Abstract
The biological activity of nanoparticle-directed therapies critically depends on cellular targeting. We examined the subtumoral fate of Particle Replication in Non-Wetting Templates (PRINT) nanoparticles in a xenografted melanoma tumor model by multi-color flow cytometry and in vivo confocal tumor imaging. These approaches were compared with the typical method of whole-organ quantification by radiolabeling. In contrast to radioactivity based detection which demonstrated a linear dose-dependent accumulation in the organ, flow cytometry revealed that particle association with cancer cells became dose-independent with increased particle doses and that the majority of the nanoparticles in the tumor were associated with cancer cells despite a low fractional association. In vivo imaging demonstrated an inverse relationship between tumor cell association and other immune cells, likely macrophages. Finally, variation in particle size nonuniformly affected subtumoral association. This study demonstrates the importance of subtumoral targeting when assessing nanoparticle activity within tumors.
Keywords: PRINT, Nanomedicine, Nanoparticle, Flow Cytometry, Cancer
Graphical abstract
Background
Appreciation of the Enhanced Permeation and Retention (EPR) effect for nanocarrier-mediated drug delivery in oncology has resulted in a focus on the accumulation of particles in whole tumors.1 A range of methods to determine the fraction of the injected dose of the carrier or cargo that accumulates in a whole organ or tumor has driven the assessment of nanoparticle targeting to solid tumors.2–13 However, tumors are composed of a variety of cell types, such as fibroblasts and endothelial cells and macrophages and neutrophils, in addition to cancer cells. The relative distribution of these cell types varies between tumors.14–17 Whole organ approaches are unable to discriminate between accumulation in the intended target, typically cancer cells, and other cells or the extracellular space. For cargo with an intracellular mechanism of action, such as nucleic acids and proteins, delivery to specific cell types is crucial to assessing nanoparticle efficacy and optimizing targeting.
Methods for the identification of subtumoral cellular components include microscopy and flow cytometry. Confocal microscopy has been used to determine particle internalization in vivo by analyzing multiple sections of an organ.18 However, meaningful quantification can be challenging. Flow cytometry permits concurrent cellular identification and nanoparticle quantification. Previous studies that have used flow cytometry to examine nanoparticle targeting to organs have not explored the effects of particle characteristics (composition, shape, etc.) or dose on the accumulation in specific cell populations and do not correlate their findings with whole organ assessment.14,19–25 Studies that account for both nanocarrier properties as well as intra-organ or intra-tumor distribution have the potential to best inform nanoparticle design and delivery.
PRINT is a top-down fabrication strategy that relies on precision molds, offering the advantage of reproducible production of monodisperse particles. This reproducibility eliminates large variation in particle sizes (i.e. PDI) that could influence the association of a subset of the particles with one cell population over another confounding data interpretation. In addition, PRINT also affords homogeneity in the composition of the particles and flexibility in the composition of the desired nanoparticle material.
Using flow cytometry, whole organ assessment and live animal in vivo confocal microscopy, we analyzed the cell type-specific distribution of PRINT nanoparticles. We identified wide variation in subtumoral cellular association and identify dose and particle properties that influence cellular targeting.
Methods
Materials
Poly(ethylene glycol) diacrylate (Mw 700) (PEG700DA), 2-aminoethyl methacrylate hydrochloride (AEM), diphenyl (2,4,6-trimethylbenzoyl)-phosphine oxide (TPO), and sucrose were purchased from Sigma-Aldrich. Thermo Scientific Dylight 488 maleimide, dimethylformamide (DMF), triethylamine (TEA), pyridine, borate buffer (pH 8.6), acetic anhydride, and methanol were obtained from Fisher Scientific. Conventional filters (2 µm) were purchased from Agilent and poly(vinyl alcohol) (Mw 2000) (PVOH) was purchased from Acros Organics. PRINT molds (80 nm×80 nm×320 nm) were obtained from Liquidia Technologies. Tetraethylene glycolmonoacrylate (HP4A) was synthesized in-house as previously described.26 Methoxy-PEG(5k)-succinimidyl carboxy methyl ester (mPEG5k-SCM) was purchased from Creative PEGWorks. Typsin, DPBS, and cell culture media were purchased from Gibco.
PRINT nanoparticle fabrication and characterization
The PRINT particle fabrication technique has been described previously in detail.27,28 The pre-particle solution was prepared by dissolving 3.5 wt% of the various reactive monomers in methanol. The preparticle solution was comprised of 67.75 wt% HP4A, 20 wt% AEM, 10 wt% PEG700DA, 1 wt% TPO and 1.25 wt% Dylight 488 maleimide. Stock particle concentrations were determined by thermogravimetric analysis (TGA) on both an aliquot of the stock and a centrifuged sample of the stock, to account for any mass due to PVOH, using a TA Instruments Q5000. Particles were visualized by scanning electron microscopy (SEM) using a Hitachi S-4700 SEM. Prior to imaging, SEM samples were coated with 3.5 nm of gold-palladium alloy using a Cressington 108 auto sputter coater. Particle size and zeta potential were measured by dynamic light scattering (DLS) on a Zetasizer Nano ZS (Malvern Instruments, Ltd.).
Particles were PEGylated and acetylated following a previously described method.27 Post-acetylation, particles were analyzed by TGA, DLS and SEM and stored at 4°C. To radiolabel the nanoparticles, high specific activity 64Cu (14000±7600 Ci/mmol or 518±28 TBq/mmol) was obtained from the Washington University School of Medicine (St. Louis, MO, USA). 64Cu was produced on a CS-15 biomedical cyclotron by the 64Ni(p,n)64Cu nuclear reaction using previously established methods29, with a half-life of 12.7 hours. Following PEGylation (described above), particles were characterized as described above by TGA and reacted in 0.1 M Na2CO3 buffer (pH 9) with 2-(4-isothiocyanatobenzyl) 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (p-SCN-Bn-DOTA) at 5 mg/mL (2:1 DOTA:AEM molar ratio). A conversion from positive to negative zeta potential indicated that the reaction went to completion. Particles were then incubated with 64CuCl2 for 30 minutes at 65 °C in 0.1 M ammonium acetate, washed 3 times by centrifugation with deionized water, and resuspended for injection in 9.25% sucrose.
Cells, cell culture, and spheroid injections
Cells were cultured as previously described in high glucose DMEM (Gibco) with 10% FBS (Gemini Bio-products) and 1% Penicillin/Streptomycin (Gibco).30 Hanging droplet spheroids were generated by trypsinization of the cells and resuspension to 2 × 106 cells/mL. Twenty microliters of the cell suspension was pipetted into wells of a 60-well minitray (Nunc, Thermofisher scientific). The minitray was inverted and placed in a humidified 150 mm dish. Cells were incubated at 37 °C and 5% CO2 for 4–6 days. Individual spheroids were harvested and verified visually. Athymic nude mice were anesthetized by inhalation with 2% isoflurane. A mouse ear was affixed onto a conical tube with double-sided tape. The tumor spheroid were injected as described30. Successful spheroid injection was verified by epifluorescence macroscopic imaging (Supplemental Figure S1A).
Mice and Particle Injections
All animals were handled according to the NIH Guide for the Care and Use of Laboratory Animals. All Procedures were approved by the University of North Carolina-Chapel Hill Institutional Animal Care and Use Committee (IACUC), protocol #11–154.0. 4–8 week old, male Foxn1nu (athymic; C57BL/6J background) nude mice were purchased from the UNC Animal Services Core. Nanoparticles were suspended in isotonic 9.25% sucrose and injected intravenously via the tail vein at a maximum of 300 µl per animal. Tumors were of equal volumes between groups and ranged from 100–300mm3, as determined using caliper measurements using the formula V=(L/2)×(W/2)×(H/2)×(4/3×π) (Supplemental Figure S1A). Particles were resuspended at different concentrations (1.45, 4.65, 14.65 mg/mL) to deliver similar injection volumes of approximately 300 µl.
Tumor Dissociation and Flow Cytometry
Tumors were harvested as described with slight modifications.31 Briefly, tumors were dissociated for 3 hours in 5 mL of DMEM with 10% FBS containing 1500 U collagenase and 500 U hyaluronidase (10× collagenase/hyaluronidase, StemCell Technologies). After centrifugation (600 ×g for 5 minutes), cell pellets were resuspended in a 1:4 mixture of HEPES Buffered Saline containing 2% FBS (HF solution) and RBC Lysis Buffer (0.8% NH4Cl and 0.1 mM EDTA, Stem Cell Technologies), recentrifuged (450 × g for 5 minutes) then resuspended in RBC lysis buffer. After centrifugation, cells were resuspended in 0.05% Trypsin/EDTA solution (Gibco) and incubated 5 minutes at 37 °C. Cells were pelleted and resuspended in Dispase (1 U/mL) and DNase I (0.1 mg/mL). After 30 minute incubation at 37 °C, 10 mL of HF solution was added, and cells were passed through a 40 µm cell strainer (Fisher), centrifuged and resuspended in HF solution. Cells were then washed and counted with a hemocytometer. Live-Dead Fixable Blue (Invitrogen) was then added at a concentration of 1 µl/4×106 cells in 1 mL PBS and incubated on ice for 15 minutes. Cells were washed with PBS and then resuspended in 100 µl PBS. Fc Block (BD Biosciences) was incubated with the sample for 5 minutes on ice followed by an antibody (Biolegend) mixture consisting of PE-Cy7 CD31 (clone 390), APC F4/80 (clone BM8), Alexa 700 Ly6G (clone 1A8), and Pacific Blue CD45 (clone 30-F11). After 1 hour on ice, cells were washed with PBS and fixed in 500µl of 4% paraformaldehyde for 15 minutes at room temperature. Cells were washed twice with FACS buffer (1% FBS in PBS), then resuspended in a final volume of 500 µl of FACS Buffer and stored at 4°C until data acquisition (LSRII, BD Biosciences). Data analysis of FCS3 files was performed using FACSDiva version 10.6 (BD Biosciences). Live Dead Blue (Invitrogen) was used to gate on the living cells. All surface markers were compared to their fluorescence minus one (FMO) controls to set appropriate gates. Particle association was determined by comparison to control sucrose injected animals to establish appropriate gating.
Two-photon Microscopy
Tumor-bearing animals with tumors between 10–30mm3 were imaged as previously described.30 Animals were anesthetized with isofluorane, and tumors were imaged before and after administration of particles. All imaging was performed at 910 nm with an Olympus FV1000MPE mounted on an upright BX-61WI microscope, using a 25×, 1.05 N.A. (2 mm W.D.) water immersion objective with optical imaging gel to capture images. Olympus Fluoview software and microscope settings were consistent for all acquired images (the laser power was at 14% and each channel’s PMT voltage was 580, 635, and 600, respectively). The laser unit (MaiTai DeepSee) is tunable from 690–1040 nm with a pulse width < 100 fs. Three channel non-descan detectors were used: Ch1 (420–460 nm) BFP, Ch2 (495–540nm) GFP, Ch3 (575–630 nm) RFP.
Statistics
All data, unless otherwise noted, were analyzed using a one-way ANOVA and Tukey’s post-hoc analysis. Mean fluorescence data was analyzed using a two-way ANOVA and Tukey’s post-hoc analysis.
Results
PRINT fabrication
To explore the influence of particle characteristics on nanoparticle association, we generated tumor spheroids from LKB498 mouse melanoma cells that stably express the red fluorescent protein tdTomato.30 Tumor cell spheroids were implanted intradermally and allowed to grow to a size of ~100 mm3 (Supplemental Figure S1A–B). Nanoparticles were fabricated using the Particle Replication in Non- Wetting Templates (PRINT) technique.27,28,32 PRINT generates nanoparticles of highly consistent and precise size, shape, and composition, eliminating variables which could influence cellular targeting. 80 nm×80 nm×320 nm PRINT nanoparticles were composed of a covalently cross-linked hydrogel matrix and were PEGylated as previously described.27 Depending on the assay, fluorescent moieties were incorporated, or copper-64 (64Cu) was chelated to the particles. Particles were characterized by DLS and SEM and demonstrated a similar negative zeta potential and narrow size distribution (Supplemental Figure S1C).
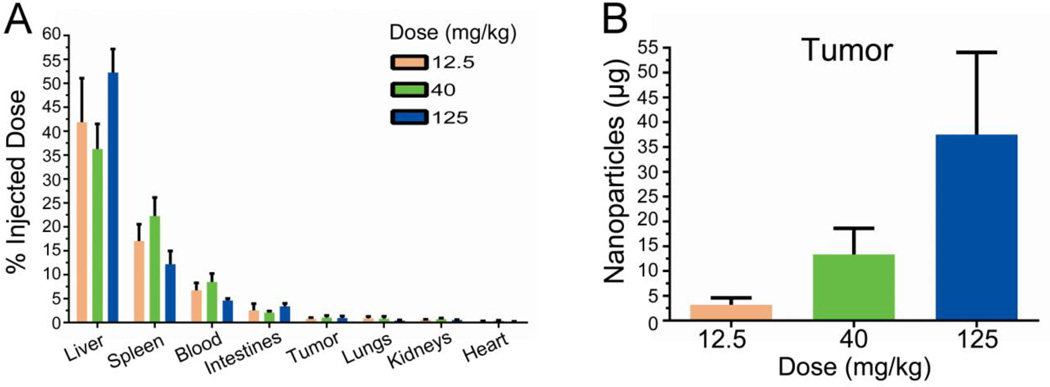
To determine nanoparticle accumulation using a whole-organ based approach, 64Cu radiolabeled PRINT nanoparticles were administered IV at several doses into tumor bearing mice. Eighteen hours following particle administration, animals were sacrificed and organs were harvested. Gamma emission was measured from several organs and the tumor accounting for ~70% of the injected dose. Radioactive signal demonstrated a wide distribution, ranging from ~45% of particles associated with the liver to ~0.2% of particles associated with the heart (Figure 1A). This particle distribution is consistent with other types of nanoparticles2,12,13,18. The relative fraction of the injected dose detected in each organ was unaffected by dose (Figure 1A). However, the total amount of particles contained within the tumor as well as in each organ increased in a dose-dependent manner (Figure 1B and Supplemental Figure S2).
Figure 1.
Particle accumulation measured by gamma radiation. (A) Radioactive emission was determined for each tissue 18 hours after intravenous administration. Fractional association for all tissues examined is shown. (B) Calculated mass of nanoparticles contained within tumor organ. Error bar indicates SD of n = 4.
Sub-organ Particle Accumulation
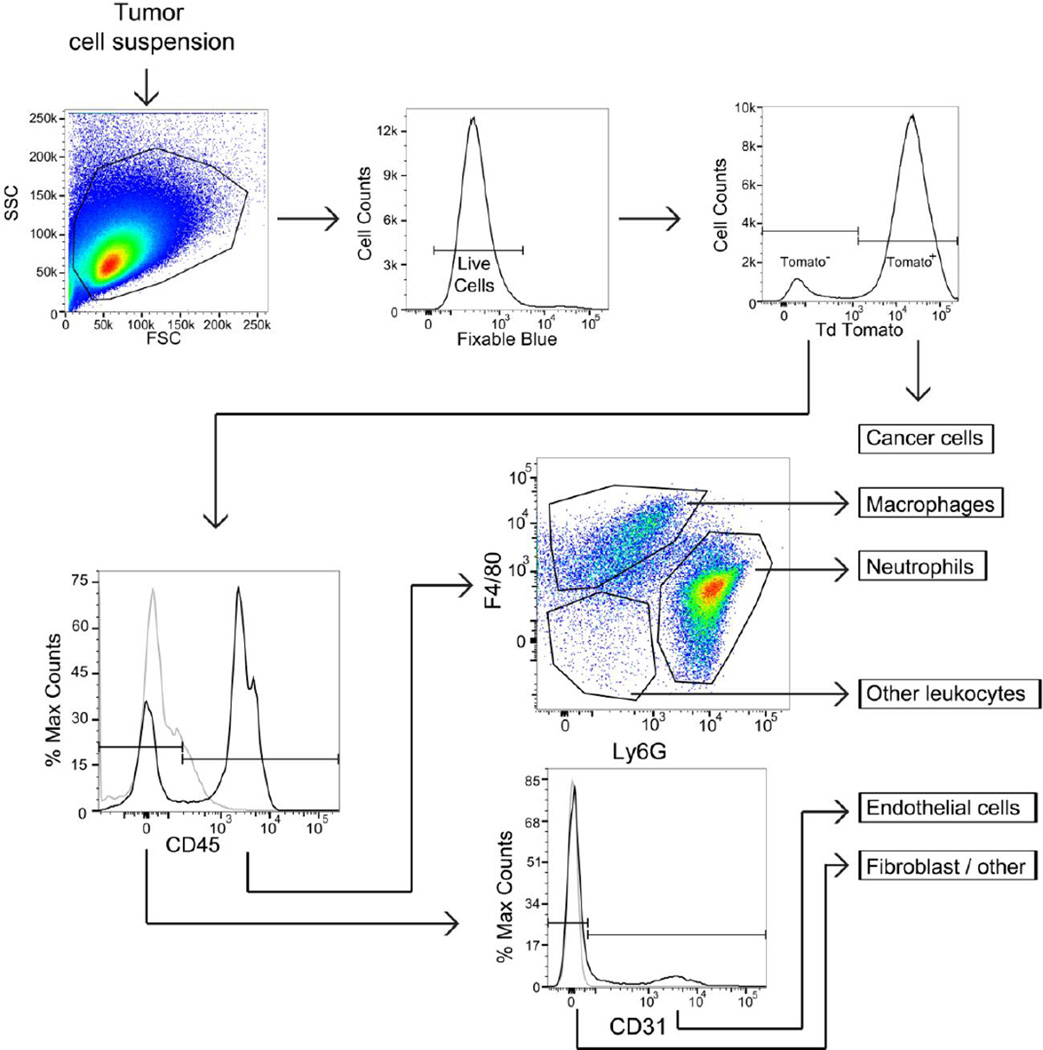
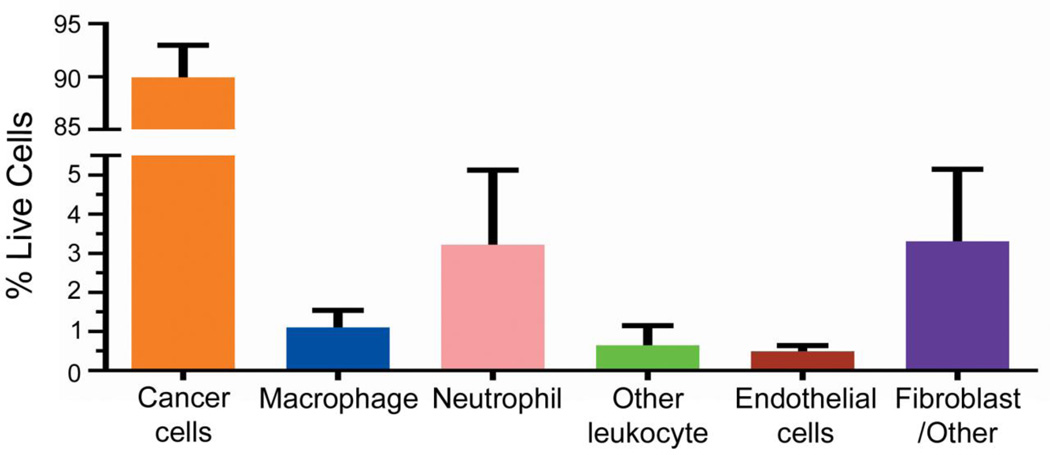
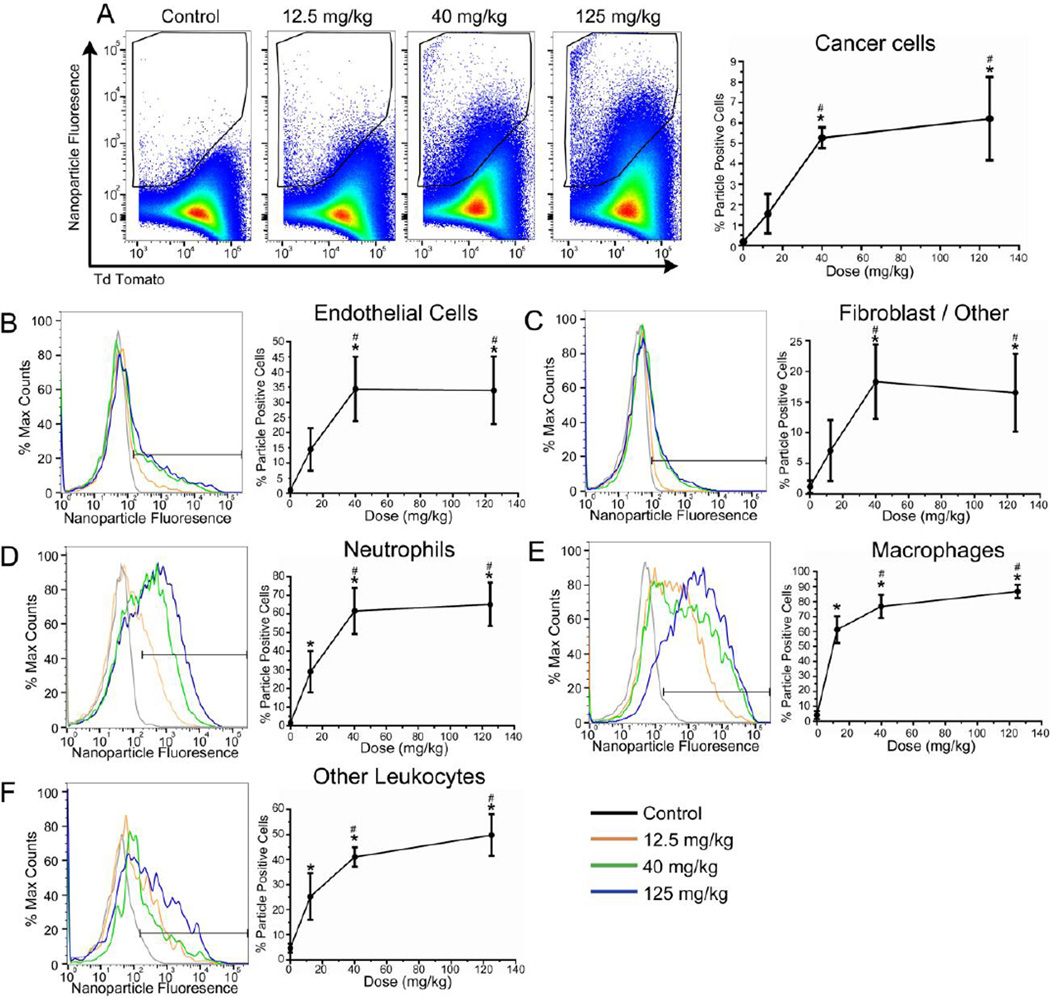
We then examined the cellular composition of our tumor model. Tumors were enzymatically dissociated to create a single cell suspension and analyzed using multi-color flow cytometry (Figure 2). After gating on viable cells, tdTomato fluorescence identified the melanoma cells. To distinguish these cells from the other cell compartments that comprise the entirety of the tumor, we refer to these cells as “cancer cells” hereafter. The remaining non-cancer cells were separated into an immune cell population based on CD45 expression (leukocyte marker). CD45+ cells were further subdivided by the expression of F4/80, which marks mouse macrophages, or Ly6G, which marks neutrophils. CD45+ F4/80− Ly6G− cells were classified as “other leukocytes” representing a mixture of cells that includes natural killer cells, dendritic cells, and B cells. CD31 surface expression in the CD45− cell population identified endothelial cells, with the remaining CD45− and CD31− populations classified as fibroblast/other. Surface markers were compared to their fluorescence minus one (FMO) controls to ensure appropriate population gating. The tumor cell suspension reproducibly contained 89.9% cancer cells, 3.2% neutrophils, 1.1% macrophages, 0.64% other leukocytes, 0.49% endothelial cells, and 3.3% fibroblast/other cells (Figure 3). This gating strategy accounted for virtually all cells (98.6%) isolated from the tumor.
Figure 2.
Flow cytometry gating strategy used to identify subtumoral compartments. To determine relative cell populations, dissociate tumor was stained with Fixable Blue, CD45, CD31, Ly6G and F4/80. Fluorescence minus one controls (grey lines) are used to distinguish significant staining (black). Representative plots are shown.
Figure 3.
Fractional composition of LKB498 xenografted tumors. Two groups of 16 animals were processed, with the data collected and analyzed separately then aggregated for the analysis. Error bars represent SD (n = 32).
Mice were injected with fluorescent 80 × 80 × 320 nanoparticles at several doses or with a sucrose control. 18 hours after treatment, tumors were dissected from mice, dissociated, and analyzed by flow cytometry as described. Cancer cells demonstrated a dose-dependent association at doses between 12.5 to 40 mg/kg, yielding 1.5% or 5.2% particle positive cells, respectively (Figure 4A). At the 125 mg/kg dose, 6.2% association was observed. These data indicate a dose dependent increase in fractional association with an upper limit that becomes independent of dose. In vitro association data also revealed a lack of particle association at high doses (Supplemental Figure S3). Since tdTomato fluorescence could obscure or artifactually contribute to particle signal, the fractionation experiment was repeated using non-fluorescent cancer cells. These data demonstrated that the relationship between fluorescence-based quantitation of particle association and dose was unaffected by the presence of tdTomato or by compensation-associated corrections, both in vitro and in vivo, although there was a slight shift in the gated population (Supplemental Figure S3 and S4).
Figure 4.
Nanoparticle association differs across cells comprising the tumor. Flow cytometric quantification of particle-positive cells. (A) LKB498 cancer cells, (B) Macrophages, (C) Neutrophils, (D) Other leukocytes (E) Endothelial cells, (F) Fibroblast/other cells. Error bar represents SD (n = 4 per dose group). * = p < 0.05 vs control, # = p < 0.05 vs 12.5 mg/kg.
Non-cancer cell populations showed a greater fractional particle association. Approximately 15–35% of endothelial cells were positive for particle association (Figure 4A), whereas fibroblast/other cells exhibited the lowest fractional association of 5–20% (Figure 4C). 20–60% of neutrophils, 60–80% of macrophages, and 20–60% other leukocytes were associated with PRINT particles (Figure 4D–F). Strikingly, in contrast to macrophages and cells in the other leukocyte classes which demonstrated a sustained dose-dependent increase in nanoparticle association, cancer cells, endothelial cells, and cells in the fibroblast/other class, showed no increase in fractional association above the 40 mg/kg dose. These data suggest a limit to nanoparticle uptake, possibly based on the number of cells sufficiently proximal to blood vessels to contact particles or the capacity of these cells to take up the particles.
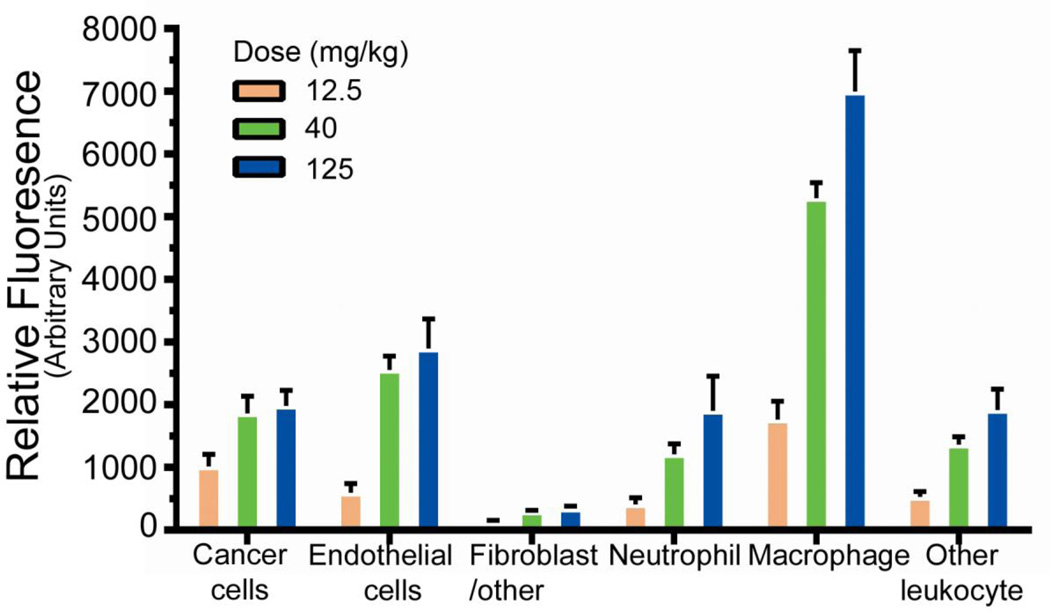
We then quantified fluorescence for individual cells. Among the cells analyzed, macrophages demonstrate the greatest mean fluorescence intensity (MFI) (Figure 5). MFI for macrophages was approximately 3.5 fold greater than that of the cancer cells, indicating that macrophages associate with a greater number of particles per cell. Similar to fractional association, fluorescence signal per cell increased in a dose-dependent fashion at the low doses for cancer cells as well as immune cells. However, signal in cancer cells plateaued despite increased dose. These data suggest that a subset of cells are able to associate with particles and, further, that these cells have a biological limitation on their ability to take up particles. Using the mean fluorescence data, the relative fluorescence in each cell compartment was calculated. These data demonstrated that although a low fraction of cancer cells are particle-positive, after adjusting for the fraction of cancer cells within the sample, cancer cells represent the cell type with the largest relative accumulation of particles (Supplemental Figure S5).
Figure 5.
Mean fluorescence intensity of the particle-positive cells. Background fluorescence from the uninjected group was subtracted. Error bars represent SD (n = 4 per dose group).
We then explored the fate of nanoparticles in the tumors of live mice by intravital two-photon microscopy. Mice harboring tdTomato+ LKB498 tumors were anesthetized and imaged before and after intravenous injection of 40mg/kg Dylight488 conjugated nanoparticles. The use of an excitatory wavelength of 910nm enabled simultaneous imaging of the tumor (tdTomato), the fluorescent nanoparticles (Dylight488), and bundled collagen (via second harmonic generation signal, SHG). As early as 10 minutes after nanoparticle injection, we observed the presence of nanoparticles within the tumor, which was demonstrated by the colocalization of red and green fluorescence suggesting the presence of particles associated with tumor cells (Figure 6A, Supplemental Figure S6). We also observed, however, a cell population with greater fluorescence intensity. Based on flow cytometry, we suspect that these bright cells are immune cells such as macrophages. We examined this pattern over several days holding imaging settings constant. In regions exhibiting few of these bright cells, colocalization between tdTomato+ tumor cells and Dylight488 nanoparticles was increased compared to regions of high cell infiltration (Figure 6B). This inverse relationship suggests regional variation within the tumor such that cancer cell particle association is more robust in macrophage-free areas of the tumor We also performed two-photon microscopy on normal ear dermis to explore particle accumulation in normal dermal tissue. We observed that particle fluorescence remains detectable only in apparent vascular spaces (Supplemental Figure S7). These data may indicate the role of tumor vasculature and EPR in PRINT nanoparticle delivery to the cells that make up the tumor.
Figure 6.
Nanoparticles preferentially concentrate in immune cells and decrease in tumor mass over time. (A) Two-photon images of tdTomato+ LKB498 melanoma spheroid tumor in murine ear dermis at 5 minutes before and 10 minutes, 24 hours, and 4 days after intravenous injection of Dylight488 conjugated nanoparticles. Maximum intensity projection, FV1000MPE, 14% power, 910 nm, 25× objective. Scale bar = 50 µm. (B) Two-photon images of tdTomato-positive cancer and infiltrating cells 24 hours postinjection of Dylight488 conjugated nanoparticles. Maximum intensity projection; FV1000MPE, 14% power, 910 nm, 25× Objective, 3× Zoom. Scale bar = 50 µm.
Effect of Particle Size on Association
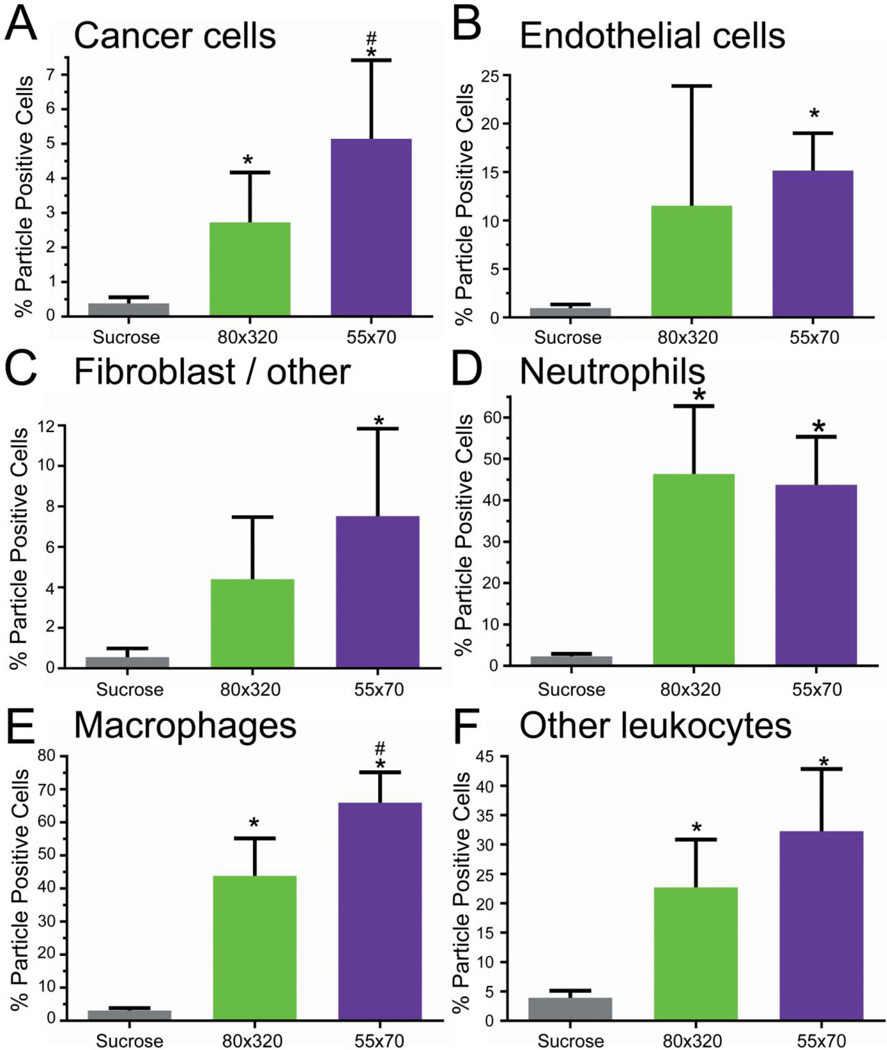
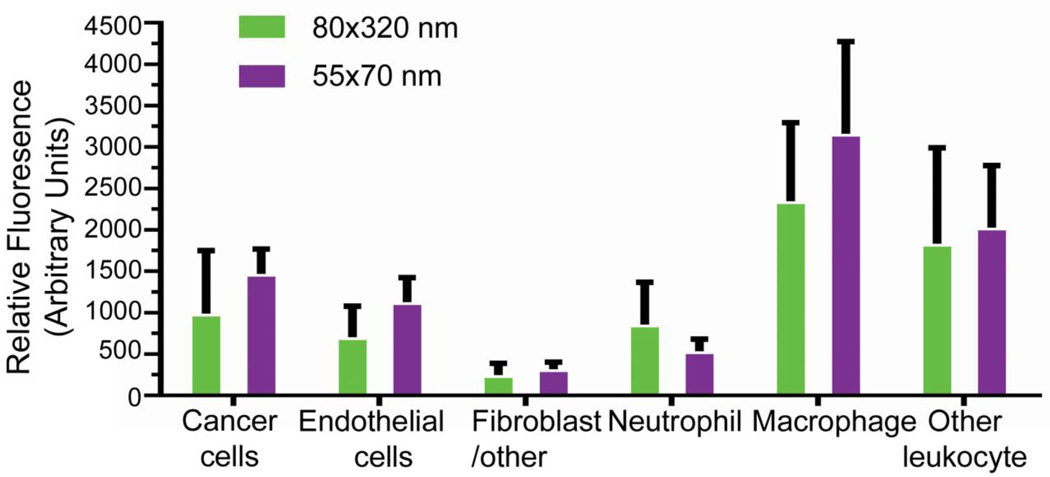
We then asked whether altering particle properties would selectively influence subtumoral cellular association. Focusing on size, 55 × 70 nm particles were fabricated and functionalized as before (Supplemental Figure 8). Compared with the 80 × 320 nm particles, the 55 × 70 nm particles demonstrated a significant increase in cancer cell (4.5 % vs 2.5%) and macrophage association (65% vs 37%) (Figure 7A,E). However, this effect was not universal. Fractional associations for neutrophils, other leukocytes, endothelial cells, and fibroblast/other cells were not affected (Figure 7B–D,F). The mean fluorescence intensity of the particle-positive cells was similar across particle sizes (Figure 8), indicating that the cells associate with a similar particle mass. Relative distribution of the cells as calculated from fluorescence suggests that smaller particle size increases relative cancer cell association (Supplemental Figure S9). These data indicate that varying particle size can shift subtumoral targeting enhancing cancer cell deposition.
Figure 7.
Nanoparticle size influences particle uptake in non-immune cells. Particle association for each cell type population was determined by flow cytometry. (A) LKB498 cancer cells, (B) Macrophages, (C) Neutrophils, (D) Other leukocytes (E) Endothelial cells, (F) Fibroblast/other cells. Error bar represents SD (n = 6 per group). * = p<0.05 vs Sucrose, # = p<0.05 vs 80×320.
Figure 8.
Mean fluorescence intensity was determined for particle-positive cells. Values were background corrected and error bars represent SD (n = 6 per dose group).
Discussion
The accurate quantification of nanoparticle association with tumors poses many challenges. Commonly, tumors are regarded as a homogeneous mass without attention to the range of cellular components and regional variation.14–17 To understand subtumoral distribution of nanoparticles, we took advantage of highly uniform PRINT nanoparticles to examine the single-cell fate of particles in an orthotopic spheroid tumor model. We observed a striking discordance between whole-tumor and individual cell analysis. Whole-organ quantification demonstrated dose-dependent accumulation. In contrast, cell-specific analysis revealed that delivery becomes dose-independent at higher doses for cancer cell population. We observed that macrophages associate with more particles per cell as compared to cancer cells. Although the relationship between tumor cell and macrophage number will vary between tumors, in this model cancer cells associated with the greatest number and mass of nanoparticles. Since flow cytometry is unable to discriminate between internalized and surface bound particles, intravital two-photon imaging was used to examine the fate of nanoparticles in an intact animal. These data, supported flow cytometry results, suggest that local immune cells disproportionately accumulate nanoparticles and may regionally influence cancer cell uptake. Flow cytometry also demonstrated the non-uniform influence of particle properties on subtumoral particle distribution. Reduced particle size increased the association with cancer cells.
Nanomedicines can be used to passively accumulate small molecule cargo (e.g. chemotherapy) in tumors via the EPR effect. However, biologically active, membrane-impermeable cargo (e.g. siRNA) requires intracellular and cell-specific delivery. Single-cell assessment offers lineage-specific quantification of nanoparticle targeting. The discrepancy between whole organ and single cell assessment limits the way radioactivity-based accumulation experiments can be interpreted. It is possible that at higher doses, particles accumulate predominantly in the extracellular space as interactions due to charge33,34 or particle size5,35 affect the volume available for nanoparticle diffusion36,37. Given these physical constraints and limited capacity for cellular association with particles, excess particles may accumulate in the extracellular space or be taken up by phagocytic cells. Radioactivity-based assessment of nanoparticle accumulation may not be proportional to cancer cell association.
The highly uniform size distribution of PRINT nanoparticles enabled examination of the influence of particle size. Reduction in particle size increased relative cancer cell association. Smaller sized particles have been shown to increase accumulation in tumors5,35,38 and may penetrate the extracellular matrix more efficiently enabling contact with a larger number of cancer cells. Alternatively, limitations based on the mechanism for cancer cell uptake may favor smaller particles. These data support our hypothesis that particle-specific features influence subtumoral distribution and that these features can be optimized to target specific cell populations in tumors and organs.
In conclusion, by quantifying nanoparticle uptake across the specific cellular compartments that constitute a tumor we have been able to explore the relative and absolute distribution of nanoparticles as well as identify the impact of altered particle properties. The analysis of nanoparticle accumulation on a cellular level provides important insights into the fundamental behavior of nanoparticles and the interplay between nanomedicines and the different physiological environment present within tumors and between tumors of different types. Cellular-level quantification is critical when assessing the function of impermeable cargo that depends on an intracellular mechanism of action.
Supplementary Material
Acknowledgments
The authors thank Charlene Santos, Mark Ross, and the UNC Lineberger Animal Studies Core. We acknowledge Amar Khumbhar of CHANL, Nancy Fisher and the UNC Flow Cytometry Core for their advice and assistance. We also thank Dr. William Zamboni and members of the Davis and DeSimone labs for helpful discussions.
Funding Sources
This work is supported in part by Liquidia Technologies, an NIH Director’s Pioneer Award (1DP1OD006432), the Carolina Center for Nanotechnology Excellence (U54CA151652), and the UNC University Cancer Research Fund. IJD is supported by grants from the NIH (R01CA166447), the Corn-Hammond Fund for Pediatric Cancer Research, the Wide Open Charitable Foundation, and the V Foundation for Cancer Research. The UNC Flow Cytometry Core Facility is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Materials and methods, supplemental data figures, supplemental movie figures are available via the Internet.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript
The authors declare the following competing financial interest(s): J.M.D. is a founder and maintains a financial interest in Liquidia Technologies.
References
- 1.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy : mechanism of tumoritropic accumulation of proteins and the antitumor agent Smancs. Cancer Res. 1986;46(12):6387–6392. [PubMed] [Google Scholar]
- 2.Poon Z, Lee JB, Morton SW, Hammond PT. Controlling in vivo dtability and biodistribution in electrostatically assembled nanoparticles for systemic delivery. Nano Lett. 2011;11(5):2096–2103. doi: 10.1021/nl200636r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Tseng Y-C, Huang L. Biodistribution dtudies of nanoparticles using fluorescence imaging: a qualitative or quantitative method? Pharm Res. 2012;29(12):3273–3277. doi: 10.1007/s11095-012-0818-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crayton SH, Elias DR, Al Zaki A, Cheng Z, Tsourkas A. ICP-MS analysis of lanthanide-doped nanoparticles as a non-radiative, multiplex approach to quantify biodistribution and blood clearance. Biomaterials. 2012;33(5):1509–1519. doi: 10.1016/j.biomaterials.2011.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Ven AL, Kim P, Haley O, et al. Rapid tumoritropic accumulation of systemically injected plateloid particles and their biodistribution. J Control Release. 2012;158(1):148–155. doi: 10.1016/j.jconrel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psimadas D, Georgoulias P, Valotassiou V, et al. Molecular nanomedicine towards cancer:111In-labeled nanoparticles. J Pharm Sci. 2012;101(7):2271–2280. doi: 10.1002/jps.23146. [DOI] [PubMed] [Google Scholar]
- 7.Chu KS, Hasan W, Rawal S, et al. Plasma, tumor and tissue pharmacokinetics of docetaxel delivered via nanoparticles of different sizes and shapes in mice bearing SKOV-3 human ovarian carcinoma xenograft. Nanomedicine. 2013;9(5):686–693. doi: 10.1016/j.nano.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo C-L, Chou M-H, Lu P-L, et al. The effect of PEG-5K grafting level and particle size on tumor accumulation and cellular uptake. Int J Pharm. 2013;456(2):424–431. doi: 10.1016/j.ijpharm.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 9.Ueno T, Dutta P, Keliher E, et al. Nanoparticle PET-CT detects rejection and immunomodulation in cardiac allografts. Circ Cardiovasc Imaging. 2013;6(4):568–573. doi: 10.1161/CIRCIMAGING.113.000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark DP, Ghaghada K, Moding EJ, et al. In vivo characterization of tumor vasculature using iodine and gold nanoparticles and dual energy micro-CT. Phys Med Biol. 2013;58(6):1683–1704. doi: 10.1088/0031-9155/58/6/1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernsting MJ, Murakami M, Roy A, et al. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. J Control Release. 2013;172(3):782–794. doi: 10.1016/j.jconrel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesh S, Iyer AK, Gattacceca F, et al. In vivo biodistribution of siRNA and cisplatin administered using CD44-targeted hyaluronic acid nanoparticles. J Control Release. 2013;172(3):699–706. doi: 10.1016/j.jconrel.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toita R, Nakao K, Mahara A, et al. Biodistribution of vaccines comprised of hydrophobically-modified poly(γ-glutamic acid) nanoparticles and antigen proteins using fluorescence imaging. Bioorg Med Chem. 2013;21(21):6608–6615. doi: 10.1016/j.bmc.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Movahedi K, Laoui D, Gysemans C, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Hu H, Ma H, et al. The anti-tumor effect and increased tregs infiltration mediated by rAAV-SLC vector. Mol Biol Rep. 2013;40(10):5615–23. doi: 10.1007/s11033-013-2663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, Rodriguez BL. Molecular targeting of liposomal nanoparticles to tumor microenvironment. Int J Nanomedicine. 2013;8:61–71. doi: 10.2147/IJN.S37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 18.Xiao K, Li Y, Luo J, et al. The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials. 2011;32(13):3435–3446. doi: 10.1016/j.biomaterials.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida JPM, Lin AY, Langsner RJ, et al. In vivo immune cell distribution of gold nanoparticles in naïve and tumor bearing mice. Small. 2014;10(4):812–819. doi: 10.1002/smll.201301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blank F, Stumbles PA, Seydoux E, et al. Size-dependent uptake of particles by pulmonary antigen-presenting cell populations and trafficking to regional lymph nodes. Am J Respir Cell Mol ecular Biol. 2013;49(1):67–77. doi: 10.1165/rcmb.2012-0387OC. [DOI] [PubMed] [Google Scholar]
- 21.Kirpotin DB, Drummond DC, Shao Y, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66(13):6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 22.Kourtis IC, Hirosue S, de Titta A, et al. Peripherally administered nanoparticles target monocytic myeloid cells, secondary lymphoid organs and tumors in mice. PLoS One. 2013;8(4):e61646. doi: 10.1371/journal.pone.0061646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolova V, Flace A, Bauer M, et al. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38(5):1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 24.VanHandel M, Alizadeh D, Zhang L, et al. Selective uptake of multi-walled carbon nanotubes by tumor macrophages in a murine glioma model. J Neuroimmunol. 2009;208(1–2):3–9. doi: 10.1016/j.jneuroim.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Zheng M, Librizzi D, Kılıç A, et al. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials. 2012;33(27):6551–6558. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 26.Guzmán J, Iglesias MT, Riande E, et al. Synthesis and polymerization of acrylic monomers with hydrophilic long side groups. Oxygen transport through water swollen membranes prepared from these polymers. Polymer (Guildf) 1997;38(20):5227–5232. [Google Scholar]
- 27.Perry JL, Reuter KG, Kai MP, et al. PEGylated PRINT Nanoparticles: theimpact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett. 2012;12(10):5304–5310. doi: 10.1021/nl302638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enlow EM, Luft JC, Napier ME, et al. Potent engineered PLGA nanoparticles by virtue of exceptionally high chemotherapeutic loadings. Nano Lett. 2011;11(2):808–813. doi: 10.1021/nl104117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kume M, Carey PC, Gaehle G, et al. A semi-automated system for the routine production of copper-64. Appl Radiat Isot. 2012;70(8):1803–1806. doi: 10.1016/j.apradiso.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Chan KT, Jones SW, Brighton HE, et al. Intravital imaging of a spheroid-based orthotopic model of melanoma in the mouse ear skin. Intravital. 2013;2(2):1–8. doi: 10.4161/intv.25805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharpless NE. Preparation and immortalization of primary murine cells. In: Celis JE, editor. Cell Biology. A laboratory handbook. 3rd. London: 2006. pp. 223–228. 3rd ed. [Google Scholar]
- 32.Chu KS, Schorzman AN, Finniss MC, et al. Nanoparticle drug loading as a design parameter to improve docetaxel pharmacokinetics and efficacy. Biomaterials. 2013;34(33):8424–8429. doi: 10.1016/j.biomaterials.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellian M, Yuan F, Trubetskoy VS, et al. Vascular permeability in a human tumour xenograft: molecular charge dependence. Br J Cancer. 2000;82(9):1513–1518. doi: 10.1054/bjoc.1999.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stylianopoulos T, Poh M-Z, Insin N, et al. Diffusion of particles in the extracellular matrix: the effect of repulsive electrostatic interactions. Biophys J. 2010;99(5):1342–1349. doi: 10.1016/j.bpj.2010.06.016. doi:10.1016/j.bpj.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith BR, Kempen P, Bouley D, et al. Shape matters: intravital microscopy reveals surprising geometrical dependence for nanoparticles in tumor models of extravasation. Nano Lett. 2012;12(7):3369–3377. doi: 10.1021/nl204175t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krol A, Maresca J, Dewhirst MW, et al. Available volume fraction of macromolecules in the extravascular space of a fibrosarcoma: implications for drug delivery. Cancer Res. 1999;59(16):4136–4141. [PubMed] [Google Scholar]
- 38.Huo S, Ma H, Huang K, et al. Superior penetration and retention behavior of 50 nm gold nanoparticles in tumors. Cancer Res. 2013;73(1):319–330. doi: 10.1158/0008-5472.CAN-12-2071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.