Abstract
Gene promoters are essential regions of DNA where the transcriptional molecular machinery to produce RNA molecules is recruited. In this process, DNA epigenetic modifications can acquire a fundamental role in the regulation of gene expression. Recently, in a previous work of our group, functional features and DNA methylation involved in the ovine HSP90AA1 gene expression regulation have been observed. In this work, we report a combination of methylation analysis by bisulfite sequencing in several tissues and at different developmental stages together with in silico bioinformatic analysis of putative regulating factors in order to identify regulative mechanisms both at the promoter and gene body. Our results show a “hybrid structure” (TATA box + CpG island) of the ovine HSP90AA1 gene promoter both in somatic and non-differentiated germ tissues, revealing the ability of the HSP90AA1 gene to be regulated both in an inducible and constitutive fashion. In addition, in silico analysis showed that several putative alternative spliced regulatory motifs, exonic splicing enhancers (ESEs), and G-quadruplex secondary structures were somehow related to the DNA methylation pattern found. The results obtained here could help explain the differences in cell-type transcripts, tissue expression rate, and transcription silencing mechanisms found in this gene.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0668-6) contains supplementary material, which is available to authorized users.
Keywords: In silico analysis, HSP90AA1, Sheep, Methylation pattern, Regulatory motifs, Structural features
Introduction
Gene expression varies among tissues and even different cell types but also in response to specific signals (physiological, environmental, etc.). The main mechanism of transcriptional regulation is orchestrated by proteins called transcription factors (TFs), which promote (as activators) or block (as repressors) the recruitment of the RNA polymerase II (Pol II complex).
A promoter is a DNA region where the transcription of a particular gene is initiated. Two main types of promoters have been described (Carninci et al. 2006): TATA box-enriched initiated at a single transcriptional start site (TSS) and 5′-CpG-3′ dinucleotide (CpG)-rich promoters containing multiple TSS. In TATA box-enriched promoters, the initiation of transcription by the RNA polymerase II machinery encompasses several sequence motifs including the initiator (INR), the TFIIB recognition element (BRE), or the downstream promoter element (DPE) (Butler and Kadonaga 2002). Moreover, TATA box-enriched promoters in conjunction with enhancers, repressors, and insulators define combinatorial codes that specify gene expression patterns (Kim et al. 2005; Juven-Gershon et al. 2008). However, CpG-rich promoters usually lack the regulatory elements and display dispersed initiation patterns (Deaton and Bird 2011). Approximately 65–70 % of promoters have associated CpG islands (CGIs) and are generally hypomethylated, while promoters containing low CpG density are usually hypermethylated (Saxonov et al. 2006; Weber and Schubeler 2007). Furthermore, CpG-rich promoters often contain multiple Sp1 transcription factor-binding sites (Carninci et al. 2006). They are more “plastic” because in addition to TFs, their transcriptional activity is subjected to epigenetic control (Carninci et al. 2006). Recently, the term “hybrid” promoter has appeared to define those ones characterized by the presence of both a CGI and a TATA box structure (Carninci et al. 2006; Deaton and Bird 2011).
Methylation is a major biochemical alteration that governs multi-tiered epigenetic regulation of gene expression. In mammals, methylation is mainly found in CpG dinucleotides at the C5 position of cytosines and is generally associated to CpG-rich promoters. CpGs are methylated in bulk DNA with the exception of called CGIs (Bird 1986). CGIs consist of multiple CpG sites ranging from 200 to 3000 nucleotides (Deaton and Bird 2011) and are frequently associated with promoter regions, 5′ ends of housekeeping and many tissue-specific genes and 3′ ends of some tissue-specific genes (Gardiner-Garden and Frommer 1987). Methylation of CGIs is associated with a repressed chromatin state and inhibition of gene expression (Bird and Wolffe 1999). For most genes, hypermethylation of CGIs is linked to transcriptional silencing. DNA methylation silences transcription through three different mechanisms: (1) sterical obstacle by which methylation prevents the binding of TFs in their specific target sites (Watt and Molloy 1988), (2) specific recognition of methyl-CpG binding factors (MBPs) that can trigger a potentially repressive response (Klose and Bird 2006), and (3) the repressed condition of chromatin modifications (Medvedeva et al. 2013).
Genome-wide analyses of DNA methylation have shown distinct profiles of DNA methylation associated with different cell types and tissues, suggesting that methylation is important for cellular identity (Bernstein et al. 2007). In mammalian development, the global DNA methylation profile of the genome is dynamically reprogrammed during gametogenesis and early embryogenesis (Reik 2007). During embryogenesis, DNA is passively demethylated along early cell divisions. Subsequently, de novo DNA methylation of dividing cells establishes the CpG methylation marks that guide restriction of gene expression patterns associated with tissue-specific lineages (Smith et al. 2012). In adult tissues, CpG methylation marks must also be maintained by DNA methyl-transferases during DNA replication to preserve the identity and function of differentiating cell types and for self-renewal of adult stem cell populations (Trowbridge and Orkin 2010; Bocker et al. 2011; Berdasco and Esteller 2010).
Taking only into account the information content of alternatively spliced sites (ASSs) with consensus sequences like 5′ splice sites, 3′ splice sites, and branch sites is insufficient for splice site selection (Cartegni et al. 2003). Some factors are responsible for splicing regulation, which are essential to ensure a correct gene expression, CGI methylation, exonic splicing enhancers (ESEs), or G-quadruplex (G4) structures. The role of CGI methylation is well known, but its impact on alternative splicing is still unclear (Anastasiadou et al. 2011). Previous studies have shown that a specific DNA structure may influence the alternative splicing, in particular, DNA methylation of cytosine at exons compared to introns (Shukla et al. 2011). There is evidence that a DNA-binding protein like the CCCTC-binding factor (CTCF) can promote inclusion of exons. However, the presence of methylation on CpGs can compromise its binding to DNA, thus contributing to tissue-specific alternative splicing by alternative promoters (Shukla et al. 2011; Wan et al. 2013). The study of tissue-specific differentially methylated region (T-DMR) allows the identification of transcription factor-binding motifs containing CpG dinucleotides. These T-DMR analyses are associated with binding motifs with either exon inclusion (positive regulation), exon exclusion (negative regulation), or both (Wan et al. 2013).
ESEs are other regulatory splicing factors that detect cis-elements, and they act like splicing enhancers and splicing silencers (Cartegni et al. 2003). These splicing regulatory elements can be affected by mutations (Cartegni et al. 2003) and DNA methylation. The recognition of ESEs’ CpG-containing sites by splicing regulatory factors (SR proteins) can also be altered by methylation (Anastasiadou et al. 2011).
A well-characterized secondary structure that has a great impact on translation is the G4 structure. These structures are guanine-rich nucleic acid sequences that can fold into a non-canonical tetrahelical structure that is very stable and has the ability to strongly repress translation (Beaudoin and Perreault 2010). The sequence motif G≥3NxG≥3NxG≥3NxG≥3 confers the ability to form a four-stranded (G-quadruplex or G4) structure in which interactions among strands are stabilized by G-quartets. G4 motifs are enriched at the TSS, the 5′-UTR, and the 5′ end of the first intron and depleted in coding regions (Maizels and Gray 2013). In the promoters of several eukaryotic and prokaryotic genes, G-rich sequences with potential to form G4 have been identified (Rankin et al. 2005; Rawal et al. 2006; Stevens et al. 2014). The potential for G4 structures to inhibit DNA synthesis has long been recognized (Han et al. 1999; Boán et al. 2004), as has its ability to act as a transcriptional repressor (Siddiqui-Jain et al. 2002; Lin et al. 2013). An early work described stabilization of G4 through cytosine-cytosine base pairing, an effect that was greatly enhanced by cytosine methylation (Hardin et al. 1993). Moreover, an exceptional ability to methylate unusual DNA structures by DNA methyltransferase has been pointed out (Smith et al. 1991). G4 motifs can occur at methylated sites. For example, the CpG island of the oncogene BCL2 contains G-rich regions capable of forming a G4 structure, and it has been shown that methylation of cytosines within the G4 motif markedly stabilizes the G-quadruplex (Lin et al. 2013).
Heat shock (HS, also known as heat stress and hyperthermia) is one of the primary organism and cellular stressors. The transcription of more than 100 genes, such as encoding factors participating in protein folding, degradation, transport, RNA repair, and metabolic pathways, is upregulated under HS conditions (Gasch et al. 2000; Tabuchi et al. 2008; Richter et al. 2010). The best known genes involved in the heat shock response are those encoding the heat shock proteins (HSPs) which are synthesized in response not only to HS but also to other types of cellular stressors (Csermely et al. 1998). The HSP90AA1 gene encoding the inducible form of the HSP90, hsp90α, is ubiquitously expressed, but brain and testes are the tissues in which the highest transcription levels are found (Sreedhar et al. 2004). Hsp90α is necessary for spermatogenesis and meiotic progression in mammalian testis (Grad et al. 2010). The ovine HSP90AA1 gene has been studied thoroughly by our group since 2008 (Marcos-Carcavilla et al. 2008, 2010a, b; Oner et al. 2012; Salces-Ortiz et al. 2013, 2015a, b). A recent functional analysis of the ovine HSP90AA1 gene (Salces-Ortiz et al. 2015c) has demonstrated that two linked polymorphisms, a cytosine insertion (g.667_668insC; rs397514115) and a C to G transversion (g.660G>C; rs397514116), both located at the promoter region, are mainly responsible for the observed differences in the expression rate of the gene in response to heat stress events. Also in this work, an allele-specific methylation was found for the adjacent cytosine of the g.660G > C SNP (rs397514116), which however has not been unequivocally linked with the observed differences at the transcription level.
The aim of the present study was (1) to characterize the structure and the regulation motifs of the HSP90AA1 promoter, (2) to study the methylation pattern of the HSP90AA1 promoter and gene body in several tissues and at different developmental stages using DNA bisulfite techniques, and (3) in silico exploration of putative mechanisms involved in its epigenetic regulation pattern.
Methods
Ethics statement
The current study was carried out under a Project License from the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) Scientific Ethic Committee. Animal manipulations were performed according to the Spanish Policy for Animal Protection RD 53/2013, which meets the European Union Directive 86/609 about the protection of animals used in experimentation. We hereby confirm that the INIA Scientific Ethic Committee (IACUC) specifically approved this study.
Animal material and nucleic acid isolation
Samples from different tissues, ages, and ovine breeds were obtained for this specific study. Sample identification, tissue, breed, age, genotype, and extraction kit method are described in Supplemental Table 1. Moreover, blood samples were those used in Salces-Ortiz and co-workers (Salces-Ortiz et al. 2015b).
PCR and genotyping
Polymerase chain reaction was performed and the resulting PCR fragments were sequenced as in Salces-Ortiz et al. (2013). A promoter fragment of 499 bp was obtained, containing 11 polymorphisms (Salces-Ortiz et al. 2015c). Primers used in the PCR and PCR conditions were previously described in Salces-Ortiz et al. (2013) (Supplemental Table 1).
DNA methylation analysis by sequencing
DNA methylation status was determined using sodium bisulfite treatment (Hajkova et al. 2002). Bisulfite treatment was performed with ≤2 μg of 32 samples of genomic DNA from different tissues using Epitect Plus Bisulfite Conversion Kit (Qiagen, Valencia, CA, USA) and EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) following manufacturer’s guidelines.
Genomic DNA and DNA bisulfite-treated concentrations were determined using a NanoDrop ND-1000 UV/Vis spectrophotometer (NanoDrop Technologies Inc., DE, USA).
Primers, PCR of bisulfite-treated DNA, amplicon sizes, Tm, PCR kits, analysis of fragments, and its potential artifacts are previously described in Salces-Ortiz et al. (2015c).
To elucidate if epigenetic mark patterns were different among tissues and ages, we have compared brain, liver, testicle, and sperm from adult rams and heart, testicle, and ovary from young animals (90 days) (Supplemental Table 1).
To confirm allele-specific methylation, the reverse strand was analyzed separately with allele-specific sequencing primers amplifying two fragments (511 and 418 bp) to encompass the promoter region (Hajkova et al. 2002).
Detection and resolution of artifacts in bisulfate sequencing: re-sequencing and combined bisulfite restriction analysis
Bisulfite sequencing has been associated with technical difficulties and potential artifacts. Accordingly, we resolved it firstly by re-sequencing and secondly by RFLP analysis with restriction enzymes to analyze CpG sites where methylation was uncertain in PCR products of DNA treated with sodium bisulfite (Xiong and Laird 1997).
Prediction of CpG island of ovine HSP90AA1 gene
The sequence of the ovine HSP90AA1 gene (DQ983231.1) was analyzed regarding the length and frequency of CpG dinucleotides. Data were obtained from “The Sequence Manipulation Suite Software: CpG islands” (http://www.bioinformatics.org/sms/cpg_island.html) (Gardiner-Garden and Frommer 1987).
Tissue-specific transcripts and RNAseq expression in ovine and human
The source data used in the study of tissue-specific transcripts and RNAseq expression of ovine and human HSP90AA1 gene is fully available online (http://www.ensembl.org/Ovis_aries; http://www.ensembl.org/Homo_sapiens). Transcript graphics of the tissues studied were extracted from the Ensembl database. The differences in RNAseq expression between tissues were calculated as the ratio of total alignments (number of alignments + alignments omitted) relative to total alignments of liver (tissue with the least number of alignments).
Prediction of putative TFs that bind predicted CpGs affecting the regulation of alternative splicing
Analyses of T-DMRs were based on different sequenced genomic regions (three exons and two introns) and several sequence motifs containing CpGs. The predicted consensus T-DMRs were enumerated and compared with the HSP90AA1 sequence using a motif comparison tool (meme.nbcr.net/meme/cgi-bin/tomtom.cgi). Those TFs that have a similar structure to CTCF (class zinc coordinating and family ββα–ZF) were chosen to be candidates for alternative splicing (AS) regulation (Lorincz et al. 2004).
Distribution of ASSs and putative ESEs
To identify patterns regulating tissue-specific AS, we looked for ASSs. Also, we looked for any relationship between the epigenetic mark distribution in putative ESEs (Anastasiadou et al. 2011). To achieve these objectives, human alternatively spliced sites and human putative ESEs were identified in the ovine HSP90AA1 gene sequence using ESEfinder 3.0 software (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi) (Anastasiadou et al. 2011; Cartegni et al. 2003).
Prediction of G4 in ovine HSP90AA1 gene
QGRS Mapper software (Kikin et al. 2006) was used to predict composition and distribution of putative G4 in the HSP90AA1 gene (DQ983231.1). Prediction of G4 structures was made in both strands.
HSP90AA1 gene sequence showed a great potential to form G4 structures because it has a G-rich sequence both in the promoter region and in the gene body. However, to know the potential contribution of G4 between non-template (in the reverse strand) and template strand sequences (in the forward strand), both were tested separately (Eddy and Maizels 2008).
Results
HSP90AA1 promoter structure and associated CpG island
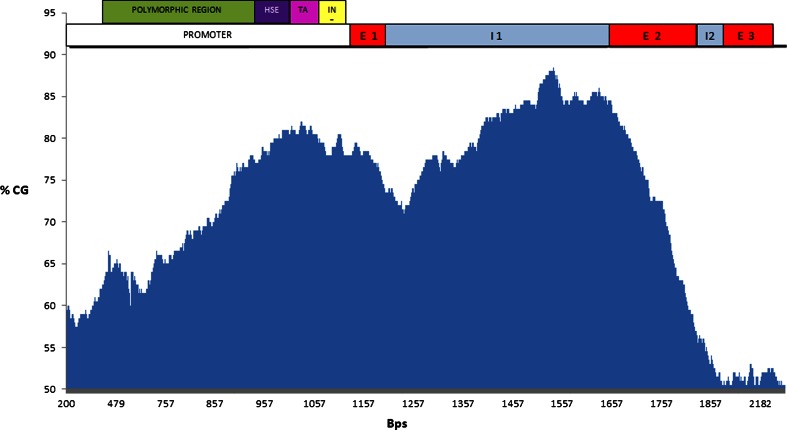
In the present work, we have confirmed the presence of a 5′ CGI along the HSP90AA1 gene by predictive analysis. We have described graphically the location of the CGI extended through the promoter region, exons, and introns of the associated gene (Fig. 1). However, this result is insufficient to define the boundaries of the CGI. Bisulfite sequencing analysis allowed us to establish the limits of the CGI in a tissue-specific manner (see Fig. 2).
Fig. 1.
Graphic representation of the ovine HSP90AA1 CpG island (based on the reference sequence from the NCBI database, DQ983231.1; 5917 bp). The X axis represents the base pairs, while the Y axis represents the percentage of GC (data obtained from the Sequence Manipulation Suite http://www.bioinformatics.org/sms/cpg_island.html). The putative CpG island spans through the core promoter including the heat shock element (HSE), TATA box, and initiator element, also the exon 1,intron 1, exon 2, and intron 2 (see top of the figure)
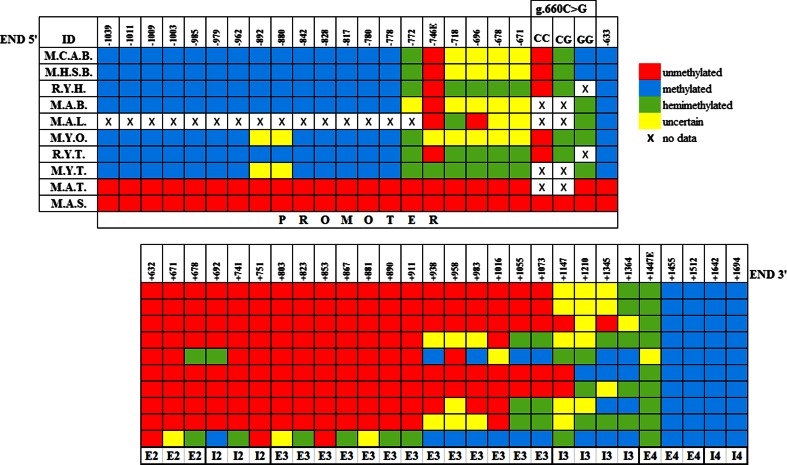
Fig. 2.
Methylation pattern representation of the HSP90AA1 CGI across different tissues. Identifications (IDs) from tissues are as follows: M.C.A.B. blood samples from non-stressed adults of Manchega breed, M.H.S.B. blood samples from heat-stressed adults of Manchega breed, R.Y.H. heart samples from young animals of Rasa Aragonesa breed, M.A.B. brain sample from an adult animal of Manchega breed, M.A.L. liver sample from an adult of Manchega breed, M.Y.O. ovary samples from young animals of Manchega breed, R.Y.T. testicule samples from young animals of Rasa Aragonesa breed, M.Y.T. testicule sample from a young animal of Manchega breed, M.A.T. testicle sample from an adult animal of Manchega breed, and M.A.S. sperm samples from adult animals of Manchega breed. Numbers on the top of the figure indicate the position of the CpG cytosine relative to TSS. CpGs from −582 to +632 positions relative to TSS are un-methylated (not shown in the figure). The signs at the bottom of the figure indicate positions at the promoter region of the gene, exons (E), and introns (I). CpG at position −778 represents the 5′ boundary of the CGI in somatic and non-differentiated germ tissues. Differentiated germ tissues have no identified CGI border in 5′. CpG at position +1455 (3′ end) is methylated in all tissues. Patterns of epigenetic marks are specific of each tissue. The rs397514116 (g.660G > C) mutation creates an ASM in blood and an “allele-specific hemi-methylation” in somatic and non-differentiated germ tissues. Differentiated germ tissues have no ASM
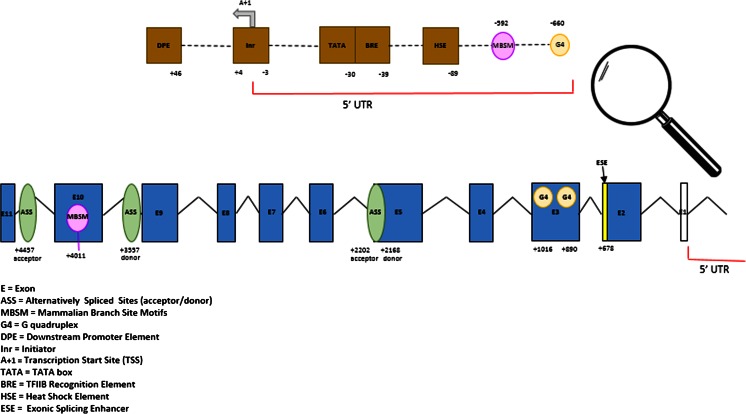
The CGI includes core promoter elements like a regulatory heat shock element (HSE), a consensus BRE motif, which is followed by a canonical TATA box (first described by Marcos-Carcavilla et al. (2008)) and an Inr consensus motif which includes one TSS (A+1). They are located precisely at positions −89, −39, and −30 relative to TSS, respectively. Furthermore, the CGI is enriched with putative consensus motif-binding sites such as nine Sp1 (M6 GGGCGGR) at positions −151, −147, −45, −23, −12, +99, +127, +156, and +364; two ELK-1 (M3 SCGGAAGY) at positions −271 and −85 (last one coincides with the HSE sequence); and one M22 (TGCGCANK) at position −134 (all positions described relative to the TSS position). Finally, a putative downstream core promoter element (DPE) at position +46 relative to TSS (position in the Inr) was found (Supplemental Figs. 1 and 3).
Epigenetic mark patterns
Allele-specific methylation
In our previous work, we had shown that the rs397514116 (g.660G > C) mutation created an allele-specific methylation (ASM) in blood (Salces-Ortiz et al. 2015c). In the present work, we have observed on additional tissues studied an allele-specific hemi-methylation associated with the GG genotype of the rs397514116 (g.660G > C) mutation except in adult testicle and sperm tissues that are not methylated (Fig. 2).
Differences among tissues
According to the bisulfite sequencing results, the same CpG island 5′ border as in blood was found in heart, brain, and ovary. Regarding epigenetic marks on the gene body, germ cells like Manchega young ovary and Rasa Aragonesa young testicle were free of epigenetic marks in exon 2, intron 2, and exon 3. Conversely, Manchega young testicle, Manchega adult testicle, and somatic tissues (liver and brain) showed epigenetic marks that extended part of exon 3 (Fig. 2).
Different CGI epigenetic mark patterns between somatic tissues of the same individual and similarly in identical somatic tissue types from different individuals have also been observed as in human (Grunau et al. 2000).
Differences between developmental stages (young vs adult)
To determine if differences in the epigenetic mark pattern exist at different ages, we compared testicle tissue from adult and young (90 and >90 days) animals and sperm from adults. A progressive loss of promoter methylation with cell differentiation was observed. Moreover, all epigenetic marks disappeared at the promoter of both adult testicle tissue (primarily composed by mature germ cells) (Bellve et al. 1977; Oakes et al. 2007) and mature sperm from the ejaculate. In addition, a progressive increase of epigenetic marks in the gene body of these tissues was addressed (Fig. 2).
Finally, we also observed a similar pattern across CGI from different individuals of equal age and a different CGI epigenetic mark pattern between samples at different ages.
Detection and resolution of artifacts in bisulfate sequencing: re-sequencing and combined bisulfite restriction analysis
CpG sites at positions −772, −746, −718, −696, −678, and −671 preceding to rs397514116 (g.660G > C) and site at position +1447 preceding to 5′ CGI boundary have doubtful patterns of methylation marks (see Fig. 2). We tried to solve these doubts with a restriction enzymes assay. CpGs at position −746 (only restriction target found and called −746E) and at position +1447 (called +1447E) have restriction targets to BstBI enzyme and to TaqI enzyme, respectively.
Based on combined bisulfite restriction analysis (COBRA), we have observed that the CpG at position +1447E was hemi-methylated in all tissues analyzed except for liver. The CpG at position −746E was not methylated in any somatic or mature sperm tissue analyzed, while it was hemi-methylated in young testis. However, we could not obtain conclusive results about the methylation status of this CpG on young ovary samples (Fig. 2).
In silico study of the putative regulating factors
Tissue-specific transcripts and RNAseq expression in ovine
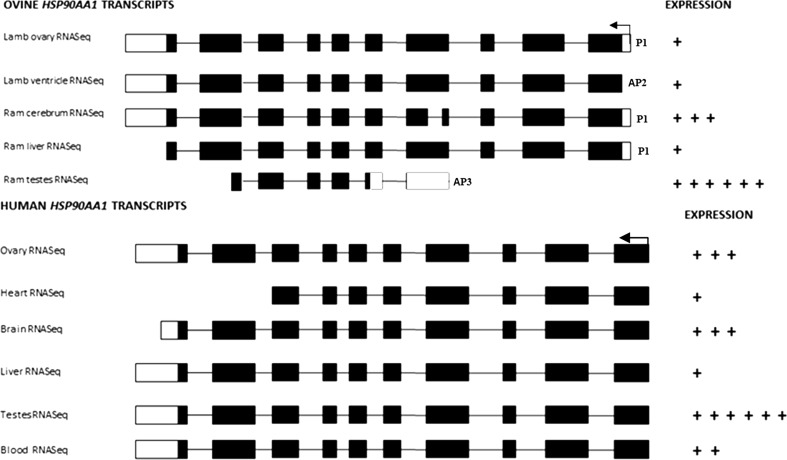
On the basis of the results of tissue-specific transcripts of the ovine HSP90AA1 gene fully available on public data sources, we identified a promoter (P1) (described above) and two alternative promoters (AP2 and AP3). P1 and AP2 have different TSSs and identical open reading frames (ORF). P1 is active in ovary, cerebrum, and liver, whereas AP2 is active exclusively in heart (from tissue data available) and AP3 is only active in testes. Moreover, AP3 exclusively contains a TSS in exon 5 and the ORF in the middle of exon 6 (Fig. 4), which defines a transcript of different size in testes compared to those from other tissues. Furthermore, splicing differences in the transcript structure of brain tissue (exon 5) and testes tissue (exon 6 and exon 10) were found (see Fig. 4).
Fig. 4.
Tissue-specific transcripts from the ovine and human HSP90AA1 genes. Only tissues analyzed in the present work and their reverse strands are shown. The arrow indicates the direction of transcription. P1 (promoter), AP2, and AP3 (alternative promoters 2 and 3) indicate the type of promoter. Graphical representation by crosses of the each expression ratio is based on total alignments of RNAseq data (number of alignments + alignments omitted) with respect to the total alignments of liver (which is the tissue with the least number of alignments) (available at http://www.ensembl.org/Ovis_aries and http://www.ensembl.org/Homo_sapiens)
On the other hand, according to in silico RNAseq ratio between tissues, we have seen the greatest expression both in testis and brain, relatively similar expression rates in heart and ovary, and the lowest expression in liver (see Fig. 4).
Prediction of putative TFs that bind to the predicted CpG-containing motifs and that affect the regulation of alternative splicing
T-DMRs are associated with intragenic regions included within CGIs (Wan et al. 2013). Most of the detected T-DMRs were located in both introns and exons of the sequence studied here, exon 2, intron 2, exon 3, and intron 3 (Figs. 2 and 4). Supplemental Table 2 shows putative transcription factors involved in positive/negative gene splicing regulation associated with DNA methylation pattern.
Detection of ASS sequences in the HSP90AA1 gene
Two canonical splice signal sequence motifs were found in the ovine HSP90AA1 gene. One of them was located in intron 5 with a “donor splice motif” straddling the 3′ end of exon 5 and the beginning of intron 5 (at position + 2168 relative to TSS). An “acceptor splice motif” was located at the end of intron 5 (at position +2202). Two “mammalian branch site motifs” (component in the effective splicing of pre-mRNA) were located in promoter region (at position −592) and in exon 10 (at position +4011). The other donor splice motif was located in intron 9 (at position +3557) and an acceptor splice motif straddling intron 10 and exon 11 (at position +4457 relative to TSS) (Supplemental Figs. 2 and 3).
Detection of ESEs in the CpG-containing motifs
Methylation of CpGs affect the binding of SR proteins to the ESE containing CpGs. In the HSP90AA1 sequence, the following nine putative ESEs were predicted in silico: two SF2/ASF IgM-BRCA1 (at positions −255 and +3133 relative to TSS), three SRp55 (at positions +10, +23, and +4457), one SC35 (at position +1695), two SRp40 (at positions +1318 and 3262), and one SF2/ASF SR protein (at position +674). Three of them include CpG-containing sites in ESEs regions, two SRp55 in exon 1 (+10 and +23) (TGCGTC) and one SF2/ASF SR protein in exon 2 (+674) (CAGACGT) (Supplemental Figs. 2 and 3).
Only in SF2/ASF SR, protein differences were found in the epigenetic mark patterns in different tissues. A CpG cytosine at position +678 detected in the SF2/ASF was found to be hemi-methylated in exon 2 only in sperm (Figs. 2 and 3).
Fig. 3.
Graphic representation of ovine HSP90AA1 gene and their putative regulatory motifs related with transcription and splicing processes found in our in silico study. Every site is numbered relative to the TSS
Identification of methyl-CpG for G4 in the HSP90AA1 gene
Our results showed that upstream and downstream regions of the TSS are G-rich in the reverse strand (non-template strand), where 56 G4s with a maximum length of 40 bp were predicted (Supplemental Table 3). Only two of them, at positions 2024 and 2142 (Supplemental Table 3), showed a hemi-methylated CpG in exon 3 (at position +890 relative to TSS) and a methylated CpG in exon 3 (at position +1016 relative to TSS) in sperm (Figs. 2 and 3).
In the forward strand (template sequence), 26 G4 sequences with a maximum length of 30 bp were predicted. Two of them had the highest G scores, at position 4556 and at position 5431 (Supplemental Table 4). One of them, at position 5431, encompasses the g.660G > C (rs397514116) mutation (promoter region at position −660 relative to TSS). The CpG site created by the presence of the G allele of this mutation is methylated in DNA extracted from blood samples and hemi-methylated and non-methylated in DNA from other tissues (Figs. 2 and 3).
Discussion
The frequency of CpG dinucleotides in human genomes is 1 %—less than one quarter of the expected frequency. Scarano et al. proposed that the CpG deficiency is due to an increased vulnerability of methyl-cytosines to spontaneously deaminate to thymine in genomes with CpG cytosine methylation (Scarano et al. 1967). The majority of CpG cytosines of the genome are methylated with the exception of those located inside the CGIs in normal adult somatic tissues (Bird et al. 1985; Lorincz et al. 2004). The prediction of CGIs in the genome is based on the expected frequency of CpGs. The most commonly used CGI detection algorithms (Gardiner-Garden and Frommer 1987; Takai and Jones 2002) however, are not useful for searching either the exact position of the CGI boundaries or for the analysis of mature germ tissues. Bisulfite sequencing analysis allowed us to establish the limits of the CGI in a tissue-specific manner. 5′ end of CGI reaches up to promoter in somatic and young germinal tissues, and 3′ end of CGI reaches up to exon 4 in blood (Salces-Ortiz et al. 2015c) and heart tissues. It is noteworthy that the methylation marks disappear in promoter region in mature germinal tissues, whereas these methylation marks appear in exon 2 in mature sperm (see Fig. 2). Thus, a study of methylation patterns of the CpGs constitutes a useful tool not only for a better mapping of CGIs but also for elucidating its transcriptional regulatory role in the different regions of the genes (Deaton and Bird 2011). In this work, evidence of the CGI associated to the 5′ end of the ovine HSP90AA1 gene in the tissues studied has allowed us to learn some aspects of the dynamic role of germ tissue methylation as a result of their epigenetic reprogramming during gametogenesis (Preveti et al. 2009). This last is revealed as differences of the methylation patterns in a tissue-specific way (see Fig. 2).
CGIs are transcriptional regulatory structures (Deaton and Bird 2011) where the transcription is initiated from multiple transcription-binding sites (Carninci et al. 2006) in contrast to the transcription at TATA core promoter that occurs from a single site or localized cluster of sites (Butler and Kadonaga 2002). A hybrid structure of the HSP90AA1 gene promoter has been confirmed since it contains a TATA box and a CGI. These structures provide promoters with a dual transcriptional behavior. The TATA box+ Inr (synergistic configuration) and the HSE complex can direct a “strong transcription” initiation in response to heat shock events. The CGI may also function in concert with the “basal transcription” factors to mediate the induced and constitutive expressions of the gene (Butler and Kadonaga 2002). TATA-containing genes are often highly regulated by biotic or stress stimuli, while TATA-less promoters with or without CGI are frequently involved in basic housekeeping (HK) processes (Basehoar et al. 2004; Kimura et al. 2006; Yang et al. 2007). This dual behavior could explain the fact that the HSP90AA1 gene is more inducible than its constitutive counterpart, the HSP90AB1, which has non-canonical core promoter elements associated with a 5′ end CGI (ENSOARG00000009343). In fact, HSP90AB1 is more stable under thermo-neutral and heat stress conditions than HSP90AA1 and several commercial HK tested in previous works of our group (Salces-Ortiz et al. 2013; Serrano et al. 2011).
In a previous work (Salces-Ortiz et al. 2015c), an ASM caused by a C to G transversion (rs397514116) in DNA extracted from blood samples was detected. In this current work, other somatic and immature germ tissues were analyzed. A general allele-specific “hemi-methylation” and “no methylation” of mature germ tissues was assessed (Fig. 2). These results confirm that the allele-specific methylation is also a tissue-specific event, and therefore, the different patterns observed could play an important role in the gene expression regulation (Chuang et al. 2013). Additional information could be provided by the CpGs at positions −772, −746, −718, −696, −678, and −671 preceding the rs397514116 (g.660G > C) and the CpG at position −633. However, the results of the COBRA assay (Zhenggang and Laird 1997), in particular at the CpG site (−746), led us to think that the methylation patterns observed are potential artifacts of the bisulfite sequencing technique. This can be due to the fact that some preparations can be more resistant to bisulfite conversion than others, even though the same DNA isolation protocol was used, or due to different residual amounts of protein in the DNA samples (Warnecke et al. 2002). Furthermore, some sequences within a fully converted region are often resistant to a “powerful methylation,” in particular cytosines located immediately 5′, in our case, the CpG at position −633 that is always methylated both in somatic and immature germ tissues. This type of non-conversion sequence may involve the formation of a stable secondary structure around the methylated CpG site, creating a localized region of dsDNA and preventing the access by bisulfite (Fig. 2; Warnecke et al. 2002).
There are several genome-wide studies that endorse the relationship between T-DMRs and gene expression, both in differentiated tissues (Song et al. 2005; Rakyan et al. 2008; De Bustos et al. 2009) and in pluripotent cells (Meissner et al. 2008). Other investigators have focused on CGI methylation in the gene body. These works suggest that gene body methylation has the potential to suppress transcriptional noise or repress spurious transcription (Huh et al. 2013), to repress the activity of intragenic promoters (Maunakea et al. 2010), to initiate the formation of chromatin structure that affects the transcription elongation (Lorincz et al. 2004), and to promote transcription pausing (Shukla et al. 2011). Recent evidence also suggests that gene body methylation is involved in splicing (Jones 2012). Maunakea and colleagues surveyed DNA methylation patterns across the genome in human brain tissue. They found that 34 % of CGIs in gene bodies are methylated compared with 16 % of all CGIs and just 2 % of CGIs at 5′ promoters. Importantly, the tissue-specific DNA methylation is much more common at CGIs in gene bodies than those in 5′ promoters (Maunakea et al. 2010).
Gene body methylation patterns detected here (Fig. 2) provide us information that could help explain the role of epigenetic marks in the differences in expression rate and in the type of transcripts across the tissues observed (Fig. 4). First, the markedly different gene body methylation patterns in heart and ram testes correlate with the lowest and highest HSP90AA1 transcriptional activities, respectively (both ovine and human expression online data; Fig. 4). Second, the presence of alternative transcripts supports the role for gene body methylation in regulating the use of tissue-specific alternative promoters (AP2 and AP3 detected for HSP90AA1) (Maunakea et al. 2010) which contribute to regulate the transcriptional complexity of the gene (Kimura et al. 2006; Landry et al. 2003). And third, five transcript isoforms and three protein isoforms were found in a tissue-specific manner (Fig. 4). Although HSP90AA1 has its highest expression in brain and testis (Csermely et al. 1998), we cannot consider it a tissue-specific gene (Meissner et al. 2008) due to its ubiquitous expression (Liu et al. 2014). Instead, when the CGI promoter is active, gene expression regulation is tissue-specific favored by the connection between alternative promoters, alternative splicing, and DNA epigenetic marks (Huang et al. 2010; Manley and Tacke 1996; Previti et al. 2009). This effect is revealed as a tissue-specific alternative splicing being, in our case, the differences observed in testicular and brain transcripts (Fig. 4). These findings could support the evidence that alternative promoters influence alternative splicing processes shown in other species (Pecci et al. 2001).
Alternative splicing is an efficient, accurate, and essential co-transcriptional process (Pandya-Jones and Black 2009) to ensure proper gene expression (Cartegni and Krainer 2003). This process may be affected by the impact of CpG methylation on the conformational structure of the DNA sequence (Anastasiadou et al. 2011). At this point, one of the aims of this work was to study the effect of methylation on the binding of splicing factors and splicing regulatory elements in the ovine HSP90AA1 gene body.
On the one hand, comparative analysis of T-DMRs at the HSP90AA1 gene revealed groups of binding motifs and putative TFs associated with negative regulation or exon exclusion, positive regulation, or exon inclusion and both mechanisms in sperm cells (Supplementary Table 2). If we consider that the predominant cells of adult testicle are spermatozoa (Bellve et al. 1977; Oakes et al. 2007), then these results may confirm that differential DNA methylation gives rise to negative regulation of alternative splicing in testicle. Moreover, it would support the role of intragenic epigenetic marks in regulating AP and AS in the HSP90AA1 gene body. Therefore, the regulation of transcription in testis is very different from other tissues, as observed here, precisely because of the use of an AP (AP2) (Kimura et al. 2006).
On the other hand, the information provided by the detection of canonical alternative splicing sites (Supplementary Fig. 2)—two donor splice sites (in brown), two acceptor splice sites (in green), and two branch sites (in blue)—would be consistent with alternative splicing events in spermatic tissue (Figs. 3 and 4).
Finally, SR proteins are a conserved family of proteins, involved in both constitutive and alternative splicing, that act like splicing enhancers and/or silencers (Manley and Tacke 1996) recognizing cis-elements or ESEs with their corresponding putative sequence motifs containing CpGs. They are particularly important when AS is involved (Cartegni et al. 2003). In our work, we found a putative ESE in exon 2 (Supplemental Fig. 2) that contains a differentially hemi-methylated cytosine at position +678 relative to TSS in sperm (Figs. 2 and 3). This finding could be consistent with gene regulation by introducing a mark that influences binding of a SR protein, SF2/ASF, that recognizes CpG-containing ESE. Therefore, SF2/ASF might have a regulatory role in alternative splicing synergistically with DNA methylation in spermatic tissue (Shukla et al. 2011; Wan et al. 2013). In summary, in silico results reveal that the distribution of CpG methylation and sequence motifs could be associated with splicing events in sperm tissue.
The formation of G4 within gene promoters can modify/influence their expression regulation (Eddy and Maizels 2008). Regions with a potential to form G4 have been identified within the promoters of several proto-oncogenes, including c-MYC and c-KIT (Siddiqui-Jain et al. 2002; Rankin et al. 2005). The presence of G4 motifs in promoters of oncogenes, such as MYC and RAS, fueled efforts to develop small-molecule ligands that would bind to a postulated G4 and downregulate gene expression (Balasubramanian et al. 2011). Moreover, G4 structures could be involved in transcription inhibition by the presence of close DNA methylation. Hardin and colleagues (Hardin et al. 1993) showed that G4-containing methylated CpGs had higher stability in vitro. A more recent in vitro analysis suggests that CpG methylation can greatly increase the thermal stability of a G4 formed in the P1 promoter of the Bcl-2 oncogene (Lin et al. 2013). In our in silico study, the upstream region of TSS in the template strand (Supplemental Fig. 4) showed one potential G4 at position 5431 (Supplemental Table 4) which includes the g.660G > C (rs397514116) mutation (Fig. 3). This putative G4 structure should be stabilized when the cytosine of the CpG is methylated (Lin et al. 2013). This result could explain, in addition to the regulatory methylation mechanism itself, the expression differences observed between g.660G > C alleles (Salces-Ortiz et al. 2015c). If G4 motifs do contribute to the epigenetic regulation by enabling epigenetic marks to be reset upon replication, then genes that respond rapidly to external stimuli would be predicted to be G4 enriched (Maizels and Gray 2013). This should be the case of the HSP90AA1 gene in which a quick and high upregulation is induced as response to heat stress events.
Moreover, a G4 structure within the gene body could play a role in regulation of splicing events (Kikin et al. 2006). Factors associated with RNA processing, including hnRNP D, CBF, the UP1 derived from hnRNP A1, and the nucleolin, bind G4 DNA through their conserved RNA-recognition motif and RNA-binding domain (RRM/RBD) regions (Dempsey et al. 1999; Zhang et al. 2006). This raises the possibility that G4 conformations in RNA transcripts may enhance binding by factors that promote RNA processing and thus stimulate splicing or transport of specific transcripts (Eddy and Maizels 2008). The HSP90AA1 non-template strand is G4-rich downstream of the TSS (Supplemental Fig. 3), but only in two of them, and in sperm cells, epigenetic marks have been detected. A G4 containing a hemi-methylated CpG in E3 at position +890 and a G4 including a methylated CpG in E3 at position +1016 in sperm have been predicted (Fig. 3). Therefore, G4 and methylation would constitute another regulatory mechanism of splicing events.
To summarize, we can confirm that a CGI has been detected in the ovine HSP90AA1 gene, which confers its promoter with a particular constitutive and inductive transcriptional behaviors to cope, in an optimal way, with the large number of roles in which this gene is involved. The methylation patterns of the CGI assessed have led us to speculate about the possible role of epigenetic marks in the regulation of the gene expression and alternative splicing in several tissue types.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Description of the CpG island motifs located at the HSP90AA1 promoter region. The following motifs are highlighted: HSE (purple) at position −89; BRE (blue) at position −39; TATA box (pink) at position −30; TSS (A+1) (grey and underlined) at the Inr (yellow); M6 consensus motifs (Sp1) (i.e., GC boxes) (grey) at positions −151, −147, −45, −23, −12, +99, +127, +156, and +364; M3 consensus motifs (ELK-1) (green) at positions −271 and −85; M22 consensus motif (red) at position −134; and DPE motif (olive green) at position +46 (all positions described here are relative to TSS). (GIF 285 kb)
Representation of alternative spliced sites (ASSs) at the ovine HSP90AA1 gene: exons are grey shaded, mammalian branch sites are in blue (at positions −592 and +4011), donor splice sites highlighted in brown (at positions +2168 and +3557), and acceptor splice sites in green (at positions +2202 and +4457). Exonic splicing enhancers (ESE): eight ESEs are highlighted in pink, two of them include a CpG which are in underlined (at positions −255, +10, +23, +1318, +1695, +3133, +3262, and +4457), and finally, one ESE which contains a tissue-specific differentially methylated CpG is in red shaded (at position +674) (all positions described here are relative to TSS). (DOCX 15 kb)
Representation of a potential G4 at HSP90AA1 non-template strand (reverse sequence) DQ983231.1 (5917 bp). Exons are grey shaded. G4 with G scores of 46–61 are in red. G4s with G scores of 30 and 31 are in yellow. G4s with G scores of 29–20 are in pink, while G4s with G scores of 9–5 are in green. Two G4s at positions +890 and +1016 relative to TSS contain tissue-specific differentially methylated CpGs which are circled. (DOCX 23 kb)
A potential G4 at the HSP90AA1 template strand (forward sequence; DQ983231.1, 5917 bp) is represented. Exons are grey shaded. G4s with G scores of 37–42 are highlighted in red. G4s with G scores of 20 and 21 are in yellow. G4s with G scores of 18–11 are in pink. G4s with G scores of 6–0 are in green. The G4 at position −660 relative to TSS contains a polymorphic CpG. The SNP (rs397514116) associated is circled. (DOCX 23 kb)
Sample identification, tissue, breed, age, genotype, and DNA extraction methods are summarized. Identifications (IDs) from tissues are as follows: M.C.A.B. blood samples from non-stressed adults of Manchega breed, M.H.S.B. blood samples from heat-stressed adults of Manchega breed, R.Y.H. heart samples from young animals of Rasa Aragonesa breed, M.A.B. brain sample from an adult animal of Manchega breed, M.A.L. liver sample from an adult of Manchega breed, M.Y.O. ovary samples from young animals of Manchega breed, R.Y.T. testicle samples from young animals of Rasa Aragonesa breed, M.Y.T. testicle sample from a young animal of Manchega breed, M.A.T. testicle sample from an adult animal of Manchega breed, and M.A.S. sperm samples from adult animals of Manchega breed. The DNA extraction procedures used in this study are the following: salting out protocol (Miller et al. 1988), Gentra Puregene DNA Purification Kit protocol (Gentra, Minnesota, USA), and MasterPure DNA Purification Kit protocol (Epicentre, Wisconsin, USA). M.C.A.B., M.H.S.B., and M.B.T. data are extracted from Salces-Ortiz et al. (2015c). (DOCX 13 kb)
Summary of putative transcription factors involved in positive/negative gene-splicing regulation associated with DNA methylation pattern. The detection of tissue-specific differentially methylated regions (T-DMRs) were enumerated and compared with the HSP90AA1 sequence using a motif comparison tool (meme.nbcr.net/meme/cgi-bin/tomtom.cgi). Most T-DMRs were located both in introns and exons of the sequence studied here, E2, I2, E3, and I3 in sperm tissue with respect to the other tissues. Putative TFs that bind to differentially methylated motifs containing CpGs and that can affect the regulation of alternative splicing were predicted. Here, it has been defined as “negative” regulation when a relative increase of epigenetic marks seems to be associated with the exclusion of several exons (E2, E3, E4, E5, and part of the E6) in sperm tissue and defined as “positive” regulation or inclusion of several exons when it is compared to the rest of the tissues analyzed; in this case, methylation marks seem to be associated with inclusion of exons. The exon 1 is not transcribed. (XLSX 14 kb)
Extract of an output from G-quadruplex analysis tool (QGRS). Position, length, QGRS sequences, and G score of non-template strand are shown. In the reverse strand sequence of the ovine HSP90AA1 gene (DQ983231.1), 56 G4s with a maximum length of 40 bp were predicted. The positions obtained are not related to TSS. (XLSX 15 kb)
Extract of an output from G-quadruplex analysis tool (QGRS). Position, length, QGRS sequences, and G score of template strand are shown. In the forward strand sequence of the ovine HSP90AA1 gene (DQ983231.1), 26 G4s with a maximum length of 30 bp were predicted. The positions obtained are not related to TSS. (XLSX 13 kb)
Acknowledgments
This work was supported by the RTA2009-00098 INIA project. We thank AGRAMA breeders association and CITA (Centro de Investigación y Tecnología Agroalimentaria de Aragón) for providing the biological samples.
Abbreviations
- AP
Alternative promoter
- AS
Alternative splicing
- ASM
Allele-specific methylation
- ASS
Alternative splice site
- BRE
TFIIB recognition element
- CpG
5′-CpG-3′ dinucleotide
- CGI
CpG island
- DPE
Downstream promoter element
- ESEs
Exonic splicinge
- G4
G-quadruplex
- hnRNP A
Heterogeneous nuclear ribonucleoprotein A
- hnRNP D
Heterogeneous nuclear ribonucleoprotein D
- HS
Heat shock
- HSE
Heat shock element
- HSF1
Heat shock factor 1
- HSFs
Heat shock transcription factors
- HSP
Heat shock protein
- hsp90α
90-KDa heat shock protein alpha
- Inr
Initiator
- MBP
Methyl-binding protein
- ORF
Open reading frame
- PCR
Polymerase chain reaction
- Pol II complex
RNA polymerase complex
- RNAseq
RNA sequencing
- RBD
RNA-binding domain region
- RRM
RNA-recognition motif and region
- SP
Specific protein
- SR
Protein serine/arginine splicing factor
- T-DMR
Tissue-specific differentially methylated region
- TF
Transcription factor
- TSS
Transcriptional start site
References
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30:267–275. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/S0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Beaudoin JD, Perreault JP. 50-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;30:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bird A. GpG-rich islands and the function of DNA methylation. Nature. 1986;321:219–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A, Wolffe A. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Boán F, Blanco MG, Barros P, González AI, Gómez-Márquez J. Inhibition of DNA synthesis by K+ stabilised G-quadruplex promotes allelic preferential amplification. FEBS Lett. 2004;571:112–118. doi: 10.1016/j.febslet.2004.06.062. [DOI] [PubMed] [Google Scholar]
- Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LC, Kao CF, Shih WL, Kuo PH. Pathway analysis using information from allele-specific gene methylation in genome-wide association studies for bipolar disorder. PlosOne. 2013 doi: 10.1371/journal.pone.0053092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/S0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, Eichler EE, Hsu L, Henikoff S, Dumanski JP, Trask BJ. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin. 2009 doi: 10.1186/1756-8935-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey LA, Sun H, Hanakahi LA, Maizels N. DNA binding by LR1 and its subunits, nucleolin and hnRNP D: a role for G-G pairing in immunoglobulin switch recombination. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad I, Cederroth CR, Walicki J, Grey C, Barluenga S, et al. The molecular chaperone Hsp90alpha is required for meiotic progression of spermatocytes beyond pachytene in the mouse. PLoS ONE. 2010;5:e15770. doi: 10.1371/journal.pone.0015770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C, Hindermann W, Rosenthal A. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum Mol Gene. 2000;9:2651–2663. doi: 10.1093/hmg/9.18.2651. [DOI] [PubMed] [Google Scholar]
- Hajkova P, el-Maarri O, Engemann S, Oswald J, Olek A, Walter J. DNA-methylation analysis by the bisulfite-assisted genomic sequencing method. Methods Mol Biol. 2002;200:143–154. doi: 10.1385/1-59259-182-5:143. [DOI] [PubMed] [Google Scholar]
- Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27(2):537–42. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin CC, Corregan MBrown BA, II, Frederick LN. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32(22):5870–80. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh I, Zeng J, Park T, Yi SV (2013) DNA methylation and transcriptional noise Epigenetics & chromatin 6:9. doi:10.1186/1756-8935-6-9 [DOI] [PMC free article] [PubMed]
- Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. doi:10.1038/nrg3230 [DOI] [PubMed]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) Nature 436:876–80. doi:10.1038/nature03877 [DOI] [PMC free article] [PubMed]
- Kimura K, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Lin J, Hou JQ, Xiang HD, Yan YY, Gu YC, Tan JH, Li D, Gu LQ, Ou TM, Huang ZS. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation. Biochem Biophys Res Commun. 2013;433(4):368–73. doi: 10.1016/j.bbrc.2012.12.040. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang L, Pu Y, Liu Z, Zhao Y, Qin S. Cloning and expression of a cytosolic HSP90 gene in Chlorella vulgaris. BioMed Res Int. 2014;2014:487050. doi: 10.1155/2014/487050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. Plos Genet. 2013;9(4):e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Calvo JH, González C, Moazami-Goudarzi K, Laurent P, Bertaud M, Hayes H, Beattie AE, Serrano C, Lyahyai J, Martín-Burriel I, Serrano M. Structural and functional analysis of the HSP90AA1 gene: distribution of polymorphisms among sheep with different responses to scrapie. Cell Stress Chaperones. 2008;13:19–29. doi: 10.1007/s12192-007-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Mutkainen M, González C, Calvo JH, Kantanen J, Sanz A, Marzanov NS, Pérez-Guzmán MD, Serrano M. A SNP in the HSP90AA1 gene 5′ flanking region is associated with the adaptation to differential thermal conditions in the ovine species. Cell Stress Chaperones. 2010;15(1):67–81. doi: 10.1007/s12192-009-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Moreno C, Serrano M, Laurent P, Cribiu EP, Andréoletti O, Ruesche J, Weisbecker JL, Calvo JH, Moazami-Goudarzi K. Polymorphisms in the HSP90AA1 5′ flanking region are associated with scrapie incubation period in sheep. Cell Stress Chaperones. 2010;15(4):343–9. doi: 10.1007/s12192-009-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva YA, et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2013;15:119. doi: 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215 [DOI] [PMC free article] [PubMed]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci U S A. 2007;104:228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oner Y, Calvo JH, Serrano M, Elmaci C. Polymorphisms at the 5′ flanking region of the HSP90AA1 gene in native Turkish sheep breeds. Livest Sci. 2012;150:381–385. doi: 10.1016/j.livsci.2012.07.028. [DOI] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. Rna. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecci A, Viegas LR, Baranao JL, Beato M. Promoter choice influences alternative splicing and determines the balance of isoforms expressed from the mouse Bcl-X gene. J Biol Chem. 2001;276:21062–21069. doi: 10.1074/jbc.M008665200. [DOI] [PubMed] [Google Scholar]
- Previti C, Harari O, Zwir I, del Val C. Profile analysis and prediction of tissue-specific CpG island methylation classes. BMC Bioinformatics. 2009;10:116. doi: 10.1186/1471-2105-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S, Rezska AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal P, Kummarasetti VBR, Ravindran J, Kumer N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40(2):253–66. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Salces-Ortiz J, Gonzalez C, Martinez M, Mayoral T, Calvo JH, Serrano M. Looking for adaptive footprints in the HSP90AA1 ovine gene. BMC Evol Biol. 2015;15:7. doi: 10.1186/s12862-015-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Gonzalez C, Bolado-Carrancio A, Rodriguez-Rey JC, Calvo JH, Muñoz R, Serrano M. Ovine HSP90AA1 gene promoter: functional study and epigenetic modifications. Cell Stress Chaperones. 2015 doi: 10.1007/s12192-015-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Ramón M, Gonzalez C, Perez-Guzman MD, Garde JJ, Calvo JH, Serrano MM. Differences in the ovine HSP90AA1 gene expression rates caused by two linked polymorphisms at its promoter affect rams sperm DNA fragmentation under environmental heat stress conditions. Plos One. 2015;10(2):e0116360. doi: 10.1371/journal.pone.0116360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Gonzalez C, Moreno-Sanchez N, Calvo JH, Perez-Guzman MD, Serrano MM. Ovine HSP90AA1 expression rate is affected by several SNPs at the promoter under both basal and heat stress conditions. PLoS ONE. 2013;8(6):e66641. doi: 10.1371/journal.pone.0066641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;2006(103):1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarano E, Iaccarino M, Grippo P, Parisi E. The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos. Proc Natl Acad Sci U S A. 1967;57(5):1394–400. doi: 10.1073/pnas.57.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M et al (2011) Use of maximum likelihood-mixed models to select stable reference genes: a case of heat stress response in sheep Bmc Molecular Biology 12. doi:10.1186/1471-2199-12-36 [DOI] [PMC free article] [PubMed]
- Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bears DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99(18):11593–8. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SS, Baker DJ, Kaplan BE, Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5) methyltransferase. J Mol Biol. 1991;217(1):39–51. doi: 10.1016/0022-2836(91)90609-A. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102(9):3336–41. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedhar AS, Kalmár E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562(1–3):11–5. doi: 10.1016/S0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- Stevens AJ, Stuffrein-Roberts S, Cree SL, Gibb A, Miller AL, Doudney K, Aitchison A, Eccles MR, Joyce PR, Filichev VV, Kennedy MA. G-quadruplex structures and CpG methylation cause drop-out of the maternal allele in polymerase chain reaction amplification of the imprinted MEST gene promoter. Plos One. 2014;9(12):e113955. doi: 10.1371/journal.pone.0113955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi Y, Takasaki I, Wada S, Zhao QL, Hori T, Nomura T, Ohtsuka K, Kondo T. Genes and genetic networks responsive to mild hyperthermia in human lymphoma U973 cells. Int J Hyperthermia. 2008;24(8):613–22. doi: 10.1080/02656730802140777. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA (2002) Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A 99(6):3740–3745. doi:10.1073/pnas.052410099 [DOI] [PMC free article] [PubMed]
- Trowbridge JJ, Orkin SH. DNA methylation in adult stem cells: new insights into self-renewal. Epigenetics. 2010;5:189–193. doi: 10.4161/epi.5.3.11374. [DOI] [PubMed] [Google Scholar]
- Wan J, Oliver VF, Zhu H, Zack DJ, Qian J, Merbs SL. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013;41:8503–8514. doi: 10.1093/nar/gkt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ (2002) Identification and resolution of artifacts in bisulfite sequencing. 27(2):101–7. doi:10.1016/S1046-2023(02)00060-9 [DOI] [PubMed]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2(9):1136–43. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Current Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12:1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucl Acids Res. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the CpG island motifs located at the HSP90AA1 promoter region. The following motifs are highlighted: HSE (purple) at position −89; BRE (blue) at position −39; TATA box (pink) at position −30; TSS (A+1) (grey and underlined) at the Inr (yellow); M6 consensus motifs (Sp1) (i.e., GC boxes) (grey) at positions −151, −147, −45, −23, −12, +99, +127, +156, and +364; M3 consensus motifs (ELK-1) (green) at positions −271 and −85; M22 consensus motif (red) at position −134; and DPE motif (olive green) at position +46 (all positions described here are relative to TSS). (GIF 285 kb)
Representation of alternative spliced sites (ASSs) at the ovine HSP90AA1 gene: exons are grey shaded, mammalian branch sites are in blue (at positions −592 and +4011), donor splice sites highlighted in brown (at positions +2168 and +3557), and acceptor splice sites in green (at positions +2202 and +4457). Exonic splicing enhancers (ESE): eight ESEs are highlighted in pink, two of them include a CpG which are in underlined (at positions −255, +10, +23, +1318, +1695, +3133, +3262, and +4457), and finally, one ESE which contains a tissue-specific differentially methylated CpG is in red shaded (at position +674) (all positions described here are relative to TSS). (DOCX 15 kb)
Representation of a potential G4 at HSP90AA1 non-template strand (reverse sequence) DQ983231.1 (5917 bp). Exons are grey shaded. G4 with G scores of 46–61 are in red. G4s with G scores of 30 and 31 are in yellow. G4s with G scores of 29–20 are in pink, while G4s with G scores of 9–5 are in green. Two G4s at positions +890 and +1016 relative to TSS contain tissue-specific differentially methylated CpGs which are circled. (DOCX 23 kb)
A potential G4 at the HSP90AA1 template strand (forward sequence; DQ983231.1, 5917 bp) is represented. Exons are grey shaded. G4s with G scores of 37–42 are highlighted in red. G4s with G scores of 20 and 21 are in yellow. G4s with G scores of 18–11 are in pink. G4s with G scores of 6–0 are in green. The G4 at position −660 relative to TSS contains a polymorphic CpG. The SNP (rs397514116) associated is circled. (DOCX 23 kb)
Sample identification, tissue, breed, age, genotype, and DNA extraction methods are summarized. Identifications (IDs) from tissues are as follows: M.C.A.B. blood samples from non-stressed adults of Manchega breed, M.H.S.B. blood samples from heat-stressed adults of Manchega breed, R.Y.H. heart samples from young animals of Rasa Aragonesa breed, M.A.B. brain sample from an adult animal of Manchega breed, M.A.L. liver sample from an adult of Manchega breed, M.Y.O. ovary samples from young animals of Manchega breed, R.Y.T. testicle samples from young animals of Rasa Aragonesa breed, M.Y.T. testicle sample from a young animal of Manchega breed, M.A.T. testicle sample from an adult animal of Manchega breed, and M.A.S. sperm samples from adult animals of Manchega breed. The DNA extraction procedures used in this study are the following: salting out protocol (Miller et al. 1988), Gentra Puregene DNA Purification Kit protocol (Gentra, Minnesota, USA), and MasterPure DNA Purification Kit protocol (Epicentre, Wisconsin, USA). M.C.A.B., M.H.S.B., and M.B.T. data are extracted from Salces-Ortiz et al. (2015c). (DOCX 13 kb)
Summary of putative transcription factors involved in positive/negative gene-splicing regulation associated with DNA methylation pattern. The detection of tissue-specific differentially methylated regions (T-DMRs) were enumerated and compared with the HSP90AA1 sequence using a motif comparison tool (meme.nbcr.net/meme/cgi-bin/tomtom.cgi). Most T-DMRs were located both in introns and exons of the sequence studied here, E2, I2, E3, and I3 in sperm tissue with respect to the other tissues. Putative TFs that bind to differentially methylated motifs containing CpGs and that can affect the regulation of alternative splicing were predicted. Here, it has been defined as “negative” regulation when a relative increase of epigenetic marks seems to be associated with the exclusion of several exons (E2, E3, E4, E5, and part of the E6) in sperm tissue and defined as “positive” regulation or inclusion of several exons when it is compared to the rest of the tissues analyzed; in this case, methylation marks seem to be associated with inclusion of exons. The exon 1 is not transcribed. (XLSX 14 kb)
Extract of an output from G-quadruplex analysis tool (QGRS). Position, length, QGRS sequences, and G score of non-template strand are shown. In the reverse strand sequence of the ovine HSP90AA1 gene (DQ983231.1), 56 G4s with a maximum length of 40 bp were predicted. The positions obtained are not related to TSS. (XLSX 15 kb)
Extract of an output from G-quadruplex analysis tool (QGRS). Position, length, QGRS sequences, and G score of template strand are shown. In the forward strand sequence of the ovine HSP90AA1 gene (DQ983231.1), 26 G4s with a maximum length of 30 bp were predicted. The positions obtained are not related to TSS. (XLSX 13 kb)