Abstract
Membrane proteins are assembled through balanced interactions among protein, lipids and water. Studying their folding while maintaining the native lipid environment is necessary but challenging. Here we present methods for analyzing key elements in membrane protein folding including thermodynamic stability, compactness of the unfolded state and folding cooperativity under native conditions. The methods are based on steric trapping which couples unfolding of a doubly-biotinylated protein to binding of monovalent streptavidin (mSA). We further advanced this technology for general application by developing versatile biotin probes possessing spectroscopic reporters that are sensitized by mSA binding or protein unfolding. By applying these methods to an intramembrane protease GlpG of Escherichia coli, we elucidated a widely unraveled unfolded state, subglobal unfolding of the region encompassing the active site, and a network of cooperative and localized interactions to maintain the stability. These findings provide crucial insights into the folding energy landscape of membrane proteins.
Understanding the free energy landscape of protein folding requires determination of the free energy levels and conformations of states populated during folding as well as analysis of energy barriers to reach the native conformation1. Experimentally, this task has been carried out by equilibrium and kinetic folding studies using denaturants that can readily shift population distribution between folded and unfolded states2. However, the overall shape of the folding energy landscape substantially changes in the presence of denaturants, and certain short-lived higher energy states may not be detected in denaturing conditions3,4. Thus, studying protein folding under native conditions is necessary for a full survey of the folding energy landscape. For water-soluble proteins, methods such as hydrogen-deuterium exchange (HDX), NMR relaxation dispersion and proteolysis have revealed the dynamic and multi-state nature of the native conformational ensemble4-8 which is critical to protein function9-11. However, such features remain largely unexplored for membrane proteins. The poor accessibility of solvent water to the interior of micelles and bilayers and the large sizes of protein-micellar and protein-liposomal complexes have made it difficult to apply these methods for characterizing the native ensemble of membrane proteins12,13.
Steric trapping is a promising tool for investigating thermodynamic stability and folding of membrane proteins directly under native conditions. The method couples unfolding of a target protein labeled with two biotin tags to competitive binding of bulky monovalent streptavidin molecules (mSA, MW=52 kD)14-18 (Fig. 1a for detailed description).
Figure 1. Principle of steric trapping and steric-trapping probes developed in this study.
(a) Steric trapping principle for measuring thermodynamic stability (ΔGoU) of proteins. After conjugation of biotin tags to two specific residues that are spatially close in the folded state but distant in the amino acid sequence, the first mSA binds unhindered to either biotin label with intrinsic binding affinity (ΔGoBind). Due to the steric hindrance with pre-bound mSA, the second mSA binds only when the native tertiary contacts between biotinylated sites are unraveled by transient unfolding. Coupling of mSA binding to unfolding leads to attenuation of the apparent binding affinity of the second mSA relative to that of the first mSA, whose degree is correlated with the protein stability. Thus, thermodynamic stability of the target protein can be determined by fitting of the second binding phase (see equations (2)–(4) in Online Methods). Overall, protein unfolding is driven by the affinity and concentration of mSA without perturbing the native solvent condition. Folding reversibility is tested upon addition of excess free biotin by which bound mSA molecules are released by competition. (b) Thiol-reactive biotin derivatives possessing a spectroscopic reporter group developed in this study. BtnPyr-IA (1): biotin (red shaded)-pyrene (green shaded)-iodoacetamide (blue shaded) conjugated to a lysine template, and BtnRG-TP (2): biotin (red shaded)-1-oxyl-2,2,5,5-tetramethylpyrroline spin label (green shaded)-thiopyridine (blue shaded) conjugated to a cysteine template.
Although promising, it is yet difficult to apply steric trapping to various types of membrane proteins. The method requires two essential features: two site-specifically conjugated biotin labels on a target protein and a probe to monitor mSA binding or protein unfolding. Site-specific biotinylation has been achieved by labeling of engineered cysteine residues with thiol-reactive biotin derivatives14,15,18. For detection of unfolding, widely-used tools such as tryptophan fluorescence and circular dichroism cannot be used due to large signal interferences from mSA molecules. A method for direct detection of mSA binding has not been developed yet. Thus, the application has been limited to proteins possessing convenient unfolding readouts such as absorbance of a conformation-sensitive intrinsic chromophore (e.g. retinal in bacteriorhodopsin15) and enzymatic activities (e.g. dihydrofolate reductase14 and diacylglycerol kinase18). In this study, we developed a generalized steric trapping strategy which utilizes novel thiol-reactive biotin probes containing spectroscopic reporter groups for sensitive detection of mSA binding and protein unfolding. This strategy was applied to analyze the thermodynamic stability, compactness of the unfolded state and folding cooperativity of the six-helical bundle intramembrane protease GlpG of E. coli.
GlpG is a member of the rhomboid protease family widely conserved in all kingdoms of life. Rhomboid proteases are involved in diverse biological processes by activating membrane-bound signaling proteins or enzymes via cleavage of a specific peptide bond near the membrane19-22. Due to the functional importance of rhomboid proteases and detailed structural information available (28 PDB entries, http://www.rcsb.org/), GlpG has emerged as an important model for studying the folding of helical membrane proteins. Critical regions for the stability have been identified using heat and sodium dodecylsulfate (SDS) denaturation tests of 151 variants23. A kinetic folding study using SDS as a denaturant has suggested the existence of a compact folding nucleus in the folding transition state24. A single-molecule tweezers study has shown that GlpG largely unfolds cooperatively at a constant tension25.
Here, using steric trapping, we provide new insights into the folding energy landscape of GlpG in the absence of heat, chemical denaturants or pulling force. We elucidated an expanded heterogeneous conformational ensemble of the unfolded state, a structural region that undergoes subglobal unfolding, and an intricate network of cooperative and localized interactions to maintain the stability of GlpG.
RESULTS
Design and synthesis of new steric-trapping probes
Our steric-trapping probes are highlighted by three features that are integrated into one molecular tag (Fig. 1b): (1) a biotin group for binding mSA, (2) a thiol-reactive group for conjugation to engineered cysteine residues on a target protein, and (3) a fluorescent or paramagnetic reporter group whose spectroscopic signal is sensitized by mSA binding or protein unfolding. Each probe was synthesized by stepwise substitutions of building blocks possessing characteristic features to a lysine or cysteine template (Supplementary Results, Supplementary Notes). BtnPyr-IA (1) is a pyrene-based fluorescent sensor to detect mSA binding. When used to doubly label a target protein, pyrene fluorescence is remarkably sensitive to binding of quencher-labeled mSA by Förster resonance energy transfer (FRET). BtnRG-TP (2) is a paramagnetic sensor possessing a 1-oxyl-2,2,5,5-tetramethylpyrroline spin label to detect protein unfolding. The spin labels allow distance measurements in the native and steric-trapped unfolded state using double electron-electron resonance spectroscopy (DEER).
Steric trapping controls reversible folding of GlpG
To prove the principle of our steric trapping strategy employing the new probes, we used GlpG as a model and its proteolytic activity as a folding indicator. Here all studies were performed in dodecylmaltoside (DDM) micelles, in which a majority of functional and folding studies of GlpG have been carried out23,26-29, and with the isolated transmembrane (TM) domain (residues 87–276) for which all structures of GlpG have been solved. For precise and efficient measurement of GlpG activity with membrane-bound substrates, we developed a fluorescence-based assay that can be transformed into a high-throughput format (Supplementary Fig. 1). The activity of the TM domain of GlpG was indistinguishable from that of the full-length protein (Supplementary Fig. 2).
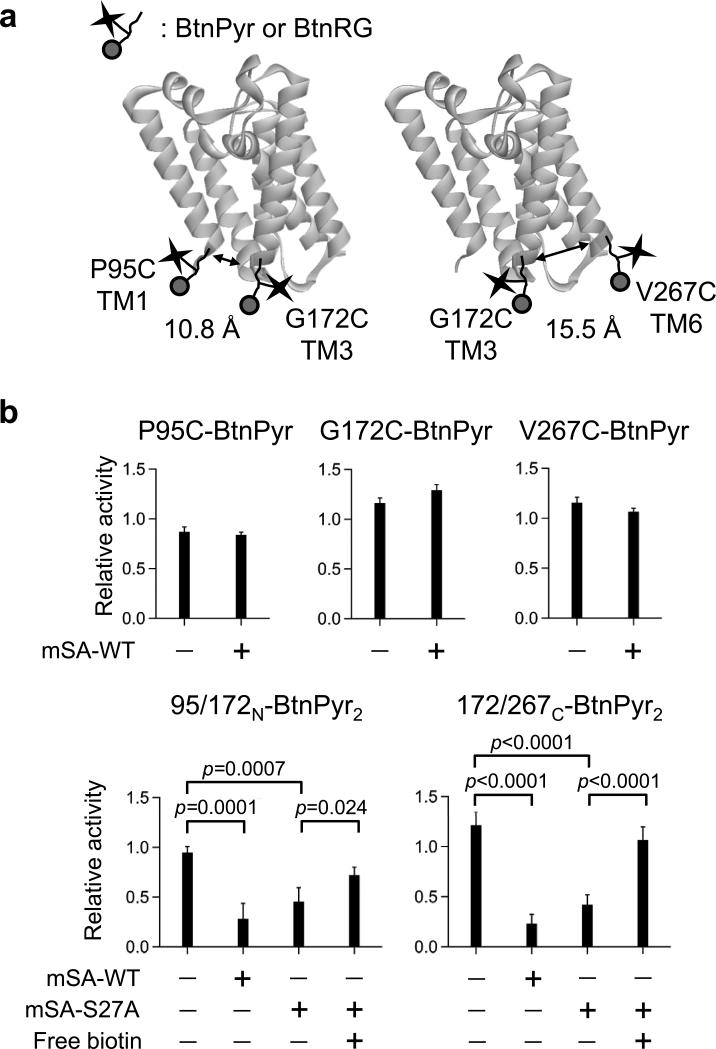
For steric trapping, we first identified optimal residue pairs for cysteine substitution to conjugate thiol-reactive biotin labels. After testing of multiple single- and double-cysteine variants, two double-cysteine variants, P95C/G172C and G172C/V267C were selected (Fig. 2a). The biotin pair conjugated to P95C/G172C is located in the approximate N-terminal half of GlpG (95/172N hereinafter) while the biotin pair to G172C/V267C is located in the C-terminal half (172/267C). Individual single-cysteine variants P95C, G172C and V267C labeled with fluorescent BtnPyr-IA maintained the wild-type activity level (Fig. 2b, top) and this activity level was not significantly altered after binding of wild-type mSA (mSA-WT). Michaelis-Menten analysis of the proteolytic activity showed that KM, kcat and kcat/KM of the single-biotin variants bound with mSA were indistinguishable from those of unbound forms demonstrating that binding of one mSA molecule to each biotin site did not perturb the structure and function of GlpG (Supplementary Fig. 2). The wild-type activity level was also maintained after labeling of double-cysteine variants. In marked contrast, saturated binding of mSA to two biotin labels on each variant induced a substantial loss of activity implying GlpG was trapped in the unfolded state (Fig. 2b, bottom).
Figure 2. GlpG reversibly unfolds by double-binding of mSA.
(a) Locations of two different biotin pairs for steric trapping in the structure of GlpG (PDB code: 3B4549) and their Cα-Cα distances. (b) Reversible control of GlpG folding tested by the proteolytic activity as a folding indicator. The second TM domain of the lactose permease50 of E. coli fused to staphylococcal nuclease was used as a model substrate throughout this study (Supplementary Fig. 1). All activity levels were normalized relative to the activity of wild-type GlpG. Error bars denote mean ± s. d. (n=5 for the data without mSA and n=3 for the data with mSA). Top panels: binding of wild-type mSA (mSA-WT) to individual single-cysteine variant labeled with BtnPyr did not affect the activity. Bottom panels: saturated binding of mSA-WT to each double-cysteine variant labeled with BtnPyr led to an inactivation of GlpG (the second bar from the left in each panel). To test folding reversibility, double-biotin GlpG variants were first inactivated with mSA-S27A possessing a weaker biotin binding affinity (Kd,bioin=1.4×10–9 M) for 2–5 days (the third bar). Next, excess free biotin was added to induce competitive dissociation of bound mSA (the fourth bar). All p-values obtained from Student's t-test were smaller than the threshold significance level (α=0.05), indicating that the activity changes for the unfolding and refolding reactions were significant.
Next, we tested if the steric-trapped unfolded state can refold after dissociation of bound mSA. Wild-type mSA binds biotin with an enormously high affinity (Kd,biotin≈10−14 M) and slow dissociation rate (koff≈days)30. Thus, we used mSA-S27A variant with a weaker biotin affinity (Supplementary Fig. 3) to facilitate dissociation of bound mSA by addition of excess free biotin (Fig. 1a)31. Both double-biotin variants which were inactivated with mSA-S27A significantly regained the activity upon addition of free biotin (Fig. 2b, bottom). For 95/172N-BtnPyr2, 50–70% of activity was regained while >90% was regained for 172/267C-BtnPyr2. Thus, we achieved reversible folding of GlpG without using denaturants by steric trapping.
Steric-trapped unfolded state is widely unraveled
So far, protein unfolding by steric trapping has been tested by the loss of enzymatic activity14,18, decrease of retinal absorbance15, or increased susceptibility to proteolysis14,15. Although those features indicate unfolding, a possibility remains that the protein conformation trapped with mSA molecules is only locally distorted or still compact with significant residual tertiary interactions. Therefore, to elucidate the conformation of the steric-trapped unfolded state as well as to gain insights into the unfolded state ensemble of membrane proteins under non-denaturing conditions, we used a thiol-reactive biotin derivative possessing a spin label (BtnRG-TP) (Fig. 1b). By labeling of double-cysteine variants of GlpG with this probe, we have the advantage of both trapping the unfolded state and measuring the distances between spin labels using DEER. DEER allows for measurements of long-range (15–60 Å) inter-spin distances32. It provides not only the most probable distance but also distance distribution, which is of great interest in characterization of the unfolded state33,34. Here, we obtained inter-spin distances for 95/172N-BtnRG2 and 172/267C-BtnRG2 in their native, SDS-induced unfolded and steric-trapped unfolded states (Fig. 3). In the native states, the distance distributions between BtnRG labels were overall similar to those between well-characterized R1 spin labels35 (Supplementary Fig. 4) demonstrating that our BtnRG label is capable of distance mapping of protein conformation.
Figure 3. DEER suggests steric trapping induce wide separation of two biotinylated sites.
Background-subtracted dipolar evolution data and their fits (left) and inter-spin distances (right) for the native (dashed lines), SDS-induced unfolded (gray solid lines, SDS mole fraction =[SDS]/([DDM]+[SDS])>0.8, in which the unfolded fraction exceeded 0.9), and steric-trapped (black solid lines) unfolded states for (a) 95/172N-BtnRG2 GlpG and (b) 172/267C-BtnRG2 GlpG. The approximate upper limit of the reliable mean distance was ~53 Å32.
For both variants, SDS induced substantial broadening of the inter-spin distance distribution over the range from the native-like distances (15–35 Å) up to ~60 Å (Fig. 3, right panels), which indicates a heterogeneous conformational ensemble of the unfolded state in SDS. Interestingly, in non-denaturing DDM micelles, the steric-trapped unfolded states also exhibited similarly broad inter-spin distance distributions. The increase of the most probable distance from ~25 Å in the native state to ~55 Å in the steric-trapped unfolded state corresponds to a ~30 Å expansion of each half of the polypeptide chain covered by the respective biotin pair. This increased dimension is comparable to the whole diameter of native GlpG. Thus, our DEER data for GlpG rules out a compact unfolded state under non-denaturing conditions, which has been observed for several water-soluble proteins36. We note that, because of the detection limit of DEER, even longer distance components (>60 Å) may have existed but not been detected. Addition of dithiothreitol to break the disulfide bond between GlpG and the biotin label bound with mSA led to a regain of activity (>70%) indicating that a majority of the unfolded conformations were able to refold (Supplementary Fig. 5).
Steric repulsion between bound mSA molecules may have biased the conformational ensemble of the unfolded state. However, during the selection of optimal biotin pairs, we found that saturated binding of mSA to the biotin pairs conjugated to G94C/G172C and G172C/N271C, whose Cα-Cα distances were similar to those of 95/172N and 172/267C, completely retained the activity and therefore did not induce unfolding (Supplementary Fig. 6). This result implies that bound mSA molecules are allowed to coexist within close distances probably also in the steric-trapped unfolded state. Therefore, steric repulsion may not fully explain the expanded unfolded state.
We further characterized the conformational features of the steric-trapped unfolded state using proteolysis by chymotrypsin, which primarily targets aromatic residues prevalent throughout GlpG (Supplementary Fig. 7). While the unfolded state bound with two mSA molecules was gradually proteolyzed over ~30 min, either native GlpG or GlpG bound with one mSA molecule was not significantly proteolyzed. As a control, we tested proteolysis of casein which dominantly exists in random coil conformation in aqueous solution37. Casein was rapidly proteolyzed within 1 min. Thus, we speculate that the steric-trapped unfolded state was significantly protected by secondary structures and micelles, but possessed more dynamic features than the native state.
DEER and proteolysis results demonstrate that steric trapping induced a true unfolded state, which was an ensemble of expanded dynamic and heterogeneous conformations. This work also represents the first measurement of the physical dimension of a helical membrane protein in its unfolded state under non-denaturing conditions.
Stability of GlpG determined by steric trapping
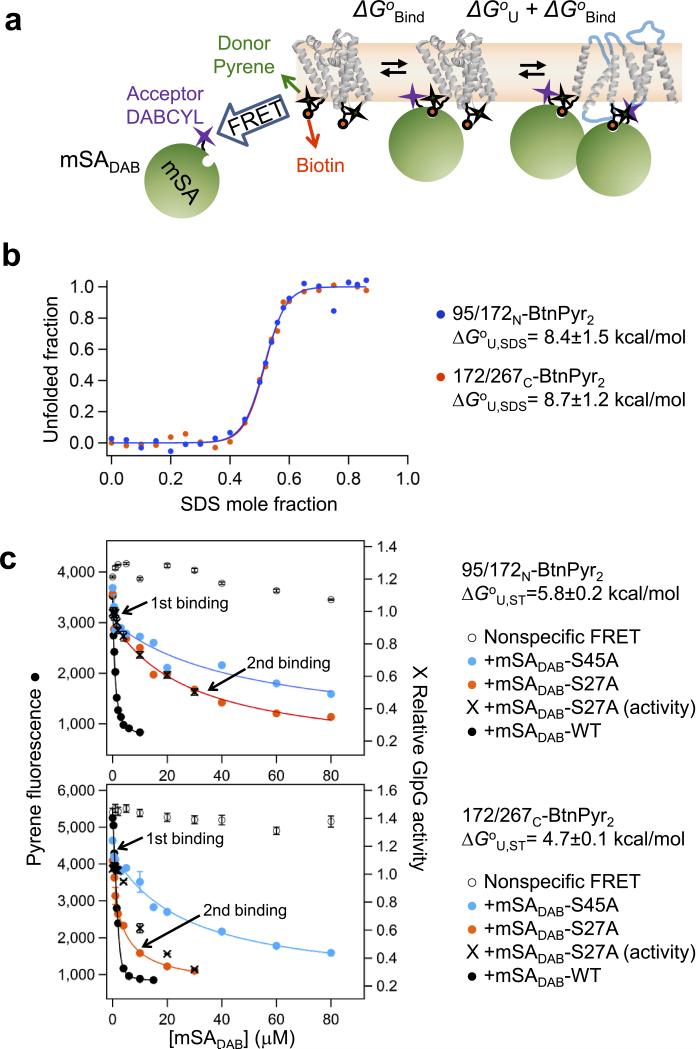
To develop a general steric-trapping strategy that does not depend on specific characteristics of a target protein, ideally the spectroscopic signal from the reporter group in our probe (Fig. 1b) should sensitively change upon either mSA binding or protein unfolding. Here we achieved a highly sensitive detection of mSA binding by employing FRET between pyrene on BtnPyr label and non-fluorescent quencher DABCYL on mSA (mSADAB) (Fig. 4a).
Figure 4. Thermodynamic stability of GlpG using steric trapping and SDS denaturation.
(a) Steric-trapping strategy using FRET between fluorescent pyrene (donor) in BtnPyr labeled on GlpG and non-fluorescent quencher DABCYL (acceptor) thiol-specifically labeled near the biotin binding pocket (Y83C) of the active subunit of mSA (mSADAB). (b) Equilibrium unfolding of GlpG variants 95/172N-BtnPyr2 and 172/267C-BtnPyr2 as a function of SDS mole fraction measured by FRET between Trp residues (donor) of GlpG and pyrene (acceptor) on BtnPyr labels (Supplementary Fig. 8). Errors in ΔGoU,SDS values denote mean ± s. d. from fitting. (c) Binding isotherms of 95/172N-BtnPyr2 and 172/267C-BtnPyr2 with three mSA variants mSADAB-WT (black circles, Kd,biotin=~10−14 M), mSADAB-S27A (red circles, Kd,biotin=1.4×10−9 M) and mSADAB-S45A (blue circles, Kd,biotin=9.0×10−9 M) (Supplementary Fig. 3). The activity change for each double-biotin variant (crosses, right y-axis) was measured at an increasing concentration of mSADAB-S27A. The thermodynamic stability (ΔGoU,ST) of each variant was obtained by fitting of the second mSA-binding phase to equation (4) in Online Methods. Procedures to obtain nonspecific FRET (open circles) are described in Online Methods. Errors in fluorescence denote mean ± s. d. (n=4). Errors in activity denote ± s. d. from fitting. Errors in ΔGoU,ST values denote mean ± s. d. (n=3).
SDS denaturation and linear extrapolation of the denaturation data to zero-SDS mole fraction yielded the same stability (ΔGoU,SDS) for 95/172N-BtnPyr2 (8.4±1.5 kcal/mol) and 172/267C-BtnPyr2 (8.7±1.2 kcal/mol) (Fig. 4b and Supplementary Fig. 8), which was similar to that of the full-length wild type (8.2±1.4 kcal/mol)24. This result indicates that the two double-biotin variants possessed the same global stability as wild type GlpG. By design, steric trapping specifically captures transient unfolding of native interactions between a biotin pair. Thus, probing the stability with two biotin pairs located in different regions (Fig. 2a) provides a novel opportunity to test the folding cooperativity of GlpG.
Using FRET, we obtained high-quality binding isotherms between each double-biotin variant and three mSADAB variants with different biotin affinities (Fig. 4c). mSADAB binds the biotin label at each site with a similar affinity (Supplementary Fig. 3a). An essential element of steric trapping to determine protein stability is choosing an mSA variant whose binding to a biotin label (ΔGoBind) optimally competes with folding (ΔGoU) to yield attenuated second binding in a desirable [mSA] range (Fig. 4a). Among mSA variants tested, mSADAB-S27A yielded an optimal separation of the first tight and second weaker binding phases (Fig. 4c). Parallel activity measurements showed that, for each GlpG variant, the second binding coincided with the activity loss (i.e. unfolding), which validated the unfolding-binding coupling. The same coupling was observed with a high-affinity variant, mSADAB-WT, further confirming that the activity loss strictly depended on the second binding of mSADAB (Supplementary Fig. 9).
Fitting of the second binding phases yielded the thermodynamic stability (ΔGoU,ST, ST: steric trapping), 5.8±0.2 kcal/mol for 95/172N-BtnPyr2 and 4.7±0.1 kcal/mol for 172/267C-BtnPyr2 (equation (4) in Online Methods) in non-denaturing DDM micelles (Fig. 4c). Both ΔGoU,ST's were significantly lower than the extrapolated stability from SDS denaturation (8.4–8.7 kcal/mol) but higher than the stability in a bicelle (6.5 kBT equivalent to ~4 kcal/mol) extrapolated to zero force from single-molecule tweezers study25. If GlpG unfolded cooperatively, the same ΔGoU,ST would be expected regardless of the position of the biotin pair. However, while SDS denaturation yielded the same global stability for the two double-biotin variants, their stabilities obtained by steric trapping were comparable but significantly different by 1.1±0.2 kcal/mol.
Subglobal unfolding of GlpG near the active site
To track down the origin of the discrepancy between the stability obtained by steric trapping (ΔGoU,ST) under a non-denaturing condition and the extrapolated stability obtained by SDS denaturation (ΔGoU,SDS), we directly measured the stability of the two double-biotin variants using steric trapping in the range of SDS mole fraction (XSDS=0–0.4), where a major fraction of GlpG existed in the folded state (folded fraction >0.9) (Fig. 4b). The ΔGoU,ST vs XSDS plot (Fig. 5) revealed two major features that clearly deviated from the behavior predicted from linear extrapolation of SDS denaturation data. First, rather than following a linearly-decreasing trend, ΔGoU,ST of both variants exhibited an upward curvature as XSDS increased. Second, while ΔGoU,ST of 95/172N-BtnPyr2 were overall larger than that of 172/267C-BtnPyr2, they remarkably converged at XSDS≈0.4 where the main unfolding transition by SDS began, and this convergence was maintained up to XSDS=0.5. This result confirms that the two variants possessed the same global stability.
Figure 5. Dependence of thermodynamic stability of GlpG on SDS mole fraction.
The plot containing ΔGoU,ST's (diamonds) obtained by steric trapping and ΔGoU,SDS's (squares) obtained by SDS denaturation as a function of SDS mole fraction (XSDS) for 95/172N-BtnPyr2 and 172/267C-BtnPyr2. To fit ΔGoU,ST, we accounted for the changes in the biotin affinity of mSADAB variants which depended on XSDS (Supplementary Fig. 3d). Errors in ΔGoU,ST denote ± s. d. from fitting. Solid lines are the linear-regression fits of ΔGoU,ST in the range of XSDS=0.2–0.4 and dashed lines indicate the extrapolation lines of ΔGoU,SDS to zero XSDS from equilibrium SDS denaturation. The slope in the ΔGoU vs XSDS plot represents the m-value. For 95/172N-BtnPyr2, m=16±3 (blue dashed line) from SDS denaturation and m=14±2 (blue solid line) from steric trapping. For 172/267C-BtnPyr2, m=17±2 (red dashed line) from SDS denaturation and m=8±1 (red solid line) from steric trapping. Errors in the m-values denote ± s. d. from fitting.
The overall nonlinearity of ΔGoU,ST against XSDS implies a complex interaction between GlpG and DDM/SDS micelles. A similar disagreement between steric trapping and SDS denaturation has been reported for bacteriorhodopsin in DMPC/CHAPSO/SDS bicelles15. In the case of GlpG, ΔGoU,ST of both variants reached a maximum at XSDS≈0.2 but linearly decreased at higher XSDS (Fig. 5). Notably, in the range of XSDS=0.2–0.4, the m-value of 95/172N-BtnPyr2 (14±2 kcal/mol/XSDS), which represents the slope of ΔGoU,ST against XSDS, was significantly larger than that of 172/267C-BtnPyr2 (8±1 kcal/mol/XSDS) but similar to those obtained by SDS denaturation (16–17 kcal/mol/XSDS).
For water-soluble proteins, the m-value is correlated with the hydrophobic surface area exposed upon unfolding38. Although the physical meaning of the m-value in SDS denaturation is still under debate39, it is most likely related to the difference in the affinity of SDS for different states of the protein, hence to the degree of exposure of buried stabilizing interactions upon unfolding40. Therefore, from the different denaturant sensitivities of the two double-biotin variants, we conclude that trapping of the unfolded state with the biotin pair 95/172N-BtnPyr2 led to substantial exposure of the buried surfaces throughout the protein, while trapping with the biotin pair 172/267C-BtnPyr2 mainly occurred through subglobal unfolding which exposed less buried-surface area9. Steric trapping of 172/267C-BtnPyr2 detects transient separation between TM3 and TM6 to which biotin labels are conjugated, and TM6 contains a biotin label (V267C-BtnPyr) as well as His254 of the catalytic dyad (Fig. 6a). Thus, subglobal unfolding should directly involve disruption of the active site.
Figure 6. Cooperativity map reveals a network of clustered cooperative and localized interactions for the stability of GlpG under a native micellar condition.
(a) Scheme for quantifying the cooperativity of interactions of a specific side chain. The stability changes (ΔΔGoU) induced by the same mutation (black star) were probed with two biotin pairs, 95/172N-BtnPyr2 and 172/267C-BtnPyr2 located in the N- and C-terminal regions, respectively, and compared to each other to yield ΔΔΔGoU using equation (1). The cyan-backbone region designates subdomain I (TM1-L1-TM2-TM3-L3198), which ends at residue 198 in the L3 loop (marked with a magenta wedge) and the yellow-backbone region (L3199-TM4-TM5-L5-TM6) indicates subdomain II. The uncertainty of the subdomain-division point was ±20–30 residues around residue 198. The detailed strategy for the subdomain dissection and the estimation of the uncertainty is described in Supplementary Fig. 10. Catalytic dyad composed of Ser201/His254 is shown as spheres. (b) Cooperativity map at a side-chain resolution. The map shows the “cooperative” (green, |ΔΔΔGoU|≤RT=0.6 kcal/mol) and “localized” (|ΔΔΔGoU|>RT) side-chain interactions. Localized interactions were further divided using additional cut-off energy values, 2RT≥|ΔΔΔGoU|>RT (“moderately-localized” interactions) and |ΔΔΔGoU|>2RT (“highly-localized” interactions). Each side chain was color-coded based on these criteria for ΔΔΔGoU as shown in the figure. Interactions mediated by residues G261 and A265 (denoted with stars) were “over-propagated”. Errors in individual ΔGoU were ±0.1−±0.2 kcal/mol (mean ± s. d. from fitting) and errors in ΔΔΔGoU ranged from ±0.1−±0.4 kcal/mol, which were calculated using the propagation of errors in ΔGoU (Supplementary Table 1).
Subglobal unfolding has been frequently observed from HDX studies of water-soluble proteins9,41,42, but not been reported for membrane proteins. Besides the different m-values, subglobal unfolding of GlpG is further supported by the lower stability of 172/267C-BtnPyr2 from steric trapping (Fig. 4c) and the reproducibly higher refolding yield of 172/267C-BtnPyr2 (>90%) than that of 95/172N-BtnPyr2 (50–70%) (Fig. 2b), implying different unfolded states.
We also note that ΔGoU,ST was larger than ΔGoU,SDS after it crossed the extrapolation lines at XSDS≈0.1 and this discrepancy became increasingly pronounced up to 2.8 kcal/mol at XSDS=0.4 (Fig. 5). We reason that the larger ΔGoU,ST was primarily due to the conformational difference between the steric-trapped unfolded state and the SDS-induced unfolded state. Our DEER result supports this argument (Fig. 3). The steric-trapped unfolded state on average exhibited larger inter-spin distances than the SDS-induced unfolded state. Thus, steric trapping appears to induce more unraveled conformations than SDS at least for the interactions between the biotinylated sites. However, we are cautious with this direct comparison because the compactness of the SDS-induced unfolded state may change as a function of XSDS due to the effects of SDS on the size and shape of mixed micelles43.
Strategy to identify cooperative interactions
The higher stability and more substantial unfolding obtained with 95/172N-BtnPyr2 indicates that the native interactions between this biotin pair in the N-terminal region are critical to the conformational integrity of the whole protein. On the other hand, the lower stability and subglobal unfolding obtained with 172/267C-BtnPyr2 indicates that the C-terminal region possesses differential folding properties from the N-terminal region. This result suggests complex energetic coupling between different regions in GlpG. To clarify this complexity, we developed a method to identify cooperative and localized interactions that contribute to the protein stability at a side-chain resolution (Fig. 6a).
First, we dissected GlpG into two subdomains: (1) the more stable N-terminal subdomain I encompassing TM1-L1-TM2-TM3-L3198 (ending at residue 198 in the L3 loop) whose unfolding was trapped with 95/172N-BtnPyr2, and (2) the less stable C-terminal subdomain II consisting of L3199-TM4-TM5-L5-TM6 (starting from residue 199) whose subglobal unfolding was trapped with 172/267C-BtnPyr2 (Supplementary Fig. 10 for dissection procedures). The uncertainty of the division point was ±20–30 residues.
Second, we made a single mutation (typically to alanine) in either subdomain to perturb a specific side-chain interaction in the background of 95/172N-BtnPyr2 and 172/267C-BtnPyr2. We referred to these background double-biotin variants as “wild type (WT)” because the wild-type native interactions were equally preserved in both as shown by SDS denaturation (Fig. 4b). We referred to two double-biotin variants possessing the same mutation as “mutants (Mut)”. Next, we probed the stability changes induced by the mutation with two different biotin pairs using steric trapping. We quantified the differential effect of the same mutation on the stability of each subdomain (ΔΔΔGoU) using equation (1) containing the stabilities of four variants:
| (1) |
ΔΔGoU,95/172N-BtnPyr2(WT-Mut) and ΔΔGoU,172/267C-BtnPyr2(WT-Mut) designate the stability changes caused by the same mutation in the backgrounds of 95/172N-BtnPyr2 and 172/267C-BtnPyr2, respectively. Thus, ΔΔΔGoU represents the difference in the stability changes that are probed with two different biotin pairs upon the same mutation.
If a mutation causes a similar degree of destabilization for both double-biotin variants with a difference within thermal fluctuation energy (|ΔΔΔGoU|≤RT=0.6 kcal/mol; R: gas constant; T=298K), the mutated site engages in a “cooperative” interaction. That is, the perturbation by the mutation similarly propagates to both subdomains. Among the cases where |ΔΔΔGoU|>RT, if a mutation preferentially destabilizes the subdomain containing it, the perturbed interactions are “localized” within that subdomain. If mutation of a residue, which makes its side-chain contacts only with the subdomain containing it, preferentially destabilizes the other subdomain, we classified the perturbation as “over-propagated”.
Cooperativity network in GlpG
We targeted 20 residues covering key packing regions23 and analyzed their roles in the folding cooperativity of GlpG (Supplementary Table 1). The stability changes upon mutation ΔΔGoU(WT-Mut) obtained by steric trapping were reasonably well correlated with the changes in melting temperature ΔTm(WT-Mut)23, which validated our approach (Supplementary Fig. 11). 20 ΔΔΔGoU values were distributed over a wide range from −1.8 to 2.0 kcal/mol and their individual errors ranged from ±0.1 to ±0.4 kcal/mol, smaller than RT (Supplementary Fig. 12). We applied four cut-off values, ΔΔΔGoU= −2RT, −RT, RT and 2RT (i.e. five sets of the cooperativity profile) to account for the wide distribution of ΔΔΔGoU as well as to more precisely resolve the degree of cooperativity of each side-chain interaction.
We mapped the effects of mutations onto the structure, which we called the “cooperativity map” (Fig. 6b). Surprisingly, we observed significant clustering of cooperative and localized interactions in defined regions in the GlpG structure and divided their spatial distributions into four distinct groups. First, cooperative interactions of five residues, Met100, Leu161, Leu174, Thr178 and Ser201, clustered in the buried region which was surrounded by subdomain I and the subdomain interface near the center of the membrane. This cooperative cluster overlapped with one of the key packing regions previously identified23 and partially with the folding nucleus formed between TM1 and TM2 in the folding transition state24.
Second, all tested residues located in the folded L1 loop (Tyr138, Thr140 and Leu143) and the residue packed against L1 (Cys104) in subdomain I engaged in moderately (RT<ΔΔΔGoU≤2RT) or highly (2RT<ΔΔΔGoU) localized interactions in subdomain I. This region is known to form non-native interactions in the folding transition state24. Third, Leu225 (ΔΔΔGoU<−2RT) and Gln226 (−2RT≤ΔΔΔGoU<−RT) in TM5 in subdomain II, which were located at the subdomain interface and exposed to the water-micelle interface, respectively, were both classified as localized in subdomain II. TM5 is not tightly packed against the rest of the protein and does not significantly contribute to the thermostability23.
In the fourth cluster, interestingly, mutation of residues at the TM4/TM6 interface (Ala253, Gly261, Ala265 and Asp268) in subdomain II preferentially destabilized subdomain I, not the subdomain containing them. Particularly, Gly261 and Ala265 make their side-chain contacts entirely with the residues in subdomain II, but perturbing these interactions exerted larger impacts on the stability of subdomain I. Thus, we classified these residues as over-propagated. The TM4/TM6 interface harbors the catalytic dyad and plays a pivotal role in both stability and function of GlpG23. Especially, Gly261 and the dyad are absolutely conserved among rhomboid proteases44. Our result suggests that these conserved residues are also critical to the energetic coupling between different regions of GlpG. Breakage of the interactions near the C-terminus and its propagation towards the N-terminus is known to be the primary mechanism of the force-induced unfolding of GlpG25.
It should be noted that 5 among 20 tested mutations completely inactivated GlpG (Supplementary Table 1). Thus, our steric trapping strategy allowed stability measurements of not only functional but also non-functional variants, which had been difficult under the original steric-trapping framework. An ANOVA test of the five sets of ΔΔΔGoU including the upper and lower standard-deviation limits of each ΔΔΔGoU indicates that differences among these sets were statistically significant (Supplementary Fig. 12). While the two double-biotin variants bearing the same mutation exhibited differential stability change in DDM micelles, they possessed the same global stability from SDS denaturation (Supplementary Fig. 13). Therefore, we conclude that the networked side-chain interactions revealed in this work is a novel phenomenon that occurs under native conditions.
DISCUSSION
Here we presented a new steric-trapping strategy to investigate thermodynamic stability of membrane proteins and conformation of their unfolded state under native conditions by employing novel thiol-reactive biotin tags. Fluorescent BtnPyr allowed determination of the thermodynamic stability of GlpG through high-quality binding isotherms obtained by FRET. Paramagnetic BtnRG enabled characterization of the unfolded state based on the distance measurements using DEER. Because this combined strategy is not limited by either target-specific unfolding readout or specific lipid environments, it is applicable to other types of membrane proteins including nonfunctional and misfolded variants whose folding characterization is difficult under native conditions.
Unfolded state of proteins has gained significant interest because it determines thermodynamic stability with the folded state, directs folding mechanisms, and serves as a target for chaperoning and degradation45. However, conformation of the unfolded state of membrane proteins was difficult to study under native conditions due to its transient nature preventing biophysical analysis. By combining DEER and steric trapping, we elucidated a largely-unraveled dynamic and heterogeneous conformational ensemble of the unfolded state of GlpG in non-denaturing micellar solution. It is still an open question to what extent trapping would affect the protein conformation beyond the region containing the biotin pair. Investigating the conformation of the unfolded state in a lipid bilayer, which provides a more defined hydrophobic environment than micelles, will be a crucial future task to understand thermodynamics and mechanisms of membrane protein folding in cell membranes.
We identified subglobal unfolding of the C-terminal region which encompasses the active site. This asymmetric stability profile of GlpG is analogous to the highly-polarized folding transition state possessing a compact folding nucleus in TM1–TM2 and largely unstructured TM3–TM624. Single-molecule tweezers study has also identified TM3–TM6 or TM5–TM6 as a flexible region25. Although we defined the region that underwent subglobal unfolding as the approximate C-terminal half, it would be more reasonable to interpret it as an ensemble-averaged event which involved unfolding of a various number of the C-terminal helices. Our work is unique in that we demonstrated partial unfolding even under a non-denaturing condition, which reflects intrinsic conformational malleability of the region that encompasses the active site. Although it is not clear if subglobal unfolding is necessary during the catalytic cycle of GlpG, we speculate that this malleability is adequate for conformational changes required for substrate interaction and catalytic mechanism. Further supporting this idea, disordering of the L5 loop, partial unfolding of TM5 and tilting of TM6 have been observed from crystal structures of GlpG in apo and inhibitor-bound forms27,46,47.
Our unprecedented cooperativity analysis suggests that the helical-bundle architecture of GlpG is maintained through a network of cooperative and localized interactions. Although the cooperativity network and its role in protein stability and function have been analyzed for water-soluble proteins4,7,9,41,48, such aspects have not been investigated for membrane proteins. Our experimentally-determined cooperativity map indicates that the degree of cooperativity was the largest for the buried residues near the center of the membrane and faded out towards the lipid- and water-contacting regions. This positional dependence of the cooperativity profile suggests that complex environmental constraints for stabilizing membrane proteins, i.e. protein-protein, protein-lipid and protein-water interactions, play an important role in the organization of the interaction network. Our general steric trapping strategy and steric trapping-based approaches will serve as powerful tools for exploring the folding energy landscape of membrane proteins in native lipid bilayers, which still remains as a far-reaching goal.
ONLINE METHODS
Synthesis of BtnPyr-IA and BtnRG-TP
Synthesis schemes and characterizations are shown in Supplementary Notes.
Preparation of GlpG DNA constructs
GlpG gene was amplified from chromosomal DNA of E. coli strain MG1655 (Coli Genetic Stock Center at Yale University) using primers containing NdeI and BamHI restriction sites. The amplified gene was ligated into pET15b vector with an N-terminal His6-tag. Site-directed mutagenesis for introducing amino acid substitutions was performed using the QuikChange Site-Directed Mutagenesis Kit (Agilent).
Expression and purification of GlpG
GlpG was expressed in E. coli BL21(DE3) RP strain. Cells were grown at 37 °C until OD600 = 0.6 was reached. Protein expression was induced with 0.5 mM isopropyl β-thiogalactopyranoside (IPTG, GoldBio), followed by additional cultivation at 15 °C for 16 h. GlpG was purified from the total membrane fraction obtained by ultracentrifugation (Beckman Coulter, Type 45 Ti rotor, 50,000 g, for 2 h) using Ni2+-NTA affinity chromatography (Qiagen) after solubilization with 2% n-dodecyl-β-D-maltoside (DDM, Anatrace).
Labeling of GlpG and determination of labeling efficiency using SDS-PAGE gel shift assay
For labeling, purified cysteine variants (0.2% DDM, 50 mM Tris-(hydroxymethyl) aminomethane hydrochloride (TrisHCl), 200 mM NaCl and pH 8.0) were diluted to less than 100 μM and incubated with a ten-fold molar excess Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl, Pierce) for 1 h at room temperature. 40 times molar excess of BtnPyr-IA or BtnRG-TP dissolved in dimethyl sulfoxide (DMSO) (~20 mg/ml) was added to the mixture while vortexing. Labeling reaction was allowed to proceed at room temperature overnight in the dark. Excess free labels were removed by extensive washing of the proteins bound to Ni2+-NTA affinity resin using 0.2% DDM, 50 mM TrisHCl, 200 mM NaCl and pH 8.0 solution. Labeled GlpG was dialyzed against 0.02% DDM, 50 mM TrisHCl, 200 mM NaCl, pH 8.0 buffer to remove imidazole.
Typically, the labeling efficiency of BtnPyr-IA and BtnRG-TP ranged from 1.5–2.2 as estimated from SDS-PAGE gel shift assay or comparison of the concentration of BtnPyr determined by pyrene absorbance (ε346nm=43,000 M·cm−1) and the concentration of GlpG determined by DC protein assay (Bio-Rad). SDS-PAGE was employed utilizing the facts that mSA maintains its tetrameric structure and the biotin-mSA complex is resistant to dissociation in the presence of SDS. SDS-PAGE gel shift assay (Supplementary Fig. 14) was carried out as follows: 10 μl of 5 μM of labeled GlpG was incubated with 10 μl of 2% SDS sample-loading buffer with 10% (v/v) β-mercaptoethanol for 30 min. Then, wild-type monovalent streptavidin (mSA-WT) was added to labeled GlpG (GlpG:mSA-WT molar ratio of 1:3) and the mixture was incubated at room temperature for 30 min before SDS-PAGE without sample heating. The gel box was incubated in ice during electrophoresis to prevent heat-induced dissociation of mSA-WT bound to biotin label on GlpG. Labeling efficiency was determined by comparing the intensities that correspond to single-mSA bound GlpG and double-mSA bound GlpG after accounting for the molecular mass of GlpG and mSA (AlphaImager, ProteinSimple). GlpG with no label was not considered because this species does not bind mSA, thus not participating in steric trapping. mSA was prepared as described previously30,51.
Expression and purification of GlpG substrate SN-LacYTM2
As a folding indicator for GlpG, we used its proteolytic activity mediating specific cleavage of a transmembrane (TM) substrate, the second TM domain of the lactose permease of E. coli50 fused to staphylococcal nuclease (SN-LacYTM2) (Supplementary Fig. 1). The DNA construct for LacYTM2 was amplified from a DNA template containing full length lactose permease using primers containing XmaI and XhoI restriction sites, which was then ligated into a pET30a vector containing SN domain16, TEV protease recognition site, and C-terminal His6-tag (SN-TEV-LacYTM2-His6). In the LacYTM2 region, the position which was five residues upstream from the scissile bond (P5 position) was substituted with cysteine for labeling with thiol-reactive, environment-sensitive fluorophore iodoacetyl-7-nitrobenz-2-oxa-1,3-diazol (IA-NBD amide, Setareh Biotech). SN-TEV-LacYTM2-His6 containing the substituted cysteine was expressed in BL21(DE3) RP E. coli strain. The protein was expressed, purified and labeled using the protocol for SN-GpATM-His6 described previously51.
Fluorescence-based high-throughput activity assay for GlpG
Activity assay (Supplementary Fig. 1) was initiated by addition of 10 times molar excess of NBD-labeled SN-LacYTM2 to purified GlpG. Time-dependent changes of NBD fluorescence was monitored in 96-well plate using SpectraMax M5e plate reader (Molecular Devices) with excitation and emission wavelengths of 485 nm and 535 nm, respectively. Fluorescence change was normalized to a control sample containing NBD-SN-LacYTM2 alone. The effect of mSA binding on the activity of single- and double-biotin variants of GlpG were tested by addition of excess mSA-WT (20 μM). The GlpG-mSA mixture was incubated overnight (single-biotin variants), for 2 days (172/267C-BtnPyr2) or for 5 days (95/172N-BtnPyr2). Folding reversibility was tested using the following steps: Each double-biotin GlpG variant was first inactivated with mSA-S27A variant (20 μM) possessing a weaker biotin binding affinity (Kd,bioin=1.4 nM) for the same incubation time as with mSA-WT. Next, excess free biotin (2 mM) was added to induce competitive dissociation of bound mSA. The activity of refolded GlpG was measured after incubation overnight. The statistical significance of the activity changes upon unfolding and refolding were evaluated using Student's t-test (n=3–5).
Double electron-electron resonance EPR spectroscopy (DEER-EPR)
DEER-EPR measurements were performed on a Bruker Elexsys 580 spectrometer with Super Q-FTu Bridge, Bruker ER 5107DQ resonator and 10 W Q-band amplifier at 80 K. The spin-labeled samples ranging from 80 to 160 μM GlpG were flash-frozen in quartz capillaries using a liquid nitrogen bath immediately prior to data collection. For data collection, 36-ns π-pump pulse was applied to the low field peak of the nitroxide absorption spectrum, and the observer π/2 (16 ns) and π (32 ns) pulses were positioned 17.8 G (50 MHz) upfield, which corresponded to the nitroxide center resonance. A two-step phase cycling (+x, −x) was carried out on the first (π/2) pulse from the observer frequency. The time domain signal collected for each sample varied from 2.3 to 2.5 μs. Based on the collection time, the reliable inter-spin distance range was ~15−~60 Å. DEER data were analyzed using the program LongDistances, which was written in LabVIEW by Christian Altenbach (http://www.biochemistry.ucla.edu/biochem/Faculty/Hubbell/).
Time-dependent proteolysis of GlpG using chymotrypsin
GlpG variants (5 μM) were incubated for 7 days (95/172N-BtnPyr2: residual activity 40%) or 4 days (172/267C-BtnPyr2: residual activity 20%) in the presence and absence of 25 μM mSA-WT in 20 mM sodium phosphate (pH 7.5), 20 mM DDM, 200 mM NaCl, 1 mM TCEP buffer. In the presence of mSA-WT, the residual activities were ~40% for 95/172N-BtnPyr2 and ~20% for 95/172N-BtnPyr2 relative to those without mSA before proteolysis. Proteolysis of 5 μM casein (from bovine milk, Sigma) was performed in the same buffer condition but without prolonged incubation. For all protein samples, proteolysis was initiated by the addition of 2.5 μM chymotrypsin (bovine chymotrypsin-α: sequencing grade, Sigma) to 10 μL aliquots, and quenched at specified time by the addition of 10 mM permethylsulfoxide. Time-dependent proteolysis was monitored by SDS-PAGE (Supplementary Fig. 7).
Construction of binding isotherms to determine thermodynamic stability of GlpG by steric trapping using FRET
1 μM of GlpG labeled with BtnPyr was titrated with mSA specifically labeled with DABCYL-plus-maleimide (AnaSpec) at Y83C-position of the active subunit (mSADAB) in 5 mM DDM, 0.25 mM TCEP, 20 mM sodium phosphate and 200 mM NaCl (pH 7.5). The titrated samples were transferred to a 96-well UV-compatible microplate, sealed with a polyolefin tape, and incubated for 5 days (for 95/172N-BtnPyr2) or 2 days (for 172/267C-BtnPyr2) at room temperature. Binding was monitored by the decrease of pyrene-monomer fluorescence at 390 nm with an excitation wavelength of 345 nm using SpectraMax M5e plate reader. Data were averaged from four readings. Nonspecific FRET was obtained by measuring the fluorescence intensity of double-biotin GlpG variants which were pre-saturated with 10 μM of the high-affinity mSA-WT (without DABCYL-label) at an increasing concentration of the lower-affinity variant mSADAB-S45A. In this condition, mSADAB-S45A cannot compete for biotin label and only diffuses around in the solution.
Fitting of binding isotherm to obtain thermodynamic stability of GlpG
Fitting equation to obtain thermodynamic stability of GlpG using steric trapping was based on the following reaction scheme14:
| (2) |
| (3) |
Fitting equation for the second mSA binding phase was:
| (4) |
where F is measured fluorescence intensity, and F0 and F∞ are the fluorescence intensities from GlpG labeled with BtnPyr at [mSA]=0 and at the saturated bound level, respectively. [mSA] is the total mSA concentration, Kd,biotin is the dissociation constant for unhindered biotin binding affinity of mSA (Supplementary Fig. 3), and KU is the equilibrium constant for unfolding of GlpG. After obtaining the fitted KU, the thermodynamic stability was calculated using the equation ΔGoU,ST =−RTlnKU.
Determination of biotin affinity (Kd,biotin) of mSA variants by FRET
Biotin binding affinity of a weaker binding mSA variant mSA-W79M was measured by titration of 50 nM GlpG single cysteine variants labeled with BtnPyr (FRET donor) with mSADAB-W79M (FRET acceptor) in 5 mM DDM, 0.25 mM TCEP, 20 mM sodium phosphate and 200 mM NaCl (pH 7.5). The titrated samples were transferred to a 96-well UV-compatible microplate, sealed with a polyolefin tape, and incubated for 24 h at room temperature. Binding was monitored by the decrease of pyrene-monomer fluorescence at 390 nm with an excitation wavelength of 345 nm using SpectraMax M5e plate reader. Data were averaged from four readings. For fitting of binding data to obtain Kd,biotin of mSADAB-W79M, the following equation was used:
| (5) |
where F is measured fluorescence intensity, PT is total GlpG concentration, [mSA] is the total mSA concentration (variable), Kd,biotin is the dissociation constant for biotin binding affinity of mSADAB, A1 is the net fluorescence change, A2 is the fluorescence level without mSADAB. Fitted values include Kd,biotin, A1 and A2; other known values were fixed.
To determine Kd,biotin of tight-binding mSA variants and mSA variants lacking DABCYL quencher label, a FRET-based competition assay was employed. 1 μM GlpG was pre-equilibrated with 2–5 times excess of mSADAB variant for 1 h at room temperature. In this condition, pyrene fluorescence was suppressed. Weaker-affinity unlabeled mSA variant was titrated into the sample. Here either DABCYL-labeled or unlabeled mSA variant had known Kd,biotin. The titrated samples were transferred to a 96-well UV-compatible microplate, sealed with a polyolefin tape, and incubated for 24 h at room temperature. Resultant dissociation of mSADAB by competition was monitored by the increase of pyrene-monomer fluorescence at 390 nm with an excitation wavelength of 345 nm using SpectraMax M5e plate reader. Data were averaged from four readings. For fitting of competition data to obtain unknown Kd,biotin, the following equation was used:
| (6) |
where F is measured fluorescence intensity, PT is the total GlpG concentration, CT is the total mSADAB concentration, [mSA] is the total unlabeled mSA concentration (variable), Kunlabel is the Kd,biotin for mSA without a DABCYL label, Kdabcyl is the Kd,biotin for mSADAB, A1 is the amplitude of binding, and A2 is the initial fluorescence level. Fitted values include the unknown Kunlabel or Kdabcyl, A1 and A2; all other values are fixed.
SDS-denaturation of GlpG variants labeled with BtnPyr
0.4 uM GlpG doubly-labeled with BtnPyr was titrated with SDS in 5 mM DDM, 20 mM Na2HPO4, 200 mM NaCl, pH 7.5, ranging from 0–0.9 SDS mole-fraction (XSDS=[SDS]/([SDS]+[DDM]). Samples were incubated overnight at room temperature. The detailed scheme for fluorescence spectroscopy to monitor SDS-induced equilibrium unfolding is described in Supplementary Fig. 8. The unfolding curves were constructed using the average of three measurements. To determine thermodynamic stability of GlpG from SDS denaturation (ΔGoU,SDS), the unfolding curves were fitted to the following two-state Santoro-Bolen equation52:
| (7) |
| (8) |
F is the net fluorescence change. Baselines for the pre- and post-transition regions were determined by the fitted parameters: FlF, the fluorescence value for fully folded GlpG; FlU, the fluorescence value for fully unfolded GlpG; mF, the slope of the fully folded baseline; and mU, the slope of the fully unfolded baseline. mSDS is the slope of the transition region against XSDS and XSDS,1/2 is the transition midpoint. Fitted values include FlF, FlU, mF, mU, mSDS, and XSDS.1/2. ΔGoU,SDS represents thermodynamic stability in the absence of denaturant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by start-up fund from Michigan State University (H.H.), Hunt for a Cure grant for basic cystic fibrosis research (H.H), National Cancer Institute R01CA149451 (X.H), National Eye Institute R01EY05216 (W.L.H.) and the Jules Stein Professor endowment (W.L.H.). The authors thank John McCracken for general advice on EPR and helping with CW-EPR measurements and Christian Altenbach for DEER analysis program. We thank Erin Deans and Caleb Arthur for helping with protein preparations. We also thank Robert Jefferson, Nicholas Woodall, Yu-Chu Chang, David Weliky, Lee Kroos, John McCracken and James Bowie for critical reading of the manuscript. We appreciate suggestions from anonymous reviewers, which greatly helped improving the quality of this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
R.G., K.G., M.K. and H.H. designed experiments, expressed and purified proteins, and performed steric trapping and other biochemical experiments; R.G., X.H. and H.H. designed new steric trapping probes; R.G., S.S. and X.H. synthesized and characterized the new probes; Z.Y. and M.K. performed EPR measurements; Z.Y., M.K., H.H. and W.L.H. interpreted DEER data; All authors contributed to writing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 3.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain AK, Handel TM, Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat Struct Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci U S A. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. J Mol Biol. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 8.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 9.Bai Y, Englander SW. Future directions in folding: the multi-state nature of protein structure. Proteins. 1996;24:145–151. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 12.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid alpha-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc Natl Acad Sci U S A. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blois TM, Hong H, Kim TH, Bowie JU. Protein unfolding with a steric trap. J Am Chem Soc. 2009;131:13914–13915. doi: 10.1021/ja905725n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YC, Bowie JU. Measuring membrane protein stability under native conditions. Proc Natl Acad Sci U S A. 2014;111:219–224. doi: 10.1073/pnas.1318576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. Proc Natl Acad Sci U S A. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong H, Bowie JU. Dramatic destabilization of transmembrane helix interactions by features of natural membrane environments. J Am Chem Soc. 2011;133:11389–11398. doi: 10.1021/ja204524c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferson RE, Blois TM, Bowie JU. Membrane proteins can have high kinetic stability. J Am Chem Soc. 2013;135:15183–15190. doi: 10.1021/ja407232b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 20.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 21.Stevenson LG, et al. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci U S A. 2007;104:1003–1008. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 23.Baker RP, Urban S. Architectural and thermodynamic principles underlying intramembrane protease function. Nat Chem Biol. 2012;8:759–768. doi: 10.1038/nchembio.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paslawski W, et al. Cooperative folding of a polytopic alpha-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc Natl Acad Sci U S A. 2015;112:7978–7983. doi: 10.1073/pnas.1424751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min D, Jefferson RE, Bowie JU, Yoon TY. Mapping the energy landscape for second-stage folding of a single membrane protein. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosyka O, et al. Activity-based probes for rhomboid proteases discovered in a mass spectrometry-based assay. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2472–2477. doi: 10.1073/pnas.1215076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoll S, et al. Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. Embo Journal. 2014;33:2408–2421. doi: 10.15252/embj.201489367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherratt AR, Blais DR, Ghasriani H, Pezacki JP, Goto NK. Activity-Based Protein Profiling of the Escherichia coli GlpG Rhomboid Protein Delineates the Catalytic Core. Biochemistry. 2012;51:7794–7803. doi: 10.1021/bi301087c. [DOI] [PubMed] [Google Scholar]
- 29.Arutyunova E, et al. Allosteric regulation of rhomboid intramembrane proteolysis. Embo Journal. 2014;33:1869–1881. doi: 10.15252/embj.201488149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klumb LA, Chu V, Stayton PS. Energetic roles of hydrogen bonds at the ureido oxygen binding pocket in the streptavidin-biotin complex. Biochemistry. 1998;37:7657–7663. doi: 10.1021/bi9803123. [DOI] [PubMed] [Google Scholar]
- 32.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 33.Dockter C, et al. Refolding of the integral membrane protein light-harvesting complex II monitored by pulse EPR. Proc Natl Acad Sci U S A. 2009;106:18485–18490. doi: 10.1073/pnas.0906462106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnamani V, Hegde BG, Langen R, Lanyi JK. Secondary and tertiary structure of bacteriorhodopsin in the SDS denatured state. Biochemistry. 2012;51:1051–1060. doi: 10.1021/bi201769z. [DOI] [PubMed] [Google Scholar]
- 35.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 36.Mok YK, Kay CM, Kay LE, Forman-Kay J. NOE data demonstrating a compact unfolded state for an SH3 domain under non-denaturing conditions. J Mol Biol. 1999;289:619–638. doi: 10.1006/jmbi.1999.2769. [DOI] [PubMed] [Google Scholar]
- 37.Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 38.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renthal R. An unfolding story of helical transmembrane proteins. Biochemistry. 2006;45:14559–14566. doi: 10.1021/bi0620454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otzen DE. Folding of DsbB in mixed micelles: A kinetic analysis of the stability of a bacterial membrane protein. Journal of Molecular Biology. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]
- 41.Bedard S, Mayne LC, Peterson RW, Wand AJ, Englander SW. The foldon substructure of staphylococcal nuclease. J Mol Biol. 2008;376:1142–1154. doi: 10.1016/j.jmb.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llinas M, Gillespie B, Dahlquist FW, Marqusee S. The energetics of T4 lysozyme reveal a hierarchy of conformations. Nat Struct Biol. 1999;6:1072–1078. doi: 10.1038/14956. [DOI] [PubMed] [Google Scholar]
- 43.Dutta A, et al. Characterization of membrane protein non-native states. 2. The SDS-unfolded states of rhodopsin. Biochemistry. 2010;49:6329–6340. doi: 10.1021/bi100339x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dill KA, Shortle D. Denatured states of proteins. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 46.Xue Y, Ha Y. Large lateral movement of transmembrane helix S5 is not required for substrate access to the active site of rhomboid intramembrane protease. J Biol Chem. 2013;288:16645–16654. doi: 10.1074/jbc.M112.438127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z, et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 48.Liu T, Whitten ST, Hilser VJ. Functional residues serve a dominant role in mediating the cooperativity of the protein ensemble. Proc Natl Acad Sci U S A. 2007;104:4347–4352. doi: 10.1073/pnas.0607132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104–1113. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–1037. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 51.Hong H, Chang YC, Bowie JU. Measuring transmembrane helix interaction strengths in lipid bilayers using steric trapping. Methods Mol Biol. 2013;1063:37–56. doi: 10.1007/978-1-62703-583-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988;27:8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.