Abstract
Proton radiotherapy is becoming more common since protons induce more precise DNA damage at the tumor site with reduced side effects to adjacent normal tissues. However, the long-term biological effects of proton irradiation in cancer initiation compared to conventional photon irradiation are poorly characterized. In this study, using a human familial adenomatous polyposis syndrome susceptible mouse model, we show that whole body irradiation with protons are more effective in inducing senescence-associated inflammatory responses (SIR) which are involved in colon cancer initiation and progression. After proton irradiation, a subset of SIR genes (Troy, Sox17, Opg, Faim2, Lpo, Tlr2 and Ptges) and a gene known to be involved in invasiveness (Plat), along with the senescence associated gene (P19Arf) are markedly increased. Following these changes loss of Casein kinase Iα (CKIα) and induction of chronic DNA damage and TP53 mutations are increased compared to x-ray irradiation. Proton irradiation also increases the number of colonic polyps, carcinomas and invasive adenocarcinomas. Pretreatment with the non-steroidal anti-inflammatory drug, CDDO-EA, reduces proton irradiation associated SIR and tumorigenesis. Thus, exposure to proton irradiation elicits significant changes in colorectal cancer initiation and progression that can be mitigated using CDDO-EA.
Keywords: Proton radiotherapy, Colorectal cancer, Senescence-associated inflammatory response (SIR), CPC, Apc mouse
Introduction
Charged particles, such as protons and heavy ions, lose energy by continuous interactions with electrons, and through occasional nuclear events within the absorbing materials (1). Thus, unlike photons, such as x-ray and γ-ray, which are absorbed by exponential attenuation, charged particles have a finite range and deposit energy in an inverse relationship with their velocity (1). Proton radiation produces a rapid fall-off of absorbed dose at the distal stopping edge, and a low level of lateral scatter, which results in sharp lateral beam edges (Bragg peak) (1–4). Proton therapy generally uses an energy range between 60 to 250 MeV/n so that tumors seated at 3 cm to 30 cm depth can be treated (5). For this reason, cancer radiotherapy treatments using protons has become much more common worldwide. However, the long-term risk and biological effects of protons in normal tissues remain to be more fully characterized.
Risk assessment is usually determined by the relative biologic effectiveness of various parameters for proton particles compared to photon (e.g. x-rays or γ-rays) exposures. Photon radiation risk estimates have been used to extrapolate to proton radiation but the reliability of the models for relative biologic effectiveness determination has uncertainties (6–7). High energy (250 MeV/n) protons have been found to have a similar biologic effectiveness to γ-rays for incidence of both simple chromosome damage and complex chromosome exchanges in human lymphocytes when irradiated with a high dose-rate (70 cGy/min) (8). However, protons increase chromosome damages in human lymphocytes when they are delivered at a lower dose-rate (0.125 cGy/min) compared to low dose-rate of γ-rays exposure (8). While a low dose-rate of protons increases chromosomal damage in vitro, cytogenetic effects of mouse bone marrow cells exposed to protons or γ-rays (both delivered at 1 cGy/min) are similar in vivo (9). While the application of scaling factors is generally accepted to be the only practical approach to human cancer risk estimation for protons, a testable hypothesis is that qualitative and quantitative differences between proton and photon effects are maintained across species, such as from mouse to human. Understanding how to scale such risks in model systems will provide the best possible framework for undertaking the same scaling of risks in humans. In order to use this approach, collection of relevant quantitative data for oncogenic and pre-oncogenic endpoints in “relevant” animal model systems are needed. In addition, little is known about the comparative molecular mechanisms involved in charged particle-induced carcinogenic pathways (10–12).
The gastrointestinal track is one of the highly sensitive tissues to radiation-induced damage, and colorectal cancer is a leading cause of cancer-related death in much of the industrialized world (13). Apc mutations play a critical initiating role in adenoma development in the inherited setting of familial adenomatous polyposis (FAP) syndrome and in sporadic tumor development (13). Mice carrying the ApcMin (Multiple intestinal neoplasia) mutation which have a nonsense mutation at codon 850 of the Apc gene appears to be responsible for predisposition to the development of multiple intestinal tumors in a mouse genetic model (14). The ApcMin mouse develops ~50 adenomas and infrequent carcinomas in the small intestine and usually die by 140 days of age. Because the vast majority of gastrointestinal tumors in human FAP patients are developed in the distal colon and rectum instead of small intestine, the ApcMin mouse has notable limitations as a model for human colorectal carcinogenesis. Thus, conditional transgenic mice in which Apc has been targeted for somatic inactivation by Cre recombinase under the control of CDX2 homeobox gene promoter (CPC;Apc mouse) has been developed (15). CPC;Apc mice developed adenomas and carcinomas mainly in the distal colon and rectum. In addition, morphologic and molecular studies of the mouse tumors reveal a striking similarity to human colorectal tumors (15).
Exposure to ionizing radiation induces intrinsic DNA damage and cellular senescence responses in human normal epithelial cells and normal fibroblasts (16). Intrinsic DNA damage and cellular senescence have recently been implicated as a major barrier against tumor initiation and progression (17–18). While senescence may initially impede tumor progression, opposing functions of senescence in tumorigenesis have also been studied. Accordingly, senescent cells secret cytokines, chemokines, and growth factors, which can facilitate tumor cell growth (19). Casein kinase I alpha (CKIα) plays a role in the Wnt signaling pathway. Ablation of CKIα triggers extensive Wnt hyper-induction and low-grade senescence-associated inflammatory responses (SIR) in the mouse intestine but homeostasis is still maintained along with wild-type p53 activity (17, 20). However, with additional alterations, such as mutations in p53, SIR loses its growth control capacity and leads to accelerated tumorigenesis and invasiveness in colorectal cancer along with overexpression of a set of invasiveness genes (termed the p53-suppressed invasiveness signature, PSIS) (17, 20). In this study, we characterized the biological effects of low dose-rate proton exposures (e.g. similar to solar particle event (SPE) simulations, sSPE) using the CPC;Apc mouse as compared to higher dose-rate protons and photon exposure. To elucidate the mechanism of low dose rate proton-induced increases in the tumor initiation and progression, the para-inflammation driving gastrointestinal track homeostasis/tumorigenesis model (17) was examined. Since protons with an energy range between 60 to 250 MeV/n (human therapeutic range) completely penetrate through a much smaller mouse body, in the present studies we used 50 MeV/n proton (which has ~2 cm of Bragg peak region). This energy would be predicted to deposit most of the energy inside a mouse, simulating what might be occurring in human patients treated with slightly higher dose rates of proton therapy.
Results
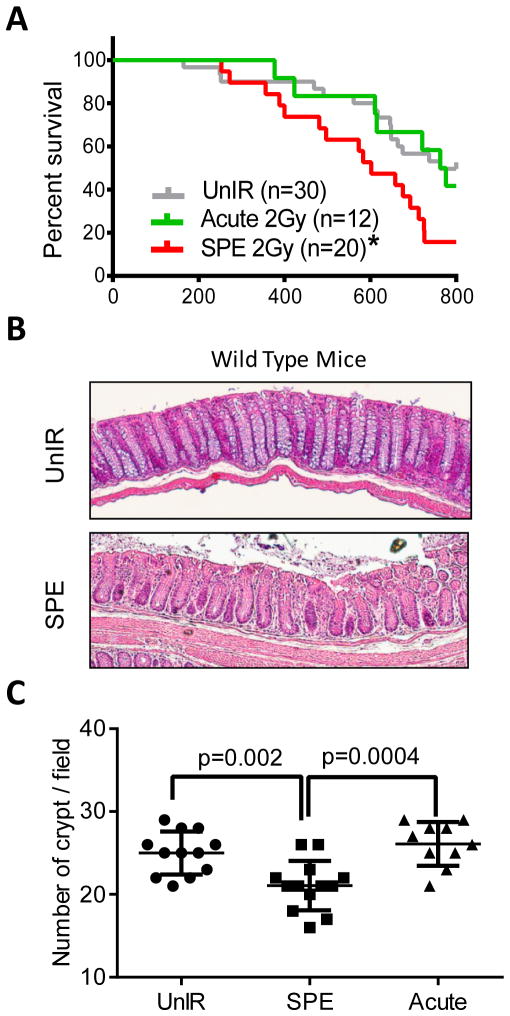
Low Dose-Rate Simulated Solar Particle Event (sSPE) Exposure Reduces Lifespan and Crypt Number in Wild Type Mice
While exposure to 2 Gy acute protons (50 MeV/n) with high dose-rate (20 cGy/min) did not change the lifespan of wild type C57BL6/J mice compared to unirradiated control mice, 2 Gy protons (SPE simulation, sSPE) with low dose-rate (1.67 cGy/min) caused markedly decreased lifespan relative to acute proton radiation. The median survival of the unirradiated control and acute proton groups were 763 day (n=30) and 769 day (n=12), respectively. In contrast, the sSPE irradiated mice showed significantly shortened median survival of 602 day (n=20) (Figure 1A). When colon histology was examined at the time of death in wild type mice, neither hyperplasias nor polyps were detected in both acute proton and sSPE irradiated mice (data not shown). However, a significant decrease in the crypt number in sSPE irradiated wild type mice compared to unirradiated or acute proton irradiated groups was observed (Figure 1B). The mean difference of crypt number per field (10× magnification) between unirradiated control and sSPE was 3.9±1.1 SD (standard deviation) (p=0.002, n=12), and the difference between acute proton and sSPE was 5.0±1.2 SD (p=0.0004, n=12) (Figure 1C).
Figure 1. Effect of low dose-rate protons (SPE simulation) on lifespan and crypt number in wild type mice.
(A) Kaplan-Meier survival plot of unirradiated or irradiated wild type mice demonstrating that irradiation with only 2 Gy sSPE significantly decreases survival compared with acute 2 Gy proton or unirradiated controls. *, P<0.05 in the log-rank test. (B) Representative images of H&E-stained colon crypts in unirradiated and sSPE-irradiated wild type mice demonstrating less crypt numbers with sSPE irradiation. (C) Quantification of crypt numbers on the 3 fields of vision with a magnification of 10X from 4 mice per group. Significant difference was evaluated by two-way ANOVA with Multiple comparisons.
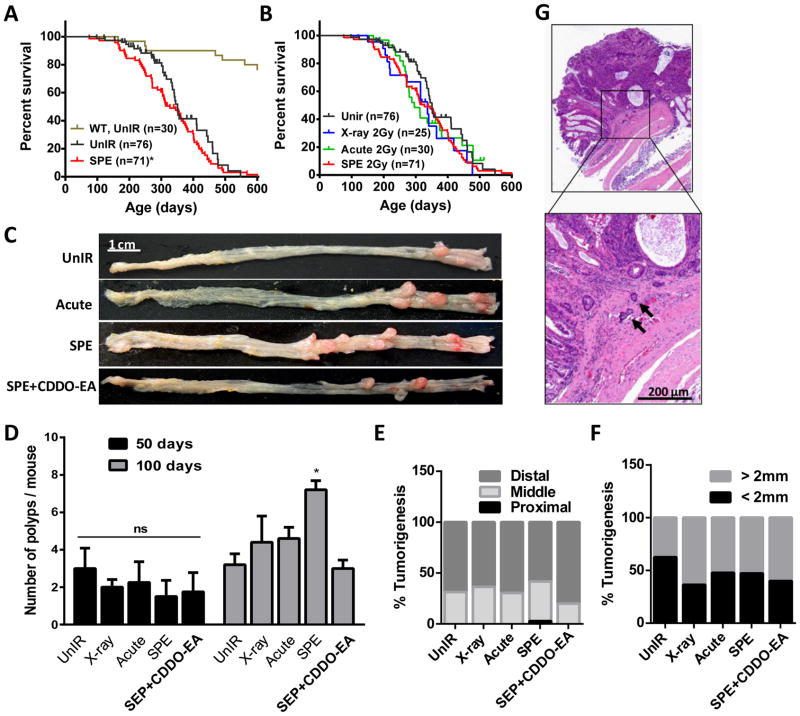
Low dose-rate sSPE Exposure Induces Higher Colonic Tumorigenesis Compared to Acute Proton or X-ray in Colon Cancer Susceptible CPC;Apc Mice
To determine the effects of proton exposure in a colon cancer susceptible mouse model, CPC;Apc mice were exposed to 2 Gy of x-ray (photon irradiation), acute high dose-rate protons, or low dose-rate protons (sSPE) radiation. While median survival of unirradiated wild type mice was calculated as 763 day, the median survival of unirradiated and sSPE exposed to colon cancer susceptible CPC;Apc mice was 351 day (n=76) and 327 day (n=71) respectively. Exposure to sSPE caused a slight decrease in median lifespan (p=0.032 in Gehan-Breslow-Wilcoxon test) in CPC;Apc mice but similar maximum lifespan was observed compared to the unirradiated control group (Figure 2A). Compared to sSPE-irradiated mice, similar survival curves were observed with acute protons (n=30) and x-ray (n=25) exposure (Figure 2B). To investigate the effects of acute proton or sSPE proton irradiation in colonic tumorigenesis, CPC;Apc mice (n=6 per study group) were observed 50 days post irradiation. However, no significant difference was observed in tumor initiation or progression at this time point post-irradiation (Figure 2D). However, 100 days after irradiation, the average number of polyps in the unirradiated control group was 3.2±1.3 SD (standard deviation), the number of polyps in 2 Gy x-ray, acute proton and sSPE-irradiated mice was 4.4±3.1 SD, 4.6±1.3 SD, and 7.2±1.1 SD respectively (Figure 2C–D). To determine if mice could be protected from sSPE-induced damage with the radioprotector, CDDO-EA (a synthetic triterpenoid) (21–25), groups of mice were fed a diet containing CDDO-EA (400 mg/kg diet) for 3 days continuously prior to sSPE exposure. Tumorigenesis of CDDO-EA treatment mice was significantly reduced to unirradiated control levels (3.0±1.0 SD) 100 days after sSPE exposure (Figure 2C–D).
Figure 2. Tumorigenic effect of simulated SPE in CPC;Apc mice.
(A–B) Kaplna-Meier survival plot of unirradiated or irradiated CPC;Apc mice. Unirradiated wild type mice (brown); unirradiated CPC;Apc mice (black); sSPE-irradiated CPC;Apc mice (red); x-ray irradiated CPC;Apc mice (blue); acute proton-irradiated CPC;Apc mice (green). *, P=0.0322 in Gehan-Breslow-Wilcoxon test compared to unirradiated CPC;Apc mice survival. (C) Representative images of polyps 100 days after irradiated or age-matched CPC;Apc mice colon. Scale bar, 1 cm. Quantification of polyp number (D), segmental distribution (E), and size (F) difference of the CPC;Apc mice 100 days after irradiation or age-matched unirradiated control. (n=6 mice per group). *, P<0.05 in the Student’s t-test compared with unirradiated control. (G) Representative image for invasion foci (black arrows) in sSPE-irradiated CPC;Apc mice. Scale bar, 200 μm.
In the segmental distribution of tumors 100 days after irradiation, unirradiated control mice showed colon tumors mostly in the distal region (68.9%) compared to the middle region (31.1%) with no tumors in the proximal region. While a significant difference was not detected in the segmental distribution of colon tumors in the x-ray and acute proton exposed group compared to the unexposed control, exposure to sSPE radiation showed higher tumorigenesis in the middle colon (38.9%) with 2.8% tumors in the proximal region (Figure 2E). When colonic tumors in each group were categorized into two groups depending on their size, there were higher numbers of larger tumors (> 2 mm) in irradiated mice relative to unirradiated control mice but there were no significant differences tumor sizes between acute proton and sSPE exposure (Figure 2F).
Histopathological analysis of the colon tumors was performed at the biological end-point and showed that 25.8% (n=62) of mice exposed to 2 Gy sSPE showed invasive adenocarcinoma. In contrast, only 6.3% (n=48) of unirradiated mice developed invasive adenocarcinoma. 2 Gy x-ray and acute proton exposed mice showed 8.0% (n=25) and 11.1% (n=27) of invasive adenocarcinoma incidence respectively. Pretreatment of CDDO-EA prior to sSPE exposure markedly decreased the incidence of invasive adenocarcinoma from 25.8% to 15.2% (n=33) (Table 1). Representative images of the invasive foci penetrating through the muscularis mucosa are shown in Figure 2G.
Table 1.
Effect of Radiation Exposure to CPC;Apc Mice
| Radiation Type | Energy (MeV/n) | Dose rate (cGy/min) | Total dose (cGy) | Median survival | Avg. polyp number (n=5) | Invasive foci incidence |
|---|---|---|---|---|---|---|
| Unirradiated Control | 0 | 0 | 0 | 351 | 3.2 | 6.3% (n=48) |
| X-ray 2 Gy | 250 kVp | 20 | 200 | 337 | 4.4 | 8.0% (n=25) |
| Proton 2 Gy (Acute) | 50 | 20 | 200 | 296 | 4.6 | 11.1% (n=27) |
| Proton 2 Gy (sSPE) | 50–150 | 1.7 | 200 | 327 | 7.2 | 25.8% (n=62)*#$ |
| Proton 2 Gy (sSPE+CDDO) | 50–150 | 1.7 | 200 | 341 | 3.0 | 15.2% (n=33) |
P<0.05 in 2-way contingency table analysis compared to unirradiated control.
P<0.05 compared to X-ray and
P=0.068 compared to Acute proton irradiated group.
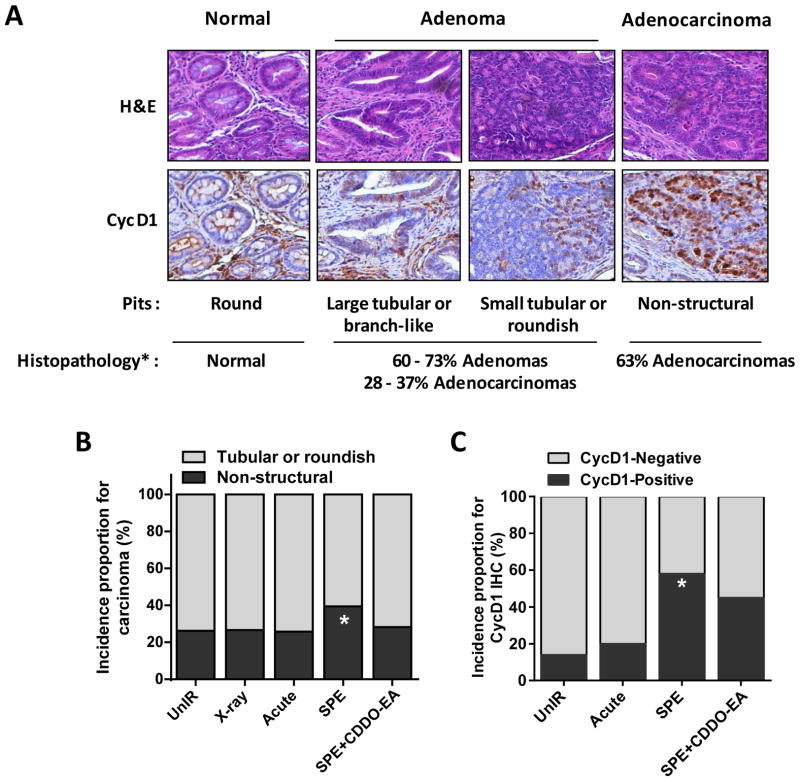
Exposure to sSPE Results in an Increased Incidence of Higher-grade Colorectal Adenocarcinomas
Human colorectal cancer can be classified by pit’s morphology through histopathological analysis (26). While round shape pits are observed in most normal colon tissues, tubular, roundish or branch-like pits are mainly found in adenomas. High grade adenocarcinomas show non-structured pit shapes (26). In addition, to better classify adenocarcinomas, immunohistochemistry of cyclin D1 was performed since others have reported a significant correlation between cyclin D1 expression and poor prognosis in colorectal cancer patients (27). Strong expression of cyclin D1 is observed in large and invasive carcinomas, and correlate with poor survival (27). In this study, we observed strong positive staining of cyclin D1 in highly progressed adenocarcinomas which had non-structured pits (Figure 3A). When colon sections collected 100 days after irradiation were scored by pit shape, exposure to sSPE radiation showed a markedly higher incidence of non-structured pit tumors (39.5%, p<0.05) relative to unirradiated (26.3%), x-ray (27.6%), or acute proton (25.8%) irradiated control groups (60–200 tumors from 30–40 mice/group were observed; Figure 3B). CDDO-EA pretreatment prior to sSPE exposure showed a decreased number of non-structured pits. Colon tissues were also categorized by immunohistochemistry of cyclin D1. In contrast to unirradiated or acute proton-irradiated control, sSPE-exposed mice had more cyclin D1-positive tumors (58%, p<0.05 compared to unirradiated control). Mice were also protected from sSPE radiation with pretreatment of CDDO-EA and showed reduced cyclin D1-positive tumors (7–12 tumors from 3 mice/group were examined) (Figure 3C).
Figure 3. Tumor grade in irradiated CPC;Apc colons comparable with unirradiated controls 100 days after irradiation.
(A) Modified classification of colorectal cancer based on pit pattern and Cyclin D1 expression (26–27, 50). (B) Quantification of overall incidences proportion for non-structural adenocarcinoma in colons of unirradiated or irradiated CPC;Apc mice demonstrates increased incidence of adenocarcinoma with sSPE exposure. (C) Quantification of overall incidence proportion for cyclin D1-positive tumors in colons demonstrating that irradiation with only 2 Gy sSPE significantly increase incidence of cyclin D1-positive carcinomas compared with unirradiated controls. *, P<0.05 compared with unirradiated control.
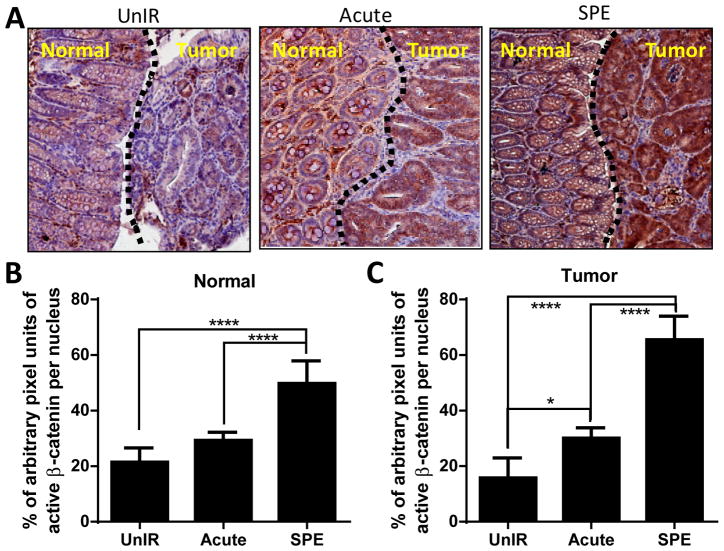
Simulated SPE Induces Prolonged Activation of β-catenin
Immunohistochemistry analysis for active-β-catenin showed markedly increased staining in both tumors and tumor-adjacent normal areas of colonic tissue in the CPC;Apc mice 100 days after sSPE irradiation (Figure 4A). Quantification showed significantly higher staining in the tumor-free normal area in sSPE-irradiated mice relative to tumor-free areas in unirradiated control and acute proton-irradiated groups (p<0.0005) (Figure 4B). Along with normal tissues, tumors in sSPE-irradiated mice also showed higher active-β-catenin staining (p<0.0005) compared with unirradiated control and acute proton radiation groups (Figure 4C). Tumors in acute proton-irradiated mice also showed an increased staining of active-β-catenin (p<0.05) relative to unirradiated control. Quantification analysis was performed in six fields of vision from the tumor-free and the tumor-bearing areas captured in each section. Average data from 4 mice are graphically presented (Figure 4B–C).
Figure 4. Prolonged activation of β-catenin in normal colons 100 days after sSPE radiation.
(A) Representative images of β-catenin (brown) stained tumor free (normal) or tumor areas of colon sections from unirradiated control, acute proton, and sSPE irradiated CPC;Apc mice. Hematoxylin (blue) was used for counterstaining. (B) Quantification of total β-catenin staining in tumor free (normal) areas of control, acute proton, and sSPE irradiated sections. (C) Quantification of total β-catenin staining in tumor areas of control, acute proton, and sSPE irradiated sections. Intensity of β-catenin staining was normalized with intensity of hematoxylin staining. At least six fields of vision from the normal and tumor areas were captured in each section and were analyzed. Average data from 4 mice are presented graphically. *, P<0.05 and ****, P<0.001 in two-way ANOVA with Multiple comparisons.
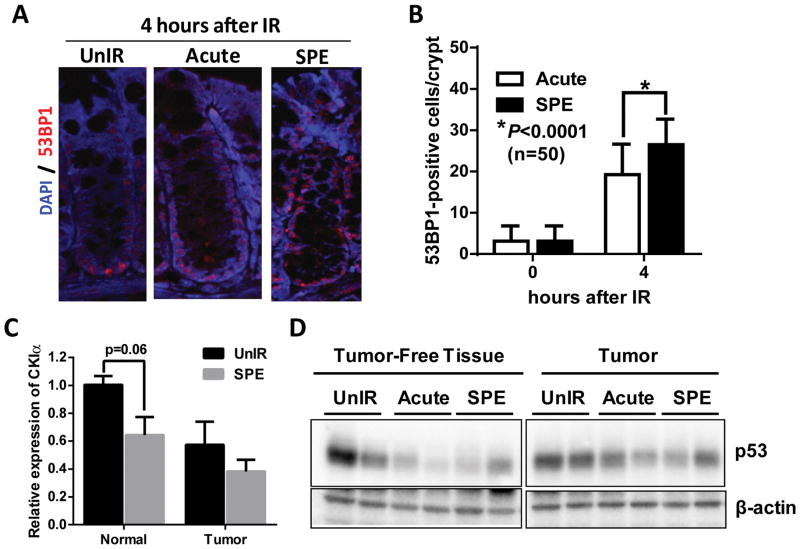
Irradiation with sSPE Increases Expression of Senescence-associated Inflammatory Response (SIR) Genes in Tumor-free Colon
To elucidate the mechanism of sSPE-induced increases in tumor initiation and progression in CPC;Apc mice, the para-inflammation driving gut homeostasis/tumorigenesis model (17) was examined 100 days after irradiation. Loss of Casein Kinase I alpha (CKIα) and/or persistent DNA damage responses trigger Senescence-associated Inflammatory Responses (SIR), a form of para-inflammation, an intermediate between basal homeostasis and chronic inflammation in epithelial cells. SIR results in a breach of homeostasis, hyperproliferation, tumorigenesis and invasive carcinogenesis with additional mutations in the tumor suppressor TP53 (17). CPC;Apc mice exposed to sSPE showed increased DNA damage responses (Figure 5A) and a significantly higher number of 53BP1-positive cells in normal colonic crypts (p<0.0001) relative to acute proton irradiated tissue 4 hours after exposure (Figure 5B). Persistent DNA damage observed up to 4 days after sSPE irradiation (Figure S1) may be due to increase of chronic oxidative stress. To test this we observed higher and persistent oxidative damage in sSPE irradiated tumor-free colon tissues compared to unirradiated or acute proton exposed tissues 100 days after irradiation (Figure S2). Casein kinase I alpha (CKIα), a component of the β-catenin-destruction complex, is involved in a number of cellular processes including DNA repair, cell division, and the Wnt signaling pathway (20). Along with low expression of CKIα in colon tumors, lower expression of CKIα in the sSPE-exposed tumor-free normal tissues was observed relative to unirradiated control (Figure 5C).
Figure 5. sSPE radiation triggers DNA damage, oxidation, and loss of CKIα and p53 expression.
(A) Representative images of 53BP1 immunostaining in normal colonic crypts 4 hours after irradiation. anti-53BP1 (red); DAPI (blue). (B) Quantification of 53BP1-positive cells per crypt demonstrating exposure to sSPE showed higher 53BP1-positive cells 4 hours after irradiation relative to acute proton irradiated groups. n=50 crypts were counted from 3 mice per group. (C) Quantitative RT-PCR shows lower expression of CKIα in normal tissue as well as tumor 100 days after sSPE irradiation relative to unirradiated control. (n=3 mice) (D) Tumor-free normal tissues irradiated with proton radiation show lower expression of p53 relative to unirradiated control.
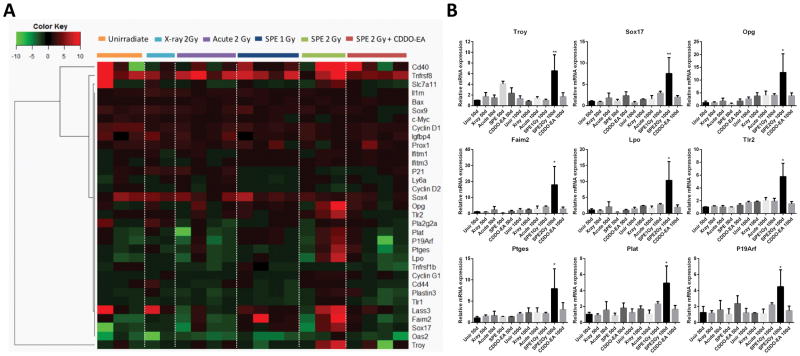
We further examined the expression of senescence-associated inflammatory genes that changed after exposure to sSPE radiation. Tumor-adjacent normal colon tissues were analyzed with qRT-PCR analysis 100 days after radiation exposure. Expression of a small subset of genes was higher in mice irradiated with 2 Gy sSPE exposure relative to unirradiated control. However, these same genes were not expressed at higher levels in 2 Gy x-ray or acute proton irradiated groups (Figure 6A). A subset of SIR genes (Troy, Sox17, Opg, Faim2, Lpo, Tlr2 and Ptges) and one PSIS gene (Plat), along with the senescence associated gene (P19Arf) were markedly increased 100 days after 2 Gy sSPE irradiation (Figure 6A–B). However, higher expression of these genes was not observed in 50 days post 2 Gy sSPE exposure group nor 100 days post 1 Gy sSPE exposure group (Figure S3). Importantly, induction of these para-inflammatory genes was dramatically suppressed by pretreatment of CDDO-EA (Figure 6).
Figure 6. Quantitative RT-PCR analysis of tumor-free distal colons reveals activation of a set of senescence-associated inflammatory response (SIR) genes after sSPE irradiation.
(A) Hierarchical clustering and associated heatmap demonstrating capacity of 33 SIR genes to segregate experimental cohorts. Color bar indicates relative fold change. (B) 9 SIR genes showing significant increase in expression 100 days after sSPE irradiation. (*, P<0.05, ANOVA).
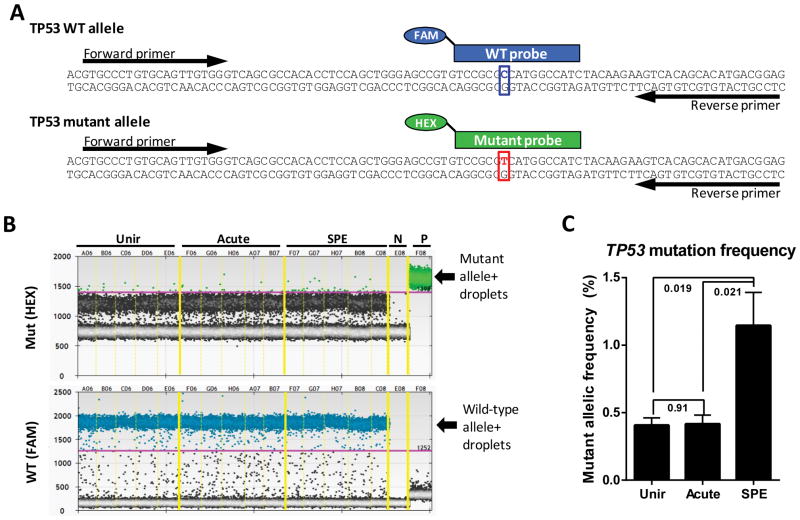
Because the combined alteration of CKIα and p53 triggers high-grade adenocarcinomas with series of gene activations (the p53-suppressed invasiveness signature, PSIS) (20), p53 expression level were assessed in tumors or tumor-free normal tissues. Compared to unirradiated control, both acute proton and sSPE irradiated groups showed lower expression of p53 in tumor-free and also tumor tissues 100 days after irradiation (Figure 5D). To address the long-term effects of sSPE irradiation on TP53 mutagenesis, we examined a specific mouse TP53 mutation using droplet digital PCR (ddPCR) (28), which can detect as little as 0.01% of a TP53 single nucleotide mutation in the wild-type background (Figure S4). Because the mouse TP53 A156V mutation (corresponding to human codon A159) has been reported at the deeply invasive colon adenocarcinoma in this CPC;Apc mouse model (15), TP53 A156V mutation was examined to determine the effects of sSPE irradiation on colorectal cancer progression. We designed a common primer pair and allele-specific TaqMan probes conjugated with different fluorophores that distinguish between the wild-type and A156V mutant TP53 sequences to screen for the mutagenesis events (Figure 7A and Figure S4). We analyzed 5 tumors per group and found that the frequency of the mutant allele differed in each group (Figure 7B). Calculated frequency of the TP53 mutant allele after sSPE exposure showed a significant (P= 0.02) 2.8-fold higher TP53 mutant frequency compared to unirradiated or acute proton irradiated control (Figure 7C).
Figure 7. ddPCR-base detection of a single-base mutation of TP53 in colon tumors.
(A) Strategy for detection of mouse TP53 mutation (p.A156V/c.467C>T) showing a common primer pair and allele-specific probes conjugated with different fluorophores (FAM and HEX) are indicated. (B) ddPCR for the TP53 mutant and wild-type allelic discrimination was performed with colon tumors from unirradiated (Unir), acute proton irradiated (Acute), and sSPE irradiated (SPE) groups. Green and blue dots represent droplets containing the mutant and the wild-type alleles, respectively. Note that while the probes weakly cross-detected the wild-type and mutant alleles, this cross-hybridization can be excluded from quantitative analysis in ddPCR by setting the threshold (pink line) of detection high enough to discriminate true-positive droplets from false-positive droplets, as shown here. N, negative control (no template); P, positive control (mutant template only). (C) Quantificatioin of ddPCR shows the average frequency of the TP53 mutant allele. P values calculated by Student’s t- test (n=5) are shown on the plot.
Discussion
Protons are the most common particle type in space and considered as a potential serious damaging factor for increased risks of cancer and other diseases in astronauts. However, due to unique dosimetric characteristics which can permit a finite range in tissues without an exit dose (3), protons have become widely used for external beam radiation therapy in the treatment of lung cancers, head and neck cancers, prostate cancers, sarcomas, brain tumors, and gastrointestinal cancers (29–33). While clinic-based proton therapy is widely being utilized, the biological long-term side effects/risks of protons have not been as carefully examined. We show, for the first time, the pro-tumorigenic effects of low dose-rate proton radiation in human FAP syndrome susceptible mouse model (CPC;Apc mice). We used 2 Gy dose of proton because radiation therapy of human cancer commonly employs 2 Gy daily fractions of ionizing radiation to achieve the planned total dose (34). In addition, crew members on the International Space Station could receive doses of 2 Gy of proton radiation or more during a solar flare event, even though shielding may partially but not completely reduce these doses (35–36). In this study, SPE simulation (sSPE) was delivered at a low dose-rate (at an average dose rate of 1.67 cGy/min) and acute protons and x-rays were delivered at a higher dose-rate (20 cGy/min). It has been reported that ionizing radiation at low dose-rate exposure can lead to higher DNA damage and cellular toxicity compared to high dose-rate exposure in vitro (37). Here, we observed that sSPE exposure decreased both the number of colonic crypts and overall maximum survival compared to acute proton exposure in wild-type mice (Figure 1). In colon tissues of the CPC;Apc mice, low dose-rate sSPE exposure showed higher acute DNA damage (Figure 5A–B) as well as chronic (100 days post irradiation) oxidative stress (Figure S2) compared to high dose-rate proton or photon exposure. When considering the typical daily dose-rate for proton therapy is about 200 cGy/min (38), we propose that lower daily dose-rate of proton may provide better tumor toxicity during proton therapy.
In addition to acute risk, mid-term (100 days post exposure) or long-term (time of death) risks of low dose-rate proton exposure were determined in the CPC;Apc mice. Exposure to sSPE radiation showed the development of a higher number of polyps compared to unirradiated or acute proton irradiated groups 100 days after exposure. While unirradiated and irradiated mice showed similar numbers of polyps at 50 days post-irradiation, sSPE irradiated mice at 100 days increased the number of polyps more than 2-fold relative to unirradiated control (Figure 2D). sSPE exposure may increase damage in normal non cancerous colonic tissues in addition to increasing the damage to pre-existing hyperplastic foci, but such damage was undetectable at 50 days post-irradiation. However, sSPE exposed mice show an increased cancer burden by 100 days post-irradiation. Besides an increase in the number of tumors (i.e. tumor initiation), tumor size and segmental distribution (i.e. tumor promotion) were changed by sSPE radiation. Exposure to sSPE radiation showed higher tumorigenesis in the middle and proximal colon region, and showed higher numbers of large tumors relative to unirradiated or acute proton/x-ray irradiated groups 100 days after exposure (Figure 2E–F). Cancer progression was accessed by histopathology at different time points, including 100 days after irradiation and at time of death. Higher numbers of adenocarcinomas were observed by both histopathological pit structure and immunohistochemistry analysis in sSPE irradiated mice. Exposure to x-ray and acute proton radiation did not show increased tumor progression 100 days after irradiation (Figure 3B–C). However, at the time of death, a slight increase incidence of invasive adenocarcinoma was observed with x-ray and acute proton exposure, along with significant increase of invasive adenocarcinoma with sSPE exposure (Table 1).
To elucidate the mechanism of sSPE-induced increases in tumor initiation and progression in CPC;Apc mice, para-inflammation driving tumorigenesis (17) was examined. The reduction or absence of CKIα and/or chronic stress triggers low-grade senescence-associated inflammatory responses (SIR) in the mouse intestine (17, 20). With further mutations in TP53, senescent cells bypass growth inhibition checkpoints leading to accelerated colon cancer tumorigenesis and invasiveness in combination with activation of the p53-suppressed invasiveness signature (PSIS) (17, 20). In this study, we demonstrated that exposure to low dose-rate proton (sSPE) resulted in more prolonged DNA damage (Figure 5A–B and Figure S1) and oxidative stress (Figure S2) in the colon tissues relative to acute high dose-rate proton radiation. We also observed that exposure to sSPE decreased expression of CKIα (Figure 5C) and increased the frequency of TP53 mutations in the colon tumors (Figure 7C) (A156V, which has been found in invasive colon carcinomas in this specific genetically engineered mouse model, CPC;Apc (15)). Therefore, a set of SIR genes (Troy, Sox17, Opg, Faim2, Lpo, Tlr2 and Ptges) and one PSIS gene (Plat), along with the senescence associated gene (P19Arf) were observed to be increased in colon tissues exposed to sSPE 100 days post-irradiation (Figure 6). Overexpression of these genes were also observed in colon tumors (data not shown). These results imply that sSPE increases tumorigenesis by inducing SIR and PSIS in the mouse intestine. In contrast to 2 Gy sSPE, acute 2 Gy proton, 2 Gy x-ray and 1 Gy sSPE did not activate these genes (Figure S3), suggesting that these genes might be a novel biomarker of dose- and dose-rate-dependent proton irradiation.
Charged particles are also considered to be one of the major risk factors for humans in space, and have emerged as a critical issue to be resolved for safe long-term missions both orbital and interplanetary. During Solar Particle Events (SPE), significant spikes in the energy and fluence of proton particles from a solar flare increase the risk of astronaut exposure to higher doses of protons. Typically SPEs develop rapidly and may last a few hours to several days emitting low dose-rate irradiation. Moreover, the occurrence, duration and size of individual events are currently unpredictable (39). In the interplanetary space environment, the radiation field consists of 2% electrons, 85.3% protons, 11.8% alpha particles (helium nuclei), and less than 1% of high charge (Z) and energy (E) (HZE) particles. Although HZE particles only account for a small amount of the GCR particle fluxes, they significantly contribute to the biological effects of space radiation (40). While protons may have less biological effects compared to HZE particles, their abundance in the space radiation field needs to be examined much more closely. In addition, HZE particles may also have important implications for carcinogenesis. To address this possibility, we compared the carcinogenic effects of Silicon (28Si) particle with sSPE exposure. Exposure to acute 2 Gy 28Si (600 MeV/n) with a dose-rate of 20 cGy/min showed similar tumor initiation, invasiveness and decreased lifespan as with 2 Gy sSPE irradiated CPC;Apc mice (manuscript in preparation). Though exposure to 2 Gy 28Si showed similar tumorigenic/invasive activity as 2 Gy sSPE, SIR/PSIS gene expression which were induced by sSPE exposure were not activated with 2 Gy 28Si exposure in tumor-free colon tissues, indicating that different types of radiation exposure activate different mechanisms to induce tumorigenesis/invasiveness in vivo (manuscript in preparation). Previously fractionated exposure of 56Fe-particle was reported to enhance lung cancer progression compared to acute 56Fe irradiation (41). In this study, however, fractionated 28Si irradiation reduced cancer progression in the colon. Colonic epithelial cells grow faster compared to bronchial epithelial cells and their turn over time is about 3–4 days. Therefore, our data can be interpreted to indicate that different type of tissues have different mechanisms to suppress or activate tumorigenesis in response to single- or multiple-fractionated radiation exposures.
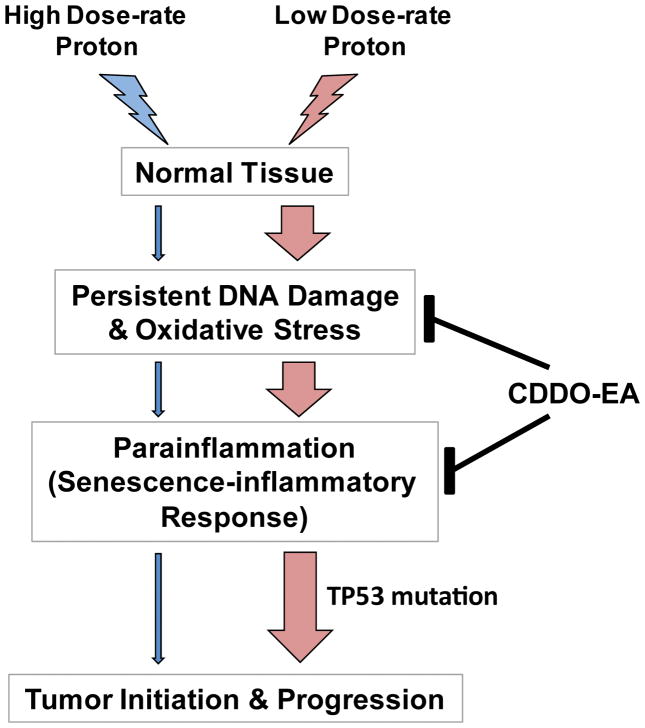
In this study, we demonstrated tumorigenic effects of proton radiation in a colorectal cancer susceptible mouse model with dose- and dose-rate-dependent effects. Exposure to low dose-rate proton (SPE simulation) increased the initiation as well as progression of colorectal cancer by inducing a set of SIR/PSIS genes expression (Figure 8). The SIR genes induced by the low dose-rate proton was different when compared with high dose-rate proton or 28Si-induced tumorigenesis indicating that there is a radiation quality dependency in radiation-induced tumorigenesis mechanisms. While proton therapy in patients is delivered in a highly localized manner, our studies used whole-body irradiation to determine tumorigenic effects. Thus, it is important to note that these present results cannot be extrapolated to the side effects of localized proton therapy in patients. Previously, we have reported that CDDO-EA reduces inflammation and DNA damage against photon irradiation by Nrf2 activation (22). Here, we also demonstrated that pretreatment with CDDO-EA protects mice and reduces tumor initiation and progression against sSPE exposure. CDDO-EA enhanced DNA repair and inhibited induction of SIR/PSIS gene expression after low dose-rate proton exposure. A new synthetic triterpenoid derivative (known as RTA408, Reata Pharmaceuticals) also has been studied as a radioprotector (42) and is currently in clinical trial for preventing radiation-induced dermatitis in patients receiving radiotherapy (NCT02142959). Collectively, these studies imply that exposure to low dose-rate protons activate SIR/PSIS gene expression by prolonged DNA damage and cellular senescence, which induces tumor initiation/progression. A synthetic triterpenoid (CDDO-EA) is a potent radioprotector against proton cancer therapy as well as a potential radioprotector and perhaps mitigator of space radiation.
Figure 8. A schematic model of low dose-rate proton-induced tumorigenesis through para-inflammation.
Exposure to low dose-rate proton induces persistent DNA damage responses and oxidative stress, which triggers senescence-inflammatory response (SIR). In p53-mutated tissues, SIR results in a breach of homeostasis, hyperproliferation, invasion, and carcinogenesis. Nonsteroidal anti-inflammatory drug, CDDO-EA, treatment moderates DNA damage, oxidative stress, and para-inflammation, thus helping to regain homeostasis.
Materials and Methods
Animal husbandry and handling
CDX2P APCflox/+ (CPC;Apc)(15) and C57BL6/J mice were used for the present studies. The colon cancer genetically engineered mouse model was kindly provided by Dr. Eric Fearon (U. of Michigan School of Medicine, Ann Arbor, MI), and a breeding colony was maintained in-house at UT Southwestern Medical Center (UTSW, TX) facility. All animal handling procedure was approved by the Institutional Animal Care and Use Committees (IACUC) of UTSW and Brookhaven National Laboratory (BNL, NY). The mice were housed 1–5 mice per cage and provided with Teklad Global 18% Protein Rodent Diet (#2018). All male and female animals, 5 to 8 weeks of age, were shipped from UTSW to BNL and were acclimated for up to 1 week before irradiation. A subset of control WT and CPC;Apc mice were shipped to BNL to account for stress in shipping and handling. All animals were transported via World Courier’s (New Hyde Park, NY) overnight delivery and returned to UTSW within 1 week after irradiation.
Synthetic triterpenoid
The synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid (CDDO) along with chemically modified derivatives, CDDO-Me (methyl ester, also known as bardoxolone methyl, BARD) and CDDO-EA (ethyl amide) are non-cytotoxic, highly multifunctional and orally available drugs that have been studied as anti-inflammatory and anti-oxidation agents in vivo and in vitro (43–45). Because ethyl amide derivative of a synthetic triterpenoid, CDDO-EA, has been reported to have enhanced pharmacodynamic activity in mouse assays compared to CDDO-Me (46), CDDO-EA (400 mg/kg diet) (provided from Reata Pharmaceuticals, Irving Texas, and Dr. Michael Sporn, Dartmouth Medical School, NH, USA) was first dissolved in an oily vehicle and prepared into chow pellets (Lab Diet #5002) by Purina (Purina-Mills, Richmond, IN, USA) (47). Group of mice were continuously fed a CDDO-EA diet or Control diet (Lab Diet #5002) for 3 days prior to radiation exposure.
Radiation exposure
Mice were irradiated at the NASA Space Radiation Laboratory (NSRL) at BNL and proton dosimetry was calculated by the NSRL physics team. Mice were housed individually in rectangular plastic cuboids boxes (#530C, AMAC Plastic Products, Petaluma, CA) and then whole-body irradiated with 2 Gy of simulated SPE (low dose-rate) or acute 50 MeV/n (high dose-rate) protons. The assembly of 25 animal cubicles was collectively arranged in an array (5 × 5) within the 20 cm × 20 cm beam area. The energy spectrum for the SPE simulation was adapted to mouse dose levels and consisted of 91.67% of the proton dose at energy of 50 MeV/n to 0.14% of the proton dose at 150 MeV/n as described previously (21) (Table S1). The 2 Gy of simulated SPE (low dose-rate proton) was delivered over 2 hours (average dose rate was about 1.67 cGy/min) and acute high dose-rate proton (50 MeV/n) was delivered with a dose-rate of 20 cGy/min. To assure uniform beam penetration through the mice, the sample assembly was rotated 180° in the vertical axis after completion of 25% of the desired dose. Groups of mice were irradiated using the X-RAD 320 irradiator (Precision X-ray, Inc) at UTSW. Mice were held in ventilated 50 ml conical Falcon tubes and placed 50 cm from the radiation source (SSD). A 5 cm diameter brass collimator was placed in the collimator holder. Mice were irradiated with a dose rate of 20 cGy/min.
Polyp counts and sampling
Irradiated mice and age matched sham-irradiated control mice were euthanized 50 or 100 days after radiation exposure. The colon was surgically removed and was flushed gently with phosphate-buffered saline (PBS) at room temperature. The colon was placed on a plate and cut open longitudinally. Polyps were counted by two independent observers. Data from multiple experiments were pooled to achieve statistical significance. Scale was used to measure the polyp size. Polyps were then carefully dissected, flash frozen in liquid nitrogen, and stored at −80°C for further DNA, RNA or protein assays. Adjacent tumor-free normal colon tissues were also dissected. A subset of colonic tissue was fixed overnight in 10% buffered formalin. Fixed tissues were embedded in paraffin and 5-μm thick longitudinal sections were prepared and used for hematoxylin and eosin (H&E) and immunohistochemistry staining.
Invasive adenocarcinoma counts
Irradiated mice and age matched sham-irradiated control mice were sacrificed at the experimental end points. The colon was surgically removed and was flushed gently with phosphate-buffered saline (PBS) at room temperature. The colon was fixed overnight in 10% buffered formalin. Fixed tissues were embedded in paraffin and two longitudinal sections were prepared with a 100 μm interval. H&E staining was performed using standard protocols. A board certified pathologist accessed all sections in each study group for invasive adenocarcinoma classified by penetrating neoplasia through the muscularis mucosa (48).
Immunohistochemistry
Unstained and deparaffinized colon sections were used for immunohistochemistry. Briefly, immunostaining was performed after antigen retrieval using sodium citrate buffer (pH 6.0) in a microwave for 20 min. After quenching endogenous peroxidase activity using 3% hydrogen peroxide for 5 min, the sections were blocked and exposed to the respective primary antibodies. Vectastain Universal ABC kit (Vector Laboratories, Burlingame, CA) and ImmPACT™ DAB kit (Vector Laboratories) were used for signal detection and color development. To determine specificity of the staining, appropriate controls were run in parallel with the experimental sections. Images were captured using bright field microscopy (Axioskop2 Plus, Carl Zeiss, Gottingen, Germany) at a magnification of 20× with 7–12 tumors from 3 mice and were analyzed using ImmumoRation plug-in of ImageJ v1.47 software (National Institutes of Health, Bethedsa, MD). Mean data of average arbitrary pixel unit of DAB color intensity per 20× field were normalized with arbitrary pixel unit of hematoxylin intensity. The following antibodies were used: Active-β-catenin (Cat#05-665, dilution 1:1000, Millipore); Cyclin D1 (Cat# 2978s, dilution 1:100, Cell Signaling); 53BP1 (Cat# ab36823, dilution 1:100, Abcam).
Quantitative RT-PCR
Frozen colon tumors or tumor-free normal colon tissues were homogenized using a liquid nitrogen-cooled mortar (Bel-Art™ Scienceware™, Fisher Scientific) and total RNA was extracted using the RNeasy Mini kit (Qiagen, MD). RNA (1 μg) was subjected to reverse transcription using the First strand cDNA synthesis kit (Roche Molecular Biochemicals, IN) and mRNA expression levels were measured by qRT-PCR using SsoFast™ EvaGrenn® Supermix (Bio-Rad, CA) in a LightCycler 480 II (Roche, IN). Primers for senescence-associated inflammatory response genes are used as described previously (17). Sequences of RT-PCR primers are listed in Table S2. Relative quantities of gene transcripts were calculated and normalized to geometric mean of UBC, TBP, GUSB, and GAPDH transcripts.
Western Blot Analysis
Frozen colon tumors or tumor-free normal colon tissues were homogenized using liquid nitrogen-cooled mortar (Bel-Art™ Scienceware™, Fisher Scientific) and were lysed in ice-cold lysis buffer (50mM Tris pH7.5, 120mM NaCl, 0.5% NP-40, and 1mM EDTA) containing protease and phosphatase inhibitor cocktails (Roche). Lysates were centrifuged and supernatants used for protein assays. Equal amounts of protein was further mixed with 5× SDS-Laemmli buffer and boiled at 95°C for 10 min, followed by a brief centrifugation. Proteins were separated by 4–15% Mini-PROTEAN TGX™ Precast Gel (Bio-Rad, CA), and transferred to a PVDF membrane using the Trans-Blot® Turbo™ Transfer System (Bio-Rad, CA). The following antibodies were used: p53 (Cat# ab26, dilution 1:1,000, Abcam); β-actin (Cat# A1978, dilution 1:20,000, Sigma Aldrich). HRP-conjugated goat anti-mouse or anti-rabbit (Jackson ImmunoResearch) were used as secondary antibodies at 1:5000 dilution and detected with the SuperSignalWest Femto Chemiluminescent Substrate Kit (dilution 1:5, Thermo Scientific) and the gel documentation system, G:BOX (Syngene, Frederick, MD).
Detection of point mutations by droplet digital PCR (ddPCR)
We manually designed the primers and probe using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/). The melting temperature (Tm) of both primers are equal and TaqMan probes Tm is designed 10°C higher than the primer Tm. Optimal primer pair and custom TaqMan probes were purchased from Sigma-Aldrich (Table S3). For optimization of the PCR annealing temperature, we mixed the following reagents: 10 μl 2× ddPCR Supermix for Probes (Bio-Rad), 50 nM FAM probe, 50 nM HEX probe, 500 nM forward primer, 500 nM reverse primer and 10 nM template of a wild-type allele or a mutant allele (20 μl total volume). Droplet generation using a QX100 Droplet Generator (Bio-Rad) was performed as described previously (49). The reaction was transferred into a 96-well PCR plate for gradient PCR on a C1000 Thermal Cycler (Bio-Rad). The thermal cycling program was: step 1, 95°C 10 min; step 2, 95°C 30 s; step 3, 50°C–65°C gradient 30 s; step 4, 72°C 30 s; repeat steps 2–4 39 times; step 5, 98°C 10 min. After the PCR was complete, the droplets were analyzed using a QX200 Droplet Reader (Bio-Rad). We chose the optimal annealing temperature that gave the best separation between negative, FAM-positive and HEX-positive populations. The optimal annealing temperature for the TP53 mutation was 60°C.
To detect point mutagenesis in TP53, we mixed the following reagents in 8-tube PCR strips: 10 μl 2× ddPCR Supermix for Probes (Bio-Rad), 50 nM FAM probe, 50 nM HEX probe, 500 nM forward primer, 500 nM reverse primer and 10 ng genomic DNAs (20 μl total volume). Droplets were generated, which was followed by PCR at the optimal temperature using T100 Thermal Cycler (Bio-Rad); the droplets were analyzed to calculate mutagenesis frequencies. Mutagenesis frequency (%) was calculated as follows:
Cross-hybridization was excluded from quantitative analysis in ddPCR by setting the threshold of detection high enough to discriminate true-positive droplets from false-positive droplets. Our pilot experiment to measure sensitivity of ddPCR to detect single nucleotide mutagenesis using template mixtures showed that we could detect as little as 0.01% of the TP53 mutant allele in the wild-type background (Figure S4).
Supplementary Material
Acknowledgments
We thank the support team at Brookhaven National Laboratory (BNL) and NASA Space Radiation Laboratory (NSRL) (Upton, NY) for helping with the Proton and HZE particles delivery to animals. We thank Dr. Michael Sporn (Hanover, NH) and Reata Pharmaceuticals (Irving, Texas) for providing CDDO-EA reagent, Summer Barron (UT Southwestern, Dallas, TX) for mouse colony maintenance and Gail Fasciani (UT Southwestern, Dallas, TX) for histological processing. This work was performed in laboratories constructed with support from NIH grant C06 RR30414. This work was supported by NASA Grants # NNX15AI21G, NNX11AC15G, NNJ05HD36G and NNX09AU95G to JWS.
Footnotes
Competing interests
JWS is on the SAB of Reata Pharmaceuticals (Irving, Texas).
References
- 1.Verhey LJ, Munzenrider JE. Proton beam therapy. Annu Rev Biophys Bioeng. 1982;11:331–57. doi: 10.1146/annurev.bb.11.060182.001555. [DOI] [PubMed] [Google Scholar]
- 2.Suit HD, Goitein M, Tepper J, Koehler AM, Schmidt RA, Schneider R. Explorotory study of proton radiation therapy using large field techniques and fractionated dose schedules. Cancer. 1975 Jun;35(6):1646–57. doi: 10.1002/1097-0142(197506)35:6<1646::aid-cncr2820350626>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 3.D’angio GJ, Lawrence JH. Medical research with high-energy heavy particles. Nucleonics. 1963;21:56–61. [Google Scholar]
- 4.Lawrence JH, Tobias CA, Born JL, Linfoot JA, Kling RP, Gottschalk A. Alpha and Proton Heavy Particles and the Bragg Peak in Therapy. Trans Am Clin Climatol Assoc. 1964;75:111–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Moravek Z, Bogner L. Analysis of the physical interactions of therapeutic proton beams in water with the use of Geant4 Monte Carlo calculations. Zeitschrift fur medizinische Physik. 2009;19(3):174–81. doi: 10.1016/j.zemedi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Dicello JF. How do we get from cell and animal data to risks for humans from space radiations? J Radiat Res. 2002 Dec;43( Suppl):S1–6. doi: 10.1269/jrr.43.s1. [DOI] [PubMed] [Google Scholar]
- 7.Williams JR, Zhang Y, Zhou H, Osman M, Cha D, Kavet R, et al. Predicting cancer rates in astronauts from animal carcinogenesis studies and cellular markers. Mutat Res. 1999 Dec 6;430(2):255–69. doi: 10.1016/s0027-5107(99)00137-2. [DOI] [PubMed] [Google Scholar]
- 8.George K, Durante M, Willingham V, Wu H, Yang TC, Cucinotta FA. Biological effectiveness of accelerated particles for the induction of chromosome damage measured in metaphase and interphase human lymphocytes. Radiat Res. 2003 Oct;160(4):425–35. doi: 10.1667/rr3064. [DOI] [PubMed] [Google Scholar]
- 9.Rithidech KN, Honikel LM, Reungpatthanaphong P, Tungjai M, Golightly M, Whorton EB. Effects of 100MeV protons delivered at 0.5 or 1cGy/min on the in vivo induction of early and delayed chromosomal damage. Mutat Res. 2013 Aug 30;756(1–2):127–40. doi: 10.1016/j.mrgentox.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004 Aug;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 11.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002 Jan 25;108(2):135–44. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 12.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990 Jun 1;61(5):759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 13.Arnold CN, Goel A, Blum HE, Boland CR. Molecular pathogenesis of colorectal cancer: implications for molecular diagnosis. Cancer. 2005 Nov 15;104(10):2035–47. doi: 10.1002/cncr.21462. [DOI] [PubMed] [Google Scholar]
- 14.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992 May 1;256(5057):668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 15.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007 Oct 15;67(20):9721–30. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 16.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008 Dec 2;6(12):2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pribluda A, Elyada E, Wiener Z, Hamza H, Goldstein RE, Biton M, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 2013 Aug 12;24(2):242–56. doi: 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006 Nov 30;444(7119):633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 19.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011 Feb 17;470(7334):409–13. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 21.Kim SB, Zhang L, Barron S, Shay JW. Inhibition of microRNA-31-5p protects human colonic epithelial cells against ionizing radiation. Life Sciences in Space Research. 2014 Apr 30;:1:67–73. doi: 10.1016/j.lssr.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, et al. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):E2949–55. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SB, Ly P, Kaisani A, Zhang L, Wright WE, Shay JW. Mitigation of radiation-induced damage by targeting EGFR in noncancerous human epithelial cells. Radiat Res. 2013 Sep;180(3):259–67. doi: 10.1667/RR3371.1. [DOI] [PubMed] [Google Scholar]
- 24.Kim SB, Lu Z, Shay JW. Oxygen and Silicon Ion Particles Induce Neoplastic Transformation in Human Colonic Epithelial Cells. Gravitational and Space Research. 2014 Aug 29;2(1):32–41. [Google Scholar]
- 25.Eskiocak U, Kim SB, Roig AI, Kitten E, Batten K, Cornelius C, et al. CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res. 2010 Jul;174(1):27–36. doi: 10.1667/RR2155.1. [DOI] [PubMed] [Google Scholar]
- 26.Kudo S. Endoscopic mucosal resection of flat and depressed types of early colorectal cancer. Endoscopy. 1993 Sep;25(7):455–61. doi: 10.1055/s-2007-1010367. [DOI] [PubMed] [Google Scholar]
- 27.Bahnassy AA, Zekri AR, El-Houssini S, El-Shehaby AM, Mahmoud MR, Abdallah S, et al. Cyclin A and cyclin D1 as significant prognostic markers in colorectal cancer patients. BMC Gastroenterol. 2004 Sep 23;4:22. doi: 10.1186/1471-230X-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nature methods. 2014 Mar;11(3):291–3. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simone CB, 2nd, Rengan R. The use of proton therapy in the treatment of lung cancers. Cancer J. 2014 Nov-Dec;20(6):427–32. doi: 10.1097/PPO.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 30.Ahn PH, Lukens JN, Teo BK, Kirk M, Lin A. The use of proton therapy in the treatment of head and neck cancers. Cancer J. 2014 Nov-Dec;20(6):421–6. doi: 10.1097/PPO.0000000000000077. [DOI] [PubMed] [Google Scholar]
- 31.Pugh TJ, Lee AK. Proton beam therapy for the treatment of prostate cancer. Cancer J. 2014 Nov-Dec;20(6):415–20. doi: 10.1097/PPO.0000000000000083. [DOI] [PubMed] [Google Scholar]
- 32.Keole S, Ashman JB, Daniels TB. Proton therapy for sarcomas. Cancer J. 2014 Nov-Dec;20(6):409–14. doi: 10.1097/PPO.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 33.Eaton BR, Yock T. The use of proton therapy in the treatment of benign or low-grade pediatric brain tumors. Cancer J. 2014 Nov-Dec;20(6):403–8. doi: 10.1097/PPO.0000000000000079. [DOI] [PubMed] [Google Scholar]
- 34.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 35.Parsons JL, Townsend LW. Interplanetary crew dose rates for the August 1972 solar particle event. Radiat Res. 2000 Jun;153(6):729–33. doi: 10.1667/0033-7587(2000)153[0729:icdrft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Townsend LW, Shinn JL, Wilson JW. Interplanetary crew exposure estimates for the August 1972 and October 1989 solar particle events. Radiat Res. 1991 Apr;126(1):108–10. [PubMed] [Google Scholar]
- 37.Collis SJ, Schwaninger JM, Ntambi AJ, Keller TW, Nelson WG, Dillehay LE, et al. Evasion of early cellular response mechanisms following low level radiation-induced DNA damage. J Biol Chem. 2004 Nov 26;279(48):49624–32. doi: 10.1074/jbc.M409600200. [DOI] [PubMed] [Google Scholar]
- 38.Breuer H, Smit BJ. Proton Therapy and Radiosurgery. Springer Science & Business Media; 2013. [Google Scholar]
- 39.Wilson JW, Cucinotta FA, Shinn JL, Simonsen LC, Dubey RR, Jordan WR, et al. Shielding from solar particle event exposures in deep space. Radiat Meas. 1999 Jun;30(3):361–82. doi: 10.1016/s1350-4487(99)00063-3. [DOI] [PubMed] [Google Scholar]
- 40.Hellweg CE, Baumstark-Khan C. Getting ready for the manned mission to Mars: the astronauts’ risk from space radiation. Naturwissenschaften. 2007 Jul;94(7):517–26. doi: 10.1007/s00114-006-0204-0. [DOI] [PubMed] [Google Scholar]
- 41.Delgado O, Batten KG, Richardson JA, Xie XJ, Gazdar AF, Kaisani AA, et al. Radiation-enhanced lung cancer progression in a transgenic mouse model of lung cancer is predictive of outcomes in human lung and breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014 Mar 15;20(6):1610–22. doi: 10.1158/1078-0432.CCR-13-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisman SA, Lee CY, Meyer CJ, Proksch JW, Sonis ST, Ward KW. Topical application of the synthetic triterpenoid RTA 408 protects mice from radiation-induced dermatitis. Radiat Res. 2014 May;181(5):512–20. doi: 10.1667/RR13578.1. [DOI] [PubMed] [Google Scholar]
- 43.Thimmulappa RK, Fuchs RJ, Malhotra D, Scollick C, Traore K, Bream JH, et al. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid Redox Signal. 2007 Nov;9(11):1963–70. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petronelli A, Pannitteri G, Testa U. Triterpenoids as new promising anticancer drugs. Anticancer Drugs. 2009 Nov;20(10):880–92. doi: 10.1097/CAD.0b013e328330fd90. [DOI] [PubMed] [Google Scholar]
- 45.Vannini N, Lorusso G, Cammarota R, Barberis M, Noonan DM, Sporn MB, et al. The synthetic oleanane triterpenoid, CDDO-methyl ester, is a potent antiangiogenic agent. Mol Cancer Ther. 2007 Dec;6(12 Pt 1):3139–46. doi: 10.1158/1535-7163.MCT-07-0451. [DOI] [PubMed] [Google Scholar]
- 46.Liby K, Royce DB, Williams CR, Risingsong R, Yore MM, Honda T, et al. The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res. 2007 Mar 15;67(6):2414–9. doi: 10.1158/0008-5472.CAN-06-4534. [DOI] [PubMed] [Google Scholar]
- 47.Neymotin A, Calingasan NY, Wille E, Naseri N, Petri S, Damiano M, et al. Neuroprotective effect of Nrf2/ARE activators, CDDO ethylamide and CDDO trifluoroethylamide, in a mouse model of amyotrophic lateral sclerosis. Free Radic Biol Med. 2011 Jul 1;51(1):88–96. doi: 10.1016/j.freeradbiomed.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003 Mar;124(3):762–77. doi: 10.1053/gast.2003.50094. [DOI] [PubMed] [Google Scholar]
- 49.Ludlow AT, Robin JD, Sayed M, Litterst CM, Shelton DN, Shay JW, et al. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic acids research. 2014 Jul;42(13):e104. doi: 10.1093/nar/gku439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutter MD. A practical guide and review of colonoscopic surveillance and chromoendoscopy in patients with colitis. Frontline Gastroenterology. 2010 doi: 10.1136/fg.2010.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.