Abstract
Introduction
In order to further advance research and development on the Clinical Data Interchange Standards Consortium (CDISC) Operational Data Model (ODM) standard, the existing research must be well understood. This paper presents a methodological review of the ODM literature. Specifically, it develops a classification schema to categorize the ODM literature according to how the standard has been applied within the clinical research data lifecycle. This paper suggests areas for future research and development that address ODM’s limitations and capitalize on its strengths to support new trends in clinical research informatics.
Methods
A systematic scan of the following databases was performed: (1) ABI/Inform, (2) ACM Digital, (3) AIS eLibrary, (4) Europe Central PubMed, (5) Google Scholar, (5) IEEE Xplore, (7) PubMed, and (8) ScienceDirect. A Web of Science citation analysis was also performed. The search term used on all databases was “CDISC ODM.” The two primary inclusion criteria were: (1) the research must examine the use of ODM as an information system solution component, or (2) the research must critically evaluate ODM against a stated solution usage scenario. Out of 2,686 articles identified, 266 were included in a title level review, resulting in 183 articles. An abstract review followed, resulting in 121 remaining articles; and after a full text scan 69 articles met the inclusion criteria.
Results
As the demand for interoperability has increased, ODM has shown remarkable flexibility and has been extended to cover a broad range of data and metadata requirements that reach well beyond ODM’s original use cases. This flexibility has yielded research literature that covers a diverse array of topic areas. A classification schema reflecting the use of ODM within the clinical research data lifecycle was created to provide a categorized and consolidated view of the ODM literature. The elements of the framework include: (1) EDC (Electronic Data Capture) and EHR (Electronic Health Record) infrastructure; (2) planning; (3) data collection; (4) data tabulations and analysis; and (5) study archival. The analysis reviews the strengths and limitations of ODM as a solution component within each section of the classification schema. This paper also identifies opportunities for future ODM research and development, including improved mechanisms for semantic alignment with external terminologies, better representation of the CDISC standards used end-to-end across the clinical research data lifecycle, improved support for real-time data exchange, the use of EHRs for research, and the inclusion of a complete study design.
Conclusions
ODM is being used in ways not originally anticipated, and covers a diverse array of use cases across the clinical research data lifecycle. ODM has been used as much as a study metadata standard as it has for data exchange. A significant portion of the literature addresses integrating EHR and clinical research data. The simplicity and readability of ODM has likely contributed to its success and broad implementation as a data and metadata standard. Keeping the core ODM model focused on the most fundamental use cases, while using extensions to handle edge cases, has kept the standard easy for developers to learn and use.
Keywords: ODM, Define-XML, CDISC, interoperability, clinical trial, EHR
Graphical Abstract
1.0 Introduction
Clinical research is essential for advancing medicine and improving patient quality of life. The expansive scope of clinical research combined with the pervasiveness of technology has given rise to increasing volumes of data, and strategies are needed to process and exchange it effectively. As clinical trials continue to grow in complexity, systematic mechanisms to collect, process, analyze, and integrate data across systems and organizational boundaries have become increasingly important. Clinical research can no longer be considered an isolated venture and is increasingly conducted in network structures where seamless data exchange is critical to operational efficiency and effectiveness. Clinical data standards improve the efficiency and quality of clinical research and more broadly of healthcare delivery in general.
Within the realm of healthcare informatics there exists a broad array of data standards that meet a variety of needs. The Clinical Data Interchange Standards Consortium (CDISC) creates data standards for clinical research that complement, and in a growing number of cases, interact with a variety of healthcare standards. The CDISC Operational Data Model (ODM) standard is a document and exchange standard created specifically to support the needs of clinical research.
The ODM standard [1] plays a key role in clinical research informatics, including areas such as data exchange, archival, U.S. Food and Drug Administration (FDA) submission, and interoperability with healthcare data. Within the highly data-centric domain of clinical research, the XML-based ODM is the standard exchange format for case report form (CRF) data and metadata [2]. Interest in ODM as a research topic has grown significantly over the last several years with increasing interest in the CDISC data standards from regulatory authorities such as the FDA [3, 4] and the Japanese Pharmaceutical and Medical Devices Agency (PMDA), as well as from the considerable resources being allocated to healthcare data interoperability [5, 6]. The FDA has stated that, “improving the efficiency and effectiveness of medical product development is a national priority” [7]. Regulatory electronic submissions have grown more complex with the average submission now a staggering 3.4 million pages, an increase of 1,423% since 2005 [8]. With this scale, inefficiencies in the clinical research data lifecycle add significant time and expense to new medical product development. Increasing efficiency requires that the networked organizations participating in clinical development exchange data seamlessly. The 2014 CDISC business case claims that using CDISC standards from the beginning of the process can save approximately $180 million per submission [1].
The ODM standard was originally published for review as v0.8 in early 2000, and at that time was called the CDISC DAIS (Data Acquisition and Interchange Standard) model. The original objective when work started in 1999 was to address the data interchange and study archival use cases. Kubick et al. [9] described ODM as established to support the essential information needs of electronic data capture (EDC) systems and paper CRF data entry systems. Other key requirements included a 21 CFR Part 11 compliant audit trail, and long-term data archival support [10].
ODM was not originally developed based on an existing clinical research or healthcare data model, but instead was designed using a bottom-up approach to meet the established data interchange, archival, and audit trail requirements. The initial focus was on a general, vendor neutral structure and syntax; industry level data models and semantics were given little consideration. For example, an effort was made to align ODM with the Biomedical Research Integrated Domain Group (BRIDG) model, but this was long after ODM was originally published. In another example, converting openEHR’s Archetype Definition Language (ADL) to ODM has been demonstrated [11], but has not been a consideration in ODM’s requirements. ODM was designed in relative isolation to meet the needs of the CDISC community, and ODM’s relationship to clinical research data models has come from usage rather than from an explicit effort to design or generate the XML from an existing model.
The first production version of ODM was published in October 2000 and was demonstrated in two Connectathon events in 2001 [12]. The current ODM version, v1.3.2, was published in December of 2013. ODM, now based on XML schema, remains under active development by the CDISC XML Technologies Team, and while the original ODM requirements remain highly relevant, use of the standard has extended well beyond the original design.
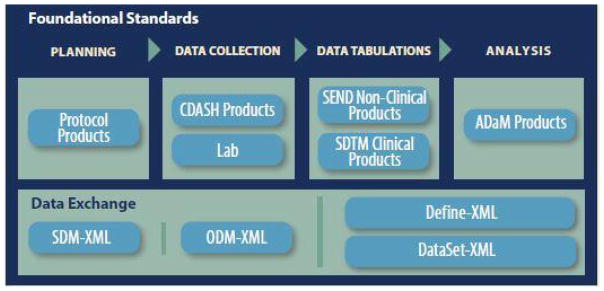
In response to increasing demands for interoperability, ODM has been extended over the years to cover a broad range of data and metadata needs [13]. This versatility has yielded research literature that reflects a diverse array of topic areas. The base ODM standard itself can be used to address a number of use cases, but standardized extensions have also been published including: (1) Define-XML for dataset metadata [14], (2) Dataset-XML for dataset data [15], (3) SDM-XML for Study Design Model [16], (4) CT-XML for Controlled Terminology [17], and (5) Analysis Results Metadata [18] for Define-XML v2. Figure 1 highlights the CDISC foundational standards covered by ODM, and standardized extensions such as Clinical Data Acquisition Standards Harmonization (CDASH) that describes the basic data collection fields for domains, the Study Data Tabulation Model (SDTM) that describes a standard structure for study data tabulations, and the Analysis Data Model (ADaM) that describes metadata models and examples for analysis datasets [1]. In addition to the extensions listed above, numerous proprietary extensions have been proposed in the research literature or implemented by practitioners. ODM’s useful form hierarchy structure, and its use of extensions to expand its applicability across a broad range of use cases has increased interest in ODM as a research topic.
Figure 1.
ODM-based standards supporting the CDISC foundational standards content
In order to further advance research on the ODM standard, the existing research must be well understood. This paper presents a systematic review of the ODM literature and uses a classification schema to organize and analyze the literature. It serves as a focal point for future ODM oriented research. Due to the number and size of the XML examples demonstrating ODM’s features and limitations, it was not feasible to include them in this paper. Instead, a website has been established as a repository for the example files at http://www.odm-review.com.
2.0 Material and methods
2.1 Research method
The literature search started with a systematic scan of online academic and conference databases in the information systems and healthcare domains performed by the lead author. The following databases were searched: (1) ABI/Inform, (2) ACM Digital, (3) AIS eLibrary, (4) Europe Central PubMed, (5) Google Scholar, (5) IEEE Xplore, (7) PubMed, and (8) ScienceDirect. A backward and forward reference search using Web of Science citation analysis for papers that passed screening was also performed with contributions from all authors. The search term used on all databases was “CDISC ODM.”
Two primary inclusion criteria were used in the selection of ODM research for examination within the proposed framework: (1) the research must examine the use of ODM as an information system solution component, or (2) the research must critically evaluate ODM against a stated solution usage scenario. Research papers not meeting these criteria were excluded from this analysis. Articles not written in the English language were also excluded from the search.
Out of 2,686 articles identified, 266 were included in a title level review resulting in 183 articles. An abstract review followed resulting in 121 remaining articles. After full text scanning of the 121 articles, 69 met the inclusion criteria and were included in the final analysis.
After reviewing the articles, a gap was identified in the literature regarding Define-XML, an ODM standard widely used in practice, but not well described in the academic literature. To address this gap an archive of 26,659 practitioner articles maintained at lexjansen.com was searched for “CDISC Define-XML.” Inclusion criteria were added to narrow the relevant articles to only those presented in 2013 – 2014 that provided a detailed description of the process for creating Define-XML files. Four additional articles were added to the final analysis based on this search.
As the articles where selected for inclusion, they were also categorized by the clinical research data lifecycle phase(s) addressed by the ODM solutions discussed in the article. The authors collaboratively developed this classification schema represented by the process diagram shown in Figure 2 and described in the next section. A consensus-based analysis process, with input from all authors, was used to review and update a categorized list of articles produced by the lead author. Each article included in the final analysis was assigned to at least one data lifecycle category, with 26% of the articles addressing multiple categories.
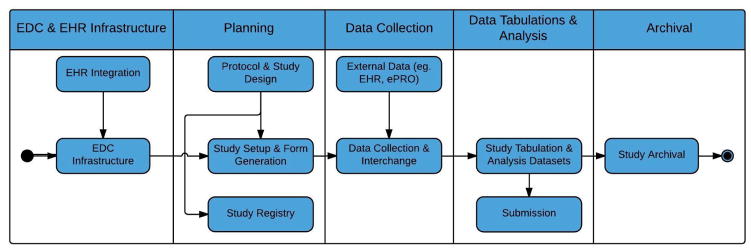
Figure 2.
Phases in the clinical research data lifecycle used by the ODM classification schema
2.2 Classification schema created for the methodological review
The purpose of establishing an ODM classification schema was to provide a categorized and consolidated view of the ODM research literature. The authors used Figure 1 as the basis for the classification schema. They consolidated the ODM use cases described in the literature into the different clinical research data lifecycle phases to yield a manageable number of categories and expanded Figure 1 to include the missing phases shown in Figure 2: EDC and EHR infrastructure, and study archival.
Each phase in the clinical research data lifecycle shown in Figure 2 represents a category in the classification schema. The categories of the proposed classification schema reflect the diverse array of use cases found in the ODM literature. A brief description of each of the five clinical research data lifecycle categories follows. Subsequent sections expand upon the applications of ODM in these five categories with details from the referenced literature.
The EDC and EHR Infrastructure phase of the lifecycle focuses on setting up the EDC data collection system and the EHR integration infrastructure to support future clinical research studies. This phase occurs once, and the infrastructure may be reused across multiple studies. After the EDC and EHR integration infrastructure has been setup, each of the remaining phases is executed for each clinical research study executed. The Planning phase covers creating a study protocol and representing it in a machine-readable format, formulating a study design, submitting a study to clinical trial registries (such as ClinicalTrials.gov), setting up a study within an EDC or other clinical data management system (CDMS), creating CRFs, defining a study schedule of events, and importing CRFs from form libraries. The Data Collection phase of the lifecycle focuses on the data collection and interchange that occurs during study execution and represents an original ODM use case [19]. The Data Tabulations and Analysis phase in the lifecycle combines the third and fourth phases shown in Figure 1 and focuses on the generation of datasets in support of standardized tabulations, analysis datasets, reporting and regulatory submissions. Study Archival is the final phase of the data lifecycle and focuses on archiving the study data and metadata such that it complies with the federal regulations [10]. It represents another original ODM use case [19].
3.0 Results
3.1 EDC and EHR infrastructure
3.1.1 ODM usage in EDC and EHR infrastructure
Though not an original ODM use case, the ODM literature in EDC and EHR infrastructure [1, 2, 20–44] identifies a surprising number of projects using ODM as a means of integrating Electronic Health Record (EHR) systems with clinical research systems [25]. Single Source, or collecting data once electronically with multiple uses in healthcare and clinical research, uses ODM to reduce the data capture burden at the investigator sites by bridging HL7 CDA and the EDC system [26, 27]. In Single Source, clinical care data flows into the EHR database, while the clinical trial data is sent to an EDC system in a parallel data flow [26] eliminating redundant data entry, reducing source data verification, and making the data available in a more timely fashion [28, 29].
The Single Source proof-of-concept Starbrite study conducted at the Duke Clinical Research Institute [27, 28, 30] captured data via the HL7 CDA, provided automated integration with ODM, and showed nearly a 75% overlap between the two. Others, such as El Fadly et al. [31] found that the overlap was only 30–50%, and in El Fadly et al. [32] it was only 13.4%. Interoperability between HL7 CDA and ODM has also been demonstrated by [32–36] and has been particularly effective for lab, demographic, medication, and vital signs data.
The x4T (exchange for Trials) system discussed in [37–39] is another Single Source implementation. The use of the x4T as a mediator between ODM and the EHR data reduced documentation time by 70% while increasing completed mandatory data elements from 82% to 100% [39]. The “Extraction and Investigator Verification” scenario identified in the Electronic Source Data Interchange (eSDI) document [29] inspired the x4T architecture [39] that is based on technologies such as the eXist XML database, XQuery, XForms, and ODM. Integrating x4T into routine patient care has improved data quality, as well as the data collection and documentation workflow [39].
The CDISC Healthcare Link Initiative, in conjunction with IHE QRPH (Integrating the Health Enterprise Quality, Research and Public Health) has produced the Retrieve Form for Data-capture (RFD) integration profile as a means of integrating EHR and clinical research data. When used with ODM, RFD establishes an automated method to capture form-based EHR data for secondary uses including clinical research, disease registries, and safety reporting [40]. RFD pre-populates ODM-based CDASH [24] CRFs with data extracted from an EHR system in HL7 CCD format [35]. This approach supports eSource by saving both the HL7 CCD and ODM XML to a regulatory compliant archive.
The RE-USE project used ODM metadata templates to support data interchange between HL7-based EHR data and clinical research data [32, 41] using a CDA/ODM mediator [31]. RE-USE included the use of an ODM metadata message to create the CRF in the EHR, and the use of an ODM data message created using the ODM mediator to exchange data [31]. In RE-USE ODM supported the use of healthcare terminologies such as SNOMED-CT [42], as well as binding data elements to concepts such as SNOMED 3.5 VF [32]. Combining ODM and HL7 using a semantic interoperability framework has enabled data interchange between EHR and EDC systems [23, 32, 35].
The SALUS (Scalable, Standard based Interoperability Framework for Sustainable Proactive Post Market Safety Studies) project addresses the lack of unified “models of use” in healthcare and clinical research by creating a common “model of meaning”. SALUS has developed a large knowledge base containing 4.7 million triples representing BRIDG, ODM, HL7 CDA, the CDISC content standards, and the relevant terminologies [21]. SALUS uses this knowledge base as the foundation of a semantic framework for achieving interoperability between clinical research systems using ODM, and EHR systems using HL7 CDA.
A large medical forms library chose ODM as its standard for representing form metadata [33]. This ODM-based medical forms repository used the BRIDG model to harmonize data elements to facilitate integration with HL7 CDA documents [33].
3.1.2 ODM enhancement opportunities identified in the literature for EDC and EHR Infrastructure
While ODM has been used to successfully integrate clinical research systems with EHR-based HL7 CDA and other medical record data formats in a limited number of cases [42], most note that their research projects used manual mapping to accomplish interoperability at the syntactic level [25, 27, 34]. Kush et al. [27] state that a general, reusable mapping between ODM and CDA would have been challenging in their project as each of the standards approaches semantics differently.
The lack of a common underlying reference model between routine healthcare and clinical research makes achieving semantic interoperability difficult [34, 35]. Semantic interoperabilityenables a clinical observation to be represented using different information models while maintaining the same meaning. Information models for representing clinical observations include HL7 Reference Information Model (RIM), CEN/ISO 13606, and BRIDG [22, 35]. CDISC standards like ODM and SDTM also represent clinical data models.
In addition to semantic differences, structural differences between standards can also impede interoperability. For example, ODM forms support 3 levels of depth, while HL7 CDA’s nested observations can be unlimited in number. This disparity is at least partially a reflection of the difference between protocol-driven clinical research and the event-driven healthcare domain.
Clinical research and healthcare lack a common set of terminologies making ODM/EHR integration challenging as similar data items may have values expressed using different vocabularies [34, 45]. HL7 CDA and ODM also represent controlled terminologies differently. The HL7 CDA standard uses the HL7 RIM to provide an external semantics source [27]. ODM tends to define its own codes without explicitly accounting for semantics [27, 45, 46].
3.2 Planning
3.2.1 ODM usage in planning
The ODM literature on planning activities covers a broad range of topics [2, 11, 13, 20–23, 45, 47–68]. The SDM-XML study design ODM extension, covering a portion of the protocol requirements, is currently available for use [16]. Despite being limited in scope, SDM-XML has been successfully applied in a number of projects. Aerts [47] showed how SDM-XML could be used to generate a caBIG Patient Study Calendar. Nepochatov [48] used study design metadata combined with core ODM metadata to generate study setups that included not only CRFs, but also a study calendar for each subject. ODM extended with SDM-XML was used by the IMI (Europe’s Innovative Medicines Initiative) EHR4CR project to support the patient recruitment process [20]. The SALUS project [21] used SDM-XML annotated with CDASH variables to demonstrate the automatic completion of research CRFs with EHR data.
Willoughby et al. [49] stated that registrations can be submitted to the World Health Organization (WHO) registry, International Clinical Trial Registry Platform (ICTRP), using an ODM extension [48]. Huser and Cimino [51] mapped the ClinicalTrials.gov schema to ODM and implemented an extension to address the gaps. In 2015, a draft ODM-based CTR-XML standard was published to represent information for submissions to clinical trial registries, such as ClinicalTrials.gov, EUDRACT, or WHO ICTRP [1].
In addition to representing elements of protocol, ODM metadata transformations can be used to generate CRFs, setup EDC systems, configure decision support systems, and drive integration with other clinical systems as described in [2, 45, 52–56]. ODM’s expressive metadata language makes it the language of choice for describing CRF content [52]. The CDISC CDASH standard [24] CRF content has been published in ODM, and can be exported from the CDISC SHARE metadata repository [69] as ODM.
Some clinical data software depends completely on ODM metadata for study setup and maintenance [2, 53]. Kuchinke et al. [58] demonstrated the transfer of a complete clinical study designed in a system at one research center, exported as an ODM file, and imported by a different system at a different research center without using any additional software tools. In addition to the basic CRF elements, ODM includes partial support for cross-element validation and derivations [59]. ODM’s vendor extension mechanism provides the means to add support for proprietary study setup and data collection features [52].
ODM’s ability to represent a broad range of CRF types was demonstrated in Bruland et al. [60] where all Clinical Data Elements (CDEs), including their semantic concepts, from 3,012 forms in the National Cancer Institute’s (NCI) Cancer Data Standards Registry and Repository were effectively mapped to ODM. The semantic concepts were captured using ODM Alias elements [60]. ODM semantic alignment is often achieved by using the Alias Name attribute to contain the code, and the Context attribute to contain the name of the code system [14, 61, 62], but in other cases implementers might choose to use a proprietary extension that more explicitly represents specific semantic annotations [11, 17, 22].
ODM is capable of supporting the requirements of a CDE repository model by adding the necessary features using vendor extensions [45]. Eli Lilly and Company demonstrated the use of ODM as a repository [63]. Dugas and Briel [64] use ODM for a large EHR and clinical research data model repository consisting of 3,320 medical forms with 102,677 data elements [23, 62, 64]. ODM has also been shown to align with the ISO 11179 metadata registry standard, with the exception of a standard representation for ISO 11179’s Data Element Concept (DEC) [13].
3.2.2 ODM enhancement opportunities identified in the literature for planning
The main limitation restricting the use of ODM to support protocol has been the lack of a complete implementation model, despite the availability of the BRIDG-based PRM (Protocol Representation Model) content model published in 2010 [49, 70]. The draft CTR-XML trial registries standard will expand ODM’s ability to represent study level elements including the WHO 20-item minimum dataset [71].
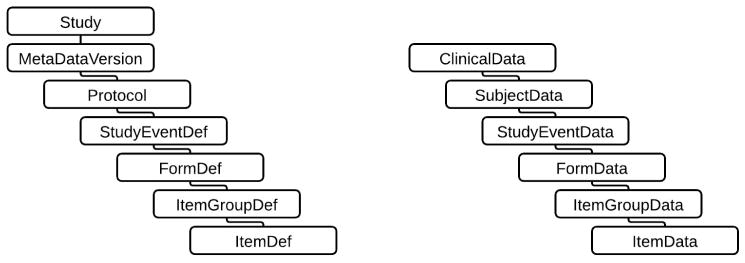
As previously described, ODM has made broad use of the Alias element to capture semantic information, but ODM lacks a formal mechanism for capturing semantics. ODM does not specify the use of specific clinical items such as CDASH, nor does it provide a specific mechanism for pointing to the source of a particular CDE. Instead, ODM provides a hierarchical container for defining clinical data items according to the needs of a given study [65], as shown in Figure 3. The lack of a source definition for a clinical data item was highlighted when the ISO 11179 DECs were found missing from the ODM model [13]. Studies that use CDASH CRFs achieve semantic alignment through a shared data standard, rather than through specific semantics [66]. ODM does not provide a mechanism for capturing the logical relationships between data elements used on different CRFs [66, 67]. Again, Alias can be used to address this shortcoming, but Alias currently relies on a common naming strategy to be effective.
Figure 3.
ODM metadata (left) and data hierarchies
By design, ODM does not include the presentation metadata needed to direct the visual rendering of CRFs [52]. Vendor extensions can be used to add the needed presentation metadata in ODM, but creating a single, standard extension to support a diverse set of clinical research software systems would be challenging [52]. This lack of metadata to describe the visual representation of CRF data fields can significantly alter a user’s experience with a CRF that is created in one system and then transferred for use in another [58]. ODM also lacks the metadata to represent the content ownership and context of use metadata as is found in copyrighted data collection instruments [45].
ODM supports multi-language CRFs, but simply applying xml:lang, a standard XML attribute for identifying language, to all ODM elements, instead of using the ODM specific TranslatedText within just five elements, broadens ODM’s multi-language support while eliminating an unnecessary level in the XML hierarchy [59].
De Melo et al. [52] cite a number of technically related ODM shortcomings, including a lack of support for arrays, that could be beneficial for capturing the value of repeating data elements. The ODM form hierarchy only supports two levels, Items within ItemGroups, as shown in Figure 3, but repeating data element CRF processing requirements could demand at least one additional layer in the hierarchy [52]. ODM also lacks a standard language to express computations and validations in the ConditionDef and MethodDef elements [45, 72].
3.3 Data collection
3.3.1 ODM usage in data collection
Data interchange, a key process in the data collection phase, is an original ODM use case, was the focus of ODM v1.0, and has been covered broadly in the literature [2, 13, 26, 35, 43, 45, 46, 52, 53, 57, 58, 60, 62–64, 73–85]. ODM’s basic hierarchical structure is particularly well suited for data capture [84]. In addition to its maturity, other noted ODM strengths include its relative simplicity and adaptability afforded by its vendor extension mechanism. Generating ODM exports to support data interchange is reasonably straightforward [83]. A large number of clinical research software vendors support the ODM-based exchange of clinical study data and metadata [2, 46, 58, 75]. ODM has also been used to support forms outside of clinical research, as was demonstrated by the ODM-based exchange of forensic autopsy data [57]. Most systems that support ODM data interchange do so via a file export and import mechanism [58, 76, 86], but some use a more modern ODM-based web services approach [53, 77, 78]. As a data interchange standard, ODM has demonstrated its usefulness as both a document and a message format. As the use of ODM to exchange data, data and metadata, or metadata alone has grown, the need for tools that validate ODM content, such as the ODM Checker [58], has increased in importance.
ODM has been used as the basis for data transformation systems in support of data integration. In Dugas and Dugas-Breit [62], ODM was the basis for an automated transformation tool to convert study data models into different formats including office documents, and statistical datasets. In this case, using semantically annotated ODM helped drive automated transformations while preserving the original semantics. The open-source compareODM tool in Dugas et al. [81] compared semantically enriched ODM forms and was able to automatically derive the differences between two versions of a form including identical, matching, or similar data items [43]. ODM has been used to integrate clinical research data into the i2b2 (Informatics for Integrating Biology and the Bedside) data model including both ontology and fact data [26, 46]. Leroux and Lefort [82] used ODM to drive the creation of RDF data cubes from longitudinal study data.
The base ODM standard has also been considered for use as a submission standard. In 2007 the FDA announced a pilot to test electronic CRF submissions in ODM instead of the currently required PDF format [87]. The FDA commented that PDF CRFs did not meet their needs, and that a suitable replacement would provide access to the CRF data, metadata, and audit trail [87]. That pilot was put on hold prior to completion.
3.3.2 ODM enhancement opportunities identified in the literature for data collection
ODM’s relative simplicity has at times also been a limiting factor impacting all aspects of interoperability including data mapping, representing semantics, data types, and terminology support. The ODM hierarchical structure, based on the elements shown in Figure 3, most clearly expresses CRF-oriented data [82, 84], and in the cases of the Define-XML and Dataset-XML, extensions have been used to represent tabular datasets [14, 15].
Despite its usefulness as an interchange standard, ODM provides limited support for data mapping information due to the lack of semantics associated with the data elements [45]. The use of the CDASH standard [24] CRF data elements in ODM format helps by providing standard naming conventions, and ultimately BRIDG [88] was intended to provide the missing semantics. However, applications using BRIDG to support data interchange semantics have been limited, and BRIDG does not include the bindings to controlled terminologies [85].
ODM does not provide the formal mechanisms needed to capture the semantics necessary for automated interchange [35]. As noted previously, Alias can be used to represent semantics, but since Alias can contain any content its usefulness in support of automated interpretation is limited [60]. Vendor extensions can be developed to represent semantics in ODM, but these extensions are not part of the normative ODM standard [2, 52].
3.4 Data tabulations and analysis
3.4.1 ODM usage in data tabulations and analysis
The Define-XML standard is an ODM extension that provides metadata to describe tabular datasets that, when used within the context of the CDISC content standards, typically describes all the SDTM, ADaM, or SEND datasets for a study [9, 89–93]. Define-XML plays a key role in establishing traceability in regulatory submission datasets [94]. The FDA added Define-XML to its Study Data Specifications in March of 2005 [3, 9], and in December 2016 it will become a requirement for submissions to the FDA [3, 14]. The FDA requirement to use the relatively archaic SAS V5 XPORT format for submission data has necessitated that the metadata be provided in a separate document [9]. To support the submission process, the ability to validate the Define-XML metadata has become increasingly important necessitating the development of validation rules and the tools that apply them, such as OpenCDISC [95] or the CDISC Define.xml Checker [96]. Kubick et al. [9] also note that the type of rich metadata provided in Define-XML is necessary for supporting integrated clinical research data repositories.
As a metadata standard, Define-XML can be used to support process automation much like the ODM metadata has been used to generate CRFs and aid in study setup. However, in practice it is often generated post-hoc to satisfy the needs of a submission. A more modern approach is to create the Define-XML metadata before the datasets are created and use it as a specification that drives the creation of study datasets. Maddox [90, 91] describes how sponsors create Define-XML files as a specification for the service providers that will produce the SDTM submission [89–92].
Dataset-XML is a new CDISC standard that represents tabular datasets in an ODM-based format and provides an alternative to the SAS V5 XPORT format. Despite being a new standard, the FDA has elected to pilot Dataset-XML as a possible alternative for submissions [4]. Dataset-XML provides a truly vendor neutral dataset exchange format without the limitations inherent in the older SAS V5 XPORT format.
3.4.2 ODM enhancement opportunities identified in the literature for data tabulations and analysis
Kubick, Ruberg, et al. [9] note that Define-XML, though a significant improvement over define.pdf, still maintains an unnatural separation of metadata and data for clinical study datasets. The availability of Dataset-XML to complement Define-XML provides opportunities to further advance the available tools and subsequently improve reviewer productivity [9, 15].
3.5 Study archival
3.5.1 ODM usage in study archival
The lifespan of regulated clinical trial data can extend to 50 years, and requires audit trails and investigator signatures. Archival in a proprietary format demanded that the data collection software and its operating environment, which often included the hardware, be archived to provide a validated platform to work with the data. ODM provides a non-proprietary means to archive data that meets the US federal regulations, and does not require the archival of proprietary software to support the use of the data [74, 97].
As an XML standard, ODM is well suited as a long-term archival solution that maintains the integrity of the data and metadata as captured from the original systems in a system-neutral, open format. ODM maintains the clinical data collected for a study, a full audit trail, electronic signatures, and the basic information needed for 21 CFR Part 11 and good clinical practice compliance [10, 73]. Kuchinke, Aerts, et al. [97] describe ODM as a structure that organizes the archived metadata and data together into a hierarchy based on the “CRF metaphor”. An increasing number of EDC systems directly export to ODM facilitating its use for archival [86].
3.5.2 ODM enhancement opportunities identified in the literature for study archival
While ODM provides the means to function as an archive for the study data and metadata, this represents one component of a complete study archive that would include other artifacts such as the study master file [97]. As noted previously, since ODM does not maintain presentation metadata it must be maintained in an extension, style sheet, or some other application capable of visually rendering the CRFs. File size has also been noted as an issue in ODM archives with file sizes ranging up to 2 GB per study [97].
Kuchinke, Aerts, et al. [97] note that using ODM as the source document archive to permit the destruction of the original paper data requires a legally compliant electronic signature. The ODM standard does not specify the level at which signatures should be created, and legally valid electronic signatures can be challenging to administer and maintain [97].
4.0 Discussion
Originally, ODM was collaboratively developed by a small CDISC team to meet the specific needs of clinical research data collection and management systems. From a data modeling perspective, ODM covered the fundamental elements of form-based data to meet the needs of regulated clinical research. ODM usage has expanded to cover new phases of the clinical research lifecycle, and now covers tabular data models in addition to the original hierarchical form-based model. As a standard authored by clinical research and XML experts, ODM has maintained an ease of understanding and use that has eluded XML standards generated from models, such as those generated using the HL7 RIM. The HL7 RIM put the needs of modelers ahead of the needs of implementers [98] making HL7 v3 difficult to implement, while ODM focused more directly on the needs of implementers. The ODM form-based model represents CRF data elements in a relatively simple manner, and limits the use of modeling abstractions favored by many of the healthcare information models, such as the HL7 RIM or BRIDG. ODM has itself been used as a model by software implementers creating solutions for clinical research. ODM has more in common with the relatively new HL7 Fast Healthcare Interoperability Resources (FHIR) standard, as the two share a number of design principles such as making use of extensions for edge cases and human readability.
4.1 Categorizing the literature by clinical research data lifecycle phase
The ODM literature reference counts by lifecycle phase for articles that met the inclusion criteria for this paper are shown in Table 1.
Table 1.
Article references by lifecycle phase
A total of 85 literature articles address the initial 3 phases of the clinical research data lifecycle, while only 8 articles address the last 2 phases. Table 2 and Table 3 in the Appendix provide a succinct synopsis of the most salient points identified for each lifecycle phase. The literature provides more coverage for study metadata than for clinical data. Almost no academic articles cover Define-XML, despite the fact that it is the most widely used ODM-based standard due to its position as a component of FDA regulatory submissions [3, 14]. The disparity between practitioner use and academic research may stem from the fact that academic institutions rarely pursue drug commercialization directly, but typically partner with bio-pharmaceutical companies to manage that phase of drug development when applicable. Many academic research projects are limited to a single site or integrated delivery network and therefore have had no need to transfer data between sites or to a regulator. This is rapidly changing with new data sharing perspectives and requirements [99].
4.2 Recommendations for future research and development
The recommendations for future ODM research and development were developed from (1) gaps in the literature, (2) ODM enhancement opportunities identified in the literature and, (3) new clinical research trends. These recommendations will be provided as inputs into a formal requirements analysis for the next version of ODM planned by the CDISC XML Technologies team to begin in 2016. All authors provided input into the recommendations, and an informal consensus process was used to determine the following list.
1. Add support for RESTful web services
In current practice clinical research data transfers predominantly occur in periodic batches, often times by sending the data for a full study in each exchange. Although the web-services based CDISC SHARE API returns ODM-based content and some vendors have implemented ODM-based web services [100], ODM lacks explicit support for modern exchange mechanisms like RESTful web services. The ODM specification does not include the means to automatically retrieve information using web services, as is the case in HL7 FHIR [101]. Leroux et al. [102] made a comparison and mapping between HL7 FHIR and CDISC ODM with the goal of achieving semantic interoperability between clinical research and healthcare. They present an approach to integrating ODM with FHIR leading to a mapping of hierarchical ODM ClinicalData elements to a set of FHIR resources [102]. Adding RESTful web services support to ODM and specifying a standardized web services API for the incremental exchange of ODM content would advance the state of practice today. Future ODM research and development would benefit by an examination of the HL7 FHIR approach to supporting RESTful interfaces, document-oriented aggregation, and semantic interoperability [98, 102].
2. Extend ODM to enable full lifecycle data traceability
Data fitness is fundamental to FDA submissions, and validation rules for the CDISC standards continue to grow in their importance as a key to applying the standards effectively [94]. As the use of validation has grown for FDA submissions, validating CDISC datasets has also grown into a common practice for Contract Research Organizations performing data services for sponsoring organizations. Inquiries into data quality within the context of ODM-based standards and the optimal means of ensuring quality, both before and during the validation step, would benefit the clinical research community. The FDA has identified a lack of traceability as one of the top 7 data standards issues [103]. Today no tools exist capable of tracing a data element from the protocol through to the clinical study report tables, listings, and figures [104]. No query capability exists to easily assess traceability within the context of the CDISC standards. Given ODM’s role in representing study metadata, machine-readable traceability represents another opportunity for further development of ODM-based standards [94].
3. Extend and evaluate ODM as an end-to-end standard representing all phases of the clinical research data lifecycle
ODM has a cogent, stable underlying data model that, through extensions, has also proven quite versatile [13] with regard to its ability to represent study setup and CRF metadata. New Protocol-XML and CTR-XML extensions will broaden the existing lifecycle coverage provided by extensions, including Define-XML, Dataset-XML, SDM-XML, and CT-XML. This list of extensions represents an increasingly comprehensive set of metadata that supports the automated generation of a growing number of study artifacts. Research highlighting ODM’s use in an end-to-end context from protocol through submission, where each state in the data lifecycle has an explicit relationship to the next, could help identify gaps or innovations that would improve its effectiveness to support clinical research process automation.
Study data archival is a strength of ODM and when linked to SDTM datasets, ADaM datasets, and EHRs it can become part of an end-to-end archive. Define-XML and Dataset-XML could aid in the archival of datasets, making it possible to archive the study datasets along with the ODM-based CRF data. New Protocol-XML and CTR-XML extensions will further extend the types of information that ODM can archive. The archival of study data is a time consuming and expensive proposition. Research into extensions that enhance ODM’s ability to function as an end-to-end data archive that captures a broader set of study artifacts would make a useful addition to the knowledge base.
Several efforts are underway to progress the development of a standardized protocol [105]. These efforts will form the foundation of a Protocol-XML standard scheduled to begin development in late 2016. A project to enhance CTR-XML to include study results is also planned to commence in late 2016. As these standards are released, an academic examination of their relative strengths and weaknesses could expedite their maturation. With the availability of a Protocol-XML standard, research into the tools and techniques for building studies based on a structured protocol would improve the quality and efficiency of the clinical research data lifecycle.
4. Extend ODM’s ability to represent semantics in a standardized way
Despite the fact that the use of semantically annotated ODM was a clear trend in the research literature, scalable semantic interoperability within healthcare and clinical research remains an unsolved problem [61]. The need to provide a standardized way to link to external code systems that does not rely on naming conventions was mentioned by a number of articles [23, 32, 60, 61]. ODM and healthcare standards like HL7 CDA provide the means to develop structural and syntactic alignment, but comparing or integrating data elements across different standards models requires the addition of semantics provided by external code systems [61]. Currently, the semantics used by most EHR and EDC systems are localized to the specific system [32]. Future research recommending an optimal approach to coding ODM data elements and CodeListItems using external code systems, such that the semantics are interpretable by software within a stated context, will accelerate the Healthcare Link agenda [33, 60, 61].
5. Extend ODM to more completely represent MDR metadata
ODM-based standards will play an increasing role in clinical research metadata repositories (MDR), and several research articles have highlighted the use of ODM in MDRs, including its alignment with ISO 11179 [13, 60]. As the number and use of MDRs grows, there will be opportunities to extend ODM to include the additional metadata needed to better support metadata libraries as demonstrated by the Eli Lilly ODM library [63]. The CDISC SHARE MDR [69] currently exports CDASH metadata in ODM and SDTM, ADaM, and SEND metadata in Define-XML. However, new ODM features are required to represent the additional metadata managed by the MDR, such as identifying the relationships between CDASH and SDTM variables, the CDASH prompt, CRF completion instructions, CDASH and SDTM core, and the CDISC notes. ODM would benefit from research that explores the additional metadata features needed to expand ODM’s role as a standard for representing metadata library content.
6. Examine the use of ODM in the submission context to complement Define-XML
Research exists on the automated analysis of CRFs [43, 81], but none have analyzed the CRF from a regulatory perspective. The benefits proposed in the 2007 FDA pilot [87] could be explored. For example, examining mechanisms for analyzing the ODM audit trail for fraud detection would be highly relevant. An analysis of how end-to-end traceability could be enhanced using ODM CRFs together with Define-XML would improve the case for submitting CRF data as ODM.
7. Examine the use of Define-XML to drive automation
Define-XML’s broad use by industry has been driven by the FDA’s requirement that “a properly functioning define.xml file is an important part of the submission of standardized electronic datasets and should not be considered optional” [106]. Define-XML can be used to specify metadata in support of the automated generation of datasets, and it is being used in this manner by the FDA’s Janus Clinical Trial Repository. As Define-XML’s role in driving the automation of end-to-end data management, data analysis, and aggregation increases, an evaluation of Define-XML’s adequacy to support these use cases would help drive future development of the standard.
Using Define-XML together with the new Dataset-XML standard creates a number of opportunities to enhance how datasets are organized and represented for regulatory submission and data interchange [4]. Combining Dataset-XML’s support for tabular data structures with ODM’s hierarchical structures could prove useful to capture the growing blend of heterogeneous data structures found in a future where a broad array of third party devices are used to capture patient data. Dataset-XML was designed to complement Define-XML, but as a new standard no literature exists analyzing the fitness of Dataset-XML as a dataset standard.
8. Create a set of standard validation rules to validate ODM-based standards across the clinical research data lifecycle
Now that datasets can be represented using an ODM-based extension, opportunities exist to create a standardized language for validating Define-XML and Dataset-XML content. A W3C standard language like XQuery has been used to process ODM-based content [2], and when combined with the schema and schematron rules might provide a standardized, executable language that works across all ODM-based standards. Additional research is needed to determine a vendor neutral formalism for expressing validation logic for verifying the structure and content of ODM-based CRFs as well as Dataset-XML datasets. Validation rules have become a prerequisite for the use of a standard in a regulatory submission context, and expressing unambiguous validation rules that can easily be converted to a computable format would advance the acceptance of the standards.
9. Extend ODM to better support data transformations and model-driven development
Research contributing to an optimal solution for incorporating data mapping metadata into ODM, as noted in [45], would benefit the development of ODM as a standard that not only represents data and metadata states within the clinical research data lifecycle, but also describes how to transition from state-to-state in support of an end-to-end data standard.
ODM is used as much for study metadata as it is for data
ODM’s flexibility and relative simplicity make efficient software based generation of data structures and CRFs possible [27]. The feasibility of model-driven architecture (MDA) in clinical research has been significantly improved by the availability of XML models like ODM. XML models support MDA’s ability to generate useful software applications [54]. De Melo, Nagler-Ihlein [52] used ODM metadata in an MDA application to generate user interfaces for mobile devices. More research exploring ways to extend ODM to support code generation and other model driven approaches to software development could help improve both the productivity and quality of automated clinical research data processing.
10. Add support for sharing de-identified data in support of secondary use
Open science, data sharing, and transparency are topics currently being explored by the clinical research community, but so are topics such as patient privacy and informed consent compliance. Standards like ODM benefit data sharing and open science because diverse, non-standard ways of collecting and representing clinical research data make it overly complex to integrate, pool, or share data for secondary analysis [107]. The FDA has stated that making de-identified and masked clinical data available for study by external experts could aid in the development of innovative new medical products, while also meeting the expectations of the patients by maximizing the use of their data [7]. Sharing undecipherable data, or data not supported by existing tools, does not advance the cause of openness and transparency. Thus, the CDISC standards must support a standard means of de-identifying datasets, including the ODM-based standards. Additional research demonstrating how ODM can be effectively used and extended in support of secondary use and big data would be of keen interest to the academic and practitioner communities. Improved ODM data citation support, to include an extension to better support the Dublin Core, would make attribution easier and encourage data sharing [108].
5.0 Conclusions
The relative simplicity and human readability of ODM has undoubtedly contributed to its success and broad implementation as a data and metadata standard. Maintaining a coherent core ODM model that accommodates most use cases, while supporting extensions to handle edge cases, has kept the standard easy for developers to learn and use. The selection of ODM for use in cases outside of clinical research, such as for representing medical forms or exchanging autopsy data, is a testament to the utility of its healthcare forms-oriented design. However, updates to ODM are needed both to address limitations as well as to keep it relevant as new automation and interoperability opportunities arise in both clinical research and healthcare. Based on the applications of ODM described in the literature, the future success of ODM may depend on how well it links to: (1) content described by other healthcare standards, (2) controlled terminologies, (3) externally defined semantics identifying ODM content, and (4) other ODM-based standards representing the different phases of the clinical research data lifecycle from protocol through analysis results.
Highlights.
ODM is used in study design, setup, execution, analysis, submissions, and archival
Much of ODM the literature addresses integrating EHR and clinical research data
ODM has been used as much as a study metadata standard as it has for data exchange
ODM’s simplicity and readability have contributed to its success
Future ODM success may depend on improved links to external standards and semantics
Acknowledgments
This research was supported in part (VH) by the Intramural Research Program of the National Institutes of Health (NIH)/National Library of Medicine (NLM)/Lister Hill National Center for Biomedical Communications (LHNCBC). We wish to thank Wayne Kubick, Rebecca Kush, and Landen Bain of CDISC, and Sally Cassells of Next Step Clinical Systems for their insightful comments on a draft version of this work. Sam Hume and Sally Cassells co-lead the CDISC XML Technologies Team.
6.0 Appendix: Summary of Findings
Table 1.
Summary of ODM Strengths
| Lifecycle Phase | Summary of Key ODM Strengths |
|---|---|
| EDC and EHR Infrastructure | ODM’s flexibility and relative simplicity have enabled it to be successfully applied across a number of EHR integration and single source projects. The Retrieve Form for Data Capture Profile (RFD) is a technical framework available to support EHR and ODM integration. ODM is broadly accepted for data collection in clinical research making it the logical target for EHR integration. ODM is increasingly listed alongside HL7 CDA as a supported XML format for healthcare informatics applications. Several European projects are currently working on EHR and EDC integration, including semantic interoperability. |
| Planning | ODM metadata has become the language of choice for describing CRFs, and has been used broadly for generating user interfaces. The ability to design a study in one system, export it in ODM, and import it into another system has been demonstrated. A study design ODM extension, SDM-XML v1.0, is currently available, and a draft CTR-XML clinical trial registry extension has been released for public review. |
| Data Collection | The ODM standard is mature, stable, relatively simple to work with, and supported by a broad number of data capture software vendors. The ODM model includes the information necessary to support the clinical research data collection process, including audit trail and digital signatures. Clinical research data web services using ODM have been demonstrated, and ODM has been shown to work as a document or a message. |
| Data Tabulations and Analysis | Define-XML supports flexibility in structural representations, and provides a rich set of metadata describing the clinical research datasets. Define-XML is broadly used in practice due to its position as a required component of a FDA regulatory submission. Dataset-XML is a new standard for representing data as tabular datasets, such as SDTM or ADaM, that complements the Define-XML metadata. |
| Study Archival | ODM maintains all the clinical data collected for a study together with the study metadata, and also contains the full audit trail, including electronic signatures. A significant number of data capture and management systems directly export to ODM as an out-of-the-box feature. |
Table 2.
Summary of ODM Limitations
| Lifecycle Phase | Summary of Key ODM Limitations |
|---|---|
| EDC and EHR Infrastructure | Most of the health IT integration successes have accomplished interoperability at the syntactic level. As long as the mapping needed for interoperability between healthcare and clinical research remains a manual process, this will prevent the large scale adoption of the ODM/CDA integration. Integration between terminologies is particularly challenging since HL7 uses terminologies differently than ODM. Integrating semantics typically relies on ODM’s Alias element which relies on a common naming strategy rather than a more formal mechanism. |
| Planning | By design, ODM does not include presentation information. ODM does not include a standard language for expressing computation and validation logic, including regular expression support. The main limitation restricting the use of ODM support for protocol has been the lack of a completed content model. |
| Data Collection | A standardized approach to using ODM with web services has not emerged. ODM does not include the metadata to capture the semantics or mapping information needed to facilitate computable semantic interoperability with other systems. |
| Data Tabulations and Analysis | Define-XML, though a significant improvement over define.pdf, still maintains an unnatural separation of metadata and data for clinical study datasets. Minimal academic research has investigated Define-XML despite its importance to practitioners. |
| Study Archival | ODM is not a comprehensive study archive that represents or provides links to the study documentation that must be archived in addition to the study data. Since ODM does not maintain presentation oriented metadata a style sheet or some other mechanism for recreating the visual aspects of the clinical data system must be maintained. File size has sometimes been noted as an issue with XML and ODM archives. |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sam Hume, Email: swhume@gmail.com.
Jozef Aerts, Email: jozef.aerts@fh-joanneum.at.
Surendra Sarnikar, Email: surendra.sarnikar@dsu.edu.
Vojtech Huser, Email: vojtech.huser@nih.gov.
References
- 1.CDISC. Clinical Data Interchange Standards Consortium. 2014 cited 2014; Available from: http://www.cdisc.org/
- 2.Forster C, Vossen G. Exploiting XML Technologies in Medical Information Systems. BLED 2012. 2012 [Google Scholar]
- 3.FDA. Study Data Specifications v1.5.1. FDA: FDA; 2010. [Google Scholar]
- 4.FDA; D.o.H.a.H. Services, editor. United States Government: Federal Register. 2013. Transport Format for the Submission of Regulatory Study Data; Notice of Pilot Project; pp. 70954–70955. [Google Scholar]
- 5.ONC. The Standards and Interoperability Framework. 2015 Jan 27;2015 Available from: http://wiki.siframework.org/ [Google Scholar]
- 6.Blumenthal D. Launching HITECH. New England Journal of Medicine. 2010;362(5):382–385. doi: 10.1056/NEJMp0912825. [DOI] [PubMed] [Google Scholar]
- 7.FDA; D.o.H.a.H. Services, editor. United States Government: Federal Register. 2013. Availability of Masked and De-identified Non-Summary Safety and Efficacy Data; Request for Comments; pp. 33421–33423. [Google Scholar]
- 8.Getz K. Technology, Regulation, Consolidation and Study Volunteer Trends. 2013 [Google Scholar]
- 9.Kubick WR, Ruberg S, Helton E. Toward a comprehensive CDISC submission data standard. Drug information journal. 2007;41(3):373–382. [Google Scholar]
- 10.FDA; FDA, editor. Guidance for Industry Part 11, Electronic Records; Electronic Signatures — Scope and Application. 2003 FDA.gov.
- 11.Garde S, et al. Can openEHR archetypes empower multi-centre clinical research? Studies in health technology and informatics. 2005;116:971–976. [PubMed] [Google Scholar]
- 12.Hume S. Applied Clinical Trials. UBM Advanstar; 2001. Connecting at the Connectathon. [Google Scholar]
- 13.Ngouongo S, Löbe M, Stausberg J. The ISO/IEC 11179 norm for metadata registries: Does it cover healthcare standards in empirical research? Journal of Biomedical Informatics. 2013;46(2):318–327. doi: 10.1016/j.jbi.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 14.CDISC. Define-XML Specification Version 2.0. CDISC; 2013. [Google Scholar]
- 15.CDISC. CDISC Dataset-XML Specification Version 1.0. Clinical Data Interchange Standards Consortium; 2014. [Google Scholar]
- 16.CDISC. CDISC Study Design Model in XML (SDM-XML) Clinical Data Interchange Standards Consortium; 2011. [Google Scholar]
- 17.CDISC. Representing Controlled Terminology in ODM. CDISC; 2011. [Google Scholar]
- 18.CDISC. Analysis Results Metadata Specification Version 1.0 for Define-XML Version 2 -Draft. Clinical Data Interchange Standards Consortium: CDISC; 2014. [Google Scholar]
- 19.CDISC. Overview of the CDISC Operational Data Model for Clinical Data Acquisition and Archive (based on CDISC ODM DTD 1.1 DRAFT) CDISC; 2001. [Google Scholar]
- 20.De Moor G, et al. Using electronic health records for clinical research: The case of the EHR4CR project. Journal of Biomedical Informatics. 2014 doi: 10.1016/j.jbi.2014.10.006. In Press. [DOI] [PubMed] [Google Scholar]
- 21.Laleci GB, Yuksel M, Dogac A. Providing semantic interoperability between clinical care and clinical research domains. Biomedical and Health Informatics, IEEE Journal of, 2013. 17(2):356–369. doi: 10.1109/TITB.2012.2219552. [DOI] [PubMed] [Google Scholar]
- 22.Bruland P, Dugas M. Transformations between CDISC ODM and openEHR Archetypes. Stud Health Technol Inform. 2014;205:1225. [Google Scholar]
- 23.Breil B, Dugas M. Analyses of medical data models-identifying common concepts and items in a repository of medical forms. Studies in health technology and informatics. 2012;192:1052–1052. [PubMed] [Google Scholar]
- 24.CDISC. Clinical Data Acquisition Standards Harmonization (CDASH) Clinical Data Interchange Standards Consortium; 2011. [Google Scholar]
- 25.Daniel C, et al. Standard-based integration profiles for clinical research and patient safety. AMIA Summits on Translational Science Proceedings; 2013; 2013. p. 47. [PMC free article] [PubMed] [Google Scholar]
- 26.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Methods Inf Med. 2009;48(1):45–54. [PubMed] [Google Scholar]
- 27.Kush R, et al. Implementing Single Source: the STARBRITE proof-of-concept study. Journal of the American Medical Informatics Association. 2007;14(5):662–673. doi: 10.1197/jamia.M2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastasiou A, Ifeachor E, Zajicek J. Data modeling methods in clinical trials: experiences from the clinical trial methods in neurodegenerative diseases project. Trials. 2011;12(Suppl 1):A15. [Google Scholar]
- 29.CDISC. Leveraging the CDISC Standards to Facilitate the use of Electronic Source Data within Clinical Trials. CDISC eSDI Group; 2006. [Google Scholar]
- 30.Alschuler L, Bain L, Kush R. Improving data collection for patient care and clinical trials. Sci Career Mag. 2004 Mar;26 [Google Scholar]
- 31.El Fadly AN, et al. CDA Template for eCRFs REUSE Project. International HL7 Interoperability Conference; 2009; Kyoto, Japan. [Google Scholar]
- 32.El Fadly AN, et al. Integrating clinical research with the healthcare enterprise: from the RE-USE project to the EHR4CR platform. Journal of Biomedical Informatics. 2011;44:S94–S102. doi: 10.1016/j.jbi.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Breil B, et al. Multilingual medical data models in ODM format–a novel form-based approach to semantic interoperability between routine health-care and clinical research. Appl Clin Inf. 2012;3:276–289. doi: 10.4338/ACI-2012-03-RA-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Fadly A, et al. Electronic Healthcare Record and clinical research in cardiovascular radiology. HL7 CDA and CDISC ODM interoperability. AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2007. [PMC free article] [PubMed] [Google Scholar]
- 35.Erturkmen GBL, et al. Building the Semantic Interoperability Architecture Enabling Sustainable Proactive Post Market Safety Studies. 2011 [Google Scholar]
- 36.Fadly A, et al. Aligning HL7 CDA Templates and CDISC ODM: An Experiment in Cardiovascular Radiology. Medinfo 2007: Proceedings of the 12th World Congress on Health (Medical) Informatics; Building Sustainable Health Systems; IOS Press; 2007. [Google Scholar]
- 37.Bruland P, Forster C, Dugas M. x4T-EDC: A Prototype for Study Documentation Based on the Single Source Concept. 24th International Conference of the European Federation for Medical Informatics; 2012. [Google Scholar]
- 38.Dziuballe P, et al. The single source architecture x4T to connect medical documentation and clinical research. Studies in health technology and informatics. 2010;169:902–906. [PubMed] [Google Scholar]
- 39.Bruland P, et al. Does single-source create an added value? Evaluating the impact of introducing x4T into the clinical routine on workflow modifications, data quality and cost–benefit. International journal of medical informatics. 2014;83(12):915–928. doi: 10.1016/j.ijmedinf.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 40.CDISC. CDISC Healthcare Link Initiative. 2014 [cited 2014 18-Dec-2014]; Available from: http://www.cdisc.org/healthcare-link.
- 41.El Fadly A, et al. The REUSE project: EHR as single datasource for biomedical research. Stud Health Technol Inform. 2010;160(Pt 2):1324–8. [PubMed] [Google Scholar]
- 42.Bernhard B, et al. HIS-based Kaplan-Meier plots-a single source approach for documenting and reusing routine survival information. BMC Medical Informatics and Decision Making. 2011;11 doi: 10.1186/1472-6947-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krumm R, et al. The need for harmonized structured documentation and chances of secondary use–Results of a systematic analysis with automated form comparison for prostate and breast cancer. Journal of Biomedical Informatics. 2014 doi: 10.1016/j.jbi.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Marcos M, et al. Interoperability of clinical decision-support systems and electronic health records using archetypes: a case study in clinical trial eligibility. Journal of Biomedical Informatics. 2013;46(4):676–689. doi: 10.1016/j.jbi.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Richesson RL, Nadkarni P. Data standards for clinical research data collection forms: current status and challenges. Journal of the American Medical Informatics Association. 2011;18(3):341–346. doi: 10.1136/amiajnl-2011-000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganslandt T, et al. Unlocking Data for Clinical Research–The German i2b2 Experience. Applied clinical informatics. 2011;2(1):116. doi: 10.4338/ACI-2010-09-CR-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aerts J. Generating a caBIG Patient Study Calendar from a Study Design in ODM with Study Design Model Extension. CDISC Journal. 2011 [Google Scholar]
- 48.Nepochatov NS. Translational research web application framework. Translational research. 2009 [Google Scholar]
- 49.Willoughby C, et al. A standard computable clinical trial protocol: The role of the BRIDG model. Drug information journal. 2007;41(3):383–392. [Google Scholar]
- 50.Karam G. International Clinical Trials Registry Platform. CDISC European Interchange 2014; 2014; Paris, France. [Google Scholar]
- 51.Huser V, CJ . NIH Research Festival. Bethesda, MD: NIH; 2014. Using CDISC Standards to Create Formal and Computable Representations of Human Clinical Research Protocols. [Google Scholar]
- 52.De Melo GM, Nagler-Ihlein J, Weber M. Generating User Interfaces from CDISC ODM for Mobile Devices. Mobile Business, 2006. ICMB’06. International Conference on; IEEE; 2006. [Google Scholar]
- 53.Tröger E, et al. Ophthabase: a generic extensible patient registry system. Acta Ophthalmologica. 2008;86(s243):0. [Google Scholar]
- 54.Crichton C, et al. Metadata-driven software for clinical trials. Proceedings of the 2009 ICSE Workshop on Software Engineering in Health Care; IEEE Computer Society; 2009. [Google Scholar]
- 55.Domínguez E, et al. Model-driven development based transformation of stereotyped class diagrams to XML schemas in a healthcare context. Advances in Conceptual Modeling–Foundations and Applications. 2007:44–53. [Google Scholar]
- 56.Fu LL, Chen T. Web-Based Case Report Form Design for Clinical Trial. Applied Mechanics and Materials. 2011;39:19–24. [Google Scholar]
- 57.Kiuchi T, et al. Legal Medicine Information System using CDISC ODM. Legal Medicine. 2013;15(6):332–334. doi: 10.1016/j.legalmed.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Kuchinke W, et al. Extended Cooperation in Clinical Studies through Exchange of CDISC Metadata between Different Study Software Solutions. Methods of information in medicine. 2006;45(4):441. [PubMed] [Google Scholar]
- 59.Yeom J, Jung S, Kim H. Language Supports for Multinational Clinical Trials in CDISC Platform. 2013 [Google Scholar]
- 60.Bruland P, et al. Interoperability in Clinical Research: From Metadata Registries to Semantically Annotated CDISC ODM. Studies in health technology and informatics. 2012;180:564. [PubMed] [Google Scholar]
- 61.Breil B, et al. Semantic enrichment of medical forms-semi-automated coding of ODM-elements via web services. Studies in health technology and informatics. 2011;180:1102–1104. [PubMed] [Google Scholar]
- 62.Dugas M, Dugas-Breit S. Integrated Data Management for Clinical Studies: Automatic Transformation of Data Models with Semantic Annotations for Principal Investigators, Data Managers and Statisticians. PloS one. 2014;9(2):e90492. doi: 10.1371/journal.pone.0090492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E.L.a. Company, editor. LillyODMLibrary. 2012 http://lillyodmlibrary.codeplex.com/ (No longer available)
- 64.DUGAS M, BREIL B. Open Medical Data Models: A Prototype of an Exchange Platform for Medical Forms. 2012 [Google Scholar]
- 65.Shabo A, Rabinovici-Cohen S, Vortman P. Revolutionary impact of XML on biomedical information interoperability. IBM systems journal. 2006;45(2):361–372. [Google Scholar]
- 66.Abler D, et al. Models for forms. Proceedings of the compilation of the co-located workshops on DSM’11, TMC’11, AGERE!’11, AOOPES’11, NEAT’11, & VMIL’11; ACM; 2011. [Google Scholar]
- 67.Davies J, et al. The cancergrid experience: Metadata-based model-driven engineering for clinical trials. Science of Computer Programming. 2013 [Google Scholar]
- 68.Nadkarni PM, Kemp R, Parikh C. Leveraging a clinical research information system to assist biospecimen data and workflow management: a hybrid approach. J Clinical Bioinformatics. 2011;1:22. doi: 10.1186/2043-9113-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.CDISC. CDISC SHARE. 2015 Jan 27;2015 Available from: http://cdisc.org/cdisc-share. [Google Scholar]
- 70.CDISC. CDISC Protocol Representation Model Version 1.0. CDISC; 2010. cdisc.org. [Google Scholar]
- 71.Huser V, et al. Standardizing Data Exchange for Clinical Research Protocols and Case Report Forms: An Assessment of the Suitability of the Clinical Data Interchange Standards Consortium (CDISC) Operational Data Model (ODM) Journal of Biomedical Informatics. 2015;(57):88–99. doi: 10.1016/j.jbi.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.CDISC. Specification for the Operational Data Model. Clinical Data Interchange Standards Consortium; 2013. [Google Scholar]
- 73.Lastic P-Y. The Convergence of Healthcare and Clinical Research Standards. 2007 [Google Scholar]
- 74.Souza T, Kush R, Evans JP. Global clinical data interchange standards are here! Drug discovery today. 2007;12(3):174–181. doi: 10.1016/j.drudis.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 75.Bickel J, Gregoric J. i2b2 Community: ODM to i2b2 importer. 2014 Nov 29;2014 Available from: https://community.i2b2.org/wiki/display/ODM2i2b2/Home. [Google Scholar]
- 76.Jiang G, et al. A collaborative framework for representation and harmonization of clinical study data elements using semantic MediaWiki. AMIA Summits on Translational Science Proceedings; 2010; 2010. [PMC free article] [PubMed] [Google Scholar]
- 77.Deserno TM, et al. SPIE Medical Imaging. International Society for Optics and Photonics; 2013. Integrating image management and analysis into OpenClinica using web services. [Google Scholar]
- 78.Haak D, et al. SPIE Medical Imaging. International Society for Optics and Photonics; 2014. OC ToGo: bed site image integration into OpenClinica with mobile devices. [Google Scholar]
- 79.Lingli F, Sheng D, Tao C. Clinical Data Management System. Biomedical Engineering and Computer Science (ICBECS), 2010 International Conference on; 2010. [Google Scholar]
- 80.Wang SA, et al. Performance of using Oracle XMLDB in the evaluation of CDISC ODM for a clinical study informatics system. Computer-Based Medical Systems, 2004. CBMS 2004. Proceedings. 17th IEEE Symposium on; IEEE; 2004. [Google Scholar]
- 81.Dugas M, et al. Automated UMLS-based comparison of medical forms. PloS one. 2013;8(7):e67883. doi: 10.1371/journal.pone.0067883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leroux H, Lefort L. Using CDISC ODM and the RDF Data Cube for the Semantic Enrichment of Longitudinal Clinical Trial Data. SWAT4LS; 2012; Citeseer. [Google Scholar]
- 83.Brandt CA, et al. Metadata-driven creation of data marts from an EAV-modeled clinical research database. International Journal of Medical Informatics. 2002;65(3):225–242. doi: 10.1016/s1386-5056(02)00047-3. [DOI] [PubMed] [Google Scholar]
- 84.Lefort L, Leroux H. Design and generation of Linked Clinical Data Cubes. 1st International Workshop on Semantic Statistics (SemStats); 2013. [Google Scholar]
- 85.Fridsma DB, et al. The BRIDG project: a technical report. Journal of the American Medical Informatics Association. 2008;15(2):130–137. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.CDISC. ODM Certified Products. 2014 [cited 2014; Available from: http://cdisc.org/odm-certified-products.
- 87.FDA; D.o.H.a.H. Services, Editor, editor. Federal Register. 2007. Electronic Case Report Form Submission; Notice of Pilot Project; pp. 11370–11371. [Google Scholar]
- 88.BRIDG. BRIDG Model Release 3.2 User’s Guide. BRIDG Semantic Coordination Committee: NCI; 2012. [Google Scholar]
- 89.Wheeldon M, Burges K. Discover Define-XML. PharmaSUG 2014; 2014; San Diego, CA. [Google Scholar]
- 90.Maddox J. Round Trip Ticket – Using the Define.xml file to Send and Receive your Study Specifications. PhUSE 2013; 2013; Brussels, Belgium. [Google Scholar]
- 91.Maddox J. Round Trip Ticket – Using the Define.xml file to Send and Receive your Study Specifications. PhamaSUG 2014; 2014; San Diego, CA. [Google Scholar]
- 92.Lightfoot G, Jansen L. Two-way Ticket, Please… All aboard the SAS® Clinical Standards Toolkit 1.5 Express. PhUSE 2013; Brussels, Belgium. 2013. [Google Scholar]
- 93.LEROUX H, et al. A method for the semantic enrichment of clinical trial data. Health Informatics: Building a Healthcare Future Through Trusted Information: Selected Papers from the 20th Australian National Health Informatics Conference (HIC 2012); IOS Press; 2012. [PubMed] [Google Scholar]
- 94.FDA; C. CDER, editor. Study Data Technical Conformance Guide. FDA; 2014. [Google Scholar]
- 95.OpenCDISC. 2014 [cited 2014; Available from: http://www.opencdisc.org/
- 96.Aerts J. XML4Pharma. 2014 [cited 2014; Available from: http://www.xml4pharma.com/
- 97.Kuchinke W, et al. CDISC standard-based electronic archiving of clinical trials. Methods of information in medicine. 2009;48(5):408. doi: 10.3414/ME9236. [DOI] [PubMed] [Google Scholar]
- 98.Grieve G, Kramer E, McKenzie L. HL7 Working Group Meeting. Baltimore, MD: HL7; 2012. Introduction to HL7 FHIR. [Google Scholar]
- 99.IOM. Sharing clinical trial data: Maximizing benefits, minimizing risk. Institue of Medicine; Washington, DC: 2015. [Google Scholar]
- 100.Newbigging A. PhUSE 2010. PhUSE; Berlin, Germany: 2010. Using web service technologies for incremental, real-time data transfers from EDC to SAS. [Google Scholar]
- 101.Raths D. Healthcare Informatics. Vendome Healthcare Media; 2014. Top Ten Tech Trends: Catching FHIR. http://www.healthcare-informatics.com/ [Google Scholar]
- 102.Leroux H, Metke-Jimenez A, Lawley M. ODM on FHIR: Towards Achieving Semantic Interoperability of Clinical Study Data. SWAT4LS International Conference; 2015; Cambridge, England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chhatre D, Malla A. CDER/CBER’s Top 7 CDISC Standards Issues. FDA; 2012. [Google Scholar]
- 104.Dootson A. PhUSE 2011. PhUSE; Brighton, UK: 2011. Tracing Data Elements through a Standard Data Flow. [Google Scholar]
- 105.Kubick W. 2015 CDISC Technical Plan. CDISC; 2015. cdisc.org. [Google Scholar]
- 106.FDA. CDER Common Data Standards Issues Document Version 1.1. FDA; 2011. [Google Scholar]
- 107.Kush R, Goldman M. Fostering Responsible Data Sharing through Standards. The New England journal of medicine. 2014;370(23):2163. doi: 10.1056/NEJMp1401444. [DOI] [PubMed] [Google Scholar]
- 108.Hoyle L, et al. Comprehensive Citation Across the Data Life Cycle Using DDI. 2014 [Google Scholar]