Abstract
Neuroinflammation is a hallmark of acute and chronic neurodegenerative disorders. Activated microglia and secreted factors such as tumor necrosis factor-alpha (TNFα) are key mediators of neuroinflammation and may contribute to neuronal dysfunction. The main aim of this study was to evaluate the therapeutic efficacy of intranasal cationic nanoemulsions encapsulating an anti-TNFα siRNA, for potential anti-inflammatory therapy, tested in an LPS induced model of neuroinflammation. The strategy of developing a cationic nanoemulsion system for silencing the TNFα gene was to efficiently provide neuroprotection against inflammation. TNFα siRNA nanoemulsions were prepared and characterized for particle size, surface charge, morphology, and stability and encapsulation efficiency. Qualitative and quantitative intracellular uptake studies by confocal imaging and flow cytometry, respectively, showed higher uptake compared to Lipofectamine® transfected siRNA. Nanoemulsions showed significantly lower (p<0.01) levels of TNFα in LPS-stimulated cells. Upon intranasal delivery of cationic nanoemulsions almost 5 fold higher uptake was observed in the rat brain compared to non-encapsulated siRNA. More importantly, intranasal delivery of TNFα siRNA nanoemulsions in vivo markedly reduced the unregulated levels of TNFα in an LPS-induced model of neuroinflammation where TNFα functions as a signaling molecule, which aggravates inflammation. These results indicate that intranasal delivery of cationic nanoemulsions encapsulating TNFα siRNA offered an efficient means of gene knockdown and this approach has significant potential in prevention of neuroinflammation.
Keywords: Neuroinflammation, tumor necrosis factor-alpha, gene silencing, small interfering RNA, cationic nanoemulsion, brain delivery
1. BACKGROUND
Neuroinflammatory signaling has long been known to be promote the pathogenesis of various neurodegenerative diseases, such as Alzheimer's disease (AD), amyotrophic lateral sclerosis (ALS), Huntington’s disease, Parkinson's disease (PD), head trauma, epilepsy, and stroke [1–5]. These disorders are devastating and expensive, with an annual cost currently exceeding several hundred billion dollars in the United States alone, and current palliative treatments are inadequate [6]. Neuroinflammation may be initiated in response to a variety of stimuli, including infection, traumatic brain injury, toxic metabolites, or autoimmunity. In the central nervous system (CNS), including the brain and spinal cord, microglia are the resident innate immune cells that are activated in response to these cues. Evidence supports that the unregulated activation of microglia in response to environmental toxins, endogenous proteins, and neuronal death results in the production of toxic factors that propagate neuronal injury. Upon stimulation by endogenous or exogenous factors, microglia produce a large number of factors (e.g., IL-1, TNFα, NO, PGE2, superoxide) that are toxic to neurons [7–9]. If the initial stimulus that elicited microglial activation is not resolved, a self-sustaining cycle of neuroinflammation can ensue and such a chronic inflammatory environment is likely to provoke neurotoxic processes in several neurodegenerative conditions. Therefore, timely delivery of anti-inflammatory regimens in patient populations identified to be at risk may afford neuroprotective effects, especially in chronic neurodegenerative diseases, such as AD and PD.
One of the strong candidate trigger proteins in neuroinflammation, and thus a potential target for therapeutic manipulation, is the potent pro-inflammatory/pro-apoptotic cytokine tumor necrosis factor-alpha (TNFα). It has been demonstrated to play a major role in central nervous system (CNS) neuroinflammation-mediated cell death in AD [10–13], PD [14, 15] and HIV-associated dementia [16] as well as several other CNS complications. Recently, agents that modulate the levels of circulating peripheral TNFα have been shown to be worthwhile biological therapeutic agents with the use of etarnecept (Enbrel®) and infliximab (Remicade®), both of which display beneficial properties against rheumatoid arthritis and other peripheral inflammatory diseases. Unfortunately, these agents are largely unable to penetrate the blood-brain barrier (BBB), which severely limits their use in the setting of neuroinflammation in the CNS. Mostly, the current research has been focused on understanding the mechanism(s) and pathological pathways underlying the various neurodegenerative diseases however, delivery of therapeutic molecules at targeted sites in CNS still remains to be a major clinical advancement that has yet to be achieved.
RNA interference (RNAi) technology shows great potential for the targeted modulation of gene expression in neuroinflammation. RNAi therapy utilizes short interfering RNA (siRNA) duplexes, usually composed of 20–25 nucleotides, and is involved in post-translational gene-silencing mechanisms. When delivered efficiently to the CNS and to the cytosol of the microglial cells, the antisense strand of the siRNA will interact with the RNA inducing silencing complex (RISC) and breakdown the complementary mRNA sequence with the action of the Argonaut enzymes. Ultimately, this approach will block the expression of specific genes, such as TNFα overexpressed in neuroinflammation. The exquisite selectivity and high potency of appropriately designed siRNA sequences affords an excellent therapeutic opportunity for prevention of neuroinflammation without associated toxicity. However, for translating RNAi from an experimental approach to a clinically viable therapeutic strategy that can benefit patients, there is a critical need for a safe and effective delivery system that can package the labile payload, afford stability during transport, permeability through tissue and cell membrane barriers, as well as cytosolic bioavailability for efficient gene silencing. For diseases of the CNS, opportunity for systemic RNAi therapy is further dampened by the formidable BBB, blood-CSF barrier and presence of efflux transporters, which limits transport of up to 98% of small molecule therapeutics and 100% of large molecular weight hydrophilic compounds, such as proteins and nucleic acid constructs [17].
In the last few years, intranasal (I.N.) administration has emerged as a non-invasive delivery option, which bypasses the BBB and allows for direct access of various biologics-based drugs to the CNS [18]. In addition, this route circumvents drug elimination by the liver and gastrointestinal tract, and enzymatic degradation in serum. This route does offer enhanced patient compliance when compared to the parenteral route. However, the nasal epithelium has some limitations such as poor absorption, enzymatic degradation, and poor permeation and retention of specifically hydrophilic drugs.
There is a need for a safe and effective delivery system that could provide penetration through the nasal olfactory epithelium and could help in overcoming the intracellular delivery barrier for siRNA. In this study, we formulated siRNA against TNFα, in a cationic nanoemulsion of omega-3 fatty acids and investigated the delivery through the intranasal route to overcome some of the associated challenges and to improve CNS delivery for down regulating the target gene in the CNS. The hypothesis for this approach is that siRNA complexed with cationic lipids in a nanoemulsion will be protected from enzymatic degradation in the nasal mucosa. Additionally, the cationic nanoemulsion formulation will provide higher mucosal residence time for higher exposure and lastly, the intranasal route of administration will provide direct access to the brain to show down-regulation of TNFα in a surgical lipopolysaccharide (LPS)-induced experimental neuroinflammation model established in female Sprague-Dawley rats.
2. MATERIALS AND METHODS
2.1. Materials
Extra pure high omega-3 fatty acid-containing flaxseed oil was kindly provided by Jedwards International (Quincy, MA). Lipoid E80® was purchased from Lipoid GMBH (Ludwigshafen, Germany). Tween 80® and stearylamine were purchased from Sigma Chemicals, Inc. (St. Louis, MO). (N-[1-(2, 3-dioleoyloxy) propyl]-N, N, N-trimethylammoniumsalt) (DOTAP) cationic lipid was purchased from Avanti polar lipids, Inc. (Alabaster, AL). A 21-base pair siRNA oligonucleotide duplex for silencing TNFα gene with the following sequence ucuucuGucuAcuGAAcuudTsdT (sense strand 5'-3') and AAGUUcAGuAGAcAGAAGAdTsdT (antisense Strand 5'-3') was kindly provided by Alnylam Pharmaceuticals (Cambridge, MA). SYBR® Gold dye and RNase enzyme were obtained from Invitrogen (Carlsbad, CA). All of the primer sequences and PCR reagents were purchased from Invitrogen (Carlsbad, CA). Lipopolysaccharide (LPS) (Escherichia coli 0111:B4) and endotoxin-tested/sterile phosphate buffered saline (PBS) were purchased from Sigma Chemicals (St. Louis, MO). A Harvard infusion pump setup was generously provided by Dr. Barbara Calderone’s laboratory at the Harvard Neuro Discovery Center (Boston, MA). A stereotaxic surgical apparatus was obtained from Dr. Craig Ferris’s laboratory in the Center for Translational Neuro Imaging (CTNI) at Northeastern University (Boston, MA).
2.2. Preparation and Characterization of siRNA-Encapsulated Nanoemulsion Formulations
Flaxseed oil-containing cationic nanoemulsion formulations encapsulating a TNFα-silencing siRNA duplex were prepared by using the cationic lipid DOTAP, which is essential in pre-condensing the negatively-charged oligonucleotide duplex and facilitates inclusion in the oil phase of the nanoemulsion for enhanced stability of the oligonucleotide duplexes. Briefly, DOTAP was first dissolved in ethanol and the siRNA stock in distilled water was added to the DOTAP solution at a specific nitrogen-to-phosphorous (N/P) ratio of 3.72:1. After complexation of the siRNA duplex with DOTAP, flaxseed oil was gradually added to the mixture to help DOTAP integration in the oil phase followed by complete evaporation of the ethanol by bubbling nitrogen. Aqueous phase containing Lipoid® E80 and Tween® 80 was then subsequently added dropwise to the oil phase. The resultant mixture was stirred for 2 minutes using a Silverson homogenizer (Model: L4RT-A, Silverson Machines, Dartmouth, MA) at 6,000 rpm and ultra-sonicated for 10 minutes using the Vibra Cell® VC 505 probe sonicator (Sonics and Material Inc., Newtown, CT) at 22% amplitude and 50% duty cycle. Blank nanoemulsions were prepared similarly without the addition of siRNA duplexes.
The blank and siRNA-encapsulated nanoemulsions (SNE) were characterized for particle size, surface charge, morphology, encapsulation efficiency, and stability of the siRNA payload. Particle size, surface charge, and polydispersity index (PDI) of freshly prepared nanoemulsion formulations were measured using a ZetaSizer Nano ZS (Malvern Instruments, Worcestershire, UK). Each sample was analyzed in triplicate at 25 °C, and the size and zeta-potential were reported as means ± SD. The morphological characterization of the oil droplets in the nanoemulsion formulations was carried out with transmission electron microscopy (TEM) using a JEOL JEM-1000 transmission electron microscope (JEOL, Tokyo, Japan). Ten microliters of the nanoemulsion was first diluted hundred times with distilled water and was placed on Formwar-coated copper grids (EM Sciences, Hatfield, PA, USA); excess fluid was drained using whatman filter paper, and the sample was negatively-stained at room temperature with 50 µL of 1.5% (w/v) phosphotungstic acid (10 minutes) prior to the observation.
2.3. siRNA Encapsulation Efficiency and Stability
Quantitative siRNA encapsulation efficiency in the cationic nanoemulsion formulations was determined using a SYBR® Gold nucleic acid reagent (Life Technologies, Carlsbad, CA) using a Bio-Tek Instruments microplate reader (Winooski, VT). A standard curve based on fluorescence emission generated from the binding of SYBR-Gold to known concentrations of double stranded siRNA was created, and the loading in the nanoemulsion was determined by treating the nanoemulsion formulation with and without Triton X-100 detergent. Total siRNA loading was calculated by subtracting the amount of free siRNA (obtained from without Triton treatment) from total siRNA (obtained from with Triton treatment). Encapsulation efficiency was defined as the ratio percent of siRNA encapsulated in nanoparticles to the total amount of siRNA added.
Qualitative assessment of encapsulated siRNA was also performed using an agarose gel retardation assay based on the principle that if siRNA is efficiently bound to DOTAP in the nanoemulsion, migration into the gel will be completely retarded. Further siRNA nanoemulsions, once treated with a 10% Triton X-100 surfactant will release the entrapped siRNA which will move freely in the gel and will be detected as free bands. The nanoemulsion formulation was prepared, as previously described, at a concentration of 0.5 mg/mL in a N/P ratio of 3.72:1 were subjected to the following different conditions: (1) no treatment, where 10uL SNE was incubated for 15 minutes with 10 µL phosphate buffer at pH 7.4; (2) low Triton® X-100 treatment, where 10 µL of SNE was incubated for 15 minutes with 10 µL of 4% Triton® X-100; and (3) high Triton treatment, where 10 µL of SNE was incubated for 15 minutes with 10 µL of 10% Triton® X-100. Resulting mixtures were applied to a 4% E-Gel (Invitrogen, Carlsbad, CA), and electrophoresis was carried out for 15 minutes using the E-Gel iBase system. Free siRNA was applied as a reference standard. The resulting gel was imaged on a ChemiDoc system (Bio Rad, Waltham, MA) using the imaging software equipped with the instrument.
Lastly, we investigated the stability of siRNA encapsulated in the nanoemulsion against RNAse-A digestion. The nanoemulsion used was prepared as previously described and in an N/P ratio of 3.72:1. The siRNA encapsulated nanoemulsion (SNE) formulations were subjected to the following conditions: (1) SNE sample (10 µL) was incubated with 10 µL RNAse-A (20 µg/mL) enzyme at 37°C for 15 minutes followed by dilution with 10 µL of phosphate buffer; (2) SNE sample (10 µL) was incubated with 10µL RNAse-A (20 µg/mL) enzyme at 37°C for 15 minutes followed by dilution with 10 µL of phosphate buffer containing 10% Triton® X-100 to release the entrapped siRNA; (3) SNE 10 µL was incubated for 15 minutes with 10 µL of phosphate buffer; and (4) SNE 10 µL was incubated with 10µL of phosphate buffer for 15 minutes followed by dilution with 10 µL of 10% Triton® X-100 to release the entrapped siRNA. Lastly, free siRNA was used as reference. The extract from each of the above samples was loaded in the well of a 4% pre-stained E-Gel (Invitrogen, Carlsbad, CA), and electrophoresis was carried out for 15 minutes using the E-Gel iBase system. The resulting gel was imaged on ChemiDoc System (Bio Rad, Waltham, MA) and digitized data was analyzed with accompanying software.
2.4. Cellular Uptake Studies and Cytotoxicity Evaluations
In order to evaluate and compare uptake and anti-inflammatory efficacy of TNFα siRNA formulation, we have used J774A.1 adherent murine macrophages. The cells were purchased from American Type Culture Collections (ATCC, Rockville, MD) and were cultured in DMEM medium from Invitrogen/Life Technologies (Carlsbad, CA) at 37 °C under 5% CO2. Cells were seeded into culture flasks for 7 days with medium supplemented with 10% fetal bovine serum.
Fluorescence confocal microscopy studies were performed in order to assess the qualitative cellular internalization of the nanoparticles. Cells (200k cells/well) were seeded on coverslips in 6-well plates and were allowed to adhere for 24 hours. To evaluate the uptake and cellular internalization of siRNA, firstly TNFα specific siRNA duplex was labeled with Cy3 dye using the silencer siRNA labeling kit from Life Sciences (Carlsbad, CA) according to manufacturer’s specifications. Nanoemulsion formulations containing Cy3 label siRNA were prepared according to the previously described method. Cells were incubated with 200 nMolar concentration of either Cy3-labeled siRNA complexed with Lipofectamine®, a cationic lipid-based transfection reagent, or with Cy3-labeled TNFα siRNA nanoemulsion (SNE). After incubation for specific periods of time, cells were washed with cold phosphate buffered saline (PBS 1× p H 7.4), fixed with 4% paraformaldehyde for 20min, washed with cold PBS, and stained with the fluorescent DNA-binding dye Hoechst® 3342 (Invitrogen, Carlsbad, CA) (1ug/mL) for 5 min. Digital images were captured by LSM 700® confocal microscope (Carl Zeiss, Gottingen, Germany) and analyzed using the NIH Image-J software. All setting parameters for fluorescence detection and images analyses were held constant to allow consistency in imaging of the sample for comparison.
TNFα siRNA conjugated with Cy3-label dye as previously mentioned. For flow cytometry 200k cells/well were seeded in 6-well plates and left to adhere for 24 h. Cells were then transfected with fluorescently labeled siRNA (Cy3-TNFα silencing) Lipofectamine® complexed and Cy3-siRNA encapsulated in nanoemulsion (SNE). After specific times, the transfection solutions was removed, cells were washed with PBS and prepared for analysis by flow cytometry. The time dependent cellular uptake of siRNA nanoemulsion was quantitatively analyzed using a Becton Dickinson FACS-Calibur® 4 color flow cytometer (BD Biosciences, Franklin Lakes, and NJ). The results obtained were analyzed using Cell-Quest Pro-software cytometer (BD Biosciences, Franklin Lakes, and NJ). The relative amounts of intracellular siRNA were evaluated based on FL-2 channel (585/42 emission). The percentage of cellular uptake was calculated based on the geometric mean (Gm) using the following formula: percent uptake = (Gmexp)/ (Gmctrl)*100 where Gmexp is the Gm of cells in the different experimental conditions and Gmctrl is the Gm of cells in the control condition, without treatment. The data presented are the mean fluorescent signals from a total of 10,000 events within a gated region. The data is presented as percent uptake compared to control untreated cells.
The acute tolerability of the siRNA-loaded nanoemulsions was assessed using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Cells were seeded at a density of 10,000 cells/well in 96-well plate; after 24 hours, cells were rinsed and incubated with different concentrations (2, 10, 50 and 100nM) of siRNA-encapsulated nanoemulsion diluted with the cell medium. Treatment with cell media was used as a negative control and treatment with poly(ethylene imine) (PEI, M.wt. 10kDa), a cationic cytotoxic polymer, was used as a positive control. Following 6 h and 19 h of treatment time, the cells were rinsed with fresh media and 50 uL of MTT reagent (Vybrant® MTT Cell Proliferation Assay Kit, Life Technologies, Carlsbad, CA) was added. Two hours after incubation, the medium was replaced by 200 µL of dimethyl sulfoxide (DMSO) to stop the reaction and lyse the cells. During the incubation period, the colorless MTT dye was converted to formazan by the live cells. After shaking the plates for few minutes, absorbance of the solution was measured at 490 nm wavelength using a BioTek Synergy® HT microplate reader. The percent cell viability was calculated from the absorbance values relative to those of untreated cells. The samples were tested with n = 8 replicates.
2.5. In Vitro Gene Silencing in LPS-Stimulated J774A.1 Cells
In order to evaluate and compare the anti-inflammatory effect of TNFα silencing small interfering RNA (siRNA) formulations, J774A.1 adherent murine macrophage stimulated with LPS were used as an in vitro model of activated microglia. Bacterial LPS has been extensively used in models studying inflammation as it mimics many inflammatory effects of cytokines, such as up-regulation of cytokines such as TNFα, IL-1β, and IL-6 [19, 20]. LPS is the most abundant component within the cell wall of gram-negative bacteria. It can stimulate the release of inflammatory cytokines in various cell types, leading to an acute inflammatory response towards pathogens. We evaluated the gene silencing efficacy of TNFα silencing siRNA in J774A.1 macrophages before we could evaluate the in vivo distribution and knockdown in an LPS induced model of neuroinflammation in rats. J774A.1 cells were plated in 6-well microplates at 200k density and on day 2, they were treated with siRNA nanoemulsions at various concentrations (10 nM, 50 nM, and 100 nM) and were compared to treatment with siRNA administered with Lipofectamine® complexation. We have used a non-specific (scrambled) siRNA incorporated in the nanoemulsions as a negative control (SNE-NTC). After 24 hours of pre-treatment, cells were exposed to 100 ng/mL LPS for 6 hours. Following 6 hours of stimulation, the supernatant was collected and cells were washed and collected by centrifugation at 5,000g for 10 min. The cell pellet was used for RNA extraction using the manufactures protocol (Roche, Indianapolis, IN). An RNA quantitation and integrity test was performed before the use of RNA in the RT step. Gene quantitation was performed in two steps; reverse transcription (RT) step- cDNA is reverse transcribed from 100 ng of RNA, followed by PCR step where PCR products are quantitatively synthesized from 5µL cDNA samples using the TaqMan Gene Expression master mix according to TaqMan gene expression assay kit instructions (Invitrogen, Carlsbad, CA). The Taqman probe/primer for target gene TNFα was ordered from Invitrogen with a FAM label and β-actin endogenous gene probe/primer set was ordered with the VIC probe label for distinction from the target gene detection. Samples were analyzed by relative quantitation method by calculating ΔΔCt values for each treatment.
2.6. Nose-to-Brain siRNA Delivery in Sprague-Dawley Rats
2.6.1. Experimental Animals
All of the experiments with female Sprague-Dawley rats described in this publication were approved by the Northeastern University’s Institutional Animal Care and Use Committee and the University’s Biological Safety Committee. Sprague-Dawley rats, weighing between 190 and 210 g, were purchased from Charles River Laboratories (Cambridge, MA) and housed under a 12 h light/dark cycle with food and water provided ad libitum. The experimental animal protocols were carried out in accordance with institutional guidelines and animals were allowed to acclimate for at least 48 hours prior to initiation of any experiments.
2.6.2. Quantitative siRNA Levels in the Brain and Plasma upon I.N. Administration
In order to determine if I.N. administration of cationic nanoemulsion formulations can successfully achieve higher uptake in the brain compared to non-formulated siRNA, the duplexes were encapsulated siRNA in cationic charged nanoemulsions as described previously. Sprague-Dawley rats were anesthetized with an intraperitoneal injection of ketamine/xylazine hydrochloride. Body temperatures were maintained at 37 °C using a heat lamp. The animals were randomly divided into five groups (n=3); animals in the first group were administered TNFα siRNA (STNF-6h) in saline solution and scarified after 6 hours from the first administered dose, those in the second group were administered TNFα siRNA (STNF-24h) in saline solution and sacrificed after 24 h from first administered dose, and animals in the third and fourth group were administered siRNA nanoemulsions (SNE-6h) and sacrificed after 6 h (SNE-6h) and after 24 h (SNE-24) respectively. The last group was administered vehicle (PBS) alone as a control group.
Dosing was performed by administration of 5 µL/nostril of the siRNA nanoemulsion (SNE) or siRNA solution using a micropipette (Eppendorf P-20), with a hold time of 1 minute between each dose. A drop was placed at the opening allowing the animal to snort the drop into nasal cavity. After the first administration, second dosing was performed under isoflurane to minimize the anesthesia overdose. The siRNA-encapsulated nanoemulsion (SNE) or siRNA solution controls in saline were injected every 2 h for 6 h and for 24 h dosing, formulation was injected 2 times on day 1 and day 2 according to the protocol below. The total siRNA-TNFα dose administered to each rat was 3mg/kg.
Thirty-minutes after the last injection, blood samples and the whole brain were collected. The brain tissues and dissected brain sections were quickly rinsed with saline and blotted with filter paper to remove the blood taint as much as possible. Plasma was separated from whole blood after centrifuging at 2,000g for 10 minutes at 4°C. The brain tissue was then homogenized using a Qiagen Tissue Lyser (Qiagen, Germantown, MD) to prepare tissue lysates. The homogenized tissue lysates were subsequently diluted at 1:500 to prepare dilute samples. Using the appropriate reverse primer, anti-primer and the tissue lysate, the annealing step was run initially followed by RT-PCR. The TNFα siRNA sequence, reverse primer, forward primer, and an anti-primer sequence used in the assay are listed below.
Reverse: GGAAGCCGACAAGGCGTAA
Forward: /56-FAM/ACTCCCTCCCTCGATTTAAGTTCAGTAGACA
Anti-primer: AAATCGAGGGAGGGAG/3BHQ_1/
As a first step, the siRNA was denatured and annealed to the RT primer (6 µL diluted siRNA and 18 µL reverse primer, 100 nM). The siRNA duplex was denatured by incubating at 95 °C for 5 minutes. Primers were then annealed by 2 minutes incubation at 80°C, 70°C, 60°C and 45 °C with 4 °C hold. Then the reverse transcription reaction was carried out as follows. A master mix was made by mixing the following components: RT-PCR buffer (6.25 µL), forward primer (10 µM, 0.12 µL), reverse primer (10 µM, 0.12 µL), anti-primer (100 µM, 0.12 µL), 25 × RT-PCR enzyme (0.5 µL) and water (1.5 µL). A total of 17 µL of this master mix was then mixed with 7 µL of sample and ran the PCR at the following listed conditions: 50 °C (10 minutes), 95 °C (10 minutes), 40 cycles, 95 °C (15 s), 45 °C (60 s). The amplified siRNAs were finally detected and quantitated by running a standard curve using lysate from untreated vehicle rat tissue or plasma and spiked with known siRNA concentrations ranging from 2,000 ng/mL to 0.002 ng/mL. For rat plasma, samples were first diluted 1000-fold with PBS and quantitation was performed similar to the brain samples. Statistical analysis was performed using the two-tailed unpaired t-test to compare the mean values standard errors and to determine if there was any significant difference between delivery systems; p values of <0.05 were considered to be statistically significant.
2.7. LPS-Induced Neuroinflammation Model and In Vivo Gene Silencing
Due to the central role of inflammation in various diseases, the need for purely inflammation-driven animal’s model has emerged. We have utilized an in vivo LPS-induced PD model that was initially developed by Castano, et al. [21]. The LPS-induced PD model has been widely accepted and used for understanding the pathogenesis of PD and for testing various anti-inflammatory treatment therapies for targeting neuroinflammation [22–25].
Although the combination of various factors is thought to contribute to the neurodegenerative processes both in cell culture systems and animal models, the precise mechanisms of action remain to be elucidated. In order to evaluate the potential protective effect of anti-inflammatory TNFα siRNA in LPS-induced degeneration of nigral dopaminergic neurons, we compared the gene silencing effect of I.N. dosed, formulated and non-formulated TNFα siRNA, on various pro-inflammatory cytokines. Briefly, 1 mg/kg of TNFα silencing siRNA duplex was dosed intranasal 16 hours before LPS injection either as a nanoemulsion formulation (SNE) or as saline solution (STNF). Microinjection of LPS in the substantia nigra was performed using a stereotaxic apparatus. Rats were deeply anesthetized first using isoflurane inhalation. The rat’s head was shaved and swabbed with 70% isopropyl alcohol and betadine. Rats were then placed in the stereotaxic instrument. Body temperature was maintained throughout the procedure at 37 °C using a heating pad (Fintronics). A sterile scalpel was used to create a 1–2 cm rostral to caudal incision on the scalp, and to expose the bregma. Tissue overlying the suture lines was scraped away and the skull was dried using a dryer. A drill (Dremel®) was then used to create a burr-hole at the following stereotaxic coordinates: 4.8 mm posterior to bregma, 1.7 mm lateral to the midline, and 8.2 mm ventral to the surface of the skull [26] for injection into the substantia nigra region. LPS stock solution (2mg/ml) prepared in sterile phosphate buffer saline (PBS) was filled in a 5µL Hamilton syringe. The needle Hamilton syringe was then lowered −8.8 mm ventral to the surface of the skull and 1 uL of LPS was injected using a motorized microinjection Harvard apparatus infusion pump at a rate of 0.5 uL/minutes. After the injection, the needle was kept in place for 2 minutes and then slowly pulled out to minimize efflux. Each rat received an injection of LPS into right side of the brain. The contralateral side was left untreated and served as an internal control and was analyzed separately.
Rat brain tissue samples were harvested 6 hours post LPS injection from injection site and the contralateral site. After micro-dissection of substantia nigra from each rat brain, tissue was stored in RNAse/DNAse free conical tubes and stored in −80°C refrigerator. The following day, the tissue samples obtained were homogenized along with 1mL of Qiazol™ (Invitrogen, Carlsbad, CA) in 12 ml sterile homogenization tubes. Total RNA was isolated and purified from brain tissue using RNeasy lipid tissue mini kit (Qiagen, Valencia, CA) according to supplier’s instruction. Gene quantitation was performed using TaqMan gene expression master mix and TaqMan gene expression assays from Applied Biosystems (Foster, CA).
The cDNA synthesis was carried out as per the guidelines provided in the TaqMan® reverse transcription kit (Invitrogen, Carlsbad, CA) using 1 ug of total RNA amount. Quantitative PCR (qPCR) procedure was carried out by using TaqMan® gene expression master mix (Invitrogen, CA). β-actin was used as the internal control in the PCR experiments. The rat specific assay primer for TNFα, IL-6, iNOS, and β–actin were obtained from Invitrogen. Fluorescence generated due to reaction amplification of the target gene was determined on a Light cycler® 480 PCR detection system. The samples were run in triplicates and data was analyzed using a comparative threshold cycle (Ct) method by calculating ΔΔCt values for each treatment and results were expressed as percent relative expression compared to β-actin as endogenous control.
2.8. Preliminary Tolerability Studies
To examine the acute safety profile, the following analysis was conducted for each treatment group as solution and as nanoemulsion with controls: body weight monitoring, histopathological evaluation of nasal mucosa and histopathological evaluation of liver sections.
2.8.1. Body Weight Changes
Periodic measurements of the body weight were performed upon injecting the control, solution and nanoemulsion based formulation containing TNFα siRNA via intranasal route of administration on day 0 to day 3. Frequent body weight measurements were made through the course of the study. The results were plotted as percent change in body weight as a function of day’s pre-treatment administration for all the treatment groups (n=2)
2.8.2 Nasal Tissue Histopathology
To evaluate the toxic effect on the nasal mucosa, rats were first dosed with different siRNA-encapsulated TNFα nanoemulsion (SNE) or aqueous solution (STNF) and after exsanguination at 6 hour time point post administration, the head was removed from the carcasses. The tissues samples were preserved in formalin fixative until histopathological processing. After fixation and decalcification, four tissue slices were processed at the following sections: (1) immediately posterior to the upper incisor teeth; (2) at the incisive papilla or the anterior nasal cavity; (3) premolar or middle part of the nasal cavity; and (4) at the middle of the first molar teeth or posterior part of the nasal cavity. The nasal tissues were processed in a conventional manner. Paraffin embedded tissues were cut into 5 µm sections and mounted on glass slides. Tissue sections were dried and de-paraffinized using a xylene substitute followed by decreasing concentrations of ethanol down to purified water. Sections were incubated in hematoxylin, rinsed with water, and incubated with 1% acid alcohol (clearing reagent). Sections were rinsed and incubated with 4% ammonia solution (bluing reagent). Sections were then incubated with eosin followed by dehydration by two changes each in 95% ethanol and 100% ethanol followed by a final change of xylene substitute. Tissues were cover slipped and a digital image was captured using a light microscopy (n=2/treatment).
2.8.3. Liver Tissue Histopathology
Liver tissues samples were collected for histopathological analysis from rats after 3 day of post treatment of control PBS, siRNA solution and siRNA nanoemulsions. These tissue samples were preserved in formalin before analysis. Paraffin embedded tissues were cut into 5 µm sections and mounted on glass slides. After processing, tissues were cover slipped and digital images were captured using a light microscopy (n=1/treatment). Blinded analysis of the toxicological profile and tissue damage, if any, was carried out by Dr. Jerry Lyon, a certified veterinary pathologist, at the Tufts University Veterinary School in Grafton, MA.
3. RESULTS
3.1. siRNA-Encapsulated Nanoemulsion Formulations
The siRNA encapsulated nanoemulsions (SNE) prepared using the cationic phospholipid DOTAP through the sonication method resulted in a homogeneous milky white emulsion. We optimized the processing conditions and found that 5 minutes ultra-sonication (energy 21%, duty cycle of 25%) resulted in a nanoemulsion with particle sizes less than 400 nm. Composition of various excipients is shown in Table 1.
Table 1.
Formulation composition for siRNA encapsulated nanoemulsion
| Composition | % wt/volume |
|---|---|
| Flaxseed Oil | 10 |
| Tween 80 | 0.8 |
| Lipoid E80 | 2.32 |
| DOTAP C.L | 0.38 |
| siRNA | 0.05 |
Nanoemulsions for siRNA were screened for optimum N/P ratios. Optimization of the N/P ratio was performed based on the particle size, zeta potential, encapsulation of the final nanoemulsion and loading capacity to achieve a minimum concentration of 1 mg/ml. The factor that was varied was the amount of DOTAP lipid and the siRNA amount was kept constant at 1 mg/ml. Results in Table 2 summarize the characterization data for the study. There was no correlation between particle size and polydispersity with change in DOTAP ratio. However, we found that there was a positive correlation between the increase in the N/P ratio (increasing DOTAP amount) and positive charge on the particles. The lower N/P ratio formulations had a big impact on encapsulation, mostly due to the fact that the amount of cationic lipid was not optimum in those formulations. The formulation with an N/P ratio of 3.72:1 was selected for further investigations as it demonstrated the best encapsulation of siRNA with a good particle size distribution (<400nm).
Table 2.
Particle size, polydispersity index, zeta potential values, and siRNA encapsulation efficiency of DOTAP/siRNA complexed nanoemulsion (N/P ratio optimization).
| Formulation | N/P Ratio | Size (nm) ± SD | PDI ± SD | Zeta Potential (mV) ± SD | % Encapsulated ± SD | % Recovery ± SD |
|---|---|---|---|---|---|---|
| SNE#1 | 7.44 | 447 ±29 | 0.24 | 45±1.80 | 82.0±5 | 98.7±05 |
| SNE#2 | 3.72 | 390.2 ±28 | 0.16 | 44.2±0.60 | 70.1±10 | 102.2±10 |
| SNE#3 | 1.86 | 386.7 ±26 | 0.52 | 3.30±0.50 | 0 | 20.8±18 |
| SNE#4 | 1.86 | 415.3 ±54 | 0.28 | 9.47±0.30 | 7.9±12 | 62.3±08 |
| SNE#5 | 0.93 | 511 ±84.2 | 0.362 | −7.01±0.60 | 7.4±06 | 116.2±05 |
| SNE#6 | 0.465 | 328.1 ±23.6 | 0.209 | −10.2±0.90 | 0 | 122.0±03 |
Each value represents mean +/− standards deviation, n=3.
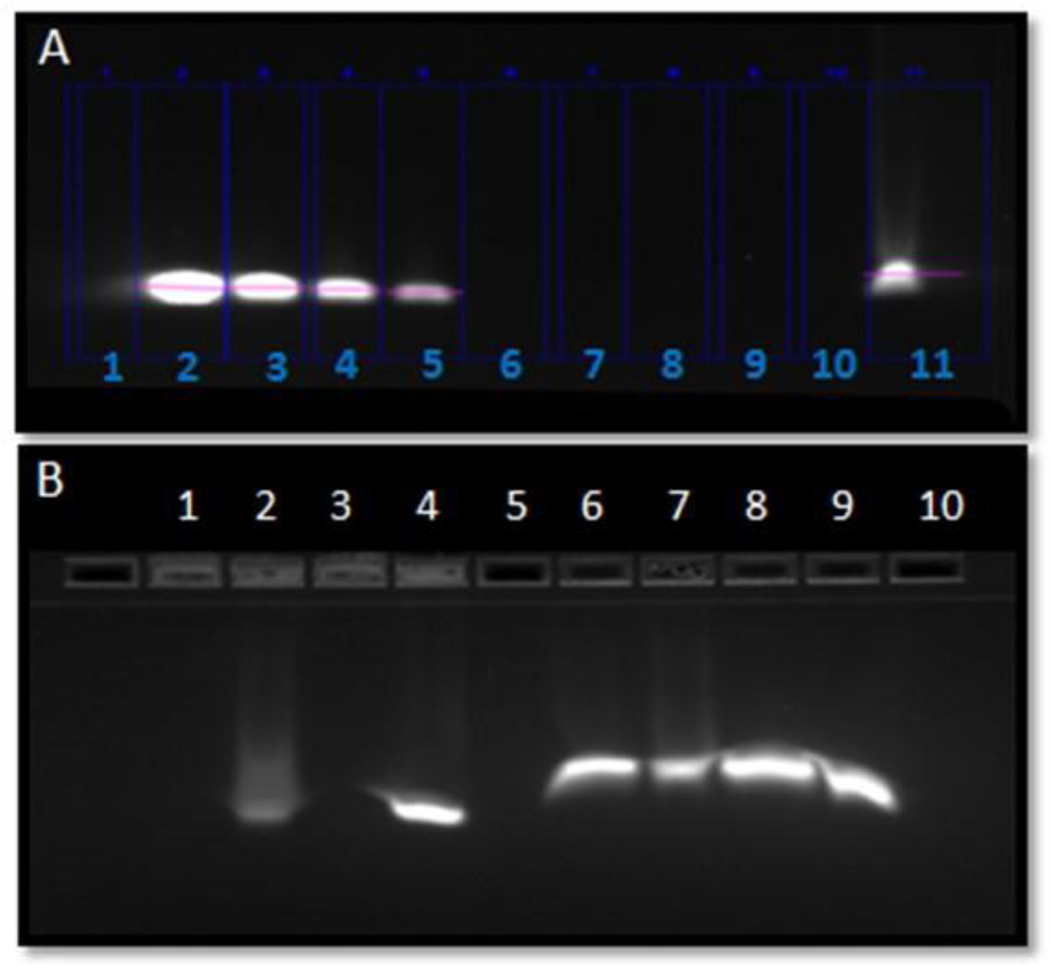
Morphology determination was performed using TEM and showed the size range from 69 nm to 168 nm for the optimum nanoemulsions with a smooth and spherical surface of particles (Supplementary Figure 1). Particle size on DLS was found to be bigger mostly because DLS measures and reports hydrodynamic diameter and bigger particle size would have higher impact on the average values. Formulations were also characterized for encapsulation efficiency based on the SYBR gold assay which showed an encapsulation efficiency of about 70±10% was achieved with good percent recovery of the total siRNA loaded in the nanoemulsion. Further, based on the E-gel retardation result siRNA was found to be completely complexed within the nanoemulsion particles as reflected by the complete retardation of siRNA in the 4% agarose gels and further it was found that when SNE was treated with a decomplexing agent (Triton-X100), siRNA was released from the particles and was able to travel through the gel and was finally visualized as a free band (Figure 1A).
Figure 1.
Electrophoretic retardation analysis of TNFα silencing siRNA encapsulation in cationic nanoemulsion (SNE). The release of intact siRNA by 10% Triton® X100 was shown for SNE in lane 11(1A) Lane 1:Empty, Lane 2: siRNA standard 250ug/mL, Lane 3: siRNA standard 125ug/mL, Lane 4: siRNA standard 62.5ug/mL, Lane 5: siRNA standard 31.25ug/mL, Lane 6 to Lane 8: Empty, Lane 9: siRNA nanoemulsion (SNE) 250ug/mL with no Triton® X100, Lane 10: SNE 250ug/mL with 2% Triton, Lane 11: SNE 250ug/mL with 10% Triton. Figure 1B shows the electrophoretic retardation analysis of siRNA stability when formulated as siRNA nanoemulsion; Lane 1: Empty, Lane 2: SNE incubated with RNAse for 15min followed by PBS dilution, Lane 3: SNE incubated with RNAse followed by Triton® X100 treatment, Lane 4: SNE incubation with PBS and dilution with PBS, Lane 5: SNE incubated with Triton and PBS dilution, Lane 6: Empty, Lane 7: siRNA incubated with RNAse and diluted with PBS, Lane 8: siRNA incubated with RNAse and Triton treatment, Lane 9: siRNA diluted with PBS only, Lane 10: siRNA incubated with PBS and treatment with Triton® X100.
siRNA degradation is one of the important barriers for siRNA intracellular delivery as it is associated with loss of therapeutic effect. Considering this, we have evaluated the protective effect of nanoemulsions against RNAse digestion. SNE after incubation with RNAse enzymes showed that siRNA in nanoemulsions is protected from enzymatic degradation as the siRNA band was found intact after siRNA was released from particles using Triton X-100. Further, we also verified that the SNE particles were physically stable and there was no phase separation during storage at 4°C for 24 hours (Figure 1B).
3.2. Cellular Uptake and Cytotoxicity
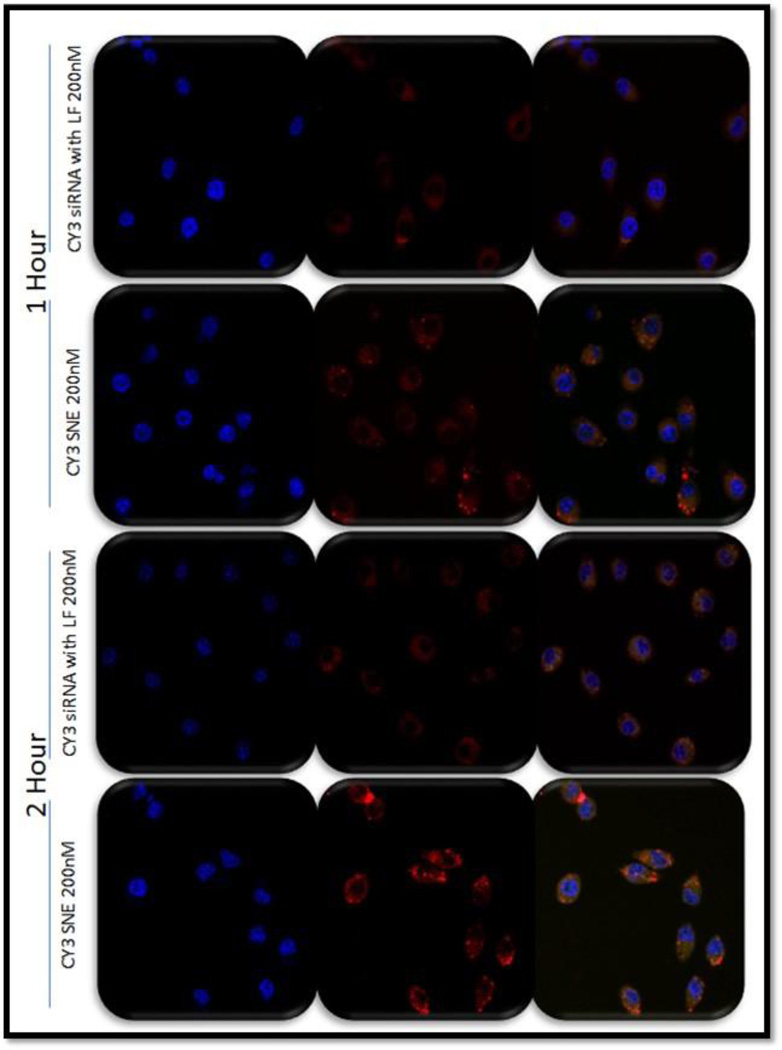
Fluorescence confocal microscopy was employed to visualize the qualitative intracellular uptake of non-formulated and nanoemulsion formulated siRNA in J774A.1 as a function of time. Intracellular uptake was analyzed at 1 h and 2 h time points after incubation of labeled siRNA particles at a siRNA concentration of 200 nM. The confocal images shown in Figure 2 show higher uptake of siRNA encapsulated in nanoemulsions as compared to siRNA delivered using Lipofectamine® (a commercially available cationic lipid transfection reagent). The higher uptake from SNE was found consistent at both the 1-hour and 2 hour time points.
Figure 2.
The uptake and trafficking of siRNA-encapsulated cationic nanoemulsion (SNE) in J774A.1 cells. Fluorescence microscopy images showing the blue (nucleus), red (siRNA) and overlay images for siRNA complexed in Lipofectamine® as compared to SNE at 200 nM concentrations. The images were taken at 40× original magnification.
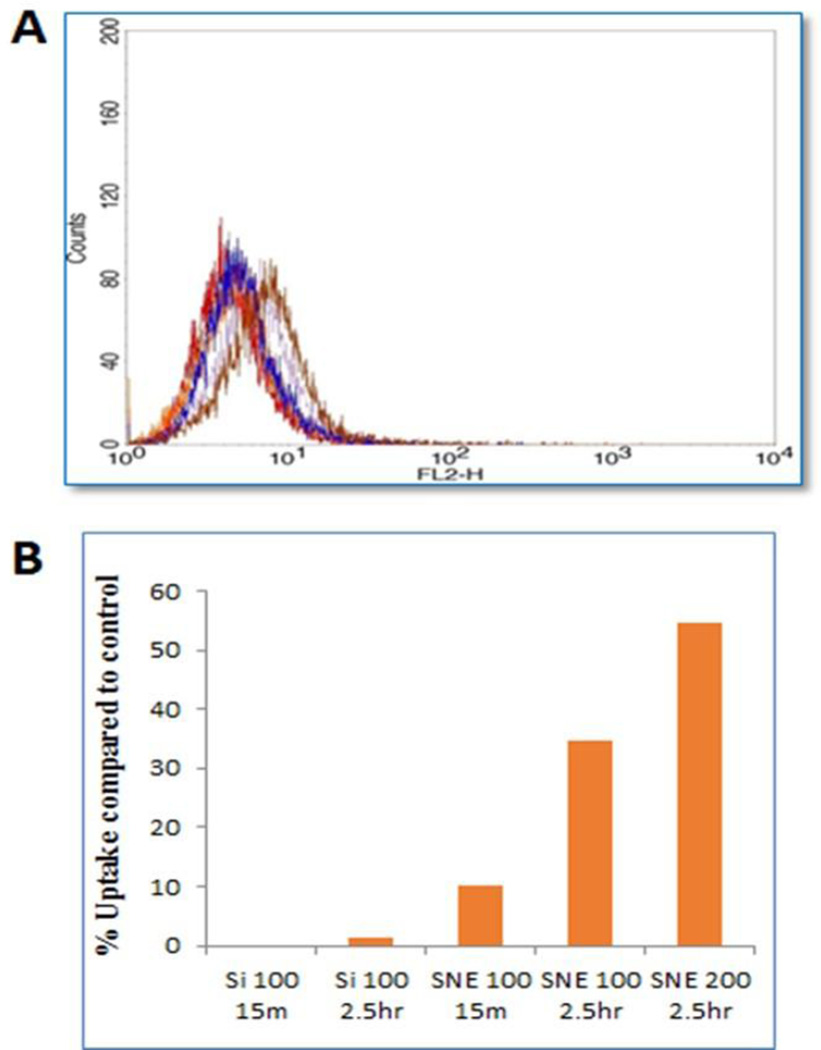
In order to ascertain the trend of nanoemulsion uptake shown by confocal images and to get a quantitative estimate of intracellular uptake, J774A.1 cells treated with Cy3-flourescently labeled nanoemulsions were analyzed by flow cytometry in a time dependent manner. The fluorescence intensity was normalized to the untreated control cells. Figure 3A and 3B shows the quantitative relative uptake of Cy3-labeled siRNA with time. SNE formulation showed higher uptake compared to the Lipofectamine® transfected samples. There was almost 10-fold higher uptake of siRNA nanoemulsion at 15 minutes time point and almost 25-fold higher uptake at later time point of 2.5 h, when compared to Lipofectamine® transfected control group. Further, when the concentration of siRNA (in SNE) was increased to 200 nM, there was a further 1.5 fold increase in fluorescence intensity showing there is a dose dependent effect for endocytosis. The flow cytometry results further confirmed the nanoemulsion internalization results of fluorescence microscopy images.
Figure 3.
Flow cytometry results showing the FL-2 channel (585/42 emission) which was used to detect the cells containing CY3-label siRNA containing particles (A). Flow cytometry data presented as percent fluorescence intensity/uptake for siRNA alone (siRNA) and nanoemulsion (SNE) compared to control untreated cells (B).
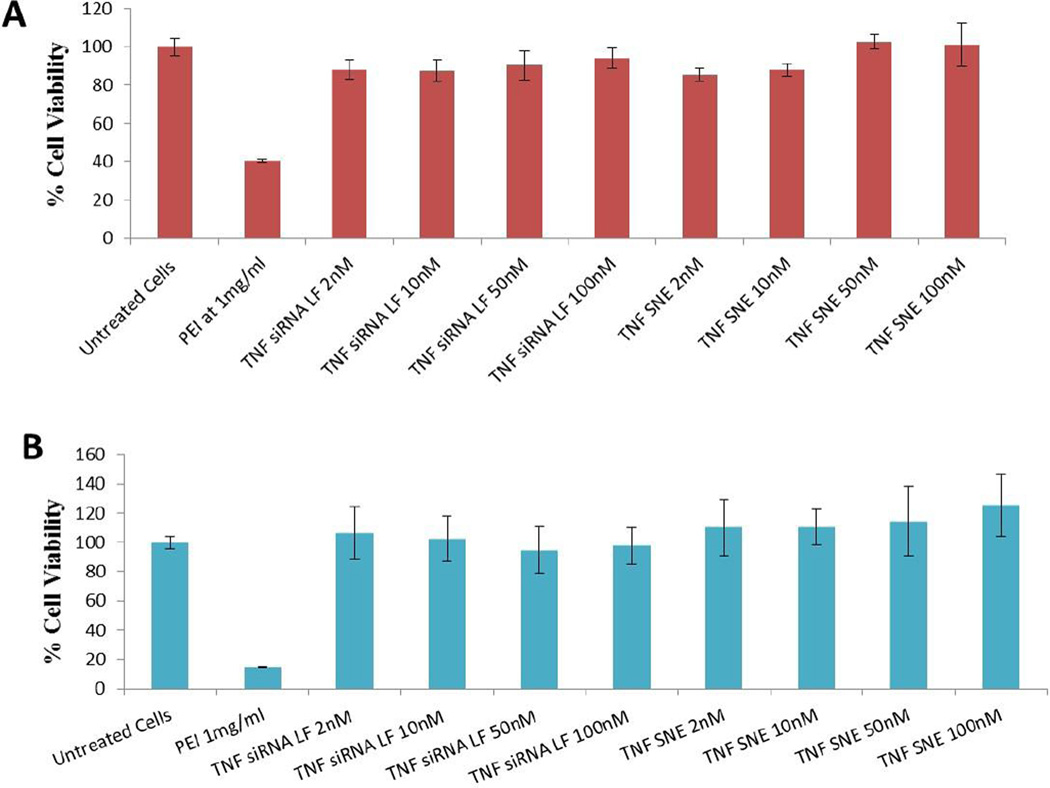
To assess potential cytotoxicity, if any, with the control and siRNA-encapsulated DOTAP nanoemulsions, the formulations were incubated at different concentrations with J774A.1 macrophages. In viable cells, the enzymes convert the water-soluble yellow MTT reagent to an insoluble purple formazan product that has absorbance at 490 nm. The cell viability results as shown in Figure 4, confirm that neither the siRNA transfected with Lipofectamine® nor the siRNA DOTAP nanoemulsions induced any significant cytotoxicity when added to cells at 2 nM to 100 nM concentration. The cytotoxicity results from SNE were found to be similar at both the early time point of 2 h (Figure 4A) and the later time point of 19 h (Figure 4B).
Figure 4.
Cytotoxicity of siRNA encapsulated cationic nanoemulsion (SNE) at 2 hours (A) and 19 hour time point (B) post-administration in J774A.1 macrophages. The cell viability of the untreated cells was considered to be 100%, and the values obtained in the rest of the treatment groups were normalized to control values and presented in percentage form. The values reported are mean ± SD (n = 8).
3.3. TNFα Silencing in LPS-Stimulated Macrophages
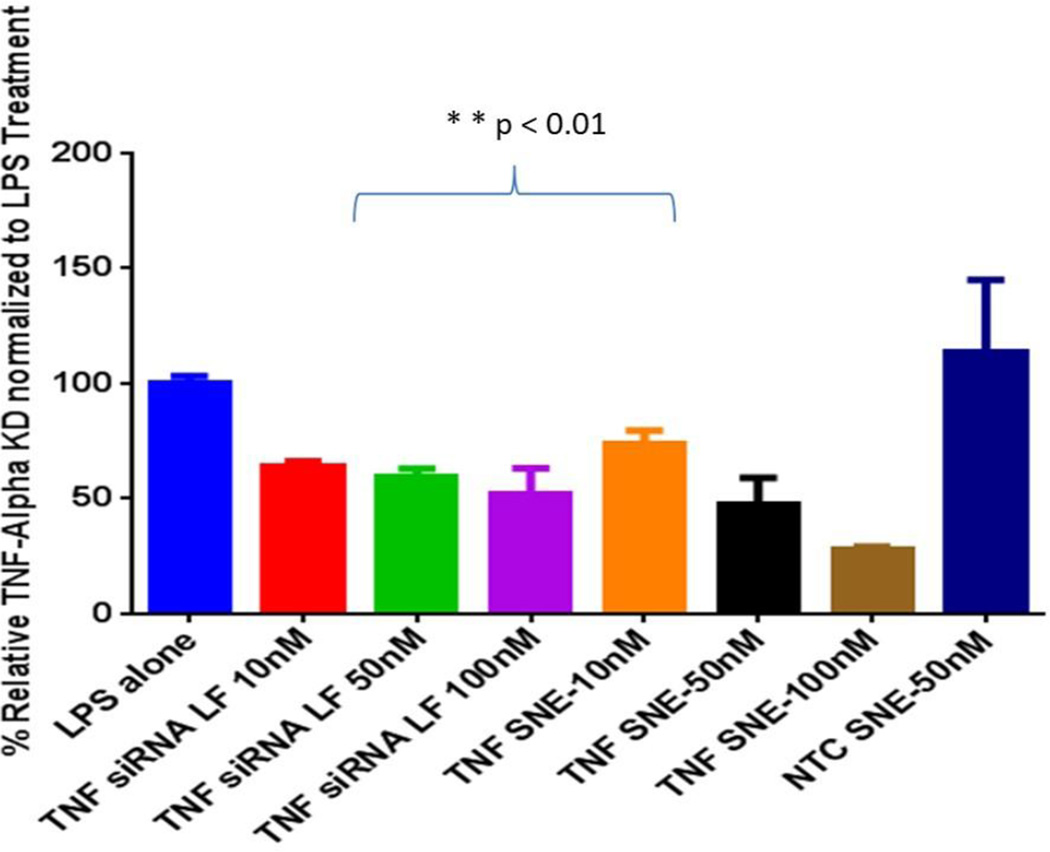
J771A.4 murine macrophages cells upon stimulation with LPS resulted in substantial up regulation of TNFα cytokine. SNE and siRNA control with Lipofectamine® were dosed at three different concentrations (10, 50 and 100nM) and after 24h of transfection cells were exposed to LPS toxin. Previously, it was shown that J774A.1 cells showed the highest stimulation for TNFα cytokine at the 6 h time point after exposure to LPS [27] and, therefore, we have selected the 6 h time point post LPS stimulation to evaluate the gene silencing effect of siRNA. Using anti-TNFα siRNA formulations, gene knockdown was determined by measuring the level of TNFα gene expression using a RT-qPCR method. As shown in Figure 5, the cationic nanoemulsions containing TNFα silencing siRNA showed dose dependent silencing efficiency which was found to be statistically higher compared to siRNA delivered with the highest concentration of Lipofectamine® transfection agent (p<0.01).
Figure 5.
TNFα gene knockdown result in lipopolysaccharide (LPS) stimulated macrophages showing inhibition of TNFα mRNA expression in the J774A.1 macrophage cell line. The data was plotted by considering the TNFα expression as 100% in LPS stimulated cells. The values reported are mean ± SD (n = 3) with **p<0.01 showing significant difference between groups
3.4. In Vivo Evaluations of siRNA Delivery in the Brain
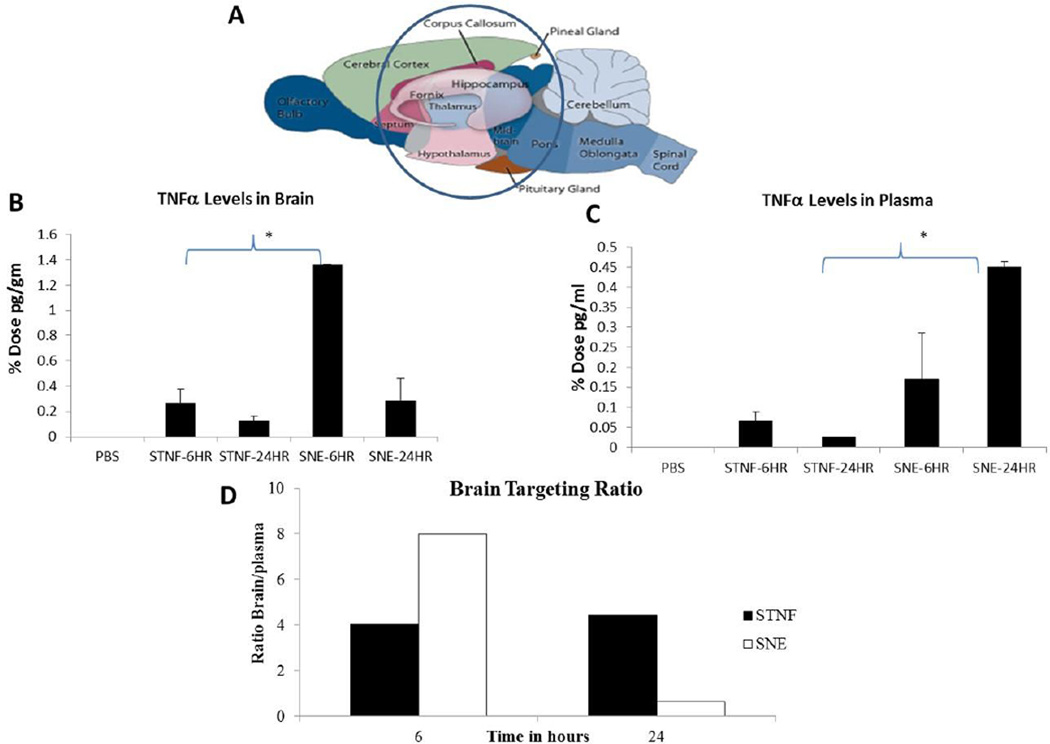
Since we were able to demonstrate previously that we were able to show higher gene silencing efficiency with siRNA nanoemulsions, we were interested in studying the distribution of the same siRNA formulation in the rat brain and plasma after intranasal delivery. For the siRNA quantification study, the TNFα siRNA was encapsulated in identical DOTAP nanoemulsions as previously described but at higher concentration of 3.8 mg/ml. SNE showed a particle size of 350 nm, zeta potential of +42 mV and 80% encapsulation efficiency. Since the substantia nigra in the brain is one of the major targets for neuroinflammation diseases, we selected middle-brain region at bregma coordinates of 4.70 mm to −7.80 mm (according to Paxinos and Watson atlas) to study the uptake of therapeutic siRNA. This region together with substantia nigra (at −4.80 mm) includes other regions like cerebral cortex, corpus callosum, hippocampus, thalamus, hypothalamus and mid-brain.. As intranasal delivery could also potentially lead to distribution in blood through the respiratory mucosa, we further assessed siRNA uptake in plasma as well. After three times dosing of siRNA in a day for a total dose of 3 mg/kg through the nasal route, blood and tissue samples were collected for siRNA quantitation at 6 h and 24 h post first administered dose (Supplementary Figure 2).
For the accurate siRNA quantitation, the anti-primer quenching based real time PCR method was utilized [28, 29] as described in the experimental section. In this method, a fluorescently labeled PCR primer was designed to anneal to the template RNA and to a universal anti-primer. Following the initial PCR, the temperature was lowered to allow the anti-primer to bind to the unincorporated primer to quench its fluorescence. Since this will not bind to the double stranded PCR product, there will be an increased fluorescent signal detected. The PCR quantitation method employed for tissue and plasma siRNA quantitation showed good linearity for the standard curve prepared in the respected biological samples (Supplementary Figure 3A and 3B). The TNFα siRNA was quantitated in middle-brain region (Figure 6A) and the percent input dose per brain region was calculated based on the starting siRNA dose. TNFα siRNA delivered intranasal in saline solution showed 0.2% injected dose at the 6 hour time point followed by a 0.12% injected dose at later time point of 24 hour. On the other hand, SNE (siRNA nanoemulsion) showed higher 1.38% injected dose in the middle-brain region at 6 hour time point. Nanoemulsions also showed reduced, 0.28% of the injected dose at the later time point of 24 hours in the brain. SNE showed significantly higher (p<0.05) accumulation compared to siRNA in solution (STNF) at 6 hour time point in middle brain region (Figure 6B). There was higher accumulation from nanoemulsions at later time points as well, but the difference was not found to be significant which could be due to high variability in the small group of animals. Interestingly, SNE at the 24 hour time point was able to provide exposure similar to the siRNA solution at 6 hour time point.
Figure 6.
Results showing comparison for the percent administered dose in the brain (B) region highlighted (A) and plasma (C) following siRNA dosed in solution (STNF) versus siRNA dosed in cationic nanoemulsion (SNE) formulation. Brain targeting ratio comparing the targeting efficiency of siRNA using either solution versus nanoemulsion is shown (D). The values reported are mean ± SEM (n = 3); *p < 0.05.
Overall, the uptake in plasma was found to be lower than the uptake in brain with either siRNA in saline solution (STNF) or siRNA encapsulated in nanoemulsion (SNE) when delivered through the intranasal route. The siRNA aqueous solution (STNF) showed only 0.06% injected dose compared to siRNA nanoemulsion, which was found to be 0.17% of the injected dose at 6 hour time point. At the later time point of 24 hour, siRNA solution delivered only 0.02% injected dose whereas siRNA nanoemulsion delivered 0.45% injected dose which was found to be statistically significantly (p<0.005). Plasma exposure overtime seems to be increasing with the nanoemulsion which could be due to the fact that emulsion due to its high positive charge surface in being able to permeate slowly through the respiratory mucosa and into the surrounding blood vasculature.
Further, the brain targeting ratio was calculated to assess and compare the advantage of both strategies delivered intranasal (Figure 6D). The ratio is calculated based on the concentration in plasma and brain at each time point. The brain targeting ratio of siRNA was found to be almost 2-fold higher for SNE (nanoemulsions) compared to STNF (solution) at the early time point of 6 hour. At the later time point of 24 h, the ratio for solution was found to be higher compared to nanoemulsions although it was still lower than the ratio observed at the early time point.
3.5 In Vivo Gene Silencing Efficacy in LPS-Induced Neuroinflammation Model
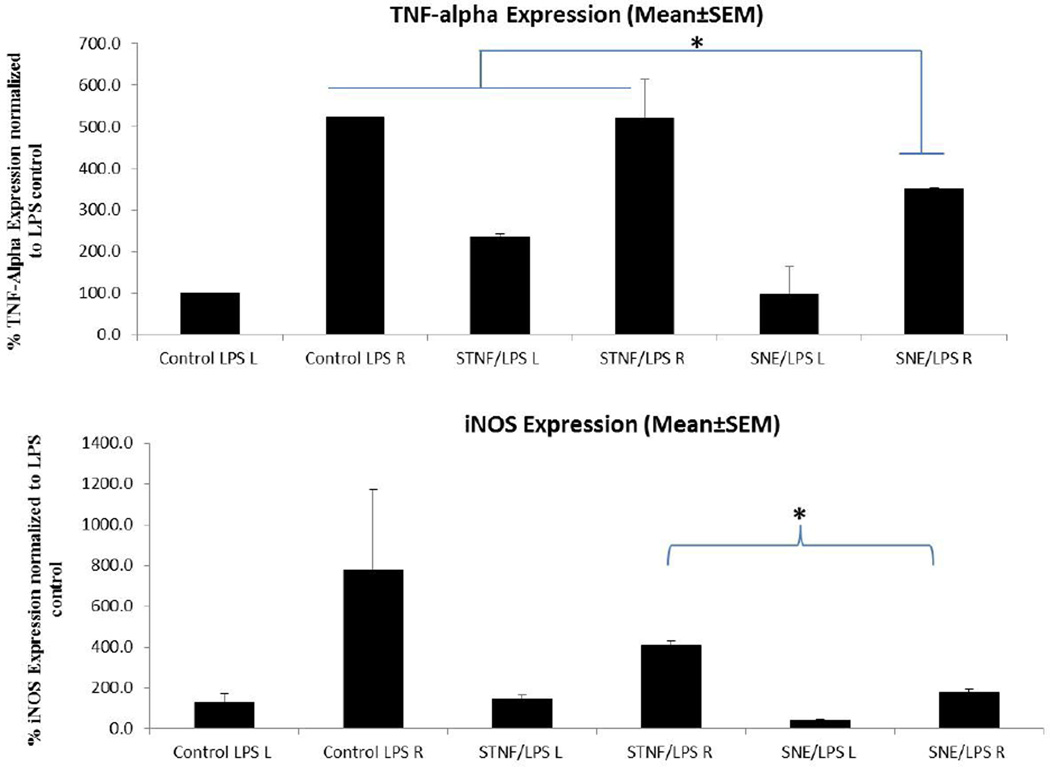
In order to assess the efficacy of TNFα siRNA nanoemulsions in vivo, we have utilized an LPS model of neuroinflammation, a commonly used model to study anti-inflammatory effects of therapeutics for neurodegenerative diseases involving inflammation. In this study, we first established the in vivo LPS induced neuroinflammation model in rats by studying the dose dependent and time dependent effect of LPS on the stimulation of various cytokines in the substantia nigra region of the brain using intra-nigral micro-injection into brain. Based on preliminary evaluations of inflammation markers (data not shown), we found that LPS-induced neuroinflammation in the rat SN was concentration- and time-dependent and we selected a dose of 2 µg LPS and a time point of 6 hour post-LPS to study the effect of anti-TNFα siRNA formulations. We evaluated the knockdown effect of TNFα siRNA upon intranasal delivery in the LPS-induced model of neuroinflammation. Rats were pre-treated with 1.5 mg/kg dose of siRNA in saline solution (STNF) or siRNA-nanoemulsion (SNE). Intranasal dosing was performed as described previously and after 24 h rats received a single intranigral injection of LPS (2 µg). After recovery from surgery rats were returned to their cages and finally brain tissues were harvested at 6 h post LPS injection. Based on the qPCR results for gene expression it was found that TNFα siRNA when delivered in solution form (STNF) showed almost no inhibition when compared to TNFα siRNA delivered as nanoemulsions (SNE). SNE statistically (p<0.05) reduced levels of TNFα from approximately 523% relative expression to 351% after 16 hours of pretreatment with siRNA (Figure 7). Left substantia nigra was used as a contralateral control for all three groups. The time frame for pre-treatment was defined based on the high distribution of siRNA in the brain from the 6 hour to 24 hour time points. Limited knockdown effects observed for the saline solution could be attributed to the limited siRNA exposure in the brain by saline solution.
Figure 7.
Inflammatory marker TNFα and iNOS specific mRNA results showing transcript expression in the substantia nigra region of rat brain. Graph comparing the pre-treatment with TNFα siRNA in solution (STNF) and siRNA-loaded cationic nanoemulsion (SNE). Data was plotted by considering the expression of control saline group as 100%. The values reported are mean ± SEM (n = 3); *p < 0.05.
Further, we also studied the effect of TNFα knockdown on other cytokines due to the possibility of crosstalk involved between different cytokines during inflammation. Intranasal dosing of TNF-siRNA in saline solution and SNE showed some down-regulation of iNOS cytokines when compared to the non-treated group, however only SNE showed statistically significant difference in levels of iNOS levels (p<0.05). Although up-regulation was observed for IL-6 cytokine after LPS toxin stimulation, both STNF and SNE showed some downregulation of IL-6 cytokine, however the results did not reach statistical significance for any of the treatment groups. As evident, upon intranasal delivery, siRNA nanoemulsions were able to provide an inhibitory effect, in the LPS-induced model of neuroinflammation. Furthermore, it was interesting to observe the crosstalk between TNFα and other cytokine down-regulation.
3.6 Preliminary Tolerability Evaluations
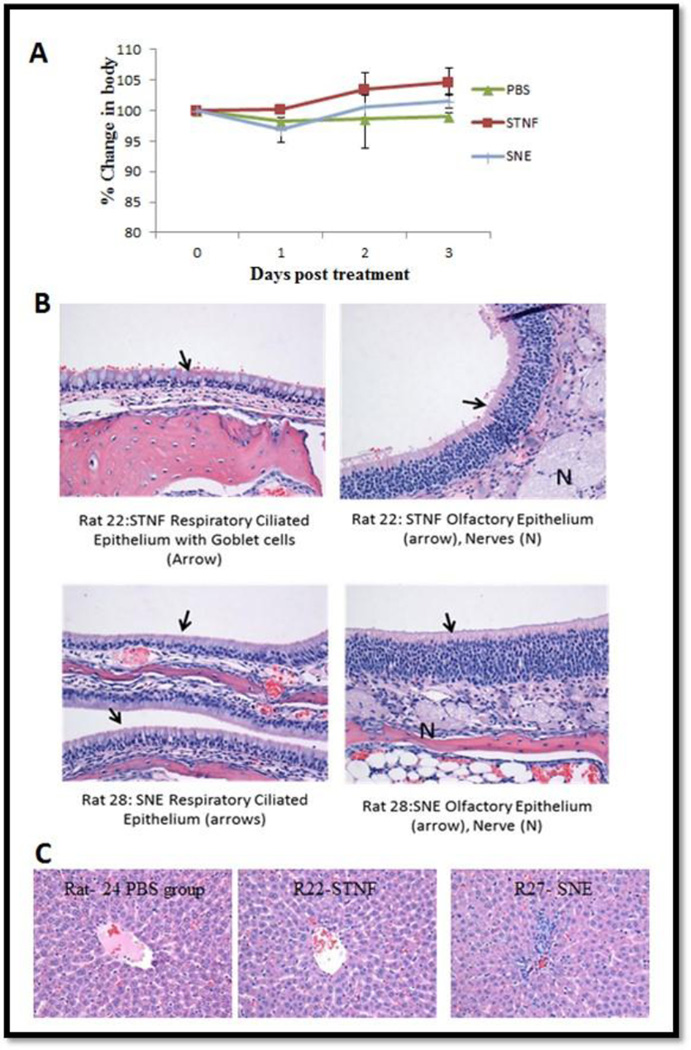
Comparing the body weights for the three groups: saline treated (control-non treated), TNFα siRNA solution (STNF) and siRNA nanoemulsions (SNE), only slight differences in body weight were seen at day 1 and all of these groups started to gain weight after 1 day (Figure 8A). These results suggest that the nanoemulsion formulations were all well tolerated in rats. Based on the histopathological report, all the normal structures of the nasal cavities were identified and no significant pathology is identified in any of the sections for any of the animals. Upon a closer view of the respiratory and the olfactory mucosal lining the nasal cavity, there were no significant changes found in both the epithelial cell lining the nasal cavity (Figure 8B). As shown in Figure 8C, there was moderate periacinar and diffuses hepatocellular vacuolation. Occasional multifocal aggregates of cells were present in portal areas consistent with extra-medullary hematopoiesis (EMH). Mild extra-medullary hematopoiesis and lipid vacuolation are considered as common incidental findings in the liver and are not thought to be related to the treatment as they were also observed in control naïve animals (R24). Hence, the liver tissues histopathological findings for treated groups are consistent with what is regarded as within normal limits.
Figure 8.
Body weight measurements to determine safety/tolerability profile upon single intranasal administration of the control and siRNA encapsulated cationic nanoemulsion (SNE) (A). Histology of nasal respiratory, olfactory epithelium (A) and liver (C) after intranasal dosing of siRNA. The values are reported as percent change in body weight as a function of pre-treatment weight of rats.
4. DISCUSSION
In order to enhance siRNA delivery for efficient TNFα silencing in vivo, we have developed a cationic lipid based nanoemulsion system for the delivery of anti-TNFα siRNA to the brain. Apart from the use of biodegradable flaxseed oil as a component, we have preferred using DOTAP lipid, due to its cationic nature which helps in higher loading capacity and also because it can complex with negatively charged biomolecules for greater intracellular uptake. These particles have shown evidence of higher cell transport based on the qualitative and quantitative cell uptake studies. DOTAP nanoemulsions showed higher uptake both at early time points and at later time point when compared to siRNA transfected with Lipofectamine®. Further advantage of using cationic particles is that there is a potential of surface modifications due to availability of functional groups when targeting is desired. An in vitro investigation for gene silencing effect was conducted using LPS-stimulated J774A.1 macrophages before this strategy could be transferred to in vivo setting. We found that SNE showed a higher gene silencing effect for TNFα transcript in LPS stimulated cells. The higher level of knockdown compared to Lipofectamine® is again contributed by the enhanced intracellular uptake of these particles by endocytosis.
Delivery of biologics to the CNS is challenging due to the presence of the BBB, blood-CSF barrier, and presence of efflux transporters. The intranasal route of administration on the other hand provides a non-invasive method of by-passing the BBB to potentially deliver biologics to the CNS. The intranasal route has been explored for successful delivery of various neuropeptides; however, lower membrane permeability acts as a significant limitation to the nasal absorption of polar high molecular weight biologics drugs like siRNA. Attempts have been made to improve the nasal absorption of polar drugs by co-administration of absorption-enhancing agents that work by modifying the tight junctions and/or act as an inhibitor of enzymatic degradation [30]. However, the use of such enhancers could be detrimental to the nasal mucosa, so the choice of the absorption-enhancer for a nasally delivered drug that is not easily absorbed must be carefully considered, especially in terms of nasal and systemic toxicity. We have prepared cationic nanoemulsion with particle sizes <400nm, as smaller particle size will provide a higher surface area for interaction and absorption in olfactory epithelium. It is known that nasal delivery offers very short residence time for molecules. For both liquid and powder formulations that are not bio-adhesive, the half-life for clearance is of the order of 15–30 minutes [31]. The advantage of using nanoemulsions for intranasal delivery is evident from the distribution study conducted in rats. In this study, a sensitive RT-PCR based quantitative method was utilized to determine the real therapeutic siRNA amounts in brain and plasma. When comparing the siRNA uptake through the intranasal route, nanoemulsions encapsulating siRNA showed higher brain uptake when compared to simple solution of siRNA in saline. Almost five fold higher percent dose was found at the 6 hour time point in middle-brain region, a region target for therapeutics [32]. Interestingly, the percent dose in brain regions at 24 h with nanoemulsion (SNE) was similar to the amount of drug found using saline solution of siRNA at 6 h. The higher amount at later time points could be due to the multifunctional potential of nanoemulsion to serve as a mucoadhesive and to enhance endocytosis due to smaller particle size. Our study compares to another recent study by Kim et al, where the HMGB1 siRNA delivery to the brain was studied using a PAMAM dendrimer as a gene carrier. Fluorescent tagged siRNA was detected in the various regions of brain within 1 hour and signal was found to be present until 12 h [33]. The study was qualitative based on imaging and hence it is hard to compare the percent dose reaching the brain with PAMAM dendrimers versus nanoemulsions, however we found measurable levels of siRNA in the brain up to a longer time point of 24 h with nanoemulsions. Further, overall lower plasma exposure was obtained via intranasal delivery, which is beneficial if specific target gene regulation is required as in case of neuroinflammation.
Additional investigations were performed to explore the capacity of the delivered siRNA to silence gene expression in an inflammation model. Due to the central role of inflammation in various diseases, the need for purely inflammation-driven animal’s model has emerged. An in vivo LPS-induced PD model was developed by Castano, et al. [21]. Subsequently, an LPS induced PD model has been widely accepted and used for understanding the pathogenesis of PD and for testing various anti-inflammatory treatment therapies for targeting neuroinflammation. In this study, we utilized an LPS-induced model of neuroinflammation to study the therapeutic effect of siRNA-nanoemulsions. Our in vitro experiments results translated well to animal studies and we showed that intranasal delivery of nanoemulsions incorporating TNFα siRNA were capable of significantly down-regulating the stimulated levels of TNFα when compared to siRNA delivered as simple saline solution. The site specific knockdown observed in this study entails an important point of not having nonspecific knockdown in other regions. By using the contralateral site as an inbuilt control, we showed that TNF- stimulation due to LPS was site specific. Further the level of downregulation of stimulated TNF-levels was more pronounced towards the injection or affected side. McApline and coworkers have previously shown that inhibition of soluble TNFα signaling can prevent lipopolysaccharide-induced Aβ plaque deposition in a transgenic AD mouse model [34]. This hypothesis is further supported by the findings from anti-TNFα approaches in AD patients where weekly perispinal etanercept (a brain TNFα fusion protein) was able to sequester excess TNFα and showed a sustained and significant improvement in cognition [35]. Further, in case of PD, genetic ablation of TNFα or treatment with a TNF-synthesis inhibitor (thalidomide) attenuated MPTP-induced dopamine depletion and reduced striatal tyrosine hydroxylase fiber density [36]. Thus, neutralization or downregulation of over expressed proinflammatory cytokines (TNFα and iNOS) as observed in our study would have clinical significance as it would help in slowing the progression or severity of the neuroinflammatory disease. At the clinically acceptable dose used in this study (1.5mg/kg), effective siRNA delivery and gene silencing could be achieved and this level of downregulation would be able to provide neuronal protection from apoptosis as shown recently in a study by Kim, et al. [37].
In addition to evaluating delivery and therapeutic efficacy, it is of utmost importance to monitor the safety and tolerability of formulations delivered through the intranasal route. It is important that intranasal delivery of particulate systems does not have any toxic effect on the sensitive nasal mucosa. When formulations were dosed in rats, we found no morphological differences in tissue histology of nasal cavity or liver in any of the treatment groups suggesting that intranasal delivery of cationic nanoemulsions of TNFα siRNA does not cause any detrimental effects to these tissues at the measured doses. A higher degree of systemic toxicity evaluation would be needed to completely understand the peripheral effects.
5. CONCLUSIONS
In this study, we have developed a non-viral cationic lipid based nanoemulsion system for down-regulating the TNFα gene transcript levels, which has implications in the prevention of neuroinflammation. We showed that this system exhibited higher cell uptake and efficiently knocked down TNFα in LPS-stimulated macrophages. Further, we have shown that intranasal delivery of TNFα siRNA nanoemulsions in rats quantitatively achieved higher uptake of therapeutic siRNA in brain compared to distribution into blood. This offers a major advantage as systemic exposure of the anti-inflammatory drugs could lead to unwanted side effects. Additionally, siRNA nanoemulsions showed site specific down regulation of TNFα cytokines when tested in LPS induced model of neuroinflammation. Overall, we have shown feasibility of using cationic lipid based nanoemulsions as a safe and effective drug delivery strategy to enhance delivery of biologics such as TNFα siRNA through the nasal route to target brain. The current findings demonstrate the utility of this approach for prevention of neuroinflammation diseases where higher CNS uptake and limited peripheral or blood compartment uptake is critical. Further investigations are needed to validate the utility of intranasal delivery of siRNA to down-regulate multiple genes that are overexpressed in complexed pathologies associated with neurodegenerative diseases both at the RNA level and protein level.
Supplementary Material
Acknowledgments
We deeply appreciate the assistance of Dr. Amit Singh with the transmission electron microscopy, flow cytometry, and confocal microscopy analyses that was performed at Northeastern University (Boston, MA). We would also like to thank Ms. Amy Dell and Mr. Michael Beverly at the Novartis Institute for Biomedical Research (Cambridge, MA) for assistance with the in vivo siRNA quantitation method. We appreciate Ms. Grishma Pawar’s assistance with the neuroinflammation model development and in vivo experiments. We also would like to thank Dr. Lara Milane for providing critical review of the manuscript.
Financial Support: This study was partially supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health through a grant R21-NS066984.
ABBREVIATIONS
- LPS
lipopolysaccharide
- TNFα
tumor necrosis factor-alpha
- siRNA
small interfering ribonucleic acids
- CNS
central nervous system
- SNTF
TNFα silencing siRNA in aqueous solution
- SNE
TNFα silencing siRNA-encapsulated in cationic nanoemulsion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Barchet TM, Amiji MM. Challenges and opportunities in CNS delivery of therapeutics for neurodegenerative diseases. Expert opinion on drug delivery. 2009;6:211–225. doi: 10.1517/17425240902758188. [DOI] [PubMed] [Google Scholar]
- 2.Glass JD, Johnson RT. Human immunodeficiency virus and the brain. Annual review of neuroscience. 1996;19:1–26. doi: 10.1146/annurev.ne.19.030196.000245. [DOI] [PubMed] [Google Scholar]
- 3.Raine CS. Multiple sclerosis: immune system molecule expression in the central nervous system. Journal of neuropathology and experimental neurology. 1994;53:328–337. doi: 10.1097/00005072-199407000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Rogers J, Luber-Narod J, Styren SD, Civin WH. Expression of immune system-associated antigens by cells of the human central nervous system: relationship to the pathology of Alzheimer's disease. Neurobiology of aging. 1988;9:339–349. doi: 10.1016/s0197-4580(88)80079-4. [DOI] [PubMed] [Google Scholar]
- 5.McGeer PL, Itagaki S, Boyes BE, McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson's and Alzheimer's disease brains. Neurology. 1988;38:1285–1291. doi: 10.1212/wnl.38.8.1285. [DOI] [PubMed] [Google Scholar]
- 6.Dorsey ER, George BP, Leff B, Willis AW. The coming crisis: obtaining care for the growing burden of neurodegenerative conditions. Neurology. 2013;80:1989–1996. doi: 10.1212/WNL.0b013e318293e2ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gayle DA, Ling Z, Tong C, Landers T, Lipton JW, Carvey PM. Lipopolysaccharide (LPS)-induced dopamine cell loss in culture: roles of tumor necrosis factor-alpha, interleukin-1beta, and nitric oxide. Brain research Developmental brain research. 2002;133:27–35. doi: 10.1016/s0165-3806(01)00315-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim WG, Mohney RP, Wilson B, Jeohn GH, Liu B, Hong JS. Regional difference in susceptibility to lipopolysaccharide-induced neurotoxicity in the rat brain: role of microglia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:6309–6316. doi: 10.1523/JNEUROSCI.20-16-06309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao HM, Jiang J, Wilson B, Zhang W, Hong JS, Liu B. Microglial activation-mediated delayed and progressive degeneration of rat nigral dopaminergic neurons: relevance to Parkinson's disease. Journal of neurochemistry. 2002;81:1285–1297. doi: 10.1046/j.1471-4159.2002.00928.x. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez A, Cacabelos R, Sanpedro C, Garcia-Fantini M, Aleixandre M. Serum TNF-alpha levels are increased and correlate negatively with free IGF-I in Alzheimer disease. Neurobiology of aging. 2007;28:533–536. doi: 10.1016/j.neurobiolaging.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Fillit H, Ding WH, Buee L, Kalman J, Altstiel L, Lawlor B, et al. Elevated circulating tumor necrosis factor levels in Alzheimer's disease. Neuroscience letters. 1991;129:318–320. doi: 10.1016/0304-3940(91)90490-k. [DOI] [PubMed] [Google Scholar]
- 12.Paganelli R, Di Iorio A, Patricelli L, Ripani F, Sparvieri E, Faricelli R, et al. Proinflammatory cytokines in sera of elderly patients with dementia: levels in vascular injury are higher than those of mild-moderate Alzheimer's disease patients. Experimental gerontology. 2002;37:257–263. doi: 10.1016/s0531-5565(01)00191-7. [DOI] [PubMed] [Google Scholar]
- 13.Tarkowski E, Blennow K, Wallin A, Tarkowski A. Intracerebral production of tumor necrosis factor-alpha, a local neuroprotective agent, in Alzheimer disease and vascular dementia. Journal of clinical immunology. 1999;19:223–230. doi: 10.1023/a:1020568013953. [DOI] [PubMed] [Google Scholar]
- 14.Mogi M, Harada M, Riederer P, Narabayashi H, Fujita K, Nagatsu T. Tumor necrosis factor-alpha (TNF-alpha) increases both in the brain and in the cerebrospinal fluid from parkinsonian patients. Neuroscience letters. 1994;165:208–210. doi: 10.1016/0304-3940(94)90746-3. [DOI] [PubMed] [Google Scholar]
- 15.Nagatsu T, Mogi M, Ichinose H, Togari A. Changes in cytokines and neurotrophins in Parkinson's disease. Journal of neural transmission Supplementum. 2000:277–290. doi: 10.1007/978-3-7091-6301-6_19. [DOI] [PubMed] [Google Scholar]
- 16.Wesselingh SL, Power C, Glass JD, Tyor WR, McArthur JC, Farber JM, et al. Intracerebral cytokine messenger RNA expression in acquired immunodeficiency syndrome dementia. Annals of neurology. 1993;33:576–582. doi: 10.1002/ana.410330604. [DOI] [PubMed] [Google Scholar]
- 17.Pardridge WM. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frey WH. Neurologic agents for nasal administration to the brain. Google Patents. 1997
- 19.Ruud TE, Gundersen Y, Wang JE, Foster SJ, Thiemermann C, Aasen AO. Activation of cytokine synthesis by systemic infusions of lipopolysaccharide and peptidoglycan in a porcine model in vivo and in vitro. Surgical infections. 2007;8:495–503. doi: 10.1089/sur.2006.083. [DOI] [PubMed] [Google Scholar]
- 20.Chirumbolo S, Franceschetti G, Zoico E, Bambace C, Cominacini L, Zamboni M. LPS response pattern of inflammatory adipokines in an in vitro 3T3-L1 murine adipocyte model. Inflammation research : official journal of the European Histamine Research Society [et al] 2014;63:495–507. doi: 10.1007/s00011-014-0721-9. [DOI] [PubMed] [Google Scholar]
- 21.Castano A, Herrera AJ, Cano J, Machado A. Lipopolysaccharide intranigral injection induces inflammatory reaction and damage in nigrostriatal dopaminergic system. Journal of neurochemistry. 1998;70:1584–1592. doi: 10.1046/j.1471-4159.1998.70041584.x. [DOI] [PubMed] [Google Scholar]
- 22.Herrera AJ, Castano A, Venero JL, Cano J, Machado A. The single intranigral injection of LPS as a new model for studying the selective effects of inflammatory reactions on dopaminergic system. Neurobiology of disease. 2000;7:429–447. doi: 10.1006/nbdi.2000.0289. [DOI] [PubMed] [Google Scholar]
- 23.Tomás-Camardiel M, Rite I, Herrera AJ, de Pablos RM, Cano J, Machado A, et al. Minocycline reduces the lipopolysaccharide-induced inflammatory reaction, peroxynitrite-mediated nitration of proteins, disruption of the blood–brain barrier, and damage in the nigral dopaminergic system. Neurobiology of disease. 2004;16:190–201. doi: 10.1016/j.nbd.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Liu J, Sun T, Zhang X, Yao J, Kai M, et al. Anti-cancer small molecule JP-8g exhibits potent in vivo anti-inflammatory activity. Sci Rep. 2014;4 doi: 10.1038/srep04372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Jiang JW, Wilson BC, Du L, Yang SN, Wang JY, et al. Systemic infusion of naloxone reduces degeneration of rat substantia nigral dopaminergic neurons induced by intranigral injection of lipopolysaccharide. The Journal of pharmacology and experimental therapeutics. 2000;295:125–132. [PubMed] [Google Scholar]
- 26.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4th. Academic Press; 1998. [DOI] [PubMed] [Google Scholar]
- 27.Jain S, Amiji M. Tuftsin-modified alginate nanoparticles as a noncondensing macrophage-targeted DNA delivery system. Biomacromolecules. 2012;13:1074–1085. doi: 10.1021/bm2017993. [DOI] [PubMed] [Google Scholar]
- 28.Stratford S, Stec S, Jadhav V, Seitzer J, Abrams M, Beverly M. Examination of real-time polymerase chain reaction methods for the detection and quantification of modified siRNA. Analytical Biochemistry. 2008;379:96–104. doi: 10.1016/j.ab.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Makrigiorgos GM. Anti-primer quenching-based real-time PCR for simplex or multiplex DNA quantification and single-nucleotide polymorphism genotyping. Nat Protocols. 2007;2:50–58. doi: 10.1038/nprot.2007.11. [DOI] [PubMed] [Google Scholar]
- 30.Illum L. Transport of drugs from the nasal cavity to the central nervous system. Eur J Pharm Sci. 2000;11:1–18. doi: 10.1016/s0928-0987(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 31.Soane RJ, Frier M, Perkins AC, Jones NS, Davis SS, Illum L. Evaluation of the clearance characteristics of bioadhesive systems in humans1. International Journal of Pharmaceutics. 1999;178:55–65. doi: 10.1016/s0378-5173(98)00367-6. [DOI] [PubMed] [Google Scholar]
- 32.Sulzer D, Surmeier DJ. Neuronal vulnerability, pathogenesis, and Parkinson's disease. Movement disorders : official journal of the Movement Disorder Society. 2013;28:41–50. doi: 10.1002/mds.25095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim I-D, Shin J-H, Kim S-W, Choi S, Ahn J, Han P-L, et al. Intranasal Delivery of HMGB1 siRNA Confers Target Gene Knockdown and Robust Neuroprotection in the Postischemic Brain. Mol Ther. 2012;20:829–839. doi: 10.1038/mt.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAlpine FE, Lee JK, Harms AS, Ruhn KA, Blurton-Jones M, Hong J, et al. Inhibition of soluble TNF signaling in a mouse model of Alzheimer's disease prevents pre-plaque amyloid-associated neuropathology. Neurobiology of disease. 2009;34:163–177. doi: 10.1016/j.nbd.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobinick E, Gross H, Weinberger A, Cohen H. TNF-alpha Modulation for Treatment of Alzheimer's Disease: A 6-Month Pilot Study. Medscape General Medicine. 2006;8:25-. [PMC free article] [PubMed] [Google Scholar]
- 36.Ferger B, Leng A, Mura A, Hengerer B, Feldon J. Genetic ablation of tumor necrosis factor-alpha (TNF-alpha) and pharmacological inhibition of TNF-synthesis attenuates MPTP toxicity in mouse striatum. Journal of neurochemistry. 2004;89:822–833. doi: 10.1111/j.1471-4159.2004.02399.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim SS, Ye C, Kumar P, Chiu I, Subramanya S, Wu H, et al. Targeted delivery of siRNA to macrophages for anti-inflammatory treatment. Mol Ther. 2010;18:993–1001. doi: 10.1038/mt.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.