Abstract
While mutations in the KRAS oncogene are amongst the most prevalent in human cancer, there are few successful treatments to target these tumors. It is also likely that heterogeneity in KRAS-mutant tumor biology significantly contributes to the response to therapy. We hypothesized that presence of commonly co-occurring mutations in STK11 and TP53 tumor suppressors may represent a significant source of heterogeneity in KRAS-mutant tumors. To address this, we utilized a large cohort of resected tumors from 442 lung adenocarcinoma patients with data including annotation of prevalent driver mutations (KRAS, EGFR) and tumor suppressor mutations (STK11 and TP53), microarray-based gene expression and clinical covariates including overall survival (OS). Specifically, we determined impact of STK11 and TP53 mutations on a new KRAS mutation-associated gene expression signature as well as previously defined signatures of tumor cell proliferation and immune surveillance responses. Interestingly, STK11, but not TP53 mutations, were associated with highly elevated expression of KRAS mutation-associated genes. Mutations in TP53 and STK11 also impacted tumor biology regardless of KRAS status, with TP53 strongly associated with enhanced proliferation and STK11 with suppression of immune surveillance. These findings illustrate the remarkably distinct ways through which tumor suppressor mutations may contribute to heterogeneity in KRAS-mutant tumor biology. In addition, these studies point to novel associations between gene mutations and immune surveillance that could impact the response to immunotherapy.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. Despite therapeutic advances over the last several decades, the overall 5-year survival remains only 16% (1). Mutations in the KRAS gene occur frequently in non-small cell lung cancer (NSCLC), especially in adenocarcinoma (~30%) though less common in squamous cell carcinoma (about 7%) (2–4). Although mutationally activated KRAS tumors were originally identified in 1982 (5), to date there are no successful treatment strategies that target these mutations (3). However, key pathways activated by KRAS and mutation-associated vulnerabilities may be therapeutically targetable including the MEK, phosphoinositide 3-kinase (PI3K), GSK-3α and RAL/TBK1 pathways (6–11). Mutations in tumor suppressor genes TP53 and STK11 are also common in lung adenocarcinoma and often co-occur with KRAS mutations (2–4, 12). While much is known about tumor promotion mechanisms of TP53, less is known about STK11 function and impact on disease progression and patient survival. The STK11 gene encodes a serine/threonine protein kinase known as liver kinase β1 (LKB1) (13). The most common STK11 mutations are deletion or inactivating mutations (14–21), which, along with murine studies provide strong evidence for a tumor suppressor function for this gene (19).
Recent studies have defined gene expression changes triggered by RAS (22–24). For example, a RAS signature associated with MEK pathway activation is also associated with sensitivity to MEK inhibitors (MEKi) (24). Gene expression studies have also defined signatures associated with enhanced tumor progression and reduced patient survival. Examples of such signatures include the malignancy risk signature reported by our group, which is rich in cell cycle regulating genes and therefore associated with highly proliferative tumors (25, 26). It is now well established that a functional immune system is crucial in controlling tumor growth (27–29). Consequently, T cell presence in tumors is associated with immune surveillance and improved patient survival (28, 30–35). It is important to note that benefit from immunotherapy, including T cell checkpoint blockade, is also commonly associated with high tumor expression of immuno-stimulatory genes and T cell infiltration (36–38). Thus, activation of key immune regulatory pathways such as JAK-STAT and NF-κB pathways (38–41) in tumor cells or tumor infiltrating non-malignant cells likely enhances the response to immunotherapy. We recently identified a gene expression signature of NF-κB regulated genes that is associated with an immune-active tumor microenvironment (42). The role and potential association between common lung cancer mutations and the immune surveillance response has however not been investigated. The goal of studies described here was to better define molecular heterogeneity in KRAS mutant tumors, especially as it relates to effects of co-occurring mutations in STK11 and TP53 tumor suppressors in shaping KRAS mutant tumor biology, proliferative and immune surveillance responses in tumors.
Results and Discussion
Study population and prevalence of mutations
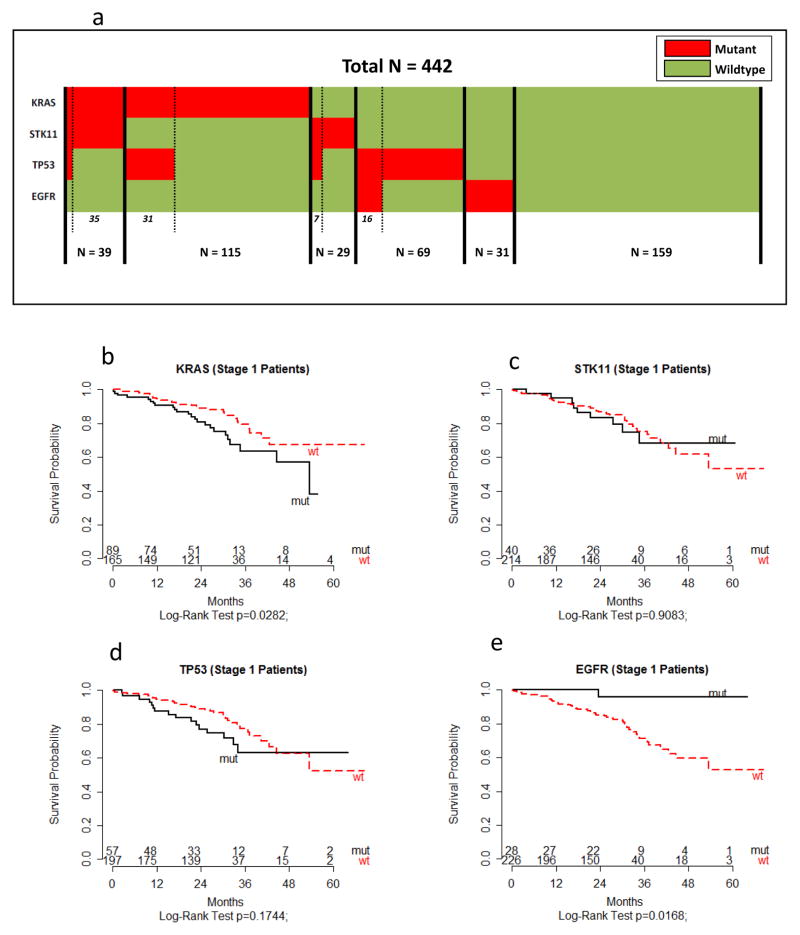
Study population characteristics and mutational status of the 442 adenocarcinoma lung cancer patients are summarized in Supplementary Table 1. Overall, the majority of patients were over 70 years of age (53.2%), female (54.3%), white (95.6%), self-reported ever smokers (91%), and early stage (stage I: 64%). In comparison of individual gene mutations to their wildtype counterpart, the overall prevalence was 34.8% for KRAS, 10.6% for EGFR, 15.3% for STK11, and 25.1% for TP53 (Supplementary Table 1). Of the 442 tumors, 159 did not harbor a mutation in any of these four genes (Fig. 1a). Key variables including demographic and clinical information associated with mutations in these 4 genes are provided in Supplementary Table 1. As expected (4), KRAS and EGFR mutations were mutually exclusive while STK11 mutations were significantly associated with KRAS mutations (p<0.0001) (Fig. 1a and Supplementary Table 1). In addition, co-occurrence of STK11 and TP53 mutations in KRAS mutant tumors was rare (n=4; Fig. 1a).
Fig. 1. Co-occurring and mutually exclusive mutations in KRAS, STK11, TP53 and EGFR in 442 human lung adenocarcinoma samples.
(a) Tumors with specific mutations are indicted in red while tumors without mutations are in green. To demonstrate co-occurring and exclusive mutations, the samples were sorted by KRAS mutations, then SKT11 mutations, then TP53 mutations, and then EGFR mutations. (b–e) Association of oncogene and tumor suppressor gene mutations with OS among stage I lung adenocarcinoma patients (n=265). Kaplan–Meier survival curves by mutation status of (b) KRAS, (c) STK11, (d) TP53, and (e) EGFR are shown. A two-sided log-rank test was used to assess statistically significant differences by mutational status. The number of patients at risk is listed below the survival curves. Additional methodology is provided in Supplemental Information.
Overall the results of sequencing analysis yielded findings similar to those in the literature and in public datasets (COSMIC) (43). Briefly, KRAS alterations occurred at codon positions 12, 13 and 61 which are each well-characterized gain-of-function positions (43). The most common mutations in EGFR have also been well-characterized (3). Although the focus of the present study was not on EGFR mutations, we found 23 L858R point mutations and 21 in frame indels in codon 19 in this cohort. Consistent with a loss-of-function mutation pattern, deletions or inactivating mutations were commonly found in TP53 and STK11. Criteria used to identify and remove germline variants in TP53 and STK11 are described in Supplementary Information which included filtering against the 1000 Genomes Project and the Exome Sequencing Project (ESP) dataset. In addition, we ensured that mutational events detected in these genes were previously reported in The Cancer Genome Atlas (TCGA) dataset, and if not, that they resulted in frame-shifted/truncated proteins (Supplementary Information). The role of KRAS mutations as a prognostic factor in NSCLC is presently unclear. However, a recent meta-analysis of 41 studies concluded that KRAS mutations are associated with poor prognosis in patients with NSCLC, especially in patients with adenocarcinoma and early stage NSCLCs (44). In the present cohort, we also found that KRAS mutations were associated with poor survival compared to wildtype among stage I patients (Fig. 1b). Conversely EGFR mutations were associated with significantly better OS compared to wildtype, while STK11 and TP53 were not significantly associated with OS (Fig. 1c–e).
Impact of STK11 and TP53 mutations on a novel KRAS mutation-associated gene expression signature
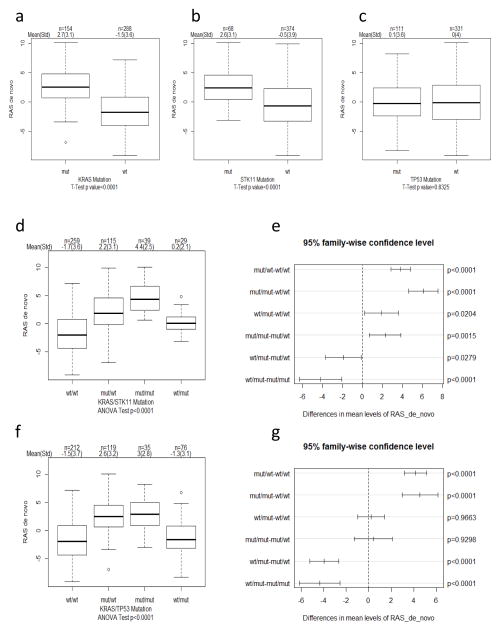
With the goal of determining impact of STK11 and TP53 mutations on KRAS-associated gene expression responses, we generated a de novo signature of differentially expressed genes in KRAS mutant versus KRAS wildtype tumors. We identified 58 probe sets encoding for 43 distinct genes that were differentially expressed in KRAS mutant tumors (p < 0.05 with a 1.5-fold change) (Supplementary Table 2 and Supplemental Information). Principal component analysis was used to evaluate activity of this signature as previously described (42). As expected, our KRAS de novo signature activity was highly elevated in KRAS mutant tumors (Fig. 2a; p<0.0001). Interestingly, activity of this signature was also substantially elevated in STK11 (Fig. 2b; p<0.0001) but not in TP53 mutant tumors (Fig. 2c; p=0.832). Importantly, signature activity was not only enhanced in KRASmut/STK11mut versus KRASmut/STK11wt tumors (Fig. 2d,e; p=0.0015), but also in KRASwt/STK11mut tumors compared to KRASwt/STK11wt tumors (Fig. 2d,e; p=0.02). Thus, STK11 mutations not only further elevate expression of these genes in KRAS mutant tumors but can also independently increase their expression.
Fig. 2. Impact of STK11 and TP53 mutations on KRAS mutation-associated gene expression.
(ac) Boxplots indicating KRAS mutation-associated (RAS de novo) signature activity (PC1) within each gene group (a) KRAS, (b) STK11, and (c) TP53. T-test was used to determine significance in difference in signature activity between mut and wt groups indicated. Sample size (n), mean and standard deviation (std) is indicated on top of each figure. (d) Boxplots and (e) pairwise comparison plots indicating RAS de novo signature activity in indicated co-occurring and exclusive mutations in KRAS and STK11. ANOVA was used to determine overall significant difference in RAS de novo signature activity among indicated groups and Tukey honest significant difference method was used to adjust for p value for pairwise comparison. (f) Boxplots and (g) pairwise comparison plots indicating RAS de novo signature activity in indicated co-occurring and exclusive mutations in KRAS and TP53. Additional methodology is provided in Supplemental Information.
To provide independent validation of KRAS signature association with above gene mutations, we performed studies on TCGA dataset. The results shown here are based upon data generated by the TCGA Research Network at: http://cancergenome.nih.gov/. Normalized RNAseq data was utilized using this dataset of 483 lung adenocarcinoma. Importantly, not only KRASmut (n=145) but also STK11mut (n=75) were very significantly associated with high KRAS signature activity (Supplemental Fig. 1; p<0.0001). In contrast, activity of this signature was not associated with TP53 mutations (Supplementary Fig. 1). Remarkably, and in complete concordance with our 442 dataset, KRAS signature activity was not only significantly enhanced in KRASmut but also in KRASwt/STK11mut tumors compared to KRASwt/STK11wt tumors (Supplementary Fig. 1; p=0.0015). Therefore, mutations in KRAS and STK11 are independently associated with upregulation of KRAS signature genes. To define the underlying biology of the KRAS signature, three analyses were performed using the following: Gene Ontology Biological process enrichment, GeneGO Pathway Map enrichment, and MSigDB pathway/signature enrichment. However, we were not able to reproducibly associate genes in this signature with a specific biological pathway. Nonetheless, several of these genes have been previously shown to be involved in RAS pathway function, including DUSP4, RASGRF1/CDC25 and HRASLS5. Interestingly, DUSP4 expression was also reported to be associated with STK11 mutations (45), suggesting that mutations in KRAS and STK11 may result in expression of at least some common genes.
Distinct association of TP53 mutations with tumor proliferative responses
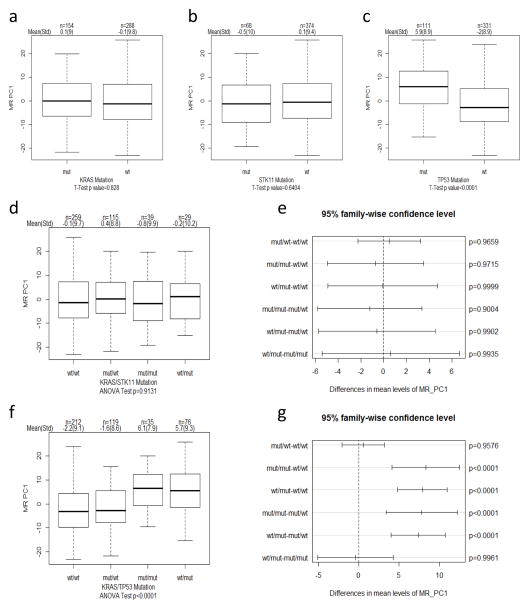
Increased tumor cell proliferation is a main driver of malignancy and known to be strongly associated with poor patient survival in multiple tumor types (25, 26). We next determined whether proliferative responses were impacted by KRAS and tumor suppressor gene mutations. To this end, we used a previously defined malignancy risk (MR; gene list in Supplementary Table 3) signature that is significantly correlated with tumor cell proliferation (25, 26). Importantly, no significant difference in MR activity was observed in KRAS mutant or STK11 mutant tumors compared to wildtype tumors (Fig. 3a,b,d,e). In contrast, TP53 mutations, either individually or with KRAS mutations, were significantly associated with higher MR activity (Fig. 3c,f,g). These findings therefore indicate that TP53 and STK11 tumor suppressor mutations have distinct association with tumor cell proliferation.
Fig. 3. Distinct association of TP53 mutations with tumor proliferative responses.
(a–b) Boxplots indicating MR signature activity (PC1) within each gene group (a) KRAS, (b) STK11, and (c) TP53. T-test was used to determine significance in difference in MR activity between mut and wt groups indicated. Sample size (n), mean and standard deviation (std) is indicated on top of each figure. (d) Boxplots and (e) pairwise comparison plots indicating MR signature activity in indicated co-occurring and exclusive mutations in KRAS and STK11. ANOVA was used to determine overall significant difference in MR activity among indicated groups and Tukey honest significant difference method was used to adjust for p value for pairwise comparison. (f) Boxplots and (g) pairwise comparison plots indicating MR signature activity in indicated co-occurring and exclusive mutations in KRAS and TP53.
Suppression of immune surveillance in STK11 mutant tumors
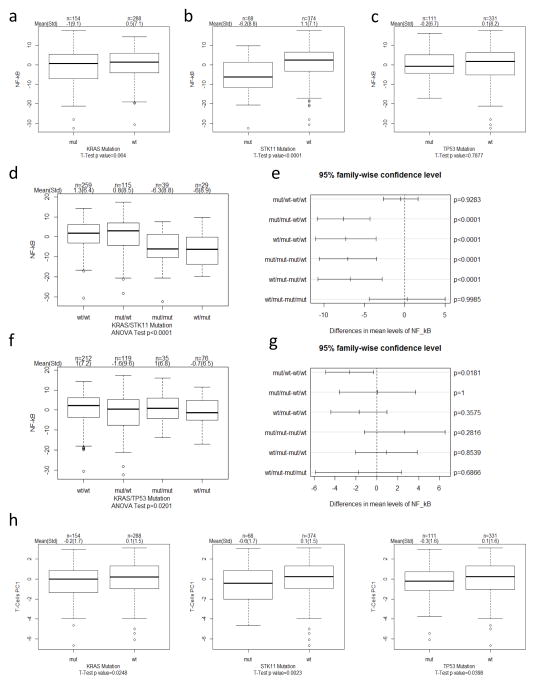
T cell mediated immune surveillance is crucial in controlling tumor growth (27–29). We recently identified a gene expression signature of NF-κB regulated genes (gene list in Supplementary Table 3) that is associated with an immune-active tumor microenvironment and T cell presence (42). Using this signature, we next determined potential association of different mutations with an immune-active tumor microenvironment. Intriguingly, only STK11 mutations were associated with significantly lower activity of this signature (Fig. 4a–c; p<0.0001). Furthermore, STK11 mutations either individually or with KRAS mutations were strongly associated with lower NF-κB signature activity (Fig. 4d–e), while no such association was seen with TP53 mutations (Fig. 4f–g). To determine more directly the impact of STK11 mutations on T cell immune surveillance, we examined T cell infiltration by using T cell receptor α and β chain expression as previously described (42). Importantly, STK11 mutations were the most significantly associated with reduced T cell presence in tumors (Fig. 4h; p=0.002), although KRAS and TP53 also showed reduced T cell presence. Overall, these findings indicate that TP53 and STK11 tumor suppresser genes may promote tumor progression by different mechanisms: while TP53 mutations lead to greater proliferative responses, STK11 mutations appear to be associated with suppression of the tumor immune surveillance response. To the best of our knowledge, these findings provide amongst the first evidence of a potential association between a common cancer gene mutation and the immune surveillance response.
Fig. 4. Suppression of immune surveillance in STK11 mutant tumors.
(a–c) Boxplots indicating NF-κB signature activity (PC1) within each gene group (a) KRAS, (b) STK11, and (c) TP53. T-test was used to determine significance in difference in NF-κB activity between mut and wt groups indicated. Sample size (n), mean and standard deviation (std) is indicated on top of each figure. (d) Boxplots and (e) pairwise comparison plots indicating NF-κB signature activity in indicated co-occurring and exclusive mutations in KRAS and STK11. ANOVA was used to determine overall significant difference in NF-κB activity among indicated groups and Tukey honest significant difference method was used to adjust for p value for pairwise comparison. (f) Boxplots and (g) pairwise comparison plots indicating NF-κB signature activity in indicated co-occurring and exclusive mutations in KRAS and TP53. (h) Boxplots indicating TCR gene expression PC1 activity. T-test was used to determine significance in difference in TCR expression between indicated mut and wt groups.
The primary goal of this study was to define molecular heterogeneity in KRAS-mutant tumors resulting from co-occurring STK11 and TP53 tumor suppressor mutations. Towards that goal, a key finding reported here is that STK11 mutations can positively impact the activity of a novel KRAS mutation-associated gene expression signature. Thus, mutations in STK11 may enhance KRAS associated signaling responses, both independently and concurrently with KRAS mutations. While the association of the KRAS signature with underlying tumor cell biology remains to be defined, our results suggest that STK11 mutations may potentiate KRAS-induced signaling and gene expression responses that drive tumorigenesis. Indeed, mouse studies demonstrate acceleration of KRAS induced tumorigenesis and increased metastasis in the presence of concurrent STK11 null mutations (19).
A key finding reported here is that tumors with TP53 and STK11 mutations are associated with distinct proliferative versus immune surveillance responses. Specifically, our results indicate that TP53 mutations are strongly associated with enhanced tumor cell proliferation, a finding consistent with prior studies of this key tumor suppressor. In contrast, STK11 mutations were not associated with differences in proliferation but strongly associated with suppression of the immune surveillance response. The relative lack of co-occurrence of STK11 and TP53 mutations is also noteworthy (Fig. 1a), and indicates that distinct tumor-promoting mechanisms resulting from these mutations dominate in different tumors. Immune suppression appears to be a specific feature of STK11 mutations, which may enhance tumor progression in addition to activation of SRC-like kinases as reported previously (46). Since the response to immunotherapy is typically associated with an immune-active tumor microenvironment (36–38), our results further suggest that STK11 mutant tumors may be less responsive to immunotherapy.
In conclusion, these findings not only provide novel insights into how KRAS-mutant tumor biology is shaped by co-occurring mutations, but may also provide insights for therapeutic targeting of lung cancers with distinct tumor suppressor mutations. Specifically, these studies illustrate the potentially significant effect that mutations in tumor suppressor genes could have on therapeutic strategies, especially immunotherapy. These findings also necessitate additional studies to understand specifically how STK11/LKB1 impacts KRAS mutation-associated gene expression as well as the tumor immune surveillance response. Interestingly, recent studies showed reduced PI3K pathway activity, including activity of NF-κB activating kinase PDK1, in STK11 mutant human lung adenocarcinoma (45). Therefore, an interesting possibility is that STK11 mutations directly impact activity of NF-κB and potentially other pathways involved in immune surveillance. Future studies should therefore be directed not only towards understanding mechanisms through which STK11 mutations promote tumorigenesis through enhancement of KRAS induced responses but also by mediating suppression of immune surveillance. Finally, we believe that the extensive dataset described here will prove to be a valuable resource for cancer researchers, especially for interrogating gene expression networks prevalent in tumors.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health SPORE Grant (P50 CA119997). We would also like to thank Mr. Andrew ‘Ross’ Myers, Anastasia Belock, Mercedes Rodriguez, Edward T. Chwieseni, Marek Wloch, Hiba Gohar, and Moffitt’s Tissue Core facility, Moffitt’s Cancer Registry (Director: Karen A. Coyne), Research Information Technology (IT) group, and the Data Management and Integration Technology (DMIT) group. Total Cancer Care® is enabled, in part, by the generous support of the DeBartolo Family, and we thank the many patients who provided data and tissue to the Total Cancer Care Consortium. Our study also received valuable assistance from the following Core Facilities at the Moffitt Cancer Center: Biostatistics and Cancer Informatics, Tissue, and Molecular Genomics. This work has been supported in part by a Cancer Center Support Grant (CCSG grant P30-CA76292) at the H. Lee Moffitt Cancer Center and Research Institute, a National Cancer Institute-designated Comprehensive Cancer Center.
Footnotes
The authors declare no financial disclosures.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. Epub 2013/01/22. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–80. doi: 10.1016/S1470-2045(10)70087-5. Epub 2011/02/01. [DOI] [PubMed] [Google Scholar]
- 3.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–75. doi: 10.1038/nature07423. Epub 2008/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–8. doi: 10.1038/nature06358. Epub 2007/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1(1):2–27. doi: 10.4161/sgtp.1.1.12178. Epub 2011/06/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitin N, Rossman KL, Der CJ. Signaling interplay in Ras superfamily function. Curr Biol. 2005;15(14):R563–74. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 8.Bang D, Wilson W, Ryan M, Yeh JJ, Baldwin AS. GSK-3alpha promotes oncogenic KRAS function in pancreatic cancer via TAK1-TAB stabilization and regulation of noncanonical NF-kappaB. Cancer discovery. 2013;3(6):690–703. doi: 10.1158/2159-8290.CD-12-0541. Epub 2013/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker NM, Yee Chow H, Chernoff J, Der CJ. Molecular Pathways: Targeting RAC-p21-Activated Serine-Threonine Kinase Signaling in RAS-Driven Cancers. Clin Cancer Res. 2014;20(18):4740–6. doi: 10.1158/1078-0432.CCR-13-1727. Epub 2014/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–66. doi: 10.1016/j.cell.2013.09.041. Epub 2013/11/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119(6):1727–40. doi: 10.1172/JCI37127. Epub 2009/05/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Molecular cancer research : MCR. 2014;12(1):3–13. doi: 10.1158/1541-7786.MCR-13-0539. Epub 2014/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–75. doi: 10.1038/nrc2676. Epub 2009/07/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62(13):3659–62. Epub 2002/07/05. [PubMed] [Google Scholar]
- 15.Shah U, Sharpless NE, Hayes DN. LKB1 and lung cancer: more than the usual suspects. Cancer Res. 2008;68(10):3562–5. doi: 10.1158/0008-5472.CAN-07-6620. Epub 2008/05/17. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Cespedes M. The role of LKB1 in lung cancer. Fam Cancer. 2011;10(3):447–53. doi: 10.1007/s10689-011-9443-0. Epub 2011/04/26. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Ge G, Ji H. LKB1 in lung cancerigenesis: a serine/threonine kinase as tumor suppressor. Protein Cell. 2011;2(2):99–107. doi: 10.1007/s13238-011-1021-6. Epub 2011/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26(40):5911–8. doi: 10.1038/sj.onc.1210418. Epub 2007/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. doi: 10.1038/nature06030. Epub 2007/08/07. [DOI] [PubMed] [Google Scholar]
- 20.O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67(19):8975–9. doi: 10.1158/0008-5472.CAN-07-1328. Epub 2007/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Xiao Q, Ma H, Li L, Liu J, Feng Y, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc Natl Acad Sci U S A. 2010;107(44):18892–7. doi: 10.1073/pnas.1004952107. Epub 2010/10/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 23.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen DT, Hsu YL, Fulp WJ, Coppola D, Haura EB, Yeatman TJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. Journal of the National Cancer Institute. 2011;103(24):1859–70. doi: 10.1093/jnci/djr420. Epub 2011/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen DT, Nasir A, Culhane A, Venkataramu C, Fulp W, Rubio R, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast cancer research and treatment. 2010;119(2):335–46. doi: 10.1007/s10549-009-0344-y. Epub 2009/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nature reviews Immunology. 2006;6(11):836–48. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–70. doi: 10.1126/science.1203486. Epub 2011/03/26. [DOI] [PubMed] [Google Scholar]
- 29.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–11. doi: 10.1038/35074122. Epub 2001/04/27. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. The New England journal of medicine. 2003;348(3):203–13. doi: 10.1056/NEJMoa020177. Epub 2003/01/17. [DOI] [PubMed] [Google Scholar]
- 31.Fridman WH, Galon J, Pages F, Tartour E, Sautes-Fridman C, Kroemer G. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res. 2011;71(17):5601–5. doi: 10.1158/0008-5472.CAN-11-1316. Epub 2011/08/19. [DOI] [PubMed] [Google Scholar]
- 32.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. The New England journal of medicine. 2005;353(25):2654–66. doi: 10.1056/NEJMoa051424. Epub 2005/12/24. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. Epub 2009/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nature reviews Immunology. 2007;7(1):41–51. doi: 10.1038/nri1995. Epub 2006/12/23. [DOI] [PubMed] [Google Scholar]
- 35.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annual review of immunology. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. Epub 2011/01/12. [DOI] [PubMed] [Google Scholar]
- 36.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer immunology, immunotherapy : CII. 2012;61(7):1019–31. doi: 10.1007/s00262-011-1172-6. Epub 2011/12/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(19):2388–95. doi: 10.1200/JCO.2012.44.3762. Epub 2013/05/30. [DOI] [PubMed] [Google Scholar]
- 38.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–22. doi: 10.1038/ni.2703. Epub 2013/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34(2):67–73. doi: 10.1016/j.it.2012.10.004. Epub 2012/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gajewski TF, Schumacher T. Cancer immunotherapy. Curr Opin Immunol. 2013;25(2):259–60. doi: 10.1016/j.coi.2013.03.008. Epub 2013/04/17. [DOI] [PubMed] [Google Scholar]
- 41.Gajewski TF, Woo SR, Zha Y, Spaapen R, Zheng Y, Corrales L, et al. Cancer immunotherapy strategies based on overcoming barriers within the tumor microenvironment. Curr Opin Immunol. 2013;25(2):268–76. doi: 10.1016/j.coi.2013.02.009. Epub 2013/04/13. [DOI] [PubMed] [Google Scholar]
- 42.Hopewell EL, Zhao W, Fulp WJ, Bronk CC, Lopez AS, Massengill M, et al. Lung tumor NF-kappaB signaling promotes T cell-mediated immune surveillance. J Clin Invest. 2013;123(6):2509–22. doi: 10.1172/JCI67250. Epub 2013/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. Epub 2010/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng D, Yuan M, Li X, Chen L, Yang J, Zhao X, et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer. 2013;81(1):1–10. doi: 10.1016/j.lungcan.2013.03.019. Epub 2013/04/24. [DOI] [PubMed] [Google Scholar]
- 45.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, et al. LKB1 Loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2014;9(6):794–804. doi: 10.1097/JTO.0000000000000173. Epub 2014/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer cell. 2010;17(6):547–59. doi: 10.1016/j.ccr.2010.04.026. Epub 2010/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.