoding gene sequencing of stool for percent of 17 Clostridal strains in total GI community on day −1 and day +35, relative to allogeneic (C57BL/6J → BALB/c) BMT on day 0. Vehicle or 17-strain cocktail were administered to recipients every other day via intragastric gavage beginning day −14 and continued through day +21, relative to BMT. ND = not detected. (b)-(c) Recipients of allogeneic BMT gavaged with vehicle or colonized with 17-strain cocktail were sacrificed 21 days following BMT. (b) Intestinal luminal contents (stool) and (c) intestinal tissue were harvested and analyzed via GC/MS. (d) GVHD clinical score and (e) survival following syngeneic (BALB/c → BALB/c) and allogeneic (C57BL/6J → BALB/c) BMT with 17-strain administration, compared to vehicle control; syngeneic n=6, n=12 mice per allogeneic group. (f)-(g) Obligate anaerobes of C57BL/6J (H-2b) mice were targeted with antibiotic mixture (ampicillin 5 mg, metronidazole 4 mg, clindamycin 5 mg, and vancomycin 5 mg) by intragastric gavage for 6 days followed by colonization with cocktails of E. faecium or 17-strains, 4 and 6 days later and subsequently used as recipients of allogeneic BMT (B10.BR → C57BL/6J). 17-strain Clostridial cocktail used at the University of Michigan and Memorial Sloan Kettering was identical. (f) 16S gene sequencing was performed on stool collected from recipients of allogeneic BMT on day −1, relative to BMT. (g) Survival of allo-BMT recipients of intragastric gavage of 17 Clostridial strains cocktail; Recipients of E. faecium n=9 , 17-strain n=10 mice. *P < .05; **P < .01; ***P < .0001 of students t-test a – c, f; Mantel-Cox log-rank test d, e, g. Bars and error bars represent the means and standard errors of the mean, respectively.
Because microbiota variations are known to occur in different colonies of mice, we also determined whether mice housed at another institution (Memorial Sloan Kettering Cancer Center, USA) and treated with the same 17 Clostridial strains would also mitigate GVHD. Additionally, because clinical BMT patients are often treated with antibiotics and as an alternative approach to colonizing the indigenous microbiota, we treated C57BL/6J mice with an antibiotic cocktail (ampicillin 5mg, metronidazole 4mg, clindamycin 5mg, vancomycin 5mg) daily by intragastric gavage for 6 days to target obligate anaerobes. The mice were then colonized 4 and 6 days later with either human Enterococcus faecium or the same cocktail of 17 strains of human Clostridia17 by intragastric gavage. Once again, we characterized the fecal microbiota by 16S gene sequence analysis on day −1, relative to BMT. Upon analysis, we observed increased presence of Clostridia species in recipients that received the cocktail of 17 Clostridial strains (Fig. 6f), compared to recipients of E. faecium. The animals were next used as recipients of a MHC-mismatched B10.BR (H-2k) BMT and followed for survival. Animals that were treated with antibiotics, but were not recolonized with bacteria, died significantly faster (P<0.0001) than mice not treated with the antibiotic mixture (Supplementary Fig. 6b). These data demonstrate that antibiotic treatment eliminated beneficial microbiota similar to previous report6. More importantly, we once again observed significantly increased survival in the animals treated with the cocktail of 17 Clostridial strains (Fig. 6g). These results suggest that altering the indigenous microbiota with 17 rationally selected strains of Clostridia, known to produce high amounts of butyrate17,34, can decrease the severity of GVHD and improve survival across multiple institutions with strain independent results.
DISCUSSION
The community structure of the microbiota is altered following allo-BMT4,5. Our results now provide a novel perspective on microbial metabolites and their impact on GVHD. Our study revealed that the only significantly decreased SCFA, butyrate, is diminished in the intestinal tissue after allo-BMT. Reduction of butyrate in allo-BMT IECs decreased acetylation of histones while increasing butyrate via intragastric gavage restored acetylation of histone H4 and GVHD. An important observation was the lack of changes in luminal (stool) butyrate, despite a documented shift in the microbiome species that produce less butyrate after allo-BMT4. We posit that this may be because of reduced uptake into IECs due to decreased butyrate transporter, thus leaving overall butyrate levels not significantly reduced in the lumen because less is being taken into the IECs despite a decreased production by the shift in the microbiota after allo-BMT.
The reasons for decreased transporter proteins after allo-BMT are intriguing. Previous reports observed a decrease in SLC5A8 following alterations in the microbiota38. Thus, our findings that SLC5A8 and GPR43 are decreased in IECs following allo-BMT are consistent with previous reports4,5. Furthermore we demonstrate that exposure of IECs to inflammatory cytokines leads to reduction in the expression of butyrate transporters. These data suggest that the inflammatory milieu early after allo-BMT reduces the butyrate transporter SLC5A8 leading to its reduction in the IECs and further reducing transporter expression and butyrate intake in a feedback mechanism.
Administered butyrate is more rapidly metabolized as shown by the greater incorporation of carbon from butyrate into the TCA cycle. These data point to a novel observation on the role of energy requirements of IECs in the context of inflammation and GVHD. Our data collectively provide new insights into the role and interactions of the microbiome-derived metabolite, butyrate after allo-BMT.
Previous studies have shown that 17 rationally selected strains of Clostridia, which produce high amounts of butyrate, increase Treg cells in the intestines17. Although we did not observe an increase in Treg cells in the intestine or dependence on donor Treg cells for GVHD reduction following butyrate treatment, we cannot formally rule out the contribution of butyrate-mediated effects on Treg cells (directly or indirectly via macrophages) for decreased GVHD when they are present in the donor inoculum. Furthermore, it is possible that the beneficial effect of the administration of the 17 Clostridial strains may be secondary to factors other than butyrate. Nonetheless, when taken in light of the data demonstrating GVHD protection by direct intragastric administration of exogenous butyrate, the results suggest that butyrate is sufficient for GVHD protection.
Our data suggest that butyrate has direct salutary effects on IECs. Butyrate altered the ratio of the expression of anti-apoptotic to pro-apoptotic molecules and increased the expression of proteins relevant for junctional integrity. Nonetheless, the direct physiological relevance of only one of these mechanisms, apoptosis or junctional integrity, cannot be ascertained from our data. Furthermore, our data does not directly address the impact of butyrate on various other cells that make up GI epithelium besides IECs. Clearly, butyrate and HDACi also have a multitude of effects on immune cells and our data cannot directly measure their impact on GVHD protection. However, a growing body of evidence indicates that GVHD pathophysiology can be regulated by manipulation of the host response to injury, and not just by suppressing the donor immune system27,39,40. Our results extend these observations and suggest that direct modulation of target tissues could be an additional important strategy to decrease GVHD without resorting to further global immunosuppression.
ONLINE METHODS
Reagents
RPMI, penicillin and streptomycin, and sodium pyruvate were purchased from Gibco (Grand Island, NY); FCS from GemCell (Sacramento, CA); 2-ME from Sigma (St. Louis, MO); murine GM-CSF from Peprotech (Rocky Hill, NJ). All antibodies (Abs) used for FACS were purchased from eBioscience (San Diego, CA). DMSO and butyrate was obtained from Sigma (St. Louis, MO), and lipopolysaccharide (LPS) from InvivoGen (San Diego, CA).
Mice
Female C57BL/6J (H-2b; CD45.2+), BALB/c (H-2d) mice were purchased from National Cancer Institute and FoxP3.DTR (DREG) (H-2b) mice and C3H.sw (H-2b) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The age of mice used for experiments ranged between 7 and 12 weeks. All animals were cared for under regulations reviewed and approved by the University Committee on Use and Care of Animals of the University of Michigan, based on University Laboratory Animal Medicine guidelines.
Cell isolation and cultures
Primary intestinal epithelial cells (IECs) were obtained from C57BL/6J mice as described previously41. Briefly, luminal contents of intestine were flushed with CMF solution. Intestine was then minced into 0.5cm pieces, washed with CMF four times, transferred to CMF/FBS/EDTA, and incubated at 37 °C for 60 minutes (shaking tubes every 10 minutes). Supernatant containing IECs was then transferred through 100 µM cell filter followed by incubation on ice for 10 minutes to allow sedimentation. Supernatant was again transferred through a 75 µM cell filter. CD326+ IECs were next purified utilizing either FACS or anti-APC magnetic microbeads (Miltenyi Biotec Ltd., Auburn, CA) and an autoMACs (Miltenyi Biotec).
For viability assay, CD326+ IECs were seeded on gelatin coated 100mm culture dishes and treated in the presence of absence of indicated butyrate concentrations overnight. Cells were then subjected to or withheld from irradiation (6 Gy) and cultured for another 24 hours.
Bone Marrow Transplantation (BMT)
BMTs were performed as previously described15,42. Briefly, syngeneic (BALB/c → BALC/c or C57BL/6J → C57BL/6J) and allogeneic (C57BL/6J → BALB/c or C3H.sw → C57BL/6J) recipients received lethal irradiation. On day −1, BALB/c recipients received a total of 800 cGy of irradiation (split dose separated by 3 hours) and B6 animals received a single dose of 1000 cGy. Donor splenic CD90.2+ T cells were magnetically separated using an autoMACs (Miltenyi Biotec; Bergisch Gladbach, Germany) and 0.5 × 106 to 2.5 × 106 T cells were transferred to BALB/c and C57BL/6J recipients. 5 × 106 donor whole or TCD bone marrow was transferred to all recipients. Survival was monitored daily and the recipient body weight and GVHD clinical scores were determined weekly, as described previously42. Histopathologic analysis of the gastrointestinal (GI) tract was performed as described42. Animals received vehicle (sterile PBS, 0.0004g Na+ per dose) or sodium butyrate (10mg/kg, 0.00004g Na+ per dose) by flexible 20G-1.5” intragastric gavage needle daily for 1 week, then every other day thereafter.
For BMTs performed at MSKCC, C57BL/6J mice were treated with an antibiotic cocktail to target obligate anaerobes (ampicillin 5 mg, metronidazole 4 mg, clindamycin 5 mg, and vancomycin 5 mg) gavaged daily for 6 days (BMT days −18 to −13) followed 4 and 6 days later by oral gavage with indicated bacteria (BMT days −9 and −7) or PBS. Clostridial bacteria were cultured individually on plates and resuspended in anaerobic PBS at a final OD (600nm) of 0.02 to 0.06. One week later, mice were irradiated (12 Gy single dose) and transplanted with B10.BR BM (5 × 106) and T cells (0.5 × 106 CD5 MACS), and followed for development of clinical GVHD.
Gas chromatography mass spectrometry (GC/MS)
To determine targeted fatty acid quantitation, samples (plasma, spleen, liver, intestine, and intestinal fecal content) from mice 7d and 21d post-transplant were harvested, homogenized, and snap-frozen in liquid N2. Equal volumes of plasma and homogenized tissue were utilized, while fecal content was weighed at necropsy. Samples were dispersed in acidified water spiked with stable isotope-labeled SCFA standards and extracted with diethyl ether. The ether layer was immediately analyzed by gas chromatography/mass spectroscopy using a Phenomenex ZB-WAX column on an Agilent 6890 GC with a 5973MS detector. Quantitation was performed by calibration to internal standards. The tissue levels were normalized by protein concentration of the homogenized tissue. Heatmap data was generated using GenePattern software from the Broad Institute (Cambridge, MA). Metabolomic data, mass spectral analytical parameters and spectral raw data from the study and meta data is available in the National Institutes of Health Metabolomics Data Repository Coordinating Center (DRCC) at the University of California San Diego (http://www.metabolomicsworkbench.org/data/index.php).
Metabolic flux analysis (MFA) assessing label incorporation into luminal and intestinal tissue butyrate pools
Animals were intragastrically gavaged with a bolus (2 g/kg) of either labeled 13C2-Butyrate or non-labeled 12C-Butyrate. The small and large intestines were then harvested 6 hours later and prepared for analysis as above. The incorporation of 13C2 labeled Butyrate (Sodium butyrate-1, 2-13C2) in the butyrate pools in the lumen and the intestinal tissue were measured using GC/MS as described previously43,44 and unlabeled (m/z 145) and labeled butyrate which is 2 a.m.u higher (m/z 147) were detected. The ratio of the labeled to unlabeled peak areas were adjusted to incorporate natural distribution of 13C label and expressed as percentage of 13C incorporation into the butyrate pool in the luminal contents (Lumen) and intestinal tissue.
MFA assessing label incorporation into tricarboxylic acid (TCA) metabolite pools
The incorporation of 13C2 labeled Butyrate (Sodium butyrate-1, 2-13C2; Sigma) into the TCA cycle metabolites in the intestinal tissue were measured using LC/MS performed an Agilent 6520 high resolution Q-TOF (quadrupole-time of flight instrument) coupled with an Agilent 1200 HPLC system (Agilent Technologies, New Castle, DE), equipped with an electrospray source. The extract was subjected to hydrophilic interaction chromatography using Phenomenex Luna NH2 column (particle size 3 µm; 1 × 150mm) at a flow rate of 0.07 mL/min. Solvent A was 5 mM ammonium acetate with pH 9.9 and solvent B was acetonitrile. The column was equilibrated with 80% solvent B. The gradient was: 20-100% solvent A over 15 min; 100% solvent A over 5 min; 20% solvent B for 0.1 min 20% solvent A for 15.9 min. Liquid chromatography electrospray ionization (LC/ESI) MS in the negative mode was performed by Q-TOF instrument using the following parameters: spray voltage 3000 V, drying gas flow 10 L/min, drying gas temperature 350°C, and nebulizer pressure 20 psi. Fragmentor voltage was 150 V in full scan mode. Mass range between m/z 100 to 1500 was scanned to obtain full scan mass spectra. Two reference masses at m/z 121.050873 and m/z 922.009798 were used to obtain accurate mass measurement within 5 ppm. All chromatograms and corresponding spectra of TCA metabolites: citrate, succinate and malate and their corresponding 13C labeled counterparts were extracted and deconvoluted using the MassHunter software (Agilent Technologies, New Castle, DE). Retention time consistency were manually rechecked and compared to authentic compounds that were injected under similar chromatographic conditions. For tissue extracts, metabolite concentrations were normalized to protein content, which was determined by the Bradford-Lowry method. For the flux analyses, peak area of the labeled compounds were normalized to natural abundance of the label and represented as ratios to the total compound peak area.
16S deep sequencing
On indicated days, fecal pellets were collected from mice and stored at −80°C. DNA was extracted and purified with phenol-chloroform following bead-based lysis. Sequencing and analysis was performed as described45. Briefly, the V4 region of the 16S rRNA gene was sequenced using Ilumina MiSeq technology. Sequencers were trimmed and analyzed using Mothur35. The 16S rRNA gene sequence from each strain in the 17 strain cocktail was added to the version 9 trainset sequences from the Ribosomal Database Project46. The resulting sequences were classified by comparing to described trainset with the requirement that the confidence score is 100%.
MSKK 16S experiment were analyzed using the Illumina MiSeq platform to sequence the V4-V5 region of the 16S rRNA gene. Sequence data were compiled and processed using mothur version 1.3435, screened and filtered for quality36, then classified to the species level47 using the Greengenes reference database37.
17 strain mixture
The 17 strain mix was prepared as described17. Briefly all strains were grown in 5 ml of EG media for 24 hours at 37°C under anaerobic conditions. Each strain was grown to confluence, with the exception of St 3,8,13,26, and 29. Cells were scraped from EG agar plates and added to the 5 ml culture to obtain the same approximate optical density as the other strains. All cultures were then mixed and glycerol was added to a final concentration of 20%. Aliqouts (1 ml) were individually frozen and stored at −80°C.
Transmission Electron Microscopy (TEM)
Samples were stained as previously described25. Briefly, the intestines from mice that received butyrate or vehicle treatment were harvested 7 days and 21 days following syngeneic and allogeneic BMT and flushed with 0.1 M Sorensen’s phosphate buffer (pH 7.4) to remove luminal contents using a 20G needle. The intestines were next gently flushed with 0.1% ruthenium red (RR) containing 2.5% glutaraldehyde fixative in Sorensen’s buffer and immediately placed in a dish containing the stain/fixative. Cross sections were immediately sliced (2mm wide) from the duodenum, jejunum, and illium. The tissue was then rinsed three times with Sorensen’s buffer, containing 0.1 percent RR and post-fixed for one hour in one percent osmium tetroxide in the same buffer containing RR. The samples were again rinsed with Sorensen’s buffer containing RR. Next, the tissue was dehydrated in ascending concentrations of ethanol, treated with propylene oxide, and embedded in Epon epoxy resin. Semi-thin sections were stained with toluidine blue for tissue identification. Selected regions of interest were ultra-thin sectioned (70 nm thick) mounted on copper grids, and post stained with uranyl acetate and lead citrate. The samples were examined using a Philips CM100 electron microscope at 60 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA).
FITC-dextran assay
Food and water was withheld from all mice for four hours on day 21 post bone marrow transplant. FITC-dextran (Sigma-Aldrich; St. Louis, MO) was administered by 20G-1.5” flexible intragastric gavage needle (Braintree Scientific; Braintree, MA) at a concentration of 50mg/ml in PBS. BMT recipients received 800mg/kg (~16 mg/mouse). Four hours later, serum was collected from peripheral blood, diluted 1:1 with PBS, and analyzed on a plate reader at excitation/emission wavelength of 485nm/535nm. Concentrations of FITC-dextran experimental samples were determined based on a standard curve.
Western Blot
CD326+ purified IECs were harvested from animals that received syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) BMT. Whole cell lysates were next obtained and protein concentrations determined with Pierce BCA Protein Assay (Thermo Scientific). Equal amounts of protein (20 µg) were separated by SDS-PAGE gel electrophoresis (120V, 1.5h) and subsequently transferred to polyvinylidene difluoride (PVDF) membrane using a Bio-Rad semi-dry transfer cell (Hercules, CA) (20V, 1h). The following antibodies were used to analyze the membranes with dilutions in accordance with the manufacturers specification sheet: α-tubulin (Cell Signaling, clone 11H10), Acetyl histone H4 (Lys5/8/12/16) (EMD Millipore, clone 3HH-4C10), SLC5A8 (abcam), GPR43 (abcam), Occludin (abcam, clone EPR8208), JAM (abcam, clone EP1042Y), and Claudin 5 (abcam). Secondary anti-rabbit antibody conjugated to HRP (Jackson ImmunoResearch, Cat No. 111-035-003) was used to detect primary antibodies, where needed. Densitometric analysis was performed using ImageJ software (National Institutes of Health; Bethesda, MA).
Quantitative PCR
mRNA was isolated from samples using RNeasy kit following manufacturers instructions (Qiagen; Venlo, Netherlands). Using 1µg of each mRNA template, cDNA was synthesized using SuperScript VILO (Invitrogen; Carlsbad, CA). qPCR primers were designed for murine targets (Supplementary Table 2). All primers were verified for the production of a single specific PCR product using a melting curve program.
Chromatin Immunoprecipitation
Primary CD326+ IECs were seeded on gelatin (Cell Biologics; Chicago, IL) coated cell culture dishes (100mm) overnight followed by treatment with butyrate 1mM for 24 hours. Cells were harvested and used for ChIP analysis using EZ-Magna ChIP kit from EMD Millipore (Billerica, MA) following the manufacturers instructions. Briefly, cells were cross-linked with 1% formaldehyde and extracted chromatin was sonicated using a Bioruptor Pico by Diagenode (Denville, NJ) to yield DNA fragments predominately in the range of 200 – 1000bp. Sonicated lysates were immunoprecipitated (IP) utilizing ChIP grade specific antibodies purchased from EMD Millipore for acetylated histone-H4 and RNA Pol II or IgG control antibody. De-crosslinked DNA was next examined by qPCR using primers targeting the promoter region of the target gene BCL-B (Bcl2l10) (Forward: CCTACTCTGCCTGGCTCTTT; Reverse: ACCCTTCTGAGTCCCTGAGA), Slc5a8 (Forward: CACAGCACAGCCTTCTTTGT; Reverse: TCCAGTTCACAGTCCAGGTC), and JAM (F11r) (Forward: TGCCGGGATTAAAAGCATGG; Reverse: ACAGGGACAGCAGGATTAGG). IP efficiency of all samples was verified by qPCR analysis of the promoter region of Gapdh (Forward: CTGCAGTACTGTGGGGAGGT; Reverse: CAAAGGCGGAGTTACCAGAG). Data analysis was determined as percent of input utilizing the equations: ΔCt[normalized ChIP] = (Ct[ChIP] − (Ct[Input] − Log2 (6.644))) and % Input = 2(−ΔCt [normalized ChIP]).
Flow Cytometry
To analyze immunophenotype surface markers, lymphocytes contained in the IEC fraction or spleen were harvested, stained using recommended dilutions indicated by manufacturer product sheets and gated on CD4-conjugated PerCP/Cy5.5 (Clone: GK1.5) or CD8-conjugated APC (Clone: 53-6.7) and configurations of the following per mouse in duplicate: CD69-PE (Clone: H1.2F3), CD62L-PE (Clone: MEL-14), CD25-PE (Clone: 3C7), CD44-PerCP/Cy5.5 (Clone: IM7), CD44-APC (Clone: IM7), FoxP3-APC (Clone: FJK-16s). Stained cells were then analyzed with an Accuri C6 Flow Cytometer (BD Biosciences).
IECs were stained with CD326-conjugated APC (Clone: G8.8) and DAPI and sorted to >98% purity using a FACSAria III (BD Biosciences) gating on live cells.
All antibodies have been validated for this species and application as found in respective 1DegreeBio validation profile. CD4, CD8, CD69, CD62L, CD25, CD44, CD326 antibodies were purchased from Biolegend, FoxP3 from eBioscience, and DAPI from Life Technologies.
CTL Assay
CD8+ T cells were isolated from Balb/c (H-2d) mice using anti-CD8 microbeads and LS columns (Miltenyi Biotec) following the manufacturers instructions. CD8+ T cells were primed in the presence of irradiated (30 Gy) C57BL/6J (H-2b) splenocytes for 6 days prior to culture with primary IECs for 6h and 16h. Primary C57BL/6J IECs were harvested and incubated overnight in the presence or absence of butyrate 1 mM in gelatin coated (Cell Biologics Inc.; Chicago, IL) 100 mm-culture dishes (Fisher Scientific).
Reproducibility
Experiments were repeated at least 2 times with 3 sample replicates, bringing the n to at least 6; sample size is indicated in figure legends. To analyze one variable in one BMT experiment, at least two groups of 3 recipients are required.
Statistics
Bars and error bars represent the means and standard errors of the mean, respectively. Non-survival analysis was performed using students unpaired t test between two groups. ANOVA was used for comparisons with more than two groups. Survival data analysis was performed using a Mantel-Cox log-rank test.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (National Heart, Lung and Blood Institute, HL-090775 (P.R.) and HL-128046 (P.R.); National Cancer Institute, CA-173878 (P.R.).
Metabolomic studies were performed through the Molecular Phenotyping Core, Michigan Nutrition and Obesity Center (DK089503 (S.P.)), and Michigan Regional Metabolomics Resource Core (DK097153 (S.P.)).
We wish to acknowledge use of the Microscopy & Image-analysis Laboratory (MIL) of the University of Michigan’s Biomedical Research Core Facilities for preparation of samples and images. Support for the MIL core is provided by The University of Michigan Cancer Center (NIH grant number CA46592) and The University of Michigan Gut Peptide Research Center (NIH grant number DK34933).
Footnotes
Accession codes
Metabolomic data, mass spectral analytical parameters and spectral raw data from the study and meta data is available in the National Institutes of Health Metabolomics Data Repository Coordinating Center (DRCC) at the University of California San Diego (http://www.metabolomicsworkbench.org/data/index.php).
Author contributions
N.D.M. designed experiments, performed experiments, analyzed data, and wrote the paper. A.V.M. performed experiments and analyzed data. R.J. designed and performed experiments. M.K. performed experiments and analyzed data. A.H. designed and performed experiments. T.T. performed experiments. K.O-W. performed experiments. S-R.W. performed experiments. Y.S. performed experiments. C.R. performed experiments. H.F. performed experiments. J.B. performed experiments. Y.S.performed experiments. C.L. performed experiments. M.C. performed experiments. T.C.S wrote the paper. K.H. provided reagents and wrote the paper. V.B.Y. wrote the paper. S.P. analyzed data and wrote the paper. M.vdB.wrote the paper. P.R. designed experiments, analyzed data, and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathewson N, Reddy P. The Microbiome and Graft Versus Host Disease. Curr Stem Cell Rep. 2015 [Google Scholar]
- 4.Jenq RR, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. Journal of Experimental Medicine. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eriguchi Y, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 6.Jenq RR, van den Brink MRM. Allogeneic haematopoietic stem cell transplantation: individualized stem cell and immune therapy of cancer. Nature Reviews Cancer. 2010;10:213–221. doi: 10.1038/nrc2804. [DOI] [PubMed] [Google Scholar]
- 7.Choi S, Reddy P. Graft-versus-host disease. Panminerva Med. 2010;52:111–124. [PubMed] [Google Scholar]
- 8.Ferrara JLM, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373:1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taur Y, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013;13:869–874. doi: 10.1016/j.coph.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Fleming LL, Floch MH. Digestion and absorption of fiber carbohydrate in the colon. Am. J. Gastroenterol. 1986;81:507–511. [PubMed] [Google Scholar]
- 12.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–121. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 13.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment. Pharmacol. Ther. 1998;12:499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 14.Reddy P, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy P, et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J. Clin. Invest. 2008;118:2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi SW, et al. Vorinostat plus tacrolimus and mycophenolate to prevent graft-versus-host disease after related-donor reduced-intensity conditioning allogeneic haemopoietic stem-cell transplantation: a phase 1/2 trial. Lancet Oncol. 2014;15:87–95. doi: 10.1016/S1470-2045(13)70512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 doi: 10.1038/nature12331. doi:10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 18.Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95:2754–2759. [PubMed] [Google Scholar]
- 19.Gao S-M, et al. Histone deacetylases inhibitor sodium butyrate inhibits JAK2/STAT signaling through upregulation of SOCS1 and SOCS3 mediated by HDAC8 inhibition in myeloproliferative neoplasms. Exp. Hematol. 2013;41:261–70.e4. doi: 10.1016/j.exphem.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23:603–611. doi: 10.1016/j.tcb.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 doi: 10.1038/nature12721. doi:10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Charney AN, Micic L, Egnor RW. Nonionic diffusion of short-chain fatty acids across rat colon. Am. J. Physiol. 1998;274:G518–24. doi: 10.1152/ajpgi.1998.274.3.G518. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, et al. Cutting edge: Negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol. 2009;182:5899–5903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noth R, et al. Increased intestinal permeability and tight junction disruption by altered expression and localization of occludin in a murine graft versus host disease model. BMC Gastroenterol. 2011;11:109. doi: 10.1186/1471-230X-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soler AP, et al. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20:1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013;70:631–659. doi: 10.1007/s00018-012-1070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanash AM, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahl K, et al. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahl K, Sparwasser T. In vivo depletion of FoxP3+ Tregs using the DEREG mouse model. Methods Mol. Biol. 2011;707:157–172. doi: 10.1007/978-1-61737-979-6_10. [DOI] [PubMed] [Google Scholar]
- 32.Topham CH, Taylor SS. Mitosis and apoptosis: how is the balance set? Curr. Opin. Cell Biol. 2013;25:780–785. doi: 10.1016/j.ceb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 34.Narushima S, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J. Gastrointest. Surg. 2010;14:449–461. doi: 10.1007/s11605-009-1045-x. [DOI] [PubMed] [Google Scholar]
- 39.Takashima S, et al. The Wnt agonist R-spondin1 regulates systemic graft-versus-host disease by protecting intestinal stem cells. J. Exp. Med. 2011;208:285–294. doi: 10.1084/jem.20101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyengar S, Zhan C, Lu J, Korngold R, Schwartz DH. Treatment with a rho kinase inhibitor improves survival from graft-versus-host disease in mice after MHC-haploidentical hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 2014;20:1104–1111. doi: 10.1016/j.bbmt.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefrançois L, Lycke N. Isolation of Mouse Small Intestinal Intraepithelial Lymphocytes, Peyer's Patch, and Lamina Propria Cells. John Wiley & Sons, Inc.; 2001. doi:10.1002/0471142735.im0319s17. [DOI] [PubMed] [Google Scholar]
- 42.Reddy P, et al. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat. Med. 2005;11:1244–1249. doi: 10.1038/nm1309. [DOI] [PubMed] [Google Scholar]
- 43.Mathew AV, Seymour EM, Byun J, Pennathur S, Hummel SL. Altered Metabolic Profile with Sodium-restricted Dietary Approaches to Stop Hypertension Diet in Hypertensive Heart Failure with Preserved Ejection Fraction. J. Card. Fail. 2015 doi: 10.1016/j.cardfail.2015.10.003. doi:10.1016/j.cardfail.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mell B, et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genomics. 2015;47:187–197. doi: 10.1152/physiolgenomics.00136.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koenigsknecht MJ, et al. Dynamics and Establishment of Clostridium difficile Infection in the Murine Gastrointestinal Tract. Infect. Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.