Abstract
Oral tolerance prevents pathological inflammatory responses towards innocuous foreign antigens via peripheral regulatory T cells (pTreg cells). However, whether a particular subset of antigen-presenting cells (APCs) is required during dietary antigen exposure to instruct naïve CD4+ T cells to differentiate into pTreg cells has not been defined. Using myeloid lineage-specific APC depletion in mice, we found that monocyte-derived APCs are dispensable, while classical dendritic cells (cDCs) are critical for pTreg cell induction and oral tolerance. CD11b− cDCs from the gut-draining lymph nodes efficiently induced pTreg cells, and conversely, loss of IRF8-dependent CD11b− cDCs impaired their polarization, although oral tolerance remained intact. These data reveal the hierarchy of cDC subsets in pTreg cell induction and their redundancy during oral tolerance development.
The peripheral immune system must maintain a balance between protective responses and tolerance. This equilibrium represents a major challenge for the mucosal surfaces, particularly the intestine, which is chronically exposed to both potentially pathogenic microbes and harmless dietary and commensal-derived antigens. Not surprisingly, several cellular and molecular mechanisms exist to ensure robust tolerance induction in the mucosae.
Peripherally-induced Foxp3+ regulatory T cells (pTreg cells) are thought to be instrumental in the induction and maintenance of peripheral tolerance1, 2, 3, 4. Innocuous antigen exposure via mucosal surfaces efficiently induces pTreg cell differentiation from naïve CD4+ T cells via a retinoic acid (RA)- and TGF-β-dependent process2, 5, 6, 7, 8. In turn, genetic loss-of-function strategies that target pTreg cells result in severe inflammatory phenotypes in the lungs and intestine 3, 4.
Antigen presenting cells (APCs), including dendritic cells (DCs) and macrophages, have been ascribed critical roles in triggering pTreg cell differentiation6, 7, 8, 9, 10. In particular, intestinal APCs expressing the fraktalkine receptor CX3CR1 take up soluble luminal antigens 11, 12 and, under certain conditions, migrate to the mesenteric lymph nodes (mLNs) where they present antigens to naïve T cells13. In addition, CX3CR1–expressing phagocytes appear to transfer antigens to neighboring migratory DCs11 and these DCs are believed to induce pTreg cell conversion after they migrate to the mLNs14, 15. Indeed, both lamina propria and mLN-derived DCs, particularly αE integrin+ (CD103+) or DEC205+ DCs, produce high amounts of RA and TGF-β and efficiently induce pTreg cells 1, 6, 7, 8, 16, 17, 18, 19. However, whether these pTreg cell-inducing APCs are also required for oral tolerance induction has not been investigated. Furthermore, because the strategies relying on cell surface markers utilized to date target multiple APC lineages, the exact nature and origin of APCs responsible for pTreg cell induction in vivo are still unclear.
We demonstrate an essential role for pre-DC–derived classical dendritic cells (cDCs) for both pTreg cell and oral tolerance induction, while macrophages and monocyte-derived cells appear dispensable. Further, we identify a hierarchical pattern in pTreg cell-inducing capacity of mLN-derived cDC subsets, whereby dietary antigen mediated pTreg cell polarization is most dependent on migratory IRF8–dependent CD11b− cDCs. Oral tolerance is intact, however, in absence of this cDC subset, highlighting robustness of the process and functional redundancy of cDCs.
Results
Systemic absence of cDCs leads to break in oral tolerance
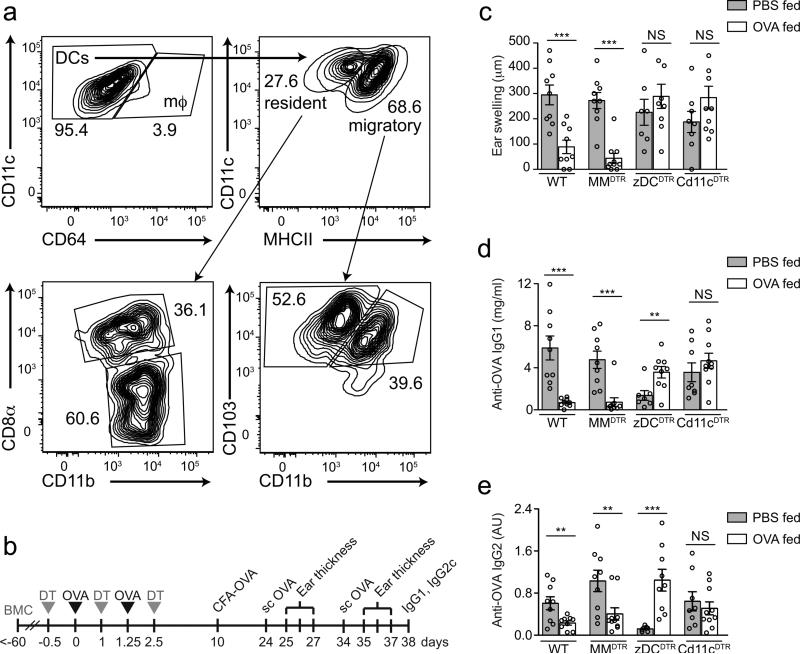
We first set out to determine whether the APCs required for induction of oral tolerance could be classified by one of the two major myeloid lineages (Supplementary Fig. 1a). We focused on the populations present in the mLNs, the major inductive sites of oral tolerance14. Macrophages were identified as Lin−MHCII+CD11c+CD64+ cells, and cDCs as Lin−MHCII+CD11c+CD64− cells (Fig. 1a)20. Within the cDCs, we distinguished between two resident MHCIIint populations, CD8α+CD11blow versus CD8α−CD11b+ and two migratory MHCIIhi populations, CD103+CD11b− versus CD103+CD11b+ (Fig. 1a). We first used a mouse model of TH1 delayed-type hypersensitivity (DTH) 9 to address whether a specific APC lineage is required for the induction phase of oral tolerance. Tolerance was assessed by measuring the cellular and humoral inflammatory immune response towards OVA in mice pre-exposed to oral ovaIbumin (OVA) or oral PBS as control and immunized with OVA in complete Freund's adjuvant (CFA) (Fig. 1b). We targeted the macrophage-monocyte lineage using mice bearing the Cre recombinase gene under the Lyz2 promoter, and the diphtheria toxin receptor (DTR) gene preceded by a loxP site-flanked stop cassette under control of the Csf1r promoter (Lyz2Cre × Csf1rLsL-DTR, herein called MMDTR mice)20 and the pre-DC-derived lineage using mice with DTR knocked in 3’ of the pre-DC lineage transcription factor Zbtb46 gene (Zbtb46DTR, herein called zDCDTR mice)21. Because DT is lethal to zDCDTR mice, due to Zbtb46 expression in vascular endothelium21, we used chimeric mice in which total bone marrow (BM) cells from zDCDTR, MMDTR or wild-type littermate control mice were transferred into lethally irradiated C57BL/6 mice. Administration of DT to MMDTR mice led to broad depletion of monocytes, monocyte– and yolk sac–derived tissue macrophages and inflammatory DCs, which lasted for at least 36h20 (Supplementary Fig. 1b, c), while DT administration to zDCDTR mice resulted in a compensatory surge of Flt3L (FMS-like tyrosine kinase 3 ligand) in the serum, and complete loss of cDCs, including migratory cDC subsets in the mLN20 (Supplementary Fig. 1c, d). To induce oral tolerance, OVA was gavaged 12 and 30 hours after the first of three DT injections given 36 hours apart, and DT and OVA administration were timed such that OVA clearance (completed after 24 hours) preceded the reappearance of APCs20 (Supplementary Fig. 1e-h). To rule out that DT administration to the bone marrow chimeric mice resulted in altered oral antigen absorption, we measured OVA uptake into different tissues at 4 hours post OVA gavage (Supplementary Fig. 1i-m). Because APCs are required for immune responses, mice were immunized with OVA+CFA 8 days after the last DT dose, when the APC niches were replenished (Fig. 1b and Supplementary Fig. 1n, o), followed by subcutaneously injection with OVA in the ear 14 and 24 days after immunization.
Figure 1. DTH responses in mice depleted of APC subsets during antigen feeding.
(a) Flow cytometry contour plots of mLN macrophages and cDC subpopulations. (b) Experimental design of oral tolerance establishment towards CFA-mediated immunity. (c) Degree of ear swelling 48 h post second subcutaneous OVA challenge in wild-type, MMDTR, zDCDTR and CD11cDTR BMCs treated with DT during oral OVA (+) or PBS (−) exposure. (d) Serum anti-OVA IgG1 and (e) anti-OVA IgG2c levels in mice described in (c) 28 days post CFA immunization. Data represent pooled values (average±SEM) from two independent experiments, n=10 per group, ns= not significant, NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test).
At 72 hours post ear injection, ear swelling, serum anti-OVA IgG1 and anti-OVA IgG2c amounts were suppressed in OVA-fed wild-type and MMDTR, but not in zDCDTR mice, when compared to their PBS-fed counterparts (Fig. 1c-e). We noticed that PBS-fed, OVA-CFA immunized zDCDTR mice produced reduced serum IgG amounts compared to wild-type or MMDTR mice, likely a consequence of the overall reduced CD4+ T cell frequency upon recent cDC depletion (Supplementary Fig. 1o). As a positive control for the APC-dependence of oral tolerance induction, we also studied chimeric mice lacking all CD11c-expressing APCs20, 22 by using BM from mice expressing DTR under control of the Itgax promoter, the gene encoding integrin CD11c (here CD11cDTR mice)20, 22. PBS-fed and OVA-fed CD11cDTR mice showed similar ear swelling and serum anti-OVA antibody responses (Fig. 1c-e), suggesting lack of tolerance to OVA. These observations indicated that monocyte-macrophage–derived APCs are dispensable for oral tolerance induction, while pre-DC–derived cells are critical.
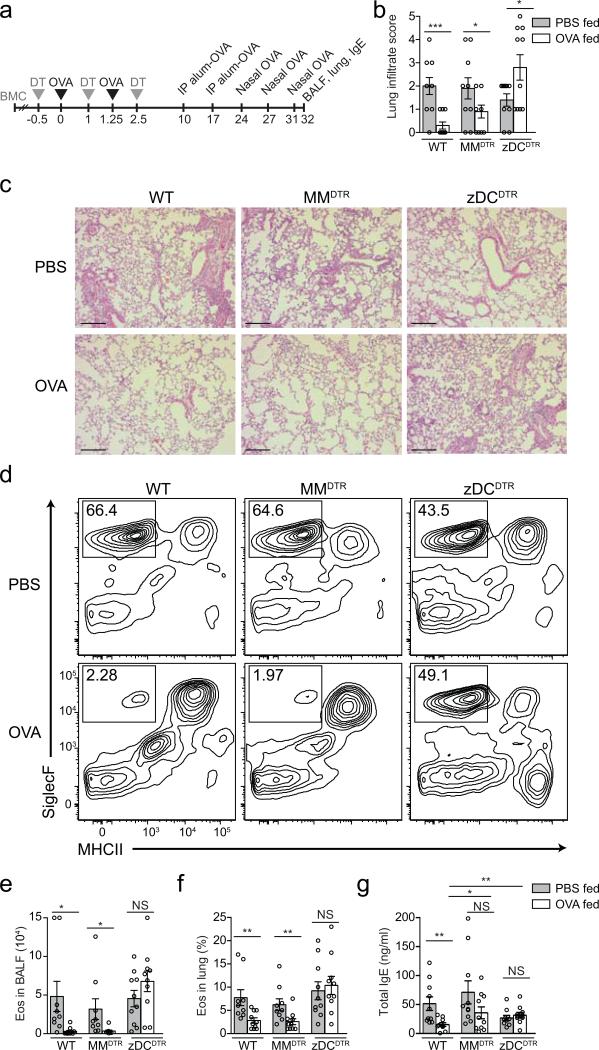
Next, we assessed the requirement for cDCs in preventing TH2 allergic responses2 (Fig. 2a). Following the same DT and oral OVA administration regimen as above, mice were immunized with OVA+Alum 8 and 15 days after the last DT injection and intranasally challenged with OVA three times in 3-day intervals. Eosinophilia in the bronchoalveolar lavage fluid (BALF) and lung, as well as total lung tissue infiltrate and serum IgE were similarly suppressed in OVA-fed wild-type and MMDTR, but not in zDCDTR mice, when compared to PBS-fed mice of their genotypes (Fig. 2b-g). These data show that the absence of cDCs during antigen feeding results into failure to establish tolerance to both TH1 and TH2 immunity.
Figure 2. Allergic airway responses in mice depleted of APC subsets during antigen feeding.
(a) Experimental design of oral tolerance establishment towards Alum-mediated immunity. (b) Histological score for inflammatory infiltrate and (c) representative hematoxylineosin staining of lung tissue from naïve (upper row) and OVA-gavaged (lower row) wild-type, MMDTR and zDCDTR mice. Scale bar, 250 μm. (d) Representative flow cytometry contour plot for SiglecF+MHCII− gating (eosinophils, eos) in the BALF, and (e) analysis of total BAL eosinophil counts and (f) relative lung tissue eosinophil frequencies among CD45+ cells in wild-type, MMDTR and zDCDTR BMCs treated with DT during oral OVA (+) or PBS (−) exposure. (g) Serum total IgE in mice described in (b-e) 21 days post Alum immunization. Data represent pooled values (average±SEM) from two independent experiments, n=10 per group, NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test).
cDCs are required for pTreg cell induction
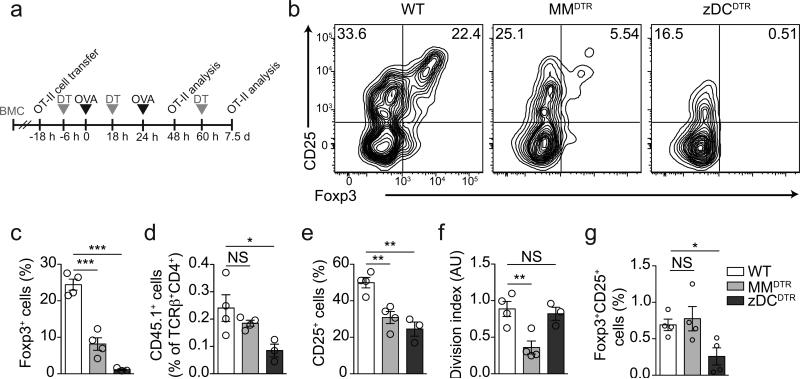
Oral tolerance is mediated by pTreg cells2, 3. We thus addressed whether the pre-DC lineage was required for pTreg cell induction upon antigen feeding by adoptively transferring sorted naïve OVA-specific CD45.1 OT-II cells into APC-depleted BM chimeric mice and assessing the appearance of CD45.1 Foxp3+ T cells in various tissues upon OVA gavage (Fig. 3a). DT administration alone did not induce OT-II cell activation, proliferation or differentiation in these chimeric mice (data not shown). 48 hours after initial administration of oral OVA MMDTR mice displayed compromised but perceptible pTreg cell induction in mLNs compared to wild-type controls (Fig. 3b, c), an effect that was accompanied by an unaltered degree of OT-II cell engraftment, but reduced overall OT-II cell activation and division (Fig. 3b-f, Supplementary Fig. 2a). In contrast, pTreg cells were virtually undetectable in OVA-fed zDCDTR mice (Fig. 3b, c) and T cell activation was decreased compared to OVA-fed wild-type mice (Fig. 3e). OT-II cell engraftment was reduced in zDCDTR mice (Fig. 3d), an observation we also made after DT administration alone (Supplementary Fig. 2b). At this time-point we did not find significant OT-II cell activation or pTreg cell induction in spleen or distal lymph nodes in any of the groups studied (data not shown). The effect of monocyte-macrophage depletion on pTreg cell induction was compensated over time, as by 7.5 days after OVA feeding the frequencies of pTreg cells in MMDTR mice were similar to those in wild-type mice (Fig. 3g), in line with the intact tolerance found in these mice. In zDCDTR mice, OT-II cell frequencies 7.5 days after OVA gavage were proportional to the CD4+ T cell compartment (Supplementary Fig. 2c), however, pTreg cell frequencies were still severely reduced in mLNs (Fig. 3g and Supplementary Fig. 2d, e), spleens and distal lymph nodes (data not shown). These data indicate that CD4+ T cell activation, proliferation and pTreg cell induction depend on cDCs and exclude an essential contribution of monocyte-derived APCs to pTreg cell generation upon oral antigen exposure.
Figure 3. Contribution of macrophage-monocyte– and pre-DC–derived cells to pTreg cell induction in vivo.
(a) Schematic representation of in vivo pTreg cell induction protocols. (b-f) Flow cytometry analysis of mLNs; (b) representative contour plots for Foxp3 and CD25 expression of TCRβ+CD4+CD45.1+ T cells; (c) Foxp3+, (d) CD45.1+, (e) CD25+ cell frequencies and (f) cell division index of adoptively transferred naïve CD45.1 OT-II cells in the mesenteric lymph nodes (mLN) of wild-type, MMDTR and zDCDTR BMCs, 48 h after first oral OVA gavage. n=4. (g) Flow cytometry analysis of CD45.1 OT-II Foxp3+ cell frequency in mLN of DT treated wild-type, MMDTR and zDCDTR BMCs, 7.5 days after first OVA gavage and 8 days post adoptive transfer of naïve CD45.1 OT-II cells. n=3. NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test). Data (average±SEM) are representative of three independent experiments.
CX3CR1+ APC contribution to pTreg cell polarization in mLN
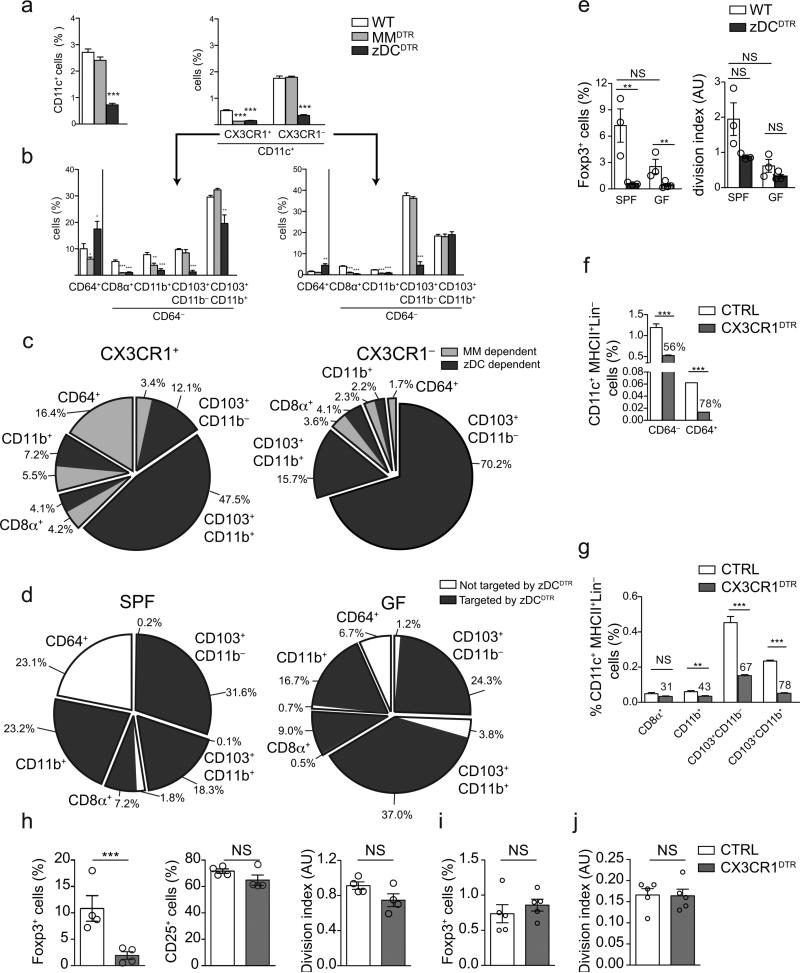
Here we distinguished APC subsets by ontogeny, but others have ascribed roles to gut-associated APCs based upon CX3CR1-targeting approaches9, 11, 13, 23. To define the lineage of origin of CX3CR1-expressing APC populations in steady-state mLNs, we generated BM chimeras from Cx3cr1+/GFP mice crossed to zDCDTR, MMDTR or wild-type control mice and characterized GFP+ cell loss upon DT injection. GFP+ cells represented roughly 25% of all CD11c+ APCs in the mLN (Fig. 4a, b). DT administration to MMDTR mice indicated that most monocyte-macrophage–derived cells were among the GFP+ cells (Fig. 4b, c). In addition to CD64+ macrophages, about half of CD8α+ and CD11b+ cells, and roughly 20% of CD103+CD11b− populations amongst the GFP+ cells were depleted by DT in MMDTR mice, indicating that part of the GFP+CD64− cells are of monocyte-macrophage origin (Fig. 4b, c). Notwithstanding, DT targeting in zDCDTR mice revealed that cDC–derived cells represented the majority of APCs among CX3CR1+ cells (Fig. 4b, c). These analyses highlight the heterogeneity of CX3CR1 expression within APC subsets.
Figure 4. Contribution of CX3CR1+ cells to APC pools and pTreg cell induction.
(a) Relative frequencies of CD11c+, CD11c+CX3CR1− and CD11c+CX3CR1+ among CD45+ cells, and APC subpopulations among (b) CX3CR1− and CX3CR1+ (CD11c+) cells in the mLNs of wild-type, MMDTR and zDCDTR BMCs, 36 h after DT administration. n=3. (c) Pie charts of relative MM (light grey) and cDC (dark grey) contributions to APC subpopulations within CX3CR1+ versus CX3CR1− cell populations. n=4, ±SD. (d) Pie chart of flow cytometry analysis of APC subpopulations frequencies in the mLN within CD45+Lin−MHCII+CD11c+ cells in SPF versus GF mice and targeting of the populations in zDCDTR mice (dark grey). n=4, ±SD. (e) Foxp3+ cell frequencies and cell division index of adoptively transferred naïve CD45.1 OT-II cells in the mLN of SPF versus GF, CX3CR1GFP/+ versus zDCDTR (CX3CR1GFP/+) BMCs, 48 h after first oral OVA gavage. n=4 (f, g) Flow cytometry analysis of APC cell frequencies in the mLN of CX3CR1lsl-DTR control versus CX3CR1DTR BMCs, 36 h post DT administration. n=4. Numbers above CX3CR1 bars depict percentage reduction upon DT administration. (h-j) Foxp3+, CD25+ cell frequencies and cell division index of adoptively transferred naïve CD45.1 OT-II cells in the mLN of CX3CR1LsL-DTR control versus CX3CR1DTR BMCs, 48 h (h) or 7.5 days (i, j) after first oral OVA gavage. n=4, NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test). Data (average±SEM) are representative of two independent experiments.
Microbiota-derived signals have been implicated in regulation of CX3CR1hi mononuclear phagocyte migration13. To better characterize these cells, we transplanted bone marrow from zDCDTR (Cx3cr1+/GFP) or wild-type Cx3cr1+/GFP control mice into germ-free (GF) and specific pathogen free (SPF) wild-type hosts. We found a selective increase of the CD103+CD11b+ population in the mLNs of GF mice compared to SPF mice (Fig. 4d, Supplementary Fig. 3a). However, the fraction of CX3CR1+ among CD103+CD11b+ cells remained the same (data not shown), indicating that this enrichment was not explained by specific CX3CR1+ cell mobilization. Depletion of cDCs in the zDCDTR (Cx3cr1+/GFP) chimeras indicated that CD11c+CD64− cells, including migratory subsets, remained pre-DC–derived in GF mice (Fig. 4d, Supplementary Fig. 3b). Finally, we asked whether the altered cDC composition in GF mice had an effect on pTreg cell induction. GF hosts displayed reduced pTreg cell induction and cell division in our OT-II transfer system, and DT administration completely prevented Foxp3 upregulation in both GF and SPF zDCDTR hosts (Fig. 4e). The reduction in CD4+ T cell numbers and OT-II engraftment in zDCDTR mice was comparable under both GF and SPF conditions (Supplementary Fig. 3c, d). These data indicate that the composition of APCs in GF mice is less favorable for pTreg cell induction and that the remaining APCs in cDC-depleted GF hosts cannot rescue the SPF phenotype.
In order to further reconcile our lineage-based approach with CX3CR1-targeting strategies and address the role of CX3CR1+ APCs in pTreg cell induction, we performed OT-II adoptive transfer experiments into Itgax-Cre × Cx3cr1LsL-DTR (CX3CR1DTR) or Cx3cr1LsL-DTR (wild-type control) BM recipients13. In CX3CR1DTR mice, Cre recombinase is under control of the Itgax promoter, while DTR is preceded by a loxP site-flanked stop cassette under the Cx3cr1 promoter, allowing DT-mediated depletion of (CD11c+) CX3CR1+ cells for at least 48 hours13. As previously reported13, DT administration to CX3CR1DTR mice led to depletion of mLN macrophages (CD11c+CD64+cells), although it also depleted CD11c+CD64− cells (Fig. 4f). The degree of depletion in each APC subset (Fig. 4g) correlated with the percentage of CX3CR1+ cells in these subsets (Fig. 4a-c). In line with this pleiotropic APC targeting, pTreg cell induction in CX3CR1DTR mice was more severely reduced than in MMDTR mice (Fig. 4h and Fig. 3c), while OT-II cell activation and division were only mildly affected and CD4+ T cell numbers and OT-II adoption were not altered when compared to wild-type controls (Fig. 4i and Supplementary Fig. 3e, f). However, similar to what was observed in MMDTR mice, pTreg cell frequencies and OT-II cell division were restored to wild-type levels by day 7.5 post OVA gavage (Fig. 4i, j). Taken together, these results suggest that CX3CR1-expressing macrophages are not essential, while cDCs are required for pTreg cell generation by dietary antigen and highlight the merits of a lineage-based approach to dissect differential APC contributions to pTreg cell induction and oral tolerance.
cDCs display differential molecular signatures
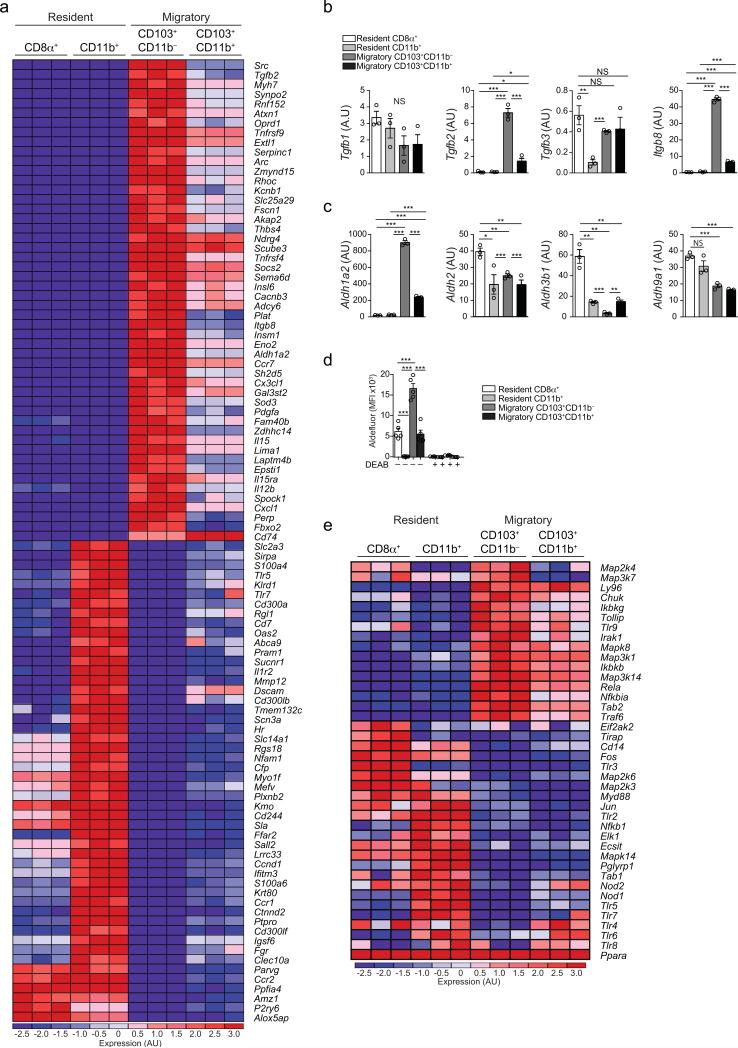
Past studies have described pTreg cell-inducing DC populations in the murine gut and gut-draining lymph nodes 6, 7, 8, 10, 16. Additionally, in recent years, large transcriptome databases for specific cDC populations have been made available24, 25, 26. However, these studies did not compare IRF4– versus IRF8–dependent (distinguishable by CD8α−CD11b+ or CD8α+CD11b− staining, respectively) among resident and migratory (CCR7− or MHCIIint versus CCR7+ or MHCIIhi) DC populations. To obtain a higher resolution of the cDC subsets and to link them with their lineage of origin, we performed RNA sequencing of cDCs present in the mLNs. We analyzed four CD11c+CD64− subsets: mLN-resident MHCIIint (CD8α+CD11blowCD103+ and CD8α− CD11b+CD103−) and migratory MHCIIhi (CD11b−CD103+ and CD11b+CD103+) subpopulations. Plasmacytoid DCs (pDCs) were not evaluated due to their very low abundance and the fact that these cells are not targeted in zDCDTR mice. Of note, we observed that the relative frequencies of cDC and lymphocyte subsets were similar in the different lymph nodes draining the proximal to distal gastrointestinal tract (Supplementary Fig. 4a-m) and therefore subjected cDCs sorted from total mLN pools to expression profiling. Unsupervised principle component analysis (PCA) revealed that resident and migratory cDC subsets were the most segregated populations (Supplementary Fig. 4n), while IRF8– and IRF4–dependent subsets followed a hierarchical pattern in their tolerance-inducing gene expression. As such, migratory, IRF8–dependent CD103+CD11b− DCs had the highest expression of retinaldehyde dehydrogenase (RALDH2) Aldh1a2, as well as Tgfb2, Itgb8 and several other genes associated with generation of pTreg cells and gut-homing. This gene signature was progressively lost by the CD8α+ and CD103+CD11b+ populations, which rather expressed other isoforms of Tgfb and Aldh; CD11b+ DCs had the lowest expression of these genes (Fig. 5a-c and Supplementary Fig. 4o, p). This hierarchy was confirmed by the activity of RALDHs in cDCs freshly isolated from total mLNs (Fig. 5d). In contrast, CD103+CD11b− DCs did not express typical pro-inflammatory genes, although Il12b (encoding IL-12p40) was preferentially expressed compared to the other DC populations. However, Il12a (encoding IL-12p35) or Il23a (encoding IL-23p19) were not present in the absence of inflammatory triggers (Supplementary Fig. 4q). While CD11b+ DCs showed the lowest pTreg cell-inducing gene signature, they had the highest expression of genes associated with pathogen sensing and initiation of inflammatory responses, including TLRs and TLR-signaling molecules (Fig. 5a, e) and a distinct cytokine expression profile (Supplementary Fig. 4q). The above expression profiling pointed to IRF8–dependent cDCs as the main tolerogenic subsets in the mLNs.
Figure 5. Characterization of mLN cDC subpopulations.
(a) Heat map of most differentially expressed genes of cDC subsets in the mLN determined by RNA-seq. (b) Arbitrary units of TGF-β synthesis and (c) retinoic acid synthesis gene cluster expression levels in mLN cDC subsets determined by RNA-seq. (a-c) n=3, representing biological replicates. (d) Flow cytometry analysis of mean fluorescence intensity (MFI) of FITC+ Aldefluor conversion product in indicated mLN cDC populations after 30 min of substrate addition, pre-incubated with or without RALDH inhibitor DEAB. n=4 (biological replicates). Data are representative of four independent experiments. (e) Heat map of expression levels of cDC subsets in the mLN determined by RNA-seq. n=3, as (a-c). Data shown as average±SEM; NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test).
Zbtb46–driven Irf8 targeting impacts pTreg cell induction
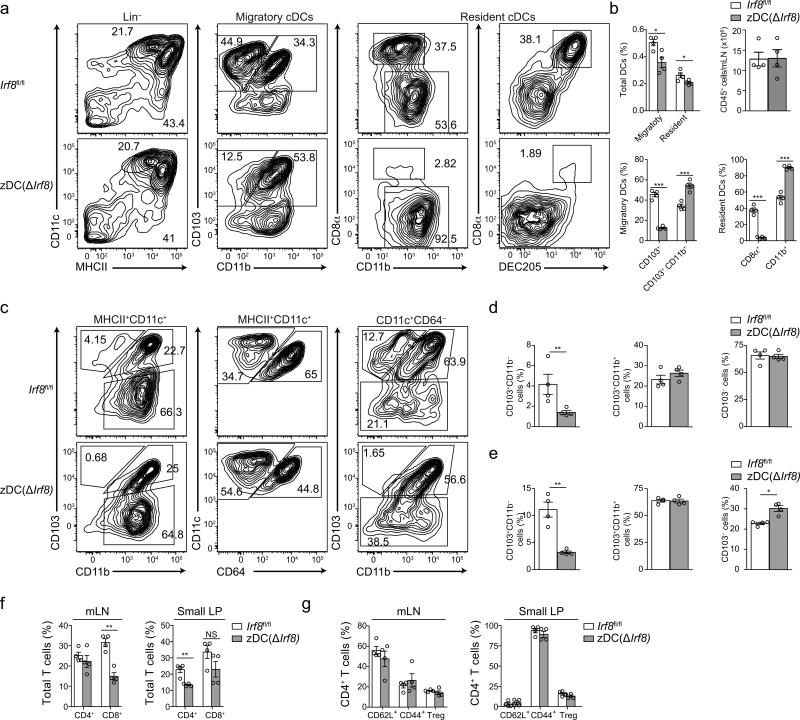
Several strategies have been devised in the past to ablate BATF3– or IRF8–dependent, or CD11b− DCs27, 28, 29, 30, but some approaches led to defects beyond those caused by loss of cDC subsets 28, 31. Recently, additional cDC-specific genetic approaches have been reported32, 33 and they revealed the reliance of T helper polarization on certain DC populations 20. However, whether a specific cDC subset is necessary for pTreg cell induction and oral tolerance has not been addressed. We therefore generated a mouse strain expressing Cre under the endogenous Zbtb46 promoter (zDCCre)34 and crossed it to mice with a loxP site flanked exon 2 of the Irf8 gene, encoding the DNA binding domain of this transcription factor (zDC(ΔIrf8) mice)35. The mice were viable, born at Mendelian ratios, and exhibited normal weights and no visible pathologies up to the age of twelve weeks. This strategy led to the loss of CD8α+ and CD11b− cDCs in mLN, lamina propria (Fig. 6a-e) and spleen (Supplementary Fig. 5a-c) of zDC(ΔIrf8) compared to Irf8fl/fl mice, with no major effects on T or B cell frequencies (Fig. 6f, g and data not shown). We noticed that excision of Irf835 led to variable loss of cDCs; however, since this property appeared conserved among BM recipients, BM transplant preserved the degree of niche depletion and permitted the generation of large and uniform cohorts of CD8α+ and CD11b− cDC-deficient mice.
Figure 6. Analysis of steady state cDC and lymphocyte populations in the mLN and lamina propria of zDC(ΔIrf8) and Irf8fl/fl mice.
(a) Representative flow cytometry contour plots, (b) relative frequencies of CD11c+, CD11c+MHCIIhi (migratory) and CD11c+MHCIIint (resident) cDCs and total CD45+ cells in the mLN of Irf8fl/fl versus zDC(ΔIrf8) mice. (c-e) Representative flow cytometry contour plots (c) and relative frequencies among MHCII+CD11c+ cells (d) or CD11c+CD64− cells (e) of small intestine lamina propria (LP) CD103+, CD103+CD11b+ and CD103− DCs in 10 weeks old Irf8fl/fl versus zDC(ΔIrf8) mice. (f) mLN and small LP CD3+CD8+ and CD3+CD4+ cells, (f) mLN and small LP CD3+CD4+Foxp3+, CD3+CD4+CD62L+ and CD3+CD4+CD44+ cells in 10 weeks old Irf8fl/fl versus zDC(ΔIrf8) mice. n=4, NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test). Data (average±SEM) are representative of two independent experiments.
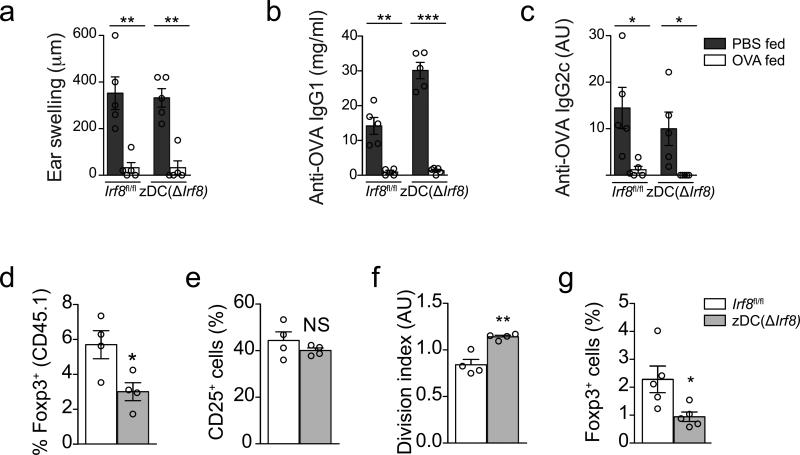
We therefore utilized zDC(ΔIrf8) BM chimeras to assess the importance of IRF8–dependent cDCs for oral tolerance (Fig. 1b, excluding DT injection). We found that ear swelling, IgG1 and IgG2c amounts in the serum were suppressed to a similar degree in zDC(ΔIrf8) and Irf8fl/f control BM recipient mice compared to the PBS-fed mice of each genotype (Fig. 7a-c). This apparently intact tolerance in zDC(ΔIrf8) mice was observed even when we immunized with OVA+CFA 24 hours after the first oral antigen encounter to blunt potential compensatory pTreg cell expansion (data not shown). Next, we directly assessed the importance of IRF8–dependent cDCs for pTreg cell polarization upon oral antigen in vivo. 48 hours after first oral OVA encounter, pTreg cell induction of adoptively transferred naïve OT-II cells was lower in zDC(ΔIrf8) than in Irf8fl/f mice, while cell division was significantly increased (Fig. 7d-f). This phenomenon persisted in mLN, spleen and other peripheral lymph nodes at 7.5 days after OVA feeding (Fig. 7g and data not shown). OT-II cells did not produce significant amounts of IFN-γ, IL-5 or IL-17a under these conditions, and this was not increased in zDC(ΔIrf8) mice (data not shown). These findings were in line with the loss of the migratory and resident mLN DC subsets with the most tolerogenic signatures. However, the impairment in pTreg cell induction was less severe than in zDCDTR mice, indicating that the remaining cDC pools could compensate for the IRF8–DC lineage loss during oral tolerance development. These results demonstrate that the targeting of IRF8–dependent cDCs partially prevents pTreg cell generation in vivo but does not affect oral tolerance, indicating that the remaining cDC pools and pTreg cells could compensate for the IRF8–DC lineage loss.
Figure 7. Assessment of oral tolerance and pTreg cell induction in zDC(ΔIrf8) and Irf8fl/fl BMCs.
(a) Degree of ear swelling 48 h post subcutaneous OVA challenge in Irf8fl/fl versus zDC(ΔIrf8) BMCs given oral OVA (+) or PBS (−) before immunization. (b) Serum anti-OVA IgG1 and (c) anti-OVA IgG2c levels in mice described in (a), 28 days post CFA immunization. n=5 per group. (d-f) Flow cytometry analysis of (d) Foxp3+, (e) CD25+ cell frequencies and (f) cell division index of adoptively transferred naïve CD45.1 OT-II cells in the mLN of Irf8fl/fl versus zDC(ΔIrf8) BMCs, 48 h after first oral OVA gavage. n=4. (g) Foxp3+ cell frequencies of adoptively transferred naïve CD45.1 OT-II cells in the mLN of Irf8fl/fl versus zDC(ΔIrf8) BMCs, 7.5 days after first oral OVA gavage. n=4. Data (average±SEM) are representative of two independent experiments each (a-c, d-f, g-h). NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test).
Hierarchical roles of cDCs in pTreg cell induction
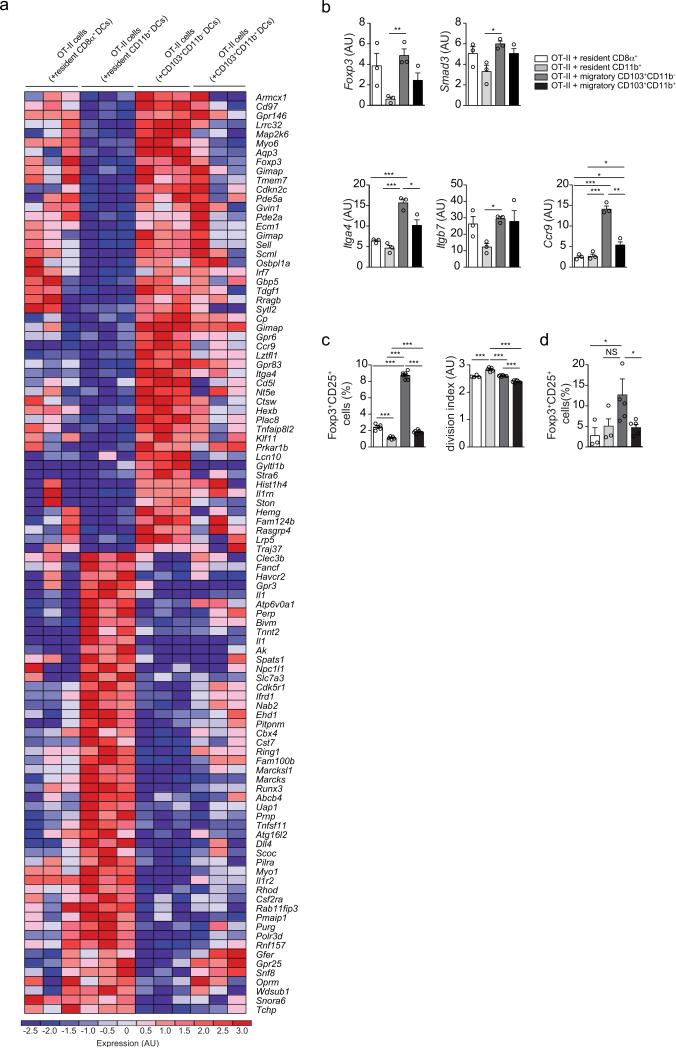
To gain insights into early changes imprinted on T cells by different DC populations, we performed RNA sequencing analyses of activated OT-II cells upon 24 hours of co-culture with mLN-derived cDC subsets and OT-II peptide. Unsupervised PCA indicated that OT-II cells stimulated by resident and migratory cDC subsets were the most segregated populations (Supplementary Fig. 6), analogous to the clustering of DC subsets (Supplementary Fig. 4n). The various cDC subsets induced a hierarchical pTreg cell pattern in OT-II cells (Fig. 8a). Consistent with the cDC gene profile (Fig. 5a-c), and with a process initiated by the TGF-β and RA pathways6, 7, 8, 16, 17, 36, naïve T cells upregulated several pTreg cell– and gut homing–associated genes when exposed to CD103+CD11b− DCs; CD103+CD11b+ and CD8α+ induced an intermediary phenotype and CD11b+ DCs were the poorest in triggering this signature (Fig. 8a, b). The pTreg cell-polarizing capacity of mLN cDCs subsets was confirmed in co-culture experiments without addition of exogenous TGF-β and RA6: CD11b−CD103+ cells were the most efficient inducers, followed by CD8α+, CD11b+CD103+ and CD11b+CD103− cells (Fig. 8c), results that closely mirrored the cDC subset and OT-II transcriptome analyses. Although the frequencies of activated OT-II cells were similar upon co-cultures with the different DC subset (data not shown), we found an inverse correlation between pTreg cell- versus proliferation-induction among cDC subsets, whereby CD8α−CD11b+ cells triggered the highest rate of cell division (Fig. 8c). Since the provision of antigen in vitro may obscure differences in antigen capture and processing that occur in vivo, we also examined pTreg cell polarization in co-cultures with cDCs sorted from OVA-fed mice. We found that CD11b−CD103+ cells consistently induced a higher frequency of Foxp3+ OT-II cells, although other cDC subsets were also able to induce Foxp3 (Fig. 8d). Overall, the differences in pTreg cell induction potential among the cDC subsets correlated with lineage but not strictly MHC class II expression: CD11b− DC subsets provided superior pTreg cell polarizing and inferior cell cycling or pro-inflammatory signals over the CD11b+ subsets, corresponding to the IRF8– and IRF4–dependent cDC lineages, respectively. In conclusion, we observed that cDC subsets play a hierarchical, but redundant role in pTreg cell induction and oral tolerance.
Figure 8. In vitro characterization of pTreg cell induction potential of cDC subpopulations in the mLN.
(a) Heat map of most differentially expressed genes in OT-II cells co-cultured with indicated mLN cDC subsets and OT-II peptide for 24 h determined by RNA-seq. n=3 (b) Treg and proliferation gene cluster expression levels of cDC subsets in the mLN determined by RNA-seq experiment shown in (a). n=3 (c) Flow cytometry analysis of Foxp3+ frequency and cell division index of sorted naïve CD45.1 OT-II cells co-cultured with indicated mLN cDC subsets, OT-II peptide, anti-IL-4 and anti-IFN-γ antibodies for 72 h. n=4-6 (n=4 for resident and n=6 for migratory DC co-culture) from three independent DC donor pools. Data is representative of three independent experiments. (d) Flow cytometry analysis of Foxp3+ frequency of sorted naïve CD45.1 OT-II cells co-cultured with indicated mLN cDC subsets obtained from mice that were fed an OVA-rich diet for 48 h, anti-IL-4 and anti-IFN-γ antibodies for 72 h. n=4-6 from three independent DC donor pools. Data shown as average±SEM; NS= not significant, *P<0.05, **P<0.01, ***P<0.005 (two-tailed t-test).
Discussion
Here we report that pre-DC– but not monocyte–derived APCs are required for both, generation of pTreg cells and oral tolerance. In addition, pre-DC–derived cDC subsets play partially-redundant roles in these processes, with IRF8–dependent cDC populations being the primary pTreg cell inducers. Our high-resolution analysis of cDCs subsets identified a pTreg cell-inducing gene signature in the IRF8-dependent, MHCIIhiCCR7hi migratory cDC population, which is consistent with the established role of CD103+ DCs6, 10 for pTreg cell polarization. By using ontogeny-based genetics, we provide evidence to reconcile previous observations defining APC requirements for pTreg cell induction using various cell surface-based targeting strategies. Because major APC subsets were depleted specifically only during oral antigen exposure, our studies do not rule out a role for monocyte-derived lineages or pDCs in the maintenance, expansion and survival of pTreg cells during later phases of oral tolerance formerly suggested9, 37.
Cell-surface molecules such as CCR2, CD11c and CX3CR1 have been used as targets to ablate APC function, development or migration in multiple studies aimed at defining a role for these cells in pTreg cell induction9, 11, 13, 15, 38, 39. Given the high expression of CX3CR1 by intestinal APCs, two main strategies targeted CX3CR1-expressing cells or Cx3cr1 itself13, 23. Receptor targeting studies using Cx3cr1GFP/GFP mice inferred that CX3CR1 is necessary for luminal antigen sampling and transfer11, 23, suggesting an essential role of fractalkine sensing for APC function and/or localization. A study using cell-transfer strategies concluded that CX3CR1 was required for macrophage-mediated pTreg cell expansion rather than pTreg cell induction9, and these results are not in contradiction to our observations. More recently, CX3CR1hi APCs were defined as lamina propria and mLN macrophages, while a fraction of DCs from pre-DC origin was reported to express low or intermediate levels of CX3CR113, 32, 40, 41, 42. A study targeting APCs by DT administration to CX3CR1DTR mice showed that while CX3CR1hi mononuclear phagocytes were efficiently deleted, CX3CR1lo/int cells (non-macrophages) remained intact in the lamina propria13. Both by CX3CR1GFP/+ reporter analysis and lineage depletion strategies, we determined the contribution of macrophages and cDC subsets to the CX3CR1+ pools in mLN and lamina propria. We observed depletion of both macrophages and CX3CR1-expressing cDCs in the mLNs of CX3CR1DTR BM recipient mice upon DT administration. Regardless of apparent targeting differences, we did not find sustained impairment of pTreg cell induction in DT-injected CX3CR1DTR, in line with our observation that monocyte-derived APCs are not strictly needed for oral antigen-mediated pTreg cell induction. However, it remains to be resolved whether CX3CR1-expressing cells play a specific role in the maintenance of pTreg cells and oral tolerance, or, alternatively, whether CX3CR1 receptor expression is required for functional and anatomical properties of cDCs.
Lineage-based intersectional genetics also provided important information regarding a hierarchy of cDC subsets in their pTreg cell induction potential. IRF8–dependent cDCs, the CD8α+ or the CD103+CD11b− cDCs populations43, 44, were highly efficient in pTreg cell generation. Our transcriptomics results confirmed and expanded previous analyses24, 25, identifying that TGF-β and RA pathways are highly enriched in migratory CD103+CD11b− DCs. Furthermore, while our data corroborate earlier observations that CD103+ DCs are efficient pTreg cell inducers via these pathways, they also identify the subpopulation amongst the three CD103+ DC populations in the mLN (CD8α+CD11blowCD103+, CD11b−CD103+ and CD11b+CD103+) governing this phenomenon. Parallel T cell and cDC RNA-seq also revealed cDC subset-specific gene signatures that closely mirror early gene programs induced in T cells, further reinforcing the role of these pathways in the generation of pTreg cells2, 4, 6, 7, 8.
In line with previous studies addressing the role of IRF4 and Notch2 in the development of cDCs26, 45, 46, 47, 48, we observed that CD11b+ DCs subsets were rather inefficient in pTreg cell induction, while they showed a superior capacity in the expansion and maintenance of CD4+ T cells and polarization towards helper T cells. The preference of particular cDC subsets in inducing pTreg cell versus effector T cell differentiation was not absolute, as demonstrated by our Irf8– targeting approach that resulted in reduced pTreg cell generation but intact oral tolerance. Our observations support recent reports that indicate a division of labor among APC subsets 13, 49. However, depletion of the microbiome, which has been shown to boost the migratory capacity of intestinal APCs13, did not rescue pTreg cell induction in mice depleted of cDCs. Additionally, although migratory IRF8–dependent cDCs appeared to have the highest capacity to induce pTreg cell differentiation in the mLN, this subset is underrepresented in the lamina propria. This observation, along with known cell-free antigen distribution through the intestinal draining lymph50, raises the possibility that antigen sampling and carriage may have only a limited impact when compared to cell-independent pathways of antigen distribution; rather, what determines the degree of pTreg cell induction is the composition of cDCs in the mLN that capture antigen in situ. Therefore, the characterization of the relative contribution APC-mediated versus cell-free antigen delivery is a logical next step for these studies.
Online Methods
Mice
C57BL/6J CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) and CD45.2 (C57BL/6J), Lyz2Cre/Cre (B6.129P2-Lyz2tm1(cre)Ifo/J), Cx3cr1GFP (B6.129P(Cg)-Ptprca Cx3cr1tm1Litt/LittJ), Cx3cr1LsL-DTR (B6N.129P2-Cx3cr1tm3(DTR)Litt/J), CD11cCre (B6.Cg-Tg(Itgax-cre)1-1Reiz/J), CD11cDTR (B6.FVB-Tg(Itgax-DTR/EGFP)57Lan/J) and Irf8fl/fl (B6(Cg)-Irf8tm1.1Hm/J) mice were purchased from the Jackson Laboratories and CD45.1 OT-II TCR-transgenic were originally purchased from Taconic Farms and maintained in our facilities. Csf1rLsL-DTR, zDCDTR and zDCCre knockin mice were generated in the Nussenzweig lab and generously shared with us20, 21, 34. These lines were interbred in our facilities to obtain the final strains described in the text. Genotyping was performed according to the protocols established for the respective strains by Jackson Laboratories. zDCCre mice were screened for Cre using PCR primers: 5’-TTACCGGTCGATGCAACGAGT-3‘ and 5‘- TTCCATGAGTGAACGAACC-3‘. Excision of the floxed Irf8 allele fragment was confirmed using Platinum DNA polymerase (Invitrogen) and primers 5‘-TTGGGGATTTCCAGGCTGTTCTA-3‘ and 5‘-TCTACACAGGAAGCAAGCCG-3‘, which give rise to ~2900 bp or ~1500 bp PCR product for the floxed or recombined locus, respectively. Mice were used at 7-12 weeks of age, and male mice were used throughout the study unless indicted otherwise. Mice were maintained at the Rockefeller University animal facilities under specific pathogen-free conditions or germ free conditions. Germ-free C57BL/6 mice were generously provided by S. Mazmanian (Caltech) and imported into germ-free flexible film isolators (Class Biologically Clean Ltd.). The mice were bred in our germ-free facility for maintenance of the strain and kept on sterilized Autoclavable Mouse Breeder Diet (5021, LabDiet, USA), which was also used for control (ex germ-free) SPF mice. Animal care and experimentation were consistent with the NIH guidelines and were approved by the Institutional Animal Care and Use Committee at the Rockefeller University.
Antibodies and cell staining
Antibodies are described in the Supplementary table 1. Aqua LIVE/DEAD® Fixable Aqua Dead Cell Stain Kit, L-34965, was purchased from Life Technologies. Horseradish peroxidase conjugated Streptavidin was purchased from Jackson ImmunoResearch Laboratories, Inc.
Lymphocyte and APC isolation from small intestine
Small intestines were separated from mesentery, and Peyer's Patches and feces were removed. Intestines were cut longitudinally and washed twice in Magnesium and Calcium free HBSS+ 1 g/l glucose (HBSS) (Gibco), and once in HBSS + 1 mM DTT (Sigma-Aldrich). Tissue was cut into 1 cm pieces and the epithelium removed by incubation in 25 ml of HBSS + 2% FCS + 1.3 mM EDTA for 15 min at 37°C at 230 rpm. Tubes were vortexed, the intestinal pieces recovered in a sieve and washed for an additional 15 min in 25 ml of HBSS + 2% FCS at 37°C at 230 rpm. Tissues were then finely chopped and digested in 8 ml of RPMI (Gibco), 2% FCS, 200 μg/ml DNaseI (Roche) and 2 mg/ml Collagenase IV (Gibco) for 2× 20 min at 37°C, 5% CO2. After the first and second 20 min, digests were taken up and resuspended 10 times. Final digests were passed through a sieve and the collagenase quenched by addition of 15 ml of cold RPMI, 2% FCS. Cell pellets were resuspended in 35% Percoll (BD Pharmigen), passed through a 100 μm mesh and separated by centrifugation in a discontinuous Percoll gradient (70%/35%) at 1000 g for 25 min at room temperature (RT). APCs and lymphocytes were isolated from the interphase, washed, and stained for FACS analysis.
Lymphocyte and APC isolation from lymph nodes and spleen
Tissues were dissected into cold HBSS supplemented with Mg2+ and Ca2+, finely chopped and incubated in 400 U/ml Collagenase D (Roche) in HBSS for 25 min at 37°C, 5% CO2. Collagenase was quenched on ice by addition of final 10% FCS. Single cell suspensions were extracted from connective tissue by taking up and resuspending the digests five times. Erythrocytes were lysed by incubation in erythrocyte lysis buffer (Sigma) for 7 min at RT.
Segmentation of mesenteric lymph nodes
Mesenteric lymph nodes draining intestinal segments were determined anatomically by following the lymphatic vessels connecting the colon, ileum and jejunum to their lymph nodes. Duodenal lymph nodes were revealed by gavaging 100 μl of olive oil (Sigma) and determining the most stomach-proximal lymph nodes surrounded by chyle, indicative of duodenal drainage, 2 h post gavage. Non-chylous stomach-proximal lymph nodes (celiac and pancreatic) were excluded from total mLN analyses.
Cell sorting
Cells were sorted using FACS Aria cell sorter flow cytometer (Becton Dickinson). For mesenteric dendritic cell sorting, cells were pre-enriched using anti-CD11c MACS beads (Miltenyi Biotec) and LS MACS Separation Columns (Miltenyi Biotec). Dendritic were sorted as Aqua−CD45+Lin−(CD3−B220−NK1.1−CD19−) CD11chi, and the subpopulations further as MHCIIintCD8α+CD11blo, MHCIIintCD8α−CD11b+, MHCIIhiCD103+CD11b− and MHCIIhiCD103+CD11b+. Naïve CD4 T cells were pre-enriched by negative selection using biotinylated antibodies against CD8α, CD25, CD11c, CD11b, TER-119, NK1.1, and B220 and anti-biotin MACS beads (Miltenyi Biotec) and sorted as Aqua−CD11c−CD8α−MHCII−Vα2+CD4+CD25−CD62hiCD44lo.
FACS analysis
Cell populations were stained with Aqua in PBS, followed by incubation with Fc block and antibodies against the indicated cell surface markers in FACS buffer (PBS, 1% BSA, 10 mM EDTA, 0.02% sodium azide). The cells were analyzed live, or fixed in 1% PFA/PBS. For Foxp3 staining analysis, surface staining was followed by permeabilization using Foxp3 Mouse Regulatory T cell Staining Kit (eBioscience). FACS data was acquired on a LSR-II flow cytometer (Becton Dickinson) and analyzed using FlowJo software package (Tri-Star). Cell division index was calculated using the FlowJo formula (http://www.flowjo.com/v765/en/proliferation.html), whereby the index represents the fraction of total cell divisions over the calculated total starting cells. Note that calculation of the division index is based on cells present at the time of analysis and cannot take into account potential lack of survival of CD45.1 cells in zDCDTR BMCs. Lineage contributions to APC pools (Fig. 4 a, b) were calculated on the assumption that DT-mediated depletion was 100% efficient: First, we determined the frequency of each APC subpopulation in DT-treated wild-type mice; next, we determined the frequency of each subpopulation in DT-treated zDCDTR (and MMDTR) mice; finally, because depletion is not 100% and every mouse has slightly different APC subpopulation ratios (routinely +/− 5% for all cDC populations), we calculated the relative contribution for cDC- (or MM-) targeted to the wild-type levels to draw pie charts.
Bone marrow chimeras
Bone marrow recipients were irradiated with two doses of 525 rad 3 h apart, and received 2-5×106 bone marrow cells by retro-orbital injection under isoflurane anesthesia. Bone marrow chimeras were reconstituted for a minimum of eight weeks after irradiation before the start of their experimental use. Germ-free bone marrow recipients were exported and irradiated in a sterile container, injected under a sterile hood and re-imported into sterile isolators. Mice tested negative for contamination 4 weeks after re-import and at the end of the experiments. For studies comparing Irf8fl/fl and zDC(ΔIrf8) bone marrow recipients, the Irf8fl/fl bone marrow was obtained from the pure Irf8fl/fl strain.
Diphtheria toxin (DT) administration
The first dose of DT (Sigma, D0564) was delivered intraperitoneally (i.p.) as 500 ng in 200 μl PBS/ 20 g body weight followed by one or two doses of 100 ng in 200 μl PBS/ 20 g body weight, as indicated. Depletion efficiency was monitored in each animal by the serum Flt3L surge upon cDC depletion (zDCDTR) and the loss of blood monocytes 24h post DT administration in MMDTR mice.
Anti-Flt3L ELISA
Serum concentrations of Flt3L were determined using an anti-Flt3L ELISA kit (R& D Systems) according to the manufacturer's protocol. Sera were diluted 1:4 to be in range of the standard curve.
Oral antigen administration
OVA (grade III, Sigma, A5378) was administered at 50 mg in 200 μl PBS by oral gavage using metal gavage needles.
125Iodine labeling of ovalbumin and 125I-OVA uptake and clearance analysis
OVA grade VII (Sigma, A7641) was labeled with 125Iodine (Perkin-Elmer) under a fume hood. Briefly, 250 μCi Na125I in 135.5 μl 0.1 N NaOH was prepared and immediately mixed with an equal volume of 0.1 N HCl containing 6.67 nmol ICl. The iodine mixture was immediately added to 1 ml of 100 μg OVA in 1M glycine, pH 9.5, 100 mM NaCl (2.26 μM OVA, equivalent to an Iodine: OVA molar ratio of 3:1), and the solution incubated at RT for 10 min. Unincorporated iodine was removed by purifying the sample on two consecutive NAP25 columns (GE Healthcare). 125I-OVA was eluted in H2O and kept at 4 °C in 1× PBS by appropriate addition of 10× PBS. Amount of labeling was determined on a Packard Cobra gamma counter and an estimated to be 95000 CPM/μg OVA. Protein purity and integrity were confirmed on a Coomassie gel and specific labeling of OVA by exposing the gel to autoradiography film. To determine OVA uptake and clearance, mice were fasted for 3 h and then gavaged at 12 pm with 50 mg OVA grade III in 200 μl PBS 300,000 CPM 25I-OVA (about 3 μg OVA). Submandibular blood and urine were collected in 5 μl aliquots; whole organs were taken at indicated time points and weighed. Radioactivity was measured using on a Packard Cobra gamma counter.
CFA immunization and subcutaneous ear challenge
Eight days after oral OVA gavage, CFA-OVA was administered subcutaneously between the shoulder blades as an emulsion of 100 μl CFA and 100 μl PBS containing 300 μg endotoxin free OVA (EndoGrade® Endotoxin-free Ovalbumin, HYGLOS, Germany, 321000) under isofluorane gas anesthesia. The first ear challenge was performed 14 days after immunization. 2.5 mg/ml endotoxin free OVA was heat aggregated in PBS at for 2 h at 65 °C, and 30 μl of OVA (left ear) or PBS vehicle (right ear) were injected under isofluorane gas anesthesia. Ear thickness was measured using a digital precision caliper (Fisher Scientific). Swelling was determined by first subtracting pre-challenge from post-challenge ear thickness, and then subtracting the swelling upon PBS injection from swelling upon OVA injection.
Alum immunization and airway challenge
Eight days after oral OVA administration, 4 ug of endotoxin-free OVA antigen adsorbed to 40 μl Imject™ Alum Adjuvant (Fisher Scientific) was injected i.p. in a final volume of 400 μl made up with PBS. Immunization was repeated after 7 days. To induce airway inflammation, mice were anesthetized and intranasally administered 10 ug of endotoxin-free OVA in 50 μl PBS (25 μl per nostril) on days 14, 17 and 21 after the first i.p. immunization.
Bronchoalveolar Lavage (BAL), lung histology and infiltrate analysis by flow cytometry
Mice were anesthetized by i.p. injection of 0.35 ml of 2.5% avertin (Sigma), the trachea was cannulated and lungs were lavaged with once with 0.5 ml and then 1.0 ml PBS. Total BAL cells were counted after erythrocyte lysis and stained for FACS analysis. Lungs were perfused via the right ventricle with 10 ml saline to wash out residual blood. One lobe was digested in 400 U/ml collagenase D/HBSS and processed for FACS analysis. Eosinophils were determined as CD45+SSAhiMHCII−CD11b+Ly6GintSiglecF+. Another lobe was fixed with 4% phosphate-buffered formalin and embedded in paraffin. Tissues sections were stained with hematoxylin/eosin (H&E) to determine cellular inflammation. Sections where assessed in a blinded fashion. An infiltration score was created whereby 0=no, 1=very mild, 2=mild, 3=locally severe, 4=uniformly severe, 5=uniformly very severe infiltration was observed in a 1 cm2 section.
Anti-OVA IgG1 and IgG2c ELISAs
High binding ELISA 96-well plates (Corning, 3690) were coated with 50 μl of 20 μg/ml OVA grade II in 100 mM Na2CO3 overnight at 4°C. Plates were washed three times PBS-0.05% Tween 20 (PBS-T, Sigma), and blocked with 150 μl 1% BSA in PBS-T for 2 h at RT. Mouse sera were obtained by submandibular bleeding and diluted in blocking buffer and added as 25 μl/ well. For anti-OVA IgG1 ELISA quantification, an anti-OVA IgG1 (Biolegend) standard curve ranging from 0.125 to 8 ng/ml was applied; for anti-OVA IgG2c ELISA quantification, a hyper-immune serum was diluted ranging from a 1:7500 (set as 1.0) to 1:480,000 (set as 0.0156) dilution and the 1:7500 concentration set as 1. Sera were measured in duplicates and at two dilutions, typically 1:500,000 and 1:2,000,000 for the anti-IgG1 ELISA, and 1:5000 and 1:20,000 for the anti-IgG2c ELISA. Standards and test sera were incubated for 1 h at RT and the plates washed three times in wash buffer. Biotinylated anti-IgG1 or anti-IgG2c applied as 50 μl at 20 ng/ml in blocking buffer, the plates incubated for 1 h and washed four times. Horseradish peroxidase linked Streptavidin (HRP-Streptavidin) was added as 50 μl at 40 ng/ml and the plates incubates for 45 min at RT and then washed five times. Fifty μl HRP substrate TMB was added and the reaction stopped by addition of 50 μl 1N H2SO4. Plates were read at A450-570nm. Absolute concentrations of anti-OVA IgG1 were determined from the standard curve; relative concentrations of anti-OVA IgG2c were determined by dividing the concentrations determined by the standard curve by 4,800.
Anti-IgE ELISA
High binding ELISA 96-well plates (Corning, 3690) were coated with 50 μl of 2μg/ml anti-IgE antibody (Invitrogen) in 100 mM Na2CO3 overnight at 4°C. Plates were washed three times PBS-0.05% Tween 20 (PBS-T, Sigma), and blocked with 1 μl 1% BSA in PBS-T for 2 h at RT. Mouse sera were diluted in blocking buffer (typically 1:2 and 1:20) and added as 25 μl/ well in duplicates. An IgE isotype control standard curve was applied at concentrations from 4 to 256 ng/ml. Standards and test sera were incubated for 2 h at RT and the plates washed three times in wash buffer. Biotinylated anti-IgE was applied as 50 μl at 33 ng/ml, the plates were incubated for 1 h and washed four times. Horseradish peroxidase linked Streptavidin (HRP-Streptavidin) was added as 50 μl at 100 ng/ml and the plates incubates for 45 min at RT and then washed five times. Fifty μl HRP substrate TMB was added and the reaction stopped by addition of 50 μl 1N H2SO4. Plates were read at A450-570nm. Absolute concentrations of serum IgE levels were determined from the standard curve.
Adoptive T cell transfer
Sorted naïve CD45.1 OT-II T cells were labeled using the Cell Trace™ Violet Cell Proliferation Kit (C-34557, Life Technologies). 2×106 or 6×106 cell were transferred by retro-orbital injection under isofluorane gas anesthesia for analyses of cells 48 h or 7.5 days after OVA gavage, respectively,.
In vitro T-cell and DC co-culture
Sorted and Cell Trace™ stained naïve OT-II cells were cultured for 3 days in flat bottom 96 well plates OT-II in the presence of sorted mLN DC subpopulations, 1 μg/ml anti-IL4 and 1 μg/ml anti-IFN-γ antibody. Antigen was either provided as 1 μg/ml of OT-II peptide (chicken OVA323-339, amino acid sequence ISQAVHAAHAEINEAGR, made by the Rockefeller University Proteomics Resource Center), or by in vivo loading of DCs by maintaining donor mice on a 20% OVA diet for 2 days and gavaging them with 50 mg OVA 4h prior to mLN isolation. Cells were co-cultured at a 5:1 T cell/DC ratio (100,000 to 20,000 or 50,000 to 10,000).
RALDH activity assay
RALDH activity was determined using the Aldefluor kit (STEMCELL™ Technologies) according to the manufacturer's protocol, with a few optimizing modifications: 3×106 cells were incubated at 1×106 cells/ml, and were pre-incubated with 30 μM DEAB (Sigma) or vehicle for 30 min at 37°C prior to the addition of 5 μl /ml Aldefluor reagent for a further 30 min.
RNA preparation and RNA sequencing
RNA from sorted cell pellets was extracted and column-purified using the Arcturus PicoPure RNA Isolation kit (Applied Biosystems). Genomic DNA was removed by on-column digest with DNAse I (Qiagen), according to the PicoPure RNA Isolation kit manual. RNA libraries were prepared using the SMARTer Ultra Low Input RNA for Illumina Sequencing kit (Clontech Laboratories) and sequenced using 50 base pair paired end reading on a HiSeq 2500 instrument (Illumina). The reads were aligned using the STAR version 2.3.0 software that permits unique alignments to Mouse Ensembl genes. Differential expression was determined by use of the Cufflinks software with default settings. Heat maps were generated by comparing the expression profile CD11b+ and CD103+CD11b− DCs, or of OT-II cells cultured with CD11b+ and CD103+CD11b− DCs. Gene clusters were determined using Gene Set Enrichment Analysis (GSEA v2.2.2) software. PCA was performed using R software.
Statistics
Statistical analysis was performed in GraphPad Prism software. Data was analyzed by applying one-way ANOVA or two-tailed unpaired Student's t-test whenever necessary. For analysis of histological scores non-parametric Mann Whitney test was used. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgements
We are indebted to M. Amoury and L. Cohn for having put thought into the project in its beginnings; to M. Nussenzweig for generously sharing resources and conceptual input; to B. Reis and V. Pedicord for assistance in mouse dissection experiments, critical discussion and help in preparing the manuscript; K. Velinzon and N. Thomas for assistance in cell sorting; The Rockefeller University Genomics Center; A. Rogoz, T. Rendon, S. Gonzalez and the Rockefeller University Comparative Bioscience Center for good animal care and genotyping; and the New York University histology core for lung histological staining. Supported by a Swiss National Foundation Advanced Postdoc fellowship (D.E.), a National Institutes of Health 1R56AI119062 grant (D.M.), and by the Leona M. and Harry B. Helmsley Charitable Trust (D.E. and D.M.).
Footnotes
Author contributions
D.E. designed the study, performed the experiments unless stated otherwise below and wrote the manuscript; J.L. generated the zDC-Cre mice, backcrossed them onto the Irf8fl/fl background, carried out their hematopoietic characterization (Fig. 7) and substantially contributed to the design of experiments; M.L. and V.J. significantly contributed to the establishment of oral tolerance protocols and to the oral tolerance experiments; T.O. performed the bioinformatic analysis of the RNA-seq experiments; D.M. initiated, designed and supervised the study, performed experiments and wrote the manuscript.
Accession Codes
All RNA-seq data are submitted to NCBI's Gene Expression Omnibus, with the accession number GEO (pending).
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von Boehmer H. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6(12):1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 2.Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115(7):1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29(1):114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostroukhova M, Seguin-Devaux C, Oriss TB, Dixon-McCarthy B, Yang L, Ameredes BT, et al. Tolerance induced by inhaled antigen involves CD4(+) T cells expressing membrane-bound TGF-beta and FOXP3. J Clin Invest. 2004;114(1):28–38. doi: 10.1172/JCI20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204(8):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 8.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204(8):1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Annacker O, Coombes JL, Malmstrom V, Uhlig HH, Bourne T, Johansson-Lindbom B, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005;202(8):1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1(+) macrophages to CD103(+) dendritic cells. Immunity. 2014;40(2):248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38(3):581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494(7435):116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203(3):519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206(13):3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21(4):527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202(8):1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Idoyaga J, Fiorese C, Zbytnuik L, Lubkin A, Miller J, Malissen B, et al. Specialized role of migratory dendritic cells in peripheral tolerance induction. J Clin Invest. 2013;123(2):844–854. doi: 10.1172/JCI65260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki S, Dudziak D, Heidkamp GF, Fiorese C, Bonito AJ, Inaba K, et al. CD8+ CD205+ splenic dendritic cells are specialized to induce Foxp3+ regulatory T cells. J Immunol. 2008;181(10):6923–6933. doi: 10.4049/jimmunol.181.10.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, et al. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med. 2013;210(10):2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209(6):1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17(2):211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307(5707):254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 24.Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol. 2014;15(1):98–108. doi: 10.1038/ni.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13(9):888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vander Lugt B, Khan AA, Hackney JA, Agrawal S, Lesch J, Zhou M, et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol. 2014;15(2):161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- 27.Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101(1):305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 28.Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC, 3rd, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196(11):1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207(4):823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322(5904):1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87(2):307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 32.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209(1):139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schraml BU, van Blijswijk J, Zelenay S, Whitney PG, Filby A, Acton SE, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154(4):843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Loschko J, Schreiber HA, Rieke G, Esterhazy D, Meredith MM, Pedicord V, et al. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J Exp Med. 2016 doi: 10.1084/jem.20160062. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J, Wang H, Shin DM, Masiuk M, Qi CF, Morse HC., 3rd. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J Immunol. 2011;186(3):1458–1466. doi: 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassani B, Villablanca EJ, Quintana FJ, Love PE, Lacy-Hulbert A, Blaner WS, et al. Gut-tropic T cells that express integrin alpha4beta7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology. 2011;141(6):2109–2118. doi: 10.1053/j.gastro.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goubier A, Dubois B, Gheit H, Joubert G, Villard-Truc F, Asselin-Paturel C, et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity. 2008;29(3):464–475. doi: 10.1016/j.immuni.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darrasse-Jeze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, et al. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J Exp Med. 2009;206(9):1853–1862. doi: 10.1084/jem.20090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36(2):276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40(5):720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, Mowat AM, et al. Intestinal CD103(−) dendritic cells migrate in lymph and prime effector T cells. Mucosal immunology. 2013;6(1):104–113. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- 42.Scott CL, Bain CC, Wright PB, Sichien D, Kotarsky K, Persson EK, et al. CCR2(+)CD103(−) intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal immunology. 2015;8(2):327–339. doi: 10.1038/mi.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O'Shea JJ, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174(5):2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto M, Kato T, Hotta C, Nishiyama A, Kurotaki D, Yoshinari M, et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PloS one. 2011;6(10):e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha -dendritic cell development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35(5):780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38(5):958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, Kc W, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14(9):937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42(5):916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Ghoshal S, Ward M, de Villiers W, Woodward J, Eckhardt E. Chylomicrons promote intestinal absorption and systemic dissemination of dietary antigen (ovalbumin) in mice. PloS one. 2009;4(12):e8442. doi: 10.1371/journal.pone.0008442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.