Abstract
Enzyme classes may contain outlier members that share mechanistic, but not sequence or structural relatedness with more common representatives. The functional annotation of such exceptional proteins can be challenging. Here, we use activity-based profiling to discover that the poorly characterized multipass transmembrane proteins AIG1 and ADTRP are atypical hydrolytic enzymes that depend on conserved threonine and histidine residues for catalysis. Both AIG1 and ADTRP hydrolyze bioactive fatty-acid esters of hydroxy-fatty acids (FAHFAs), but not other major classes of lipids. We discover multiple cell-active, covalent inhibitors of AIG1 and show that these agents block FAHFA hydrolysis in mammalian cells. These results indicate that AIG1 and ADTRP are founding members of an evolutionarily conserved class of transmembrane threonine hydrolases involved in bioactive lipid metabolism. More generally, our findings demonstrate how chemical proteomics can excavate potential cases of convergent/parallel protein evolution that defy conventional sequence- and structure-based predictions.
Introduction
Activity-based protein profiling (ABPP) uses active site-directed chemical probes to study the functions of mechanistically and/or structurally related proteins in native biological settings1-3. ABPP probes are often broad-spectrum in their reactivity such that many members of an individual enzyme class can be characterized in parallel. A prominent example is the fluorophosphonate (FP) class of probes4 that targets the serine (Ser) hydrolases, a large and diverse enzyme family that constitutes ~1% of all proteins in mammals and utilizes a conserved Ser nucleophile to hydrolyze amide, ester, and/or thioester bonds in biomolecules5. Prior work has demonstrated that FP probes provide extensive coverage of Ser hydrolases6 and also react with some members of the much smaller class of threonine (Thr) hydrolases, such as the catalytic subunits of the proteasome7. FP probes do not cross-react with other classes of hydrolytic enzymes, including cysteine, aspartyl, and metallo-hydrolases, and this selectivity has facilitated the assignment of enzymes with established substrates to the Ser hydrolase class8,9. FP-probes can also be used in a competitive ABPP format, where biological samples are pre-treated with candidate small-molecule inhibitors that may compete for binding and/or reaction with targets of FP probes. Competitive ABPP has proven to be a robust platform for the discovery, optimization, and characterization of inhibitors of Ser hydrolases2.
The Ser hydrolase family contains several sub-clans that are distantly, or even un-related to one another in terms of sequence, structure, and/or mechanism5,10. This remarkable diversity raises an intriguing question – might other, as-of-yet unassigned Ser (or Thr) hydrolases exist in the human proteome? Here, we hypothesized that reactivity with FP probes, being a near-universal feature of these enzymes, could provide a proteome-wide assay to discover cryptic members of the Ser/Thr hydrolase family that may have arisen by convergent or parallel evolution11. We evaluated human cell proteomes by quantitative, mass spectrometry (MS)-based ABPP, resulting in the discovery of a poorly characterized multipass transmembrane protein AIG1 as a highly FP-reactive protein. We show that AIG1, and the sequence-related homologous protein ADTRP, possess conserved Thr and histidine (His) residues required for FP reactivity and find that both enzymes hydrolyze the fatty-acid esters of hydroxy-fatty acid (FAHFA) class of lipids in vitro and in human cells. Taken together, these data indicate that AIG1 and ADTRP represent a mechanistically novel class of Thr-dependent transmembrane hydrolases that regulate bioactive lipid metabolism in mammals.
Results
Discovery of AIG1 as an FP-reactive protein
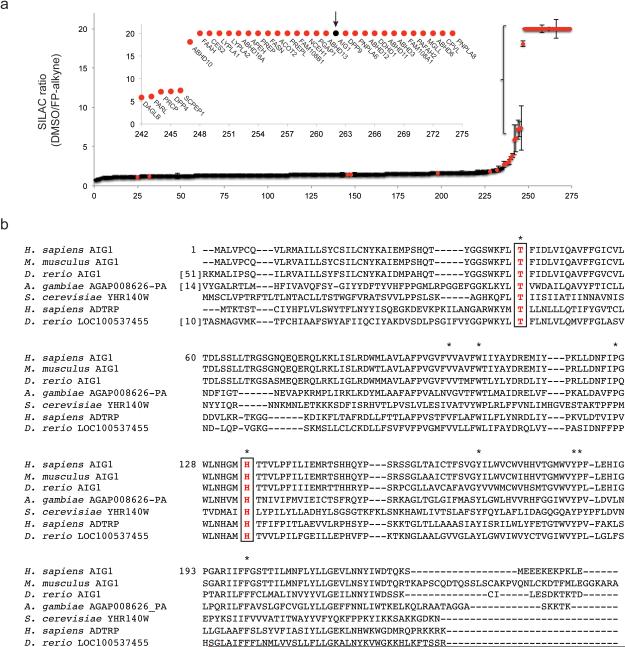
We performed a series of competitive ABPP experiments where heavy and light amino acid-labeled proteomes from a human cancer cell line (SKOV3) were pre-treated with DMSO or an FP agent (FP-alkyne12) at a concentration (20 μM) and incubation time (1 h) that, based on previous studies13, would be expected to fully label many serine hydrolases. We then exposed both DMSO-treated and FP-alkyne-blocked samples to a biotinylated FP probe (FP-biotin) and identified FP-biotin-labeled proteins by avidin enrichment and quantitative liquid chromatography (LC)-MS/MS analysis. Using this method, termed ABPP-SILAC14, we identified a group of proteins that were blocked in their reactivity with FP-biotin by pre-treatment with FP-alkyne (defined as proteins that were highly enriched in DMSO-treated compared to FP-alkyne-treated proteomes). As expected, virtually all of these proteins were annotated Ser hydrolases (Fig. 1a and Supplementary Results, Supplementary Table 1). However, within the group of FP-alkyne-sensitive proteins was one poorly characterized protein termed androgen-induced gene 1 protein (AIG1). Additional control experiments, as well as a review of legacy ABPP data sets performed in our lab, revealed that AIG1 was consistently enriched in studies that compared FP-biotin versus DMSO-treated proteomes (FP-probe-versus-no probe experiments) and showed a SILAC ratio of ~1 in probe-versus-probe control experiments where both heavy and light-labeled proteomes were treated with the same concentration of FP-biotin (Supplementary Fig. 1 and Supplementary Table 1).
Figure 1.
Discovery and characterization of AIG1 and ADTRP as FP-reactive proteins in the human proteome. (a) Competitive ABPP-SILAC analysis to identify FP-alkyne-inhibited proteins, where protein enrichment and inhibition were measured in proteomic lysates from SKOV3 cells treated with FP-alkyne (20 μM, 1 h) or DMSO using the FP-biotin probe following established protocols14. Annotated Ser hydrolases are indicated by red markers; AIG1 is marked in black and with an arrow. Data represent the median SILAC ratios ± s. d. for peptides quantified for each protein from one experiment representative of two biological replicates. (b) Sequence alignment of AIG1 orthologues from multiple species, as well as homologous ADTRP proteins (http://www.st-va.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi). Residues conserved between all sequences are marked with asterisks. The conserved Thr (Thr43 in human AIG1) and His (His134 in human AIG1) residues are boxed in red.
AIG1 is predicted to be a 28 kDa multipass transmembrane protein (with five-six predicted transmembrane domains; http://www.uniprot.org/uniprot/Q9NVV5) and was originally discovered as an androgen-induced gene product from human dermal papilla cells15 and later found to interact with the E3 ligase Pirh2 by yeast two-hybrid screening16. The biochemical functions of AIG1, however, remain unknown. BLAST and HHpred searches failed to detect any sequence homology between AIG1 and known Ser/Thr hydrolases and also revealed that there are no proteins in the Protein Data Bank (PDB) with substantial sequence-relatedness to AIG1 (Supplementary Table 2). These searches did, however, identify one protein in the human proteome that shares ~37% sequence identity with AIG1 – another poorly characterized protein termed ADTRP (androgen-dependent TFPI-regulating protein)17. We next set out to test whether AIG1 (and ADTRP) displayed biochemical properties consistent with those of a Ser hydrolase.
Conserved Thr and His residues in AIG1
We recombinantly expressed human AIG1 (hAIG1) as a C-terminal epitope-tagged protein in HEK293T cells and confirmed that this protein reacts with a rhodamine-conjugated FP probe (FP-Rh) by gel-based ABPP, which detected hAIG1 signals as 25 and 15 kDa bands in the membrane proteome of HEK293T cells (Supplementary Fig. 2a). FP-Rh-labeling of hAIG1 was time-dependent (Supplementary Fig. 2b), irreversible (Supplementary Fig. 2c), and competitively blocked by pre-treatment with FP-alkyne (Supplementary Fig. 2a). We also found that both the rat and mouse variants of AIG1 reacted with FP-Rh (Supplementary Fig. 3). Notably, when variants of AIG1 were expressed without a C-terminal epitope tag, we observed that they migrated as ~15-17 kDa proteins, which was substantially lower in molecular mass than that predicted from their full sequences (see below for further discussion). Recombinantly expressed human and mouse ADTRP were also labeled by FP-Rh and migrated as ~15 kDa proteins (Supplementary Fig. 3).
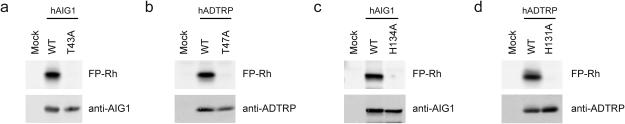
The protein sequence of hAIG1 contains 15 Ser residues, but, surprisingly, none of them were conserved across AIG1/ADTRP homologues from different species (Fig. 1b and Supplementary Fig. 4). On the other hand, we identified a single Thr residue (Thr-43 in hAIG1) that was completely conserved (Fig. 1b and Supplementary Fig. 4). We next mutated several Ser/Thr residues within the hAIG1 sequence to alanine (Ala, or A), including Thr-43 (T43), expressed these AIG1 variants in HEK293T cells, and evaluated their reactivity with FP-Rh by gel-based ABPP. Among the 15 individual Ser/Thr-to-Ala mutant proteins evaluated, only the T43A mutant showed a complete loss of FP-labeling (Fig. 2a and Supplementary Fig. 5). We also mutated the corresponding Thr residue in ADTRP (T47; Fig. 1b) and the resulting T47A mutant showed a similar loss of FP-labeling (Fig. 2b).
Figure 2.
Identification of Thr and His residues critical for FP labeling of AIG1 and ADTRP. (a-d) ABPP gels and Western blots of membrane proteomes (0.5 mg proteome mL−1) showing selective FP-Rh labeling of WT-, but not the indicated Thr-to-Ala and His-to-Ala mutants of human (h) AIG1 (a, c) and ADTRP (b, d) in transfected HEK293T cell lysates. Proteomes were treated with 1 μM FP-Rh for 30 min at 37 °C. Full images of gels and Western blots are provided in Supplementary Figure 16.
Despite extensive efforts using established LC-MS methods for mapping small-molecule probe-labeling sites in proteins18, we were unsuccessful in directly identifying Thr-43 as the site of FP-reactivity in AIG1. Thr-43 is predicted to reside within a long and hydrophobic tryptic peptide (aa 41-67), which may have complicated its detection by MS. We do note, however, that our ABPP-SILAC experiments with FP probes, in aggregate, provided 74% overall coverage of the AIG1 sequence and, in none of these experiments, was an unmodified form of the tryptic peptide bearing Thr-43 detected (Supplementary Fig. 6).
Ser/Thr hydrolases also possess conserved basic residues that activate the Ser/Thr nucleophile for catalysis. In most (but not all) Ser hydrolases, this basic residue is a histidine (His), while catalytic proteasomal subunits and other N-terminal Thr (or Ser or Cys) nucleophile-dependent enzymes use the N-terminal α-amine group as a base19,20. Review of the sequence alignment of AIG1 with homologous proteins identified a single conserved His residue His134 (Fig. 1b). Mutation of His134 to Ala in AIG1 (H134A) eliminated FP-labeling (Fig. 2c), while mutation of two other non-conserved His residues (H32A and H150A mutants) had no effect (Supplementary Fig. 5). Mutation of the corresponding conserved His residue in ADTRP to alanine (H131A) also blocked FP labeling of this protein (Fig. 2d).
We next analyzed the protein sequences for both AIG1 and ADTRP using six different transmembrane topology prediction programs (CCTOP, Phobius, PSORT II, TMHMM, TMpred, and Uniprot) and found that these programs consistently predicted that both the conserved Thr and His residues of AIG1 and ADTRP were located within transmembrane domains of these proteins (Supplementary Fig. 7).
AIG1 and ADTRP hydrolyze FAHFA lipids
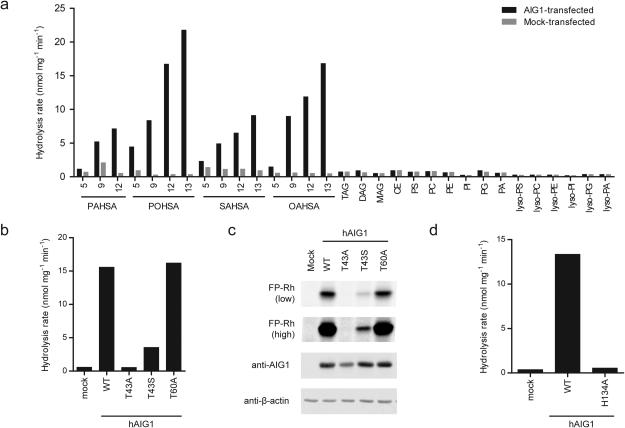
The strong reactivity displayed by AIG1 and ADTRP with FP probes and the dependency of these interactions on conserved Thr and His residues suggested that these poorly characterized proteins could represent a novel class of Thr hydrolases. The predicted multipass transmembrane structure of AIG1 and ADTRP also led us to hypothesize that one or more lipids may serve as substrates for these enzymes. We directly tested this premise by screening AIG1 and ADTRP against a panel of lipid substrates. The membrane lysates of hAIG1- and hADTRP-transfected HEK293T cells showed negligible hydrolytic activity above a mock-transfected control proteome with the majority of tested lipid substrates, including common classes of (lyso)-phospholipids and neutral lipids (e.g., tri-, di- and mono-glycerides) (Fig. 3a). In contrast, both AIG1 and ADTRP-transfected cell membrane lysates robustly hydrolyzed several fatty-acid esters of hydroxy-fatty acids (FAHFAs) (Fig. 3a and Supplementary Fig. 8a). FAHFAs are a recently identified class of bioactive lipids isolated from murine adipose tissue and their levels correlate with insulin sensitivity in both rodents and humans21. AIG1 and ADTRP displayed a preference for FAHFAs with branching distal from the carboxylate head group of the lipids (Fig. 3a).
Figure 3.
AIG1 and ADTRP are FAHFA hydrolases. (a) In vitro lipid substrate hydrolysis assays for membrane proteomes of mock and hAIG1-transfected HEK293T cells. PAHSA, palmitic acid ester of hydroxy-stearic acid; POHSA, palmitoleic acid ester of hydroxy-stearic acid; SAHSA, stearic acid ester of hydroxy-stearic acid; OAHSA, oleic acid ester of hydroxy-stearic acid; TAG, triacylglycerol (tri-C18:1); DAG, diacylglycerol (C16:0/C18:1); MAG, monoacylglycerol (C18:1); CE, cholesterol ester (C18:1); PS, phosphatidylserine (18:0, 18:2); PC, phosphatidylcholine (C17:0, C20:4); PE, phosphatidylethanolamine (C17:0, C20:4); PI, phosphatidylinositol (C16:0, C18:1); PG, phosphatidylglycerol (C16:0, C18:1); PA, phosphatidic acid (C17:0, C20:4); lyso-phospholipids (all C18:1). For each assay, 20 μg of mock or AIG1-transfected proteome was incubated with 100 μM substrate for 30 min at 37 °C. Data represent mean values for two biological replicates. (b-d) 12-OAHSA hydrolysis (b, d) and gel-based ABPP (c) assays for membrane proteomes from mock, wild-type hAIG1, and mutant hAIG1-transfected HEK293T cells. For (b, d), 20 μg of each proteome was incubated with 100 μM 12-OAHSA for 30 min at 37 °C. Data represent mean values for two biological replicates. For (c), proteomes were treated with 1 μM FP-Rh for 30 min at 37 °C, and β-actin was used as a loading control for gel experiments. Low and high exposures of the FP-Rh-labeled samples are shown to demonstrate the residual FP-labeling observed with the T43S, but not T43A mutant of hAIG1. Full images of the gel and Western blot are provided in Supplementary Figure 17.
The FAHFA hydrolase activities of AIG1 and ADTRP were abolished by mutating their putative catalytic nucleophilic residues Thr-43 and Thr-47, respectively (Fig. 3b and Supplementary Fig. 8b). Of note, mutation of Thr-43 to Ser in AIG1 impaired, but did not totally eliminate FAHFA hydrolytic activity or FP labeling of the mutant protein (Fig. 3b, c). We also tested the H134A mutant of AIG1 and H131A mutant of ADTRP and found that these proteins showed no detectable FAHFA hydrolase activity above a mock-transfected control (Fig. 3d and Supplementary Figure 8c).
Discovery and characterization of AIG1 inhibitors
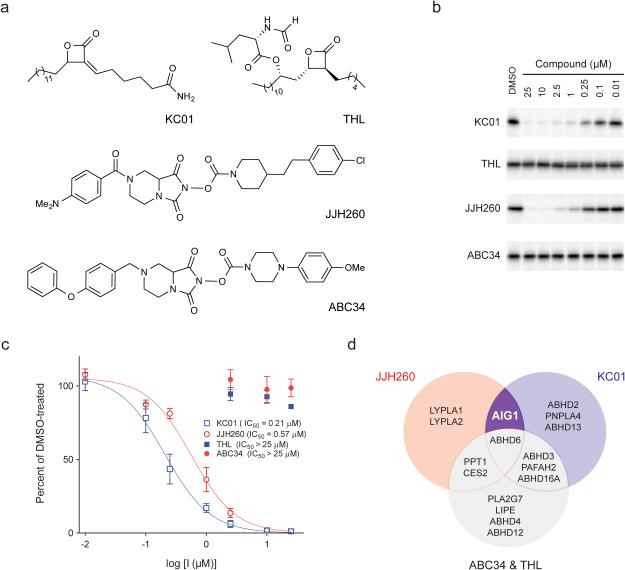
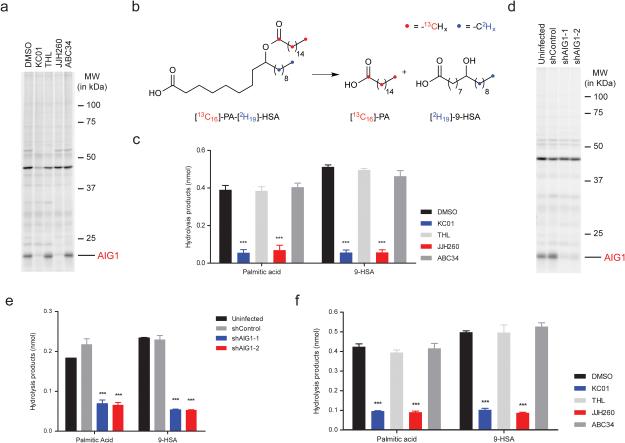
Beyond their reactivity with FPs, many Ser/Thr hydrolases are sensitive to inhibition by other types of small molecules, in particular, compounds bearing electrophilic carbonyl centers2. We performed competitive gel-based ABPP experiments with a structurally diverse library of carbamates22, β-lactones23, and activated ureas14 and identified several compounds that blocked FP-Rh labeling of hAIG1 (Supplementary Fig. 9a). Inhibitors of AIG1 included the β-lactone KC0123 and the N-hydroxyhydantoin (NHH) carbamate JJH260 (1) (Fig. 4a), which blocked FP-labeling of AIG1 with IC50 values of 0.17 ± 0.03 μM and 0.50 ± 0.14 μM, respectively (Fig. 4b and Supplementary Fig. 9b) and kobs/[I] values of 2820 ± 780 M−1s−1 and 300 ± 25 M−1s−1, respectively (Supplementary Fig. 9c). Both KC01 and JJH260 also inhibited FAHFA hydrolysis by AIG1 with IC50 values of 0.21 ± 0.08 μM and 0.57 ± 0.14 μM, respectively (Fig. 4c). We finally identified structurally related inactive control compounds – the β-lactone tetrahydrolipstatin (THL) and the NHH carbamate ABC3422 (Fig. 4a) – that did not inhibit the FP reactivity (Fig. 4b and Supplementary Fig. 9b) or FAHFA hydrolysis activity (Fig. 4c) of AIG1. KC01 and JJH260 also inhibited ADTRP as assessed by competitive gel-based ABPP, albeit with much lower potency (IC50 values of 1.3 and 8.5 μM, respectively; Supplementary Fig. 10).
Figure 4.
Discovery of inhibitors and structurally related inactive control compounds for AIG1. (a) Chemical structures of two hAIG1 inhibitors, KC01 and JJH260, and the respective control compounds, THL and ABC34. (b) ABPP gels demonstrating inhibition of FP-Rh labeling of hAIG1 by KC01 and JJH260, but not THL and ABC34. Proteomes were treated with each inhibitor at the indicated concentrations for 30 min at 37 °C, followed by FP-Rh (1 μM, 30 min, 37 °C). Full images of the gels are provided in Supplementary Figure 18. (c) Concentration-dependent inhibition of the 9-PAHSA hydrolysis activity of hAIG1-transfected HEK293T membrane proteome by KC01 (IC50 = 0.21 ± 0.08 μM) and JJH260 (0.57 ± 0.14 μM), but not THL and ABC34. For these experiments, proteomes were incubated with each inhibitor at the indicated concentration for 30 min at 37 °C. 20 μg of each proteome was then incubated with 100 μM 9-PAHSA for 30 min at 37 °C. Data represent mean values ± s. e. m. for three biological replicates. (d) Venn diagram showing overlapping and distinct target profiles for AIG1 inhibitors KC01 and JJH260 and control compounds THL and ABC34.
The activity of KC01 suggested that a fluorescent analogue WHP0123 might also serve as a tailored activity-based probe for AIG1. Consistent with this premise and with a covalent mode of inhibition of AIG1 by β-lactones, we found that wild-type hAIG1, but not the T43A mutant, was robustly labeled by WHP01 in transfected HEK293T cell membrane proteomes (Supplementary Fig. 11).
KC0123, THL24, and ABC3422 have been previously characterized for their selectivity across the Ser hydrolase class by competitive ABPP experiments. We complemented these past studies by also evaluating the selectivity profile of JJH260 by ABPP-SILAC in human cancer cell proteomes, which confirmed inhibition of endogenously expressed AIG1 and established ABHD6, LYPLA1, and LYPLA2 as additional off-targets of JJH260 (Supplementary Fig. 12a and Supplementary Table 1). PPT1, a common target of NHH carbamates22, was also found to be inhibited at low micromolar concentrations of JJH260 by gel-based ABPP (Supplementary Fig. 12b). Comparing the inhibitor selectivity profiles revealed complementary sets of off-targets, as neither LYPLA1 nor LYPLA2 were inhibited by KC0123, while the AIG1-inactive control compound ABC34 blocked both ABHD6 and PPT122 (Fig. 4d). Thus, we concluded that the function of AIG1 could be pharmacologically characterized in biological systems by evaluating the effects of both active compounds (KC01 and JJH260) compared to their inactive controls (THL and ABC34).
AIG1 is a principal FAHFA hydrolase in human cells
Little is known about how FAHFAs are inactivated in human cells, so we next asked whether AIG1 was a principal regulator of the metabolism of this lipid class in situ. RNA expression profiling indicated strong AIG1 expression in the human prostate cancer cell line (LNCaP)25, which we confirmed by ABPP-MudPIT (12 spectral counts/1 mg proteome). We were also able to detect a 15 kDa WHP01-reactive band in LNCaP cells by gel-based ABPP, and the WHP01 labeling of this protein was blocked by pre-treatment with KC01 and JJH260, but not THL and ABC34 (Fig. 5a), consistent with its designation as endogenous AIG1. Both KC01 and JJH260, but not THL or ABC34 also inhibited the FAHFA hydrolase activity of LNCaP cell lysates, which was mostly found in the membrane fraction (Supplementary Fig. 13a, b). We next established an in situ FAHFA hydrolysis assay by treating human cells with a doubly isotopically labeled substrate [13C16]-palmitic acid (PA) ester of [2H19]-9-hydroxy-stearic acid (HSA) (13C,2H-PAHSA; Fig 5b). Cells were treated with inhibitor or control compound (5 μM) for 4 h followed by the addition of 13C,2H-PAHSA (2 μM, 5 mL of media) for 2 h, and then harvested and PAHSA and its hydrolytic products –13C16-PA and 2H19-9-HSA (2H19-HSA)– extracted and analyzed by LC/MS. After first validating this experimental approach with multiple concentrations of 13C,2H-PAHSA and HEK293T cells that heterologously express AIG1 (Supplementary Fig. 13c, d), we measured 13C,2H-PAHSA hydrolysis in LNCaP cells treated with various inhibitors. KC01 and JJH260-treated cells exhibited a substantial (~70-80%) reduction in 13C16-PA and 2H19-HSA products, while cells treated with the control compounds THL and ABC34 showed negligible alterations in 13C,2H-PAHSA hydrolysis compared to DMSO-treated cells (Fig. 5c). A similar profile of inhibitor activity on 13C,2H-PAHSA hydrolysis was observed in AIG1-transfected HEK293T cells (Supplementary Fig. 13e).
Figure 5.
AIG1 inhibitors block FAHFA hydrolysis in mammalian cells. (a) ABPP gel of the membrane proteome of LNCaP cells demonstrating inhibition of endogenous AIG1 by KC01 and JJH260. Cells were treated with 5 μM of each compound for 4 h at 37 °C. lysed, and membrane proteomes labeled with WHP01 (2 μM, 30 min, 37 °C) before gel-based ABPP. (b) Structure of double-labeled 9-PAHSA ([13C16]-PA-[D19]-HSA, or 13C,2H-PAHSA) and its hydrolytic products 13C16-palmitic acid (13C16-PA) and 2H19-hydroxystearic acid (2H19-HSA). (c) 13C,2H-PAHSA hydrolysis activity of LNCaP cells treated in situ with DMSO or inhibitors (5 μM) for 4 h at 37 °C and then fed 2 μM [13C16]-PA-[D19]-HSA for 2 h. Data represent mean ± s. e. m. for four biological replicates. (d) ABPP gel of the membrane proteomes of shAIG1 and control LNCaP cell lines. Proteomes were treated with 2 μM WHP01 for 30 min at 37 °C. (e) 13C,2H-PAHSA hydrolysis activity of shAIG1 and control LNCaP cell lines. Cells were fed 2 μM 13C,2H-PAHSA for 2 h prior to analysis. Data represent mean ± s. e. m. for five biological replicates. (f) 13C,2H-PAHSA hydrolysis activity of human T-cells treated in situ with DMSO or inhibitors. Cells were treated with 5 μM of each inhibitor for 4 h at 37 °C and then fed 2 μM [13C16]-PA-[D19]-HSA for 2 h. Data represent mean ± s. e. m. for three biological replicates. ***p < 0.001 by two-sided Student’s t-test for inhibitors-treated or shAIG1 versus control cell lines.
We complemented these pharmacological studies by knocking down AIG1 expression in LNCaP cells using RNA-interference (RNAi) technology. We generated two stable shRNA-knockdown lines targeting AIG1 (shAIG1-1 and -2 lines) and a control line expressing an shRNA targeting an unrelated protein (GFP; shControl line). Substantial and selective reductions in AIG1 expression were observed in both shAIG1 lines compared to the shControl line by gel-based ABPP using the tailored and broad-spectrum probes WHP01 and FP-Rh, respectively (Fig. 5d and Supplementary Fig. 14a). Proteomic lysates from shAIG1 lines also showed significant reductions in 9-PAHSA hydrolysis compared to lysates from the shControl line (Supplementary Fig. 14b). We next treated shAIG1 and shControl cells with 13C,2H-PAHSA and observed a ~70% reduction in hydrolysis products in the shAIG1 cells compared to shControl or uninfected cell lines (Fig. 5e). Finally, we analyzed FAHFA hydrolysis in human T-cells, where we detected AIG1 by gel-based ABPP (Supplementary Fig. 15a). The FAHFA hydrolase activity of human T-cell membrane lysates was substantially reduced following treatment with KC01 or JJH260, but not THL or ABC34 (Supplementary Fig. 15b). Similar results were obtained for in situ experiments, where pre-treatment of human T-cells with KC01 or JJH260, but not THL or ABC34 blocked the cellular hydrolysis of 13C,2H-PAHSA (Fig. 5f). These results, taken together, indicate that AIG1 functions as a major FAHFA hydrolase in human cells.
Discussion
We have discovered herein using ABPP that the poorly characterized multipass transmembrane proteins AIG1 and ADTRP represent a new family of hydrolytic enzymes that degrade the FAHFA class of signaling lipids. Several lines of biochemical evidence support that AIG1 and ADTRP use conserved Thr and His residues as a catalytic nucleophile and base, respectively. That these residues are generally predicted to be embedded within transmembrane domains of AIG1 and ADTRP indicates these enzymes could have evolved to perform hydrolytic chemistry within the cell membrane environment, reminiscent of the regulated intramembrane proteolysis (RIP) class of transmembrane proteases26-29, which have also been studied by ABPP30-32. This feature, if experimentally validated, may endow AIG1 and ADTRP with a special capacity to hydrolyze FAHFAs, which, unlike most neutral- and phospho-lipids, possess ester bonds along the length of their otherwise hydrophobic acyl chains, and these bonds are presumably also buried within the lipid bilayer. It is not yet clear why AIG1 and ADTRP migrate as much lower MW proteins than that predicted from their sequence, but it is possible that these proteins are proteolytically processed in cells or migrate aberrantly by SDS-PAGE due to their high transmembrane content33. We also do not yet understand the functional differences between AIG1 and ADTRP, although we note that the cell/tissue distributions of these enzymes are quite distinct, with AIG1 being broadly expressed with highest signals in brain and macrophages, and ADTRP showing a more restricted profile of expression principally in metabolic organs, such as liver, kidney, intestine, and brown fat (biogps.org).
We have identified an initial set of pharmacological tools to study AIG1, including multiple classes of lead inhibitors (KC01, JJH260) and structurally related inactive control compounds (THL, ABC34), as well as tailored ABPP probes (WHP01) for enhanced detection of endogenous AIG1 activity in native proteomes. While further research will be required to optimize the potency and specificity of AIG1 (and ADTRP) inhibitors, we should note that NHH carbamates have been developed into selective and in vivo-active inhibitors for other Ser hydrolases22. If optimized inhibitors verify that AIG1 and/or ADTRP regulate FAHFA metabolism in vivo, these enzymes could represent new targets for treating metabolic disorders, such as type 2 diabetes21.
Finally, we offer some speculation on the existence of additional outlier members of the Ser/Thr hydrolase class. Our studies to date of FP-reactive proteins in mammalian cells have not uncovered clear examples of other proteins beyond AIG1 and ADTRP that lack designation as Ser/Thr hydrolases (e.g., see Fig. 1a), suggesting that the annotation of enzymes from this class may be near-completion in humans. The HHpred search results, however, uncovered a distinct set of uncharacterized AIG1/ADTRP-like proteins that possess the conserved Thr and His residues and are found in non-mammalian eukaryotic organisms (Panther family PTHR12242; members in insects, plants, protozoa, and other non-vertebrates). Microbial proteomes also remain much less extensively characterized, and, considering their massive evolutionary and metabolic diversity34, we would not be surprised if bacteria contain many additional classes of unassigned Ser/Thr hydrolases. Our findings underscore the power of chemical proteomic methods like ABPP for the discovery and functional characterization of such enzymes.
Online Methods
ABPP-MudPIT and competitive ABPP-MudPIT sample preparation
For ABPP-MudPIT samples, proteomes (1 mg/mL in 1 mL of PBS) were labeled with FP-biotin (5 μM) for 1 h at 23 °C while rotating. After labeling, the proteomes were denatured and precipitated using 4:1 MeOH/CHCl3, resuspended in 0.5 ml of 6 M urea in PBS, reduced using tris(2-carboxyethyl)phosphine (TCEP, 10 mM) for 30 min at 37 °C, and then alkylated using iodoacetamide (40 mM) for 30 min at 23 °C in the dark. The biotinylated proteins were enriched with PBS-washed avidin-agarose beads (100 μL; Sigma-Aldrich) by rotating at 23 °C for 1.5 h in PBS with 0.2% SDS to a final volume of 6 mL. The beads were then washed sequentially with 10 mL PBS with 0.2% SDS, 10 mL PBS (3x) and 10 mL DI H2O (3x). On-bead digestion was performed using sequencing-grade trypsin (2 μg; Promega) in 2 M urea in PBS with 2 mM CaCl2 for 12–14 h at 37 °C (200 μL). Peptides obtained from this procedure were acidified with formic acid (5%) and stored at −20 °C prior to analysis. SILAC experiments were performed using the human SKOV3 breast cancer cell line (ATC HTB-77). The isotopically labeled cell lines were generated by ≥ 5 passages in either light (100 μg/mL each of L-arginine and L-lysine) or heavy (100 μg/mL each of [13C615N4]L-arginine and [13C615N4]L-lysine) SILAC RPMI medium (ThermoScientific) supplemented with 10% dialyzed FBS (Omega Scientific) and penicillin-streptomycin (GE Life Sciences). Light and heavy cells were treated with the test compound or DMSO, respectively, at 37 °C. Cells were then washed with sterile PBS (3x), harvested, and lysed by sonication in PBS. The membrane and soluble fractions were separated by ultra-centrifugation at 100,000 g for 45 min at 4 °C. 2 mg/mL light proteome (0.5 mL) and 2 mg/mL heavy proteome (0.5 mL) were subsequently treated with FP-biotin (5 μM) for 1 h at 23 °C, combined, and processed as above for the ABPP-MudPIT protocol. For competitive ABPP experiments with FP-alkyne, the concentration of probe (20 μM) was chosen based on prior gel-based experiments demonstrating saturation of labeling for many serine hydrolases at comparable concentrations of FP probe.13 In addition, previous studies have shown effective competition of most FP-Rh labeled bands in mouse brain with 10 μM FP-alkyne.35
HHpred searches
Remote homology detection was performed using the HHpred algorithm36. Briefly, a seed alignment was generated using the CLUSTALW algorithm on the seven protein sequences shown in Fig. 1b (UniProt IDs: AIG1_HUMAN, AIG1_MOUSE, ADTRP_HUMAN, F1QYR4_DANRE, Q7Q8H9_ANOGA, YHU0_YEAST, E7FAT8_DANRE). The CLUSTALW alignment was used as input for the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred) and it was run using the default parameters to search all available HMM databases. The complete output from this analysis is provided in Supplemental Table 2.
Cell culture methods
HEK293T cells (ATCC CRL-3216) were cultured in DMEM (Gibco), supplemented with L-glutamine (2 mM), 10% FCS (Omega Scientific), and penicillin-streptomycin (GE Life Sciences) at 37 °C and 5% CO2. LNCaP cells (ATCC CRL-1740) were cultured in RPMI 1640 (Gibco), supplemented with L-glutamine (2 mM), 10% FCS, and penicillin-streptomycin at 37 °C and 5% CO2. T-cells were purified from peripheral blood mononuclear cells (PBMCs) obtained from human subjects using STEMCELL Technologies kits as per manufacturers instructions. Typically 100-150 million T-cells were obtained from 500 mL human subject derived PBMCs at > 95% purity as confirmed by flow cytometry analysis. T-cells were cultured in dye-free RPMI supplemented with 10% FCS at 37 °C and 5% CO2.
Cloning and recombinant expression of AIG1 and ADTRP
Full-length cDNA encoding human AIG1 (GE Healthcare, in pOTB7 vector) was cloned into the pCMV6-Entry vector with C-terminal Myc and DDK tags. (Sense primer: 5'-GGGCGGCCGGGAATTCGCGAACATGG-3'; Antisense primer: 3'-AAGCCTAAATTGGAAACGCGGCCGCTTTA-5'). Full-length cDNA encoding for human ADTRP in the pCMV6-XL5 vector was purchased from Origene. Full-length cDNA constructs encoding for mouse AIG1 in the pCMV-Sport6 vector, rat AIG1 in pExpress-1 vector, and mouse ADTRP in the pCMV-Sport6 vector were purchased from GE Life Sciences. To recombinantly express AIG1 or ADTRP, HEK293T cells were grown to 40% confluence in a 10 cm tissue culture plate and transiently transfected with 4 μg of the desired construct using polyethyleneimine ‘MAX’ (MW 40,000, PEI; Polysciences, Inc.) as the transfection reagent per the manufacturer’s protocol. ‘Mock’ transfected cells were transfected with 4 μg of empty vector. 48 h after transfection, cells were washed with PBS (3x), harvested by scraping, and lysed by sonication in PBS. The membrane and soluble fractions were separated by ultra-centrifugation at 100,000 g for 45 min at 4 °C. Protein concentrations were measured using the DC Protein Assay kit (Bio-Rad). Aliquots were flash-frozen and stored at −80 °C for further use.
Site-directed mutagenesis
Point mutations in both human AIG1 and ADTRP were generated by the QuikChange site-directed mutagenesis protocol (Strategene) as per the manufacturer’s instructions. For primers used for each mutation, see Supplementary Table 3. Primers were obtained from Integrated DNA Technologies. All DNA sequencing was performed by Eton Biosciences Inc.
Gel-based ABPP analysis
Tissue and cell proteomes (50 μL) were treated with either FP-rhodamine (1 μM) or WHP01 (2 μM) for 30 min at 37 °C. The reactions were then quenched by addition of 4x SDS-PAGE loading buffer (20 μL). Competitive gel-based ABPP experiments were performed as previously described.7 Samples were visualized in-gel using a ChemiDoc MP imaging system (Bio-Rad). The fluorescence from rhodamine is presented in gray scale. 2-5 s exposure times and 20-60 s exposure times were used for AIG1/ADTRP-transfected and native proteomes, respectively. Relative band intensities were quantified using ImageJ software (http://imagej.nih.gov/ij/).
Western blotting
Cell proteomes were separated by SDS-PAGE, transferred to nitrocellulose membrane (60 V for 90 min), and blocked by 5% milk in TBS-Tween. The primary antibodies used and dilutions are as follows: anti-AIG1 (Rabbit, Sigma-Aldrich, SAB1304597, 1:250), anti-ADTRP (Rabbit, Atlas, HPA048113, 1:500), and anti-β-actin (Mouse, Cell Signaling, 3700S, 1:1000). IRDye 800CW anti-rabbit and anti-mouse secondary antibodies (LI-COR, 1:10000) were used as secondary antibodies for visualization.
AIG1 and ADTRP LC/MS substrate hydrolysis assays
FAHFA substrates were purchased from Cayman Chemical Co. All non-FAHFA lipid substrates were purchased from Avanti Polar Lipids Inc. unless mentioned otherwise. 20 μg of proteome was incubated with 100 μM lipid substrate in a reaction volume of 250 μL in PBS at 37 °C with constant shaking. After 30 minutes the reaction was quenched with 400 μL of 2:1 (vol/vol) CHCl3: MeOH, doped with internal standard (0.5 nmol C17:1 heptadecenoic acid (C17:1 FFA) and 0.05 nmol 9-hydroxy-heptadecanoic acid (9-HHDA)). The mixture was vortexed and centrifuged at 2800 g for 5 min to separate the aqueous (top) and organic (bottom) phase. The organic phase was collected and dried under a stream of N2, re-solubilized in 100 μL of 2:1 (vol/vol) CHCl3: MeOH, and subjected to LC-MS analysis. A fraction of the organic extract (~ 15 μL) was injected onto an Agilent 6520 quadrupole-time-of-flight (QTOF) LC-MS and analyzed as described previously.23 All data presented for substrate hydrolysis assays is the average of 2-5 independent biological replicates, error bars represent S.D. (n = 2), and S.E.M. (n = 3-5).
Synthesis of AIG1 inhibitors and probes
FP-biotin, FP-rhodamine, and FP-alkyne were synthesized in-house as previously described.4,37,38 KC01 and WHP01 were synthesized in-house as previously described.23 ABC34 was synthesized in-house as previously described.22 Tetrahydrolipstatin (THL) was purchased from Sigma-Aldrich. [13C16]-PA-[D19]-HSA was synthesized according to the previously described method, using isotopically labeled starting materials.21 See Supplementary Note for information on the synthesis and characterization of JJH260.
9-PAHSA feeding assay
For the pharmacology studies, cells were treated in situ with 5 μM KC01, THL, JJH260, or ABC34 for 4 h at 37 °C in a volume of 5 mL in dye-free RPMI (LNCaP cells, T-cells) or dye-free DMEM (HEK293T cells) supplemented with 10% FCS. 2 μM [13C16]-PA-[D19]-HSA was then added to the media for 1 h (HEK293T cells) or 2 h (LNCaP cells, T-cells) at 37 °C. Thereafter the media was collected (5 mL) and the cells were washed with sterile PBS (3x) and harvested by scraping. The cells were re-suspended in 1 mL sterile PBS and mixed by vortexing with 3 mL of 2:1 CHCl3: MeOH with the internal standard mix (50 pmol each of C17:1 FFA, 9-HHDA, and [13C16]-PAHSA). The two phases were separated by centrifugation at 2800 g for 10 min, and the organic phase (bottom) was collected and dried under stream of N2. The lipid extracts were re-solubilized using 100 μL of 2:1 CHCl3: MeOH, and 10 μL was used for the targeted LC-MS analysis. The MRM transitions for the targeted LC-MS analysis are presented in Supplementary Table 4.
shRNA knockdown studies
AIG1 MISSION shRNA bacterial glycerol stocks were purchased, and the lentiviral-based shRNA gene knockdown was performed in LNCaP cells using the manufacturer’s protocol (Sigma-Aldrich). Briefly, 1 μg shRNA transfer vector, 0.1 μg of the VSVG envelope vector and 0.9 μg packaging vector dVPR were transfected using 6 μL of X-tremeGENE 9 (GE Life Sciences) into 1.8 ×106 HEK293T cells cultured in 5 mL DMEM with L-glutamine (2 mM) and 10% FCS and 5% CO2 to generate the lentiviral transduction particles (LTP). 0.25 × 106 LNCaP cells were infected with the LTP generated from the transfections along with 5 μg/mL polybrene (to enhance infection) cultured in 5 mL RPMI 1640 with 10% FCS and 5% CO2, and thereafter the lentiviral-infected LNCaP cells were selected on puromycin (1 μg/ml). After six rounds of puromycin selection, LNCaP cells infected with the LTP generated using the constructs TRCN0000142987 (shAIG1-1; Sigma-Aldrich) and TRCN0000422331 (shAIG1-2; Sigma-Aldrich) showed >75% knockdown of AIG1 by gel-based ABPP and substrate hydrolysis assays and were selected for further studies. As a negative control (shControl), shRNA transfer vector targeting GFP was used.
MS and data analysis
MS was performed using a LTQ (for spectral counting studies), or LTQ-Orbitrap or Orbitrap Velos (for SILAC studies), following previously described protocols (ThermoFinnigan).7,39 Peptides were eluted using a five-step multidimensional LC-MS protocol in which increasing concentrations of ammonium acetate are injected followed by a gradient of increasing acetonitrile, as previously described.40 For all samples, data were collected in data-dependent acquisition mode over a range from 400–1,800 m/z. Each full scan was followed by up to 7 or 30 fragmentation events for experiments using the LTQ and Orbitrap or Orbitrap Velos instruments, respectively. Dynamic exclusion was enabled (repeat count of 1, exclusion duration of 20 s) for all experiments. The data were searched using the ProLuCID algorithm against a human reverse-concatenated nonredundant (gene-centric) FASTA database that was assembled from the Uniprot database. ProLuCID searches specified static modification of cysteine residues (+57.0215 m/z; iodoacetamide alkylation) and required peptides to contain at least one tryptic terminus. For SILAC samples, data sets were searched independently with the following parameter files; for the light search, all amino acids were left at default masses; for the heavy search, static modifications on lysine (+8.0142 m/z) and arginine (+10.0082 m/z) were specified. For data collected on the Orbitrap mass spectrometers, precursor-ion mass tolerance was set to 50 ppm. The resulting peptide spectral matches were filtered using DTASelect (version 2.0.47), and only half-tryptic or fully tryptic peptides were accepted for identification. Peptides were restricted to a specified false positive rate of <1%. SILAC ratios were quantified using in-house CIMAGE software.18 Briefly, a 10-min retention time window was used for peak identification using 10 ppm mass accuracy and requiring a coelution R2 value greater than 0.8. Peptides detected as singletons, where only the heavy or light isotopically labeled peptide was detected and sequenced but which passed all other filtering parameters, were given a ratio of 20, which is the maximum SILAC ratio reported here.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 6 (for Mac OS X) software. Data derived from three or more replicates are shown as mean values ± s. e. m. Student’s t-test (two-tailed) was used to study statistically significant differences between study groups. A P value of <0.05 was considered statistically significant for this study.
Supplementary Material
Acknowledgments
We are grateful to K. Masuda and L. Bar-Peled for discussions on and assistance with cloning; M. Dix and K. Lum for discussions and technical expertise in designing and analyzing proteomics experiments; J. Teijaro for providing T-cells; G. Simon for assistance with the HHpred analysis; and M. Niphakis for numerous helpful discussions. This work was supported by the NIH (DA033760, DK909810), The Leona M. and Harry B. Helmsley Charitable Trust (grant #2012-PG-MED002 to A.S.), NCI Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.), a Hewitt Foundation for Medical Research Fellowship (to W.H.P.), a Chapman Charitable Trust Fellowship (to M.K.), and an Irving S. Sigal postdoctoral fellowship from the American Chemical Society (to S.S.K.).
Footnotes
Author contributions. W.H.P., M.J.K., A.S. and B.F.C. conceived the project. W.H.P., M.J.K., S.S.K., A.S., and B.F.C. designed experiments. W.H.P. performed the molecular biology and proteomics experiments. W.H.P., A.B.C., J.J.H., and A.S. synthesized compounds. M.J.K., S.S.K., and W.H.P. performed substrate assays and biochemical experiments. W.H.P., M.J.K., S.S.K., E.S., B.B.K., A.S., and B.F.C. analyzed and interpreted the data. W.H.P. and B.F.C. wrote the paper. M.J.K., S.S.K., and A.S. edited the paper.
Competing financial interests
B.F.C. is a founder and advisor to Abide Therapeutics, a biotechnology company interested in developing serine hydrolase inhibitors and therapeutics.
References
- 1.Willems LI, Overkleeft HS, van Kasteren SI. Current developments in activity-based protein profiling. Bioconjug. Chem. 2014;25:1181–1191. doi: 10.1021/bc500208y. [DOI] [PubMed] [Google Scholar]
- 2.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- 3.Berger AB, Vitorino PM, Bogyo M. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am. J. Pharmacogenomics. 2004;4:371–381. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 8.Higa HH, Diaz S, Varki A. Biochemical and genetic evidence for distinct membrane-bound and cytosolic sialic acid O-acetyl-esterases: serine-active-site enzymes. Biochem. Biophys. Res. Comm. 1987;144:1099–1108. doi: 10.1016/0006-291x(87)91425-2. [DOI] [PubMed] [Google Scholar]
- 9.Jessani N, et al. Class assignment of sequence-unrelated members of enzyme superfamilies by activity-based protein profiling. Angew. Chem. Int. Ed. Engl. 2005;44:2400–2403. doi: 10.1002/anie.200463098. [DOI] [PubMed] [Google Scholar]
- 10.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias M, Tawfik DS. Divergence and convergence in enzyme evolution: parallel evolution of paraoxonases from quorum-quenching lactonases. J. Biol. Chem. 2012;287:11–20. doi: 10.1074/jbc.R111.257329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lone AM, et al. A substrate-free activity-based protein profiling screen for the discovery of selective PREPL inhibitors. J. Am. Chem. Soc. 2011;133:11665–11674. doi: 10.1021/ja2036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 14.Adibekian A, et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo J, Kim J, Kim M. Cloning of androgen-inducible gene 1(AIG1) from human dermal papilla cells. Mol. Cells. 2001;11:35–40. [PubMed] [Google Scholar]
- 16.Wu G, Sun M, Zhang W, Huo K. AIG1 is a novel Pirh2-interacting protein that activates the NFAT signaling pathway. Frontiers in Bioscience E3. 2011:834–842. doi: 10.2741/E291. [DOI] [PubMed] [Google Scholar]
- 17.Lupu C, Zhu H, Popescu NI, Wren JD, Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118:4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends in Biochemical Sciences. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 20.Ekici OD, Paetzel M, Dalbey RE. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yore MM, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cognetta AB, 3rd, et al. Selective N-Hydroxyhydantoin carbamate inhibitors of mammalian serine hydrolases. Chem. Biol. 2015;22:928–937. doi: 10.1016/j.chembiol.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamat SS, et al. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat. Chem. Biol. 2015;11:164–171. doi: 10.1038/nchembio.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg. Med. Chem. Lett. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomura DK, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem. Biol. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe MS. Intramembrane-cleaving proteases. J. Biol. Chem. 2009;284:13969–13973. doi: 10.1074/jbc.R800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 29.Strisovsky K. Why cells need intramembrane proteases - a mechanistic perspective. FEBS J. 2015 doi: 10.1111/febs.13638. doi:10.1111/febs.13638. [DOI] [PubMed] [Google Scholar]
- 30.Sherratt AR, Blais DR, Ghasriani H, Pezacki JP, Goto NK. Activity-based protein profiling of the Escherichia coli GlpG rhomboid protein delineates the catalytic core. Biochemistry. 2012;51:7794–7803. doi: 10.1021/bi301087c. [DOI] [PubMed] [Google Scholar]
- 31.Vosyka O, et al. Activity-based probes for rhomboid proteases discovered in a mass spectrometry-based assay. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2472–2477. doi: 10.1073/pnas.1215076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen MT, Kersavond TV, Verhelst SH. Chemical tools for the study of intramembrane proteases. ACS Chem. Biol. 2015;10:2423–2434. doi: 10.1021/acschembio.5b00693. [DOI] [PubMed] [Google Scholar]
- 33.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlt JA, et al. The enzyme function initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adibekian A, et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J. Am. Chem. Soc. 2012;134:10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 38.Tully SE, Cravatt BF. Activity-based probes that target functional subclasses of phospholipases in proteomes. J. Am. Chem. Soc. 2010;132:3264–3265. doi: 10.1021/ja1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu KL, et al. DAGLβ inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat. Chem. Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.