Abstract
E-cadherin is a cell adhesion molecule best known for its function in suppressing tumor progression and metastasis. Here we show that E-cadherin promotes nucleotide excision repair through positively regulating the expression of xeroderma pigmentosum complementation group C (XPC) and DNA damage-binding protein 1 (DDB1). Loss of E-cadherin activates the E2F4 and p130/107 transcription repressor complexes to suppress the transcription of both XPC and DDB1 through activating the TGF-β pathway. Adding XPC or DDB1, or inhibiting the TGF-β pathway, increases the repair of UV-induced DNA damage in E-cadherin-inhibited cells. In mouse skin and skin tumors UVB radiation down-regulates E-cadherin. In sun-associated premalignant and malignant skin neoplasia, E-cadherin is down-regulated in association with reduced XPC and DDB1 levels. These findings demonstrate a crucial role of E-cadherin in efficient DNA repair of UV-induced DNA damage, identify a new link between epithelial adhesion and DNA repair, and suggest a mechanistic link of early E-cadherin loss in tumor initiation.
Keywords: E-cadherin, nucleotide excision repair, XPC, DDB1, TGF-β, UV, ultraviolet radiation
INTRODUCTION
Genomic integrity is constantly threatened by various endogenous and environmental insults. Fortunately, cells are equipped with specialized molecular machineries to correct specific types of DNA damage. In particular, nucleotide excision repair (NER) is a versatile DNA repair pathway that eliminates a wide variety of helix-distorting base lesions induced by environmental carcinogenic sources, including two products induced by solar ultraviolet B (UVB) radiation, cyclobutane pyrimidine dimers (CPD) and (6-4) photoproducts (6-4PP), as well as other bulky adducts induced by air pollutants (1-7).
Mammalian NER consists of two distinct subpathways: global genome NER (GG-NER), which operates throughout the entire genome, and transcription-coupled NER (TC-NER), which specifically removes lesions on the transcribed DNA strand of active genes (4, 5, 8-10). A major difference between these two pathways appears to lie in the strategies for detecting damaged bases and the phenotypes in human patients. Defects in GG-NER cause xeroderma pigmentosum (XP), an autosomal recessive disorder predisposing affected individuals to cancer development not only in the skin, but also in the brain and lungs (4, 5, 9, 11). Seven XP factors (XPA-XPG) have been identified as indispensable NER factors (4, 5, 8-10). Accumulating evidence indicates that XPC plays an essential role in GG-NER-specific damage recognition (12-14). XPC activity in NER requires polyubiquitination by the DDB E3-ligase complex that comprises the DDB1 (DNA damage-binding protein 1), DDB2, CUL4A (Cullin-family E3-ligase adaptor protein) and ROC1 (E3-ligase RING domain) subunits following UV radiation (15-17).
Recent studies from our group and others have demonstrated that the availability and the activity of these NER factors are regulated by endogenous and environmental factors to control the NER capacity (18, 19). Since these XP factors are difficult to target directly, identification of their upstream regulators will provide valuable tools for enhancing NER and therefore tumor suppression.
As a calcium-dependent cell adhesion molecule, E-cadherin is a key component of adherens junctions, structures that are crucial for the maintenance of epithelial integrity (20, 21). It is a calcium-dependent adhesion molecule that mediates homophilic interactions and the formation of catenin-containing complexes (20-22). In cancer, loss of E-cadherin is the best-characterized alteration in the activation of cancer cell invasion and metastasis in multiple cancer types (21, 23, 24), including skin cancer (25-29). However, the role of E-cadherin in DNA repair and tumor initiation remains unknown.
Using mouse models with tissue-specific E-cadherin inactivation, recent studies have shown that loss of E-cadherin induces spontaneous tumorigenesis in the liver (30), skin and mammary gland in the absence of p53 (31), and increases chemical or oncogene-induced liver tumorigenesis (30). Furthermore, ultraviolet radiation (UV) down-regulates E-cadherin in keratinocytes and melanocytes (28, 32). These findings suggest that loss of E-cadherin is an early event and may play an active role in early stage of tumorigenesis. It is possible that the early loss of E-cadherin actively contributes to tumor initiation by impairing UV-induced DNA repair, leading to increased genetic mutations and thus UV-induced tumorigenesis. Here we show that E-cadherin positively regulates UVB-induced DNA repair by promoting the transcription of XPC and DDB1 to facilitate damage recognition.
RESULTS
E-cadherin loss inhibits nucleotide excision repair and down-regulates XPC and DDB1
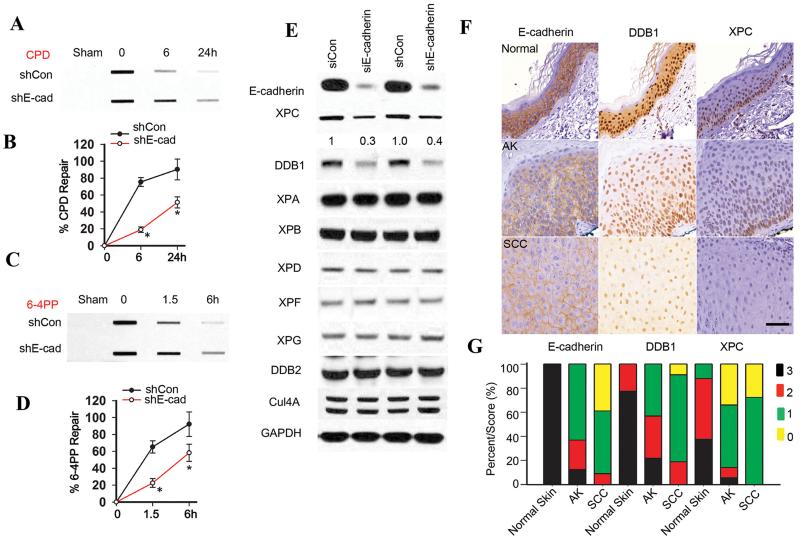
To determine whether E-cadherin has a role in the regulation of NER, we compared NER capability and NER protein levels in control and E-cadherin-inhibited cells. E-cadherin inhibition significantly reduced both CPD repair (Fig. 1A-B) and 6-4PP repair (Fig. 1C-D). As compared with the control group, E-cadherin knockdown by either siRNA or shRNA decreased the levels of XPC and DDB1, while it did not affect other NER factors, including XPA, XPB, XPD, XPF, XPG, DDB2 and Cul4A (Fig. 1E). In addition, as compared with sham-irradiated mouse epidermis, E-cadherin was down-regulated in UVB-irradiated mouse epidermis and tumors (Fig. S1A). Immunohistochemical analysis showed that, as compared with normal human skin, in actinic keratosis (AK) and human squamous cell carcinoma (SCC) the protein levels of E-cadherin, DDB1 and XPC were decreased (score 0 or 1) (Fig. 1F-G). This reduction was statistically significant as analyzed by the Mann-Whitney U test (P < 0.0005 for AK or SCC vs normal skin). The protein levels of XPC and DDB1 were positively correlated with the E-cadherin protein level in human skin and tumors (Fig. S1B) (P < 0.0001). These results indicate that E-cadherin loss inhibits NER and down-regulates XPC and DDB1.
Fig. 1. E-cadherin loss inhibits nucleotide excision repair and down-regulates XPC and DDB1.
(A and C) Slot blot analysis of the levels of CPD (A) and 6-4PP (C) in HaCaT cells transfected with vector (shCon) or shRNA targeting E-cadherin (shE-cadherin, or shE-cad) at 0, 6 and 24 h post-UVB (20 mJ/cm2) for CPD and 0, 1.5 and 6 h post-UVB (20 mJ/cm2) for 6-4PP. (B and D) Quantification of percentage (%) of CPD repair (B) from A and 6-4PP repair (D) from C. *, P < 0.05, compared with shCon group, Student’s t-test and two-way ANOVA. (E) Immunoblot analysis of E-cadherin, XPC, DDB1, XPA, XPB, XPD, XPD, XPF, XPG, DDB2, Cul4a and GAPDH in HaCaT cells transfected with shCon or shE-cadherin, or siRNA targeting E-cadherin (siE-cadherin) or control siRNA (siCon). The results were obtained from three independent experiments. (F) Representative immunohistochemical analysis of E-cadherin, XPC, and DDB1 protein levels (brown) in normal skin (n=14), actinic keratosis (AK) (n=25), and squamous cell carcinoma (SCC) (n=25). Scale bar, 50 μm. (G) Comparison of E-cadherin staining (score) in normal epidermis compared with the panel of AK and SCCs. Statistical analysis was performed using a Mann–Whitney U test.
E-cadherin regulates 6-4PP repair but not CPD repair via increasing XPC availability
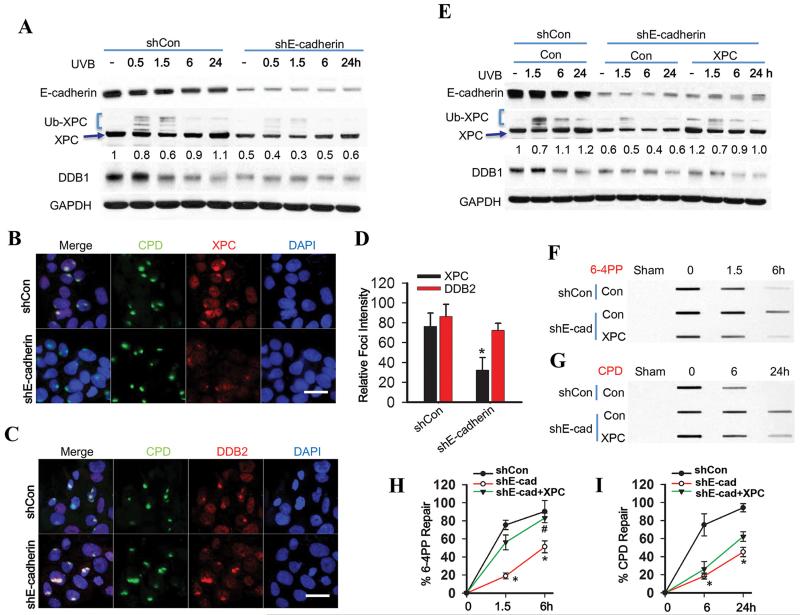
To determine the mechanism by which E-cadherin regulates the repair of CPD and 6-4PP, we assessed the functional significance of XPC and DDB1 regulation by E-cadherin. UVB down-regulated E-cadherin protein levels (Fig. 2A), consistent with previous studies (28). E-cadherin knockdown reduced UVB-induced XPC polyubiquitination (Fig. 2A), a biochemical process critical for DNA damage recognition mediated by the UV-DDB complex (15-17). E-cadherin knockdown inhibited the recruitment of XPC to subnuclear UV-induced CPD sites, while it had no effect on the recruitment of DDB2 to CPD sites (Fig. 2B-D, S2A). It is possible that the recruitment of XPC protein was due to the reduced XPC protein level or the reduced XPC ubiquitination. Our findings are in line with previous reports supporting the recruitment of DDB2 independent of XPC as well as DDB1 (33, 34). In E-cadherin knockdown cells, XPC addition by transient transfection with the pCMV-XPC construct did not affect DDB1 expression or XPC polyubiquitination (Fig. 2E). However, XPC addition increased 6-4PP repair to an extent similar to control cells but did not affect CPD repair (Fig. 2F-I). At the UVB dose used (20 mJ/cm2), UVB did not induce apoptosis, nor did the addition of XPC and/or DDB1 affect post-UVB survival (Fig. S2B). In contrast, in XPC null cells with the same DDB1 protein levels, XPC addition by lentiviral infection significantly increased the repair of both CPD and 6-4PP (Fig. S2C-G). These results demonstrate that E-cadherin regulates 6-4PP repair via increasing basal XPC levels while it may regulate CPD repair via increasing basal XPC levels and activity.
Fig. 2. E-cadherin regulates 6-4PP repair but not CPD repair via increasing XPC availability.
(A) Immunoblot analysis of E-cadherin, XPC, Ub-XPC (polyubiquitinated XPC), DDB1 and GAPDH in HaCaT cells transfected with shCon or shE-cadherin at 0, 0.5, 1.5, 6 and 24 h post-UVB (20 mJ/cm2). The results were obtained from three independent experiments. (B-C) Immunofluorescence assay of the subnuclear colocalization of CPD and XPC (B) and CPD and DDB2 (C) in HaCaT cells transfected with shCon or shE-cadherin at 0.5 h post-UVC (10 mJ/cm2) through a 5 μm micropore filter. Scale bar, 10 μm. (D) The relative intensity of XPC and DDB2 focus was calculated by analyzing 100 foci and normalized to that of CPD (n= 100, bar: SD). (E) Immunoblot analysis of E-cadherin, XPC, DDB1 and GAPDH in HaCaT cells transfected with shCon, shE-cadherin, or the combination of shE-cadherin with XPC plasmids at 0, 1.5, 6 and 24 h post-UVB (20 mJ/cm2). The results were obtained from three independent experiments. XPC protein levels in A and E were quantified using ImageJ software (below each band in arbitrary units). (F-G) Slot blot analysis of 6-4PP (F) and CPD (G) in HaCaT cells transfected with shCon, shE-cadherin, or the combination of shE-cadherin with XPC plasmids at 0, 1.5 and 6 h post-UVB (20 mJ/cm2) for 6-4PP and 0, 6 and 24 h post-UVB (20 mJ/cm2) for CPD. (H and I) Quantification of percentage (%) of 6-4PP repair from F and CPD repair from G. *, P < 0.05, compared with shCon group; #, P < 0.05, compared with shE-cadherin together with shCon groups, Student’s t-test and two-way ANOVA.
E-cadherin regulates CPD repair through increasing the levels of both XPC and DDB1
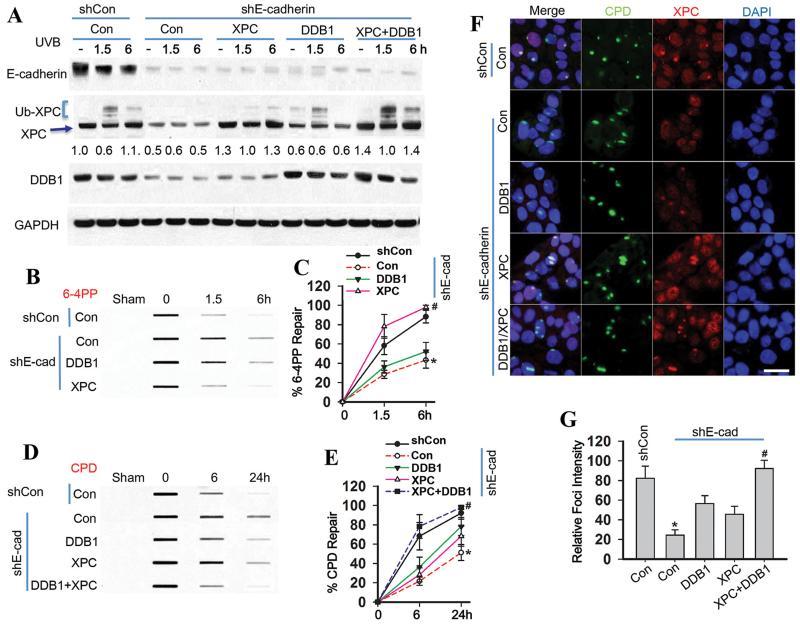
To determine how E-cadherin regulates CPD repair, we assessed the role of both DDB1 and XPC. In E-cadherin-inhibited cells, adding both XPC and DDB1 by transient transfection with the combination of the pCMV-XPC and pcDNA-DDB1 constructs increased XPC polyubiquitination (Fig. 3A) and CPD repair as well as 6-4PP repair (Fig. 3B-E). Immunofluorescence analysis demonstrated that addition of XPC alone did not affect recruitment of XPC to the subnuclear CPD sites, while addition of both XPC and DDB1 restored the recruitment of XPC to CPD sites to a level similar to control cells (Fig. 3F-G). These data demonstrate that both XPC and DDB1 are required for E-cadherin regulation of CPD repair.
Fig. 3. E-cadherin regulates CPD repair through increasing the levels of both XPC and DDB1.
(A) Immunoblot analysis of E-cadherin, XPC, Ub-XPC, DDB1 and GAPDH in HaCaT cells transfected with shCon, shE-cadherin, or the combination of shE-cadherin with vector (Con), XPC, DDB1, or the combination of XPC and DDB1 plasmids at 0, 1.5, 6 and 24 h post-UVB (20 mJ/cm2). The results were obtained from three independent experiments. XPC protein levels in A were quantified using ImageJ software (below each band in arbitrary units). (B) Slot blot analysis of 6-4PP in HaCaT cells transfected with shCon, shE-cadherin, or the combination of shE-cadherin with vector (Con), XPC, or DDB1 plasmids at 0, 1.5 and 6 h post-UVB (20 mJ/cm2). (C) Quantification of percentage (%) of 6-4PP repair from B. *, P < 0.05, compared with shCon group; #, P < 0.05, compared with shE-cadherin/Con groups. (D) Slot blot analysis of CPD in HaCaT cells treated as in B at 0, 6, or 24 h post-UVB. (E) Quantification of percentage (%) of CPD repair from D. *, P < 0.05, compared with shCon group; #, P < 0.05, compared with shE-cadherin/Con groups. (F) Immunofluorescence assay of the colocalization of XPC and subnuclear CPD in HaCaT cells transfected as in B at 0.5 h post-UVC (10 mJ/cm2) through a 5 μm micropore filter. Scale bar, 10 μm. (G) The relative intensity of XPC focus was calculated by analyzing 100 foci and normalized to that of CPD (n= 100, error bar: SD).
The TGF-β pathway but not β-catenin is required for E-cadherin loss to induce inhibition of Nucleotide Excision Repair
To determine the downstream effector pathway in E-cadherin regulation of NER, we first assessed the role of β-catenin, which has been shown to translocate from the plasma membrane to the nucleus and become activated (35). E-cadherin knockdown induced nuclear translocation of β-catenin and the activation of TCF/LEF complex, indicating the activation of the Wnt/β-catenin pathway (Fig. S3A-B). However, in E-cadherin-inhibited cells, β-catenin knockdown had no effect on XPC and DDB1 expression, XPC polyubiquitination (Fig. S3C), or the repair of CPD and 6-4PP (Fig. S3D-G), indicating that E-cadherin regulation of nucleotide excision repair does not require the Wnt/β-catenin pathway.
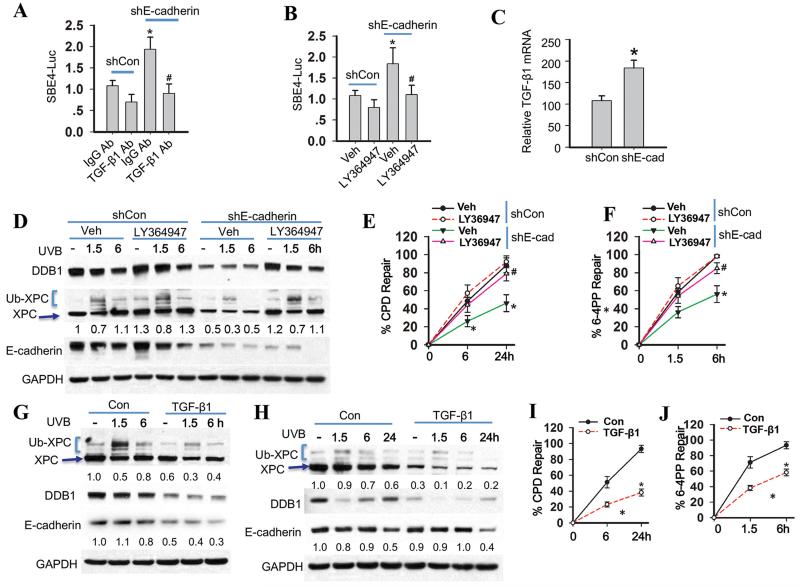
Next we determined the role of the TGF-β pathway, which has been found to be suppressed by E-cadherin (11). As compared with sun-protected normal human skin, in human SCC E-cadherin down-regulation was associated with the nuclear translocation of Smad3 (Fig. S4A), suggesting activation of the TGF-β pathway (36-38). A luciferase reporter assay of the SBE4 promoter showed that E-cadherin knockdown activated the TGF-β pathway, which was sensitive to the pan TGF-β neutralization antibody (Fig. 4A) and the TGF-β pathway inhibitor LY364947 (Fig. 4B). E-cadherin inhibition increased the TGF-β1 mRNA level (Fig. 4C). These data indicate that increased TGF-β1 expression has a role in TGF-β pathway activation by E-cadherin knockdown.
Fig. 4. The TGF-β pathway is required for suppression of nucleotide excision repair by E-cadherin loss.
(A) Luciferase reporter assay of the SBE4 luciferase reporter in HaCaT cells transfected with shCon or shE-cadherin and then treated with normal IgG or anti-TGF-β antibody for 24 h. (mean±S.D. (error bars), n=3; *, p<0.05, compared with the shCon group; #, p<0.05, compared with the shE-cadherin group). (B) Luciferase reporter assay of the SBE4 promoter in HaCaT cells transfected with shCon or shE-cadherin and then treated with vehicle or the TGF-β pathway inhibitor LY364947 (2 μM) for 24 h. (C) Real time RT-PCR analysis of TGF-β1 mRNA levels in HaCaT cells transfected with shCon or shE-cadherin. (D) Immunoblot analysis of E-cadherin, XPC, Ub-XPC, DDB1, and GAPDH in HaCaT cells transfected with shCon or shE-cadherin and then treated with vehicle or LY364947 (2 μM) for 24 h and then collected at 0, 1.5 and 6 h post-UVB (20 mJ/cm2). The results were obtained from three independent experiments. (E-F) Quantification of repair percentage (%) of CPD (E) and 6-4PP (F) in HaCaT cells transfected with shCon or shE-cadherin and then treated with vehicle or TGF-β inhibitor LY364947 (2 μM) for 24 h and collected at 0, 1.5 and 6 h post-UVB (20 mJ/cm2) for 6-4PP and 0, 6 and 24 h post-UVB (20 mJ/cm2) for CPD. *, P < 0.05, compared with shCon group; #, P < 0.05, compared with shE-cadherin/Veh groups. (G) Immunoblot analysis of E-cadherin, XPC, Ub-XPC, DDB1, and GAPDH in HaCaT cells treated with vehicle or TGF-β1 (10 ng/ml) for 48 h and then collected at 0, 1.5 and 6 h post-UVB (20 mJ/cm2). The results were obtained from three independent experiments. (H) Immunoblot analysis of E-cadherin, XPC, Ub-XPC, DDB1, and GAPDH in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 48 h and then collected at 0, 1.5 and 6 h post-UVB (20 mJ/cm2). Protein levels in D, G and H were quantified using ImageJ software (below each band in arbitrary units). (I-J) Quantification of repair percentage (%) of CPD and 6-4PP in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 48 h and then collected at 0, 1.5 and 6 h post-UVB (20 mJ/cm2) for 6-4PP and 0, 6 and 24 h post-UVB (20 mJ/cm2) for CPD. *, P < 0.05, compared with Con groups.
In E-cadherin-inhibited cells, the pan TGF-β neutralization antibody increased the levels of both XPC and DDB1 (Fig. S4B); LY364947 increased the levels of both XPC and DDB1 and polyubiquitination of XPC (Fig. 4D). LY364947 increased the recruitment of XPC to subnuclear CPD sites (Fig. S4C-D) and the repair of both CPD and 6-4PP (Fig. 4E-F). At the UVB irradiation dose used for NER analysis (20 mJ/cm2), LY364947 did not affect UVB-induced apoptosis (Fig. S4E). In HaCaT cells TGF-β1 treatment down-regulated both XPC and DDB1 (Fig. 4G). E-cadherin was also decreased by TGF-β1 treatment (Fig. 4G), consistent with previous studies in HaCaT and other cell lines detecting TGF-β1-induced E-cadherin suppression (29, 39), (40). Similarly, in normal human epidermal keratinocytes (NHEK), TGF-β1 down-regulated XPC and DDB1 (Fig. 4H) and suppressed the repair of both CPD and 6-4PP (Fig. 4I-J). However, in these cells TGF-β1 did not affect the E-cadherin level (Fig. 4H). These results demonstrate that the TGF-β pathway produces phenocopies of E-cadherin loss and is the downstream effector of E-cadherin loss in suppressing NER.
TGF-β pathway inhibits XPC and DDB1 transcription
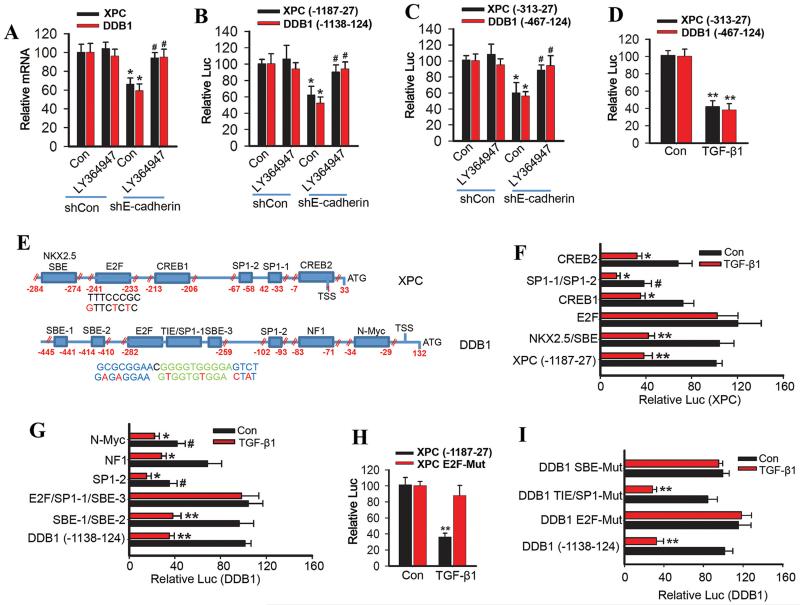
The TGF-β pathway regulates various physiological and pathological responses largely through controlling gene transcription via the Smad proteins (36-38). To determine the mechanism in the regulation of XPC and DDB1 by the E-cadherin/TGF-β pathway, we tested the hypothesis that the TGF-β pathway suppresses the transcription of XPC and DDB1 in E-cadherin-inhibited cells. Indeed, E-cadherin knockdown reduced the mRNA levels of both XPC and DDB1, which was prevented by the TGF-β pathway inhibitor (Fig. 5A).
Fig. 5. TGF-β pathway activation inhibits both XPC and DDB1 transcription.
(A) Real time RT-PCR analysis of XPC and DDB1 mRNA levels in HaCaT cells transfected with shCon or shE-cadherin and then treated with vehicle or LY364947 (2 μM) for 24 h. (B) Luciferase reporter assay of the XPC promoter (−1187-27) and DDB1 promoter (−1138-124) in HaCaT cells transfected with shCon or shE-cadherin and then treated with vehicle or LY364947 (2 μM) for 24 h. (C) Luciferase reporter assay of the XPC promoter (−313-27) and DDB1 promoter (−467-124) in HaCaT cells transfected with shCon or shE-cadherin and treated with vehicle or LY364947 (2 μM) for 24 h. (D) Luciferase reporter assay of the XPC (−313-27) and DDB1 promoter (−467-124) in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (E) Schematic representation of the deletion sites of both XPC and DDB1 promoters and the E2F site of human XPC promoter and E2F/SP1-1/SBE-3 site of human DDB1 promoter. Red nucleotides indicate mutations made in human XPC and DDB1 promoters. (F) Luciferase reporter assay of the XPC promoter with wild-type sequence (−1187-27), or deletion of the CREB2, SP1-1/SP1-2, CREB1, E2F or NKX2.5/SBE element in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (G) Luciferase reporter assay of the DDB1 promoter with wild-type sequence (−1138-124), or deletion of the N-Myc, NF1, SP1-2, E2F/SP1-1(TIE)/SBE-3 and SBE-1/SBE-2 site in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (H) Luciferase reporter assay of the XPC promoter with an intact (−1187-27) XPC or E2F site-mutated XPC in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (I) Luciferase reporter assay of the DDB1 promoter with an intact (−1138-124) DDB1 or mutation of E2F site, SP1-1(TIE) site or SBE-3 site DDB1 in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 24 h.
Next we generated the XPC promoter luciferase construct between the −1187 and 27 positions relative to the XPC TSS (transcription start site) and the DDB1 promoter luciferase construct between the −1138 and 124 positions relative to the DDB1 TSS. E-cadherin knockdown inhibited transcription of both XPC and DDB1, which was prevented by the TGF-β pathway inhibitor (Fig. 5B). Similar results were obtained from the shorter promoters of both XPC (−313-27) and DDB1 (−467-124) (Fig. 5C). TGF-β1 inhibited the transcription of the promoters of both XPC (−313-27) and DDB1 (−467-124) (Fig. 5D). Using the TRANSSERCH and TRANSFAC programs, we predicted several candidate transcription factor-binding elements in the shorter promoters of both XPC (−313-27) and DDB1 (−467-124) (Fig. S5A-B).
Next we generated promoters with deletions of the five candidate transcription factor-binding elements of the XPC promoter (CREB2, SP1-1/SP1-2, CREB1, E2F and NKX2.5/SBE) and the DDB1 promoter (N-Myc, NF1, SP1-2, E2F/SP1-1(TIE)/SBE-3 and SBE-1/SBE-2) identified. As compared with the WT XPC promoter, SP1-1/SP1-2 deletion significantly inhibited basal XPC transcription (Fig. 5E-F). However, as compared with the same WT XPC promoter, deletion of the SP1-1/SP1-2, CREB2, CREB1, or NKX2.5/SBE elements had no effect on TGF-β1-induced suppression of XPC transcription. In contrast, deletion of the E2F site completely blocked TGF-β1-induced suppression of XPC transcription (Fig. 5E-F), indicating that the E2F site in the XPC promoter is required for the regulation of XPC transcription by TGF-β1.
As compared with the WT DDB1 promoter, deletion of the SP1-2 or N-Myc element significantly reduced basal DDB1 transcription (Fig. 5E and G). However, as compared with the WT DDB1 promoter, deletion of the N-Myc, NF1, SP1-2 or SBE-1/SBE-2 elements had no effect on TGF-β1-induced suppression of DDB1 transcription. In contrast, deletion of the E2F/SP1-1(TIE)/SBE-3 site completely blocked TGF-β1-induced suppression of DDB1 transcription (Fig. 5E and G). Site-specific mutation of the XPC E2F element completely blocked TGF-β1-induced suppression of XPC transcription (Fig. 5E and H). In comparison, site-specific mutation of the DDB1 E2F or SBE-3 elements, but not the SP1-1(TIE) element, completely blocked TGF-β1-induced suppression of DDB1 transcription (Fig. 5E and I, S5C). Taken together, these data demonstrate that E-cadherin loss suppresses XPC and DDB1 transcription through activating the TGF-β pathway. TGF-β activation inhibits XPC and DDB1 transcription through the E2F element of the XPC promoter and the E2F and SBE-3 elements of the DDB1 promoter.
The TGF-β pathway promotes binding of the E2F4/P130 complex to the XPC promoter and the E2F4/Smad3/p107 complex to the DDB1 promoter
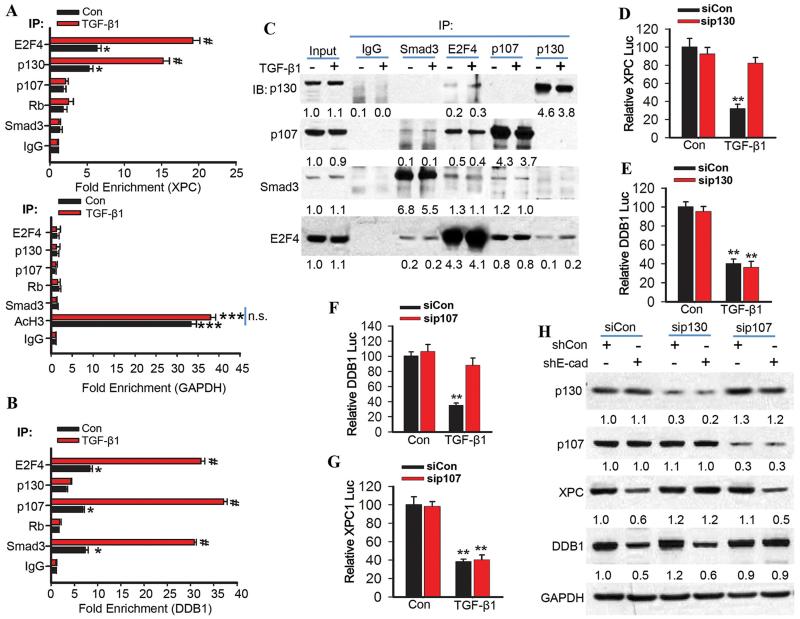
The TGF-β pathway has been demonstrated to induce the transcription-repressing activity of E2F4/5/p107/Smad via the E2F and Smad-binding sites (41). We found that E2F4 and p130 bind to the XPC promoter with the E2F element, and E2F4, p107 and Smad3 bind to the DDB1 promoter with the E2F/SP1-1(TIE)/SBE-3 element (Fig. 6A-B). TGF-β1 significantly increased these bindings (Fig. 6A-B). Immunoprecipitation analysis demonstrated that Smad3 binds with E2F4 and p107 but not p130; TGF-β1 does not affect this binding (Fig. 6C). p130 bound with E2F4 but not Smad3; TGF-β1 increased the E2F-p130 interaction (Fig. 6C).
Fig. 6. The TGF-β pathway promotes binding of the E2F4/P130 complex to the XPC promoter and the E2F4/Smad3/p107 complex to the DDB1 promoter.
(A) Chromatin immunoprecipitation (ChIP) was performed in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 6 h using the indicated antibodies. qPCR was performed with primers specific for the XPC promoter region harboring the E2F site (upper panel) and for the human GAPDH promoter region as negative and positive control (Lower panel; AcH3, acetylated histone H3). (B) Chromatin immunoprecipitations were performed in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 6 h using the indicated antibodies. qPCR was performed with primers specific for the DDB1 promoter region upstream of the E2F/SP1-1/SBE-3 binding site. (C) Immunoprecipitation was performed in NHEK cells treated with vehicle or TGF-β1 (10 ng/ml) for 6 h with the indicated antibodies, followed by immunoblot analysis of p107, p130, Smad3, and E2F4. The results were obtained from three independent experiments. (D and E) Luciferase reporter assay of the XPC (−1187-27) and DDB1 promoter (−1138-124) in NHEK cells transfected with siCon or siRNA target p130 (sip130) and then treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (F and G) Luciferase reporter assay of the DDB1 (−1138-124) and XPC promoter (−1187-27) in NHEK cells transfected with siCon or siRNA target p107 (sip107) and then treated with vehicle or TGF-β1 (10 ng/ml) for 24 h. (H) Immunoblot analysis of p107, p130, XPC, DDB1, and GAPDH in HaCaT cells transfected with shCon or shE-cadherin and then transfected with siCon, sip107 or sip130. Protein levels in C and H were quantified using ImageJ software (below each band in arbitrary units). The results were obtained from three independent experiments.
p130 knockdown blocked TGF-β1-induced suppression of XPC transcription, while it had no effect on DDB1 transcription (Fig. 6D-E). In contrast, p107 knockdown prevented TGF-β1-induced suppression of DDB1 transcription, while it had no effect on XPC transcription (Fig. 6F-G). p130 knockdown also prevented E-cadherin-loss-induced down-regulation of XPC but not DDB1, whereas p107 knockdown blocked E-cadherin-loss-induced down-regulation of DDB1 but not XPC (Fig. 6H). Taken together, these data demonstrate that the TGF-β pathway promotes the binding of E2F4 and p130 to the E2F element in the XPC promoter and thereby suppresses XPC transcription, and enhances the binding of the E2F4/Smad3/p107 complex to the E2F/SP1-2(TIE)/SBE-3 element in the DDB1 promoter and thereby suppresses DDB1 transcription.
DISCUSSION
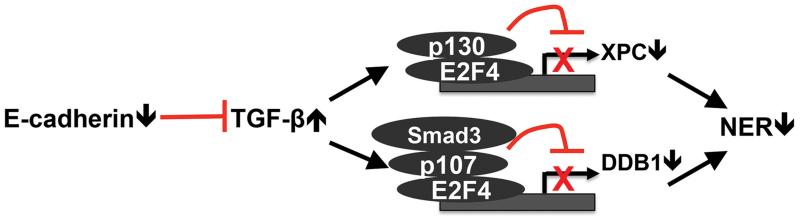
Loss of E-cadherin is the best-studied hallmark in cancer progression and metastasis (21, 23-29). Here we show that E-cadherin is critical for the efficient repair of UVB-induced DNA damage. Loss of E-cadherin impairs UV-induced DNA repair by suppressing the transcription of XPC and DDB1. In human AK and SCC specimens, E-cadherin down-regulation is associated with reduced levels of XPC and DDB1, as compared with normal human skin samples. The activation of the TGF-β pathway mimics E-cadherin loss and is required for the suppression of XPC and DDB1 expression and UV-induced DNA damage repair in E-cadherin-inhibited cells. Our findings elucidate an unexpected role of E-cadherin in DNA damage repair and suggest a molecular mechanism for E-cadherin’s suppressive role in tumor initiation.
Nucleotide excision repair is regulated at multiple levels by extracellular cues and intracellular signaling pathways through regulating the expression or activity of DNA repair factors (18, 19). In this study we found that two NER factors, XPC and DDB1, are transcriptionally regulated by E-cadherin via regulation of different E2F4 transcription repressor complexes. E-cadherin loss increased the enrichment of the E2F4/p130 complex at the XPC promoter. In contrast, at the DDB1 promoter, E-cadherin loss elevated the E2F4/p107/Smad3 complex, a signaling effector complex found in TGF-β-induced suppression of c-Myc (41). The recruitment of Smad3 to the DDB1 promoter is TGF-β1-dependent (Fig. 6B), but its interaction with E2F4/p107 complex is TGF-β1-independent. It is possible that TGF-β1 induces the nuclear translocation of the existing Smad3/E2F4/p107 and thus the binding of this complex to the DDB1 promoter. Indeed, previous reports indicated that the XPC gene is repressed by the E2F4-p130 repressor complex (42), and that disruption of this complex by the tumor suppressor ARF (alternative reading frame) or SIRT1-mediated PTEN deacetylation stimulates XPC transcription (43, 44).
Using a promoter luciferase reporter assay, a recent study showed that the basal DDB1 transcription regulation involves the E2F element, as well as the Sp1, N-Myc, and NF1 elements (45). The E2F sites in the promoters of XPC and DDB1 are required for TGF-β-induced suppression of XPC and DDB1 transcription, whereas the SBE site is only required for DDB1 promoter transcription. Together with previous studies on TGF-β-induced c-Myc suppression, which requires the SBE element (46), our findings suggest a gene-specific regulatory mechanism for TGF-β signaling. In addition to E2F4, E2F5 may also play an important role, as detected in TGF-β-induced c-Myc repression (41). Further investigation can elucidate the specific E2F family members required for XPC and DDB1 regulation by TGF-β. Together with the recent report showing that XPC also positively regulates E-cadherin (47), XPC and E-cadherin may form a positive feedback loop to suppress tumorigenesis and tumor progression.
Although the DDB2 promoter contains a functional E2F element as does the DDB1 promoter (45), DDB2 expression was not affected by E-cadherin loss, suggesting that E-cadherin specifically regulates DDB1. Furthermore, it is noteworthy that E-cadherin promotes 6-4PP repair through regulating XPC, while it promotes CPD repair via regulating the expression of both XPC and DDB1. These distinct mechanisms are supported by the specific requirement of NER factors for 6-4PP and CPD: XPC is required for the repair of both 6-4PP and CPD, whereas the UV-DDB complex including DDB1 is required for efficient repair of CPD, but not 6-4PP (33, 48). Our findings demonstrate a critical molecular link of E-cadherin to nucleotide excision repair.
E-cadherin interacts with multiple proteins and regulates related downstream pathways. E-cadherin is known to recruit β-catenin to the cell membrane and thus prevent β-catenin’s nuclear localization and transactivation (35). However, we found that regulation of nucleotide excision repair by E-cadherin loss does not require β-catenin nuclear translocation and its transactivation (Fig. S3A-G). Instead, we found that E-cadherin inhibition suppresses nucleotide excision repair through activating the TGF-β pathway. E-cadherin knockdown increased the TGF-β1 mRNA level in keratinocytes (Fig. 4C), consistent with the findings in liver cells (49).
TGF-β signaling has a pleotropic and context-dependent role in tumorigenesis and tumor progression (36-38), including in the skin (50). Recent work has shown that deletion of Smad2 or Smad4 increases skin tumorigenesis (51, 52), and Smad4 loss reduces UV-induced DNA repair (52). In contrast, Smad3 deletion inhibits skin tumorigenesis (53). It is possible that suppression of CPD repair by TGF-β1/Smad3 may play an active role in tumor initiation, since CPD but not 6-4PP drives skin tumorigenesis (54). Although TGF-β1 promotes non-homologous end-joining repair and protects cells from ionizing radiation (55), TGF-β1/Smad3 has been shown to inhibit the BRCA1-dependent repair of double-strand breaks induced by mitomycin C (56). In the chemical carcinogenesis model, TGF-β1 enhances early skin tumorigenesis in mice (57); UVB radiation induces TGF-β biosynthesis and activation (58); and TGF-β1 signaling promotes UVB-induced skin tumorigenesis (59), suggesting that TGF-β signaling is oncogenic in the UVB response. Further studies can assess the precise role of the TGF-β signaling and its individual molecular components in the UVB-induced skin tumorigenesis model. Nevertheless, our findings provide a molecular mechanism for TGF-β signaling linking E-cadherin loss to suppression of nucleotide excision repair and suggest a crucial role of the E-cadherin/TGF-β axis in tumor initiation following UVB damage.
We primarily focused on UVB irradiation for the majority of these experiments and only used UVC for the localized UV irradiation experiments, since (1) both UVC and UVB cause the formation of CPD and 6-4PP as the primary DNA damage type, and (2) UVB is clinically relevant to UV-associated human skin damage. However, we were unable to detect subnuclear formation of CPD following local UVB irradiation. It is possible that the Millipore filters block UVC while they may not sufficiently block UVB. Therefore all the localized UV irradiation experiments were performed using UVC but not UVB, as in previous reports (12, 60-64). Further investigation in greater molecular detail will clarify whether the NER process is regulated differently in response to UVC and UVB irradiation.
In summary, our findings demonstrate that E-cadherin positively regulates nucleotide excision repair by promoting the transcription of XPC and DDB1. E-cadherin loss causes the activation of TGF-β signaling, which induces transcription repression of both XPC and DDB1. Our findings may shed light on the previously unrecognized positive role of E-cadherin in DNA repair and tumor initiation.
MATERIALS AND METHODS
All human specimens were studied after approval by the University of Chicago Institutional Review Board. All animal procedures have been approved by the University of Chicago Institutional Animal Care and Use Committee. HaCaT cells (human keratinocytes and epithelial cells, kindly provided by Dr. Fusenig) were used. Detailed descriptions of all methods are provided in SI Methods.
Supplementary Material
Fig. 7. Schematic diagram of E-cadherin regulation of nucleotide excision repair (NER).
ACKNOWLEDGMENTS
We are grateful to Dr. Altaf Wani for his helpful suggestions. We thank Terri Li for immunohistochemistry and Dr. Ann Motten for a critical reading of the manuscript. This work was supported by the NIH/NIEHS grant ES016936 and ES024373 (YYH), the American Cancer Society (ACS) grant RSG-13-078-01 (YYH), the University of Chicago Cancer Research Center (P30 CA014599), the CTSA (UL1 TR000430), and the University of Chicago Friends of Dermatology Endowment Fund.
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest.
REFERENCES
- 1.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. PubMed PMID: 15189136. [DOI] [PubMed] [Google Scholar]
- 2.Niggli HJ, Rothlisberger R. Cyclobutane-type pyrimidine photodimer formation and induction of ornithine decarboxylase in human skin fibroblasts after UV irradiation. J Invest Dermatol. 1988 Dec;91(6):579–84. doi: 10.1111/1523-1747.ep12477095. PubMed PMID: 3192953. eng. [DOI] [PubMed] [Google Scholar]
- 3.Vink AA, Berg RJ, de Gruijl FR, Roza L, Baan RA. Induction, repair and accumulation of thymine dimers in the skin of UV-B-irradiated hairless mice. Carcinogenesis. 1991 May;12(5):861–4. doi: 10.1093/carcin/12.5.861. PubMed PMID: 2029750. eng. [DOI] [PubMed] [Google Scholar]
- 4.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005 Jul;5(7):564–73. doi: 10.1038/nrc1652. PubMed PMID: 16069818. Epub 2005/08/03. eng. [DOI] [PubMed] [Google Scholar]
- 5.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009 Nov;10(11):756–68. doi: 10.1038/nrg2663. PubMed PMID: 19809470. Epub 2009/10/08. eng. [DOI] [PubMed] [Google Scholar]
- 6.Braithwaite E, Wu X, Wang Z. Repair of DNA lesions induced by polycyclic aromatic hydrocarbons in human cell-free extracts: involvement of two excision repair mechanisms in vitro. Carcinogenesis. 1998 Jul;19(7):1239–46. doi: 10.1093/carcin/19.7.1239. PubMed PMID: 9683183. [DOI] [PubMed] [Google Scholar]
- 7.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010 Mar 12;37(5):702–13. doi: 10.1016/j.molcel.2010.02.003. PubMed PMID: 20227373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001 May 17;411(6835):366–74. doi: 10.1038/35077232. PubMed PMID: 11357144. Epub 2001/05/18. eng. [DOI] [PubMed] [Google Scholar]
- 9.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012 Mar;132(3 Pt 2):785–96. doi: 10.1038/jid.2011.426. PubMed PMID: 22217736. Pubmed Central PMCID: 3279615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugasawa K. Xeroderma pigmentosum genes: functions inside and outside DNA repair. Carcinogenesis. 2008 Mar;29(3):455–65. doi: 10.1093/carcin/bgm282. PubMed PMID: 18174245. eng. [DOI] [PubMed] [Google Scholar]
- 11.Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet. 2011 Mar;48(3):168–76. doi: 10.1136/jmg.2010.083022. PubMed PMID: 21097776. Pubmed Central PMCID: 3235003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001 Jul;8(1):213–24. doi: 10.1016/s1097-2765(01)00281-7. PubMed PMID: 11511374. Epub 2001/08/21. eng. [DOI] [PubMed] [Google Scholar]
- 13.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998 Aug;2(2):223–32. doi: 10.1016/s1097-2765(00)80132-x. PubMed PMID: 9734359. eng. [DOI] [PubMed] [Google Scholar]
- 14.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. Embo J. 2003 Oct 1;22(19):5293–303. doi: 10.1093/emboj/cdg489. PubMed PMID: 14517266. Epub 2003/10/01. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005 May 6;121(3):387–400. doi: 10.1016/j.cell.2005.02.035. PubMed PMID: 15882621. eng. [DOI] [PubMed] [Google Scholar]
- 16.Sugasawa K. UV-DDB: a molecular machine linking DNA repair with ubiquitination. DNA Repair (Amst) 2009 Aug 6;8(8):969–72. doi: 10.1016/j.dnarep.2009.05.001. PubMed PMID: 19493704. eng. [DOI] [PubMed] [Google Scholar]
- 17.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006 May;7(5):323–34. doi: 10.1038/nrm1908. PubMed PMID: 16633336. [DOI] [PubMed] [Google Scholar]
- 18.Kim I, He YY. Ultraviolet radiation-induced non-melanoma skin cancer: regulation of DNA damage repair and inflammation. Genes and Diseases. 2014 doi: 10.1016/j.gendis.2014.08.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah P, He YY. Molecular regulation of UV-induced DNA repair. Photochem Photobiol. 2015 doi: 10.1111/php.12406. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003 Feb 21;112(4):535–48. doi: 10.1016/s0092-8674(03)00108-9. PubMed PMID: 12600316. [DOI] [PubMed] [Google Scholar]
- 21.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008 Nov;65(23):3756–88. doi: 10.1007/s00018-008-8281-1. PubMed PMID: 18726070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J Cell Biol. 1994 Dec;127(6 Pt 2):2061–9. doi: 10.1083/jcb.127.6.2061. PubMed PMID: 7806582. Pubmed Central PMCID: 2120290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. PubMed PMID: 21376230. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008 Jun;14(6):818–29. doi: 10.1016/j.devcel.2008.05.009. PubMed PMID: 18539112. [DOI] [PubMed] [Google Scholar]
- 25.Margulis A, Zhang W, Alt-Holland A, Crawford HC, Fusenig NE, Garlick JA. E-cadherin suppression accelerates squamous cell carcinoma progression in three-dimensional, human tissue constructs. Cancer Res. 2005 Mar 1;65(5):1783–91. doi: 10.1158/0008-5472.CAN-04-3399. PubMed PMID: 15753375. [DOI] [PubMed] [Google Scholar]
- 26.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006 Sep 5;45(35):10739–46. doi: 10.1021/bi060423z. PubMed PMID: 16939226. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro P, Gomez M, Pizarro A, Gamallo C, Quintanilla M, Cano A. A role for the E-cadherin cell-cell adhesion molecule during tumor progression of mouse epidermal carcinogenesis. J Cell Biol. 1991 Oct;115(2):517–33. doi: 10.1083/jcb.115.2.517. PubMed PMID: 1918152. Pubmed Central PMCID: 2289150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brouxhon S, Kyrkanides S, O’Banion MK, Johnson R, Pearce DA, Centola GM, et al. Sequential down-regulation of E-cadherin with squamous cell carcinoma progression: loss of E-cadherin via a prostaglandin E2-EP2 dependent posttranslational mechanism. Cancer Res. 2007 Aug 15;67(16):7654–64. doi: 10.1158/0008-5472.CAN-06-4415. PubMed PMID: 17699770. [DOI] [PubMed] [Google Scholar]
- 29.Qiang L, Zhao BZ, Ming M, Wang N, He TC, Hwang S, et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci U S A. 2014;111:9241–6. doi: 10.1073/pnas.1322913111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Nakagawa H, Hikiba Y, Hirata Y, Font-Burgada J, Sakamoto K, Hayakawa Y, et al. Loss of liver E-cadherin induces sclerosing cholangitis and promotes carcinogenesis. Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):1090–5. doi: 10.1073/pnas.1322731111. PubMed PMID: 24395807. Pubmed Central PMCID: 3903249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006 Nov;10(5):437–49. doi: 10.1016/j.ccr.2006.09.013. PubMed PMID: 17097565. [DOI] [PubMed] [Google Scholar]
- 32.Jamal S, Schneider RJ. UV-induction of keratinocyte endothelin-1 downregulates E-cadherin in melanocytes and melanoma cells. J Clin Invest. 2002 Aug;110(4):443–52. doi: 10.1172/JCI13729. PubMed PMID: 12189238. Pubmed Central PMCID: 150409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, et al. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006 Sep 1;66(17):8590–7. doi: 10.1158/0008-5472.CAN-06-1115. PubMed PMID: 16951172. eng. [DOI] [PubMed] [Google Scholar]
- 34.Oh KS, Imoto K, Emmert S, Tamura D, DiGiovanna JJ, Kraemer KH. Nucleotide excision repair proteins rapidly accumulate but fail to persist in human XP-E (DDB2 mutant) cells. Photochem Photobiol. 2011 May-Jun;87(3):729–33. doi: 10.1111/j.1751-1097.2011.00909.x. PubMed PMID: 21388382. Pubmed Central PMCID: 3082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999 Apr;112(Pt 8):1237–45. doi: 10.1242/jcs.112.8.1237. PubMed PMID: 10085258. [DOI] [PubMed] [Google Scholar]
- 36.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012 Oct;13(10):616–30. doi: 10.1038/nrm3434. PubMed PMID: 22992590. Pubmed Central PMCID: 4027049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010 Jun;10(6):415–24. doi: 10.1038/nrc2853. PubMed PMID: 20495575. [DOI] [PubMed] [Google Scholar]
- 38.Fleisch MC, Maxwell CA, Barcellos-Hoff MH. The pleiotropic roles of transforming growth factor beta in homeostasis and carcinogenesis of endocrine organs. Endocr Relat Cancer. 2006 Jun;13(2):379–400. doi: 10.1677/erc.1.01112. PubMed PMID: 16728569. [DOI] [PubMed] [Google Scholar]
- 39.Kao YC, Wu LW, Shi CS, Chu CH, Huang CW, Kuo CP, et al. Downregulation of thrombomodulin, a novel target of Snail, induces tumorigenesis through epithelial-mesenchymal transition. Mol Cell Biol. 2010 Oct;30(20):4767–85. doi: 10.1128/MCB.01021-09. PubMed PMID: 20713448. Pubmed Central PMCID: 2950553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelmann R, Nguyen-Tat MD, Giehl K, Adler G, Wedlich D, Menke A. TGFbeta-induced downregulation of E-cadherin-based cell-cell adhesion depends on PI3-kinase and PTEN. J Cell Sci. 2005 Oct 15;118(Pt 20):4901–12. doi: 10.1242/jcs.02594. PubMed PMID: 16219695. [DOI] [PubMed] [Google Scholar]
- 41.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002 Jul 12;110(1):19–32. doi: 10.1016/s0092-8674(02)00801-2. PubMed PMID: 12150994. [DOI] [PubMed] [Google Scholar]
- 42.Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004 Nov 5;16(3):399–411. doi: 10.1016/j.molcel.2004.09.037. PubMed PMID: 15525513. Epub 2004/11/05. eng. [DOI] [PubMed] [Google Scholar]
- 43.Dominguez-Brauer C, Chen YJ, Brauer PM, Pimkina J, Raychaudhuri P. ARF stimulates XPC to trigger nucleotide excision repair by regulating the repressor complex of E2F4. EMBO Rep. 2009 Sep;10(9):1036–42. doi: 10.1038/embor.2009.139. PubMed PMID: 19644500. Epub 2009/08/01. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22623–8. doi: 10.1073/pnas.1010377108. PubMed PMID: 21149730. Pubmed Central PMCID: 3012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols AF, Itoh T, Zolezzi F, Hutsell S, Linn S. Basal transcriptional regulation of human damage-specific DNA-binding protein genes DDB1 and DDB2 by Sp1, E2F, N-myc and NF1 elements. Nucleic Acids Res. 2003 Jan 15;31(2):562–9. doi: 10.1093/nar/gkg152. PubMed PMID: 12527763. Pubmed Central PMCID: 140516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol. 2004 Mar;24(6):2546–59. doi: 10.1128/MCB.24.6.2546-2559.2004. PubMed PMID: 14993291. Pubmed Central PMCID: 355825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui T, Srivastava AK, Han C, Yang L, Zhao R, Zou N, et al. XPC inhibits NSCLC cell proliferation and migration by enhancing E-Cadherin expression. Oncotarget. 2015 Apr 30;6(12):10060–72. doi: 10.18632/oncotarget.3542. PubMed PMID: 25871391. Pubmed Central PMCID: 4496340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol. 2011 Oct;9(10):e1001183. doi: 10.1371/journal.pbio.1001183. PubMed PMID: 22039351. Pubmed Central PMCID: 3201922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho IJ, Kim YW, Han CY, Kim EH, Anderson RA, Lee YS, et al. E-cadherin antagonizes transforming growth factor beta1 gene induction in hepatic stellate cells by inhibiting RhoA-dependent Smad3 phosphorylation. Hepatology. 2010 Dec;52(6):2053–64. doi: 10.1002/hep.23931. PubMed PMID: 20890948. Pubmed Central PMCID: 3086490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glick AB. The Role of TGFbeta Signaling in Squamous Cell Cancer: Lessons from Mouse Models. J Skin Cancer. 2012;2012:249063. doi: 10.1155/2012/249063. PubMed PMID: 23326666. Pubmed Central PMCID: 3541634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008 Aug;118(8):2722–32. doi: 10.1172/JCI33713. PubMed PMID: 18618014. Pubmed Central PMCID: 2447925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitra D, Fernandez P, Bian L, Song N, Li F, Han G, et al. Smad4 loss in mouse keratinocytes leads to increased susceptibility to UV carcinogenesis with reduced Ercc1-mediated DNA repair. J Invest Dermatol. 2013 Nov;133(11):2609–16. doi: 10.1038/jid.2013.213. PubMed PMID: 23648546. Pubmed Central PMCID: 3783584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li AG, Lu SL, Zhang MX, Deng C, Wang XJ. Smad3 knockout mice exhibit a resistance to skin chemical carcinogenesis. Cancer Res. 2004 Nov 1;64(21):7836–45. doi: 10.1158/0008-5472.CAN-04-1331. PubMed PMID: 15520189. [DOI] [PubMed] [Google Scholar]
- 54.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker AP, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005 Jan 26;15(2):105–15. doi: 10.1016/j.cub.2005.01.001. PubMed PMID: 15668165. eng. [DOI] [PubMed] [Google Scholar]
- 55.Yi JY, Kim MR, Lee J, An YS, Jin YB, Park IC, et al. TGF-beta1 Protects Cells from Gamma-IR by Enhancing the Activity of the NHEJ Repair Pathway. Mol Cancer Res. 2014 Oct 15; doi: 10.1158/1541-7786.MCR-14-0098-T. PubMed PMID: 25319009. [DOI] [PubMed] [Google Scholar]
- 56.Dubrovska A, Kanamoto T, Lomnytska M, Heldin CH, Volodko N, Souchelnytskyi S. TGFbeta1/Smad3 counteracts BRCA1-dependent repair of DNA damage. Oncogene. 2005 Mar 31;24(14):2289–97. doi: 10.1038/sj.onc.1208443. PubMed PMID: 15735739. [DOI] [PubMed] [Google Scholar]
- 57.Perez-Lorenzo R, Markell LM, Hogan KA, Yuspa SH, Glick AB. Transforming growth factor beta1 enhances tumor promotion in mouse skin carcinogenesis. Carcinogenesis. 2010 Jun;31(6):1116–23. doi: 10.1093/carcin/bgq041. PubMed PMID: 20172950. Pubmed Central PMCID: 2878359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Kochevar IE. Involvement of UVB-induced reactive oxygen species in TGF-beta biosynthesis and activation in keratinocytes. Free Radic Biol Med. 2005 Apr 1;38(7):890–7. doi: 10.1016/j.freeradbiomed.2004.12.005. PubMed PMID: 15749385. [DOI] [PubMed] [Google Scholar]
- 59.Ravindran A, Mohammed J, Gunderson AJ, Cui X, Glick AB. Tumor-promoting role of TGFbeta1 signaling in ultraviolet B-induced skin carcinogenesis is associated with cutaneous inflammation and lymph node migration of dermal dendritic cells. Carcinogenesis. 2014 Apr;35(4):959–66. doi: 10.1093/carcin/bgt486. PubMed PMID: 24363069. Pubmed Central PMCID: 3977148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, et al. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J Invest Dermatol. 2001 Nov;117(5):1156–61. doi: 10.1046/j.0022-202x.2001.01540.x. PubMed PMID: 11710927. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi N, Katsumi S, Imoto K, Nakagawa A, Miyagawa S, Furumura M, et al. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001 Apr;14(2):94–102. doi: 10.1034/j.1600-0749.2001.140204.x. PubMed PMID: 11310797. [DOI] [PubMed] [Google Scholar]
- 62.Imoto K, Kobayashi N, Katsumi S, Nishiwaki Y, Iwamoto TA, Yamamoto A, et al. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J Invest Dermatol. 2002 Nov;119(5):1177–82. doi: 10.1046/j.1523-1747.2002.19514.x. PubMed PMID: 12445209. [DOI] [PubMed] [Google Scholar]
- 63.Boyle J, Ueda T, Oh KS, Imoto K, Tamura D, Jagdeo J, et al. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum Mutat. 2008 Oct;29(10):1194–208. doi: 10.1002/humu.20768. PubMed PMID: 18470933. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang QE, Han C, Zhao R, Wani G, Zhu Q, Gong L, et al. p38 MAPK- and Akt-mediated p300 phosphorylation regulates its degradation to facilitate nucleotide excision repair. Nucleic Acids Res. 2013 Feb 1;41(3):1722–33. doi: 10.1093/nar/gks1312. PubMed PMID: 23275565. Pubmed Central PMCID: 3561975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.