PREFACE
Animals assemble and maintain a diverse, yet host-specific gut microbial community. In addition to characteristic microbial compositions along the longitudinal axis of the intestines, discrete bacterial communities form in microhabitats, such as the gut lumen, colon mucus layers and colon crypts. In this Review, we examine how spatial distribution of symbiotic bacteria among physical niches in the gut impacts the development and maintenance of a resilient microbial ecosystem. We consider novel hypotheses for how nutrient selection, immune activation and other mechanisms control the biogeography of bacteria in the gut and discuss the relevance of this spatial heterogeneity to health and disease.
Humans and other mammals harbor a complex gastrointestinal microbiota, which includes all three domains of life (Archaea, Bacteria and Eukaryota). This extraordinary symbiosis, formed via a series of exposures to environmental factors, is initiated upon contact with the vaginal microbiota during birth1. Abrupt changes during the first year of life follow a pattern that corresponds to gestational age in both mice2 and humans3, which suggests that strong deterministic processes shape the composition of the microbiota during development. These population shifts may be explained by influences from diet, the developing immune system, chemical exposures, and potentially founder effects of initial colonizers. Founder effects are not well understood in the mammalian gut, but the profound changes in host gene expression that occur in response to microorganisms, and the great potential for syntrophic interactions between bacteria suggest that early colonizers may have long-term effects on the establishment of the microbiota. The immune system imposes selective pressure on the microbiota through both innate and adaptive mechanisms such as antimicrobial peptides4, secreted immunoglobulin A (IgA)5, and other contributing factors6 (see below). However, current research suggests that diet may have the greatest impact on microbiota assembly.
Prior to weaning, breast milk plays a crucial part in shaping the microbial community composition via transmission of the milk microbiota to the infant gut7, protection from harmful species by secreted maternal antibodies8, and selection for certain species by milk oligosaccharides, which can be used by microorganisms as carbon sources9. For example, in in vitro competitive growth experiments, Bifidobacterium longum benefits from its ability to use fucosylated oligosaccharides that are present in human milk to outgrow other bacteria that are usually present in the gut microbiota, such as Escherichia coli and Clostridium perfringens10. Several species of Bacteroides can also utilize fucosylated oligosaccharides as carbon sources11, suggesting that their colonization may be aided by prebiotic properties of milk. Accordingly, children of mothers with nonfunctional fucosyltransferase 2, an enzyme required for fucosylation of milk oligosaccharides, display lower levels of fecal Bifidobacteria and Bacteroides species12. The importance of diet in determining the composition of the microbial community in the gut is also highlighted by the observation that transition to solid foods coincides with establishment of an adult-like microbiota.
The adult intestinal microbiota consists of hundreds to thousands of species, dominated by the Bacteroidetes and Firmicutes phyla13. This ecosystem is distinct from that of any other microbial habitats that have been surveyed14, and includes many species that exist nowhere else in nature, indicating that coevolution of the host with its gut microbial symbionts (including commensals and mutualists) has generated powerful selective mechanisms. A recent study of how different microbial communities colonize gnotobiotic animals showed that deterministic mechanisms (presumably host-microorganism interactions) led to reproducible shaping of the microbiota regardless of the source of the input community15.
The adult intestinal microbiota is also partially stable, as a core of ~40 bacterial species (accounting for 75% of the gut microbiota in terms of abundance) persists for at least a year in individuals16. A more extensive longitudinal study found that 60% of all bacterial strains within an individual persisted for five years17. During severe perturbations such as antibiotic treatment, the fecal community is depleted to a low-diversity consortium, but after a recovery period membership and relative abundance largely resemble the pretreatment state18. Some species that are depleted to undetectable levels in stool are later recovered18, suggesting that there may be reservoirs of bacterial cells that can re-seed the intestinal lumen.
The mucus layer, crypts of the colon and appendix are examples of privileged anatomical sites, protected from the fecal stream and accessible only to certain microorganisms. In this Review, we highlight relevant features of spatial heterogeneity of bacterial species and communities in the gut microbiota, and discuss the impact of microbial localization in engendering specific and stable colonization with profound implications for health and disease.
MICROBIAL COMPOSITION OF THE GUT
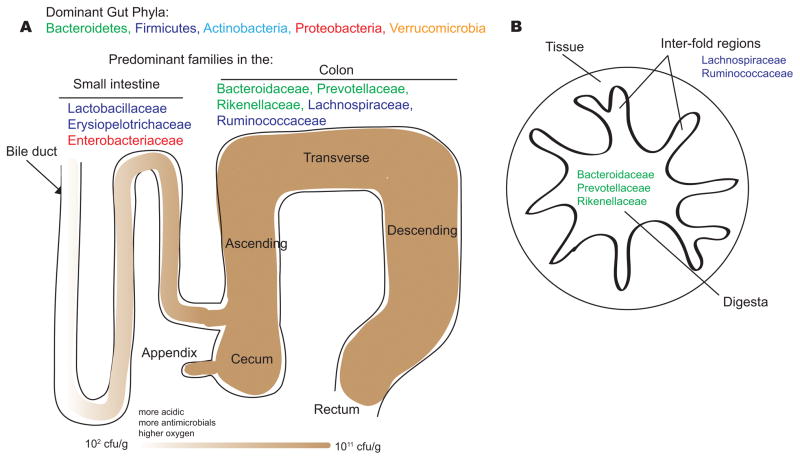
The mammalian lower gastrointestinal tract contains a variety of distinct microbial habitats along the small intestine, cecum, and large intestine (colon). Physiological variation along the lengths of the small intestine and colon include chemical and nutrient gradients, as well as compartmentalized host immune activity, which are known to influence bacterial community composition. For example, the small intestine is more acidic, and has higher levels of oxygen and antimicrobials than the colon (Figure 1A). Therefore, the small intestine microbial community is dominated by fast-growing facultative anaerobes that tolerate the combined effects of bile acids and antimicrobials, while still effectively competing for simple carbohydrates that are available in this region of the gastrointestinal tract. Bile acids, secreted through the bile duct at the proximal end of the small intestine, are bactericidal to certain species due to their surfactant properties and are known to broadly shape the composition of the microbiota, especially in the small intestine. For example, feeding mice excess bile acids generally stimulates the growth of Firmicutes and inhibits Bacteroidetes19. Additionally, the shorter transit time in the small intestine compared to colon (an order of magnitude shorter, despite the increased length of the small intestine) is thought to make bacterial adherence to tissue or mucus an important factor for persistent colonization of the small intestine.
Figure 1. Microbial habitats in the human lower gastrointestinal tract.
The dominant bacterial phyla in the gut are the Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia. The dominant bacterial families of the small intestine and colon reflect physiological differences along the length of the gut. For example, a gradient of oxygen, antimicrobial peptides (including bile acids, secreted by the bile duct), and pH limits the bacterial density in the small intestinal community, whereas the colon carries high bacterial loads. In the small intestine, Lactobacillaceae and Enterobacteriaceae dominate, whereas the colon is characterized by the presence of Bacteroidaceae, Prevotellaceae, Rikenellaceae, Lachnospiraceae and Ruminococcaceae. A cross-section of the colon shows the digesta – which is dominated by Bacteroidaceae, Prevotellaceae and Rikenellaceae – and the inter-fold regions of the lumen – which are dominated by Lachnospiraceae and Ruminococcaceae.
In ileostomy samples from humans, the small intestine was found to exhibit lower bacterial diversity than the colon, and was highly enriched in certain Proteobacteria and Clostridium species20. Furthermore, a metatranscriptomic analysis revealed that the expression of genes involved in central metabolism and in pathways responsible for import of simple sugars by facultative anaerobes was greatly enriched in ileal samples, compared to fecal samples20. In mice, Lactobacillaceae and Proteobacteria (especially Enterobacteriaceae) are enriched in the small intestine21 (Figure 1A). Although bacteria in the small intestine are potentially competing with the host for nutrients, host-derived bile acids and antimicrobial peptides limit bacterial growth to low densities in proximal regions. Only at the distal end of the small intestine (in the terminal ileum) do bacterial densities reach saturating levels similar to those found in the large intestine (Figure 1A).
The cecum and colon cultivate the most dense and diverse communities of all body habitats. Mice, like most herbivorous mammals, have a large cecum between the small and large intestine where plant fibers are slowly digested by the microbiota. Humans have a small pouch-like cecum with an attached appendix, a thin tube-like extension (Figure 1A). In the cecum and colon, microorganisms are responsible for the breakdown of otherwise ‘resistant’ polysaccharides that are not metabolized during transit through the small intestine. Lower concentrations of antimicrobials, slower transit time, and a lack of available simple carbon sources facilitate the growth of fermentative polysaccharide-degrading anaerobes, notably those of the high-abundance families Bacteroidaceae and Clostridiaceae. In the mouse, the cecum is enriched in Ruminococcaceae and Lachnospiraceae, while the colon is enriched in Bacteroidaceae and Prevotellaceae21. Rikenellaceae are prominent in both the cecum and colon21. Various host factors drive community differences over the cross-sectional axis of the gut. The entire wall of the colon folds over itself, creating compartments between folds (inter-fold regions) that are distinct from the central lumenal compartment (Figure 1B). In mouse studies that used laser capture microdissection to profile the composition of the microbial communities in discrete regions, significant differences were observed between the central lumen compartment and the inter-fold region22,23. Specifically, the Firmicute families Lachnospiraceae and Ruminococcaceae were enriched between folds while the Bacteroidetes families Prevotellaceae, Bacteroidaceae, and Rikenellaceae were enriched in the digesta22. Relative to the digesta, the inter-fold regions are likely to contain greater amounts of mucus, which can serve as a nutrient source for certain bacteria.
Gut microhabitats: mucus and colon crypts
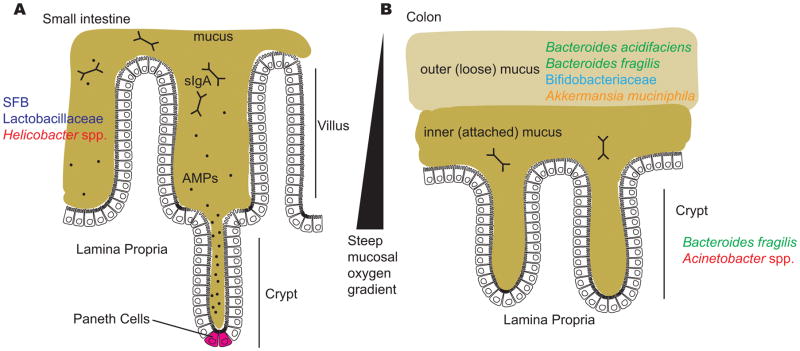
Throughout the human small intestine and colon, specialized epithelial cells called goblet cells secrete a mucus layer of varying thickness that partially or fully covers the epithelium depending on the region, creating a boundary between the gut lumen and host tissue (Figure 2A and 2B). The small intestine harbors a single, tightly-attached mucus layer (Figure 2A), whereas in the colon, mucus is organized into two distinct layers: an outer, loose layer, and an inner, denser layer that is firmly attached to the epithelium (Figure 2B). As mentioned above, bacterial densities are much higher in the colon, compared to the small intestine, and examination of the colon by fluorescence in situ hybridization (FISH) has shown that the inner mucus layer appears essentially sterile next to the densely populated outer layer24. In addition to mucus density itself serving as a physical obstacle for microorganisms, antimicrobial molecules and oxygen secreted from the epithelium accumulate higher local concentrations within the mucosa, especially in the small intestine, greatly restricting potential microbial inhabitants.
Figure 2. The mucus layers of the small intestine and colon.
Several factors limit the ability of gut bacteria to access host cells, including the mucus layers in the small intestine and the colon; antimicrobial peptides in the small intestine, including those produced by Paneth cells at the base of the crypts; secreted immunoglobulin A (sIgA) in both the small intestine and colon; and a steep oxygen gradient that influences which bacteria are capable of surviving close to the epithelial surface. A) The surface of the small intestine is shaped into villi and crypts and is colonized by certain adherent species, including segmented filamentous bacteria (SFB), Lactobacillaceae and Helicobacter spp. B) The colon has two distinct mucus structures: the outer layer is colonized by mucin-degrading bacteria and is characterized by the presence of Bacteroides acidifaciens, Bacteroides fragilis, Bifidobacteriaceae and Akkermansia muciniphila and the inner layer and crypts are penetrated at low density by a more restricted community that includes Bacteroides fragilis and Acinetobacter spp.
Mucus is continuously secreted and the outer layers are sloughed off, generating ‘islands’ of mucus that are carried into the fecal stream25. In mice, a viscosity gradient of the gel-forming mucus increases from the proximal colon (which includes the cecum and the ascending and transverse colon) to distal colonic sites (which includes the descending colon and the sigmoid colon that connects to the rectum). Accordingly, there are more mucus-associated bacteria in the proximal region26. Mucosal biofilm formation in the proximal colon is conserved from mammals to amphibians27, suggesting an ancient, evolutionarily conserved origin of this region for interactions with bacteria. Therefore, the mucus layers of the gastrointestinal tract create environments that are distinct, protected habitats for specific bacterial ecosystems that thrive in proximity to host tissue.
Divergence between the mucosal and digesta-associated colonic communities has been observed in several mammals including humans28, macaques29, mice30, cows31, and flying squirrels32. More specifically, human colon biopsy and swab samples have revealed a distinct mucosal community enriched in Actinobacteria and Proteobacteria compared to the lumen community33. Certain species are highly enriched in colon mucus, such as the mucin-degraders Bacteroides acidifaciens in mice34, Bacteroides fragilis in macaques29, and Akkermansia muciniphila in mice and humans34,35 (Figure 2B). Human mucosal communities in biopsy36–38 and lavage39 samples of the colon contain significant variability between sample locations less than one centimeter apart, suggestive of the existence of mucosal microbial populations in patches. Interestingly, an imaging study using approaches that carefully preserve the structure of feces also identified discrete patches; individual groups of bacteria were found to spatially vary in abundance from undetectable to saturating levels25. This spatial niche partitioning in feces may be reflective of aggregates of interacting microorganisms, heterogeneity of nutrient availability in plant fibers, or microenvironments in mucosally-associated communities that imprint the digesta as it transits through the gut. Therefore, microbial profiling of fecal samples, which is the most common strategy employed in microbiome studies, represents an incomplete and skewed view of even the colon, which has distinct mucosal communities and spatial heterogeneity that is lost upon sample homogenization.
Some bacteria completely penetrate the mucus and are able to associate directly with the epithelium, within the crypts of the colon. Crypt-associated microorganisms were first described using electron microscopy40,41. Many subsequent imaging studies likely failed to observe or underestimated the number of tissue-associated bacteria because common washing and fixing methods can remove mucosal biofilms42. This led to the hypothesis that the mucosal surface is largely devoid of microbial colonization in healthy individuals. However, imaging studies using Carnoy’s fixative, which is known to preserve the mucosal layer, found that there are bacteria in a significant fraction of colonic crypts in healthy mice43 and humans44. More recent work using laser microdissection and sequencing to profile mouse crypt-associated communities revealed that the community is especially dominated by Acinetobacter spp. and is generally enriched for Proteobacteria capable of aerobic metabolism23 (Figure 2B). Evasion of immune responses and particular metabolic activities are likely required for crypt occupancy by microorganisms specialized to reside in close proximity to the host. A well-characterized example of this adaptation is the ability of the human symbiont B. fragilis to enter crypts of the proximal colon of mice via a process requiring both modulation of the immune system45 and utilization of specific host-derived nutrients46 (see below). While dogma has emerged that microorganisms contact mucosal surfaces exclusively in disease states, it appears that life-long physical associations between specific members of the microbiota and their hosts represent symbioses forged over millennia of co-evolution.
MECHANISMS RESPONSIBLE FOR GUT BIOGEOGRAPHY
Several factors influence the biogeography of bacteria within the gut, including diet, antimicrobials, mucus and adherence, and the host immune system.
Diet and nutrients
Bacterial metabolism in the gut likely contributes to the localization of particular groups of microorganisms. Because fatty acids and simple carbohydrates from food are absorbed and depleted during transit through the small intestine, sustainability of the colonic bacterial ecosystem requires growth by fermentation of complex polysaccharides, the principal carbon sources that reach the colon. Best studied in this regard are Bacteroides species, which are able to catabolize polysaccharides derived from the diet and from the host47. Compared to other gut bacteria, Bacteroides have the largest number and diversity of genes involved in polysaccharide degradation48. This extensive array of polysaccharide utilization systems is dominated by those resembling the starch utilization system (Sus), originally described in Bacteroides thetaiotaomicron 49. Sus systems consist of lipid-anchored enzymes either secreted or displayed on the bacterial cell surface that can catabolize particular complex glycans into smaller oligosaccharides, which are then imported through a dedicated outer membrane transporter (Figure 3A). In the gut, Bacteroides species use Sus-like systems to break down dietary polysaccharides and host-derived mucin glycans50. The genome of B. thetaiotaomicron encodes 88 Sus-like systems presumably with different glycan specificities, providing remarkable metabolic flexibility51. Based on these findings, Bacteroides species, and B. thetaiotaomicron in particular, are sometimes referred to as “generalists,” capable of occupying a variety of metabolic niches depending on the availability of diverse polysaccharide nutrients.
Figure 3. Bacterial colonization determinants.
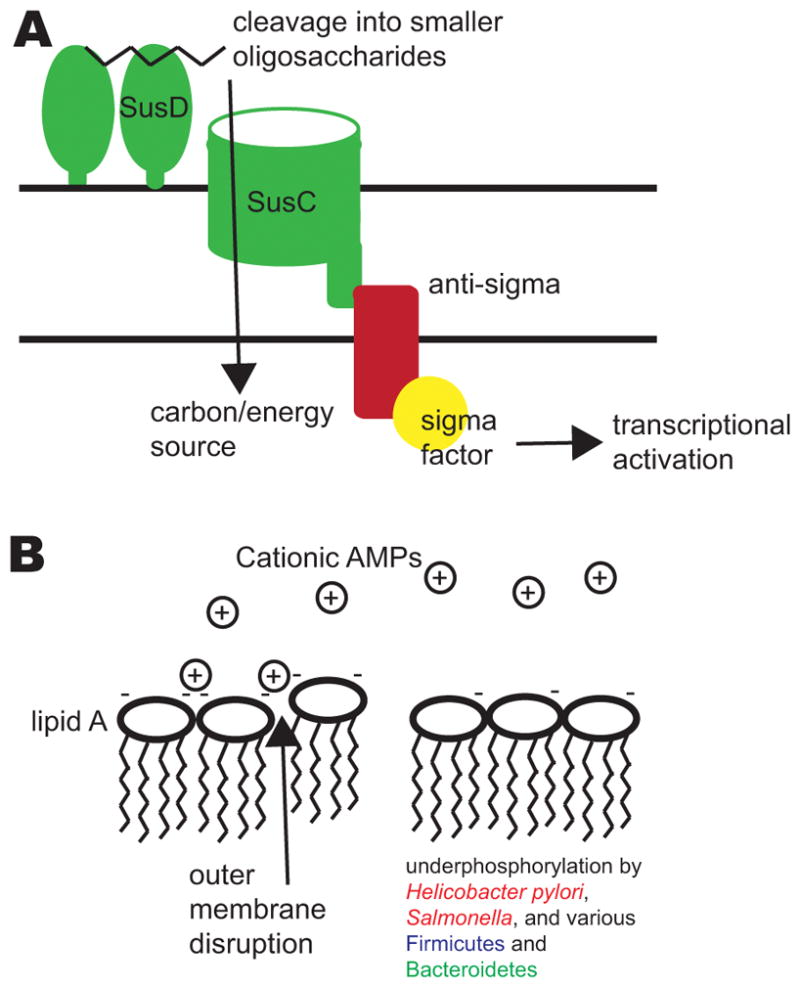
Several factors affect the localization of bacteria within the gastrointestinal tract, including the ability to utilize different glycans and to resist antimicrobial peptides (AMPs). A) Sus-like systems in Bacteroides species allow the utilization of complex polysaccharides from the diet or the host. The figure illustrates a generalized schematic of a Sus-like system. Homologues of SusD and other outer membrane lipid-anchored enzymes bind and cleave the glycans (such as starch) into smaller oligosaccharides that are then imported by the SusC-like outer membrane transporter. Interaction with the cognate glycan often leads to transmembrane signaling to activate gene regulatory mechanisms, such as a two-component system or a transmembrane anti-sigma factor which releases and activates a sigma factor. Downstream transcriptional regulation allows Bacteroides species to respond to local availability of glycans. B) Cationic AMPs in the small intestine, which also pass into the colon via the fecal stream, disrupt bacterial outer membranes by interacting with negative charges on their surface. By removing phosphate groups from lipid A of lipopolysaccharide (LPS), pathogens and commensals alike – such as Helicobacter pylori, Salmonella spp., and various Firmicutes and Bacteroidetes –reduce the negative charge on their membranes and evade attack by cationic AMPs.
Diet-derived polysaccharides control microbial community composition in the lumen of the colon. Unsurprisingly, the influence of diet is readily apparent in studies that profile the fecal community. A study of humans that completely switched between plant and animal-based diets showed that the microbiome abruptly shifts with diet52. Over small time scales this effect is reversible, suggesting that these changes represent transient ecosystem adaptations via blooms of particular species in the lumen while the mucosal reservoir remains unchanged. Many studies of Bacteroides glycan metabolism have shown that restricting the polysaccharide content of the mouse diet allows selection for species (or strains) that are capable of metabolizing the complex glycans present, such as fructans53, human milk oligosaccharides11, fucosylated mucin glycans54, and mannan55. Presumably, the variety of Sus-like systems present in the genomes of Bacteroides provides the metabolic plasticity to persist in the gut despite short and long-term changes in nutrient availability. However, even in terms of monosaccharide and disaccharide utilization, there is a hierarchy of bacteria that are more efficient consumers, which helps explain how diet can dramatically and rapidly change the composition of the fecal community. Importantly, the nutrient environment of the gut lumen may be in a dynamic state of flux due to potential meal-to-meal variability, especially in omnivorous mammals.
In contrast to the variable conditions in the gut lumen, mammals likely maintain a more consistent nutrient balance in the mucosa, which serves as a stable positive selection factor for certain species of bacteria. Mucus degradation and metabolism by gut microorganisms provides access to privileged spatial niches and therefore a competitive advantage over other species, both indigenous and invasive. For example, several studies have shown that the ability to grow in an in vitro mucus culture is generally predictive of the ability of a bacterial species to colonize the mouse gut56,57. MUC2 alone is coated with over 100 different O-linked glycan structures in humans58. These glycans differ between mice and humans59, and differences in complex glycan “preference” by various bacterial species is a suggested mechanism of host-specific selection of a characteristic microbiome profile. In agreement, computational models have shown that positive selection at the epithelium via the ability to metabolize specific nutrients can be a more powerful mechanism for shaping host-associated microbial communities than negative selection driven by antimicrobials60.
A. muciniphila, a prominent symbiont in many mammals, is one of the most effective mucin degraders in vitro35 and is consistently found at high abundance in the mucus layer in humans35 and mice34. Consumption of mucus glycans as a carbon and energy source allows A. muciniphila and other mucin-degraders to colonize the gut independently of the animal’s diet, providing a clear advantage to the bacteria during conditions of nutrient deprivation. Accordingly, levels of A. muciniphila increase in fasting Syrian hamsters61 and hibernating ground squirrels62. Similarly, during intestinal inflammation in mice, the community metatranscriptome indicates increased mucin utilization with a corresponding increase in abundance of the mucin-degrading B. acidifaciens63. In gnotobiotic mice, restriction of complex polysaccharides in the diet causes the generalist B. thetaiotaomicron to shift its metabolism to utilize mucin glycans50. Further work has revealed that mutations in Sus-like systems involved in mucin glycan utilization in B. thetaiotaomicron cause a defect in competitive colonization and in vertical transmission of bacteria from mother to pup64. Therefore, the ability to utilize mucus as a carbon and energy source contributes to the ability of some microorganisms to stably colonize the host and transfer to offspring across generations. Not surprisingly, genetic manipulation of enteric mucus production in mice changes microbial community composition54,65. In turn, gut bacteria affect transcription of mucin-encoding genes in mice66. Overall, development of a healthy mucosa is a collaborative, bi-directional event between the host and the gut microbiota, creating an environment that allows the specific members to establish persistent colonization via utilization of host-derived glycans.
In some cases, the ability of a bacterium to colonize the gut may be determined by its ability to utilize a specific, yet limiting, nutrient. Bacterial species-specific carbohydrate utilization systems termed commensal colonization factors (CCFs) have been identified in B. fragilis and Bacteroides vulgatus, and allow these bacteria to colonize saturable nutrient niches46. This discovery was made based on the observation that gnotobiotic mice colonized with a specific Bacteroides species are resistant to colonization by the same species, but not colonization by closely related species. A genetic screen revealed that a set of genes encoding the CCF system was required for this intra-species colonization resistance phenotype (Box 1), suggesting that CCFs are responsible for defining the species-specific niche. Accordingly, when the ccf genes from B. fragilis were expressed in B. vulgatus, the resulting hybrid strain gained the ability to colonize an alternate niche. The CCF system was also required for penetration of B. fragilis into the crypts of the colon and long-term resilience to intestinal perturbations such as antibiotic treatment and gastroenteritis. Collectively, these data suggest that while metabolic flexibility allows bacterial adaptation in the lumen environment, the occupation of a narrowly-defined, tissue-associated niche is likely very important for stable colonization by some bacteria.
Box 1. Colonization resistance.
One of the benefits afforded by the microbiota to the host is colonization resistance to pathogens. Invasive species of bacteria are inhibited from colonizing the gut because they are unable to displace indigenous species that have gained a strong foothold. After years of studying colonization resistance against pathogens in gnotobiotic animals in the 1960’s and 70’s, Rolf Freter theorized that the ability of a bacterial species to colonize the gut is determined by its ability to utilize a specific, limiting nutrient135. This notion has been well supported by studies showing that colonization resistance to pathogens is mediated by the availability of nutrient niches in the cases of Escherichia coli136 and Clostridium difficile137. But Freter’s hypothesis reached even further, suggesting that the relative amounts of limiting nutrients could dictate the abundance of each species in the indigenous community. Correspondingly, the variety of host-derived growth substrates could explain the stable diversity of the gut microbiota if individual species have evolved to specialize in the uptake and metabolism of specific, limiting nutrients, such as in the case of Bacteroides fragilis46. The concept of spatial niche partitioning being governed by host production of specific and scarce nutrient resources is attractive, and may help explain both long-term persistence and resilience of the microbiome, as well as colonization resistance to pathogens.
Antimicrobials
Specialized epithelial immune cells called Paneth cells reside at the base of the crypts of the small intestine, secreting an array of antimicrobials that restrict the growth of bacteria that are found near the mucosal surface4. Many of these molecules are cationic antimicrobial peptides that interact with and disrupt negatively charged bacterial membranes (Figure 3B). Modifications to lipid A, a major component of the outer membrane of gram-negative bacteria, are known to confer resistance to cationic antimicrobial peptides in several pathogens67. Interestingly, underphosphorylation of this lipid portion of LPS, a modification shared with the pathobiont Helicobacter pylori, was found to be important for resilient colonization by B. thetaiotaomicron during inflammation68 (Figure 3B).
The concentration of a variety of antimicrobials is higher toward the proximal end of the small intestine, creating a gradient that leads to a higher abundance and diversity of bacteria in distal locations (Figure 1A). For example, the lectin RegIIIγ is bactericidal to gram-positive bacteria that dominate the small intestine because it binds to and disrupts their exposed peptidoglycan layer. RegIIIγ is required to prevent massive infiltration of the mucosa and microbial invasion of the tissue69. In addition to RegIIIγ, the innate immune system deploys many other antimicrobials (such as alpha-defensins from Paneth cells and beta-defensins from neutrophils) with differing specificities to limit access to the epithelium70, and resistance to these host-derived antimicrobial peptides is a general feature of many indigenous gut species of Firmicutes and Bacteroidetes68.
In addition to these antimicrobials, gut bacteria, which are largely anaerobic, must contend with reactive oxygen species produced by aerobic host metabolism. Rapid dilution and consumption of oxygen secreted from the host tissue generates a gradient of oxygen that decreases in concentration from tissue to lumen (Figure 2). Accordingly, the mucosal community is enriched in genes required for resistance to reactive oxygen species33. Notably, although all Bacteroides species are classified as obligate anaerobes, B. fragilis can use oxygen as a terminal electron acceptor at nanomolar concentrations71. B. fragilis and tissue-associated microaerophilic Lactobacillaceae express catalase, superoxide dismutase, and other enzymes to inactivate reactive oxygen species72. Altogether, these mechanisms restrict access to the epithelium to a subset of bacterial species that not only can utilize nutrients found only at the tissue boundary, but can survive host antimicrobial strategies as well.
Mucus and adhesion
To access the epithelium, pathogens and commensals alike must contend with the mucus barrier and the immune system (Figure 4). Secreted MUC2 forms peptide crosslinks to create a viscous gel-like substance73, serving as a barrier and host defense mechanism74. In mice lacking MUC2, the crypts of the colon are filled with bacteria and the tissue is covered in biofilms24, indicating that the gel-forming mucus is the primary barrier to tissue association by the microbiota at large. However, certain bacteria are able to penetrate the mucus by swimming or eating their way through.
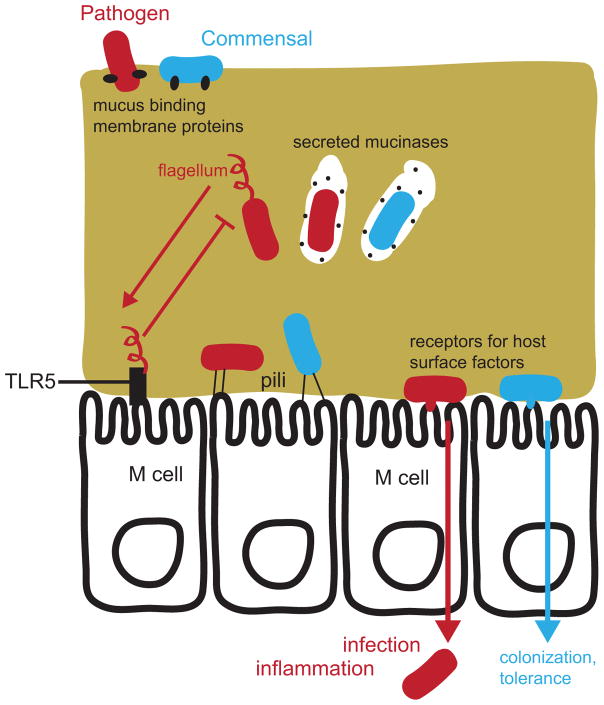
Figure 4. Bacterial access to the epithelium.
Both bacterial pathogens (red) and commensals (or mutualists; blue) have the ability to cross the mucus layer and access the gut epithelium. Lectins and other mucus-binding proteins facilitate initial interactions with the mucus layer. Mucinases and proteases are used to degrade mucus for bacteria to “eat” their way through, while some pathogens such as Salmonella spp. use flagella to swim through the viscous mucus. TLR5 sensing of flagellin effectively leads to inhibition of flagellar biosynthesis for most bacteria in the gut. Adherence to the tissue is achieved by both commensals and pathogens through pili, lectins, and other outer-membrane proteins that target ligands on the epithelial cell surface. Adherence facilitates gut colonization for both commensals and pathogens, and also allows tissue invasion by pathogenic bacteria. Microfold cells (M cells) are specialized immune sentinel epithelial cells that detect gut bacteria and are also exploited by many pathogens as a means of translocation across the epithelium.
In the gut, bacterial motility is generally restricted due to the immunogenicity of flagellin, which is a ligand for Toll-like Receptor 5 (TLR5)75, and the viscosity of mucus limits the effectiveness of swimming (Figure 4). Still, the enteric pathogen Salmonella enterica subspecies enterica serovar Typhimurium depends on flagella and chemotaxis to penetrate the mucus layer and to reach host tissue76. E. coli and close relative Shigella flexneri opt for an alternative strategy of secreting a mucin-binding serine protease, Pic, which rapidly digests mucus (Figure 4). Interestingly, Pic also causes hypersecretion of mucus, which may interfere with the ability of indigenous bacteria to compete with the pathogen77. Similarly, another family of mucus-degrading proteins, M60-like peptidases, are conserved in pathogens and commensal mucosal bacteria from the Proteobacteria, Firmicutes, Bacteroidetes, and other phyla78. In enterotoxigenic E. coli, an M60-like peptidase was required for association with villi in the mouse small intestine79.
In addition to the ability to penetrate the mucus layer, bacterial adhesion to the epithelium also influences the microbial composition of the gut, especially in the small intestine (Figure 2A). Species of Helicobacter adhere to and colonize the the stomach and small intestine tissue via adherence to epithelial surface glycans80. Further downstream in the small intestine, segmented filamentous bacteria (SFB) adhere intimately to the epithelial surface, as first described in imaging studies of mice81. Host-specific strains of SFB appear to be present in many mammals, including humans82. These bacteria were only recently cultured in vitro using tissue-cultured enterocytes as a platform to support their growth, reinforcing the idea that they are obligate symbionts with the mammalian gut tissue83. Their mechanism of attachment is still a mystery, though the attachment site is marked by accumulation of actin and leaves a visible indentation on the surface of the epithelial cell following removal of the filaments83. By virtue of intimate host association, SFB shape the host immune response84 and impact autoimmune disease in mouse models85,86.
The molecular mechanisms underlying how microorganisms attach to host tissue have been well-studied in pathogens (reviewed in Ref 87). Although all of these features were initially discovered and described in pathogens, they are found in many commensal bacteria. Bacteria adhere to mucus and epithelial surfaces by deploying outer membrane proteins, capsules, lectins, adhesins, and fimbriae (attachment pili) (Figure 4). For example, the non-invasive pathogen Vibrio cholerae forms a layer of adhered cells on the wall of the small intestine using toxin-coregulated pili (TCP) 88. V. cholerae also binds mucins using an outer membrane N-acetyl-D-glucosamine (GlcNAc)-binding protein, which may also facilitate penetration of the mucus and access to the epithelium89. Without attachment, these naturally plankton-associated marine bacteria are unable to colonize the gut, and thus are avirulent. E. coli possesses a great number of lectins with diverse sugar specificities allowing it to bind mucins as well as other glycoproteins and extracellular matrix components of epithelial cells90. Invasive pathogens also depend on adherence factors as a preceding step to penetration and infection of the tissue. Listeria monocytogenes expresses a surface protein, internalin A, which binds epithelial E-cadherin (a host cell adhesion protein) as a first step before exploiting actin to induce phagocytosis91. Studies of S. Typhimurium also reveal a critical role of apical surface attachment in inducing neutrophil-mediated inflammation, which appears to paradoxically promote infection92 by providing a competitive advantage for the pathogen over the resident microbiota93.
Beneficial microorganisms also adhere to particular regions of the epithelium and can serve to exclude adherent pathogens by occupying limited binding sites, although little is known about the underlying mechanisms or functions of this process (Box 1). Early imaging studies revealed that Lactobacillus spp. that form adherent layers on the epithelium in the rat stomach prevent yeast94 and staphylococcal95 adherence to the epithelium. Members of the family Lactobacillaceae (such as Lactobacillus and Lactoccocus) that colonize the small intestine and stomach have become model systems for studying adhesion by commensals, with exopolysaccharides, pili, and cell wall-anchored proteins found to be involved in interacting with mucus, extracellular matrix proteins, and other molecular targets on the epithelial cell surface96. Notably, cell wall-anchored mucus-binding proteins (MUBs) unique to lactobacilli are known to be involved in both adherence and aggregation97. Strain-specific diversity in adherence and aggregation factors underlies the host specificity of Lactobacillis reuteri, indicating that tissue-associated biofilm formation is fundamental to colonization by this species98. Other means of attachment involve mechanisms conserved with pathogens, such as adhesive pili in Lactobacillus rhamnosus that bind mucus99. Analogous mechanisms can be found in unrelated species such as Bifidobacterium bifidum, which uses pili to bind extracellular matrix proteins, contributing to bacterial aggregation100.
Collectively, these studies suggest that interactions with mucus and adherence to intestinal epithelial cells appear to be adaptations used by pathogens during infection, as well as strategies employed by commensals during persistent colonization (Figure 4).
Immunomodulation
In order to persist in the gut, non-pathogenic bacteria that intimately associate with host tissue must be tolerated by the immune system. The mucosa is inundated with large amounts of secreted Immunoglobulin A (sIgA) to monitor the microbiota. Many bacteria in the gut are coated in sIgA, and this subpopulation broadly resembles the mucosal population101. Certain adherent species such as Helicobacter spp. and SFB are especially highly coated in sIgA102. Binding of sIgA to bacteria may contribute to mucosal biofilm formation, which serves as a barrier to pathogen adherence103. Gnotobiotic studies with Rag1 knockout mice (which effectively have no adaptive immune system) showed that experimental coating of B. thetaiotaomicron with sIgA reduces microbial fitness but also leads to reduced inflammatory signaling and changes to bacterial gene expression5,104. Through these mechanisms, sIgA mediates homeostasis between the host and the microbiota, as well as potential pathogens at mucosal surfaces. Furthermore, natural antibodies have evolved to recognize bacterial capsular polysaccharides; while largely studied in the context of infectious agents, such antibodies may also represent an evolutionarily conserved strategy used by the host to sense indigenous bacterial species. However, examples on how the immune system can dependably distinguish between harmful and beneficial microorganisms remain limited.
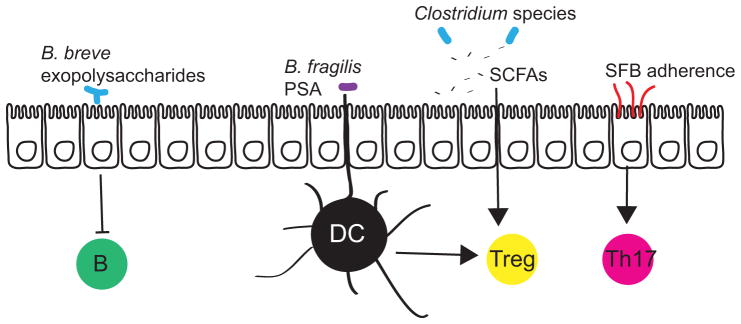
An alternate view is that the immune system is not “hard-wired” to discriminate between various classes of microorganisms, but rather that specific species have adapted to promote their own immunologic tolerance. A few examples of active, species-specific immunomodulation by beneficial microorganisms suggest that some bacteria display signals that ensure their own tolerance by the immune system (Figure 5). B. fragilis is one of the best-understood gut bacteria in terms of immunomodulation. A component of its capsule, polysaccharide A (PSA), signals through an antigen-presenting cell intermediary to stimulate production of IL-10 by an anti-inflammatory subset of immune cells, regulatory T cells105, contributing to the ability of B. fragilis to enter the mucus layer of the colon45 (Figure 5). Surface fucosylation of the bacterial capsule also contributes to B. fragilis fitness in the gut, perhaps by mimicking the host cell surface to elicit a tolerogenic immune response106. Through these specific molecular signals, B. fragilis induces an anti-inflammatory immune profile that facilitates its own colonization. Similarly, exopolysaccharides of Bifidobacterium breve promote immune tolerance by decreasing the production of pro-inflammatory cytokines and preventing a B-cell response107 (Figure 5). Through a less well-defined mechanism, B. breve also induces IL-10 production by regulatory T cells108. Notably, both B. fragilis and species of Bifidobacterium are known to closely associate with the host, which may necessitate immunomodulation to prevent an inflammatory reaction against these bacteria. Similarly, adherent SFB stimulate the development of a subset of T helper cells, Th17 cells, which are required for normal SFB colonization and also confer resistance to the pathogen Citrobacter rodentium84 (Figure 5; Box 1). Clostridia are able to induce regulatory T cells, but a population of many species is much more effective than single isolates or combinations of a few, suggesting this is a combined effect of production of different metabolites, such as short chain fatty acids, by different species (see below)109 (Figure 5). Similarly, a defined community of eight mouse gut bacterial species (including several members of families Clostridiaceae and Lactobacillaceae) referred to as the altered Schaedler’s flora110, was also shown to modulate immune responses mediated by regulatory T cells. Therefore, it is likely that many other beneficial microorganisms have co-evolved with the immune system to facilitate stable long-term colonization.
Figure 5. Immunomodulation by commensal gut bacteria.
Commensal gut bacteria induce immunomodulation via interaction with epithelial cells, antigen presenting cells (such as dendritic cells (DCs)), and via production of signaling metabolites. The exopolysaccharides of adherent Bifidobacterium breve reduce the production of inflammatory cytokines to dampen B cell responses. The capsular polysaccharide PSA of Bacteroides fragilis and short-chain fatty acids (SCFAs) produced by many species of Clostridia (and other genera) stimulate the production of the anti-inflammatory interleukin-10 (IL-10) by regulatory T cells. Segmented filamentous bacteria (SFB) intercalate between microvilli of epithelial cells and stimulate the development of Th17 cells, which are important for mucosal immunity to extracellular pathogens.
Several non-specific signals in the gut also promote tolerance towards beneficial microorganisms. Short chain fatty acids such as butyrate, propionate, and acetate are the end-products of anaerobic fermentation of sugars, which is the dominant metabolism in the colon. The development of regulatory T cells is stimulated by these molecules111,112, which could be a more general way for the immune system to recognize beneficial bacteria or to assess the total fermentative productivity of the community. Mucus is another non-specific anti-inflammatory signal. When MUC2 is taken up by dendritic cells in mice, it inhibits the expression of pro-inflammatory signals113, raising the possibility that indigenous mucin-degraders may induce host tolerance by being co-presented with mucus. Pathogens also have an arsenal of anti-inflammatory mechanisms to suppress the immune system to promote infection114. Particularly perplexing is the fact that features traditionally regarded as virulence factors in pathogens, such as capsular polysaccharides and pili, are also colonization factors in beneficial bacteria. Our notion of the defining characteristics of pathogens is likely clouded by a historical under-appreciation of similar colonization strategies used by beneficial species (Figure 4). It is not surprising that similar mechanisms of host association (mucus penetration, adherence, immune modulation) are used by pathogenic and commensal bacteria alike; however, a key distinguishing feature is that commensals have either not evolved traits resembling traditional virulence factors, or have evolved additional features or modifications to offset the host-response to such factors. This perspective suggests that commensal bacteria may have reached an immunologic and metabolic ‘truce’ with their host, enabling persistent establishment of defined microbial habitats and elaborate microbial biogeographies.
MICRO-BIOGEOGRAPHY IN HEALTH AND DISEASE
Microhabitats in the gut are likely to contribute to the development and stability of microbial communities because spatially stratified niches facilitate greater diversity. In mouse pups, the fecal microbiota is initially dominated by Proteobacteria, a signature of the small intestine, but switches following weaning to Clostridia and Bacteroides, which are characteristic of the adult colon2. The sequential development of the microbiota thus may occur from proximal to distal compartments, which makes sense as dispersal in the gut is largely unidirectional along the fecal stream. Because of this restriction on dispersal, depletion of beneficial species, especially in the colon, could be catastrophic without a mechanism to replenish the community. Therefore, protected regions that are less susceptible to variable conditions in the gut may serve as reservoirs of bacterial cells that can seed growth in the lumen, possibly after an environmental insult (Figure 6). In the case of B. fragilis, mutants that are unable to colonize the crypts of the colon are less resilient to intestinal perturbations such as antibiotic treatment and enteric infection46. This is also a proposed function of the human appendix, which has a mucus and bacteria-filled lumen contiguous with the cecum115. The appendix is protected from the fecal stream, yet harbors a diverse microbial community and a contingent of specialized immune cells. The appendix is also phylogenetically widespread and evolved independently at least twice, providing strong evidence that this is not a vestigial structure as once believed27. In the rabbit appendix, indigenous bacteria coordinate the education of B and T-lymphocytes, suggesting that these tissue-associated niches are venues for immunomodulation116. Microhabitats such as crypts, mucus, and the appendix may be crucial to facilitate immune homeostasis, to protect microbial inhabitants from competitors, and to re-populate the gut following catastrophic perturbations that alter bacterial community structure or deplete certain species from the lumen.
Figure 6. Gut microhabitats as reservoirs of bacterial diversity.
Specific niches such as crypts, the inner mucus, and the appendix may be crucial to facilitate immune homeostasis, to protect microbial inhabitants from competitors, and to re-populate the gut following perturbations that alter bacterial community structure or deplete certain species from the lumen. A) A subset of species is able to penetrate the inner mucus layer and enter crypt spaces. B) Environmental challenges such as diet perturbations, abnormalities in gastrointestinal motility, and antibiotic consumption massively alter the lumen community. However, the more stable mucosal environment and crypts protect important bacterial species. C) The crypts and mucosa serve as reservoirs to repopulate the lumen.
Micro-biogeography alterations during disease
The adverse effects of altered composition of the healthy microbiota, known as dysbiosis, on host health have long been appreciated. Increasing clinical evidence links dysbiosis with various immune, metabolic, and neurological disorders in both intestinal and extra-intestinal sites. For example, inflammatory bowel disease (IBD) is associated with changes in the gut microbiota, characterized by decreased abundance of Clostridia117–119 and overall reduction in bacterial diversity118–120. Childhood asthma is correlated with low intestinal microbial diversity during the first month of life121. The obesity-associated microbiota is characterized by reduced microbial diversity and, in some studies, an increased Firmicutes:Bacteroidetes ratio122. In recent years, the role of gut dysbiosis in the pathogenesis of chronic liver diseases 123,124, colorectal cancer (CRC) 125,126, and even neuropsychiatric dysfunctions127 has been explored in animal models and humans. For clinical applications, profiling of the fecal microbiota has been widely used as a surrogate for the gastrointestinal bacterial community due to non-invasive and straightforward sample collection; however, fecal populations may be less informative than mucosal biopsies in defining disease-associated dysbiosis128, a notion that requires additional experimental support. Below, we detail two examples that illustrate the importance of micro-biogeography alterations of the gut microbiota during disease: IBD and hepatic encephalopathy.
IBD is characterized by inflammation of the gastrointestinal tract resulting in pain, vomiting, diarrhea, and other complications including severe weight loss and behavioral changes. Generally IBD is categorized into two syndromes, Crohn disease, which may involve inflammation throughout the gastrointestinal tract (mouth to anus), and ulcerative colitis, where pathology is restricted to the large intestine. For over a decade, studies have attempted to define a pattern of dysbiosis associated with IBD and yielded inconsistent and sometimes contradicting results129. Studies that focused on fecal microbiota reported wide inter-individual differences in composition with overall microbial diversity reduced118 in Crohn disease patients compared to healthy controls. However, a study in which human gut microbiota were assessed for the ability to drive colitis pathology in mice found that bacteria contributing to the disease are highly coated in sIgA102, suggesting that the mucosal or tissue-associated population is most relevant. Human studies based on biopsy samples elucidated several consistent features in line with this hypothesis: patients had increased concentration of bacteria on the mucosal surface130; decreased microbial diversity; 119,120; decreased abundance of Clostridium species 117; and increased number of Enterobacteriaceae (especially adherent, invasive E. coli) in ileal mucosa131. Most recently, both the lumen- and mucosa-associated microbiota were profiled in a large cohort of new-onset, treatment naïve pediatric patients with Crohn disease and non-IBD controls. Analysis of the mucosal microbiota revealed a significant drop in species richness, an increase in Enterobacteriaceae, a decrease in Clostridiales, and significant changes in several other previously unidentified taxa. Importantly, these dysbiotic signatures were lost when stool samples were examined128. Intriguingly, a laser-capture microdissection study of colon crypt mucus in patients with ulcerative colitis found that they had lower levels of crypt-associated bacteria132. Overall, these studies highlight that distinguishing between fecal and mucosal microbial communities is particularly important for finding a reproducible microbial signature of IBD. Moving from correlations to a potential causal etiology of the microbiota for IBD and other disorders will require further study of mucosal communities, focusing on the interactions between the host and microbiota.
Biogeographical changes in the gut microbiota may also influence liver function. Hepatic encephalopathy is a neuropsychiatric complication of cirrhosis and direct sequelae of gut dysbiosis. As a result of impaired liver function and the presence of porto-systemic shunts (bypass of the liver by the circulatory system), toxic metabolites produced by the gut microbiota evade liver catabolism and cross the blood-brain barrier, leading to cerebral toxicity123. Interestingly, a comparison of the fecal microbiota of cirrhotic patients with and without hepatic encephalopathy showed minimal differences, whereas analyzing the microbiota composition of the colonic mucosa revealed significant changes, including lower Roseburia and higher Enterococcus, Veillonella, Megasphaera, Burkholderia, and Bifidobacterium in cirrhotic patients with hepatic encephalopathy133. The bacterial genera over-represented in the mucosa of patients with hepatic encephalopathy (Megaspheara, Veillonella, Burkholderia, and Bifidobacterium) were also correlated with poor cognition, higher inflammation, and higher clinical severity score. In summary, mucosal dysbiosis in the gut, but not in the composition of the fecal community, significantly correlates with the severity of chronic liver disease phenotypes, including hepatic encephalopathy.
CONCLUSION
We have highlighted evidence that the microbiota is biogeographically stratified within the gut on different spatial scales and axes. Progress towards a functional understanding of the microbiota necessitates increased attention to microhabitats within the gut ecosystem, and to spatial relationships between microorganisms and between microorganisms and the host. Fecal community profiling enabled by next-generation sequencing provides a valuable picture of the diversity, specificity, stability, and developmental dynamics of the gut microbiota, but focusing on measurements of abundance in feces neglects the importance of mucus and tissue-associated organisms and cannot account for spatial distributions. Similarly, studies in gnotobiotic animals allow a reductionist approach to studying host-microorganism interactions akin to methods traditionally employed by microbiologists studying pathogens, but this simplified methodology is likely to miss important contributions from interspecies interactions. The functional study of gut microbial ecology using “meta-omics” techniques enables one to account for the behaviors of the community as a whole, but attributing functions to particular microbial members remains a challenge in community-level ecology. Therefore, testing unifying hypotheses using both reductionist and ecological approaches will be essential to our understanding of the microbiota and its biological functions.
More than half a century ago, in “Microorganisms Indigenous to Man,” the microbiologist Theodor Rosebury lamented on the lack of a general theory for influences that control composition of the microbiota, the roles of individual members, and functions that affect the host134. With the true complexity of the problem revealed recently by sequencing advances, research is only now in a position to fulfill Rosebury’s call for a general theory. Rolf Freter’s nutrient niche hypothesis135, which states that limiting nutrients control the population level of species that are particularly adept at utilizing them, provides a metabolic foundation to explain some of the nascent observations in the field. But when Freter proposed his ideas, we were unaware of the role of immunomodulation by non-pathogens, which requires access to the tissue. Based on evidence outlined in this review, we propose that the host presents limiting nutrients as well as attachment sites in privileged locations. Furthermore, the immune system has an active role in allowing only beneficial species to access these locations during homeostasis. Selection for particular species close to the epithelium creates protected, stable reservoirs for microorganisms to persist in the face of rapidly changing conditions in the gut lumen. Through localized, immune-facilitated, and adherence-dependent nutrient selection, the host maintains stability of a diverse community of microbial symbionts.
Online summary.
The gut microbiota is spatially stratified along the longitudinal and cross-sectional axes of the gut. Chemical and nutrient gradients, antimicrobial peptides, and physical features of the gut contribute to differences in microbial community composition in different locations.
The mucosal and lumenal microbiota of the gut represent distinct microbial communities. On a smaller scale, patchiness within these communities suggests that they are highly spatially organized.
Diet imparts a large effect on microbial colonization and relative abundance, but some bacteria can thrive independently of dietary changes by living on host-derived nutrients such as mucin glycans. Therefore, the mucus layer can harbor a reservoir of bacteria that are maintained regardless of food intake. The appendix and colon crypts may also be examples of such microbial reservoirs.
Only a subset of gut symbionts are able to access the epithelial surface. Mucus, antimicrobial peptides, and adaptive immune activity limit tissue accessibility. Direct interfacing between the host and microbial symbionts may be important for maintenance of homeostasis.
Immunomodulation by certain symbionts allows the host to tolerate intimate relationships with potentially beneficial microorganisms. This may be a way in which commensals distinguish themselves from pathogens and prevent their elimination by the immune system.
While many diseases have been associated with dysbiosis, understanding the function of the microbiota in health and disease requires accounting for the biogeography of the community. Recent human studies have found differences specific to the mucosal community in cases of inflammatory bowel disease and hepatic encephalopathy.
Acknowledgments
Thanks to Elaine Hsiao (UCLA), Brittany Needham and Timothy Sampson (Caltech) for critical comments on the manuscript. G.P.D. is supported by an NSF Graduate Research Fellowship (DGE-1144469). Work in the Mazmanian laboratory is supported by funding from the National Institutes of Health (GM099535, DK078938, MH100556), the Emerald Foundation, and the Simons Foundation.
Glossary
- Symbionts
in ecology, these are organisms that participate in a close relationship with other organisms. The term symbionts encompasses organisms that participate in different types of relationships, including mutualists, commensals and parasites.
- Mutualists
in ecology, these are organisms that participate in a symbiotic relationship in which both parties benefit.
- Commensals
in ecology, these are organisms that participate in a symbiotic relationship in which one party benefits from the other without affecting it. Historically, commensals is also used as a term for the resident gut bacteria, though many of these may be mutualists
- Pathobiont
a symbiont with the potential to promote pathology under conditions that deviate from homeostasis, (such as in immunocompromised or nutrient deprived individuals)
- Indigenous
organisms that are native to a particular habitat (also termed autochthonous), as distinct from organisms that are simply passing through a habitat (allochthonous)
- Dysbiosis
deviations from a normal microbial community, such as imbalances in abundance, membership or localization of microorganisms.
- Gnotobiotic
usually refers to formerly germ-free animals that carry a defined microbiota. The composition of the microbiota in these animals is usually determined experimentally.
- Paneth cells
specialized epithelial cells at the base of crypts in the small intestine that secrete antimicrobial peptides.
- Goblet cells
specialized epithelial cells throughout the gastroinstestinal tract that secrete gel-forming mucins. Goblet cells can also be present in other mucosal epithelial surfaces throughout the body.
- Colonization resistance
the prevention of invasion of an exogenous species into a microbial community. In the gut, colonization resistance may be a result of resource competition, spatial exclusion or direct inhibition by commensal microorganisms or of selection mediated by host factors.
- Microaerophilic
obligate aerobic microorganisms that only thrive in environments with relatively low oxygen concentrations, such as at the epithelial surface in the gut.
- Microbiota
The microorganisms that inhabit a particular habitat
- Syntrophic
A metabolic relationship between difference species in which one feeds another
- Prebiotics
Foods that stimulate the growth of commensal or mutualistic gut bacteria
- Biofilm
An aggregation of bacteria stuck to each other and to a surface
- Digesta
The bulk of dietary fibers that is digested as it transits the gastrointestinal tract
- MUC2
The most abundant mucin protein in the human gut (also in mice, where it is referred to as Muc2).
- Secreted Immunoglobulin A (sIgA)
By far the most abundant class of antibody found in the gut
Biographies
Gregory P. Donaldson is a graduate student in the Mazmanian lab at Caltech, studying the microbial genetic and molecular basis of gut colonization in species of Bacteroides. He received his B.S. in Microbiology from the University of Maryland, College Park, where he studied biofilm regulation in Pseudomonas aeruginosa.
S. Melanie Lee is a former member of the Mazmanian lab and is currently a resident physician trainee of Psychiatry at the Neuropsychiatric Institute of UCLA. She received her B.S. in Biology from Harvey Mudd College, her Ph.D. in Biology from Caltech, and M.D. from the Keck School of Medicine of the University of Southern California.
Sarkis K. Mazmanian is the Louis and Nelly Soux Professor of Microbiology at Caltech. He received his B.S. and Ph.D. in Microbiology, Immunology and Molecular Genetics from UCLA, studying mechanisms by which Staphylococcus aureus displays surface protein adhesins during animal infection. As a fellow at Harvard Medical School, he investigated how gut bacteria influence immune system development. Dr. Mazmanian’s laboratory focuses on studying mechanisms by which the gut microbiome impacts the immune and the nervous systems.
References
- 1.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasegawa M, et al. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect Immun. 2010;78:639–650. doi: 10.1128/IAI.01043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Rosa PS, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci USA. 2014;111:12522–12527. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 5.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández L, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Rogier EW, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z-T, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013 doi: 10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu ZT, et al. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology. 2013;23:169–177. doi: 10.1093/glycob/cws138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcobal A, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis ZT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:425. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS ONE. 2013;8:e69621. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam KBMS, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 20.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu S, et al. Bacterial Community Mapping of the Mouse Gastrointestinal Tract. PLoS ONE. 2013;8:e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. Laser-capture microdissection and 16S sequencing were used to profile the microbiome of the inter-fold regions of the proximal colon, revealing a community distinct from the central lumen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pédron T, et al. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3 doi: 10.1128/mBio.00116-12. First 16S sequencing study of the colon crypt microbiome, demonstrating that the crypt community includes many aerobic bacteria and has a distinct profile relative to lumenal bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–579. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Swidsinski A, et al. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis. 2007;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- 27.SMITH HF, et al. Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. J Evol Biol. 2009;22:1984–1999. doi: 10.1111/j.1420-9101.2009.01809.x. [DOI] [PubMed] [Google Scholar]
- 28.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. Detailed investigation of differences in the lumen and mucosal communities along the GI tract of macaques shows that many taxa have preferred spatial habitats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE. 2010;5:e13607. doi: 10.1371/journal.pone.0013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malmuthuge N, Griebel PJ, Guan LL. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Applied and Environmental Microbiology. 2014;80:2021–2028. doi: 10.1128/AEM.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu HP, et al. Spatial heterogeneity of gut microbiota reveals multiple bacterial communities with distinct characteristics. Sci Rep. 2014;4:6185. doi: 10.1038/srep06185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albenberg L, et al. Correlation Between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota in Humans and Mice. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.07.020. Careful measurements of oxygen content in the gut shows a steep oxygen gradient in the mucus that is predictive of community membership based on the ability of bacteria to tolerate oxygen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry D, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1219247110. Isotope-labeling of mucosal proteins in the gut followed by nanoSIMS detection was used in conjunction with FISH to identify mucosal bacteria that were consuming host-derived proteins. This powerful method provides in situ support that certain mucin-degrading bacteria largely forage host-derived nutrients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Png CW, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 36.Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS ONE. 2011;6:e25042. doi: 10.1371/journal.pone.0025042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2013 doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6:57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong M, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS ONE. 2013;8:e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis CP, Mulcahy D, Takeuchi A, Savage DC. Location and description of spiral-shaped microorganisms in the normal rat cecum. Infect Immun. 1972;6:184–192. doi: 10.1128/iai.6.2.184-192.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savage DC, Blumershine RV. Surface-surface associations in microbial communities populating epithelial habitats in the murine gastrointestinal ecosystem: scanning electron microscopy. Infect Immun. 1974;10:240–250. doi: 10.1128/iai.10.1.240-250.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palestrant D, et al. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol. 2004;28:23–27. [PubMed] [Google Scholar]
- 43.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. Identification of a glycan binding and import system in species ofBacteroides that determines their species-specific niche, localization in colon crypts, and resilience in the face of intestinal perturbations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 49.Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. Journal of Bacteriology. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. Transcriptional profiling ofBacteroides thetaiotaomicron in the gut of gnotobiotic animals allowed the identification of genes involved in the utilization of diet-derived and host-derived nutrients. [DOI] [PubMed] [Google Scholar]
- 51.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenburg ED, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kashyap PC, et al. Genetically dictated change in host mucuscarbohydrate landscape exerts a diet-dependenteffect on the gut microbiota. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wadolkowski EA, Laux DC, Cohen PS. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988;56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gries DM, Pultz NJ, Donskey CJ. Growth in cecal mucus facilitates colonization of the mouse intestinal tract by methicillin-resistant Staphylococcus aureus. J Infect Dis. 2005;192:1621–1627. doi: 10.1086/491737. [DOI] [PubMed] [Google Scholar]
- 58.Larsson JMH, Karlsson H, Sjövall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 59.Thomsson KA, et al. Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1- and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology. 2012;22:1128–1139. doi: 10.1093/glycob/cws083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. A mathematical modeling study demonstrating that positive selection through presentation of nutrients is a more effective way for hosts to control surface bacterial communities than negative selection by antimicrobial compounds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonoyama K, et al. Response of gut microbiota to fasting and hibernation in Syrian hamsters. Applied and Environmental Microbiology. 2009;75:6451–6456. doi: 10.1128/AEM.00692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol Regul Integr Comp Physiol. 2013;304:R33–42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwab C, et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 2014;8:1101–1114. doi: 10.1038/ismej.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. Mutants in genes involved in mucin O-glycan utilization were found to be defective in colonization when animals were fed diets without plant polysaccharides and in vertical transmission from mother to pup. This demonstrates the importance of host-derived nutrients in the mucus for stable and long-term colonization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sommer F, et al. Altered mucus glycosylation in core 1 o-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS ONE. 2014;9:e85254. doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergström A, et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes. 2012;5:402. doi: 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullen TW, et al. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. This paper demonstrates the necessity of bacterial sensing by the epithelium and secretion of RegIIIgamma by Paneth cells to prevent microbial overgrowth of the epithelial surface in the small intestine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 72.Miyoshi A, et al. Oxidative stress in Lactococcus lactis. Genet Mol Res. 2003;2:348–359. [PubMed] [Google Scholar]
- 73.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. A detailed investigation of the protein content of colon mucus and distinguishing factors between the outer and inner layers. MUC2 is shown to be required to prevent bacterial overgrowth on the epithelial surface and invasion of tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pelaseyed T, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stecher B, et al. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Navarro-Garcia F, et al. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect Immun. 2010;78:4101–4109. doi: 10.1128/IAI.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS ONE. 2012;7:e30287. doi: 10.1371/journal.pone.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo Q, et al. Enterotoxigenic Escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun. 2014;82:509–521. doi: 10.1128/IAI.01106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahdavi J, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin Y, et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnupf P, et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015 doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunology. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 88.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhowmick R, et al. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect Immun. 2008;76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mouricout M. Interactions between the enteric pathogen and the host. An assortment of bacterial lectins and a set of glycoconjugate receptors. Adv Exp Med Biol. 1997;412:109–123. [PubMed] [Google Scholar]
- 91.Lecuit M, et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 92.McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Savage DC. Microbial interference between indigenous yeast and lactobacilli in the rodent stomach. Journal of Bacteriology. 1969;98:1278–1283. doi: 10.1128/jb.98.3.1278-1283.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morotomi M, Watanabe T, Suegara N, Kawai Y, Mutai M. Distribution of indigenous bacteria in the digestive tract of conventional and gnotobiotic rats. Infect Immun. 1975;11:962–968. doi: 10.1128/iai.11.5.962-968.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sengupta R, et al. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediators Inflamm. 2013;2013:237921–16. doi: 10.1155/2013/237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mackenzie DA, et al. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology (Reading, Engl) 2010;156:3368–3378. doi: 10.1099/mic.0.043265-0. [DOI] [PubMed] [Google Scholar]
- 98.Frese SA, et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013;9:e1004057. doi: 10.1371/journal.pgen.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Ossowski I, et al. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Applied and Environmental Microbiology. 2010;76:2049–2057. doi: 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turroni F, et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mathias A, Corthésy B. N-Glycans on secretory component: mediators of the interaction between secretory IgA and gram-positive commensals sustaining intestinal homeostasis. Gut Microbes. 2011;2:287–293. doi: 10.4161/gmic.2.5.18269. [DOI] [PubMed] [Google Scholar]
- 104.Peterson DA, et al. Characterizing the Interactions Between a Naturally-primed Immunoglobulin A and its Conserved Bacteroides thetaiotaomicron Species-specific Epitope in Gnotobiotic Mice. J Biol Chem. 2015 doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 107.Fanning S, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeon SG, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8:e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunology. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 111.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith PM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013 doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shan M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 115.Randal Bollinger R, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol. 2007;249:826–831. doi: 10.1016/j.jtbi.2007.08.032. This study proposes the hypothesis that the appendix harbors a protected reservoir of bacterial cells that could repopulate the cecum and large intestine. [DOI] [PubMed] [Google Scholar]
- 116.Hanson NB, Lanning DK. Microbial induction of B and T cell areas in rabbit appendix. Dev Comp Immunol. 2008;32:980–991. doi: 10.1016/j.dci.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Walker AW, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Abrahamsson TR, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 122.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 123.Garcovich M, Zocco MA, Roccarina D, Ponziani FR, Gasbarrini A. Prevention and treatment of hepatic encephalopathy: focusing on gut microbiota. World J Gastroenterol. 2012;18:6693–6700. doi: 10.3748/wjg.v18.i46.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012 doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- 126.Wu N, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 127.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 128.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. 16S sequencing analysis of fecal, ileal mucosa and rectal mucosa samples from early stage Crohn’s patients before treatment shows dysbiosis in the mucosal samples and no difference in fecal samples. Following treatment, there are unrelated differences in fecal samples, suggesting that the fecal dysbiosis previously observed may be a secondary effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Swidsinski A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 131.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 132.Rowan F, et al. Bacterial colonization of colonic crypt mucous gel and disease activity in ulcerative colitis. Ann Surg. 2010;252:869–875. doi: 10.1097/SLA.0b013e3181fdc54c. [DOI] [PubMed] [Google Scholar]
- 133.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–85. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rosebury T. Microorganisms Indigenous to Man. McGraw-Hill; 1962. [Google Scholar]
- 135.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS ONE. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]