Abstract
Proteostasis is an integral component of healthy aging, ensuring maintenance of protein structural and functional integrity with concomitant impact upon health span and longevity. In most metazoans, increasing age is accompanied by a decline in protein quality control resulting in the accrual of damaged, self-aggregating cytotoxic proteins. A notable exception to this trend is observed in the longest-lived rodent, the naked mole-rat (NMR, Heterocephalus glaber) which maintains proteostasis and proteasome-mediated degradation and autophagy during aging. We hypothesized that high levels of the proteolytic degradation may enable better maintenance of proteostasis during aging contributing to enhanced species maximum lifespan potential (MLSP). We test this by examining proteasome activity, proteasome-related HSPs, the heat-shock factor 1 (HSF1) transcription factor, and several markers of autophagy in the liver and quadriceps muscles of eight rodent species with divergent MLSP. All subterranean-dwelling species had higher levels of proteasome activity and autophagy, possibly linked to having to dig in soils rich in heavy metals and where underground atmospheres have reduced oxygen availability. Even after correcting for phylogenetic relatedness, a significant (p < 0.02) positive correlation between MLSP, HSP25, HSF1, proteasome activity, and autophagy-related protein 12 (ATG12) was observed, suggesting that the proteolytic degradation machinery and maintenance of protein quality play a pivotal role in species longevity among rodents.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0672-x) contains supplementary material, which is available to authorized users.
Keywords: Aging, Chaperones, Proteostasis, Naked mole rat, Proteasome
Introduction
Conformation states of cellular proteins are in continuous flux, spontaneously unfolding and refolding. Should protein misfolding occur, improper protein-protein interactions, protein aggregation, and subsequent cytotoxicity may result (Gidalevitz et al. 2011). Cells have complex quality control systems employing molecular chaperones to help resolve misfolded and aggregated proteins and thereby maintain protein homeostasis. There is still much debate regarding the role of molecular chaperones in longevity and if their function is attenuated or potentiated with advancing age (Morimoto and Cuervo 2009). Heat-shock factor 1 (HSF1) is the master transcription factor that regulates the expression of this class of molecular chaperones (i.e., heat shock proteins [HSPs] (Shamovsky and Nudler 2008). Both HSF1 expression and that of some of its regulated genes (i.e., constitutive heat shock protein 70 (HSC70) and inducible HSP70) have been linked to enhanced stress response, increased longevity, and/or improved health in diverse species ranging from Caenorhabditis elegans to primates (Kavanagh et al. 2011; Morley and Morimoto 2004; Salway et al. 2011a; Sarup et al. 2014).
Irreversibly damaged or misfolded proteins are transported by chaperones to the UPS and/or autophagosomes for proteolytic degradation. The UPS is responsible for normal turnover of short-lived proteins, as well as for the degradation of damaged, denatured, and oxidized proteins (Chondrogianni et al. 2014). Similarly autophagy is responsible for the degradation of relatively long-lived proteins, large protein aggregates, and for the removal of damaged or redundant organelles (Levine and Klionsky 2004). Both of these proteolytic systems, like chaperones, are upregulated in response to environmental stressors such as heat, nutrient limitation, and oxidative stress (Hohn and Grune 2014; Levine and Klionsky 2004). These all work in concert to maintain chaperone-mediated proteostasis.
The proteolytic components of the 26S proteasome (26S) require ATP to degrade ubiquitinated proteins (Demartino and Gillette 2007). 26S consists of a 20S catalytic core and 19S regulatory caps which remove ubiquitin and unfold proteins prior to catalytic cleavage by the 20S catalytic core (Demartino and Gillette 2007). The proteasome can however, act in an ubiquitin-independent manner when ATP levels are reduced. Under these circumstances, the 19S regulatory caps are either replaced by the 11S activator (PA28) or removed completely (Demartino and Gillette 2007; Freudenburg et al. 2013). The proteasome has three main proteolytic activities: (a) chymotrypsin-like (ChTL) cleaves proteins after a hydrophobic amino acid, (b) trypsin-like (TL) cuts after a basic residue, and (c) caspase like or peptidyl glutamyl peptide hydrolyzing (PGPH) incises after acidic amino acids (Gaczynska and Osmulski 2005).
Proteasome function reportedly declines with age in mice, and the resultant accrual of damaged or misfolded proteins may possibly contribute to the decline in physiological function commonly observed during aging (Dasuri et al. 2009; Rodriguez et al. 2010). Indeed, overexpression of proteasome subunits and genetic alterations aimed at enhancing proteasome activity has been shown to increase lifespan in yeast and C. elegans (Chondrogianni et al. 2015; Kruegel et al. 2011) and as such basal proteasome activities may contribute to disparate species longevity. Similarly, genetic manipulations that give rise to increased autophagy possibly via the initiation and/or formation of the autophagosome have been also implicated in lifespan extension in yeast, worms, and vertebrate models (Lapierre et al. 2013; Mai et al. 2012; Pride et al. 2015).
We have previously observed that long-lived naked mole-rats (NMRs; Rodentia, Heterocephalus glaber) when compared to laboratory mice, exhibit high expression levels of several chaperones as well as proteasome activity and autophagy (Pride et al. 2015; Rodriguez et al. 2012, 2014). We questioned if these traits were unique features of the NMR or if they are key components linked to both species longevity and health span in muscle and liver tissue. Using eight rodent species representing five different rodent families and ranging in MLSP from approximately four (golden hamster, laboratory mice) to more than 30 years (NMRs) (Fig. 1) were used in a comparative study to assess the role of these proteolytic pathways in divergent species longevity. Because across all phyla, the species body size correlates with longevity (Speakman 2005), we also examined the association between these proteins and pathways with body mass. Further, we assess whether this pattern of expression or activity of proteasome and/or levels of autophagy-related proteins and the various chaperones related to these two processes, namely, HSP70 and HSP40, could be a marker or determinant of a more robust health span and increased longevity.
Fig. 1.
Mass, maximum species life span potential (MLSP), and phylogeny of species used in this study. a Species used in this study organized by mass. Abbreviations are noted under their genus and species (column 1), and these abbreviations will be used in all subsequent figures. b Phylogenetic tree of the species used in this study determined by five nuclear genes under the generalized time-reversible model of sequence evolution as previously described (Edrey et al. 2012). Branch lengths are inferred. See also Statistical Analyses in Methods section for more details. c Comparison of mass to MLSP in the species used in this study
Methods
Animals
Naked mole-rats (Heterocephalus glaber) and Damaraland mole-rats (Fukomys damarensis) were from the well-characterized colonies of Dr. Rochelle Buffenstein housed at the University of Texas Health Science Center, San Antonio. Young 4- to 6-month-old laboratory mice (C57BL6; Mus musculus) were obtained from Jackson Laboratories (Bar Harbor, ME, USA). Young guinea pigs (Cavia porcellus), gerbils (Meriones unguiculatus), and hamsters (Mesocricetus auratus) were purchased from Harlan Laboratories (Denver, CO, USA). These animals were housed under the Association for Assessment and Accreditation of Laboratory Animal Care International standards of care. Tissues of young white-footed mice (Peromyscus eremicus) were obtained from the lab of Dr. Kristan. Spalax galili tissues were harvested from wild-caught animals kept in the laboratory of Dr. Nevo. Tissues harvested outside our laboratory were shipped to our laboratory on dry ice. See Fig. 1 for a list of all the animals used in this study, their maximum species lifespan potential (MLSP), and their average mass according to the AnAGE database (www. genomics.senesence.info/species) (deMagalhães and Costa 2009).
All animals used in these studies were young, healthy female individuals. Tissues from at least seven different individuals of each species were used for our measurements. Immediately after euthanasia, tissues were flash-frozen in liquid nitrogen and stored at −80 °C until use. All animal protocols were approved by the University of Texas Health Science Center at San Antonio Institutional Animal Care and Use Committee.
Preparation of tissue lysates
A sample of liver or muscle from at least seven individuals per species was used. The individual tissue was weighed and disrupted in a 2-mL Potter-Elvehjem homogenizer in reticulocyte standard buffer (RSB, 10 mM HEPES, pH-6.2, 10 mM NaCl, 1.4 mM MgCl2) at a weight-to-volume ratio of 1 g of tissue to 1 mL of buffer for liver or 1 g of tissue to 5 mL of buffer for muscle. One portion of the tissue was placed in RSB supplemented with the addition of 1 mM ATP, 0.5 mM DTT, and 5 mM MgCl2 to help maintain intact 26S subassemblies (Liu et al. 2006) for peptidolytic assays. To the other equal portion, one tablet/10 ml protease and phosphatase inhibitor mini tablets were added (Thermo Fisher Scientific, Waltham, MA, USA) for use in Western blots. To clear the lysate of debris, the homogenates were spun at 1000×g for 12 min, and the supernatant was then stored as 50 μl aliquots at −80 °C until further use.
Peptidolytic activity assays
In each assay, 20 μg of whole tissue lysates were incubated with 100 μM of substrate specific for the type of proteasome activity. A saturating concentration of proteasome inhibitor N-(benzyloxycarbonyl) leucinyl-leucinyl-leucinal (MG132) was added to parallel samples. The difference of the fluorescence released with and without inhibitor was used as a measure of the net peptidolytic activity of proteasome as previously described using model peptide substrates to represent cleavage after hydrophobic (chymotrypsin-like, ChTL) residues, basic residues (trypsin-like, TL), and acidic residues (post-glutamyl, peptide-hydrolizing, PGPH) (Rodriguez et al. 2010).
Western blot analyses
Tissue lysates were separated using a 4–20 % SDS-PAGE (Biorad Life Sciences) and transferred to nitrocellulose membranes (Biorad Life Sciences). The membranes were probed with antibodies against the following proteasome and chaperone proteins: HSP110 (rabbit pAb, SPA1101, 1:5 K), HSP90 (mouse mAb, SPA831, 1:20 K), HSF1 (rabbit pAb, SPA901, 1:5 K), p-HSF1 (Ser326) (rabbit pAb, SPA902, 1:5 K), HSP70/72 (mouse mAb, SPA810, 1:10 K), HSP40 (HDJ1) (rabbit pAb, SPA400, 1:2.5 K), HSP25/HSPB1 (rabbit pAb, SPA801, 1:10 K), α7 (mouse mAb, PW8110, 1:5 K), RPT5 (mouse mAb, PW8310, 1:5 K) (Enzo Life Sciences, Plymouth Meeting, PA, USA), carboxyl-terminus of HSP70-interacting protein (CHIP) (rabbit mAb(C3B6), #2080, 1:2.5 K) (Cell Signaling Technology, Inc., Danvers, MA, USA), HSC70 (goat pAb, sc-1059, 1:2 K), and PA28α (goat pAb, sc-21267, 1:2 K) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Antibodies against GAPDH (mouse mAb, G8795, 1:30 K) (Sigma-Aldrich, St. Louis, MO, USA) were used as a loading control. For autophagy proteins, the following antibodies were used all at 1:5 K dilutions: beclin-1 (rabbit mAb, #3495P), ATG12 (rabbit mAb, #4180P), LC3A/B (rabbit mAb, #4108P), ATG5 (rabbit mAb, #8540P), ATG7 (rabbit mAb, #8558P), and ATG3 (rabbit mAb, #3451P) (Cell Signaling Technology). The GAPDH antibody detailed above was used as a loading control.
Primary antibodies were detected using anti-rabbit IRDye 680LT or anti-mouse IR Dye800 CW (Li-Cor, Lincoln, NB, USA) conjugated antibodies. Secondary antibodies were incubated at 1:10 K for 2 h at room temperature, and images were captured and subsequently quantified using the Odyssey Imaging System (Li-Cor) by quantifying fluorescent signals as integrated intensities (I.I. K Counts) using the Odyssey Infrared Imaging System, Application Version 3.0 software. We used a local background subtraction method to subtract independent background values from each box, more specifically, the “median” background function with a 3-pixel width border above and below each box was subtracted from individual counts. We calculated ratios for each antibody against the GAPDH loading control using I.I. K Counts.
Statistical analyses
Multispecies comparisons for MLSP and mass were performed by using the linear regression program in GraphPad Prism 6 (GraphPad Software, Inc.) and followed by phylogenetic independent contrast (PIC), analyses using the phylogenetic tree used in Fig. 1 was previously described (Edrey et al. 2012). This phylogeny estimate was constructed using a maximum likelihood method applied to five nuclear genes. The PIC covariance matrix derived from this phylogenetic tree assumed a Brownian model of trait evolution. For the regression analysis, the MLSP variable was used in both original and log-transformed scales as the outcome variable, and the body mass and the protein levels were used as predictors. For the residual analysis, MLSP was regressed onto body mass, and the residuals of MLSP were used as the outcome in the PIC regression with protein levels as predictors. The PIC used was implemented within the Analyses of Phylogenetics and Evolution package (APE) analyses of the linear regression in R statistical software assuming a Brownian model of trait evolution (Paradis et al. 2004). To transform the various phenotypes to a standard scale, variables in the cluster analysis were mean-centered and scaled to have standard deviations of 1.0. The clustering algorithm used the Euclidean distance and the complete linkage method. The hierarchical clustering was based only on the traits measured and did not explicitly consider the phylogenetic tree. Hence, the dendrograms from the clustering were derived from the phenotypes and not the phylogenetic information. Clustering and heatmaps were implemented with R statistical software (R Development Core Team 2010).
Results
HSP25 and HSF1 protein levels correlate with rodent species longevity in both liver and quadriceps muscle tissues
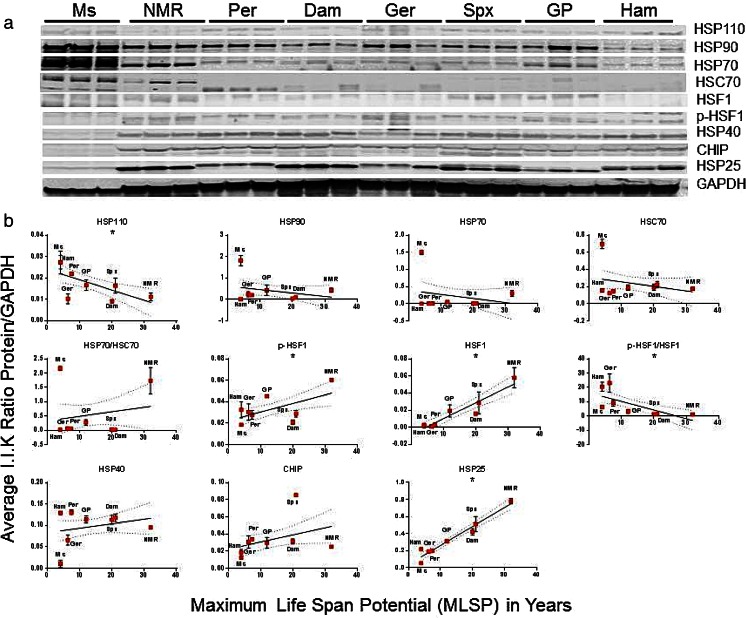
To examine if basal protein levels of HSP are linked to maximum lifespan potential, we used a comparative biology approach and evaluated protein content for a panel of selected HSPs in quadriceps muscle and liver lysates from eight rodent species using Western blotting techniques. There was a significant positive association between protein expression of HSP25, phosphorylated HSF1 (p-HSF1), and total HSF1 with MLSP (Fig. 2, Table S1) in quadriceps muscle. Conversely, HSP110 showed a negative correlation (p < 0.05) with MLSP in muscle tissue (Fig. 2, Table S1).
Fig. 2.
HSP25 and active HSF1 correlate with maximum life span potential (MLSP) in quadriceps muscle of eight rodent species. a Representative Western blots showing the heat-shock chaperone protein (HSP) species and HSF1 forms tested in this study. This analysis was repeated five times with similar results. b Quantitation and linear regression analyses (GraphPad Prism v 6.0) showed that HSP25 (R 2 = 0.89, p < 0.001), phosphorylated HSF1 (p-HSF1; R 2 = 0.30, p = 0.006) and total HSF1 (R 2 = 0.23, p = 0.03) correlated positively with MLSP in quadriceps muscle lysates. HSP110 correlated negatively (R 2 = 0.32, p = 0.004). Plots show mean ± S.E.M. (n = 4 per experiment) and *indicates significance. See also Table 1, S1
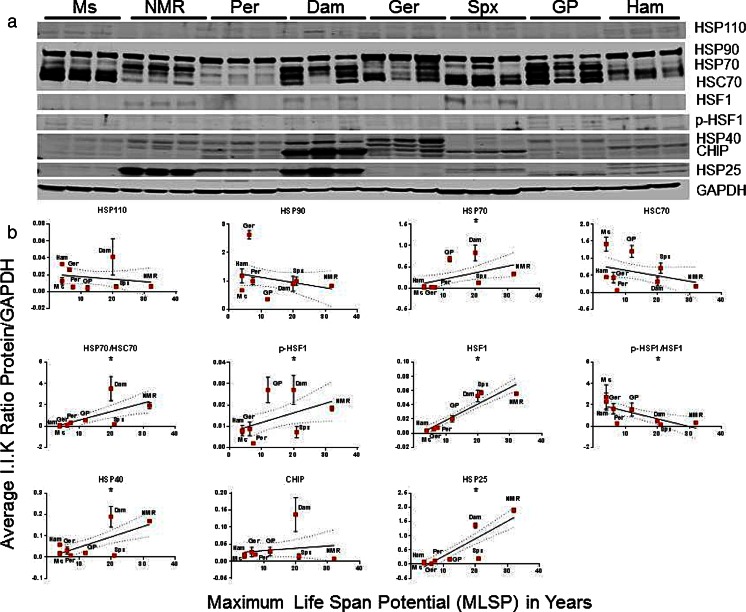
In liver lysates, protein expression of HSP25, HSF1, and p-HSF1 correlated positively (p < 0.05) with MLSP (Fig. 3, Table S1) such that for the doubling of HSP25 protein levels longevity was extended close to threefold. Furthermore, there was also a positive association (p < 0.05) between HSP40, HSP70, and HSP70 flux (the ratio of HSP70 to HSC70) in liver tissue (Figure S1). However, HSC70, HSP70, and p-HSF1 in liver tissue also showed a significant positive correlation with mass. Based on the R 2 values, these relationships with body mass were stronger than those with MLSP (Table S1). Significant associations between HSP25, HSP70 flux, and total HSF1 with MLSP were independent of mass in this tissue (Figure S2, Table S1). After removing the effects of body size, the magnitude of association between protein expression of HSP25 and HSF1 with MLSP was nevertheless robust (Table S2).
Fig. 3.
In liver tissue of eight rodent species, HSP25, HSP40, active HSF1, and HSP70 flux correlate with maximum life span potential (MLSP). a Representative Western blots showing the heat-shock chaperone protein (HSP) species and HSF1 forms tested in this study. This analysis was repeated five times with similar results. b Quantitation and linear regression analyses (GraphPad Prism v 6.0) showed that again HSP25 (R 2 = 0.76, p < 0.001), phosphorylated HSF1 (p-HSF1; R 2 = 0.17, p = 0.04), and total HSF1 (R 2 = 0.82, p < 0.001) correlated positively with MLSP, this time in liver lysates. Further, HSP40 (R 2 = 0.40, p < 0.001), HSP70 (R 2 = 0.21, p = 0.02), and the HSP70 flux (the ratio of HSP70 to HSC70; R 2 = 0.35, p = 0.003) also showed a positive correlation MLSP in these samples. Plots show mean ± S.E.M. (n = 4 per experiment) and *indicates significance. See also Table 1, S1
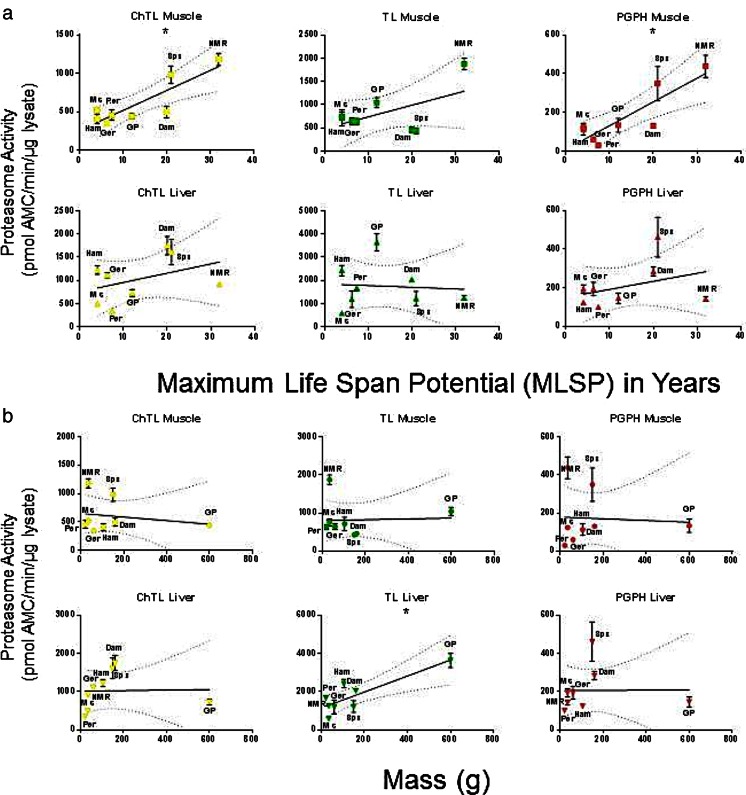
Proteasome activity correlates with longevity in muscle but not in liver
Significant positive correlations between ChTL and PGPH activities, with MLSP in quadriceps muscle were evident (Fig. 4a (top), Table S1). In contrast, none of the activities were associated with MLSP in liver lysates (Fig. 4a (bottom)). Similarly, there were no significant correlations between proteasome activities with body mass in the quadriceps muscle, although TL activity did show a positive correlation with species mass in the liver (Fig. 4b, Table S1). We evaluated if differences in proteasome activity reflected differences in specific activity or proteasome assembly population. Therefore, we tested protein expression of α7 for total proteasome, regulatory particle ATPase subunit 5 (RPT5) for 26 proteasome, and proteasome activator subunit 1 (PA28α) for immunoproteasome to measure the association between the type and amount of proteasomes and MLSP or mass. α7 did not correlate with either MLSP or mass (p > 0.05), but RPT5 and PA28α both correlated positively with mass (p < 0.05) in the liver (Figure S3, Table S1). There were no significant correlations of proteasome activity with mass or MLSP in the muscle (Figure S3). Thus, MLSP only correlated with proteasome activity, but not proteasome content, in quadriceps muscle suggesting that proteasome efficiency is more important for species longevity.
Fig. 4.
Both chymotrypsin-like (ChTL) and caspase-like (PGPH) proteasome activity correlated with maximum life span potential (MLSP) in quadriceps muscle in eight rodent species. a Linear regression analyses (GraphPad Prism v 6.0) on proteasome activity from quadriceps muscle in eight rodent species reveal that both ChTL (R 2 = 0.73, p = 0.007) and PGPH (R 2 = 0.72, p = 0.008) have significant positive correlations with MLSP. Trypsin-like (TL) activity did not. In liver lysates, there was no correlation with MLSP. b In contrast, there was no correlation between proteasome activity and mass in quadriceps muscle lysates, although in liver lysates, TL activity showed a positive correlation (R 2 = 0.72, p = 0.008). Plots show mean ± S.E.M. (n = 7). Significance is indicated by *. See also Table 1, S1
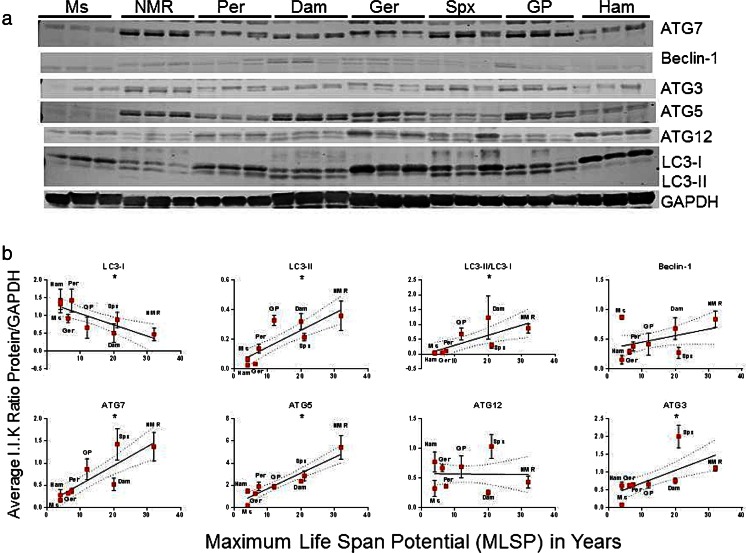
Autophagy markers show significant correlations with MLSP
Since chaperones such as HSC70 and HSP25 interact with and regulate autophagy, (Janen et al. 2010; Rout et al. 2014) we tested autophagy-related protein expression in our eight rodent species. In quadriceps muscle, microtubule-associated protein 1A/1B-light chain 3 bound to phosphatidylethanolamine (LC3-II), autophagic flux (LC3-II/LC3-I, (i.e., ratio of bound to unbound LC3A/B)), autophagy-related protein 7 (ATG7), ATG5, and ATG3 correlated positively (p < 0.05) with MLSP; whereas, LC3-I was negatively correlated (p < 0.05) with MLSP (Fig. 5, Table S1). Examination of autophagic markers in liver tissue revealed that LC3-II, ATG7, and ATG3 were positively correlated (p < 0.05) with MLSP (Fig. 6, Table S1). Further examination of correlations between mass and autophagic markers revealed that LC3-II correlated positively (p < 0.05) with mass in muscle samples though the R 2 value compared to muscle was much lower (R 2 = 0.18 vs R 2 = 0.57; Table S1). Autophagic flux is also positively correlated (p < 0.05) with increased mass in liver lysates. Interestingly, LC3-I and ATG12 exhibited significant inverse relationships with mass (Figure S4, Table S1). Hence, only ATG7 and ATG3 showed a positive correlation with MLSP in both tissues independent of mass.
Fig. 5.
Markers of autophagy show both positive and negative correlations with rodent maximum life span potential (MLSP) in quadriceps muscle. a Representative Western blots showing the various autophagy-related proteins examined in this study. This analysis was repeated three times with similar results. b Quantitation and linear regression analyses (GraphPad Prism v 6.0) in quadriceps muscle lysates showed that, LC3-II (R 2 = 0.57, p < 0.001), LC3-II/LC3-I (R 2 = 0.50, p = 0.005), ATG7 (R 2 = 0.52, p < 0.001), ATG5 (R 2 = 0.73, p < 0.001), and ATG3 (R 2 = 0.47, p = 0.003) correlated positively with MLSP. In contrast, LC3-I (R 2 = 0.34, p = 0.002) showed a negative correlation. Plots show mean ± S.E.M. (n = 3 per experiment) and *indicates significance. See also Table 1, S1
Fig. 6.
LC3-II, ATG7 and ATG3 show correlations with maximum life span potential (MLSP) in liver tissue. a Representative Western blots showing the various autophagy-related proteins examined in this study. This analysis was repeated three times with similar results. b Quantitation and linear regression analyses (GraphPad Prism v 6.0) in liver lysates revealed that LC3-II (R 2 = 0.46, p < 0.001), ATG7 (R 2 = 0.19, p = 0.03) and ATG3 (R 2 = 0.18, p = 0.04) showed positive correlations with MLSP. Plots show mean ± S.E.M. (n = 3 per experiment) and *indicates significance. See also Table S1
Phylogenetic independent contrast and residual analyses reveal potential markers of longevity
Traits from closely related species are inherently non-independent. We used phylogenetic independent contrast (PIC) to correct for phylogenetic-specific traits that could mask the linear regression relationships related to MLSP (Garland et al. 1992; Paradis et al. 2004). In muscle tissue, these analyses revealed that HSP25 (R 2 = 0.94, p < 0.001) and HSF1 (R 2 = 0.90, p < 0.001) positively correlated with MLSP (Table 1). ChTL (R 2 = 0.88, p = 0.007) and PGPH (R 2 = 0.82, p = 0.008) activities showed a similar positive relationship (Table 1). From the autophagy-related genes measured, ATG12 (R 2 = 0.72, p = 0.03) showed a positive correlation while Beclin-1 (R 2 = 0.69, p = 0.02) had a negative correlation independent of phylogeny (Table 1). In liver tissue, again HSP25 (R 2 = 0.89, p = 0.005) and HSF1 (R 2 = 0.88, p < 0.001) showed positive correlation with MLSP after PIC analyses (Table 1). Curiously, HSF1 flux showed a negative correlation (R 2 = 0.44, p < 0.04) (Table 1). HSP70 flux (R 2 = 0.57, p = 0.08) and HSP40 (R 2 = 0.83, p = 0.06) approached but fell short of significance after PIC (Table 1). There were no correlations between the various proteins and MLSP in muscle that were independent of phylogeny in relation to mass; however in the liver, both RPT5 (R 2 = 0.92, p = 0.005) and TL proteasome activity (R 2 = 0.75, p = 0.008) were still significant after PIC (Table 1; See Supplementary Material Tables S2 for full PIC and Residual analyses). Thus, after PIC analyses, only the proteins HSP25 and HSF1 showed a positive relationship to MLSP in both tissues while tests of degradative processes (proteasome and autophagy) correlated with MLSP only in muscle tissue.
Table 1.
Selected linear regression statistics after phylogenetic independent contrast (PIC) and residual analyses showing statistically significant protein content or activity as they relate to maximum species life span potential (MLSP) in years (y) and mass in grams (g)
| Protein or activity | R | R 2 | P value |
|---|---|---|---|
| Muscle MLSP (y) | |||
| HSF1 | 0.95 | 0.90 | <0.001 |
| HSP25 | 0.97 | 0.95 | <0.001 |
| ChTL activity | 0.85 | 0.73 | 0.007 |
| PGPH activity | 0.85 | 0.72 | 0.007 |
| Beclin-1 | −0.79 | 0.62 | 0.02 |
| ATG12 | 0.76 | 0.58 | 0.03 |
| Liver MLSP (y) | |||
| HSP70/HSC70 | 0.66 | 0.43 | 0.08 |
| HSF1 | 0.93 | 0.86 | 0.001 |
| p-HSF1/HSF1 | −0.72 | 0.52 | 0.04 |
| HSP40 | 0.68 | 0.47 | 0.06 |
| HSP25 | 0.87 | 0.76 | 0.005 |
| Liver Mass (g) | |||
| TL activity | 0.85 | 0.72 | 0.008 |
| RPT5 | 0.87 | 0.76 | 0.005 |
Cluster analyses show tissue diversity in regards to markers of species longevity
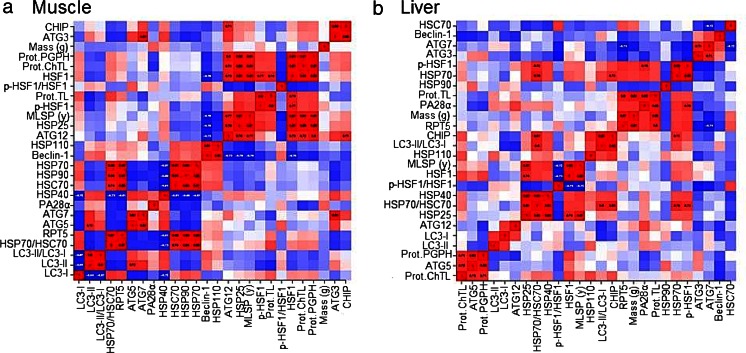
We employed cluster analyses to combine the observations of positive and negative correlations between the various protein expression patterns, proteasome activity, mass, and MLSP phenotypes between species with particular emphasis on proteins and proteasome activities that were still significant after PIC. In hierarchical clustering (Fig. 7a, b), phenotypes that were most similar were paired and treated as a unit (pair1). Next, the most similar phenotypes (including the pair1) are grouped and treated as a unit (pair2), etc. Clustering by this hierarchical method revealed that interactions between the various phenotypes were distinct for tissue types (Fig. 7a, b). In muscle tissue, MLSP strongly and positively associated with HSP25, p-HSF1, ATG12, all three proteasome activities, whereas MLSP negatively associated with beclin-1 (Fig. 7a, upper right). The most similar HSP90/HSP70 pairing correlated negatively with HSC70 and HSP40 (Fig. 7a, middle left), but positively with RPT5 and HSP70 flux (Fig. 7a, middle-bottom left). Other significant relationships were observed between the ATG3/ATG7 pair (Fig. 7a, upper right) and the interactions between the various LC3 phenotypes (Fig. 7a, lower left). In the liver, unlike in the muscle, MLSP did not associate with proteasome activity, but still correlated positively with HSP25 (Fig. 7b). The MLSP/HSP25 pairing also correlated positively with HSP70 flux, HSP40, total HSF1, and p-HSF1 (Fig. 7b, middle-lower left). Mass, however, showed positive correlation with TL proteasome activity, RPT5, and PA28α (Fig. 7b, middle-upper right).
Fig. 7.
Cluster analyses show distinct, tissue-related patterns of interactions between chaperone proteins and activities in the proteostatic network and their correlations to mass and maximum life span potential (MLSP). Positive correlations are indicated in red, while negative correlations are indicated in blue. In muscle (a), HSP25 strongly and positively associated with MLSP, p-HSF1, and all three proteasome activities (upper right cluster). Another strong positive grouping existed between HSP70 and HSP90 that correlated negatively with HSC70 (middle-lower cluster). In the liver (b), unlike in muscle, HSP25 did not cluster with proteasome activity, but instead with MLSP and HSP70 flux (middle lower left). Mass and TL proteasome activity (upper right) showed a positive relationship and HSP70 and p-HSF1 also showed a positive correlation that had a negative relationship with HSF1 flux (mid-left center)
Heatmap cluster analyses comparing the activity, protein expression, mass, and MLSP phenotypes tested against species once again revealed divergent patterns based on tissue type (Figure S5A,B). The analyses of phenotypes from muscle tissue showed that NMRs and laboratory mice were only loosely grouped, but these were not highly similar and both of these species dissimilar to the rest of the species (Figure S5A). These distinct clusters on the column tree mainly anchored by high proteasome activity, HSP25, and MLSP in the NMR and low proteasome activity and lifespan but high HSC/HSP70 and HSP40 in the laboratory mouse (Figure S5A). The analyses in the liver (Figure S5B) in contrast, divided the species very close to the phylogenetic tree (Fig. 1b) although anchored by the same phenotypes that distinguished NMR and mice in muscle. Phenotypes grouped similarly to that seen in the hierarchical clustering. For example, the HSP25/MLSP pairing clustered with ChTL and PGPH proteasome activities, HSF1, and ATG12 in the muscle (Figure S5A) while the close association of HSP40 and HSP25 sorted with HSP70 flux, MLSP, and HSF1 (Figure S5B). Therefore, while several proteins showed correlations with MLSP in both the liver and muscle, hierarchical clustering suggests that their association with other proteins and potentially their cellular targets diverge between tissues.
Discussion
In this study, we tested the hypothesis that the various components contributing to the level of chaperone-mediated protein degradation (i.e., proteasome activity, key chaperones, and autophagy) are key components of rodent longevity. Following PIC and correction for size differences, using rodents ranging in mass from 25 to 600 g and ranging in MLSP from 4 to >30 years, we observed that under these stringent criteria only, the small heat shock chaperone HSP25 and total HSF1 (Table 1) were significantly (p < 0.001) linked with MLSP. These significant relationships with MLSP but not body mass in both the muscle and liver suggest that HSP25 and HSF1 are likely to play a critical role in species longevity.
HSF1 positively correlates with lifespan
Increased HSF1 reportedly improves longevity in worms (Hsu et al. 2003). This is attributed to the fact that this transcription factor promotes the expression of several small heat shock proteins that in turn assist in the maintenance of protein structural integrity and conformation (Hsu et al. 2003). Thereby, this pathway prevents the accumulation of misfolded proteins and protein aggregates and sustains proteostasis. Strikingly, crossing HSF1-overexpressing mice with the human amyloid precursor protein (PDAPP) Alzheimer’s disease (AD) mouse model not only reduced the accrual of beta amyloid [Aβ] aggregates commonly observed in brain tissue of this AD mouse model but also improved cognitive function (Pierce et al. 2013). These findings highlight the importance of HSF1 in intracellular proteostasis. The protective nature of HSF1 in maintaining cellular integrity is also apparent in mouse fibroblasts; cells overexpressing HSF1 show enhanced resistance to protein aggregation stressors (Pierce et al. 2010). Although to date there are no data showing that overexpression of HSF1 in mice increases lifespan, these animals show enhanced protection against proteotoxic stress and well-maintained protein homeostasis (Pierce et al. 2010, 2013). Conversely and in line with the premise of a critical role of HSF1 in proteostasis, HSF1 knockout animals reportedly are more susceptible to prion disease and die more rapidly (20 % faster) than do wild-type animals inoculated with the identical quantity of prion protein (Steele et al. 2008). Upstream regulation of this key chaperone transcription factor may be more important, since total HSF1, not p-HSF1, positively correlated with MLSP in both tissues. One possible candidate for transcription regulation of HSF1 is nuclear factor-erythroid 2 p45-related factor 2 (Nrf2, also called Nfe2l2) which interacts with the HSF-1 network (Alavez et al. 2011). Nrf2 can also modulate the expression of HSP70, HSP40, and proteasome subunits (Hensen et al. 2013; Kwak et al. 2007) and recently has been shown to correlate with rodent MLSP in liver tissue (Lewis et al. 2015).
HSP25 is a critical component of proteostasis and longevity under basal and stressed conditions
Most chaperones, including HSP70 and HSP40, were differentially correlated to MLSP or mass based in a tissue-specific manner. Surprisingly, however, only HSP25 positively correlated with MLSP in both of the tissues tested. Moreover, these data were able to withstand the stringency of PIC (Figs. 1 and 2, Table 1, Table S1) as well as residual analyses to remove the effects of body size (Table S2). HSP25 serves as a chaperone to prevent misfolding during heat stress, inflammatory stress, and cell proliferation for key intracellular structural proteins such as actin and myosin (Doshi et al. 2010). Its expression correlates with increases in both autolysosomal and proteasome-mediated protein degradation (Chen et al. 2012; Parcellier et al. 2003) and is integral to the regulation of the NFκB inflammatory pathway (Parcellier et al. 2003; Salari et al. 2013). Moreover, research using Drosophila has demonstrated that overexpression of the HSP25 ortholog in flies can increase lifespan and augment resistance to oxidative stress (Morrow et al. 2004). Also, increased expression of the human HSP25 protein prevents protein aggregation and cell death in models of Parkinson’s and Huntington’s disease (Wyttenbach et al. 2002). Basal levels of HSP25 are higher in fibroblasts taken from long-lived bats and marsupials when compared to shorter-lived species within their respective taxonomic orders (Pride et al. 2015). Furthermore, expression of HSP25 is greatly enhanced after heat stress in fibroblasts from longer-lived rodents and marsupials compared to shorter-lived closely related species (Pride et al. 2015). Collectively, our data as well as those of previous research suggest that HSP25 could be a pivotal player in the regulation of protein homeostasis. As more data accumulate especially in long-living species, this may prove to be a universal mediator of extended lifespan possibly through its stress-related upregulation and response to aggregation within cells.
Proteasome activity correlates with lifespan in muscle
In this study, we found that proteasome activity correlated with longevity, independent of mass, but only in muscle tissue (Fig. 3, S3; Table 1, Table S1, S2). Ubiquitous overexpression of proteasome subunits has been shown to extend lifespan in Drosophila and C. elegans (Chondrogianni et al. 2015; Tonoki et al. 2009), but these studies lack tissue specificity. Interestingly, in Drosophila, loss of proteasome activity and assembly induced gross muscle disorganization and organ dysfunction (Haas et al. 2007), suggesting that proteasome stability and efficiency may play a critical role in muscle function (Fragala et al. 2014). However, we found that while activity correlated with MLSP in the muscle, the actual proteasome content for the 20S (α7), 26S ATPase (RPT5) proteasome, or the immunoproteasome (PA28α) (Figure S3) did not. These data imply that rather than altering proteasome levels to induce changes in its activity, there is more upstream control of substrate regulation (i.e., chaperones, E3 ligases, deubiquitinases, etc.). Such actions would lead to an improvement in proteasome-mediated degradation efficiency. Our investigations did not reveal any correlation between proteasome activity and MLSP in liver tissue, though there were significant correlations with body size and TL activity, as well as, RPT5 and PA28α levels (Fig. 3, S3, Table 1, Table S1, S2). This lack of correlation of MLSP in liver tissue is in agreement with a previous study that reported that the liver, proteasome activity did not correlate with lifespan (Salway et al. 2011b) and may reflect important differences linked specific to terminally differentiated tissues.
Proteins from the ubiquitin-like conjugation system of autophagy correlate with MLSP in muscle
While ATG7 and ATG3 showed significant correlations with MLSP in both liver and muscle lysates, only beclin-1 (negative) and ATG12 (positive) still showed a correlation with MLSP in muscle after PIC analyses (Figs. 4 and 5, Table 1, Table S1). Beclin-1 inhibition has been shown to prevent cell death in highly metabolic tissues such as cardiac myocytes and neurons during stress and injury (Diskin et al. 2005; Valentim et al. 2006). These findings suggest that an overabundance of this protein may not be beneficial to organ recovery, and in the case of muscle, may complicate regenerative processes. Increased expression of ATG12 improved lifespan in an in vitro aging model of human epithelial cells by improving mitophagy and its effects on cell homeostasis (Mai et al. 2012). ATG12 conjugates with ATG5 to initiate autophagosome formation (Yang and Klionsky 2009), an integral part of the repair mechanisms aiding in long-term health in muscle tissue. Although ATG7 and ATG3 aid in the activation of autophagosomes by facilitating conjugation between ATG5 and ATG12 (Diskin et al. 2005; Tanida et al. 1999; Yang and Klionsky 2009), the quantitation of these proteins did not correlate with MLSP after PIC analyses. This could suggest that the high levels of these proteins in NMR and Damaraland mole-rats may be specific phylogenetic traits of the African mole rats.
The role of tissue type, ecology, and phylogeny on MLSP
Heatmap analyses revealed that most rodent species clustered close to their cladogram phylogeny in the liver but not in the muscle (Fig. 7, S5). Interestingly, the two independent outliers, neither closely related, in muscle were both the long-lived NMRs and short-lived laboratory mouse (Figure S5A, B). As both the genetics of a species and the degree of protection afforded by its habitat may contribute to MLSP (Sibly and Brown 2007), tissue differences in these biomarkers of proteostasis could highlight the disparate functions of the tissues and their vulnerability to environmental influence. In the muscle, for example, HSP25 correlated with MLSP even after PIC and showed high protein expression in all three subterranean species (NMR, Damarland, and blind mole-rats) regardless of their phylogeny. This convergence may reflect that they are all subject to similar ecophysiological challenges linked to their subterranean ecological niches (Fig. 2, Table 1). Strikingly, this convergent pattern was not evident in liver tissue. Protein expression of HSP25 was lower in Spalax compared to the two bathyergid species, but nevertheless still correlated significantly with MLSP after PIC (Fig. 3, Table 1). There are several possible explanations for this: (a) it may be reflective of a strong phylogenetic component with the Bathyergidae expressing more HSP25 and having better protection or (b) this may be due to the solitary nature of Spalax compared to both of the Bathyergids in this study which live in large social groups and might therefore encounter even more stressful hypoxic/hypercapnic habitat than that of the solitary species (Buffenstein 2005; Nevo 2013). Therefore, the bathyergids might require higher levels of protein protection. Even within a single solitary species such as Spalax galili, subpopulations residing in different habitats show differential expression of markers of protein homeostasis (Li et al. 2015; Nevo 2013), highlighting the importance of environmental niche in proteostatic responses. As such, a third possibility is that proteostatic response is not only a phylogenetic trait but also is prone to intraspecific adaptation (Li et al. 2015). Spalax was wild caught whereas the other species were born in the lab. While the possibility exists that in the wild there could be more of a need for a higher proteostatic capacity compared to those reared in the lab. As such, results from lab-reared individuals could be an underestimation of the usual level of protein expression for that species in the wild, the data still suggest that these underground-originated species generally have higher proteostatic capacity compared to the other animals. If protein expression and measures of proteolytic activity are higher in wild NMRS or Damaraland mole-rats, then the correlations with MLSP might be further strengthened especially in cases where Damaraland mole-rats have lower protein expression or proteolytic activities compared to the Spalax.
Although body mass across all phyla correlates with MLSP (Sibly and Brown 2007; Speakman 2005), in our rodent subset, we found no significant correlations between the proteins measured and proteolytic activities tested in muscle and body mass, yet several of these variables were strongly correlated with MLSP. While this may reflect our small sample size, analyses of biochemical or molecular characteristics in muscle tissue nevertheless may be a better reflection of health span or longevity markers because mass (and its variation in captive sedentary animals) can be a confounding factor. As such, the differences observed with hierarchical clustering (Fig. 7a, b) may reflect tissue-specific prioritization of responses to protein homeostatic challenge. In muscle, a mostly terminally differentiated tissue, perhaps there is a higher priority for protein turnover of damaged proteins via proteasome activity or autophagy so that damaged proteins do not form aggregates that could lead to apoptosis, muscle loss, and ultimately sarcopenia. An enhancement of this protein and cellular maintenance could preserve long-term organ function.
Conclusions
In this study, we see a significant positive correlation between rodent species longevity and the various components of chaperone-mediated protein degradation pathways, especially in the muscle. Further, we identified an integral chaperone, the small heat shock chaperone HSP25, and its putative transcription factor HSF1, that appear to be critical component of protein homeostasis in both proliferating and post mitotic tissues. In the muscle, this chaperone’s upregulation is associated with both proteasomal degradation and autophagosomal initiation perhaps to better facilitate substrate presentation to these proteolytic machineries. In the liver, the association of HSP25 with HSP70 flux suggests that HSP25 enhances rescue of misfolded proteins, prior to degradation. Overexpression of HSF1 has previously been shown to extend lifespan, and this study reinforces that hypothesis. It is possible that these longer-lived species may have HSF1 at higher levels primed to respond to proteostatic challenge. However, while major transcription factors have large effects on lifespan by controlling the activities of multiple downstream effector molecules, the two effector molecules elucidated here HSP25 and ATG12 will allow us to determine the specific molecular targets with which these proteins interact. Discovering the mechanisms behind these interactions can aid us in understanding the process of functional decline and loss of protein quality control during aging.
Electronic supplementary material
Below is the link to the electronic supplementary material.
None of the HSPs tested correlated significantly with mass in the quadriceps muscle of seven rodent species. Quantitation and linear regression analyses against mass were performed on the same set of proteins examined in Fig. 1. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table S1. (JPG 49 kb)
Several HSPs also correlate with mass. Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Fig. 2. HSP70 (R2 = 0.31, p = 0.005), HSC70 (R2 = 0.20, p = 0.03), and phosphorylated HSF1 (p-HSF1; R2 = 0.36, p = 0.004) showed positive correlation versus mass in liver lysates. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table 1, S1. (JPG 54 kb)
Markers of proteasome content correlate with mass in liver tissue. (A) Representative Western Blots showing the constitutive 20S proteasome protein α7, the 26S ATPase protein RPT5, and PA28α immunoproteasome subunit in the tested rodent species. This analysis was repeated five times with similar results. (B) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against MLSP and Mass for quadriceps muscle and liver lysates. In muscle, there were no association with mass or MLSP from any of the proteins tested. In liver RPT5 (R2 = 0.68, p < 0.001) and PA28α (R2 = 0.30, p =0.005) showed a positive correlation with mass. α7 did not show a correlation with mass or MLSP in either of the tissues. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table S1. (JPG 84 kb)
Autophagic markers also correlate strongly with mass. (A) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Figure 4. Only LC3-II correlated significantly with mass (R2 = 0.18, p = 0.04). (B) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Figure 5. In liver, LC3-I (R2 = 0.19, p = 0.03), LC3-II/LC3-1 (R2 = 0.69, p <0.001), and ATG12 (R2 = 0.21, p = 0.02) showed significant correlations with mass. Plots show mean ± S.E.M. (n = 3 per experiment) and * indicates (p < 0.05) significance. See also Table S1. (JPG 66 kb)
Species map differently based on tissue type. Heat maps (A, B) show phenotypes as columns and rodent species as rows. The phenotypes are centered and scaled so that they have mean 0 and standard deviation (SD) of 1. This means that the colors represent the number of SD above the average of all species. While the muscle map (A) showed that naked mole-rats (NMR) and mice (Ms) are distinct from the other species, in the liver map (B) the clusters grouped very similar to the phylogenetic tree (C). (JPG 65 kb)
(DOCX 199 kb)
(XLSX 26 kb)
Acknowledgments
We thank Megan Smith, Vivian Cerritos, and Lillian Cerritos for the excellent care of the naked mole-rat and Damaraland mole-rat colonies. This work was supported by grants from the American Federation for Aging Research (AFAR), the Glenn Foundation for Medical Research, and the NIH/National Institute on Aging (NIA R21 AG043912-01A1) (R.B.). K.A.R. was supported by NIA training grant T32 AG021890 and an AFAR postdoctoral fellowship and now by a K99/R00 Pathway to Independence Award (AG049940-01A1) from the NIA. The comparative biology core of the Nathan Shock Center (UTHSCSA) and the Glen foundation partially contributed to the costs associated with animal husbandry.
Compliance with ethical standards
All animal protocols were approved by the University of Texas Health Science Center at San Antonio Institutional Animal Care and Use Committee.
References
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. The naked mole-rat; a new long-living model for human aging research. J Gerontol Biol Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- Chen SF, et al. Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci. 2012;19:52. doi: 10.1186/1423-0127-19-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrogianni N, et al. Protein damage, repair and proteolysis. Mol Asp Med. 2014;35:1–71. doi: 10.1016/j.mam.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Georgila K, Kourtis N, Tavernarakis N, Gonos ES. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J. 2015;29:611–622. doi: 10.1096/fj.14-252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasuri K, et al. Comparison of liver and brain proteasomes for oxidative stress induced inactivation: influence of aging and dietary restriction. Free Radic Res. 2009;43:28–36. doi: 10.1080/10715760802534812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deMagalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Bio. 2009;22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Development Core Team R. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Diskin T, Tal-Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma. 2005;22:750–762. doi: 10.1089/neu.2005.22.750. [DOI] [PubMed] [Google Scholar]
- Doshi BM, Hightower LE, Lee J. HSPB1, actin filament dynamics, and aging cells. Ann N Y Acad Sci. 2010;1197:76–84. doi: 10.1111/j.1749-6632.2010.05191.x. [DOI] [PubMed] [Google Scholar]
- Edrey YH, et al. Sustained high levels of neuregulin-1 in the longest-lived rodents; a key determinant of rodent longevity. Aging Cell. 2012;11:213–222. doi: 10.1111/j.1474-9726.2011.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragala MS, et al. Strength and function response to clinical interventions of older women categorized by weakness and low lean mass using classifications from the foundation for the National Institute of Health Sarcopenia Project. J Gerontol A Biol Sci Med Sci. 2014 doi: 10.1093/gerona/glu110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenburg W, et al. Reduction in ATP levels triggers immunoproteasome activation by the 11S (PA28) regulator during early antiviral response mediated by IFNbeta in mouse pancreatic beta-cells. PLoS One. 2013;8:e52408. doi: 10.1371/journal.pone.0052408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaczynska M, Osmulski PA. Small-molecule inhibitors of proteasome activity. Methods Mol Biol. 2005;301:3–22. doi: 10.1385/1-59259-895-1:003. [DOI] [PubMed] [Google Scholar]
- Garland T, Harvey PH, Ives AR. Procedures for the analysis of comparative data Using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. doi: 10.1093/sysbio/41.1.18. [DOI] [Google Scholar]
- Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3:a009704. doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KF, Woodruff E, 3rd, Broadie K. Proteasome function is required to maintain muscle cellular architecture. Biol Cell. 2007;99:615–626. doi: 10.1042/BC20070019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensen SM, Heldens L, van Enckevort CM, van Genesen ST, Pruijn GJ, Lubsen NH. Activation of the antioxidant response in methionine deprived human cells results in an HSF1-independent increase in HSPA1A mRNA levels. Biochimie. 2013;95:1245–1251. doi: 10.1016/j.biochi.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Hohn TJ, Grune T. The proteasome and the degradation of oxidized proteins: Part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Janen SB, Chaachouay H, Richter-Landsberg C. Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia. 2010;58:1766–1774. doi: 10.1002/glia.21047. [DOI] [PubMed] [Google Scholar]
- Kavanagh K, Flynn DM, Jenkins KA, Zhang L, Wagner JD. Restoring HSP70 deficiencies improves glucose tolerance in diabetic monkeys. Am J Physiol Endocrinol Metab. 2011;300:E894–E901. doi: 10.1152/ajpendo.00699.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U, et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2007;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc Natl Acad Sci U S A. 2015;112:3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, et al. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc Natl Acad Sci U S A. 2015;112:11905–11910. doi: 10.1073/pnas.1514896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-W, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai S, Muster B, Bereiter-Hahn J, Jendrach M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy. 2012;8:47–62. doi: 10.4161/auto.8.1.18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Ger Biol Sci Med Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Nevo E. Stress, adaptation, and speciation in the evolution of the blind mole rat Spalax, in Israel. Mol Phylogenet Evol. 2013;66:515–525. doi: 10.1016/j.ympev.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parcellier A, et al. HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol Cell Biol. 2003;23:5790–5802. doi: 10.1128/MCB.23.16.5790-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Wei R, Halade D, Yoo SE, Ran Q, Richardson A. A Novel mouse model of enhanced proteostasis: full-length human heat shock factor 1 transgenic mice. Biochem Biophys Res Commun. 2010;402:59–65. doi: 10.1016/j.bbrc.2010.09.111. [DOI] [PubMed] [Google Scholar]
- Pierce A, et al. Over-expression of heat shock factor 1 phenocopies the effect of chronic inhibition of TOR by rapamycin and is sufficient to ameliorate Alzheimer’s-like deficits in mice modeling the disease. J Neurochem. 2013;124:880–893. doi: 10.1111/jnc.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride H, et al. Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem Biophys Res Commun. 2015;457:669–675. doi: 10.1016/j.bbrc.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Rodriguez KA, Gaczynska M, Osmulski PA. Molecular mechanisms of proteasome plasticity in aging. Mech Ageing Dev. 2010;131:144–155. doi: 10.1016/j.mad.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KA, Edrey YH, Osmulski P, Gaczynska M, Buffenstein R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS One. 2012;7:e35890. doi: 10.1371/journal.pone.0035890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KA, Osmulski PA, Pierce A, Weintraub ST, Gaczynska M, Buffenstein R. A cytosolic protein factor from the naked mole-rat activates proteasomes of other species and protects these from inhibition. Biochim Biophys Acta. 2014;1842:2060–2072. doi: 10.1016/j.bbadis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout AK, Strub MP, Piszczek G, Tjandra N. Structure of transmembrane domain of lysosome-associated membrane protein type 2a (LAMP-2A) reveals key features for substrate specificity in chaperone-mediated autophagy. J Biol Chem. 2014;289:35111–35123. doi: 10.1074/jbc.M114.609446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari S, et al. Extracellular HSP27 acts as a signaling molecule to activate NF-kappaB in macrophages. Cell Stress Chaperones. 2013;18:53–63. doi: 10.1007/s12192-012-0356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salway KD, Gallagher EJ, Page MM, Stuart JA. Higher levels of heat shock proteins in longer-lived mammals and birds. Mech Ageing Dev. 2011;132:287–297. doi: 10.1016/j.mad.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Salway KD, Page MM, Faure PA, Burness G, Stuart JA. Enhanced protein repair and recycling are not correlated with longevity in 15 vertebrate endotherm species. Age (Dordr) 2011;33:33–47. doi: 10.1007/s11357-010-9157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarup P, Sorensen P, Loeschcke V. The long-term effects of a life-prolonging heat treatment on the Drosophila melanogaster transcriptome suggest that heat shock proteins extend lifespan. Exp Gerontol. 2014;50:34–39. doi: 10.1016/j.exger.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibly RM, Brown JH. Effects of body size and lifestyle on evolution of mammal life histories. Proc Natl Acad Sci U S A. 2007;104:17707–17712. doi: 10.1073/pnas.0707725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. Body size, energy metabolism and lifespan. J Exp Biol. 2005;208:1717–1730. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- Steele AD, et al. Heat shock factor 1 regulates lifespan as distinct from disease onset in prion disease. Proc Natl Acad Sci U S A. 2008;105:13626–13631. doi: 10.1073/pnas.0806319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentim L, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo AP, Rubinsztein DC. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
None of the HSPs tested correlated significantly with mass in the quadriceps muscle of seven rodent species. Quantitation and linear regression analyses against mass were performed on the same set of proteins examined in Fig. 1. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table S1. (JPG 49 kb)
Several HSPs also correlate with mass. Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Fig. 2. HSP70 (R2 = 0.31, p = 0.005), HSC70 (R2 = 0.20, p = 0.03), and phosphorylated HSF1 (p-HSF1; R2 = 0.36, p = 0.004) showed positive correlation versus mass in liver lysates. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table 1, S1. (JPG 54 kb)
Markers of proteasome content correlate with mass in liver tissue. (A) Representative Western Blots showing the constitutive 20S proteasome protein α7, the 26S ATPase protein RPT5, and PA28α immunoproteasome subunit in the tested rodent species. This analysis was repeated five times with similar results. (B) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against MLSP and Mass for quadriceps muscle and liver lysates. In muscle, there were no association with mass or MLSP from any of the proteins tested. In liver RPT5 (R2 = 0.68, p < 0.001) and PA28α (R2 = 0.30, p =0.005) showed a positive correlation with mass. α7 did not show a correlation with mass or MLSP in either of the tissues. Plots show mean ± S.E.M. (n = 4 per experiment) and * indicates significance (p < 0.05). See also Table S1. (JPG 84 kb)
Autophagic markers also correlate strongly with mass. (A) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Figure 4. Only LC3-II correlated significantly with mass (R2 = 0.18, p = 0.04). (B) Quantitation and linear regression analyses (GraphPad Prism v 6.0) against mass were performed on the same set of proteins examined in Figure 5. In liver, LC3-I (R2 = 0.19, p = 0.03), LC3-II/LC3-1 (R2 = 0.69, p <0.001), and ATG12 (R2 = 0.21, p = 0.02) showed significant correlations with mass. Plots show mean ± S.E.M. (n = 3 per experiment) and * indicates (p < 0.05) significance. See also Table S1. (JPG 66 kb)
Species map differently based on tissue type. Heat maps (A, B) show phenotypes as columns and rodent species as rows. The phenotypes are centered and scaled so that they have mean 0 and standard deviation (SD) of 1. This means that the colors represent the number of SD above the average of all species. While the muscle map (A) showed that naked mole-rats (NMR) and mice (Ms) are distinct from the other species, in the liver map (B) the clusters grouped very similar to the phylogenetic tree (C). (JPG 65 kb)
(DOCX 199 kb)
(XLSX 26 kb)