Abstract
Accumulation of amyloid β (Aβ) in the brain is a key pathological hallmark of Alzheimer’s disease (AD). Because aging is the most prominent risk factor for AD, understanding the molecular changes during aging is likely to provide critical insights into AD pathogenesis. However, studies on the role of miRNAs in aging and AD pathogenesis have only recently been initiated. Identifying miRNAs dysregulated by the aging process in the brain may lead to novel understanding of molecular mechanisms of AD pathogenesis. Here, we identified that miR-186 levels are gradually decreased in cortices of mouse brains during aging. In addition, we demonstrated that miR-186 suppresses β-site APP-cleaving enzyme 1 (BACE1) expression by directly targeting the 3′UTR of Bace1 mRNA in neuronal cells. In contrast, inhibition of endogenous miR-186 significantly increased BACE1 levels in neuronal cells. Importantly, miR-186 overexpression significantly decreased Aβ level by suppressing BACE1 expression in cells expressing human pathogenic mutant APP. Taken together, our data demonstrate that miR-186 is a potent negative regulator of BACE1 in neuronal cells and it may be one of the molecular links between brain aging and the increased risk for AD during aging.
Keywords: aging, microRNA, miR-186, BACE1, Aβ, Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is the most common form of dementia. AD is characterized with progressive loss of memory and cognitive decline (Hardy & Selkoe 2002). Although details of AD pathogenesis still remain elusive, abnormal accumulation of amyloid-β (Aβ) peptide in the brain is hypothesized to trigger pathogenic cascades that lead to AD (Haass & Selkoe 2007). Therefore, strategies to reduce Aβ accumulation in the brain are actively being pursued as AD therapies.
Aβ is generated through sequential cleavages of amyloid precursor protein (APP) by β-site APP-cleaving enzyme 1 (BACE1) and γ-secretase complex (De Strooper 2010). Given its critical role in Aβ production, BACE1 has been one of the prime therapeutic targets for AD. In fact, several studies unequivocally demonstrated that inhibition of BACE1 by RNAi-mediated knock-down or genetic deletion ameliorates Aβ-associated pathologies and cognitive deficits in mouse models of Aβ-amyloidosis (Laird et al. 2005, Luo et al. 2001, McConlogue et al. 2007, Ohno et al. 2004, Singer et al. 2005). Previous studies consistently reported that expression and activity of BACE1 are increased during aging in the brains mice, rats and humans (Boissonneault et al. 2009, Che et al. 2014, Fukumoto et al. 2004, Rossner et al. 2006), raising the possibility of a link between aging-associated alterations in BACE1 and the increased risk for AD. Nonetheless, little is known about the molecular mechanism of how BACE1 expression is regulated by the aging process. Therefore, identifying the regulatory mechanisms of BACE1 expression in the brain by aging may provide new therapeutic opportunities for AD.
Recently, microRNAs (miRNAs) have emerged as key post-transcriptional regulators of protein coding genes (Krol et al. 2010, Bartel 2009). Genome-encoded small non-coding miRNAs regulate gene expression by binding to their target messenger RNAs (mRNAs), leading to translational repression and/or degradation of their target mRNAs (Huntzinger & Izaurralde 2011). Expression patterns of a subset of miRNAs often show disease-specific signatures, as is the case in neurodegenerative diseases (Lau & de Strooper 2010, Lau et al. 2013, Wang et al. 2011, Delay et al. 2012). Growing evidence suggests that dysregulation of miRNAs may contribute to the onset and/or progression of various diseases by regulating disease-associated genes (Delay et al. 2012, Schonrock et al. 2012, Esteller 2011).
Aging is the most prominent risk factor for AD. Given the critical roles of Aβ in AD pathogenesis and strong correlation between brain Aβ levels and age (Fukumoto et al. 2004, Lesne et al. 2013), understanding how the Aβ level is regulated during aging is likely to provide critical insights into AD pathogenesis. Growing evidence suggests that AD pathology, such as cerebral Aβ accumulation, probably begins one to two decades before onset of clinical symptoms (Holtzman et al. 2011, Perrin et al. 2009). Therefore, understanding the molecular changes that occur through the aging process, in particular, at middle age may provide critical insights into early pathogenesis of AD. Interestingly, emerging evidence also suggests that in humans the aging processes, such as proinflammation and cognitive decline, begin in midlife(Ferrucci et al. 2005, Belsky et al. 2015). Genome wide expression profiling studies have identified several genes and biological pathways affected by aging (Bishop et al. 2010, Lopez-Otin et al. 2013, Kenyon 2010, Yankner et al. 2008). However, studies on the roles that miRNAs play in brain aging and AD pathogenesis have only recently been initiated (Inukai et al. 2012, Persengiev et al. 2011, Somel et al. 2010, Eda et al. 2011). Here we found that miR-186 levels are gradually decreased in mouse cortices during aging. We demonstrated that overexpression of miR-186 suppresses BACE1 expression in neuronal cells. In addition, inhibition of miR-186 significantly increases BACE1 expression. Taken together, our data suggest that miR-186 may be one of the putative molecular links between aging and AD development.
Materials and methods
Mice and cells
All procedures with animals followed the guidelines approved by the Animal Studies Committee at Washington University and Mayo Clinic College of Medicine. Wild-type C57BL/6J mice (Jackson lab, 000664) were sacrificed at different ages as indicated. Then, different brain regions and organs were immediately dissected out and processed according to the experimental purposes. Female mice were used to measure miR-186 expression in different organs. All other experiments were performed with male mice.
Cortical primary astrocytes were obtained from P2 pups of C57B6 mice, as described previously (Kim et al. 2009). Cortical primary neurons were prepared from E16 embryos and fully differentiated before harvesting cells at DIV14, as described previously (Kim et al. 2009). The purity of primary cells was monitored by neuron-specific tubulin, beta 3 class III (βIII-tub) and astrocyte-specific glial fibrillary acidic protein (GFAP). Neuro-2a cells and 7PA2 cells, Chinese hamster ovary (CHO) cells overexpressing human APP770 containing Indiana mutation (V717F) (Podlisny et al. 1995), were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10 % fetal bovine serum (FBS) and 1 % penicillin/streptomycin at 37°C in a humidified 5 % CO2 incubator. Transient transfections were performed with Lipofectamine™ 2000 (Invitrogen) to Neuro-2a cells and with Lipofectamine™ RNAiMAX (Invitrogen) to 7PA2 cells according to the manufacturer’s guides. Synthetic miR-186 (5′ - CAAAGAAUUCUCCUUUUGGGCU - 3′) and negative control (5′ - UUCUCCGAACGUGUCACGUTT - 3′) were from Insight Genomics. Locked nucleic acid (LNA)-based anti-miR-186 (5′ - AGCCCAAAAGGAGAATTCT - 3′) and negative anti-control (5′ - GTGTAACACGTCTATACGCCCA - 3′) were from EXIQON.
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNAs were extracted using TRIzol® Reagent (Invitrogen) and reverse transcribed with High Capacity cDNA Reverse Transcription kit (Applied Biosystems) for mRNAs or with Mir-X™ miRNA First-Strand Synthesis Kit (Clontech) for miRNAs. Quantitative PCR was performed with Power SYBR Green PCR Master Mix, ABI 7500, and ABI 7900 (Applied Biosystems) using default thermal cycling program. Except U6 Primer and universal reverse primer (Clontech), all other primers were purchased at Sigma Life Science (Sigma). Endogenous Gapdh, U6, or sno202 was used as normalization controls for mRNAs and miRNAs, as indicated. Relative levels of mRNAs and miRNAs were calculated by comparative Ct method using ABI 7500 software (version2.0.5) and GeneEx 5.3.2 (Multid analyses). The primer sequences used were: Bace1, 5′-CAGTGGGACCACCAACCTTC-3′ and 5′-GCTGCCTTGATGGACTTGAC-3′; Gapdh, 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′. miR-186 mature sequence and universal reverse primer were used for miR-186.
Luciferase reporter assay
psiCHECK vector harboring the entire 3′ UTR of mouse Bace1 mRNA downstream of the Renilla luciferase ORF (psiCHECK-mBace1 3′ UTR) was a kind gift from Dr. Patrick Provost (Université Laval, Quebec, Canada) (Boissonneault et al. 2009). This vector also contains a constitutively expressed firefly luciferase gene for the normalization of transfection efficiency. pmirGLO vectors harboring the partial 3′ UTRs (~ 40 nt) including the predicted binding sites of mouse Bace1 mRNA downstream of the firefly luciferase ORF (pmirGLO-mBace1 MRE) were cloned in our laboratory. pmirGLO vector also contains a constitutively expressed Renilla luciferase gene for the normalization of transfection efficiency. Neuro-2a cells were plated at a density of 4 × 104 cells/well in a 96-well plate a day before transfection. 0.12 μg of luciferase reporter vector was cotransfected with miR-186 or negative control at a final concentration of 75 nM for 24 h. Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Each experiment was independently repeated twice or more in duplicate or triplicate.
Aβ secretion assay
7PA2 cells were plated in a 24-well plate at a density of 9 × 104 cells/well. 24 h later, cells were transfected with miR-186 or negative control. 48 h post-transfection, media was changed to Opti-MEM®I with 1 % FBS. 6 h after media change, media were collected on ice and centrifuged at 6,000 x g for 10 min to remove cell debris. Cells were washed with PBS buffer and lysed on ice for 30 min with cell lysis buffer (1% Triton X-100, 150mM NaCl, 50mM Tris-HCl, pH 8) containing protease inhibitor cocktail (Roche). Cell lysates were centrifuged at 17,000 x g for 10 min and supernatants were collected for Western bot analyses. The levels of APP, BACE1, APP C-terminal fragment α (CTFα), and CTFβ were normalized with corresponding GAPDH levels. The levels of soluble APPα (sAPPα), soluble APPβ (sAPPβ), and Aβ were normalized with corresponding cellular APP levels. Experiments included duplicates or triplicates and were repeated independently twice or more.
Western blot analysis
Western blots were performed as previously described (Kim et al. 2012). Protein concentration of cell lysates was determined by BCA protein assay (Thermo Fisher). Equal amounts of total protein for each lysate were separated on 4–20 % TGX™ (Tris-Glycine eXtended) gels (Bio-Rad) and transferred to 0.2 μm pore size nitrocellulose membranes (Bio-Rad). For Aβ detection, same volumes of media were run on 16.5% Tris-Tricine gel (Bio-Rad). After transferring, membranes were boiled for 10 minutes in PBS, as previously reported (McGowan et al. 2005). All membranes were blocked with 4% non-fat dry milk in TBS/T (Tris buffered saline with 0.125% Tween-20). Blots were probed with rabbit anti-APP antibody (Invitrogen), rabbit anti-BACE1 antibody (D10E5, Cell Signaling Technology), mouse anti-Aβ antibody (82E1, IBL International), rabbit anti-sAPPβ antibody (IBL International), rabbit anti-GAPDH antibody (FL-335, Santa Cruz Biotechnology), or mouse anti-actin antibody (Sigma) at room temperature for 1 h. Anti-APP antibody (Invitrogen) was used for CTFα and CTFβ detection. 6E10 (Covance) antibody was used for sAPPα detection. After secondary antibody incubation, membranes were developed using Lumigen TMA-6 ECL detection kit (Lumigen).
Statistical analysis
To determine the statistical significance (*p<0.05, **p<0.01, ***p<0.001), we first tested whether our data sets passed the equal variance test (Levene Median test) and normality test (Kolmogorov-Smirnov test) (SigmaStat 3.5). After confirmation that the data did not violate the assumptions of parametric testing, a two-tailed Student’s t-test was used when two groups were compared (GraphPad Prism 6). In the cases of multiple comparisons, One-way ANOVA was used after post-hoc analysis as indicated. All data are shown as mean ± standard error of the mean (SEM).
Results
miR-186 expression is decreased in cortex during aging
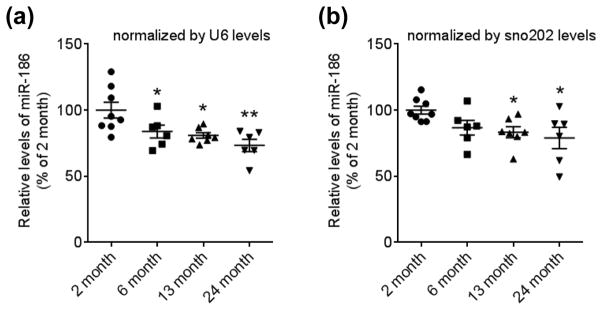
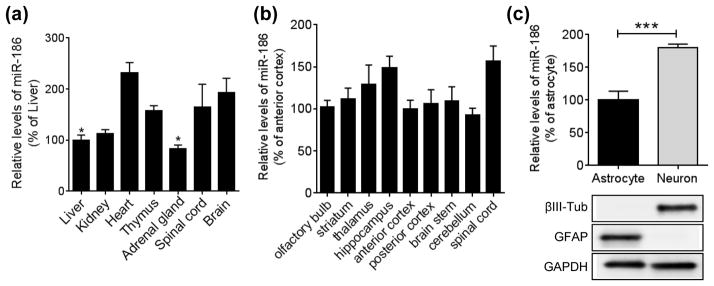
Previous studies have shown that only a small population of miRNAs accounts for a majority of miRNA pool in the brain (Shin et al. 2010, Hu et al. 2011, Landgraf et al. 2007, Berezikov et al. 2006). To identify unique miRNA signature in the aged brain, we first compared about 100 miRNA levels between young (2 month) and middle (13 month) aged brains. We identified that miR-186 expression is significantly decreased in mouse cortices at 13 month of age, compared to 2 month of age, and it shows a trend of further decrease at 24 month of age (Fig. 1, A and B). miR-186 is expressed throughout different organs (Fig. 2A). In the brain, miR-186 is broadly expressed across multiple brain sub-regions in mice (Fig. 2B). Many miRNAs are known to have unique expression patterns in different brain cell types (Jovicic et al. 2013). To study the role of miR-186 in relevant cell types, we next measured miR-186 expression in primary astrocytes and neurons isolated from mouse cortices. Of note, miR-186 showed relatively higher expression in neurons, compared to astrocytes (Fig. 2C), suggesting that miR-186 may have prominent roles in neurons.
FIGURE 1.
miR-186 expression is decreased during aging in the mouse brain cortex. Mature miR-186 levels were analyzed by qRT-PCR in mouse cortex at different ages (n= 8 for 2 month, 6 for 6 month, 7 for 13 month, 6 for 24 month). Each level was represented as a percentage of 2 month old mice. Values are mean ± SEM (*, p<0.05; **, p<0.01; One-way ANOVA compared to 2 month with post hoc Holm-Sidak’s test).
FIGURE 2.
miR-186 expression pattern in the brain. (a) Relative expression levels of miR-186 in different mouse tissues. Endogenous levels of miR-186 were analyzed by qRT-PCR in different mouse tissues of 3.5-month-old C57B6 mice (n = 3). Each level was normalized by corresponding U6 level and represented as a percentage of liver (*, p<0.05; One-way ANOVA compared to brain with post hoc Dunnett’s test). (b) Relative expression of miR-186 in the mouse brain (n = 3). Each level was normalized by corresponding U6 level and represented as a percentage of the anterior cortex (One-way ANOVA compared to anterior cortex with post hoc Dunnett’s test). (c) Relative expression levels of miR-186 in primary cells isolated from mice brains. miR-186 levels were analyzed in mouse primary neurons and astrocytes by qRT-PCR. The purity of primary culture cells was assessed by Western blotting of cell lysates with neuron-specific βIII-Tubulin and astrocyte-specific GFAP antibodies. Data are shown as a percentage of astrocytes (n = 6, ***, p<0.001, t-test). All Values are mean ± SEM.
miR-186 suppresses BACE1 expression by directly targeting 3′UTR of BACE1 mRNA
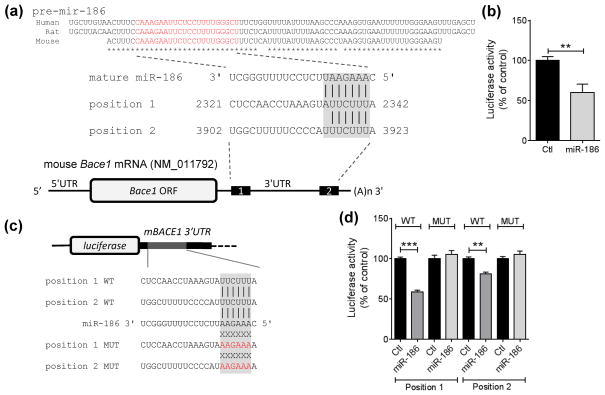
To identify direct target genes of miR-186, we first searched genes that have putative binding sites in the 3′UTR of their mRNA by utilizing several miRNA-target prediction algorithms as described previously (Kim et al. 2012). Among several candidate target genes, BACE1 was predicted commonly by several different algorithms (TargetScan, miRanda, PICTAR) (Fig. 3A). miR-186 has two putative binding sites in the 3′UTR of mouse Bace1 mRNA with complementary seed match (Fig. 3A). The first predicted position is 8mer site, which has complementary seed match at position 2 – 7 of mature miR-186 followed by a nucleotide “A” and additional match at position 8 of mature miR-186 (Fig. 3A). The second predicted position is 7mer-A1 site, which has complementary seed match at position 2 – 7 of mature miR-186 followed by a nucleotide “A” (Fig. 3A). The sequences of precursor microRNA-186 (pre-mir-186) and mature microRNA-186 (miR-186) are well conserved in mammals, such as human, mouse, and rat (Fig. 3A). Because miR-186 is well expressed in the brain and preferentially in neurons (Fig. 2), we also considered the expression profile of the predicted targets in our candidate gene selection. Among several candidates, we focused on BACE1 for two reasons: 1) Since BACE1 is predominantly expressed in neurons (Vassar et al. 1999, Rossner et al. 2001), miR-186 coexpressed in neurons may be able to locally regulate BACE1 expression. 2) Previous reports showed that BACE1 protein levels are increased in the brains of mice and rats at middle age compared to young age (Boissonneault et al. 2009, Che et al. 2014). However, up-regulation of BACE1 protein levels during aging is not due to the change of its mRNA levels (Hebert et al. 2008, Boissonneault et al. 2009). We also observed that the levels of BACE1 mRNA are not altered during aging in mouse cortices (data not shown), suggesting that BACE1 expression is regulated at the post-transcriptional level during aging. Taken together, these findings raise the possibility of a link between reduction of miR-186 levels and induction of BACE1 expression during aging.
FIGURE 3.
miR-186 directly targets the 3′UTR of Bace1 mRNA. (a) Schematic diagram of target sites of miR-186 in Bace1 mRNA. The mature sequence of miR-186 is shown in red within pre-mir-186. miR-186 has 2 potential targeting sites in the mouse Bace1 3′UTR with complementary seed match indicated with gray box. Asterisk (*) indicates conserved nucleotides. (b) miR-186 directly targets the 3′UTR of Bace1 mRNA. (n = 7 per group). Luciferase reporter assays were performed with the reporter construct containing the full-length 3′ UTR of mBace1 mRNA downstream of Renilla luciferase. Mouse Neuro-2a neuronal cells were transfected with reporter construct with 75 nM negative control (Ctl) or miR-186. Renilla luciferase activity was normalized with the corresponding firefly luciferase activity. (c) Schematic diagram of luciferase constructs with miRNA recognition sites (MREs). The reporter constructs contain the partial 3′ UTRs (~ 40 nt) including the predicted sites of mouse Bace1 mRNA downstream of the firefly luciferase ORF (pmirGLO-mBace1 MRE WT). The mutated sequences in the seed match region are shown in red (pmirGLO-mBace1 MRE MUT). (d) miR-186 directly targets both MREs in the 3′UTR of Bace1 mRNA (n = 4). Mouse Neuro-2a neuronal cells were transfected with pmirGLO-mBace1 MRE-WT or MRE–MUT construct with 75 nM negative control (Ctl) or miR-186. Firefly luciferase activity was normalized with the corresponding Renilla luciferase activity. Data are shown as a percentage of control. Values are mean ± SEM (**, p<0.01; ***, p<0.001, t-test).
To determine whether miR-186 directly targets the 3′UTR of Bace1 mRNA, we performed a luciferase assay with a reporter construct containing the entire 3′UTR of mouse Bace1 mRNA at the downstream of luciferase gene (Fig. 3B). We transfected this luciferase reporter construct with negative control or miR-186 to mouse Neuro-2a neuronal cells and then measured luciferase activities. Compared to the negative control, miR-186 markedly reduced luciferase activity (Fig. 3B). We next examined whether miR-186 targets the predicted sites in the 3′UTR of mouse Bace1 mRNA. To this end, we generated the luciferase constructs which harbor partial 3′UTR fragments containing the predicted sites with either wild-type or mutated sequence in the seed-match regions at the downstream of luciferase gene (Fig. 3C). Compared to the negative control, miR-186 decreased luciferase activities by targeting both of the predicted sites (Fig. 3D). miR-186 had no effect on luciferase activity when the seed-match sequences were mutated, demonstrating that miR-186 targets both sites through its direct binding to Bace1 mRNA by complementary seed match.
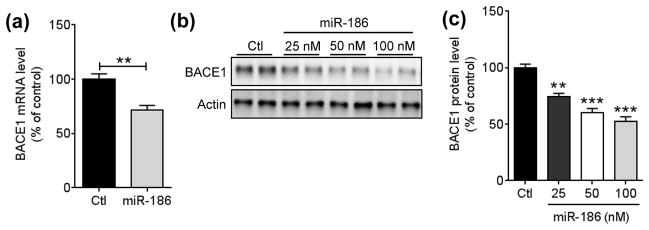
Since miRNA can suppress target expression via destabilizing mRNA and/or inhibiting translation, we next examined whether miR-186 affects Bace1 mRNA levels. Compared to the negative control, miR-186 overexpression significantly decreased BACE1 mRNA levels in Neuro-2a cells (Fig. 4A). Furthermore, miR-186 decreased BACE1 protein levels in a dose-dependent manner (Fig. 4, B and C). Taken together, these data demonstrate that overexpression of miR-186 suppresses BACE1 expression by inhibiting translation as well as destabilizing mRNA.
FIGURE 4.
miR-186 decreases both mRNA and protein expression levels of endogenous BACE1 in neuronal cells. (a) Reduction of Bace1 mRNA by miR-186 in Neuro-2a cells. 48 h post-transfection with 75 nM of negative control or miR-186, Bace1 mRNA levels were measured by qRT-PCR and normalized by corresponding Gapdh levels (n = 6, **, p<0.01; t-test). (b, c) Dose-dependent decrease of endogenous BACE1 protein levels by miR-186 in Neuro2a cells. 48 h post-transfection with indicated amount of negative control or miR-186, BACE1 protein levels were monitored by Western blot. Representative Western blot images (b) and relative BACE1 protein levels (c) are shown. BACE1 protein levels were normalize by corresponding β-actin levels (n = 4). All data are shown as a percentage of control. Values are mean ± SEM (**, p<0.01; ***, p<0.001, One-way ANOVA compared to control with post hoc Dunnett’s test).
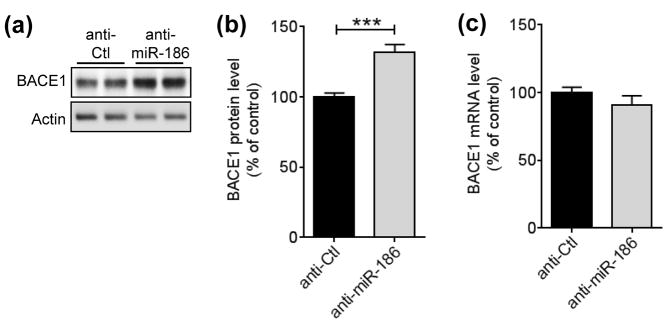
Inhibition of endogenous miR-186 increases BACE1 expression
Although overexpression of miRNA can strongly regulate its target gene, endogenous miRNA may not have a significant effect on target gene expression due to multiple reasons, such as its low expression level or competition with other miRNAs for the same targeting site. Therefore, it is crucial to assess miRNA’s function by modulating endogenous miRNA levels. Locked nucleic acid (LNA)-based anti-miRs are nuclease-resistant single-stranded antisense oligonucleotides with high binding affinity to their target miRNAs (Lennox & Behlke 2010). To determine if endogenous miR-186 is functional in regulating BACE1 expression, we transfected LNA-based anti-miR-186 or negative anti-control to Neuro-2a cells and then assessed BACE1 levels by Western blot. Inhibition of endogenous miR-186 by anti-miR-186 significantly increased BACE1 protein levels (Fig. 5, A and B). Unlike overexpression of miR-186, inhibition of miR-186 did not affect BACE1 mRNA levels (Fig. 5C), suggesting that BACE1 expression is actively suppressed by endogenous miR-186 under basal condition through translational inhibition in neuronal cells.
FIGURE 5.
Inhibition of endogenous miR-186 increases BACE1 expression in neuronal cells. (a, b) Inhibition of miR-186 increases BACE1 protein levels in Neuro-2a cells. Neuro-2a cells were transfected with 50 nM of anti-miR-186 or anti-control (anti-Ctl). 72 h post-transfection, cell lysates were applied for Western blot. A representative western blot images (a) and relative BACE1 levels (b) are shown. BACE1 protein levels were quantified as a percentage of control after normalization by corresponding β-actin levels (n = 7). (c) Inhibition of miR-186 does not affect BACE1 mRNA levels in Neuro-2a cells. Neuro-2a cells were transfected with 50 nM of anti-miR-186 or anti-control (anti-Ctl). 72 h post-transfection, Bace1 mRNA levels were measured by qRT-PCR and normalized by corresponding Gapdh levels (n = 7). Data are shown as a percentage of control. Values are mean ± SEM (***, p<0.001, t-test).
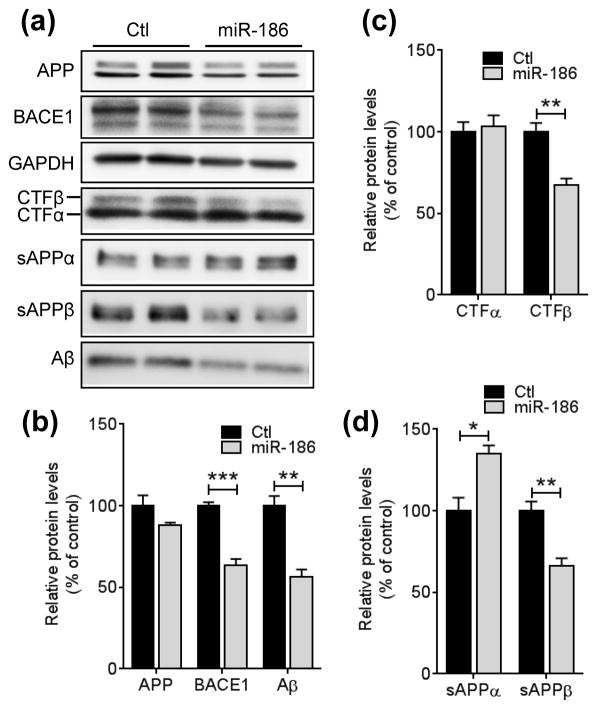
Overexpression of miR-186 decreases the secreted Aβ levels in cells expressing pathogenic mutant APP
Given the rate-limiting role of BACE1 in Aβ generation from APP, strategies to lower cerebral Aβ levels by inhibiting BACE1 have been extensively pursued as AD therapeutics (De Strooper et al. 2010). Therefore, we tested if miR-186 can repress Aβ generation. First, 7PA2 cells expressing the Indiana mutant form of human APP770 were transfected with negative control or miR-186. Then, the levels of APP and CTFs in the cell lysates and sAPPs and secreted Aβ in the media were measured by Western blot (Fig. 6A). miR-186 markedly decreased the levels of cellular CTFβ, secreted sAPPβ, and total Aβ (all species of Aβ, including Aβ40 and Aβ42) in 7PA2 cells compared to the negative control (Fig. 6B – D).
FIGURE 6.
miR-186 represses Aβ secretion by suppressing BACE1 expression. 7PA2 cells expressing Indiana mutant hAPP were transfected with 75 nM negative control or miR-186. 48 h post-transfection, cells were incubated in fresh medium for 6 h and then, cell and media were collected for Western blot analyses. Representative western blot images (a) and relative protein levels (b - d) are shown (n = 7). 82E1 anti-Aβ western blot detects total Aβ, including Aβ40 and Aβ42. Data are shown as a percentage of control. Values are mean ± SEM (*, p<0.05; **, p<0.01; ***, p<0.001, t-test).
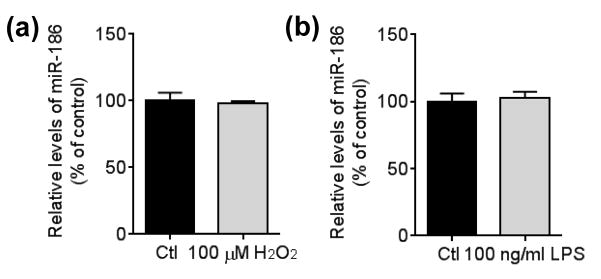
miR-186 expression is independent of oxidative stress and inflammation
Mounting evidence has shown that aging is accompanied with the increase of oxidative- stress and inflammation in the brain (Franceschi et al. 2000, Finkel & Holbrook 2000, Balaban et al. 2005, Lee et al. 2000, Guest et al. 2014, Lynch 2010). Interestingly, several studies have suggested that both oxidative stress and inflammation increase BACE1 expression (Chami & Checler 2012, Rossner et al. 2006, Tamagno et al. 2012). Because miR-186 levels were decreased during aging in the mouse brain, these findings prompted us to determine if miR-186 expression may be regulated by those stresses. When we analyzed the effects of oxidative stress induced by H2O2 and inflammation induced by LPS on miR-186 expression in Neuro-2a cells, miR-186 levels were not affected by either oxidative stress or inflammation (Figure 7 A and B).
FIGURE 7.
miR-186 expression is not affected by either oxidative stress or inflammation. Neuro-2a cells were incubated 100 μM H2O2 (a) or 100 ng/ml LPS (b) for 24 h. miR-186 levels were measured by qRT-PCR and normalized by corresponding U6 levels (n = 4, t-test).
Discussion
In this study, we found that miR-186 levels in the cortex of the mouse brain gradually decrease during aging. Moreover, we demonstrated that miR-186 is a potent negative regulator of BACE1 expression in neuronal cells. Both miR-186 and BACE1 are expressed in the brain and are particularly enriched in neurons (Fig. 2) (Vassar et al. 1999). Furthermore, overexpression of miR-186 markedly decreased Aβ level. Given the up-regulation of BACE1 expression in the brain during aging (Boissonneault et al. 2009, Che et al. 2014), it is tempting to speculate that down-regulation of miR-186 by the aging process may be involved in the induction of BACE1 expression and thereby cause the accumulation of Aβ during aging.
So far, several miRNAs have been identified as regulators of BACE1 expression (Hebert et al. 2008, Zong et al. 2011, Zhu et al. 2012, Fang et al. 2012, Wang et al. 2008b, Boissonneault et al. 2009, Long et al. 2014), suggesting that an extensive network of multiple miRNAs may regulate BACE1 expression in a coordinated manner. Some of these are known to be down-regulated in the mouse brain during aging (Boissonneault et al. 2009). Therefore, it is likely that dysregulation of multiple miRNAs along with other potential regulators may coordinate to increase the levels of BACE1 and Aβ during aging process. Although there have been several miRNA profiling studies with human samples, our understanding of the miRNA’s roles in human brain aging is still limited (Persengiev et al. 2012, Persengiev et al. 2011, Smith-Vikos & Slack 2012, Somel et al. 2010, Hu et al. 2011, Wei et al. 2015, Noren Hooten et al. 2013, Noren Hooten et al. 2010). Further studies are warranted to determine the roles of miRNAs in brain aging, in particular, the roles of miRNAs in regulating BACE1 expression during aging in the human brain.
Aging is broadly defined by a progressive decline in physiological functions, accompanied with increase of oxidative stress and inflammation in the brain (Franceschi et al. 2000, Finkel & Holbrook 2000, Balaban et al. 2005, Lee et al. 2000, Guest et al. 2014, Lynch 2010, Lopez-Otin et al. 2013). Our study suggests that miR-186 expression may not be regulated by aging-associated oxidative stress and inflammation. Although BACE1 expression has been known to be up-regulated by oxidative stress and inflammation (Chami & Checler 2012, Rossner et al. 2006, Tamagno et al. 2012), our data suggest that miR-186 does not mediate the BACE1 induction by those aging-associated stresses. Aside from oxidative stress and inflammation, aging is a multifactorial process in which many other biological pathways are involved. While we cannot address the cause of miR-186 reduction in aged brain, it will be interesting to study further which biological pathway is responsible for miR-186 dysregulation in aged brain.
BACE1 has multiple substrates involved in various processes in the brain (Vassar et al. 2009). Complete loss of Bace1 in mice are known to lead to schizophrenia-like phenotypes and deficits in cognitive function, mossy fiber long-term potentiation (LTP), synaptic plasticity, and axon guidance (Savonenko et al. 2008, Wang et al. 2008a, Petrus & Lee 2014, Hitt et al. 2012, Rajapaksha et al. 2011). Therefore, further in vivo animal studies are warranted to determine whether miR-186 regulates cognition and memory during brain aging process.
Recently, nicastrin, a subunit of γ-secretase complex, was identified as a target of miR-186 (Delay et al. 2014), suggesting that miR-186 represses Aβ generation at both β- and γ-cleavage steps. Moreover, miR-186 suppresses purinergic receptor P2X, ligand-gated ion channel, 7 (P2RX7) in epithelial cancer cells (Zhou et al. 2008). P2RX7 mediates apoptosis by activating mitochondrial caspase 9 pathway in human cervical endothelial cells (Wang et al. 2004). Intriguingly, P2RX7 levels are elevated in AD brains (McLarnon et al. 2006). P2RX7 is known to increase α-cleavage of APP in vitro (Delarasse et al. 2011). However, pharmacological inhibition of P2RX7 reduces hippocampal amyloid plaques in an Aβ-amyloidosis mouse model (Diaz-Hernandez et al. 2012). Although the effect of miR-186 on P2RX7 in the brain remains elusive, these findings, together with our results, suggest miR-186 may represent a novel therapeutic target for AD. Therefore, further in vivo animal studies are warranted to determine therapeutic effects of miR-186 for AD.
Post-mortem analyses revealed that BACE1 expression is elevated in cortices of AD patients compared to the age-matched, non-demented controls (Fukumoto et al. 2002, Yang et al. 2003, Hebert et al. 2008, Ahmed et al. 2010). Recent miRNA profiling studies showed that multiple miRNAs are dysregulated in the affected regions of AD brains (Lau et al. 2013, Hebert et al. 2008, Wang et al. 2011, Delay et al. 2012, Cogswell et al. 2008, Lukiw 2007). Among those, several miRNAs regulating BACE1 expression are known to be down-regulated in the affected regions of AD brains (Hebert et al. 2008, Nelson & Wang 2010, Long et al. 2014, Lau et al. 2013). Interestingly, one report showed that miR-186 level is decreased in the white matter of the temporal cortex in AD brain (Wang et al. 2011). On the other hand, another study reported that miR-186 is up-regulated in the grey matter of the prefrontal cortex in AD brains (Lau et al. 2013). At this point, it is unclear whether these data suggest that miR-186 levels are regulated in a brain region-specific manner in AD brains. Although the role of miR-186 in AD pathogenesis remains elusive, dysregulation of multiple miRNAs regulating BACE1 may be involved in the induction of BACE1 in AD brains and thereby exacerbating Aβ-associated pathology in AD patients.
In conclusion, we found that miR-186 gradually decreases in the mouse brain cortex during aging. In addition, we demonstrated that inhibition of miR-186 increases BACE1 expression, while overexpression of miR-186 strongly suppresses its expression in neuronal cells. Our work suggests that miR-186 might be one of the molecular links between advancing brain aging and the increased risk for AD during aging.
Acknowledgments
We are grateful to Patrick Provost (Université Laval) for psiCHECK-mBace1 3′ UTR plasmid and Dr. Koo (UCSD) for 7PA2-APP cell line. This work was supported by GHR Foundation (JK), Mayo Clinic Center for Individualized Medicine (JK), NIH grants NS069329 (J.K.), AG016574 (J.K.), AG005681 (J.K), Mayo Graduate School (H.Y), and BrightFocus Foundation A2012421 (J.K.).
Abbreviations used
- AD
Alzheimer’s disease
- Aβ
amyloid-β
- APP
amyloid precursor protein
- BACE1
β-site APP-cleaving enzyme1
- βIII-Tub
tubulin, beta 3 class III
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- LPS
lipopolysaccharide
- miR
microRNA
- MRE
miRNA recognition sites
- ORF
open reading frame
- pre-miRNA
precursor microRNA
Footnotes
The authors have no conflict of interests.
References
- Ahmed RR, Holler CJ, Webb RL, Li F, Beckett TL, Murphy MP. BACE1 and BACE2 enzymatic activities in Alzheimer’s disease. J Neurochem. 2010;112:1045–1053. doi: 10.1111/j.1471-4159.2009.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Houts R, et al. Quantification of biological aging in young adults. Proc Natl Acad Sci U S A. 2015;112:E4104–4110. doi: 10.1073/pnas.1506264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami L, Checler F. BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and beta-amyloid production in Alzheimer’s disease. Mol Neurodegener. 2012;7:52. doi: 10.1186/1750-1326-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che H, Sun LH, Guo F, et al. Expression of Amyloid-Associated miRNAs in Both the Forebrain Cortex and Hippocampus of Middle-Aged Rat. Cell Physiol Biochem. 2014;33:11–22. doi: 10.1159/000356646. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol Rev. 2010;90:465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarasse C, Auger R, Gonnord P, Fontaine B, Kanellopoulos JM. The purinergic receptor P2X7 triggers alpha-secretase-dependent processing of the amyloid precursor protein. J Biol Chem. 2011;286:2596–2606. doi: 10.1074/jbc.M110.200618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Dorval V, Fok A, et al. MicroRNAs targeting Nicastrin regulate Abeta production and are affected by target site polymorphisms. Front Mol Neurosci. 2014;7:67. doi: 10.3389/fnmol.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Mandemakers W, Hebert SS. MicroRNAs in Alzheimer’s disease. Neurobiol Dis. 2012;46:285–290. doi: 10.1016/j.nbd.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Diaz-Hernandez JI, Gomez-Villafuertes R, Leon-Otegui M, et al. In vivo P2X7 inhibition reduces amyloid plaques in Alzheimer’s disease through GSK3beta and secretases. Neurobiol Aging. 2012;33:1816–1828. doi: 10.1016/j.neurobiolaging.2011.09.040. [DOI] [PubMed] [Google Scholar]
- Eda A, Takahashi M, Fukushima T, Hohjoh H. Alteration of microRNA expression in the process of mouse brain growth. Gene. 2011;485:46–52. doi: 10.1016/j.gene.2011.05.034. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Fang M, Wang J, Zhang X, et al. The miR-124 regulates the expression of BACE1/beta-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Ferrucci L, Corsi A, Lauretani F, et al. The origins of age-related proinflammatory state. Blood. 2005;105:2294–2299. doi: 10.1182/blood-2004-07-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J, Grant R, Mori TA, Croft KD. Changes in oxidative damage, inflammation and [NAD(H)] with age in cerebrospinal fluid. PloS one. 2014;9:e85335. doi: 10.1371/journal.pone.0085335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt B, Riordan SM, Kukreja L, Eimer WA, Rajapaksha TW, Vassar R. beta-Site amyloid precursor protein (APP)-cleaving enzyme 1 (BACE1)-deficient mice exhibit a close homolog of L1 (CHL1) loss-of-function phenotype involving axon guidance defects. J Biol Chem. 2012;287:38408–38425. doi: 10.1074/jbc.M112.415505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, Guo S, Xi J, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Inukai S, de Lencastre A, Turner M, Slack F. Novel MicroRNAs Differentially Expressed during Aging in the Mouse Brain. PLoS One. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Roshan R, Moisoi N, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33:5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, et al. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 2009;64:632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Ramirez CM, et al. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Laird FM, Cai H, Savonenko AV, et al. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Bossers K, Janky R, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 2010;21:768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- Lesne SE, Sherman MA, Grant M, et al. Brain amyloid-beta oligomers in ageing and Alzheimer’s disease. Brain. 2013;136:1383–1398. doi: 10.1093/brain/awt062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Ray B, Lahiri DK. MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem. 2014;289:5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Lynch MA. Age-related neuroinflammatory changes negatively impact on neuronal function. Front Aging Neurosci. 2010;1:6. doi: 10.3389/neuro.24.006.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L, Buttini M, Anderson JP, et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, et al. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarnon JG, Ryu JK, Walker DG, Choi HB. Upregulated expression of purinergic P2X(7) receptor in Alzheimer disease and amyloid-beta peptide-treated microglia and in peptide-injected rat hippocampus. J Neuropathol Exp Neurol. 2006;65:1090–1097. doi: 10.1097/01.jnen.0000240470.97295.d3. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer’s disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PloS one. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Fitzpatrick M, Wood WH, 3rd, et al. Age-related changes in microRNA levels in serum. Aging. 2013;5:725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Sametsky EA, Younkin LH, et al. BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer’s disease. Neuron. 2004;41:27–33. doi: 10.1016/s0896-6273(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev S, Kondova I, Otting N, Koeppen AH, Bontrop RE. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol Aging. 2011;32:2316.e2317–2327. doi: 10.1016/j.neurobiolaging.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Kondova II, Bontrop RE. The Impact of MicroRNAs on Brain Aging and Neurodegeneration. Curr Gerontol Geriatr Res. 2012;2012:359369. doi: 10.1155/2012/359369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrus E, Lee HK. BACE1 Is Necessary for Experience-Dependent Homeostatic Synaptic Plasticity in Visual Cortex. Neural Plast. 2014;2014:128631. doi: 10.1155/2014/128631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlisny MB, Ostaszewski BL, Squazzo SL, et al. Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- Rajapaksha TW, Eimer WA, Bozza TC, Vassar R. The Alzheimer’s beta-secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol Neurodegener. 2011;6:88. doi: 10.1186/1750-1326-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Apelt J, Schliebs R, Perez-Polo JR, Bigl V. Neuronal and glial beta-secretase (BACE) protein expression in transgenic Tg2576 mice with amyloid plaque pathology. J Neurosci Res. 2001;64:437–446. doi: 10.1002/jnr.1095. [DOI] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer’s disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Savonenko AV, Melnikova T, Laird FM, Stewart KA, Price DL, Wong PC. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci U S A. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N, Matamales M, Ittner LM, Gotz J. MicroRNA networks surrounding APP and amyloid-beta metabolism--implications for Alzheimer’s disease. Exp Neurol. 2012;235:447–454. doi: 10.1016/j.expneurol.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- Smith-Vikos T, Slack FJ. MicroRNAs and their roles in aging. J Cell Sci. 2012;125:7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Guo S, Fu N, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagno E, Guglielmotto M, Monteleone D, Vercelli A, Tabaton M. Transcriptional and post-transcriptional regulation of beta-secretase. IUBMB life. 2012;64:943–950. doi: 10.1002/iub.1099. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song L, Laird F, Wong PC, Lee HK. BACE1 knock-outs display deficits in activity-dependent potentiation of synaptic transmission at mossy fiber to CA3 synapses in the hippocampus. J Neurosci. 2008a;28:8677–8681. doi: 10.1523/JNEUROSCI.2440-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li X, Wang L, Feng YH, Zeng R, Gorodeski G. Antiapoptotic effects of estrogen in normal and cancer human cervical epithelial cells. Endocrinology. 2004;145:5568–5579. doi: 10.1210/en.2004-0807. [DOI] [PubMed] [Google Scholar]
- Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008b;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YN, Hu HY, Xie GC, et al. Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol. 2015;16:41. doi: 10.1186/s13059-015-0608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LB, Lindholm K, Yan R, et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, Gorodeski GI. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem. 2008;283:28274–28286. doi: 10.1074/jbc.M802663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HC, Wang LM, Wang M, et al. MicroRNA-195 downregulates Alzheimer’s disease amyloid-beta production by targeting BACE1. Brain Res Bull. 2012;88:596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang H, Dong W, et al. miR-29c regulates BACE1 protein expression. Brain Res. 2011;1395:108–115. doi: 10.1016/j.brainres.2011.04.035. [DOI] [PubMed] [Google Scholar]