Abstract
The Na+-dependent glutamate transporter, GLT-1 (EAAT2), shows selective expression in astrocytes, and neurons induce expression of GLT-1 in astrocytes. In unpublished analyses of GLT-1 promoter reporter mice, we identified an evolutionarily conserved domain of 467 nucleotides ~8 kb upstream of the GLT-1 translation start site that is required for astrocytic expression. Using in silico approaches, we identified Pax6 as a transcription factor that could contribute to the control of GLT-1 expression by binding within this region. We demonstrated expression of Pax6 protein in astrocytes in vivo. Lentiviral transduction of astrocytes with exogenous Pax6 increased expression of enhanced green fluorescent protein (eGFP) in astrocytes prepared from transgenic mice that use a bacterial artificial chromosome (BAC) containing a large genomic region surrounding the GLT-1 gene to control expression of eGFP. It also increased GLT-1 protein, and GLT-1-mediated activity, while there was no effect on the levels of astroglial glutamate transporter, GLAST. Transduction of astrocytes with an shRNA directed against Pax6 reduced neuron-dependent induction of GLT-1 or eGFP. Finally, we confirmed Pax6 interaction with the predicted DNA binding site in electrophoretic mobility assays (EMSA) and chromatin immunoprecipitation (ChIP). Together, these studies show that Pax6 contributes to regulation of GLT-1 through an interaction with these distal elements and identify a novel role of Pax6 in astrocyte biology.
Keywords: glutamate transport, Pax6, astrocytes, EAAT2, GLT-1, transcriptional regulation
Graphical Abstract
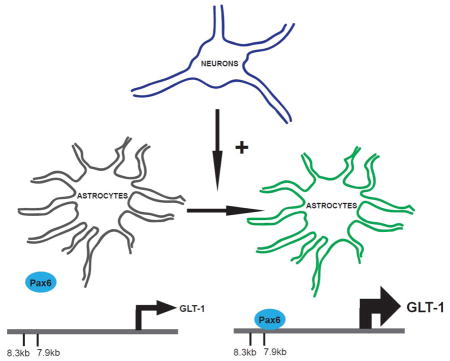
The astroglial glutamate transporter, GLT-1, shows selective expression in astrocytes and its expression can be induced by neurons. In this study we demonstrate that Pax6 is expressed in astrocytes and binds to the GLT-1 promoter in vitro and in vivo. Exogenous expression of Pax6 increases GLT-1 and enhanced green fluorescent protein (eGFP) expression in astrocytes from a transgenic mouse line that uses the GLT-1 gene to drive eGFP expression, and an shRNA directly against Pax6 attenuates neurondependent induction of GLT-1/eGFP. We therefore conclude that Pax6 contributes to the neuron-dependent induction of GLT-1.
Introduction
Extracellular glutamate is cleared by a family of Na+-dependent transporters, including GLAST (EAAT1), GLT-1 (EAAT2), EAAC1 (EAAT3), EAAT4 and EAAT5 (for reviews, see Danbolt 2001, Beart & O’Shea R 2006, Tzingounis & Wadiche 2007, Vandenberg & Ryan 2013). There are several lines of evidence to suggest that GLT-1/EAAT2 (encoded by the SLC1A2 gene in humans, slc1a2 in mice) mediates the bulk of glutamate uptake in vivo. First, the pharmacology of transport measured in crude synaptosomal membranes parallels that observed for cloned GLT-1 expressed in heterologous expression systems and is different from that observed for the other transporters expressed in heterologous systems (Robinson et al. 1991, Arriza et al. 1994 for review, see Robinson & Dowd 1997). Second, immunodepletion of GLT-1 from solubilized brain extracts reduces activity that can be subsequently reconstituted to 10% of control (Haugeto et al. 1996). Finally, genetic deletion of GLT-1 reduces transport activity in crude synaptosomes to ~5% of control (Tanaka et al. 1997). In fact knock-down of GLT-1 expression results in increased extracellular glutamate and excitotoxic cell death (Rothstein et al. 1996). Genetic deletion of GLT-1 results in premature death that is associated with spontaneous seizures (Tanaka et al. 1997).
GLT-1 transcription is regulated both in vitro and in vivo. For example, during development GLT-1 mRNA and protein levels uniquely increase during the period of synaptogenesis (Sutherland et al. 1996, Furuta et al. 1997). Little or no GLT-1 protein is found in astrocytes in vitro, but co-culturing with neurons induces expression of GLT-1 in astrocytes (Swanson et al. 1997, Schlag et al. 1998). Lesioning neurons causes a loss of GLT-1 in target areas in vivo (Ginsberg et al. 1995, Levy et al. 1995, Ginsberg et al. 1996, Hein et al. 2001) and decreases expression of GLT-1 in vitro (Schlag et al. 1998). These studies strongly suggest that neurons contribute to induction and maintenance of GLT-1 expression in vitro and in vivo. This effect of neurons is partly mediated by soluble secreted molecules (Gegelashvili et al. 1997, Schlag et al. 1998, Zelenaia et al. 2000), but contact also contributes to this effect (Yang et al. 2009). GLT-1 expression in cultured astrocytes is also controlled by other stimuli, including dbcAMP, pituitary adenylate cyclase-activating peptide, and epidermal growth factor (Swanson et al. 1997, Schlag et al. 1998 Figiel & Engele 2000, Zelenaia et al. 2000). Decreases in GLT-1 expression, both mRNA and protein, have been observed in a variety of neurologic insults, suggesting the loss of GLT-1 may contribute the pathogenesis of a variety of diseases (for reviews, see Sheldon & Robinson 2007, Kim et al. 2011). The β-lactam antibiotic, ceftriaxone, was initially identified in a high throughput screen for transcriptional activators of GLT-1 (Rothstein et al. 2005). While ceftriaxone also increases expression of system Xc- (Lewerenz et al. 2009), it delays the onset of muscle weakness, motor neuron loss, and prolonged survival in a rodent model of ALS (Rothstein et al. 2005) and also shows protective effects in a number of other animal models of disease (for review, see Soni et al. 2014). Ceftriaxone also reduces heroine relapse in a rodent model. Importantly, these effects are attenuated by antisense knock-down of GLT-1 (Shen et al. 2014). Together, these results suggest that GLT-1 expression can be dynamically regulated and that targeting transcriptional up-regulation of GLT-1 may be an interesting therapeutic strategy for a variety of neurologic and mental health disorders.
Several years ago, Su and colleagues isolated the proximal 2.5 kb of the GLT-1 promoter and characterized reporter expression in human primary human fetal astrocytes (Su et al. 2003, Sitcheran et al. 2005). They mapped domains within this 2.5 kb region that contributed to induction of reporter expression and identified nuclear factor-κB binding sites within this region that contribute to activation by dbcAMP and epidermal growth factor. They also showed that the effects of ceftriaxone are dependent upon nuclear factor-κB (NF-κB) binding sites within this region (Lee et al. 2008 for review, see Kim et al. 2011). Using many of these same constructs, we demonstrated that the effects of neurons are dependent upon binding of kappa B-motif binding protein (Yang et al. 2009) and nuclear factor-κB (Ghosh et al. 2011) to this proximal region of the promoter. Although these studies demonstrate that the proximal region of the GLT-1 promoter is important for regulation of GLT-1 expression, they do not address distal enhancer regions that may play an important role in regulating expression of GLT-1.
When 8.3 kb of the GLT-1 promoter is used to control transcription of fluorescent reporter protein in transgenic animals, expression is restricted to astrocytes (Yang et al. 2011). Interestingly, analysis of various GLT-1 promoter reporter mice employing increasing amounts of the GLT-1 promoter region, we find that 7.9 kb of the GLT-1 promoter is not sufficient to achieve expression of GLT-1 in astrocytes in vivo (unpublished observations). In the present study, we report that there is a fairly large evolutionarily conserved domain surrounding this region and that this domain contains several evolutionarily conserved ‘putative’ transcription factor binding sites. Pax6 was identified as a potential candidate for regulation of GLT-1 expression within this region. We provide evidence that Pax6 contributes to induction of GLT-1 expression through an interaction with an evolutionarily conserved binding site in this region in vitro and in vivo.
Materials And Methods
Animals
A colony of BAC GLT1 eGFP mice was maintained in the animal facility of Children’s Hospital of Philadelphia (Regan et al. 2007). All animals were housed at standard temperature- humidity-, and in a light-controlled environment with ad libitum access to food and water. Mice of either sex 1–2 days of age were used to generate astrocyte cultures. Chromatin immunoprecipitation experiments were conducted with adult male animals (~45 days of age). Timed-pregnant Sprague-Dawley rats were obtained from Charles River Laboratories (Kingston, NY, USA) for the generation of wild-type rodent cell suspensions. The care and treatment of animals in all experimental procedures followed the National Institutes of Health Guidelines for Care and Use of Laboratory Animals. These studies were approved by the institutional animal care and use committee of the Children’s Hospital of Philadelphia.
In Silico analysis of the GLT-1 gene
The NCBI housed DCODE database (http://ecrbase.dcode.org) was used to align and compare the 5′ non-coding region of GLT-1 gene from mammalian species (mouse, rat, dog, cow, monkey, chimpanzee and human). As previously described, evolutionarily conserved regions were identified using the default criteria of 70% homology for at least 100 nucleotides (Ghosh et al. 2011). The GeneChip microarray data deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), a transcriptome database for astrocytes, neurons, and oligodendrocytes, were accessed to find the mRNA expression levels of the identified transcription factors (Lovatt et al. 2007, Cahoy et al., 2008).
Primary cultures of astrocytes
Astrocyte cultures were prepared from cortices of young (1–2 days of age) BAC GLT1 eGFP mice as previously described (Li et al. 2006, Ghosh et al. 2011). Briefly, after dissociation into single cells with trypsin, cells were plated at a density of 2.5×105 cells/ml in 75 cm2 flasks and maintained in a 5% CO2 incubator at 37°C. The medium was replaced twice a week until the cells were ~90% confluent. Cultures were treated with A2B5 hybridoma supernatant (from Dr. Judy Grinspan, Children’s Hospital of Philadelphia) and Low Tox-M rabbit complement (Cedar Lane) to eliminate A2B5-positive oligodendrocyte precursors cells that express GLT-1 (Zelenaia et al. 2000). After another two days in culture, astrocytes were re-plated onto either 6-well or 12-well plates. After 3–5 d, astrocytes were transduced with lentiviral vectors. In some experiments, lentivirus infected astrocytes on 6-well plates were further overlaid with a cell suspension containing neurons and astrocytes, prepared from embryonic (day 17–18) Sprague Dawley rats as described earlier (Gonzalez et al. 2007, Ghosh et al. 2011). Cells were layered over the astrocytes at a density of 4×105 cells/ml. One-fourth of medium was exchanged with fresh medium every 3–4 d. After 7–10 d, the cultures contain a relatively high density of process-bearing neurons that overlay an intact monolayer of astrocytes (Ghosh et al. 2011). As reported earlier, there was no difference in the induction of eGFP expression with cell suspensions from either wild-type mice or rats (Ghosh et al. 2011).
Constructs
A short hairpin RNA (shRNA) directed against mouse Pax6 (GenBank Accession: NM_013627) was designed using Gene Link RNAi Explorer (Clontech). A unique nucleotide sequence between 1187 and 1210 was identified; this sequence is ~45% in GC content with a melting temperature of ~60°C. A 23 bp sequence (5′-GCGGAAGCTGCAAAGAAATAGAA-3′) corresponding to this position was used to create a hairpin-type RNAi cassette. The cassette consists of two complementary oligonucleotides, a sense strand and an antisense strand. The sense strand includes a guanosine (G) nucleotide, followed by the target sequence, a hairpin loop sequence 5′-TTCAAGAGA-3′, antisense of the target sequence, a 5–6 nucleotide long poly (T) tract, and a unique restriction site (HindIII). The oligonucleotide cassette also included BamHI and EcoRI restriction site overhangs on the 5′ end of the sense strand and on the 5′ end of the antisense strand, respectively for cloning into the lentiviral transfer vector (pTY-CMV). A control, scrambled oligonucleotide (5′-GGAGATAAAGCCGACGAATAAGA-3′) was used to prepare a non-silencing shRNA sequence; this sequence does not target any known eukaryotic gene. The oligonucleotide cassettes were synthesized by integrated DNA technology. Mouse Pax6 cDNA (Thermo Scientific; Clone ID 4008490; GenBank Accession: BC0011272) or Discosoma species (dsRED) were cloned into the multiple cloning site of pTY-CMV. The sequences of all constructs were verified prior to use.
Lentiviral vector
We used pseudotyped lentiviral vectors to express Pax6 or shRNAs that reduce Pax6 expression. These were produced according to the procedure described earlier (Li et al. 2006, Ghosh et al. 2011). Briefly, HEK-293T cells were transfected with a mixture of 3 cDNAs, including a packaging plasmid (pCMV Δr8.2, 16.8 μg), an envelope plasmid (JS-86, 5.6 μg), and a transfer plasmid (pTY-CMV, 22.5 μg) containing the transgene of interest using Ca2+-phosphate transfection kit (Clontech). Approximately 12 h after transfection, the cells were rinsed and fresh growth medium (12 ml) was added. Virus containing medium was concentrated by centrifugation at 50,000 g for 2 h at 4°C on each of the following 2 d. Concentrated virus was re-suspended in ice-cold cell culture medium (1 ml) and stored at −80°C for no more than one month prior to use. Viral titer was measured after each preparation using HIV-1 P24 ELISA assay kit (Perkin Elmer). Astrocytes were infected at a concentration of 400 ng of p24/ml in 1 mL of media per well in a 6-well plate or 0.5 mL of media per well in a 12-well plate. After 1 h, an equal volume of fresh media (no virus) was added. Twenty-four hours later, the media was exchanged for fresh media. Using this approach, we routinely observe ~80–90% transduction efficiency within 3–5 days in astrocytes transduced with fluorescent reporter proteins, and there is no effect on the morphology of the astrocytes nor is there any evidence of cell death (Li et al. 2006, Ghosh et al. 2011). When two viruses were used, the initial concentration was 200ng of p24/mL for each virus so that the total p24 concentration did not exceed 400 ng p24/mL.
Transfection and transduction of HEK 293T cells
HEK-293T cells were transiently transfected with 5 μg of Pax6 in the PTy-CMV vector or with empty PTy-CMV using the Ca2+ phosphate transfection kit (Clontech). In parallel, cells were also transduced with virus engineered to exogenously express Pax6 or with empty PTy-CMV (as described above). The cells were passaged 24 hours prior to either transfection or transduction. Cell were lysed 36 hours later in all transfections and in three transduction experiments. In one transduction, the cells were passaged 1:10 36 hours after virus application and maintained in culture for an additional 9 days (11 days total) before lysis. Cortical tissue was used as a positive control for GLT-1 immunoreactivity. Briefly, an adult BAC GLT1 eGFP mouse was anaesthetized with isofluorane and rapidly decapitated. Cortical tissue was harvested and homogenized in solubilization buffer (Dunlop et al. 2003).
Western blot analysis
Cells were lysed and harvested as described earlier (Li et al. 2006, Ghosh et al. 2011). Protein was measured using bicinchoninic acid (BCA kit; Pierce), and equal amounts of protein were resolved on 10% SDS-PAGE minigels (Bio-Rad), and transferred to polyvinylidine fluoride membrane (Immobilon-FL; Millipore) using transblot apparatus (Bio-Rad). After blocking for 1 h at room temperature in a 20 mM Tris buffer containing: 0.9% NaCl, 0.1% Tween 20, and 5% nonfat dry milk (pH 7.5), the membranes were then probed with rabbit anti GLT-1 (C-terminal-directed; 1:5000) (Rothstein et al. 1994), rabbit anti-GFP (1:5000; Sigma, cat# G1544), rabbit anti-actin (1:1000; Sigma, cat# 2066), mouse anti-GLAST (1:200; Miltenyi Biotec, cat# 130-095-822), rabbit anti-Pax6 (1:50; Covance, cat# PRB-278P), rabbit anti-Pax6 (10 μg/ml: Sigma, cat# SAB 1410 879), or in some cases a combination of these antibodies. Using lysates from transfected and untransfected HEK-293T cells, we tested the specificity and sensitivity of the anti-Pax6 antibodies and found that combining the two antibodies together improved both specificity and sensitivity (data not shown). Membranes were washed in blocking solution containing 1% nonfat dry milk and then incubated with fluorescent conjugated anti-rabbit or anti-mouse secondary antibody (1:10000; LI-COR Biosciences) and visualized using an Odyssey Infrared Imaging system (LI-COR Biosciences). As reported earlier (Haugeto et al. 1996), GLT-1 and GLAST variably form multimers that do not dissociate in solubilizing buffer. Monomers and multimers were quantified separately. As there was no evidence for differential effects, total transporter immunoreactivity was analyzed. Data were analyzed both with and without normalization to actin immunoreactivity. Both approaches yielded similar results, but normalization to actin slightly reduced variability. Therefore, this approach was used for data presented.
Glutamate uptake assay
Sodium-dependent transport activity was measured from lentivirus-transduced astrocytes on 12 well plates as described earlier (Li et al. 2006, Ghosh et al. 2011). Cells were first rinsed with Na+ or choline-containing buffer (2×1 mL). Accumulation of radioactive L-[3H]-glutamate (0.5 μM) was measured in presence or absence of 300 μM dihydrokainate (DHK; Tocris Bioscience), a selective inhibitor of GLT-1 mediated glutamate uptake (Robinson et al. 1991, Arriza et al. 1994). We previously showed that uptake is linear for at least 10 min under these conditions, and uptake was stopped after 5 min by rinsing the cells with ice-cold choline-containing buffer. After solubilization with NaOH, an aliquot of the cell extract was used for quantification of protein (Lowry et al. 1951) and a separate aliquot was used for analysis of radioactivity (Beckman LS 6500 scintillation counter). Dihydrokainate-sensitive Na+-dependent uptake was compared in control and transduced astrocytes (Ghosh et al. 2011).
Electrophoretic mobility shift assay
Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) from adult mice cortex as previously described (Ghosh et al. 2011). A double stranded oligonucleotide surrounding the putative Pax6 binding site (at -7896 upstream of translation start site) and corresponding to between -7912 and -7867 of the human GLT-1 promoter was synthesized. Similarly a mutant, control variant was made by altering the last 3 bp of Pax6 binding site from AAT to GGG. The sense strand of the wild-type probe was 5′-TCAGGTGAGGAATACATTCAAGACTGAATTCTTGGAGAAAAACGA-3′, and the sense strand of the mutant probe was 5′-TCAGGTGAGGAATACATTCAAGACTGGGGCCTTGGAGAAAAACGA-3′. Unlabeled and biotin-labeled oligonucleotides were synthesized by integrated DNA technology. Electrophoretic mobility shift assays (EMSAs) were performed according to the procedure described in Lightshift Chemiluminescent EMSA kit (Pierce) and as described earlier (Ghosh et al. 2011). Briefly, 20 μl reactions containing 2 μl of nuclear extract, 20 fmol/ml biotin-labeled oligo, 1 mg/ml poly (dI-dC), 50% glycerol, 100mM MgCl2, and 1% NP-40 were incubated at room temperature for 25 min. To test specificity of the reactions, 200-fold excess unlabeled oligo was included in parallel incubations. For supershift assays, 10 μg of rabbit anti-Pax6 (Covance) was added to the binding reaction and incubated for an additional 2 h at room temperature. The entire reaction mixture were resolved using 6% non-denaturing polyacrylamide gels at 200 V for 4 h in 1X Tris-Borate-EDTA (TBE) buffer and transferred overnight to Hybond-N+ hybridization membrane (GE Healthcare) at 30 V. Membrane and Protein-DNA complexes were cross-linked using a UV light and biotin-labeled DNA was detected by chemiluminescence (Chemiluminescent nuclic acid detection module kit; Pierce).
Chromatin immunoprecipitation (ChIP) assay
Pax6-DNA interactions were examined using the EZ-CHIP Chromatin Immunoprecipitation Kit (Millipore) as described previously (Ghosh et al. 2011). In brief, cortex, cerebellum, and kidney were harvested from adult wild-type mice, crossed-chopped, and incubated with 1.5% formaldehyde for 15 min to cross-link protein-DNA complexes. After quenching with excess glycine, tissue with rinsed with 1X PBS and triturated repeatedly using a glass Pasteur pipette. This suspension was repeatedly sonicated in SDS lysis buffer containing 1% SDS, 10 mM EDTA, 50mM Tris, pH 8.1 to obtain sheared DNA of between 100–400 nucleotides in length. After preclearing, an aliquot of each specimen (1% of precleared DNA) was saved and used for direct PCR amplification; this specimen is termed input. The remainder of the sample was incubated with a rabbit anti-Pax6 (Covance, Sigma) or control rabbit IgG (Invitrogen) overnight at 4°C. Protein-DNA-antibody complexes were isolated using agarose beads. After washing with increasing salt concentration, antibody-DNA complexes were eluted and un-cross-linked using RNase A and Proteinase K treatment. Resulting DNA was purified, extracted and PCR amplified using the following primers around the Pax6 binding site: 5′ AGTGTTTATCCAGAGGCTTGG 3′ (-8017 to -7997 relative to start codon of first exon) and 5′ CCATAAGTTCAGGAACATAAC 3′ (-7874 to -7854). Amplified products were resolved on 1% agarose gel and bands were visualized using a ChemiImager 4400 imaging system. Band intensities were quantified densitometrically using ImageJ (version 1.47d) softwere. Values were normalized to input.
Statistical Analysis
Results are expressed as mean ± standard error of mean of at least three independent experiments. Student’s t-test or one-way ANOVA followed by Bonferroni post hoc analysis were performed using Instat 3 (GraphPad Software Inc.). A p value of less than 0.05 was considered significant.
Results
Identification of minimal promoter required for transcription of GLT-1 in vivo
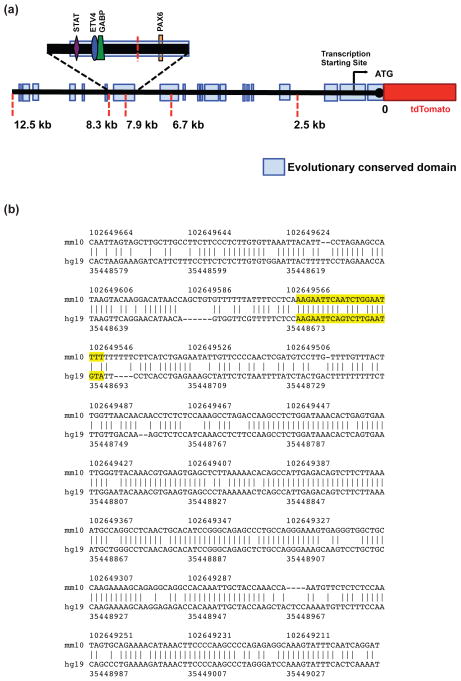
In earlier studies, we and others focused on understanding the role of transcription factors that bind to the proximal 2.5 kb of the 5′ non-coding region of GLT-1 gene and contribute to the control of expression (see Introduction). While this region is highly conserved, there are several additional evolutionarily conserved domains distal to this region, extending to almost 12.5 kb (Fig. 1). In unpublished studies, we generated mice that utilize 2.5, 6.7, or 7.9 kb of the 5′ non-coding region of the GLT-1 gene to control expression of tdTomato to identify the minimal amount of the promoter that is required to direct astrocyte-specific expression. None of these multiple transgenic mouse lines express the reporter gene in astrocytes/GLT-1 expressing cells (Rothstein, unpublished). In contrast, there is clear overlap of tdTomato with eGFP when 8.3 kb of the 5′ non-coding region is used to control expression of tdTomato (Yang et al. 2011). Based on these analyses, we hypothesized that the evolutionarily conserved 467 bp fragment that is ~8kb upstream of the translation start site is required for in vivo expression of GLT-1 in astrocytes.
Figure 1. Schematic representation of putative promoter region of GLT-1 gene (~ to scale) and the aligned human and mouse 467 bp ECR.
(a) Evolutionary conserved domains are indicated as light blue rectangular boxes, and are relative to the translation start site. Red dashed lines represent locations of restriction sites used to generate mice in which increasing lengths of the 5′ non-coding region of the human GLT-1 gene was used to control expression of a reporter protein, tdTomato (Rothstein, unpublished observations). These mice were bred with BAC GLT-1 eGFP transgenic mice to generate ‘dual’ reporter mice. The magnified portion represents the 467 bp evolutionarily conserved domain that is ~8kb upstream of the translation start site. The relative locations of ‘putative’ transcription factor binding sites for astrocyte-enriched transcription factors are depicted. (b) The human and mouse sequences from the DCODE database from the 467 bp evolutionarily conserved domain. The ‘putative’ Pax6 binding site is highlighted in yellow.
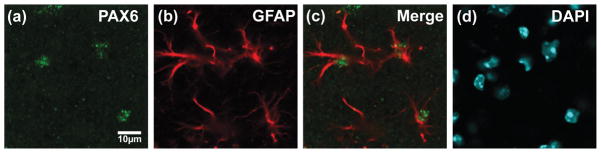
We tested for evolutionarily conserved transcription factor binding sites using the DCODE database (see methods). We identified 21 evolutionary conserved, putative transcription factor binding sites. As three ‘putative’ binding sites that were observed more than once, 18 transcription factors were implicated using this strategy (See Table 1). Some of these binding sites bind dimers of transcription factors (e.g. GABP), and some of these binding sites bind more than one factor (e.g. STAT or HSF). Of these transcription factors, Pax6 was the only mRNA that was greater than 10-fold higher in astrocytes compared to other brain cells in two different analyses of astrocyte RNA levels using expression arrays (Lovatt et al. 2007, Cahoy et al. 2008). STAT3, STAT5a, STAT5b, GABPa, GABPb2, and ETV4 (PEA3) were the only other transcription factors that were enriched in astrocytes, but either the enrichment was less than that observed with Pax6 (STATs, ETV4, & GABPa, ~2- to 8-fold) or the results were not consistent between the two sets of data (GABPb2). While our current study was underway, a third transcriptome analysis was conducted using RNA sequencing (RNA-seq) technology (Zhang et al. 2014). An examination of this database leads to a similar conclusion, but there were some differences. Based on presence of a ‘putative’ Pax6 binding site and the 10-fold enrichment in astrocytes, we hypothesized that Pax6 might contribute to the control of GLT-1 expression through an interaction with this domain. In agreement with a relatively recent study (Sakurai & Osumi 2008), we observed Pax6 immunoreactivity in cells that also stain for the astroglial protein, glial fibrillary acidic protein (GFAP) (Fig. 2). In fact, this staining was restricted to the nucleus, as identified with 4′,6-diamidino-2-phenylindole (DAPI) staining, within the cell body of GFAP+ cells, consistent with the expected location for a transcription factor.
Table 1.
Astrocytic expression of transcription factors with ‘putative’ binding sites in the domain ~8kb upstream of the translation start site in the GLT-1 gene
| Enriched in astrocytes (> 1.5-fold) | Not enriched in astrocytes | Not identified |
|---|---|---|
| ETV4 (PEA3) | E4F1 | CET168 |
| GABPα | ELK1 | CETS1P54 |
| GABPβ1 | GABPβ2 | NKX2-5B |
| PAX6 | GATA2 | PAX3 |
| STAT3 | HIC1 | |
| STAT5a | HIF1α | |
| STAT5b | HSF1 | |
| HSF2 | ||
| NF1 | ||
| PCBP1 (αCP1) | ||
| RUNX1 (AML) | ||
| RUNX2 (OSF2) | ||
| STAT1 | ||
| STAT6 |
The 467 bp, evolutionarily conserved domain that is ~8kb upstream of the translation start site in the GLT-1 gene was analyzed for evolutionarily conserved transcription factor binding sites using the ECR browser in the DCODE database. Twenty-one transcription factor binding sites were identified. The expression levels of each of the transcription factors implicated from these binding sites was initially analyzed using two databases (Lovatt et al, 2007; and Cahoy et al., 2008). While the current studies were underway, a third expression analysis was conducted (Zhang et al., 2014). The table reflects data obtained from these three different databases. “Enriched in astrocytes” was defined as >1.5-fold higher levels of mRNA in astrocytes compared to other cells in at least two of the three databases. “Not enriched in astrocytes” includes transcription factors that were <1.5 higher in astrocytes, equal levels in astrocytes and other cells, or enriched in other brain cells in at least two of the three databases. “Not identified” indicates they were not identified in at least 2 of the 3 sets of data.
Figure 2. Distribution of Pax6 in cortex.
Anti-Pax6 and anti-GFAP antibodies were used to test for co-localization in astrocytes in cortex. Nuclei were stained with DAPI. Data are representative of three independent experiments.
Effects of exogenous Pax6 and shRNA targeting Pax6 on GLT-1 expression and function
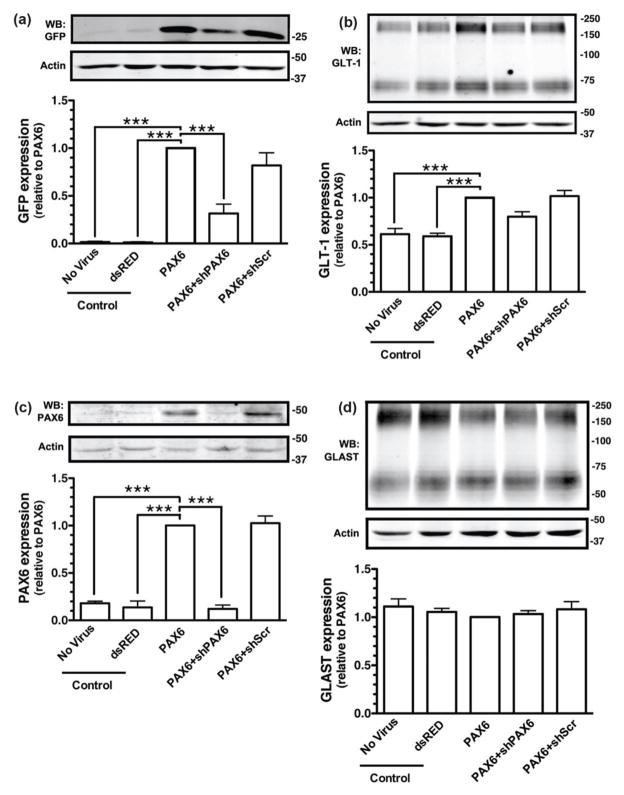
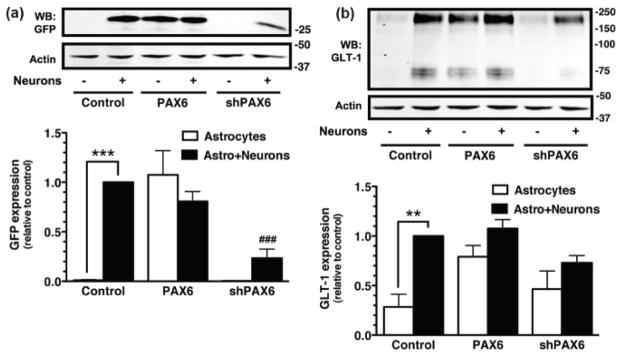
To determine if exogenous expression of Pax6 is sufficient to increase transcription of GLT-1/eGFP, astrocytes were prepared from the BAC GLT1 eGFP mice and transduced with lentiviral vectors engineered to express control protein (dsRED), Pax6, an shRNA directed against Pax6, a control scrambled shRNA, or combinations of these vectors. Exogenous expression of Pax6 resulted in a dramatic increase in the steady-state levels of eGFP, and this effect was attenuated by co-expression of the shRNA directed against Pax6 (Fig. 3a). Although the effects of Pax6 on GLT-1 protein were, as previously observed with other transcription factors (Ghosh et al. 2011, Yang et al. 2009), more modest than the effects on eGFP protein levels, Pax6 caused a significant increase in the steady state levels of GLT-1 protein (Fig. 3b). In these same experiments, the levels of Pax6 were examined by Western blot and as expected, the levels of Pax6 protein were higher in astrocytes transduced with the lentiviral vector engineered to express Pax6. This increase in Pax6 levels was specifically blocked by expression by an shRNA directed against Pax6 with no effect of expression of scrambled sequence (Fig. 3c). Finally, Pax6 had no effect on expression of the other astroglial glutamate transporter, GLAST (Figure 3d).
Figure 3. Effects of Pax6 and shRNA against Pax6 on the levels of eGFP or GLT-1 protein.
Cortical astrocytes from BAC GLT-1 eGFP transgenic mice were infected with different combinations of lentiviral vectors, including those that contain: dsRED & empty virus, Pax6 & empty virus, Pax6 and shRNA directed against Pax6, or Pax6 and a scrambled shRNA (shScr). Cells were harvested after 10 d and the levels of eGFP (a) or GLT-1 (b) were analyzed by Western blot. Fifteen μg of cell lysate was loaded to each lane. Top, Representative Western blots for eGFP or GLT-1. Bottom, Summary of eGFP or GLT-1 protein levels normalized to actin and expressed relative to the levels observed in astrocytes infected with Pax6. Cell lysates were also obtained from astrocytes that were not transduced with lentivirus and used as an additional control (No Virus). Pax6 (c) and GLAST (d) protein levels were also analyzed in these same specimens. Top, A representative Western Blot; bottom, graphical summary of data. Fifty μg of cell lysate was loaded in each lane for Western blot analysis of Pax6. No other immunoreactive bands were observed in these blots. Data are the mean ± SEM of three independent experiments. ***p < 0.001 compared to corresponding Pax6 infected astrocytes.
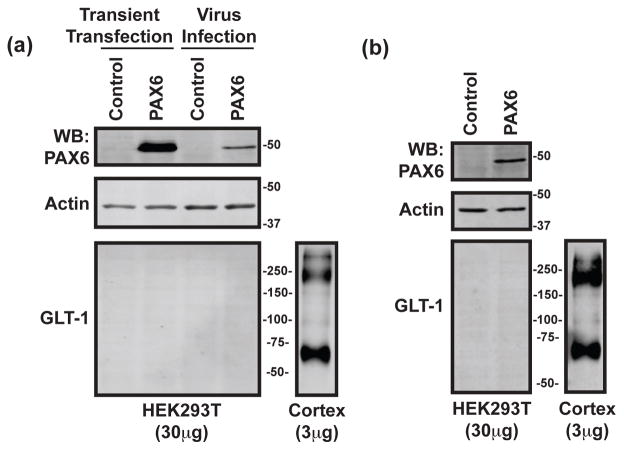
To determine if this effect of exogenous Pax6 is specific for astrocytes, we used two approaches, transient transfection or lentiviral transduction, to over-express Pax6 in HEK-293T cells. HEK-293T cells do not normally express GLT-1. We find that both approaches dramatically increase the amount of Pax6 protein in these cells, but did not result in any detectable GLT-1 (Fig. 4a). Cortical tissue was also resolved in these same experiments (on the same gels) as a positive control for our ability to detect GLT-1 immunoreactivity. These cells were harvested two days after transfection. At this stage, the HEK-293T cells were starting to grow over one-another. In one experiment, the HEK-293T cells were re-plated and maintained for 11 days after infection with lentivirus. Under these conditions, Pax6 was still clearly and dramatically increased over control, but no GLT-1 protein was detected. Together these studies show that exogenous expression of Pax6 is not sufficient to induce expression of GLT-1 in all cell types, suggesting that other factors are required.
Figure 4. Effects of exogenous expression of Pax6 in HEK-293T cells.
(a) HEK-293T cells were transiently transfected with Pax6 or control (empty PTy-CMV) and harvested for protein analysis after 36 hours. In parallel, HEK-293T cells were transduced with a lentiviral vector engineered to express Pax6 or empty PTy-CMV and also harvested after 36 hours. The samples were analyzed for Pax6, actin, and GLT-1 by Western blot. GLT-1 was never detected in these samples. These results are representative of three independent experiments. Note, cortical tissue lysates were also loaded onto these same gels; the lanes were cut for more effective presentation in these figures (same exposure). (b) In one of these experiments, the HEK-293T cells transduced with a lentiviral vector were passaged for 11 days and then harvested. Under these conditions, no GLT-1 immunoreactivity was detected.
In our previous study, we generated GLT-1 gene-based reporter constructs to study interactions of NF-κB with the promoter and neuron-dependent activation of these reporter constructs (Ghosh et al. 2011). These studies included mutant variants that lack the ‘putative’ NF-κB binding sites. We tried to generate and subclone similar reporter constructs to evaluate Pax6-dependent activation, but we were unsuccessful at subcloning these long (8.3 kb) promoter fragments combined with luciferase into the lentiviral backbone. In future studies, it will be important to examine the effects of deletion of this Pax6 binding site in vivo using CRISPR-based approaches (for reviews, see Cong et al. 2013, Gaj et al. 2013, Mali et al. 2013).
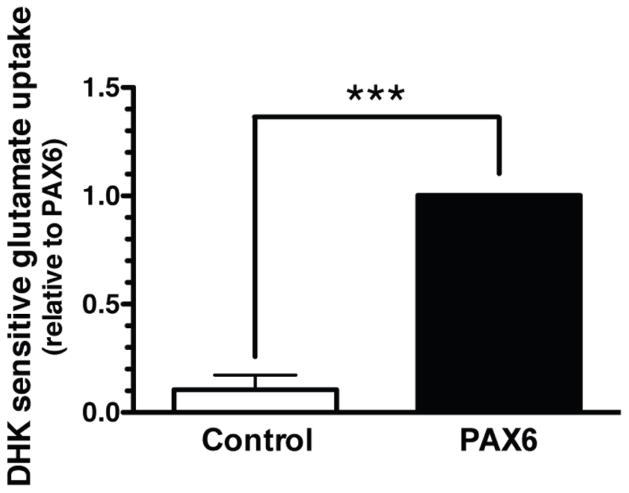
Dihydrokainate selectively inhibits GLT-1-mediated activity (Arriza et al. 1994); therefore dihydrokainate-sensitive uptake has been used by several groups including ours as a measure of GLT-1 mediated uptake (Dabir et al. 2006, Ghosh et al. 2011). To determine whether the effects of Pax6 on eGFP and GLT-1 levels are associated with an increase in functional GLT-1-mediated uptake, we examined the effect of exogenous expression of Pax6 on dihydrokainate-sensitive uptake. We observed that transduction of Pax6 had a very small effect on total Na+-dependent transport activity (0.34 ± 0.09 nmol/mg/min in control-dsRED-transduced astrocytes, 0.43 ± 0.1 nmol/mg/min in astrocytes transduced with Pax6; n = 4; P = 0.14 by paired t-test). As previously reported (Garlin et al. 1995), we observed essentially no DHK-sensitive uptake in astrocytes transduced with dsRed as a control while exogenous expression of Pax6 caused a significant (~10-fold) increase in DHK-sensitive transport (Fig. 5). Together, these studies demonstrate that exogenous expression of Pax6 is sufficient to increase GLT-1 protein and GLT-1 mediated activity. The fact that exogenous expression of Pax6 also caused an increase in expression of eGPF in astrocytes prepared from the BAC GLT-1 eGFP mice suggests that these effects are due to increased transcription and are not related to effects on translation or protein stability.
Figure 5. Effect of exogenous expression of Pax6 on dihydrokainate (DHK)-sensitive Na+-dependent glutamate uptake into astrocytes.
Cortical astrocytes from BAC GLT-1 eGFP transgenic mice were infected with lentiviral construct containing dsRED (control) or Pax6. After 10 days, glutamate (0.5 μM) uptake was measured in the presence and absence of Na+ and in the presence and absence of DHK (300 μM). Each analysis was conducted in triplicate. Data are the mean ± SEM of 4 independent experiments.
Effects of Pax6 shRNA on neuron-dependent activation of GLT-1
Co-culturing neurons with astrocytes increases astrocytic expression of GLT-1 protein (Swanson et al. 1997, Schlag et al. 1998). Similarly, co-culturing astrocytes derived from the BAC GLT1 eGFP mice with neurons from wild-type rodents (mice or rats) increases astrocytic expression of eGFP (Yang et al. 2009, Ghosh et al. 2011). To determine if neuron-dependent induction of eGFP/GLT-1 expression in astrocytes is dependent upon Pax6, astrocytes derived from BAC GLT1 eGFP mice cortex were first transduced with either exogenous Pax6, with the shRNA directed against Pax6, or with a control lentiviral vector that expresses no transgene. Five days later, a suspension of cells from embryonic cortex of wild-type rodent (rat) was added to the culture, and 7–10 days later cells were harvested for the analysis of eGFP and GLT-1 protein. As was previously observed (Swanson et al. 1997, Schlag et al. 1998, Yang et al. 2009, Ghosh et al. 2011), the levels eGFP and GLT-1 protein were significantly higher in the neuron-containing cultures compared to the cultures that only contain astrocytes (Fig. 6). It should be noted that the suspension of embryonic cortex also introduces a source of additional astrocytes that might express GLT-1; loading equal protein in each lane ensures that the percentage of total protein that comes from astrocytes is actually lower in the cultures that also contain neurons. Of course, these additional astrocytes are derived from wild-type animals so they do not contribute to the total amount of eGFP. When neurons are present, the levels of neither eGFP nor GLT-1 were significantly affected by the exogenous expression of Pax6, indicating that the effects of neurons and Pax6 are not additive. Expression of shRNA directed against Pax6 significantly reduced neuron-dependent induction of eGFP (Fig. 6a), and the mixed cultures did not express significantly higher levels of GLT-1 protein compared to astrocytes alone under these same conditions (Fig. 6b). These studies provide strong evidence that Pax6 is necessary for neuron-dependent induction of GLT-1 expression.
Figure 6. Effects of shRNA against Pax6 on neuron-dependent induction of eGFP/GLT-1.
Cortical astrocytes were infected with lentiviral particles carrying an empty expression vector (control), Pax6, or shRNA directed against Pax6 (shPAX6). Five days after transduction, a cell suspension from embryonic (E17) brain tissue containing neurons and astrocytes obtained from wild-type animals (rats) was overlaid on the top of infected astrocytes. After 10 d, the levels of eGFP (a) or GLT-1 (b) were analyzed by Western blot. Fifteen μg of cell lysate was loaded in each lane. Top, Representative Western blots for eGFP and GLT-1 protein. Bottom, summary of quantification of eGFP and GLT-1 protein levels normalized to actin and expressed relative to the levels observed in control astrocytes with neurons. In all three experiments, eGFP was never detected in the absence of neurons and was observed in all experiments with neurons. Data are the mean ± SEM of four independent experiments. **p < 0.01, ***p < 0.001 compared to corresponding astrocyte control. ###p < 0.001 compared to astrocytes infected with control vector plus neurons.
Interaction of Pax6 with the distal elements on GLT-1 promoter ex vivo
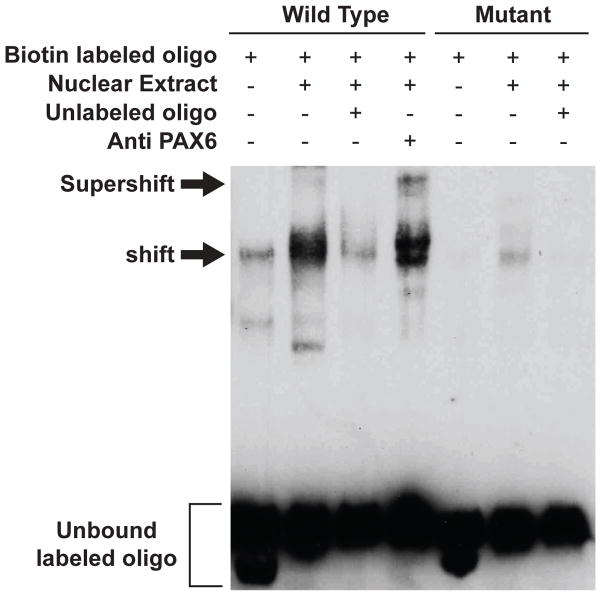
To determine if the ‘putative’ evolutionarily conserved Pax6 binding site located at -7896 relative to the translation start site is an authentic Pax6 binding site, we tested for formation of protein complexes between a synthetic biotin-labeled oligonucleotide that contains the region surrounding this site and nuclear extracts obtained from cortices of adult wild-type mice using electrophoretic mobility shift assays (EMSA). We also tested for formation of even larger complexes upon the addition of an anti-Pax6 antibody, termed supershift. Incubation of the oligonucleotide corresponding to the GLT-1 promoter region with nuclear extracts from cortex resulted in formation of complexes (labeled shift in Fig. 7). The amount of this complex was dramatically reduced by addition of excess unlabeled oligonucleotide. No binding was observed when the last three nucleotides of Pax6 binding site were changed from AAT to GGG. These controls show that the shift is specific and dependent upon the specific ‘putative’ Pax6 binding site. To determine if the complexes do in fact contain Pax6, an anti-Pax6 antibody was included in parallel incubations. The addition of anti-Pax6 antibody resulted in an additional band (see band labeled supershift in Fig. 7). We note the consistent presence of a faint band with the sample that only contained biotin labeled oligonucleotide (left lane). The simplest explanation for this band is spillover. We also note that we consistently observed a faint band with the sample that contained the mutant form of oligonucleotide and nuclear extract. This may represent lower affinity binding of Pax6 or perhaps some other protein of similar molecular weight binds to the mutated site.
Figure 7. Analysis of Pax6 binding to the ‘putative’ binding site in the evolutionarily conserved domain in the GLT-1 gene.
Biotin-labeled double-stranded oligonucleotides, identical in sequence to the region surrounding the Pax6 binding site of the GLT-1 gene (centered at -7896 upstream of the translation start site), were incubated with nuclear extracts prepared from adult mouse cortex (Wild Type). Parallel incubations included excess unlabeled oligonucleotide or anti-Pax6 antibody. A parallel set of experiments was also conduced using an oligonucleotide containing a mutation in the last three nucleotides (AAT to GGG) of the Pax6 binding site (Mutant). Shifted and supershifted oligonucleotide complexes are labeled. This analysis was conducted in four independent experiments. In one of these experiment, the bands were vertical smears. Therefore, the current figure is representative of three independent experiments.
Together, these studies suggest that Pax6 can interact with this ‘putative’ Pax6 binding site ex vivo.
Test for an interaction of Pax6 with the distal elements on GLT-1 promoter in vivo
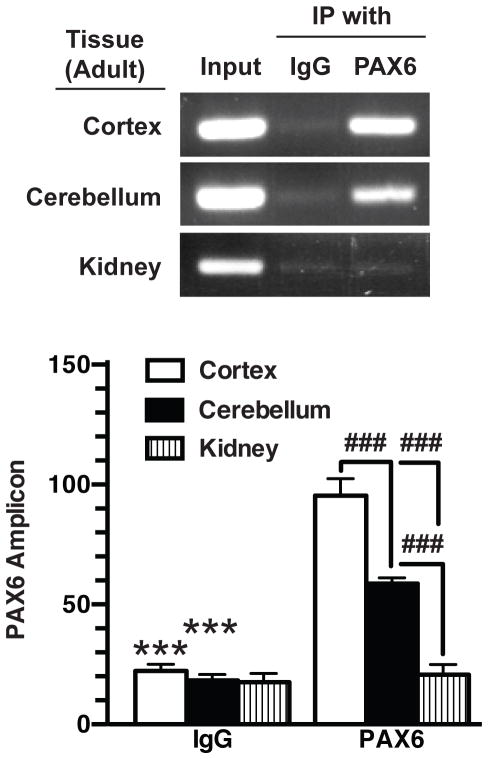
To determine if Pax6 interacts with this region of the GLT-1 promoter in vivo, anti-Pax6 antibodies were used to immunoprecipitate Pax6-DNA complexes from tissues obtained from wild-type mice. DNA sequences corresponding to the ‘putative’ Pax6 binding site were amplified from anti-Pax6 immuno-isolates of cortical tissue; this sequence was not immunoprecipitated using control antibody (Fig. 8). These ChIP studies were extended to kidney, a tissue that expresses no GLT-1 protein (Shayakul et al. 1997, Ghosh et al. 2011). There was no evidence for an interaction between Pax6 and the GLT-1 promoter in kidney. The fact that an amplified product was detected in the material used for immunoprecipitation (labeled input in Fig. 8) strongly suggests that the absence of signal is not related to inappropriate isolation of DNA from kidney. The level of GLT-1 protein is lower in cerebellum than that observed in cortex (Rothstein et al. 1994); therefore we used to ChIP to determine if there is less Pax6 binding to the promoter in cerebellar tissue. As was observed with DNA isolated from cortical tissue, a DNA fragment that includes the ‘putative’ Pax6 binding site was immunoisolated using an anti-Pax6 antibody, but the amount of interaction was lower than that observed in cortical tissue. Together these studies provide strong evidence that the evolutionarily conserved domain, that is ~8k upstream of the translation start site, contains an authentic Pax6 binding site.
Figure 8. Analysis of interaction of Pax6 with GLT-1 promoter in vivo by ChIP.
Nuclear extracts from adult, wild type mouse cortex, cerebellum, or kidney were crosslinked with formaldehyde, sheared, and immunoprecipitated with control IgG or anti-Pax6 antibodies. Eluted DNA fragments were subjected to PCR amplification with primers surrounding the putative Pax6 binding site. One percent of the sheared chromatin was quantified as input. Intensities of amplified fragments were normalized and expressed relative to that observed with input. These results are the mean ± SEM of three independent experiments. ***indicates a p value of <0.001 compared with corresponding samples isolated with anti-Pax6 antibodies. ###indicates a p value of < 0.001 for the comparisons identified.
Discussion
The proximal region of the GLT-1 promoter region is highly evolutionarily conserved (see Fig. 1) and clearly contributes to the control of GLT-1 expression (Su et al. 2003, Sitcheran et al. 2005, Yang et al. 2009, Ghosh et al. 2011), however no studies have determined if this region is sufficient to direct astrocyte expression in vivo. In unpublished studies, we show that the evolutionarily conserved domain ~8kb upstream of the translational start site is required for astrocyte-specific expression of the GLT-1. This evolutionarily conserved domain contains several evolutionarily-conserved, putative, transcription factor binding sites. In the current study, we focused on Pax6. Pax6 contains paired and homeobox DNA-binding domains and contributes to the development of a variety of tissues (for reviews, see Simpson & Price 2002, Manuel & Price 2005, Manuel et al. 2015). Humans with aniridia (absence of an iris) have mutations in Pax6 and heterozygous/homozygous deletion of Pax6 results in decreased eye size or absence of eyes in mice (for review, see Cvekl & Ashery-Padan 2014). In the nervous system, Pax6 contributes to the differential differentiation of the radial glia into neurons or astrocytes. In fact, the absence of Pax6 reduces the differentiation of radial glia into neurons (Heins et al. 2002). In agreement with an earlier study (Sakurai & Osumi 2008), we find that Pax6 is found in astrocytes and the immunoreactivity is enriched in nuclei. mRNA expression profiles show that Pax6 is expressed in astrocytes; in fact Pax6 mRNA levels are consistently ~10-fold higher in astrocytes than in other cells in the brain (Lovatt et al. 2007, Cahoy et al. 2008, Zhang et al. 2014). Relatively few studies have examined the role of Pax6 in astrocyte biology. Sakaurai and colleagues demonstrated that astrocytes isolated from Pax6 null mice display increased proliferation and higher migration potential (Sakurai & Osumi 2008). Astrocytes that lack Pax6 are also less responsive to the differentiating effects of dbcAMP (Sakurai & Osumi 2008). It is not known if the previously documented effects of dbcAMP on GLT-1 expression (Swanson et al. 1997, Schlag et al. 1998) are also dependent upon Pax6. A more recent study has demonstrated that transient ischemia causes an increase in Pax6 expression in GFAP+ cells after 24 h (Steliga et al. 2013). In the present study, we demonstrate that exogenous Pax6 increases expression of GLT-1 and does not affect expression of GLAST. As the expression of GLT-1 is a marker of astrocyte maturation, these studies suggest that Pax6 contributes to this process.
In this same evolutionarily conserved domain, there are several other evolutionarily conserved, ‘putative’ transcription factor binding sites (Table 1). The family of STAT transcription factors may be an interesting topic for future investigation. The STATs are cytoplasmic proteins that, upon activation by immune complexes or cytokines, are phosphorylated, dimerize, and translocate to the nucleus where they affect transcription of their target genes (for review, see Kanski et al. 2013). While STAT5a and STAT5b are primarily associated the mammary glands and hematopoietic cells (for review, see Grimley et al. 1999), STAT3 has been implicated in a variety of astrocytic functions, including gliogenesis, glial differentiation, and protection from reactive oxygen species (Lapp et al. 2014 for review, see Sloan & Barres 2014). Ciliary neurotrophic factor is known to activate the JAK-STAT pathway and causes a change in glycosylation of GLT-1 and redistribution to lipid rafts (Escartin et al. 2006). The JAK-STAT pathway also contributes to the regulation of glial glutamate transporter expression after hypoxia (Raymond et al. 2011). As of the date of submission of this article, pubmed searches of the other two transcription factors (GABP & ETV4 also called PEA3) when combined with the word ‘astrocyte’ reveals no publications. In other cell systems, GABP is an obligate heteromultimer with two alpha and two beta subunits. GABP and ETV4 are both members of the ets family of transcription factors that bind similar purine-rich DNA sequences. GABP is involved in many cellular functions including differentiation, cell cycle control, and cell signaling. Although it is widely expressed, it can regulate lineage-restricted genes through signaling events and protein-protein interactions (for review, see Rosmarin et al. 2004). ETV4 (PEA3) is primarily associated with oncogenesis (for review, see Oh et al. 2012) and little appears to be known of any role it plays in brain, though one study identifies a role in cortical laminar patterning (Hasegawa et al. 2004). The potential roles of these transcription factors in GLT-1 regulation will require additional studies. Furthermore, it will also be important to examine the possible interdependence of Pax6 binding with that of NF-κB and KBBP, two transcription factors that we and others have shown bind to the proximal region of the promoter and increase GLT-1 expression (see Introduction, Su et al. 2003, Sitcheran et al. 2005, Yang et al. 2009, Ghosh et al. 2011).
In the dual reporter mice that utilize the 8.3 kb promoter reporter to control expression of tdTomato and a BAC containing a very large region of the GLT-1 gene to control expression of eGFP, we find that tdTomato essentially always overlaps with eGFP (>98% tdTomato-expressing cells also express eGFP), but eGFP does not always overlap with tdTomato (Yang et al., 2011). This suggests that while 8.3 kb of the promoter is sufficient to drive expression of reporter in a subset of astrocytes, it is not sufficient to drive expression of GLT-1 in all astrocytes. This has two possible implications. First, this suggests that these two reporters may define subtypes of astrocytes. In fact, our preliminary studies with expression analyses and proteomic analyses of cells isolated using fluorescence activated cell sorting support this hypothesis (Chen et al. 2014). These data will be published as a separate study. Second, these studies suggest that these two populations of astrocytes may engage different mechanisms to control expression of GLT-1. Additional independent lines of evidence would be needed confirm this hypothesis. In addition, it will be important to understand how and why this might occur. These issues will be the subject of future investigations.
Understanding the expression of GLT-1 is likely to have an impact in two areas. First, GLT-1 expression is correlated with morphological changes that are associated with maturation of astrocytes and with maturation the nervous system (Furuta et al. 1997). It seems likely that the effects of different factors on GLT-1 expression may generalize to other markers of astrocyte differentiation. There is evidence for morphologically distinct subtypes of astrocytes (Oberheim et al. 2009) and several groups have begun to define molecular subtypes in vitro and in vivo (Bachoo et al. 2004, Imura et al. 2006, Yeh et al. 2009, Stahlberg et al. 2011, Lau et al. 2012, Benesova et al. 2012, Rusnakova et al. 2013, Kasymov et al. 2013 for reviews, see Chaboub & Deneen 2012). As the molecular subtypes of astrocytes are identified, it will be interesting to learn if Pax6 contributes to broader differences in subtypes of astrocytes. In this regard, Pax6 is differentially expressed in subpopulations of astrocytes in the spinal cord (Hochstim et al. 2008). Second, as indicated in the introduction, expression of GLT-1 is decreased in several different neurologic diseases and up-regulation of GLT-1 with the transcriptional activator, ceftriaxone, is therapeutic in several different preclinical models (for review, see Soni et al. 2014). As the field develops a better understanding of the mechanisms that contribute to control of GLT-1, it will be possible to develop drugs that more selectively target GLT-1, depending on whether the effects of transcriptional activation generalize to other astroglial proteins that may or may not be beneficial.
Acknowledgments
This work was supported by NIH grants R01 NS36465 & NS 092067 to MBR and JDR, Target ALS, Muscular Dystrophy Association, The Robert Packard Center for ALS Research. The institutional Intellectual and Developmental Disabilities Research Center (U54 HD086984) partially supports core services, including Cellular Neuroscience and Analytical Neurochemistry, that provided assistance with imaging, provision of A2B5 hybridoma supernatant, and analyses of uptake. We would also like to thank members of the Robinson laboratory for their advice and suggestions with this project.
Abbreviations
- BAC
bacterial artificial chromosome
- ChIP
chromatin immunoprecipitation
- DAPI
4′,6-diamidino-2-phenylindole
- DHK
dihydrokainate
- EAAT
excitatory amino acid transporter
- eGFP
enhanced green fluorescent protein
- GLAST
glutamate-aspartate transporter
- GLT-1
glutamate transporter-1
- JAK
janus kinase
- NF-κB
nuclear factor-κB
- STAT
signal transducer and activator of transcription
Footnotes
Conflicts of interest: None
The authors have no conflicts of interests to declare.
References
- Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. Journal of Neuroscience. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, et al. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci U S A. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beart PM, O’Shea RD. Transporters for L-glutamate: An update on their molecular pharmacology and pathological involvement. Br J Pharmacol. 2006 doi: 10.1038/sj.bjp.0706949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesova J, Rusnakova V, Honsa P, Pivonkova H, Dzamba D, Kubista M, Anderova M. Distinct expression/function of potassium and chloride channels contributes to the diverse volume regulation in cortical astrocytes of GFAP/EGFP mice. PLoS One. 2012;7:e29725. doi: 10.1371/journal.pone.0029725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub LS, Deneen B. Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Developmental neuroscience. 2012;34:379–388. doi: 10.1159/000343723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Sattler R, Ghosh M, Robinson MB, Rothstein JD. Identification and characterization of novel subtypes of astrocytes. Society for Neuroscience Abstract. 2014;305(15) [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir DV, Robinson MB, Swanson E, Zhang B, Trojanowski JQ, Lee VM, Forman MS. Impaired glutamate transport in a mouse model of tau pathology in astrocytes. J Neurosci. 2006;26:644–654. doi: 10.1523/JNEUROSCI.3861-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Progress in neurobiology. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Dunlop J, McIvain HB, She Y, Howland DS. Impaired spinal cord glutamate transport capacity and reduced sensitivity to riluzole in a transgenic superoxide dismutase mutant rat model of amytrophic lateral sclerosis. Journal of Neuroscience. 2003;23:1688–1696. doi: 10.1523/JNEUROSCI.23-05-01688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C, Brouillet E, Gubellini P, et al. Ciliary neurotrophic factor activates astrocytes, redistributes their glutamate transporters GLAST and GLT-1 to raft microdomains, and improves glutamate handling in vivo. J Neurosci. 2006;26:5978–5989. doi: 10.1523/JNEUROSCI.0302-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel M, Engele J. Pituitary adenylate cyclase-activating polypeptide (PACAP), a neuron-derived peptide regulating glial glutamate transport and metabolism. Journal of Neuroscience. 2000;20:3596–3605. doi: 10.1523/JNEUROSCI.20-10-03596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat central nervous system development. Journal of Neuroscience. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlin AB, Sinor AD, Sinor JD, Jee SH, Grinspan JB, Robinson MB. Pharmacology of sodium-dependent high-affinity L-[3H]glutamate transport in glial cultures. Journal of Neurochemistry. 1995;64:2572–2580. doi: 10.1046/j.1471-4159.1995.64062572.x. [DOI] [PubMed] [Google Scholar]
- Gegelashvili G, Danbolt NC, Schousboe A. Neuronal soluble factors differentially regulate the expression of the GLT1 and GLAST glutamate transporters in cultured astroglia. J Neurochem. 1997;69:2612–2615. doi: 10.1046/j.1471-4159.1997.69062612.x. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Yang Y, Rothstein JD, Robinson MB. Nuclear Factor-kB contributes to neuron-dependent induction of GLT-1 expression in astrocytes. Journal of Neuroscience. 2011;31:9159–9169. doi: 10.1523/JNEUROSCI.0302-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Martin LJ, Rothstein JD. Regional deafferentation down-regulates subtypes of glutamate transporter proteins. Journal of Neurochemistry. 1995;65:2800–2803. doi: 10.1046/j.1471-4159.1995.65062800.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Rothstein JD, Price DL, Martin LJ. Fimbria-fornix transections selectively down-regulate subtypes of glutamate transporter and glutamate receptor proteins in septum and hippocampus. Journal of Neurochemistry. 1996;67:1208–1216. doi: 10.1046/j.1471-4159.1996.67031208.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez MI, Krizman-Genda E, Robinson MB. Caveolin-1 Regulates the Delivery and Endocytosis of the Glutamate Transporter, Excitatory Amino Acid Carrier 1. J Biol Chem. 2007;282:29855–29865. doi: 10.1074/jbc.M704738200. [DOI] [PubMed] [Google Scholar]
- Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine & growth factor reviews. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Ashigaki S, Takamatsu M, Suzuki-Migishima R, Ohbayashi N, Itoh N, Takada S, Tanabe Y. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J Neurosci. 2004;24:8711–8719. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugeto O, Ullensvang K, Levy LM, Chaudhry FA, Honore T, Nielsen M, Lehre KP, Danbolt NC. Brain glutamate transporter proteins form homomultimers. J Biol Chem. 1996;271:27715–27722. doi: 10.1074/jbc.271.44.27715. [DOI] [PubMed] [Google Scholar]
- Hein C, Horvath E, Kugler P. Glutamate transporter expression in astrocytes of the rat dentate gyrus following lesion of the entorhinal cortex. European Journal of Neuroscience. 2001;13:1839–1848. doi: 10.1046/j.0953-816x.2001.01559.x. [DOI] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura T, Nakano I, Kornblum HI, Sofroniew MV. Phenotypic and functional heterogeneity of GFAP-expressing cells in vitro: differential expression of LeX/CD15 by GFAP-expressing multipotent neural stem cells and non-neurogenic astrocytes. Glia. 2006;53:277–293. doi: 10.1002/glia.20281. [DOI] [PubMed] [Google Scholar]
- Kanski R, van Strien ME, van Tijn P, Hol EM. A star is born: new insights into the mechanism of astrogenesis. Cellular and molecular life sciences : CMLS. 2013 doi: 10.1007/s00018-013-1435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasymov V, Larina O, Castaldo C, Marina N, Patrushev M, Kasparov S, Gourine AV. Differential Sensitivity of Brainstem versus Cortical Astrocytes to Changes in pH Reveals Functional Regional Specialization of Astroglia. J Neurosci. 2013;33:435–441. doi: 10.1523/JNEUROSCI.2813-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SG, Kegelman TP, et al. Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: opportunities for developing novel therapeutics. Journal of cellular physiology. 2011;226:2484–2493. doi: 10.1002/jcp.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp DW, Zhang SS, Barnstable CJ. Stat3 mediates LIF-induced protection of astrocytes against toxic ROS by upregulating the UPC2 mRNA pool. Glia. 2014;62:159–170. doi: 10.1002/glia.22594. [DOI] [PubMed] [Google Scholar]
- Lau CL, Perreau VM, Chen MJ, Cate HS, Merlo D, Cheung NS, O’Shea RD, Beart PM. Transcriptomic profiling of astrocytes treated with the Rho kinase inhibitor fasudil reveals cytoskeletal and pro-survival responses. Journal of cellular physiology. 2012;227:1199–1211. doi: 10.1002/jcp.22838. [DOI] [PubMed] [Google Scholar]
- Lee SG, Su ZZ, Emdad L, Gupta P, Sarkar D, Borjabad A, Volsky DJ, Fisher PB. Mechanism of ceftriaxone induction of excitatory amino acid transporter-2 expression and glutamate uptake in primary human astrocytes. J Biol Chem. 2008;283:13116–13123. doi: 10.1074/jbc.M707697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy LM, Lehre KP, Walaas SI, Storm-Mathisen J, Danbolt NC. Down-regulation of glial glutamate transporters after glutamatergic denervation in the rat brain. European Journal of Neuroscience. 1995;7:2036–2041. doi: 10.1111/j.1460-9568.1995.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Lewerenz J, Albrecht P, Tien ML, et al. Induction of Nrf2 and xCT are involved in the action of the neuroprotective antibiotic ceftriaxone in vitro. J Neurochem. 2009;111:332–343. doi: 10.1111/j.1471-4159.2009.06347.x. [DOI] [PubMed] [Google Scholar]
- Li LB, Toan SV, Zelenaia O, Watson DJ, Wolfe JH, Rothstein JD, Robinson MB. Regulation of astrocytic glutamate transporter expression by Akt: Evidence for a selective transcriptional effect on the GLT-1/EAAT2 subtype. Journal of Neurochemistry. 2006;97:759–771. doi: 10.1111/j.1471-4159.2006.03743.x. [DOI] [PubMed] [Google Scholar]
- Lovatt D, Sonnewald U, Waagepetersen HS, et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27:12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosenberg NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry. 1951;193:265–275. [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nature methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M, Price DJ. Role of Pax6 in forebrain regionalization. Brain research bulletin. 2005;66:387–393. doi: 10.1016/j.brainresbull.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Manuel MN, Mi D, Mason JO, Price DJ. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Frontiers in cellular neuroscience. 2015;9:70. doi: 10.3389/fncel.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochimica et biophysica acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Li P, Mangin JM, Huntsman M, Gallo V. Chronic perinatal hypoxia reduces glutamate-aspartate transporter function in astrocytes through the Janus kinase/signal transducer and activator of transcription pathway. J Neurosci. 2011;31:17864–17871. doi: 10.1523/JNEUROSCI.3179-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MB, Dowd LA. Heterogeneity and functional properties of subtypes of sodium-dependent glutamate transporters in the mammalian central nervous system. Advances in Pharmacology. 1997;37:69–115. doi: 10.1016/s1054-3589(08)60948-5. [DOI] [PubMed] [Google Scholar]
- Robinson MB, Hunter-Ensor M, Sinor J. Pharmacologically distinct sodium-dependent L-[3H]glutamate transport processes in rat brain. Brain research. 1991;544:196–202. doi: 10.1016/0006-8993(91)90054-y. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood cells, molecules & diseases. 2004;32:143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Martin L, Levey AI, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl RW. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Patel S, Regan MR, et al. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- Rusnakova V, Honsa P, Dzamba D, Stahlberg A, Kubista M, Anderova M. Heterogeneity of astrocytes: from development to injury - single cell gene expression. PLoS One. 2013;8:e69734. doi: 10.1371/journal.pone.0069734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Osumi N. The neurogenesis-controlling factor, Pax6, inhibits proliferation and promotes maturation in murine astrocytes. J Neurosci. 2008;28:4604–4612. doi: 10.1523/JNEUROSCI.5074-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag BD, Vondrasek JR, Munir M, Kalandadze A, Zelenaia OA, Rothstein JD, Robinson MB. Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Molecular Pharmacology. 1998;53:355–369. doi: 10.1124/mol.53.3.355. [DOI] [PubMed] [Google Scholar]
- Shayakul C, Kanai Y, Lee WS, Brown D, Rothstein JD, Hediger MA. Localization of the high-affinity glutamate transporter EAAC1 in rat kidney. American Journal of Physiology. 1997;273:F1023–F1029. doi: 10.1152/ajprenal.1997.273.6.F1023. [DOI] [PubMed] [Google Scholar]
- Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochemistry international. 2007;51:333–355. doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HW, Scofield MD, Boger H, Hensley M, Kalivas PW. Synaptic glutamate spillover due to impaired glutamate uptake mediates heroin relapse. J Neurosci. 2014;34:5649–5657. doi: 10.1523/JNEUROSCI.4564-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Price DJ. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- Sitcheran R, Gupta P, Fisher PB, Baldwin AS. Positive and negative regulation of EAAT2 by NF-kappaB: a role for N-myc in TNFalpha-controlled repression. EMBO Journal. 2005;24:510–520. doi: 10.1038/sj.emboj.7600555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Barres BA. Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr Opin Neurobiol. 2014;27:75–81. doi: 10.1016/j.conb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni N, Reddy BV, Kumar P. GLT-1 transporter: An effective pharmacological target for various neurological disorders. Pharmacology, biochemistry, and behavior. 2014;127C:70–81. doi: 10.1016/j.pbb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Stahlberg A, Andersson D, Aurelius J, Faiz M, Pekna M, Kubista M, Pekny M. Defining cell populations with single-cell gene expression profiling: correlations and identification of astrocyte subpopulations. Nucleic acids research. 2011;39:e24. doi: 10.1093/nar/gkq1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steliga A, Waskow M, Karwacki Z, Wojcik S, Lietzau G, Klejbor I, Kowianski P. Transcription factor Pax6 is expressed by astroglia after transient brain ischemia in the rat model. Folia neuropathologica/Association of Polish Neuropathologists and Medical Research Centre, Polish Academy of Sciences. 2013;51:203–213. doi: 10.5114/fn.2013.37704. [DOI] [PubMed] [Google Scholar]
- Su ZZ, Leszczyniecka M, Kang DC, Sarkar D, Chao W, Volsky DJ, Fisher PB. Insights into glutamate transport regulation in human astrocytes: cloning of the promoter for excitatory amino acid transporter 2 (EAAT2) Proc Natl Acad Sci U S A. 2003;100:1955–1960. doi: 10.1073/pnas.0136555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland ML, Delaney TA, Noebels JL. Glutamate transporter mRNA expression in proliferative zones of the developing and adult murine CNS. Journal of Neuroscience. 1996;16:2191–2207. doi: 10.1523/JNEUROSCI.16-07-02191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RA, Liu J, Miller JW, Rothstein JD, Farrell K, Stein BA, Longuemare MC. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Watase K, Manabe T, et al. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Wadiche JI. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nature reviews Neuroscience. 2007;8:935–947. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- Vandenberg RJ, Ryan RM. Mechanisms of glutamate transport. Physiol Rev. 2013;93:1621–1657. doi: 10.1152/physrev.00007.2013. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Watkins A, et al. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61:880–894. doi: 10.1016/j.neuron.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59:200–207. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TH, Lee da Y, Gianino SM, Gutmann DH. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia. 2009;57:1239–1249. doi: 10.1002/glia.20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaia O, Schlag BD, Gochenauer GE, Ganel R, Song W, Beesley JS, Grinspan JB, Rothstein JD, Robinson MB. Epidermal growth factor receptor agonists increase expression of glutamate transporter GLT-1 in astrocytes through pathways dependent on phosphatidylinositol 3-kinase and transcription factor NF-κB. Molecular Pharmacology. 2000;57:667–678. doi: 10.1124/mol.57.4.667. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]